Abstract
The predominant motor neuron disease in infants and adults is spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS), respectively. SMA is caused by insufficient levels of the Survival Motor Neuron (SMN) protein, which operates as part of the multiprotein SMN complex that includes the DEAD-box RNA helicase Gemin3/DDX20/DP103. C9orf72, SOD1, TDP-43 and FUS are ranked as the four major genes causing familial ALS. Accumulating evidence has revealed a surprising molecular overlap between SMA and ALS. Here, we ask the question of whether Drosophila can also be exploited to study shared pathogenic pathways. Focusing on motor behaviour, muscle mass and survival, we show that disruption of either TBPH/TDP-43 or Caz/FUS enhance defects associated with Gemin3 loss-of-function. Gemin3-associated neuromuscular junction overgrowth was however suppressed. Sod1 depletion had a modifying effect in late adulthood. We also show that Gemin3 self-interacts and Gem3ΔN, a helicase domain deletion mutant, retains the ability to interact with its wild-type counterpart. Importantly, mutant:wild-type dimers are favoured more than wild-type:wild-type dimers. In addition to reinforcing the link between SMA and ALS, further exploration of mechanistic overlaps is now possible in a genetically tractable model organism. Notably, Gemin3 can be elevated to a candidate for modifying motor neuron degeneration.
Subject terms: Epistasis, Amyotrophic lateral sclerosis
Introduction
Motor neuron disease (MND) encompasses a seemingly heterogeneous group of neurological conditions that are nonetheless characterised by muscle weakness and paralysis thought to arise from the selective degeneration of motor neurons. Genetic factors play a major role in disease pathogenesis and the knowledge that mutations in genes encoding RNA-binding proteins (RBPs) can lead to MND, underscores RNA dysregulation as a key contributor to motor dysfunction1–4. In infants, the predominant MND is spinal muscular atrophy (SMA), typically an autosomal recessive condition caused by inactivating mutations in the survival motor neuron 1 (SMN1) gene that are partly counteracted by the paralogous SMN2 gene. Rather than total loss of the SMN1- or SMN2-encoded SMN protein, SMA is the result of insufficient SMN levels5. SMN, operating as part of a large multiprotein complex that includes Gemins 2–8 and Unrip, is indispensable for chaperoning the assembly of spliceosomal small nuclear ribonucleoproteins (snRNPs)6,7, in addition to a possible role in the assembly and axonal trafficking of messenger ribonucleoproteins (mRNPs) in motor neurons8. In adults, the most common MND is amyotrophic lateral sclerosis (ALS), which can be inherited (~10%) but is mostly sporadic (~90%). Chromosome 9 open reading frame 72 (C9orf72), Cu/Zn superoxide dismutase 1 (SOD1), transactive response DNA binding protein (TARDBP) and fused in sarcoma (FUS), in that order, are ranked as the four most common genes causing familial ALS and mutations in these genes are increasingly detected in sporadic cases9,10. TAR DNA binding-protein 43 or TDP-43 (encoded by TARDBP), and FUS are RBPs that are involved in multiple levels of RNA processing11,12.
Although SMA and ALS are traditionally considered as separate MNDs, a notion supported by differences in genetic aetiology, disease onset and type of affected motor neurons, accumulating evidence has revealed a surprising overlap at a molecular level. First, SMN and/or SMN complex members are components of the interactomes of SOD113, TDP-4314,15, FUS16–19 or the dipeptide repeat (DPR) proteins resulting from hexanucleotide repeat expansion in the C9orf72 gene20. In addition, both TDP-43 and FUS were reported to localise to gems14,15, which are nuclear bodies enriched in SMN complexes7,21,22. Second, both diseases are characterised by disrupted RNA processing including snRNP perturbation6,15,18,23–27 and axonal transport defects8,17,28–30. Third, both ALS and SMA were found to co-occur within families31. Fourth, an abnormal change in SMN1 copy number including gene deletion or duplication increases susceptibility to sporadic ALS32–34, presumably because deviations from normal SMN protein levels render motor neurons more vulnerable to degeneration. In corroboration, SMN deficiency was found to accelerate phenotypic severity in mutant SOD1 mice13. Fifth, and most important, a depleted number of gems resulting from SMN reduction was identified as a signature feature of ALS in addition to SMA35. A follow-up study unexpectedly showed that motor neurons derived from SMA or ALS patients have heterogeneous SMN levels with those having low levels being highly susceptible to cell death36. This observation explains why increasing SMN levels was found to be beneficial not only to SMA37,38 but also to ALS, at least, in SOD139,40 and TDP-4341 mouse models. Whether SMA therapeutics elevating SMN levels are also effective in ALS patients still remains to be determined.
Known and unknown components of molecular pathways can be uncovered in an unbiased fashion via genetic approaches. Drosophila has emerged as a premier model system for this task in view of its genetic tractability42,43. Indeed, genome-wide screens in Drosophila have yielded several modifier genes that are relevant to the pathology underlying either SMA44–47 or ALS48–52. However, the overlap has been surprisingly minimal and one study even reported that overexpression or RNAi-mediated knockdown of SMN failed to modify human FUS (hFUS)-induced neurodegeneration in Drosophila eyes23. This is in contrast to earlier findings in a cell-based system showing that overexpression of SMN rescued axonal defects induced by mutant FUS17. Although a common pathway uniting SMA and ALS could have developed later in evolution, it is highly likely that the screenable phenotype used in Drosophila-based investigations was not adequate to uncover interactions between SMA- and ALS-linked proteins. Therefore, the question of whether Drosophila can be exploited to study the shared pathogenic pathway linking SMA and ALS remains. Here, we address this question by using a different approach. First, instead of SMN, we focus on Gemin3, which is a core member of the SMN complex7. Our rationale is based on accumulating evidence that has essentially shifted the limelight from SMN to its Gemin associates revealing, a previously undisclosed, starring role in the operations of the SMN complex6,53. Second, we probe for a modifying effect in muscle, a tissue that is increasingly considered as a primary site of pathogenesis in both SMA and ALS54–59.
Gemin3, also known as DDX20 or DP103, is a DEAD-box RNA helicase which is involved in multiple cellular processes60. Most documented are its roles in RNA metabolism, including snRNP biogenesis where it functions within the SMN complex. To this end, we have recently shown that, in Drosophila, Gemin3 interacts both genetically and physically with pICln and Tgs1, two fundamental players in the snRNP biogenesis cycle61. Here, we extend the functional relationship to three key proteins linked to ALS. Hence, we demonstrate that a combination of Gemin3 and TDP-43 or FUS disruption exacerbates viability defects, motor dysfunction and muscle atrophy whilst suppressing neuromuscular junction (NMJ) overgrowth. Loss of Sod1 function is also responsible for inducing a prominent motoric decline in Gemin3 mutant flies at a late stage in adult life. The likely explanation is an interference in a common pathway. Additionally, we show that Gemin3 is capable of self-binding and Gem3ΔN, a helicase domain deletion mutant, enhances the association when bound to wild-type Gemin3, an observation that offers an explanation for its dominant-negative mechanism of action. Collectively, our data reinforce the link between SMA and ALS in addition to giving impetus to further studies on the shared mechanisms in a genetically tractable model organism.
Results
Overexpression of human TDP-43 in a Gem3 mutant background induces adult lethality
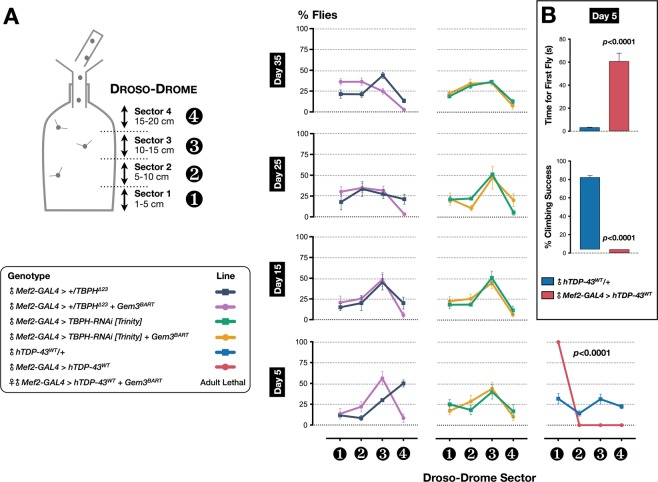
Similar to SMN45,62,63, loss of Gemin3 impacts adult viability and induces motor dysfunction64–66. In addition to Gemin3, a select number of SMN complex components, including Gemin2, Gemin4, Gemin5, Gemin8 and Unrip, are required for neuromuscular function and survival in Drosophila61,64,67. It is therefore highly plausible that SMA is triggered by any perturbation in the stoichiometry of the SMN complex. We have recently isolated Gem3BART, a hypomorphic version of the Gem3ΔN mutant, which lacks the N-terminal helicase core. Subsequently, we reported that alterations in the levels of SMN complex components precipitate the viability and motor phenotypes of Gem3BART adult flies68. A similar outcome was observed on disruption of snRNP biogenesis factors pICln and Tgs161. We wished to investigate whether a functional interaction also extends to Gemin3 and ALS-linked TDP-43. Missense mutations in this protein have been identified in 5% of familial and <1% of sporadic ALS cases10. TDP-43, an evolutionarily conserved protein, comprises of 2 RNA recognition motifs (RRMs), a nuclear localisation signal and a nuclear export sequence that mediate nuclear shuttling, as well as a C-terminal glycine-rich region where the majority of ALS-associated mutations occur11. Importantly, truncated TDP-43 is mislocalised from its predominantly nuclear location to ubiquitin-containing cytoplasmic inclusions in neurons of both sporadic and most familial forms of ALS10,69,70. Loss of TDP-43 nuclear function has been proposed as a primary mechanism linking TDP-43 proteinopathy to neuromuscular degeneration in ALS. In this context, we first asked whether decreased levels of TDP-43 can modify Gem3BART phenotypes. We note that neither haploinsufficiency (TBPHΔ23) nor RNAi-induced knockdown (TBPH-RNAi) of TBPH, the Drosophila TDP-43 homologue, had any effect on motor and viability phenotypes in flies with muscle-restricted Gem3BART expression (Fig. 1, Table 1 and data not shown).
Figure 1.
Gain-of-function identifies TDP-43 as a modifier of survival in Gem3BART-expressing flies. (A) Left: Flight performance was assessed by the Droso-Drome apparatus, where determination of flight capacity is based on which sector flies land after they are introduced at the top. Right: Removal of one copy (+/TBPHΔ23) or RNAi-mediated knockdown (TBPH-RNAi [Trinity]) of TBPH does not impair the motoric ability of flies with pan-muscular expression of Gem3BART. Indeed, at all time points, no significant differences were observed between the two groups in the number of flies that had no flight ability (sector 1) compared to those that retained the ability to fly (sectors 2–4). In contrast, ectopic expression of wild-type human TDP-43 (hTDP-43WT) in muscle impairs flight as early as day 5 post-eclosion. Flies were non-fliers, hence they all fell in sector 1. (B) Climbing success rate of flies with muscle-specific hTDP-43 overexpression was drastically reduced compared to control animals. Furthermore, assessment of the time taken for the first fly to reach a pre-set threshold determined that flies took significantly longer to attain this goal in contrast to the control genotype. In (A,B), data presented are the mean ± S.E.M. of at least 4 independent experiments, and for each time point measured, n ≥ 60 per genotype. Symbols indicate the sex of the genotype assessed: ♂ = males, ♀ = females, and ♀♂ = males + females. Significance as tested by two-way ANOVA, followed by Bonferroni’s post hoc tests (A) and the unpaired t-test (B) is indicated by the exact p-value.
