Abstract
In Brazil, cutaneous leishmaniasis is caused predominantly by L. (V.) braziliensis. The few therapeutic drugs available exhibit several limitations, mainly related to drug toxicity and reduced efficacy in some regions. Miltefosine (MF), the only oral drug available for leishmaniasis treatment, is not widely available and has not yet been approved for human use in Brazil. Our group previously reported the existence of differential susceptibility among L. (V.) braziliensis clinical isolates. In this work, we further characterized three of these isolates of L. (V.) braziliensis chosen because they exhibited the lowest and the highest MF half maximal inhibitory concentrations and were therefore considered less tolerant or more tolerant, respectively. Uptake of MF, and also of phosphocholine, were found to be significantly different in more tolerant parasites compared to the less sensitive isolate, which raised the hypothesis of differences in the MF transport complex Miltefosine Transporter (MT)-Ros3. Although some polymorphisms in those genes were found, they did not correlate with the drug susceptibility phenotype. Drug efflux and compartmentalization were similar in the isolates tested, and amphotericin B susceptibility was retained in MF tolerant parasites, suggesting that increased fitness was also not the basis of observed differences. Transcriptomic analysis revealed that Ros3 mRNA levels were upregulated in the sensitive strain compared to the tolerant ones. Increased mRNA abundance in more tolerant isolates was validated by quantitative PCR. Our results suggest that differential gene expression of the MT transporter complex is the basis of the differential susceptibility in these unselected, naturally occurring parasites.
Keywords: Leishmania braziliensis, Isolates, Miltefosine, Susceptibility, Uptake, RNAseq
Graphical abstract
Highlights
-
•
Brazilian L. (V.) braziliensis isolates vary in mitefosine susceptibility.
-
•
Diminished drug internalization was observed in more tolerant isolates.
-
•
Drug susceptibility did not correlate with SNPs in MT-Ros3 genes.
-
•
Drug efflux and compartmentalization were similar in the isolates tested.
-
•
Increased drug sensitivity is accompanied by Ros3 mRNA upregulation.
1. Introduction
Leishmania (Viannia) braziliensis is the main causative species of cutaneous leishmaniasis (CL) in Brazil with almost 20,000 new cases each year (Banuls et al., 2007; Grimaldi et al., 1989; WHO, 2018). Infections caused by this species can lead to the destructive mucocutaneous manifestation, which is potentially deadly (Burza et al., 2018). Chemotherapy for leishmaniasis in Brazil relies mainly on the pentavalent antimonial meglumine antimoniate, a drug with serious limitations due to toxicity and parenteral administration. Efficacy in CL treatment with this drug has been reported to be as low as 50% in some regions of the country (Chrusciak-Talhari et al., 2011; Machado et al., 2010). The alkylphosphocholine miltefosine (MF) was shown to be active against Leishmania parasites and is currently the most effective oral drug for leishmaniasis treatment (Sunyoto et al., 2018). Initial demonstrations of miltefosine's efficacy for the treatment of visceral leishmaniasis (VL) in India were followed over time by concerns regarding the selection of drug resistant parasites. Resistant clinical isolates have already been described (Srivastava et al., 2017). Furthermore, Leishmania susceptibility to miltefosine was shown to vary, not only between different species but also in isolates of the same species (Bhandari et al., 2012; Espada et al., 2017; Hendrickx et al., 2015; Kumar et al., 2009; Prajapati et al., 2013; Utaile et al., 2013; Yardley et al., 2005).
Resistance to MF in Leishmania has been correlated with reduction in the amount of drug inside the parasite (Deep et al., 2017; Mondelaers et al., 2016; Perez-Victoria et al., 2003a, 2006a, 2006b; Sanchez-Canete et al., 2009; Vacchina et al., 2016). The most well accepted hypothesis for this reduced intracellular drug accumulation is the presence of a defective complex Miltefosine Transporter (MT)-Ros3, the main pathway associated with MF transport (Coelho et al., 2014; Fernandez-Prada et al., 2016; Laffitte et al., 2016b; Mondelaers et al., 2016; Perez-Victoria et al., 2003b, 2006a; Seifert et al., 2007; Shaw et al., 2016). However, a reduction in MF accumulation and susceptibility has also been observed in parasites harboring a normal transport machinery, suggesting that tolerance to the drug could be the result of other mechanisms as well (Bhandari et al., 2012; Coelho et al., 2014; Deep et al., 2017; Obonaga et al., 2014). A recently published review emphasized the diversity of MF tolerance mechanisms described for Leishmania, suggesting that this phenotype could be multifactorial (Hefnawy et al., 2017). For example, MF tolerance has also been attributed to drug efflux (Perez-Victoria et al., 2006b), differences in membrane interactions such as variations in phospholipid content (phosphatidylcholine and phosphatidylethanolamine) of the parasite membrane (Rakotomanga et al., 2007) and increased detox related proteins (Deep et al., 2017).
In Brazil, MF is not yet approved for use in humans, but two clinical trials showed superior efficacy in CL treatment when compared with meglumine antimoniate (Chrusciak-Talhari et al., 2011; Machado et al., 2010). We have, in a previous study, characterized the susceptibility of L. (V.) braziliensis clinical isolates recovered from CL patients in Brazil. The half-maximal effective concentrations (EC50) against these isolates varied 6-fold and 15-fold for promastigotes and intracellular amastigotes, respectively. Interestingly, these isolates had not been exposed to miltefosine previously and were recovered from patients before treatment with other leishmanicidal drugs (Espada et al., 2017).
Aiming to identify the molecular basis involved in the variable tolerance of L. (V.) braziliensis to MF, we chose three isolates from the same geographical region (Bahia, Brazil) with the least or the highest drug tolerances, together with the reference strain of this species, to study drug uptake and compartmentalization and gene expression profiles. Global gene expression analysis revealed differences that could explain the diverse phenotypes of susceptibility and uptake.
2. Materials and methods
2.1. Chemical compounds
BODIPY labeled MF [11-(4′,4′-Difluoro-6′-ethyl-1′,3′,5′,7′-tetramethyl-4′-bora3′a,4′a-diaza-s¬-indacen-2′-yl)-undecylphosphocholine] (MT-EtBDP) was prepared as described (Hornillos et al., 2008). BODIPY-PC [(2-(4,4-Difluoro-5-Methyl-4-Bora-3a,4a-Diaza-s-Indacene-3-Dodecanoyl)-1-Hexadecanoyl-sn-Glycero-3-Phosphocholine] was purchased from Molecular Probes®. MitoTracker™ Deep Red FM was purchased from Invitrogen™ and Lysosomal Staining Reagent - NIR – Cytopainter from Abcam. Amphotericin B was purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.2. Parasites
Parasites were grown in M199 medium supplemented with 10% heat inactivated fetal calf serum (FCS), 0,25% hemin, and 2% male urine at 25 °C. L. (V.) braziliensis reference strain (MHOM/BR/1975/M2903) and three clinical isolates previously characterized for susceptibility to MF by our group (Espada et al., 2017) were used in this study: MHOM/BR/2005/LTCP16012, which is more sensitive to MF, and MHOM/BR/2006/LTCP16907 and MHOM/BR/2009/LTCP19446, which are more tolerant to the drug (Table 1).
Table 1.
Susceptibility of promastigotes and intracellular amastigotes of L. (V.) braziliensis isolates to miltefosine.
| Miltefosinea |
Amphotericin B |
||
|---|---|---|---|
| Promastigotesb |
Amastigotesc |
Promastigotesb |
|
| EC50 (μM)d | EC50 (μM)d | EC50 (nM)d | |
| MHOM/BR/1975/M2903 | 53.5 ± 6.6 | 2.7 ± 0.2 | 45.8 ± 1.4 |
| MHOM/BR/2005/LTCP16012 | 22.9 ± 3.7 | 0.8 ± 0.1 | 50.9 ± 3.0 |
| MHOM/BR/2006/LTCP 16907 | 101.2 ± 6.0 | 3.3 ± 0.4 | 39.7 ± 0.5 |
| MHOM/BR/2009/LTCP 19446 | 90.4 ± 5.2 | 4.2 ± 0.2 | 27.5 ± 1.6 |
Data previously described in Espada et al. (2017).
Activity of miltefosine or amphotericin B determined against late log phase promastigotes, through MTT conversion.
Amastigote assays performed in infected BMDM.
Effective concentrations (EC50 and EC90). Mean ± standard deviation of three independent experiments performed in triplicates.
2.3. Uptake of labeled phosphocholine and miltefosine
Log-phase promastigotes were incubated in HEPES-NaCl buffer (21 mM HEPES, 137 mM NaCl, 5 mM KCl, 0,7 mM NaH2PO4, 6 mM glucose, pH 7.05) containing 0.3% (w/v) BSA and 500 μM phenylmethylsulfonyl fluoride (PMSF) (Sigma Aldrich) for 15 min to inhibit phospholipid catabolism. Next, 10 μM BODIPY-PC or 1 μM MT-EtBDP was added and parasites were incubated for 1 h or 5 min, respectively, at 25 °C. In order to remove the non-internalized labeled molecules, parasites were washed three times with ice-cold HEPES-NaCl containing 0.3% BSA. The parasites were then suspended in PBS for flow cytometry analyses using Guava EasyCyte Mini Flow Cytometer System (Millipore) and 20,000 events per sample were evaluated. Statistical analyses were performed using one-way ANOVA analysis followed by Tukey's multiple comparison tests in Graph Pad Prism 6.0.
2.4. Confocal microscopy
For MT-EtBDP localization and co-localization assays, 5 × 106 log-phase promastigotes were incubated in the presence of 1.5 μM MitoTracker™ Deep Red FM or Lysosomal Satining Reagent - NIR for 30 min and washed twice with PBS. Parasites were then incubated with MT-EtBDP in HEPES-NaCl buffer supplemented with 0.3% BSA and 500 μM PMSF for 5 min and washed three times with HEPES-NaCl containing 0.3% BSA. Parasites were left to adhere to culture plates previously treated with 0.5% poly-lysine for approximately 1 min. Confocal images were captured in a 1024 × 1024 pixel format using a Zeiss LSM 780 confocal laser scanning inverted microscope (Carl Zeiss, Germany). Images were captured with an alpha Plan-Apochromatic 100x/1.46 Oil DIC M27 objective (Zeiss), applying a zoom factor of 2.
Alternatively, for incorporation assays, 5 × 106 log-phase promastigotes were left to adhere to CELLView™ (Greiner bio-one) culture dishes previously treated with 0.1% poly-lysine for approximately 1 min. Non-adhered parasites were removed, and 100 μL of 1 mg/mL PBS-glucose was added. After focusing, video acquisition was started and MT-EtBPD was added to a final concentration of 0.5 μM. Time-series images were acquired in a 512 × 512 pixel format with an image interval of 970 ms for 7 min and 39 s. Image quantification was performed using the ROI tool in the Zen 2011 software (Zeiss, version 11.00.190) and processing with FIJI software (Schindelin et al., 2012). Fluorescence intensity was calculated for each parasite present on focal field and background fluorescence intensity was used as control. Area under the curve calculation followed by t-test analysis were performed using GraphPad Prism 6.0.
2.5. Measurement of residual MF at different time points
Log-phase promastigotes were incubated with 1 μM MF-EtBPD in HEPES-NaCl buffer supplemented with 0.3% BSA and 500 μM PMSF for 5 min. Parasites were washed with HEPES-NaCl 0.3% BSA and 5 × 106 parasites were ressuspended in 1 mg/mL PBS-Glucose for MT-EtBDP uptake quantification. After 1, 2, 24 and 48 h, 5 × 106 parasites were pelleted and the supernatant was recovered. Parasites were washed again with HEPES-NaCl 0.3% BSA to remove non-internalized molecules and the amount of residual fluorescence inside the parasites was evaluated by flow cytometry using Guava EasyCyte Mini Flow Cytometer System. A total of 20,000 events were analyzed. Results were expressed as the fluorescence remaining inside the parasite compared to the initial fluorescence after the initial uptake.
Alternatively, for labeled MF efflux determination, the fluorescence in the recovered supernatant was measured using POLARstar® Omega microplate reader with excitation at 485 nm and emission at 530-10 nm.