Table 1.
Alleles of ALS-linked genes investigated in this study and their effect on viability when expressed either alone or in combination with Gem3BART in muscle tissue.
| ALS GENE | ALLELE | REF. | VIABILITY | |
|---|---|---|---|---|
| Mef2-GAL4> | Mef2-GAL4 > + Gem3BART | |||
| Hs: TDP-43; Dm: TBPH | LOF: +/TBPHΔ23 | 73 | Adult Viable | Adult Viable |
| LOF1: TBPH-RNAi [Trinity] | 73 | Adult Viable | Adult Viable | |
| LOF1: TBPH-RNAi [Merton] | 73 | Adult Viable | Adult Viable | |
| LOF1: TBPH-RNAi [Maudlin] | 126 | Death at P | Death at P | |
| OE2: hTDP-43WT | 72 | Adult Viable | Death at P | |
| OE1: hTDP-43WT.GFP#10 | 72 | Death at P | Death at L3 | |
| OE1: hTDP-43WT.GFP#16 | 72 | Death at P | Death at L3 | |
| OE1: hTDP-43CTF.GFP#14 | 72 | Death prior to L3 | N/A | |
| OE1: Flag.hTDP-43WT | 73 | Death at P | Death at L3 | |
| OE1: Flag.TBPHWT | 73 | Death prior to L3 | N/A | |
| OE2: TBPHWT | 75 | Death prior to L3 | N/A | |
| OE2: Venus-TBPHWT | 75 | Death at P | Death prior to L3 | |
| Hs: FUS; Dm: Caz | LOF: caz1/+* | 75 | Adult Viable | Adult Viable |
| LOF2: caz-RNAi [Kellogg] | 121,127 | Adult Viable | Adult Viable | |
| LOF2: caz-RNAi [Oriel] | N/A | Adult Viable | Adult Viable | |
| OE2: Flag.cazWT | 75 | Adult Viable | Death at P | |
| OE2: Flag.cazP398L | 75 | Death at P | Death at P | |
| OE2: Flag.hFUSWT | 75 | Adult Viable | Death prior to L3 | |
| OE2: HA.hFUSWT | 128 | Death at P | Death at P | |
| OE1: hFUSP525L-RFP.HA | 129 | Death prior to L3 | N/A | |
| OE1: hFUSWT-RFP.HA | 129 | Death prior to L3 | N/A | |
| OE1: hFUSR524S-RFP.HA | 129 | Death at P | Death at P | |
| OE2: Flag.hFUSP525L | 75 | Death at P | Death at L3 | |
| Hs: C9orf72 | OE2: G4C2-3 | 123 | Adult Viable | Adult Viable |
| OE2: G4C2-36 | 123 | Adult Viable | Adult Viable | |
| OE2: GR-36 | 123 | Adult Viable | Adult Viable | |
| OE2: GR-100 | 123 | Death prior to L3 | N/A | |
| OE2: PR-36 | 123 | Adult Viable | Adult Viable | |
| OE2: PR-100 | 123 | Adult Viable | Adult Viable | |
| Hs: SOD1; Dm: Sod1 | LOF: Sod1n1/+* | 80 | Adult Viable | Adult Viable |
| LOF2: Sod1-RNAi [Pembroke] | 122 | Adult Viable | Adult Viable | |
| LOF1: Sod1-RNAi [Hertford] | 122 | Adult Viable | Adult Viable | |
| OE1: hSOD1WT | 130 | Adult Viable | Adult Viable | |
| OE2: hSOD1WT. HA | 131 | Adult Viable | Adult Viable | |
| OE1: hSOD1G85R | 130 | Adult Viable | Adult Viable | |
| OE1: hSOD1A4V | 130 | Adult Viable | Adult Viable | |
| OE1: Sod1 | 130 | Adult Viable | Adult Viable | |
1Transgenesis: random insertion; 2Transgenesis: Φ-C31 site-specific insertion; *Heterozygote; Hs, Homo sapiens (human); Dm, Drosophila melanogaster (fruit fly); N/A = Not Applicable; L3, third instar larval stage; P, pupal stage, LOF, loss of function; OE, overexpression.
Recently, considerable attention has been given to the toxic effect of cytoplasmic TDP-43 protein aggregates71. To this end, we next queried whether gain rather than loss of TDP-43 function is a modifying factor. Expression of wild-type human TDP-43 (hTDP-43WT) in muscle leads to adult flies that have climbing defects and are entirely flightless when compared to controls (Fig. 1). Flies also have a shortened life-span, therefore surviving less than a week post-eclosion. Notably, in combination with Gem3BART, hTDP-43 induced adult lethality with flies dying at the pupal stage. Furthermore, transgenes with higher expression levels and/or epitope-tagged versions of hTDP-43 (hTDP-43WT.GFP#10, weak expression; hTDP-43WT.GFP#16, strong expression; ref. 72) were found to induce death at the earlier third instar stage (L3) when combined with Gem3BART in contrast to death at pupal stage when they were expressed alone (Table 1). This trend, which was also observed with an independently-generated line (Flag.hTDP-43WT, ref. 73; Table 1), shows that phenotypic enhancement is dependent on dose or modifications that interfere with protein structure. Muscle-directed expression of hTDP-43CTF, which mimics the major TDP-43 C-terminal fragment found in cytosolic aggregates of ALS patients, was found to induce lethality at the first instar larval stage most likely because the transgene is highly expressing72. This excluded its use for interaction analysis (Table 1). Importantly, overexpression of endogenous TBPH replicated the modifier effect of its human counterpart. Hence, whereas alone it induces death at the pupal stage, when combined with Gem3BART it enhanced survival defects with flies dying earlier than the L3 stage (Table 1). Overall, these findings are suggestive of a genetic interaction between Gemin3 and TBPH or its human homologue, TDP-43.
Knockdown or overexpression of caz/FUS enhances Gem3 mutant phenotypes
FUS is another major RBP that is mutated in both familial (5%) and sporadic (<1%) ALS cases10. FUS is a highly-conserved protein possessing an N-terminal domain rich in glutamine, glycine, serine and tyrosine residues (QGSY region), a glycine-rich region, an RRM, multiple arginine/glycine/glycine (RGG) repeats in an arginine- and glycine-rich region, and a zinc finger motif at the C-terminus. Mutations cluster in the glycine-rich region and in the extreme C-terminus where the nuclear localisation signal is likely to reside11. Similar to TDP-43, mutant FUS is mislocalised to the cytoplasm where it forms ubiquitinated aggregates74. Considering that FUS and TDP-43 function in a common pathway with FUS acting downstream of TDP-4375,76, we hypothesised that Gemin3 is likely to have a functional relationship not only with TBPH/TDP-43 but also with FUS. To this end, we first tested whether haploinsufficiency of cabeza (caz), the Drosophila homologue of FUS, can induce motor deficits when placed in a Gem3BART genetic background. Interestingly, we find a subtle yet statistically significant difference in motoric abilities at late adulthood (day 35 post-eclosion) in caz mutant heterozygous flies (caz1/+) that had muscle-restricted Gem3BART expression compared to those that did not (Fig. 2). Subsequently, we asked whether phenotypic enhancement is dose-dependent. Hence, we induced muscle-specific RNAi-mediated knockdown of caz in wild-type versus Gem3BART flies. We observed that a moderately-expressing RNAi transgene targeting the C-terminus (caz-RNAi [Kellogg], Supplementary Fig. S1) induced flight defects as early as day 15 post-eclosion with flies then exhibiting an age-dependent progressive worsening in phenotype (Fig. 2). A stronger RNAi transgene targeting the same region but based on short hairpin microRNA (shRNA) technology (caz-RNAi [Oriel], Supplementary Fig. S1) was capable of inducing motor defects at an earlier stage during adulthood (Fig. 2), further confirming that modification is dependent on Caz protein levels with a severe reduction inducing the highest impact.
Figure 2.
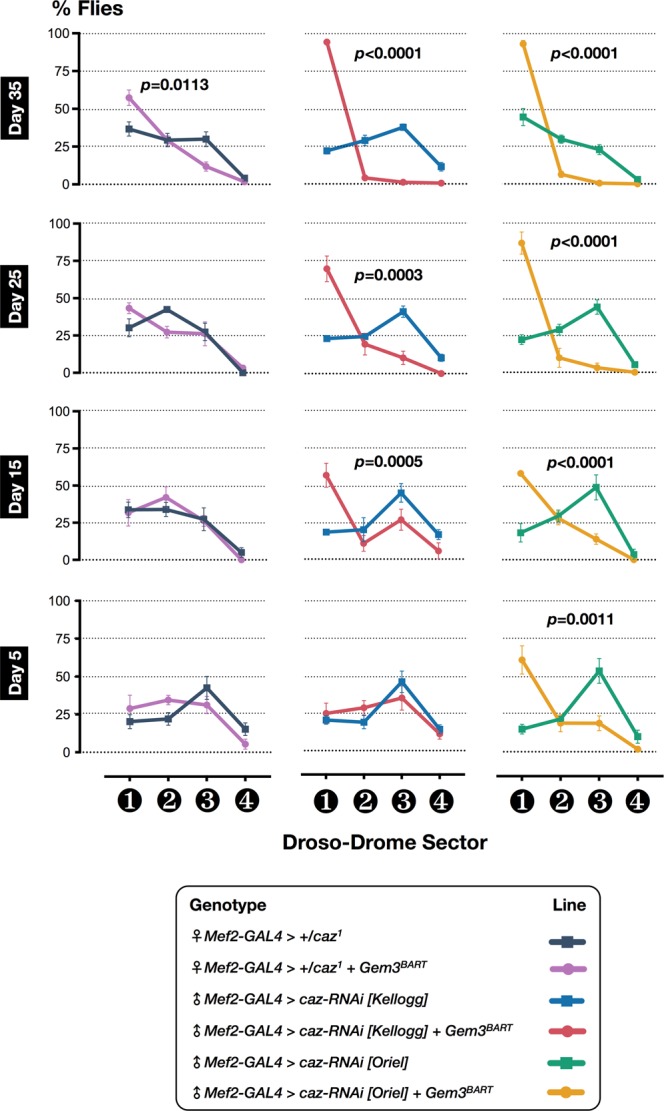
Loss of caz function impacts neuromuscular ability in Gem3 mutant flies. Left panel: In a heterozygous caz deficient background, brought about by the caz1 null mutant, Gem3BART flies develop flight defects at a late stage during adulthood. Hence, motor deficits become obvious only on day 35 post-eclosion. Middle panel: A greater reduction in Caz levels induced by a moderately-expressing RNAi transgene (caz-RNAi [Kellogg]) induced flight defects at an earlier stage (day 15 post-eclosion) and an increase in severity was observed with age. Right panel: A more pronounced age-dependent progressive decline in flight capacity can be brought about by knockdown mediated by a stronger RNAi transgene (caz-RNAi [Oriel]). Data presented are the mean ± S.E.M. of at least 4 independent experiments, and, for each time point measured, n ≥ 60 per genotype. Symbols indicate the sex of the genotype assessed: ♂ = males, and ♀ = females. Significance as tested by two-way ANOVA, followed by Bonferroni’s post hoc tests is indicated by the exact p-value.