2.6. DNA sequencing and analyses
Total genomic DNA was isolated using DNAzol reagent (Invitrogen™). Specific primers for each gene analyzed in this study (Table S1) were used for amplification with Accuprime Taq DNA polymerase system (Invitrogen™) or Platinum® Taq DNA Polymerase High Fidelity (Invitrogen™). Internal and external primers were designed using Primer 3 Software (Koressaar and Remm, 2007; Rozen and Skaletsky, 2000) (Table S1) based on the sequences of MT (LbrM.13.1380 and LbrM.13.1400) and Ros3 (LbrM.32.0580) L. (V.) braziliensis M2904 genes available at TriTrypDB (http://trytripdb.org) (Aslett et al., 2010). For MT genes, each ORF was amplified separately using the primer pair LbrM.13.1380 ORF-F and LbrM.13.1380 ORF-R for the LbrM.13.1380 gene and the pair LbrM.13.1400 ORF-F and LbrM.13.1400 ORF-R for the LbrM.13.1400 gene. Amplified products containing these ORFs were then used as template for further amplification of smaller fragments. For this purpose, an external primer specific for each gene, and an internal primer in common for both genes were used. The amplification with the primer pair LbrM.13.1380 ORF-F (or LbrM.13.1400 ORF-F) and R3 resulted in a 2.4 kb fragment of MT, which has an intersection region with the fragment amplified with LbrM.13.1380 ORF-R (or LbrM.13.1400 ORF-R) and F3 primer pair (1.2 kb fragment). PCR amplified products of each fragment were purified using PureLink® Quick Gel Extraction kit (Invitrogen™) and then each fragment was individually cloned in the pGEM®-T Easy Vector system (Promega).
Nucleotide sequences were determined with Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). Several primers annealing to sequences inside (R1, F1, R2, F2, R3, F5) and outside (M13-F, M13-R, LbrM.13.1380 ORF-F/LbrM.13.1400 ORF-F and LbrM.13.1380 ORF-R/LbrM.13.1400 ORF-R) the cloned inserts were used in order to improve sequencing quality (Table S1).
For Ros3 gene sequencing, the ORF (1092 bp) was directly cloned. Thus, Ros3 was amplified using the primer pair LbrM.32.0580 ORF-F and LbrM.32.0580 ORF-R (Table S1), cloned in pGEM®-T Easy Vector system and sequenced as described. At least 4 individual clones of each clinical isolate were sequenced for each gene.
Single nucleotide polymorphisms (SNPs) analyses were performed using novoSNP (Weckx et al., 2005) and visual inspection. For analyses performed using novoSNP, a score of 20 was chosen as a cutoff. In case of heterozygosis, only polymorphisms present in two or more clones, or in one clone and in the reference strain were validated. For visual inspection, a consensus sequence of each clone of each isolate and reference strain was obtained using Seqman (DNASTAR). These sequences were aligned using MegAlign (DNASTAR) and polymorphisms between sequences were annotated. Again, in case of heterozygosis only polymorphisms present in two or more clones, or in one clone and in the reference strain were validated. In order to obtain a consensus sequence for each isolate, consensus of clones were aligned and nucleotide sequences are available at GenBank under submission IDs: 2166810, 2166809 and 2166797.
2.7. Susceptibility of clinical isolates to amphotericin B (ampB)
The activity of ampB against promastigotes of L. (V.) braziliensis isolates and reference strains was assessed by the MTT assay (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) as previously described (Zauli-Nascimento et al., 2010). Briefly, 2 × 106 log-phase promastigotes were incubated in presence of increasing concentrations of ampB (0–700 nM). After 24 h, cell viability was determined by incubation with MTT followed by optical density (OD) absorbance at 595 nm (referenced at 690 nm). EC50 values were calculated by sigmoidal regression curves using GraphPad Prism 6.0 software.
2.8. RNA preparation
Total RNA from log-phase promastigotes was obtained using RNEasy® mini kit (Quiagen®) following manufacturer specifications. Alternatively, RNAs were extracted with Trizol® according to the manufacturer's instructions and then were purified using the RNEasy mini kit in order to improve RNA purity. RNA samples were then treated with DNAse I Amplification Grade (Invitrogen™) for DNA contamination removal according to manufacturer's instructions.
2.9. RNA sequencing and data analysis
cDNA library preparation and sequencing were performed at Oklahoma Medical Research Foundation NGS Core (Oklahoma, USA). RNA integrity was determined using the Agilent TapeStation 4200 system and Poly(A) enriched stranded cDNA libraries were obtained using Illumina TruSeq Sample Preparation kit (San Diego, CA, USA). Pair-end sequencing (2 × 150bp) was performed on the Illumina HiSeq 3000 platform.
FastQC software was used to evaluate library sequence quality (Andrews, 2010). Illumina adapter removal and trimming was done using the Cutadapt tool in pair-end mode (Martin, 2011). Reads were trimmed for removal of bases with average phred score under 20, and also for reads with less than 20 bases. Bowtie 2 (version 2.3.4) (Langmead and Salzberg, 2012) was used to map sequences against the TriTrypDB L. (V.) braziliensis M2904 genome (release 35). Two mismatches per read were allowed. Aligned libraries were sorted using Samtools (Li et al., 2009). Featurecounts program was then used to determine the number of mapped reads over each CDS coordinate on reference genome (Liao et al., 2014).
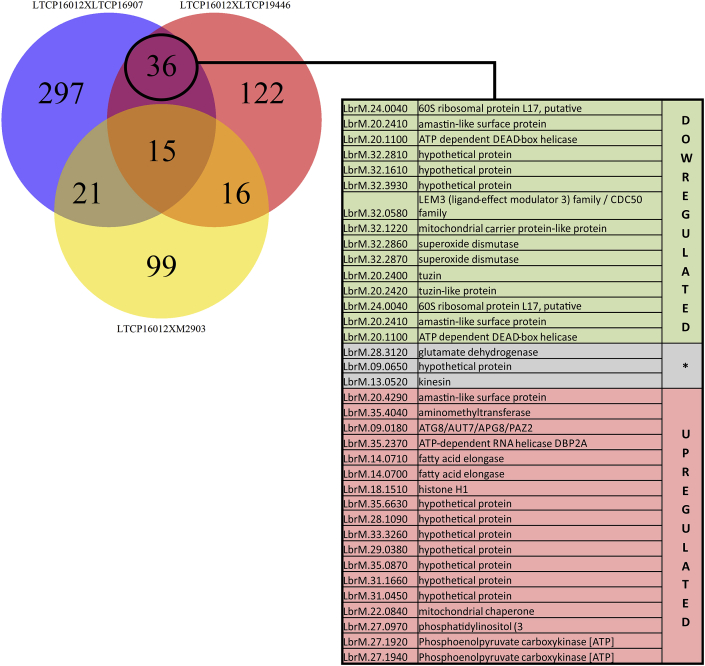
Differential gene expression was assessed using Limma R package (Bioconductor) (Ritchie et al., 2015). The analysis protocol was based on a previous study (Law et al., 2016). Briefly, count tables were loaded on RStudio and transformed to counts per million (CPM). Features harboring CPM values under 1 were filtered out of the analysis. A Multidimensional Scaling (MDS) plot was then generated in order to evaluate the replicate weight for further differential gene expression analysis. TMM (trimmed means of M-value) and voom normalization methods were then applied in order to minimize biases caused by highly expressed genes (heteroscedascity). P-value was adjusted by the Benjamini and Hocherg method (Benjamini and Hochberg, 1995) and genes were considered differentially expressed between isolates when presenting fold-change values above 2 and p-value under 0.05. Comparisons between differentially expressed genes (DEG) in sensitive and tolerant strains and in the reference strain M2903 were identified by the analysis of different pairs (LTCP16012 vs LTCP16907, LTCP16012 vs LTCP19446, LTCP16012 vs M2903). A Venn Diagram was produced indicating the number of genes in each area of intersection (Fig. 5).
Fig. 5.
RNAseq revealed genes differentially expressed in both tolerant isolates compared to the sensitive strain, but absent in comparisons with M2903. Those genes were selected as possibly related to MF tolerance in these L. (V.) braziliensis clinical isolates. The table lists the description of these 36 DEG genes. Genes marked in green are upregulated in the sensitive isolate (LTCP 16012) compared to both tolerant strains (LTCP 16907 and LTCP, 19446). Genes in red are downregulated in the sensitive strain compared to tolerant strains. Genes shown in gray were found to be differentially expressed in these contrasts but not in the same direction, being upregulated in one comparison but down in the other. The detailed list of these 36 DEG containing logFC and p-value information is available as Supplementary Table 4.
2.10. Real time PCR (RT-PCR)
Ros3 differential expression found among the isolates by RNAseq was validated by real-time RT-PCR (qPCR) of this region. RNA used as template for cDNA synthesis was the same RNA used for transcriptome sequencing. Generation of cDNA molecules from RNA was done using MuLV Reverse Transcriptase (Applied Biosystems). Briefly, 6 μg of DNAse treated RNA were incubated with 1 μg Random Primers (Invitrogen™) for 10 min at 4 °C. After this period 5X MulV-RT buffer, 0.1 M DTT, 25 mM MgCl2 and 10 mM dNTPs were added to the system and incubated at 42 °C for 2 min. Reverse Transcriptase was then added to the RT + tubes but not the RT-tubes (control of DNA absence). The reaction was incubated at 25 °C for 10 min, followed by incubation at 48 °C for 30 min and 95 °C for 5 min according to the manufacturer's instructions.
One hundred nanograms of in vitro synthesized cDNA was used as template. qPCR was performed in a StepOne™ Plus System (Applied Bios ystems) using SYBR® Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). The following program was used: 95 °C for 10 min followed by 40 cycles at 95 °C for 15s, 60 °C for 60s, and 72 °C for 20s. Primers used to amplify the target and reference genes were designed using Primer3 (Koressaar and Remm, 2007; Rozen and Skaletsky, 2000) and manufactured by IDT (Integrated DNA Technologies, Inc.). A 148 bp fragment of Ros3 gene was amplified using the primer pair Ros3-F and Ros3-R (Table S1). The housekeeping GAPDH gene was used for normalization and was amplified using the primer pair GAPDH-F and GAPDH-R, which amplifies a 147 bp GAPDH gene fragment (Table S1).
Three biological replicates for each isolate, and three technical replicates of each sample were evaluated for Ros3 and GAPDH mRNA abundance determination. The threshold cycle (Ct) obtained for Ros3 in each sample was normalized by the Ct of the GAPDH gene (reference gene). The 2−ΔΔCt equation was used to determine the relative expression of Ros3 genes in these isolates compared to the reference strain M2903 (Livak and Schmittgen, 2001). 2−ΔΔCt values were then plotted on GraphPad prism 6 and statistical analysis performed using one-way ANOVA analysis followed by Tukey's multiple comparison tests.
3. Results
3.1. Susceptibility of L.(V.) braziliensis clinical isolates to ampB
Aiming to understand if the intrinsic variation of susceptibility to MF observed was exclusive to this drug or a more widespread phenomenon, we evaluated the susceptibility of the clinical isolates used in this study to ampB. Although some degree of variability was found the differences between the isolates’ ampB EC50 were much less pronounced than the 6-fold variations found for MF, varying from 27 to 46 nM. Furthermore, the susceptibility phenotype was inversely correlated (r = −0.80 p = 0.33) for these two drugs: LTCP 16012, the most sensitive isolate for MF, was the least sensitive for ampB, and inversely, LTCP 16907 and LTCP 19446 were more susceptible to ampB (Table 1).
3.2. Uptake of MF inversely correlates to MF susceptibility
The amount of MF accumulated inside the parasite was previously shown to be strictly related to parasite sensitivity to MF, regardless of the Leishmania species and the origin of tolerance to MF (intrinsic or acquired) (Coelho et al., 2014; Deep et al., 2017; Fernandez-Prada et al., 2016; Mondelaers et al., 2016; Perez-Victoria et al., 2003a, 2006a).