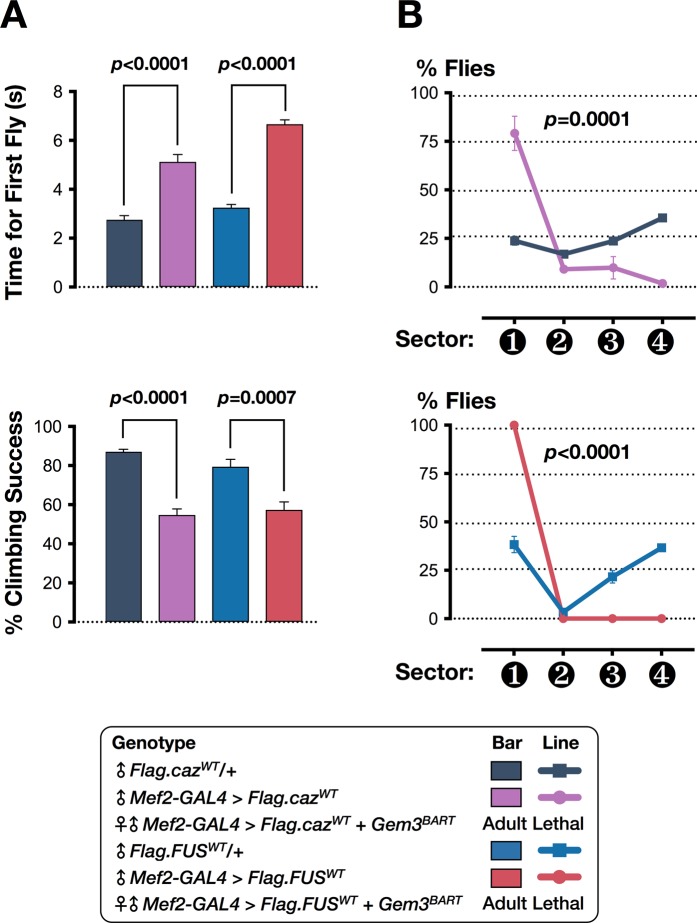
Similar to TDP-43, in addition to loss of nuclear function, a toxic gain of function due to the formation of cytoplasmic aggregates has been implicated as a predominant mechanism underpinning FUS-associated pathophysiology74,77. In this context, we questioned whether upregulation of caz or overexpression of human FUS (hFUSWT) can also act as enhancers when placed in a Gem3 mutant background. In this regard, in a wild-type background, muscle-restricted increase in Caz protein levels was sufficient to induce both climbing and flight defects as early as day 5 post-eclosion (Fig. 3). However, in Gem3BART flies, cazWT upregulation induced lethality before eclosion with the majority of flies dying during the pupal stage (Table 1). Similarly, ectopic expression of hFUSWT in muscles was enough to cause motor deficits in young adult flies when applied to a wild-type background (Fig. 3). In combination with Gem3BART, hFUSWT overexpression remarkably induced death during early development (Table 1). A trend towards reduced survival was also observed in Gem3BART flies upon expression of hFUS with a pathogenic mutation in the C-terminus (hFUSP525L), though not when expressing cazP398L, its equivalent in Drosophila (Table 1). Collectively, these findings show that either loss or gain of Caz/FUS function aggravate the motor and viability phenotypes of Gem3 mutant flies, which is highly suggestive of a genetic association between Gemin3 and caz/FUS.
Figure 3.
Gain of caz/FUS function in muscle impairs motor performance. (A) Overexpression of either caz or human FUS (hFUS) in muscle leads to adult flies with reduced mobility. Hence, on assessment, the first fly took significantly longer to reach a pre-set threshold. Furthermore, at a population level, climbing success was profoundly reduced. (B) Flies were in their majority non-fliers. Thus, when tested, a significant percentage dropped to the base (sector 1) of the Droso-Drome. Importantly, in combination with Gem3BART, muscle-specific overexpression of either caz or hFUS induced lethality prior to eclosion. Data presented are the mean ± S.E.M. of at least 4 independent experiments, and for each time point measured, n ≥ 60 per genotype. Symbols indicate the sex of the genotype assessed: ♂ = males, ♀ = females, and ♀♂ = males + females. Adult-viable flies were assessed at day 5 post-eclosion. Significance as tested by the unpaired t-test (A) and two-way ANOVA, followed by Bonferroni’s post hoc tests (B) is indicated by the exact p-value.
Expression of C9orf72 repeat expansions has no effect on Gem3 mutant flies
We next sought to broaden our investigation by determining whether Gem3 mutant phenotypes are also induced by disruption of other major ALS-linked genes. C9orf72 is the most frequently mutated gene in ALS. Enormous expansions of an intronic hexanucleotide repeat (GGGGCC, G4C2) cause a large portion of familial (25%) and sporadic (10%) ALS10. Transcripts containing repeats form intranuclear RNA foci that sequester nuclear proteins. In the cytoplasm, expanded RNA also undergoes repeat-associated non-AUG (RAN) translation to produce toxic dipeptide repeat (DPR) proteins9. Transgenic expression of a non-pathogenic repeat length (G4C2-3) in muscle tissue had no effect on neuromuscular function in either wild-type or Gem3BART flies. Thus, at all time points assessed, flies were relatively healthy and no major differences were apparent between the two groups at any stage during their adult life (Fig. S2). Although muscle-restricted expression of 36 repeats (G4C2-36), previously shown to be neurotoxic78, caused flight defects in early adulthood, the phenotype in Gem3BART flies was surprisingly identical to that of control flies with a wild-type background (Fig. S2). It is noteworthy that a similar outcome was observed in either background when assessing animals expressing two arginine-containing DPR proteins, glycine-arginine (GR-36) or proline-arginine (PR-100) (Fig. S2). In addition to confirming that repeats are damaging through the production of DPR proteins78, this result underscores that neither expanded repeats nor poly-GR/poly-PR proteins enhance Gem3 loss-of-function.
Loss rather than gain of Sod1 function enhances motor deficits in Gem3 mutant flies
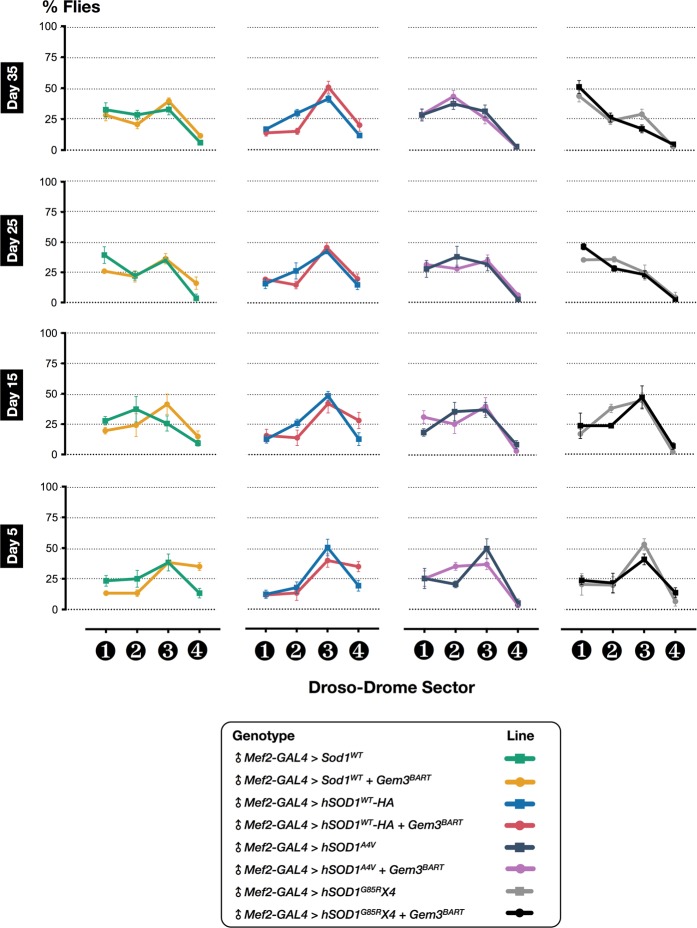
The first ALS gene to be identified was SOD1, which encodes for the Cu-Zn superoxide dismutase, an abundant ubiquitously-expressed cytoplasmic enzyme. SOD1 is the second most commonly mutated gene in ALS, contributing to 20% and 2% of familial and sporadic cases, respectively10. SOD1 performs an important antioxidant function by catalysing the conversion of highly reactive superoxide to hydrogen peroxide or oxygen. Nevertheless, neuromuscular degeneration is thought to be driven by one or more acquired toxicities of the mutant protein rather than loss of dismutase activity. Indeed, similar to TDP-43 and FUS, most ALS-causing SOD1 mutants form ubiquitinated cytoplasmic aggregates that are toxic to various cellular processes79. Against this backdrop, we investigated whether gain of Sod1 function is also capable of triggering motor dysfunction in Gem3 mutant flies. We found that neither overexpression of Drosophila Sod1 nor ectopic expression of wild-type human SOD1 (hSOD1WT) had any negative effect in either a wild-type or a Gem3 mutant background (Fig. 4). Expression of pathogenic variants including hSOD1A4V or hSOD1G85R gave a similar result, hence they were not damaging in either genetic background (Fig. 4). Interestingly, we were surprised to note that less than 50% reduction in enzymatic activity, brought about by heterozygosity for the missense allele Sod1G51S (+/Sod1G51S)80, induced a prominent decrease in neuromuscular function during late adulthood in Gem3 mutant flies, suggesting that these organisms are susceptible to oxidative stress when they get old. We confirmed this result through the use of muscle-specific RNAi-mediated loss of Sod1 function in wild-type versus Gem3BART flies. Hence, we show that an RNAi transgene targeting the C-terminus (Sod1-RNAi [Pembroke], Supplementary Fig. S1) similarly provoked flight defects in adult flies aged to day 35 post-eclosion (Fig. 5). Motor defects became apparent at an even earlier stage (day 25 post-eclosion) when we made use of a stronger RNAi transgene targeting the same region but having a longer hairpin sequence (Sod1-RNAi [Hertford], Fig. 5 and Supplementary Fig. S1). Overall, these findings demonstrate that Gem3 mutant phenotypes are hastened by Sod1 loss-of-function rather than by gain-of-function, hence allowing us to uncover a genetic interaction between Gemin3 and Sod1.
Figure 4.
Sod1 gain-of-function has no effect on motor behaviour in Gem3 mutant flies. Overexpression of either Sod1 or human SOD1 (hSOD1) is not deleterious to animals with a marginal loss of Gem3 function (left panel). Overexpression of pathogenic hSOD1 A4V or G85R variants is equally not a damaging factor to Gem3 mutant flies (right panel). Organisms did not show differences in motor function at all time points assessed. Data presented are the mean ± S.E.M. of at least 4 independent experiments, and, for each time point measured, n ≥ 60 males (♂) per genotype.
Figure 5.

Loss of Sod1 function impairs motor behaviour in Gem3 mutant flies. Placed in a Gem3BART genetic background, heterozygotes for an enzymatic null Sod1 allele (+/Sod1G51S) only induce deficits in old age (left panel). A similar outcome is achieved in Gem3BART flies with muscle-selective RNAi-mediated knockdown of Sod1 (middle panel). The use of a stronger RNAi transgene causes defects at an earlier stage of adult life (right panel). Data presented are the mean ± S.E.M. of at least 4 independent experiments, and, for each time point measured, n ≥ 60 males (♂) per genotype. Significance as tested by two-way ANOVA, followed by Bonferroni’s post hoc tests, is indicated by the exact p-value.
TDP-43 or FUS gain of function enhance muscle atrophy and suppresses neuromuscular junction overgrowth in Gem3 mutant flies
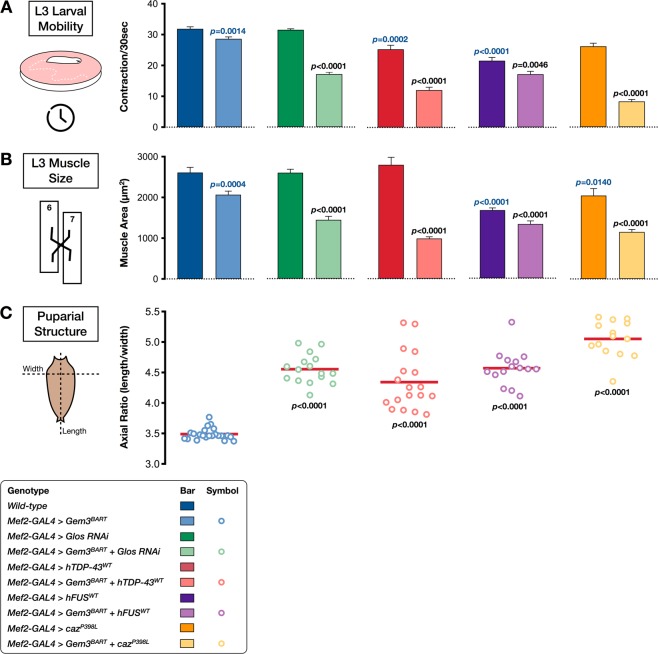
Above we showed that upregulation of caz or TBPH, in flies with Gem3 loss of function, induced adult lethality. Overexpression of the respective human homologue gave an analogous outcome. We next investigated whether the neuromuscular function of these animals is perturbed prior to their death. Gem3BART animals devoid of any genetic modifying factor(s) eclose normally and neuromuscular function is relatively unperturbed in adult flies. Surprisingly, when analysing larval crawling, we observed a slight yet significant decline in mobility in Gem3BART larvae compared to their wild-type counterparts (Fig. 6A). Notably, this difference can be explained by a substantial difference in muscle surface area between Gem3 mutant and control larvae. Thus, the former had a pronounced reduction in muscle size (Fig. 6B). We asked whether these phenotypes are amenable to modification by genetic factors. To this end, we introduced Gaulos RNAi in Gem3 mutants. Gaulos (Glos) was recently identified as the Drosophila orthologue of Gemin467,81, a putative co-factor of Gemin382. Remarkably, muscle-driven Glos reduction caused a further decline in both the locomotor ability (Fig. 6A) and muscle size of Gem3BART larvae (Fig. 6B), hence confirming that the phenotypes can be genetically enhanced. Importantly, we demonstrate that compared to the baseline provided by Gem3BART larvae, ectopic expression of either hTDP-43 or hFUS was responsible for an additional degree of sluggishness in larvae as demonstrated by their less frequent movements (Fig. 6A). Interestingly, the difference was reflected in muscle size, hence muscle atrophy was greatly enhanced upon TDP-43 or FUS gain-of-function (Fig. 6B). Upregulation of TBPH causes Gem3BART flies to die before the third instar larval stage (Table 1), hence precluding assessment of flies. Upregulation of caz in Gem3BART flies induces larval mobility defects but has no effect on muscle size (data not shown). However, overexpression of the pathogenic variant cazP398L in Gem3BART flies was found to mirror the neuromuscular phenotypes observed on TDP-43/FUS gain of function in the same genetic background (Fig. 6A,B).
Figure 6.
Overexpression of hTDP-43 or hFUS/cazP398L in Gem3 mutant flies causes early mobility defects, reduced muscle size and aberrant puparial structures. (A) Third instar larvae with muscle-specific Gem3BART expression have a significant reduction in velocity compared to wild-type controls. This deficit was profoundly enhanced on overexpression of hTDP-43WT, hFUSWT or cazP398L. A similar result was obtained on knockdown of Gaulos (Glos RNAi), which served as a positive control. In the absence of Gem3BART, only overexpression of hTDP-43WT or hFUSWT was found to induce a decrease in larval mobility compared to wild-type animals. (B) Compared to the wild-type control, Gem3BART larvae also show a significant reduction in muscle size that dramatically worsens on Glos knockdown or upon overexpression of hTDP-43WT, hFUSWT, or cazP398L, all directed to muscle. In a wild-type background, muscle size was also found reduced in larvae with Glos knockdown or those overexpressing FUS/cazP398L. (C) Sluggish larval behaviour leads to the formation of puparia that have a significantly higher axial ratio (defined as length/width) compared to the baseline offered by Gem3BART larvae. In (A–C) data presented are the mean ± S.E.M. of at least 3 independent experiments, and n ≥ 15 per genotype. Equal number of male and female larvae were used in each assay. Significance as tested by the unpaired t-test is indicated by the exact p-value, shown either in blue (comparison to wild-type larvae) or black (comparison to Gem3BART larvae).
Consequent to muscle atrophy and the subsequent decline in muscle power, all genotypes assessed failed to contract adequately during pupariation. Hence, in a Gem3BART genetic background, Glos knockdown (serving as a positive control) or overexpression of hTDP-43/hFUS/cazP398L, all induced a puparial axial ratio that was significantly higher than the baseline observed in Gem3 mutant flies devoid of any genetic manipulation (Fig. 6C). Finally, we assessed the morphology of the neuromuscular junction (NMJ). Muscle-directed expression of Gem3BART causes an appreciative expansion of the NMJ (Fig. 7A), hence, parameters including area (Fig. 7B), number of branches (Fig. 7C) and bouton numbers (Fig. 7D) were all significantly elevated compared to the wild-type control. This phenotype was similar to that previously reported for flies with homozygous Gem3 loss-of-function in all tissues65. NMJ overgrowth was also observed on Glos knockdown and upon expression of cazP398L, both of them directed to muscle (Fig. 7). No deviations from the wild-type NMJ parameters were however seen on expression of either hTDP-43 or hFUS in muscle (Fig. 7). Interestingly, in a Gem3BART background, expression of hTDP-43, hFUS and, to a slightly lower degree, cazP398L (but not Glos RNAi) suppresses the NMJ overgrowth phenotype associated with Gem3 loss-of-function, with key NMJ morphology parameters reduced to the wild-type range (Fig. 7). In sum, these observations strengthen the evidence favouring the possibility that Gemin3 acts together with TBPH/TDP-43 and caz/FUS within a genetic pathway that influences viability and neuromuscular function.
Figure 7.
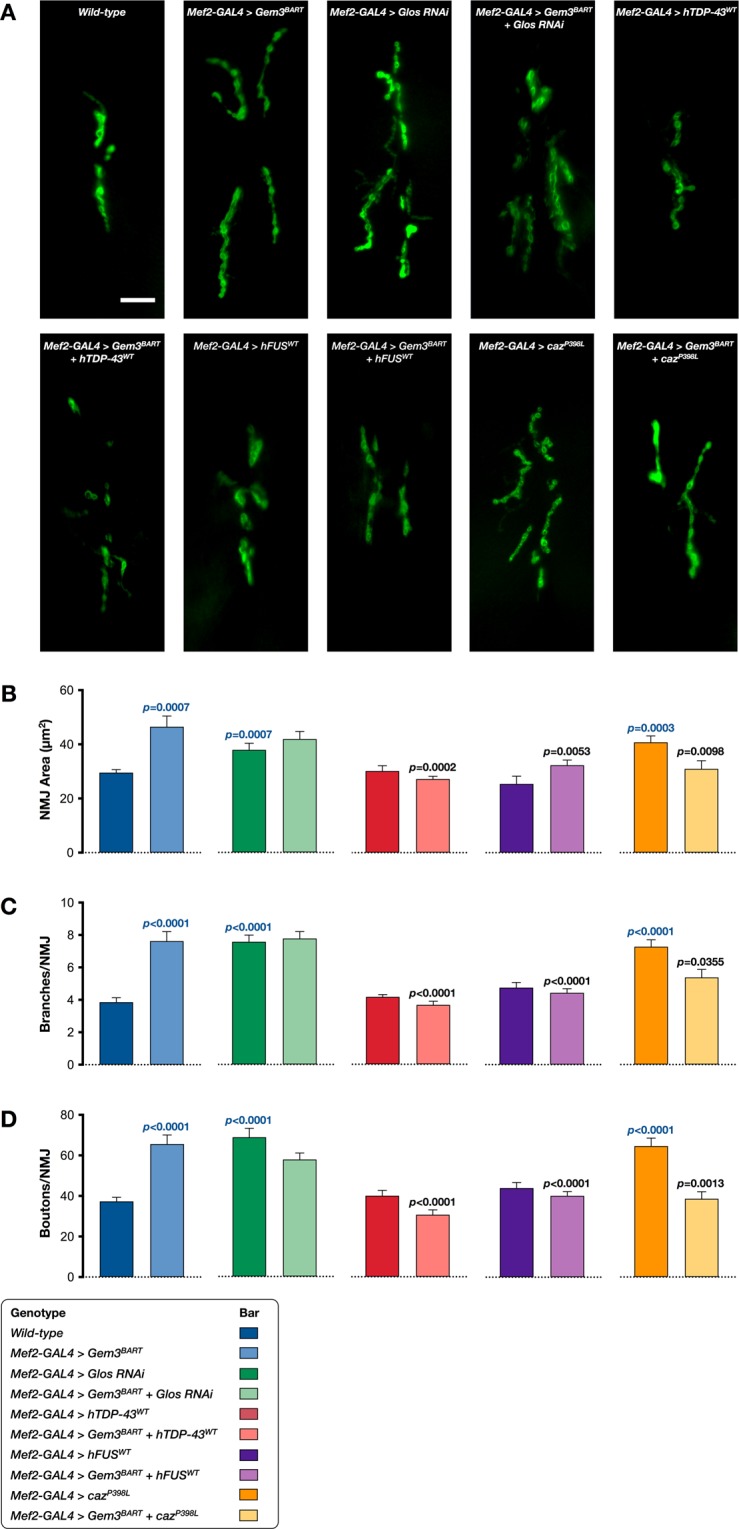
Overexpression of hTDP-43 or hFUS/cazP398L suppresses NMJ expansion in Gem3 mutant flies. (A) Representative images of NMJs innervating ventral longitudinal muscles 6 and 7 in third instar larvae stained with post-synaptic anti-DLG antibody (scale bar = 10 μm). Visual inspection reveals that compared to wild-type, muscle-directed expression of Gem3BART induces an overgrown NMJ morphology that is restored on overexpression of hTDP-43WT, hFUSWT and, to a lesser degree, cazP398L. An overgrowth phenotype is also obvious on Glos knockdown (applied singularly or combined with Gem3BART) or in flies expressing cazP398L. These observations were confirmed upon quantification of NMJ area (B), number of branches per NMJ (C) and number of boutons within a single NMJ (D). In (B–D) data presented are the mean ± S.E.M. and n ≥ 18 per genotype. Equal number of male and female larvae were assessed. Significance as tested by the unpaired t-test is indicated by the exact p-value, shown either in blue (comparison to wild-type larvae) or black (comparison to Gem3BART larvae).