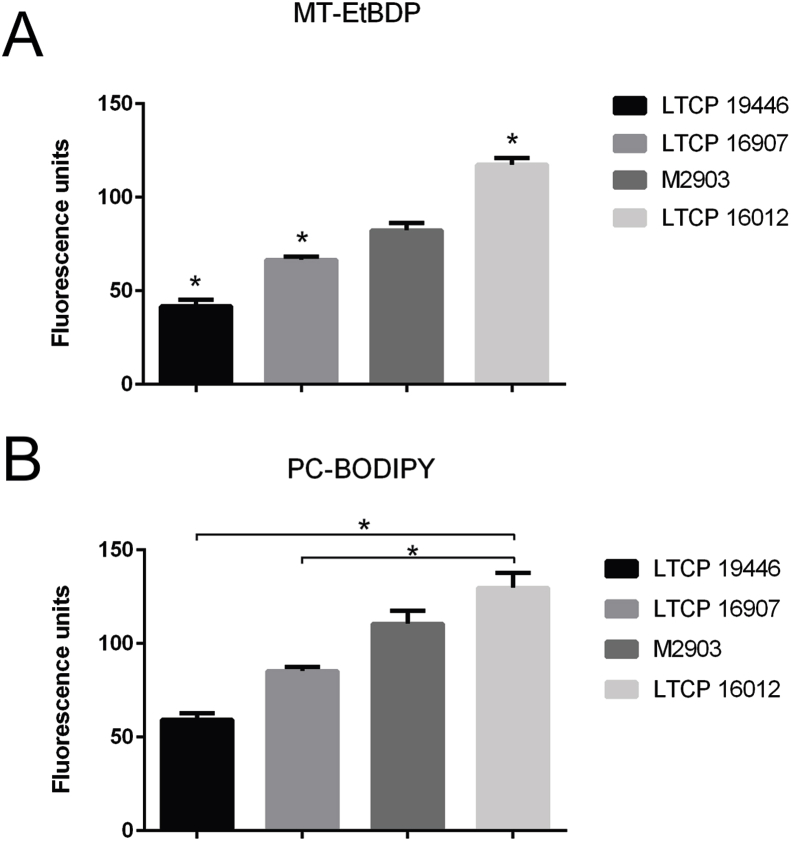
To evaluate whether MF accumulation differed in these isolates, we quantified the uptake of a BODIPY-labeled MF (MT-EtBDP) by flow cytometry in one sensitive (LTCP 16012), two tolerant isolates (LTCP 16907 and LTCP, 19446) and for the reference strain M2903. As MT is a P4-ATPase involved in phospholipid transport in Leishmania parasites (Perez-Victoria et al., 2003b), and MF (hexadecilphosphocholine) is a phosphocholine analogue, we also determined whether BODIPY-PC uptake was impaired in less sensitive isolates.
All clinical isolates and the reference strain revealed differences in MT-EtBDP uptake when compared to each other (Fig. 1). For BODIPY-PC, differential uptake was also observed for all clinical isolates. The uptake of BODIPY-PC by the reference strain was not significantly different from LTCP 16012 and LTCP 16907 (Fig. 1). The amount of MT-EtBDP and BODIPY-PC accumulated inside the parasite was inversely correlated with MF susceptibility (r = −0.80, p = 0.33 and r = −0.70, p = 0.23, respectively) so that LTCP 16012 (EC50 = 22.9 μM) retained both more BODIPY-PC and MT-EtBDP and LTPC 16907 (EC50 = 101.2 μM) and LTCP 19446 (EC50 = 90.4 μM) retained less of these labeled molecules (Fig. 1).
Fig. 1.
Uptake of labeled phosphocholine MT-EtBDP (A) and PC-BODIPY (B) by L. (V.) braziliensis isolates and the reference strain M2903. Isolates were incubated with labeled molecules and fluorescence intensity inside parasite was measured by flow cytometry. Results are representative of three independent experiments and show the mean and standard deviation of fluorescence measured for three technical replicates.
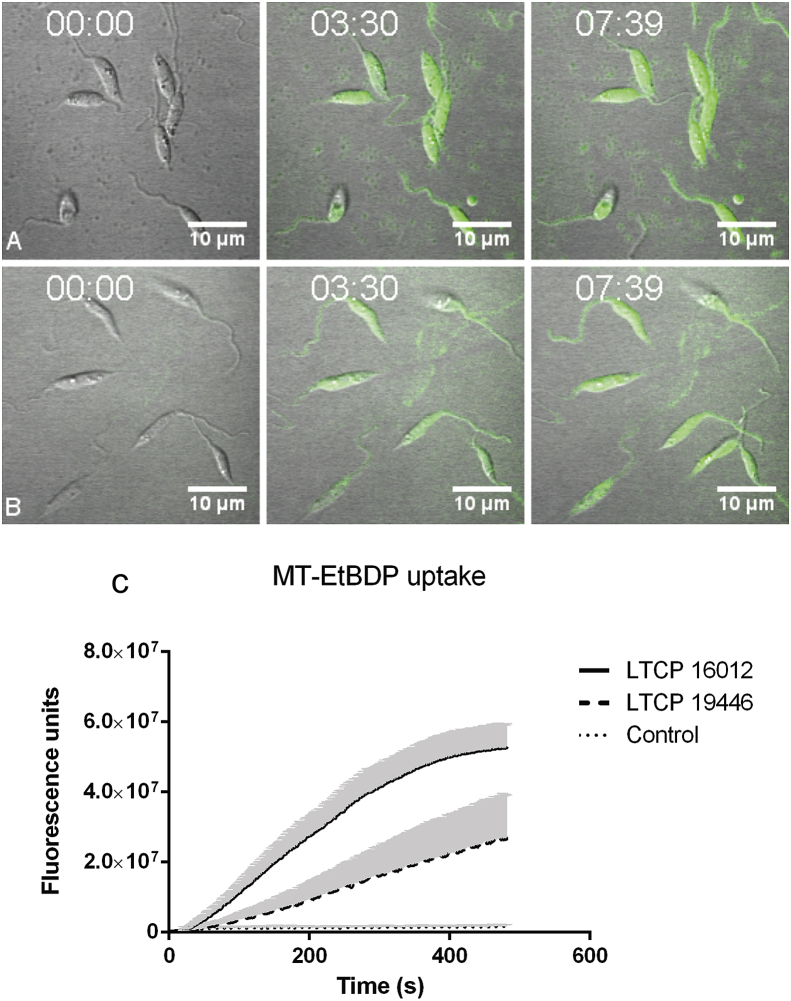
Differential uptake of MT-EtBDP was also observed by confocal microscopy. MT-EtBDP was added to parasites and the incorporation of labeled miltefosine was followed during eight minutes. Fluorescence intensity inside each parasite on the focal field over time was then measured resulting in a MT-EtBDP uptake curve. As previously observed, LTCP 19446 accumulated less MF in all time points evaluated compared to LTCP 16012 (Fig. 2). Area under the curve analyses followed by T test showed significant differences in the amount of intracellular MF and uptake kinetics so that the LTCP 19446 isolate accumulated MF slower and in lower amounts compared to LTCP 16012. Background fluorescence intensity was also measured and did not increase over time (Fig. 2).