Self-association of full-length Gemin3 explains loss of function induced by the Gem3BART mutant
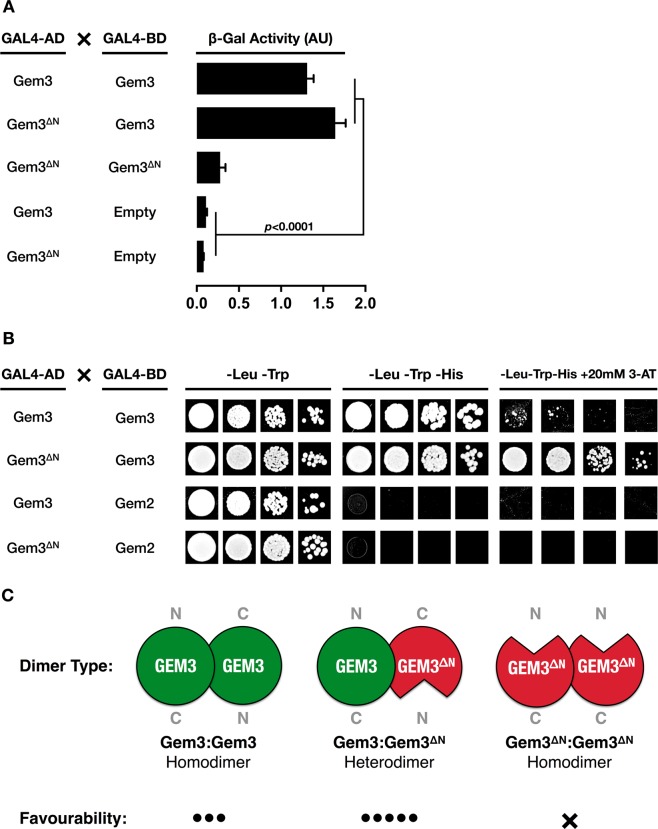
In conclusion, we wished to gain insights into the mode of action of the Gem3BART allele, a hypomorphic or weaker version of the Gem3ΔN mutant. The latter differs from wild-type Gemin3 in that it lacks the N-terminus which hosts the helicase domains68. Previously, we have shown that expression of Gem3ΔN mimics Gem3 knockdown and, together, the genetic alterations cause lethality. This allowed us to conclude that expression of Gem3ΔN in a wild-type background induces a loss of Gemin3 function64. We hypothesised that Gem3ΔN interacts with its wild-type full-length counterpart and in so doing, it interferes with its function. This model assumes that Gemin3 is capable of self-association for which the evidence is presently lacking. Through a yeast two-hybrid assay, we confirm this model, hence, we show that Drosophila Gemin3 is able to strongly interact with itself (Fig. 8A). Although this property is lost with Gem3ΔN, remarkably, we demonstrate that Gem3ΔN is capable of binding to its full-length counterpart (Fig. 8A).
Figure 8.
Full-length Gemin3 interacts not only with itself but also with Gem3ΔN. (A) Measurement of β-galactosidase activity was used to assay the expression of the GAL1-LacZ reporter gene that is produced by the combinations of the indicated proteins. Full-length Gemin3 interacts with itself or with its N-terminal truncated form (Gem3ΔN). Gem3ΔN is not capable of self-binding. An empty vector served as a negative control. Individual bars represent the mean ± S.E.M. β-galactosidase activity of 3 independent experiments. Significance as tested by one-way ANOVA is indicated by the exact p-value. (B) The reporter strain carrying yeast two-hybrid plasmids expressing the indicated proteins was spotted in serial dilutions on –Leu–Trp–His plates in the presence of 20 mM 3-AT. Under these conditions, Gem3-Gem3 interaction is unfavourable whereas the Gem3-Gem3ΔN association remains favourable. Interaction between Gem3 or Gem3ΔN with Gemin2 served as a negative control. At least 3 independent experiments were performed, and the result of one representative is shown. (C) Model for Gemin3 dimerisation. Left, Gemin3 monomers are hypothesised to bind to each other in the reverse direction. Middle, The Gem3ΔN mutant is a truncated version of Gemin3 lacking the N-terminus, which hosts the helicase domains. In the presence of Gem3ΔN, wild-type:mutant dimers are favoured more than wild-type:wild-type dimers. Right, A model predicting self-association in the opposite orientation infers that Gem3ΔN is not self-binding, hence, mutant:mutant dimers do not form. Abbreviations: N, N-terminus; C, C-terminus.
We next questioned whether the Gem3ΔN-Gem3 interaction is stronger than the one between wild-type Gemin3 monomers, hence favouring the capture of endogenous Gemin3 into non-functional oligomers. To this end, we re-performed the yeast two-hybrid assay in the presence of 3-amino-1,2,4-triazole (3-AT). 3-AT competitively inhibits imidazole glycerol-phosphate dehydratase, a histidine (His) biosynthetic enzyme, thus limiting His synthesis83,84. We demonstrate that yeast containing both the Gem3 bait and the Gem3ΔN prey were capable of growing on –Leu–Trp–His selective plates at 20 mM 3-AT (Fig. 8B), thus indicating that the two-hybrid Gem3-Gem3ΔN interaction is strong enough to overcome the growth inhibitory effect of 3-AT in the medium. In summation, these findings are first supportive of Gemin3 self-binding. Second, they allow us to postulate that the dominant-negative nature of Gem3ΔN arises from its most-favourable binding to endogenous Gemin3. This is predicted to titre wild-type Gemin3 into non-functional dimers or oligomers.
Discussion
In this study, we sought to determine whether a functional interaction exists between Gemin3, a core SMN complex component, and major ALS-associated proteins. Focusing on motor behaviour, muscle mass, NMJ structure and survival, which are all profoundly affected in motor neuron disease, we show that disruption of either TBPH/TDP-43 or Caz/FUS enhance muscle defects but are able to suppress NMJ morphology deficits, both induced by Gemin3 loss-of-function. We also found that depletion of Sod1 has an enhancing effect on neuromuscular function in old age. In addition to highlighting shared pathways most likely involving aspects of ribostasis and oxidative stress, our findings reinforce the link between SMA and ALS. Importantly, they extend our knowledge on the function of Gemin3, showing for the first time that it self-interacts, which is a property that makes it prone to loss of function.
Defective chaperoning of spliceosome assembly and/or missplicing have long been known to have a major role in the pathophysiology of motor neuron degeneration. A plethora of in vivo studies unequivocally show that snRNP assembly defects and the consequential missplicing events can induce the selective motor phenotype that is typical in SMA patients (reviewed in ref. 6). In this regard, we have recently shown that, in Drosophila, perturbation of snRNP biogenesis factors pICln or Tgs1 causes motor deficits that mirror those brought about by loss of SMN or select Gemins including Gemin361. This corroborates earlier findings demonstrating that knockdown of pICln or U1 snRNP leads to MND-like phenotypes in zebrafish85,86. Notably, by discovering an interaction between Gemin3 and either pICln or Tgs161, we underscored that these factors participate in a common pathway that most likely centres on the synthesis of snRNPs which form the backbone of the spliceosome. Here, we widen our findings by uncovering a genetic association between Gemin3 and two RBPs with important roles in diverse aspects of RNA metabolism, namely TDP-43 and FUS. Our data suggest that Gemin3, TDP-43 and FUS function in overlapping pathways that influence viability, muscle mass, NMJ morphology and motoric ability.
Considering the roles of Gemin3, TDP-43 and FUS at different points in the life of the spliceosome, we speculate that the intersecting pathways are vital for the correct splicing of mRNAs, which we believe is a main contributor to the health and optimal function of the neuromuscular system. In support, although its exact activities in snRNP assembly remain unclear, Gemin3 is indispensable for this process in vivo60. Considering TDP-43 and FUS, long before their implication in ALS, these two proteins were reported to influence pre-mRNA splicing or interact with known splicing factors87,88. Both RBPs were later shown to bind to predominantly UG-rich sequences in RNA transcripts, regulating the expression and alternative splicing of multiple yet distinct target genes, particularly those with exceptionally long introns89–93. Thus, loss of TDP-43 or FUS in cell lines or mouse brain leads to splicing defects that are mostly different for either factor89,90,92,94,95. Even splicing of snRNP components is altered based on studies in sporadic ALS patient-derived lower motor neurons that had nuclear TDP-43 depletion96 or FUS knockdown in a human cell line97. Overexpression of ALS-causing TDP-43 or FUS mutants, which cause neuromuscular phenotypes in mice, were also found to induce aberrant RNA splicing98,99. Similarly, Drosophila with knockout or overexpression of TBPH exhibit splicing alterations100. Interestingly, expression levels of several snRNAs were found significantly increased in brains of flies overexpressing human TDP-43101 and decreased in fibroblasts derived from ALS patients with FUS mutations or FUS transgenic mice19. Notably, in an ALS mouse model, an endogenous C-terminal domain mutation in TDP-43 was recently reported to induce a gain of splicing function. Hence, splicing activity of TDP-43 was modified in such a way that it leads to the excision of otherwise normally conserved exons, thereby, termed ‘skiptic exons’102. Pathogenic TDP-43 or FUS mutations are also known to affect splicing in a gain-of-toxic-function manner by mislocalising snRNPs, SMN and splicing factors (PSF and NeuN) to the cytosol18,19,86,103,104.
It is interesting to note that whereas gain of TDP-43 or FUS function were both shown to enhance Gemin3 motor deficits, we observed that loss-of-function was consequential only for FUS. This can be explained by the more intimate relationship of FUS with the SMN complex. In this respect, in addition to U1 (and U11) snRNP components19,103, SMN complex members including SMN and Gemin3 were shown to be integral members of the FUS interactome19. It is plausible that the SMN complex might collaborate with FUS in an as yet unknown snRNP-related function, which is disrupted by deviations from normal Caz/FUS levels. Thus, ALS-causative mutations in FUS were found to strengthen the interaction with SMN potentially sequestering SMN and, most probably, its associates, from their normal localisation and function19. It is important to note that we do not exclude the possibility that Gemin3 cooperates with TDP-43 and FUS in other steps of RNA metabolism including transcription or RNA transport given that all three factors are known to participate in either process11,12,28,60,105.
Gem3 mutant phenotypes were not hastened by SOD1 or C9orf72 gain-of-function. This allows us to infer that the genetic interaction between Gemin3 and TBPH/TDP-43 or caz/FUS is specific. Nonetheless, reduced levels of Sod1 brought about by heterozygosity for a Sod1 mutant or RNAi-mediated knockdown were surprisingly found to induce motor deficits in Gem3 mutant flies during late adulthood. Oxidative stress is exacerbated by age106 and paucity of Sod1107–109. Notably, snRNP assembly function of the SMN complex was found to be inhibited by oxidative stress in a dose-dependent manner110. This observation adds to the plethora of evidence showing that oxidative stress perturbs RNA metabolism (reviewed in ref. 111). Thus, it is reasonable to speculate that oxidative stress is a modifying factor for Gemin3 function in snRNP synthesis as part of the SMN complex.