Fig. 2.
Labeled miltefosine (MT-EtBDP) uptake evaluated by confocal microscopy. Parasites were adhered to poly-lysine coated plates and MF was added. Uptake was monitored during 8 min and fluorescence intensity over time was calculated. Uptake of MT-EtBDP (green) by L. (V.) braziliensis isolates LTCP 16012 (A) and LTCP 19446 (B) at the time of MT-EtBDP addition and after 3:30 and 7:39 min. The complete video is available as a supplementary file. (C) Measured fluorescence of each parasite in the focal field was plotted for area under the curve (AUC) analysis. Black lines represent the mean and gray shade represents the standard deviation of fluorescence intensity measured for each isolate population (LTCP 16012 n = 17 and LTCP, 19446 n = 11). Small dotted line represents the mean and SD of fluorescence units measured for background in different regions of the plate.
3.3. Measurement of residual intracellular MF at different time points
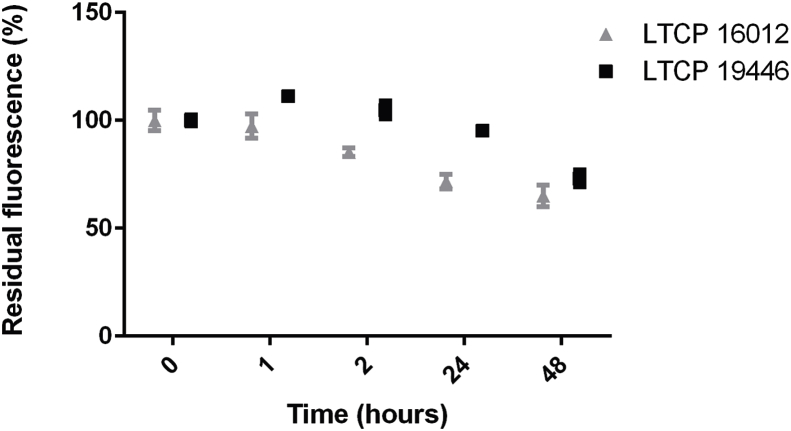
The efflux of MF has also been pointed out as a possible mechanism of resistance in Leishmania parasites (Perez-Victoria et al., 2006b). In order to evaluate MF remaining inside parasites after internalization, parasites were loaded with MT-EtBDP and successive measurements of fluorescence retained in the cell bodies were done by flow cytometry at 1 h, 2 h, 24 h and 48 h for LTCP 16012 and LTCP 19446 isolates (Fig. 3). At point 0 h, just after uptake, the isolates demonstrated the differential uptake of MF described above. During the follow-up, no significant differences in MF retention were observed. After 48 h, both isolates presented a 25% reduction in MT-EtBDP fluorescence compared to the intracellular drug detected at time 0, suggesting that not the efflux but the differential intake might be the reason for the differential susceptibility to MF (Fig. 3). Fluorescence in the culture supernatant was not detected. These data suggested that the drug metabolism was similar among these isolates.
Fig. 3.
Residual fluorescence inside L. braziliensis isolates LTCP 16012 and LTCP 19446 was assessed by flow cytometry. After initial uptake determination, fluorescence associated with cell bodies was measured after 1 h, 2 h, 24 h and 48 h. Fluorescence intensity at each point time was normalized by the initial uptake of MT-EtBDP since these isolates showed differences in drug uptake. Each point represents mean and SD of three technical replicates and the graph show a representative experiment of three.
3.4. Intracellular MF localization in L.(V.) braziliensis clinical isolates
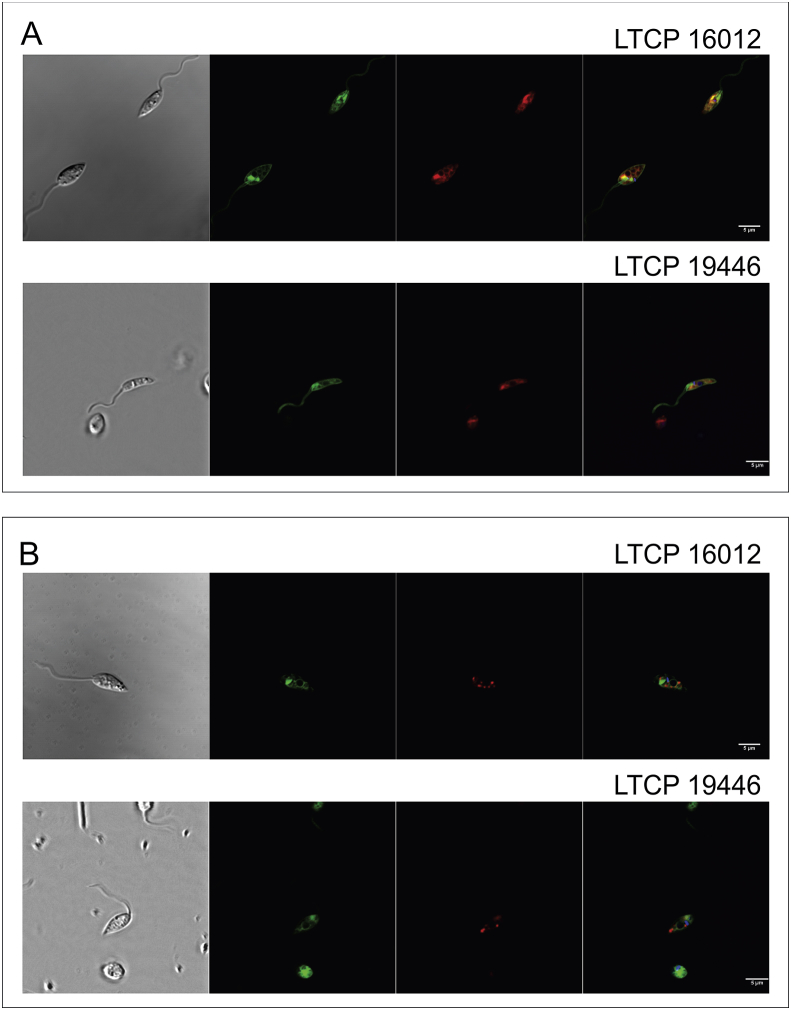
In order to evaluate if differential susceptibility could be a result of MF storage in different cell compartments in more and less sensitive isolates, MF localization inside these parasites was evaluated (Fig. 4). MT-EtBDP location was similar in both isolates (LTCP 16012 and LTCP 19446), being concentrated mainly in the anterior portion of the parasites' cell body. The labeling pattern, close to the flagellum and kinetoplast, and lateral to the nucleus suggested that MF could be retained in organelles such as the mitochondrion and multi-vesicular tubule. Labeling parasites simultaneously with Mitotracker and MT-EtBDP revealed a partial co-localization of these two fluorescent-labeled molecules, suggesting that MF partially localized to the mitochondrion (Fig. 4A). On the other hand, simultaneous labeling with Lysonir (marker for acidic compartments) and MT-EtBPD did not indicate a co-localization, suggesting that MF do not accumulate in the parasite's multi-vesicular tube (Fig. 4B).