Ectopic overexpression of the helicase core deletion mutant Gem3ΔN in a wild-type background induces phenotypes that overlap those resulting from Gemin3 loss-of-function. In this regard, either overexpression of Gem3ΔN or RNAi-mediated knockdown of Gemin3, both targeted to muscle tissue, was shown previously to disrupt motor behaviour. Applied simultaneously, these two genetic manipulations were found to cause lethality64. Thus, the evidence favours the possibility that Gem3ΔN interferes with endogenous Gemin3 to induce loss of function, hence acting as a dominant-negative mutant or a Muller’s antimorph. Here, we show that Gemin3 is capable of self-interaction and Gem3ΔN retains the ability to interact with its wild-type counterpart. Importantly, analysis of interaction strength demonstrates that wild-type:mutant dimers are favoured more than wild-type:wild-type dimers (Fig. 8C). Based on these findings, we predict that Gem3ΔN titres endogenous Gemin3 into non-functional dimers or oligomers. Sequestration of Gemin3 can perturb SMN complex stoichiometry in addition to inhibiting the participation of Gemin3 in SMN complex-related activities including snRNP assembly or recycling.
The formation of wild-type:mutant dimers or even oligomers can potentially hinder the catalytic activity of Gemin3, thus raising the question of whether dimerization of Gemin3 is a prerequisite for its ATPase-dependent RNP chaperoning activities. Self-interaction that is independent of the RNA substrate is rather unusual for members of the DEAD-box RNA helicase family. It has been reported in prokaryotes for Escherichia coli RhIB112, Thermus thermophilus Hera113, Bacillus subtilis CshA114 and cyanobacteria CrhR115. In this context, our findings add Gemin3 to the growing list of eukaryotic DEAD-box RNA helicases that also have a self-interaction property including transcriptional regulators DDX5/p68 and DDX17/p72116. Domain analysis of both T. thermophilus Hera and B. Subtilis CshA revealed that efficient dimerization is dependent on protein regions other than the those hosting the two highly-conserved RecA-like helicase domains which are required for RNA substrate binding and catalytic activity113,114. For E. coli RhIB, cyanobacteria CrhR115 and eukaryotic DDX5 or DDX17, a large part of the conserved core was required for self-association112,116, a finding that also applies for Gemin3. To this end, we show that, alone, the C-terminal domain of Gemin3 is incapable of self-binding, hence, the formation of Gem3ΔN homodimers is an unfavourable reaction (Fig. 8C). However, interaction is observed in the presence of the N-terminus, making the formation of Gem3ΔN:Gem3 dimers a highly favourable reaction. This indicates that self-interaction requires that the full-length protein is present in at least one of the two monomers, thus raising the possibility that Gemin3 monomers bind to each other in the reverse direction. This model warrants future investigation through further molecular and structural studies.
Given the functional interaction of ALS-linked TBPH/TDP-43, Caz/FUS and Sod1 with Gemin3, which itself is intimately associated with the SMA-causative SMN, our work adds to the substantial collection of evidence supporting convergence of the molecular mechanisms of two major MNDs. Although we speculate that defects in RNA metabolism might be central to the pathophysiology of ALS and SMA, further investigation of the mechanistic overlaps is now possible in a genetically-tractable model organism. Importantly, given our findings, we propose Gemin3 as a candidate for modifying motor neuron degeneration.
Materials and Methods
Flies
Flies were cultured on food consisting of sugar, corn meal, yeast, and agar in plastic vials at an incubation temperature of 25 °C under 12 hours day/night cycles. The wild-type strain was w1118. For adult-based assays, male flies were used except where indicated. In instances where females were used, flies were virgins. For larval-based assays, equal number of male and female larvae were assessed. Inducible transgenes were expressed via the bipartite GAL4/upstream activation sequence (UAS) system (reviewed in ref. 117). Muscle-exclusive expression was achieved through the use of the Mef2-GAL4 driver118. The Gem3BART allele (UAS.Gem3BART) was generated previously by transposition of the Gem3ΔN transgenic allele into a repressive region on chromosome 268. The UAS.Glos-IRDEX (Glos RNAi) transgene was described and characterised previously67. TBPHΔ23 and caz1 are small deletions that partially remove the coding and 5’ sequence of TBPH73 and caz75, respectively. They are considered as null alleles of the respective gene. The Sod1G51S mutant (also known as Sod1n108 or Sod1n1) carries an EMS-generated missense mutation in the Sod1 gene that disrupts dimer contact119. Sod1G51S homozygotes were reported to be null for superoxide dismutase activity. In Sod1G51S heterozygotes, superoxide dismutase activity was reported to be close to 40%80. The RNAi transgenic constructs, UAS.TBPH-RNAi [Trinity] (ID: 38377), UAS.TBPH-RNAi [Merton] (ID: 38379), UAS.TBPH-RNAi [Maudlin] (ID: 104401), UAS.caz-RNAi [Kellogg] (ID: 100291), UAS.caz-RNAi [Oriel] (ID: 330388), UAS.Sod1-RNAi [Hertford] (ID: 31551), and UAS.Sod1-RNAi [Pembroke] (ID: 108307) were obtained from the Vienna Drosophila Resource Center, Austria120, and were described previously73,121,122. The provenance of the various UAS transgenes for TBPH, hTDP-43, caz, hFUS, Sod1 and hSOD1 is referenced in Table 1. All C9orf72-related transgenic lines were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) at Indiana University, USA and were characterised previously123. Combination of the various genetic tools including GAL4 drivers, alleles, and transgenes was performed according to standard genetic crossing schemes.
Mobility assays
Mobility assays in larvae and adult flies were conducted as described previously67. In brief, third instar larvae were first placed on a 0.7% agar plate. Subsequently, the number of forward body wall contractions exhibited by the organism in 30 seconds were counted. Each larva was assessed three times before an average was taken. To assess climbing performance in adult flies, two empty polystyrene tubes were vertically joined by tape facing each other. Flies (15–20) were then transferred into the lower tube and allowed to acclimatize. Flies were then gently tapped down to the bottom of the vial. The time for the first fly within a group to cross an 8 cm threshold was first measured. Consequently, the number of flies per group, that can climb above the 8 cm mark by 10 seconds, was determined. For each group of flies, four trials were performed. A minimum of four groups were assayed per genotype.
Flight assay
Flight performance was assessed as detailed previously61,67. This assay made use of the Droso-Drome apparatus, which consists of a 1 L glass bottle coated with an alcohol-based sticky fluid, and divided into 4 sectors, of 5 cm each, spanning a total height of 20 cm. In short, flies first underwent a ‘warm-up’ by inducing negative geotaxis in an empty tube for 6 times. Organisms were then dropped into the Droso-Drome to induce flight. The number of flies stuck to each sector was next counted, divided by the total number of flies dropped and multiplied by 100 to generate the percentage number of flies per sector. Fight ability correlates with the height or sector in which flies are distributed, hence, fly percentages that are skewed towards the lower sectors of the Droso-Drome are indicative of reduced flight capacity.
Puparial axial ratios
Length and width of puparia were first measured from still images. As reported previously61,67, calculation of puparial axial ratios involved dividing the length by the width of the puparia.
Immunohistochemistry
The same immunohistochemistry procedures described previously65 were followed. Briefly, body wall muscles of wandering third instar larvae were dissected in phosphate buffered saline (PBS), fixed in 4% paraformaldehyde in PBS and washed in PBS + 0.1% Triton X-100 (PBT). Tissues were then stained overnight at room temperature by mouse anti-Discs large antibody (1:1000; Developmental Studies Hybridoma Bank, University of Iowa, USA). On the following day, tissues were washed in PBT and stained overnight at room temperature with anti-mouse Alexa Fluor 488-conjugated secondary goat antibody (1:50) and Alexa Fluor 546-conjugated Phalloidin (1:50). After a final wash in PBT, the samples were mounted in 90% glycerol with anti-fade. Imaging was performed with Optika B-600TiFL microscope (20x or 40x objectives).
Analysis of muscle size and NMJ morphology
ImageJ software (NIH) was used to quantify both muscle and NMJ area. The former comprised of both ventral longitudinal muscles 6 and 7 derived from abdominal segments 2–4 whereas the latter constituted the postsynaptic region on the same muscles stained by the anti-Discs large antibody. Branch number was determined by counting the number of arborisations containing at least two boutons within a single NMJ. To determine, bouton numbers, all boutons were counted within a single NMJ.
Yeast two-hybrid assays
Two-hybrid assays were performed as described previously67. Briefly, baits and preys were obtained by PCR amplification of cDNA and ligation into the pAS∆∆ and pACT2st vectors, respectively124. Primer sequences and PCR regimes are available upon request. The cDNA clones for Gemin2 (LD47479) and Gemin3 (LD05563) were obtained from the Drosophila Genomics Resource Centre (Indiana University, USA). Gem3ΔN was synthesised as described previously65. The bait pAS∆∆ construct containing the protein sequence fused in frame with the GAL4 DNA binding domain (GAL4-BD) was used to transform the CG1945 strain, which was then selected on –Trp plates. The prey pACT2st construct containing the protein sequence fused in frame with the GAL4 activation domain (GAL4-AD) was used to transform the Y187 strain, which was then selected on –Leu plates. Mating of bait and prey strains was achieved overnight on rich yeast extract peptone dextrose (YPD) plates and –Trp –Leu plates were used to select diploids containing bait/prey combinations. Protein-protein interactions were screened by spotting serial dilutions on –Trp–Leu–His plates. Where indicated, 30 mM 3-amino-1,2,4-triazole (3-AT) was added to the medium to assess interaction strength. Incubations were performed at 28° C for 3 to 5 days. The β-galactosidase assay was used to quantify yeast two-hybrid interactions. Cells were grown in –Trp–Leu selective medium to an OD600 = 0.5–1.0. Activity was measured from extracts as reported previously125.
Statistical analysis
Values are presented as means ± S.E.M. unless otherwise indicated. The unpaired t-test was used to compare measures between 2 groups whereas one-way ANOVA was applied for multiple comparisons with the control. Two-way ANOVA, followed by Bonferroni’s post hoc test, was used to determine differences between 2 groups in the percentage number of fliers (sectors 2–4) vs. non-fliers (sector 1) in the flight assay (GraphPad Prism v8.0.1). Differences were deemed statistically significant if p < 0.05, and when this is the case, the exact p-value is presented.
Supplementary information
Acknowledgements
The authors are indebted to Matthew Camilleri for unwavering technical and administrative support. Thanks also goes to Julia Camilleri for assistance with experiments. We are also very grateful to Aaron Voigt, Magalie Lecourtois, Jane Wu, Brian McCabe, and Fabian Feiguin for fly stocks. This work was supported by the University of Malta Research Fund to RJC, and the Malta Council for Science & Technology Internationalisation Partnership Award to RJC. RC was supported by the Erasmus+ programme of the EU. ML was supported by an Endeavour Scholarship (Malta), part-financed by the EU – European Social Fund under Operational Programme II – Cohesion Policy 2014–2020, “Investing in human capital to create more opportunities and promote the well-being of society”. RMB was supported by a Bjorn Formosa Scholarship for Advanced Research into ALS/MND funded by the non-profit organisation, ALS Malta Foundation, facilitated by the Research Trust (RIDT) of the University of Malta.