Fig. 4.
Localization of MT-EtBDP in L. braziliensis promastigotes was analyzed by confocal microscopy. For both isolates, the same pattern of MT-EtBDP labeling (green) was observed (A and B) being mostly concentrated in the anterior portion of parasite, near the flagellar pocket and around nucleus. For co-localization assays, parasites were initially labeled with Hoechst (blue), (A) Mitotracker Deep Red FM (red) or (B) Lysonyr (red). After incubation with MT-EtBDP co-localization was determined by overlapping images obtained for each labeling.
3.5. Nucleotide sequencing of the genes encoding the MT-Ros3 complex
Since the differential MF susceptibility found in these L. (V.) braziliensis clinical isolates was shown to be correlated to differential drug uptake, we characterized the genes encoding the MF transporter. In L. (V.) braziliensis, an extra copy of the MT gene (LbrM13.1380) is present (LbrM.13.1400). These two copies exhibit 99% nucleotide identity. Both copies of the MT and the Ros3 genes were sequenced. We found polymorphisms between the M2903 sequence obtained and data available for the M2904 type strain (http://tritrypdb.org/tritrypdb/) as well as polymorphisms between the isolates (Table S2). Analyses of LbrM.13.1380, the orthologue of L. (L.) donovani MT revealed 32 polymorphisms among the analyzed sequences, of which 9 were synonymous and 23 non-synonymous substitutions (Table S2). A similar number of polymorphisms was observed for the paralogue of this gene (LbrM.13.1400), with substitutions of which 13 were synonymous and 26 non-synonymous (Table S2). Despite the high number of non-synonymous polymorphisms found between the sequences, none of them could justify the differential susceptibility and uptake of MF found in these clinical isolates, since these variations were found simultaneously in a less susceptible isolate and a more susceptible or reference strains, or were not present in both less susceptible clinical isolates at the same time.
As an efficient MF transport in Leishmania depends not only on MT but also on the Ros3 protein (Perez-Victoria et al., 2006a), we also determined the nucleotide sequence of the LbrM.32.0580 gene. The analysis revealed 6 polymorphisms between the sequences of the three isolates, and of the M2903 and M2904 reference strains, of which 3 were synonymous and 3 were non-synonymous substitutions (Table S2). None of the variations observed could be correlated with the different phenotypes of MF susceptibility and uptake.
3.6. Transcriptome analysis reveals DEG between sensitive and tolerant isolates
In order to identify potential genes involved in the differential phenotype of susceptibility to MF, whole transcriptome was performed in log-phase promastigotes of sensitive and tolerant isolates.
After library trimming, reads were mapped to the M2904 sequence and all samples presented alignment rates above 94%. The number of reads aligning to each feature was counted and 65 genes out of the 8176 annotated were filtered out for having CPM <1. Normalized libraries were then analyzed for differential gene expression discovery. Only DEG presenting fold change above 2 and p-value under 0.05 were considered. Within these conditions, the greatest number of DEG was found for the comparison between LTCP 16907 and the reference strain M2903 followed by the comparison between LTCP 16012 and LTCP 16907, which presented 432 and 369 DEG events, respectively (Table S3). On the other hand, the least number of DEG was found in the comparison between LTCP 16012 and the reference strain M2903 with 151 genes differentially expressed. The number of DEG revealed in the other comparisons is described in Table S3. The complete data set listing the DEGs found in all pairwise comparisons is presented in Tables S4–S9.
A Venn diagram was generated with the comparisons between the more tolerant isolate LTCP 16012 with all the others. The aim was to identify the cluster differentially expressed when LTCP 16012 was compared with both LTCP 16907 and LTCP 19446 (less tolerant) but not with the type strain M2903 (Fig. 5, Table S10). We found 36 DEG clustering between sensitive and tolerant isolates. Except for three genes, the majority of DEG identified were in accordance between the comparisons, being up or downregulated in both tolerant isolates when compared to sensitive isolates (Fig. 5).
Interestingly, Ros3 mRNA was found upregulated in the sensitive isolate compared to the tolerant isolates. When we compared LTCP 16012 with both LTCP 16907 and LTCP 19446, Ros3 appeared 2.05 and 2.14-fold upregulated in the sensitive isolate when compared to LTCP 16907 and LTCP 19446 tolerant isolates, respectively.
The other 35 DEG (Table S10) observed in this cluster are under investigation for genes potentially related to MF susceptibility and uptake in these isolates.