Author contributions
R.J.C. and R.B. conceived and designed the experiments. R.C., M.L., J.C., R.M.B. and R.J.C. performed the experiments and data interpretation. R.J.C. wrote the manuscript with contributions from R.B. and N.V. The authors declare that they have no conflict of interest.
Data availability
All data generated or analysed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-53508-4.
References
- 1.Ibrahim F, Nakaya T, Mourelatos Z. RNA dysregulation in diseases of motor neurons. Annu Rev Pathol. 2012;7:323–352. doi: 10.1146/annurev-pathol-011110-130307. [DOI] [PubMed] [Google Scholar]
- 2.Baumer D, Ansorge O, Almeida M, Talbot K. The role of RNA processing in the pathogenesis of motor neuron degeneration. Expert reviews in molecular medicine. 2010;12:e21. doi: 10.1017/S1462399410001523. [DOI] [PubMed] [Google Scholar]
- 3.Ito Daisuke, Hatano Mami, Suzuki Norihiro. RNA binding proteins and the pathological cascade in ALS/FTD neurodegeneration. Science Translational Medicine. 2017;9(415):eaah5436. doi: 10.1126/scitranslmed.aah5436. [DOI] [PubMed] [Google Scholar]
- 4.Butti Z, Patten SA. RNA Dysregulation in Amyotrophic Lateral Sclerosis. Front Genet. 2018;9:712. doi: 10.3389/fgene.2018.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanfranco M, Vassallo N, Cauchi RJ. Spinal Muscular Atrophy: From Defective Chaperoning of snRNP Assembly to Neuromuscular Dysfunction. Front Mol Biosci. 2017;4:41. doi: 10.3389/fmolb.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cauchi RJSMN. and Gemins: ‘we are family’… or are we? Insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. Bioessays. 2010;32:1077–1089. doi: 10.1002/bies.201000088. [DOI] [PubMed] [Google Scholar]
- 8.Donlin-Asp PG, Bassell GJ, Rossoll W. A role for the survival of motor neuron protein in mRNP assembly and transport. Curr Opin Neurobiol. 2016;39:53–61. doi: 10.1016/j.conb.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Brown RH, Al-Chalabi A. Amyotrophic Lateral Sclerosis. N Engl J Med. 2017;377:162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 10.Taylor JP, Brown RH, Jr., Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratti A, Buratti E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J Neurochem. 2016;138(Suppl 1):95–111. doi: 10.1111/jnc.13625. [DOI] [PubMed] [Google Scholar]
- 13.Turner BJ, Parkinson NJ, Davies KE, Talbot K. Survival motor neuron deficiency enhances progression in an amyotrophic lateral sclerosis mouse model. Neurobiol Dis. 2009;34:511–517. doi: 10.1016/j.nbd.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Wang IF, Reddy NM, Shen CK. Higher order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci USA. 2002;99:13583–13588. doi: 10.1073/pnas.212483099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuiji H, et al. Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO Mol Med. 2013;5:221–234. doi: 10.1002/emmm.201202303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki T, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2:799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groen EJ, et al. ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN. Hum Mol Genet. 2013;22:3690–3704. doi: 10.1093/hmg/ddt222. [DOI] [PubMed] [Google Scholar]
- 18.Gerbino V, Carri MT, Cozzolino M, Achsel T, Mislocalised FUS. mutants stall spliceosomal snRNPs in the cytoplasm. Neurobiol Dis. 2013;55:120–128. doi: 10.1016/j.nbd.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Sun S, et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun. 2015;6:6171. doi: 10.1038/ncomms7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin S, et al. Evidence that C9ORF72 Dipeptide Repeat Proteins Associate with U2 snRNP to Cause Mis-splicing in ALS/FTD Patients. Cell Rep. 2017;19:2244–2256. doi: 10.1016/j.celrep.2017.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. Embo J. 1996;15:3555–3565. doi: 10.1002/j.1460-2075.1996.tb00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cauchi RJ. Gem formation upon constitutive Gemin3 overexpression in Drosophila. Cell Biol Int. 2011;35:1233–1238. doi: 10.1042/CBI20110147. [DOI] [PubMed] [Google Scholar]
- 23.Mirra A, et al. Functional interaction between FUS and SMN underlies SMA-like splicing changes in wild-type hFUS mice. Sci Rep. 2017;7:2033. doi: 10.1038/s41598-017-02195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishihara T, et al. Decreased number of Gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22:4136–4147. doi: 10.1093/hmg/ddt262. [DOI] [PubMed] [Google Scholar]
- 25.Workman E, Kolb SJ, Battle DJ. Spliceosomal small nuclear ribonucleoprotein biogenesis defects and motor neuron selectivity in spinal muscular atrophy. Brain Res. 2012;1462:93–99. doi: 10.1016/j.brainres.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, et al. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci USA. 2013;110:19348–19353. doi: 10.1073/pnas.1319280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boulisfane N, et al. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet. 2011;20:641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- 28.Yasuda K, Mili S. Dysregulated axonal RNA translation in amyotrophic lateral sclerosis. Wiley Interdiscip Rev RNA. 2016;7:589–603. doi: 10.1002/wrna.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldwin KR, Godena VK, Hewitt VL, Whitworth AJ. Axonal transport defects are a common phenotype in Drosophila models of ALS. Hum Mol Genet. 2016;25:2378–2392. doi: 10.1093/hmg/ddw105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corcia P, et al. Phenotypic and genotypic studies of ALS cases in ALS-SMA families. Amyotroph Lateral Scler Frontotemporal Degener. 2018;19:432–437. doi: 10.1080/21678421.2018.1440406. [DOI] [PubMed] [Google Scholar]
- 32.Blauw HM, et al. SMN1 gene duplications are associated with sporadic ALS. Neurology. 2012;78:776–780. doi: 10.1212/WNL.0b013e318249f697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corcia P, et al. SMN1 gene, but not SMN2, is a risk factor for sporadic ALS. Neurology. 2006;67:1147–1150. doi: 10.1212/01.wnl.0000233830.85206.1e. [DOI] [PubMed] [Google Scholar]
- 34.Veldink JH, et al. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology. 2005;65:820–825. doi: 10.1212/01.wnl.0000174472.03292.dd. [DOI] [PubMed] [Google Scholar]
- 35.Cauchi RJ. Gem depletion: amyotrophic lateral sclerosis and spinal muscular atrophy crossover. CNS Neurosci Ther. 2014;20:574–581. doi: 10.1111/cns.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Muela N, et al. Single-Cell Analysis of SMN Reveals Its Broader Role in Neuromuscular Disease. Cell Rep. 2017;18:1484–1498. doi: 10.1016/j.celrep.2017.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercuri E, et al. Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N Engl J Med. 2018;378:625–635. doi: 10.1056/NEJMoa1710504. [DOI] [PubMed] [Google Scholar]
- 38.Finkel RS, et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N Engl J Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 39.Turner BJ, et al. Overexpression of survival motor neuron improves neuromuscular function and motor neuron survival in mutant SOD1 mice. Neurobiol Aging. 2013;35:906–915. doi: 10.1016/j.neurobiolaging.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kariya S, et al. Mutant superoxide dismutase 1 (SOD1), a cause of amyotrophic lateral sclerosis, disrupts the recruitment of SMN, the spinal muscular atrophy protein to nuclear Cajal bodies. Hum Mol Genet. 2012;21:3421–3434. doi: 10.1093/hmg/dds174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perera ND, et al. Enhancing survival motor neuron expression extends lifespan and attenuates neurodegeneration in mutant TDP-43 mice. Hum Mol Genet. 2016;25:4080–4093. doi: 10.1093/hmg/ddw247. [DOI] [PubMed] [Google Scholar]
- 42.Aquilina Beppe, Cauchi Ruben J. Modelling motor neuron disease in fruit flies: Lessons from spinal muscular atrophy. Journal of Neuroscience Methods. 2018;310:3–11. doi: 10.1016/j.jneumeth.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Olesnicky Eugenia, Wright Ethan. Drosophila as a Model for Assessing the Function of RNA-Binding Proteins during Neurogenesis and Neurological Disease. Journal of Developmental Biology. 2018;6(3):21. doi: 10.3390/jdb6030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sen A, et al. Genetic circuitry of Survival motor neuron, the gene underlying spinal muscular atrophy. Proc Natl Acad Sci USA. 2013;110:E2371–2380. doi: 10.1073/pnas.1301738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang HC, et al. Modeling spinal muscular atrophy in Drosophila. PLoS ONE. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aquilina B, Cauchi RJ. Genetic screen identifies a requirement for SMN in mRNA localisation within the Drosophila oocyte. BMC Res Notes. 2018;11:378. doi: 10.1186/s13104-018-3496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dimitriadi M, et al. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 2010;6:e1001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freibaum BD, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casci I, Pandey UB. A fruitful endeavor: modeling ALS in the fruit fly. Brain Res. 2015;1607:47–74. doi: 10.1016/j.brainres.2014.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steyaert J, et al. FUS-induced neurotoxicity in Drosophila is prevented by downregulating nucleocytoplasmic transport proteins. Hum Mol Genet. 2018;27:4103–4116. doi: 10.1093/hmg/ddy303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhan L, Hanson KA, Kim SH, Tare A, Tibbetts RS. Identification of genetic modifiers of TDP-43 neurotoxicity in Drosophila. PLoS One. 2013;8:e57214. doi: 10.1371/journal.pone.0057214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borg R, Cauchi RJ. GEMINs: potential therapeutic targets for spinal muscular atrophy? Front Neurosci. 2014;8:325. doi: 10.3389/fnins.2014.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hua Y, et al. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes & development. 2015;29:288–297. doi: 10.1101/gad.256644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee AJ, Awano T, Park GH, Monani UR. Limited phenotypic effects of selectively augmenting the SMN protein in the neurons of a mouse model of severe spinal muscular atrophy. PloS one. 2012;7:e46353. doi: 10.1371/journal.pone.0046353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boyer JG, Ferrier A, Kothary R. More than a bystander: the contributions of intrinsic skeletal muscle defects in motor neuron diseases. Front Physiol. 2013;4:356. doi: 10.3389/fphys.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loeffler JP, Picchiarelli G, Dupuis L, Gonzalez De Aguilar JL. The Role of Skeletal Muscle in Amyotrophic Lateral Sclerosis. Brain Pathol. 2016;26:227–236. doi: 10.1111/bpa.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamilton G, Gillingwater TH. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Nash LA, Burns JK, Chardon JW, Kothary R, Parks RJ. Spinal Muscular Atrophy: More than a Disease of Motor Neurons? Curr Mol Med. 2016;16:779–792. doi: 10.2174/1566524016666161128113338. [DOI] [PubMed] [Google Scholar]
- 60.Curmi F, Cauchi RJ. The multiple lives of DEAD-box RNA helicase DP103/DDX20/Gemin3. Biochem Soc Trans. 2018;46:329–341. doi: 10.1042/BST20180016. [DOI] [PubMed] [Google Scholar]
- 61.Borg RM, Fenech Salerno B, Vassallo N, Bordonne R, Cauchi RJ. Disruption of snRNP biogenesis factors Tgs1 and pICln induces phenotypes that mirror aspects of SMN-Gemins complex perturbation in Drosophila, providing new insights into spinal muscular atrophy. Neurobiol Dis. 2016;94:245–258. doi: 10.1016/j.nbd.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 62.Chan YB, et al. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum. Mol. Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- 63.Rajendra TK, et al. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J Cell Biol. 2007;176:831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borg R, Cauchi RJ. The Gemin Associates of Survival Motor Neuron are Required for Motor Function in Drosophila. PLoS ONE. 2013;8:e83878. doi: 10.1371/journal.pone.0083878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cauchi RJ, Davies KE, Liu JL. A motor function for the DEAD-box RNA helicase, Gemin3, in Drosophila. PLoS Genet. 2008;4:e1000265. doi: 10.1371/journal.pgen.1000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shpargel KB, Praveen K, Rajendra TK, Matera AG. Gemin3 is an essential gene required for larval motor function and pupation in Drosophila. Mol Biol Cell. 2009;20:90–101. doi: 10.1091/mbc.e08-01-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lanfranco M, et al. Novel interactors of the Drosophila Survival Motor Neuron (SMN) Complex suggest its full conservation. FEBS Lett. 2017;591:3600–3614. doi: 10.1002/1873-3468.12853. [DOI] [PubMed] [Google Scholar]
- 68.Borg RM, Bordonne R, Vassallo N, Cauchi RJ. Genetic Interactions between the Members of the SMN-Gemins Complex in Drosophila. PloS one. 2015;10:e0130974. doi: 10.1371/journal.pone.0130974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 70.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 71.Lee EB, Lee VM, Trojanowski JQ. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2011;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voigt A, et al. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS One. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feiguin F, et al. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 74.Ederle H, Dormann D. TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett. 2017;591:1489–1507. doi: 10.1002/1873-3468.12646. [DOI] [PubMed] [Google Scholar]
- 75.Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lanson NA, Jr., et al. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng HX, et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann Neurol. 2010;67:739–748. doi: 10.1002/ana.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 80.Phillips JP, et al. Subunit-destabilizing mutations in Drosophila copper/zinc superoxide dismutase: neuropathology and a model of dimer dysequilibrium. Proc Natl Acad Sci USA. 1995;92:8574–8578. doi: 10.1073/pnas.92.19.8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matera AG, et al. Composition of the Survival Motor Neuron (SMN) Complex in Drosophila melanogaster. G3 (Bethesda) 2019;9:491–503. doi: 10.1534/g3.118.200874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charroux B, et al. Gemin4: a novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hilton JL, Kearney PC, Ames BN. Mode of action of the herbicide, 3-amino-1,2,4-triazole(amitrole): inhibition of an enzyme of histidine biosynthesis. Arch Biochem Biophys. 1965;112:544–547. doi: 10.1016/0003-9861(65)90093-7. [DOI] [PubMed] [Google Scholar]
- 84.Klopotowski T, Wiater A. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Arch Biochem Biophys. 1965;112:562–566. doi: 10.1016/0003-9861(65)90096-2. [DOI] [PubMed] [Google Scholar]
- 85.Winkler C, et al. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu Y, et al. U1 snRNP is mislocalized in ALS patient fibroblasts bearing NLS mutations in FUS and is required for motor neuron outgrowth in zebrafish. Nucleic Acids Res. 2015;43:3208–3218. doi: 10.1093/nar/gkv157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem. 1998;273:27761–27764. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- 88.Buratti E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rogelj B, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Colombrita C, et al. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem. 2012;287:15635–15647. doi: 10.1074/jbc.M111.333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Colombrita C, et al. From transcriptomic to protein level changes in TDP-43 and FUS loss-of-function cell models. Biochim Biophys Acta. 2015;1849:1398–1410. doi: 10.1016/j.bbagrm.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 95.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–655. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Highley JR, et al. Loss of nuclear TDP-43 in amyotrophic lateral sclerosis (ALS) causes altered expression of splicing machinery and widespread dysregulation of RNA splicing in motor neurones. Neuropathol Appl Neurobiol. 2014;40:670–685. doi: 10.1111/nan.12148. [DOI] [PubMed] [Google Scholar]
- 97.van Blitterswijk M, et al. Characterization of FUS mutations in amyotrophic lateral sclerosis using RNA-Seq. PLoS One. 2013;8:e60788. doi: 10.1371/journal.pone.0060788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arnold ES, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci USA. 2013;110:E736–745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qiu H, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hazelett DJ, Chang JC, Lakeland DL, Morton DB. Comparison of parallel high-throughput RNA sequencing between knockout of TDP-43 and its overexpression reveals primarily nonreciprocal and nonoverlapping gene expression changes in the central nervous system of Drosophila. G3 (Bethesda) 2012;2:789–802. doi: 10.1534/g3.112.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chung CY, et al. Aberrant activation of non-coding RNA targets of transcriptional elongation complexes contributes to TDP-43 toxicity. Nat Commun. 2018;9:4406. doi: 10.1038/s41467-018-06543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fratta, P. et al. Mice with endogenous TDP-43 mutations exhibit gain of splicing function and characteristics of amyotrophic lateral sclerosis. EMBO J37, 10.15252/embj.201798684 (2018). [DOI] [PMC free article] [PubMed]
- 103.Reber S, et al. Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 2016;35:1504–1521. doi: 10.15252/embj.201593791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang G, et al. Cytoplasmic mislocalization of RNA splicing factors and aberrant neuronal gene splicing in TDP-43 transgenic pig brain. Mol Neurodegener. 2015;10:42. doi: 10.1186/s13024-015-0036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Coyne AN, Zaepfel BL, Zarnescu DC. Failure to Deliver and Translate-New Insights into RNA Dysregulation in ALS. Front Cell Neurosci. 2017;11:243. doi: 10.3389/fncel.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oka S, Hirai J, Yasukawa T, Nakahara Y, Inoue YH. A correlation of reactive oxygen species accumulation by depletion of superoxide dismutases with age-dependent impairment in the nervous system and muscles of Drosophila adults. Biogerontology. 2015;16:485–501. doi: 10.1007/s10522-015-9570-3. [DOI] [PubMed] [Google Scholar]
- 107.Sahin A, et al. Human SOD1 ALS Mutations in a Drosophila Knock-In Model Cause Severe Phenotypes and Reveal Dosage-Sensitive Gain- and Loss-of-Function Components. Genetics. 2017;205:707–723. doi: 10.1534/genetics.116.190850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Phillips JP, Campbell SD, Michaud D, Charbonneau M, Hilliker AJ. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci USA. 1989;86:2761–2765. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Parkes TL, Kirby K, Phillips JP, Hilliker AJ. Transgenic analysis of the cSOD-null phenotypic syndrome in Drosophila. Genome. 1998;41:642–651. doi: 10.1139/g98-068. [DOI] [PubMed] [Google Scholar]
- 110.Wan L, Ottinger E, Cho S, Dreyfuss G. Inactivation of the SMN complex by oxidative stress. Mol Cell. 2008;31:244–254. doi: 10.1016/j.molcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bozzo F, Mirra A, Carri MT. Oxidative stress and mitochondrial damage in the pathogenesis of ALS: New perspectives. Neurosci Lett. 2017;636:3–8. doi: 10.1016/j.neulet.2016.04.065. [DOI] [PubMed] [Google Scholar]
- 112.Liou GG, Chang HY, Lin CS, Lin-Chao S. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J Biol Chem. 2002;277:41157–41162. doi: 10.1074/jbc.M206618200. [DOI] [PubMed] [Google Scholar]
- 113.Klostermeier D, Rudolph MG. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res. 2009;37:421–430. doi: 10.1093/nar/gkn947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lehnik-Habrink M, et al. The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol Microbiol. 2010;77:958–971. doi: 10.1111/j.1365-2958.2010.07264.x. [DOI] [PubMed] [Google Scholar]
- 115.Skeik, R. M. MSc Thesis: Dimerization of the DEAD-Box Cyanobacterial RNA Helicase Redox, CrhR Master of Science thesis, University of Alberta (2012).
- 116.Ogilvie VC, et al. The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res. 2003;31:1470–1480. doi: 10.1093/nar/gkg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cauchi RJ, van den Heuvel M. The fly as a model for neurodegenerative diseases: is it worth the jump? Neurodegener Dis. 2006;3:338–356. doi: 10.1159/000097303. [DOI] [PubMed] [Google Scholar]
- 118.Ranganayakulu G, Schulz RA, Olson EN. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev Biol. 1996;176:143–148. doi: 10.1006/dbio.1996.9987. [DOI] [PubMed] [Google Scholar]
- 119.Campbell SD, Hilliker AJ, Phillips JP. Cytogenetic analysis of the cSOD microregion in Drosophila melanogaster. Genetics. 1986;112:205–215. doi: 10.1093/genetics/112.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 121.Frickenhaus M, Wagner M, Mallik M, Catinozzi M, Storkebaum E. Highly efficient cell-type-specific gene inactivation reveals a key function for the Drosophila FUS homolog cabeza in neurons. Sci Rep. 2015;5:9107. doi: 10.1038/srep09107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martin I, Jones MA, Grotewiel M. Manipulation of Sod1 expression ubiquitously, but not in the nervous system or muscle, impacts age-related parameters in Drosophila. FEBS Lett. 2009;583:2308–2314. doi: 10.1016/j.febslet.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mizielinska S, Isaacs AM. C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia: gain or loss of function? Curr Opin Neurol. 2014;27:515–523. doi: 10.1097/WCO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fromont-Racine M, Rain JC, Legrain P. Building protein-protein networks by two-hybrid mating strategy. Methods in enzymology. 2002;350:513–524. doi: 10.1016/S0076-6879(02)50982-4. [DOI] [PubMed] [Google Scholar]
- 125.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 126.Kushimura Y, et al. Overexpression of ter94, Drosophila VCP, improves motor neuron degeneration induced by knockdown of TBPH, Drosophila TDP-43. Am J Neurodegener Dis. 2018;7:11–31. [PMC free article] [PubMed] [Google Scholar]
- 127.Xia R, et al. Motor neuron apoptosis and neuromuscular junction perturbation are prominent features in a Drosophila model of Fus-mediated ALS. Mol Neurodegener. 2012;7:10. doi: 10.1186/1750-1326-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miguel L, et al. Accumulation of insoluble forms of FUS protein correlates with toxicity in Drosophila. Neurobiol Aging. 2012;33:1008 e1001–1015. doi: 10.1016/j.neurobiolaging.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 129.Chen Y, et al. Expression of human FUS protein in Drosophila leads to progressive neurodegeneration. Protein Cell. 2011;2:477–486. doi: 10.1007/s13238-011-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Watson MR, Lagow RD, Xu K, Zhang B, Bonini NM. A drosophila model for amyotrophic lateral sclerosis reveals motor neuron damage by human SOD1. J Biol Chem. 2008;283:24972–24981. doi: 10.1074/jbc.M804817200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zografos L, et al. Functional characterisation of human synaptic genes expressed in the Drosophila brain. Biol Open. 2016;5:662–667. doi: 10.1242/bio.016261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.