3.7. Validation of Ros3 as a potential gene involved in MF susceptibility
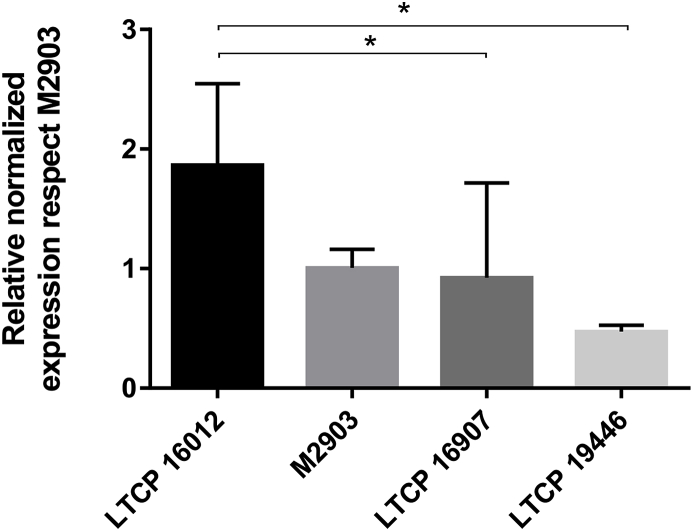
In order to validate Ros3 differential expression we quantified Ros3 mRNA by qPCR. The housekeeping GAPDH gene was used for normalization purposes. Relative expression of Ros3 mRNA was determined adopting M2903 strain as a reference. Data obtained for LTCP 16907 did not show clear differences in reference to the type strain M2903, due to sample heterogeneity. However, the profile of Ros3 expression observed in RNAseq experiments was confirmed using the qPCR approach. Ros3 mRNA abundance was higher in LTCP 16012 compared to M2903 and LTCP 16907 (approximately 2-fold). The comparison between LTCP 16012 and LTCP19446 revealed a more pronounced difference with an approximately 4-fold increase in the more sensitive isolate (Fig. 6).
Fig. 6.
Relative abundance of Ros3 mRNA normalized by GAPDH mRNA expression for each L. (V.) braziliensis isolate compared to M2903. mRNA quantification was assessed by qRT-PCR and normalized using the 2ΔΔCt method. Three biological replicates of each isolate were evaluated in three technical replicates of each sample.
4. Discussion
In this work, we investigated whether the mechanisms previously described as responsible for MF resistance were involved in the differential susceptibility to this drug observed amongst L. (V.) braziliensis clinical isolates. For this purpose, three L. (V.) braziliensis isolates were chosen based on their geographical origin and polar tolerance to miltefosine (the highest and the lowest from Bahia state in a panel of 17 previously characterized isolates). One more sensitive (LTCP 16012), two more tolerant (LTCP 1607 and LTCP, 19446) isolates and a reference strain for this species were used.
The uptake of labeled MF (MT-EtBDP) and phosphocholine (PC-BODIPY) were evaluated in these parasites by two different methodologies. Flow cytometry results showed that both, MT-EtBDP and PC-BODIPY were differentially accumulated by sensitive and tolerant parasites and this uptake was inversely correlated to MF susceptibility. The kinetics of drug entry in the isolates LTCP 16012 and LTCP 19946 led to a slower and less pronounced accumulation. Therefore, MF internalization and retention inside the parasite was strictly correlated to MF susceptibility. Similar results were previously described for Peruvian L. (V.) braziliensis isolates (Sanchez-Canete et al., 2009), which presented 4-fold differences in internalization rates, despite the less striking difference in susceptibility observed (EC50 for promastigotes varying between 80 and 140 μM) (Sanchez-Canete et al., 2009). Differences in MF internalization causing susceptibility variation were also observed in other Leishmania species such as L. (L.) donovani (Deep et al., 2017) and L. (L.) infantum (Fernandez-Prada et al., 2016; Laffitte et al., 2016a; Mondelaers et al., 2016).
Since the MT-Ros3 complex is responsible for the inward transport of phospholipids in Leishmania, the finding that PC internalization was also reduced in tolerant isolates suggested that the intrinsic differences in these parasites could be related to this complex. Variation in PC uptake was also observed in L. (L.) amazonensis selected by MF drug pressure (Coelho et al., 2014). Together, our findings showed an intrinsic difference in MF susceptibility among these isolates (since they were never exposed to MF) that was correlated to the amount of MF these parasites were able to internalize. Additionally, the differences in PC internalization suggested that the mechanism involved in these phenotypes was responsible for both, PC and MF transport in L. (V.) braziliensis.
It has already been shown that impairment of the MT-Ros3 complex causes alterations in membrane composition and symmetry (Fernandez-Prada et al., 2016; Weingartner et al., 2010). Moreover, change in membrane phospholipid composition was observed in Leishmania parasites after treatment with MF, with an increase in phosphatidylethanolamine and reduction in phosphatidylcholine levels due to a reduction in choline uptake (Rakotomanga et al., 2007). All these findings led us to consider the MT-Ros3 complex as the cause of the differences in susceptibility and uptake of MF.
MF resistance has been strongly correlated with MT and/or Ros3 gene mutations, causing significant reduction in MF transport and thus increasing the tolerance of the parasite to this drug (Coelho et al., 2014; Fernandez-Prada et al., 2016; Laffitte et al., 2016a; Mondelaers et al., 2016; Perez-Victoria et al., 2003b; Seifert et al., 2007; Shaw et al., 2016). Mutations in genes encoding this complex have even been related to a reduction in sensitivity to another leishmanicidal drug, ampB, due to changes in membrane lipid composition (Fernandez-Prada et al., 2016). The determination of MT-Ros3 coding sequences in these isolates revealed a number of polymorphisms in the MT genes but without a pattern that could be assigned to more tolerant or sensitive isolates. Moreover, the classical hotspots related to MF resistance caused by MT mutations in other Leishmania species were not observed in these isolates (Bhandari et al., 2012; Coelho et al., 2012; Cojean et al., 2012; Mondelaers et al., 2016; Perez-Victoria et al., 2003a). Therefore, we were not able to correlate differences in susceptibility and uptake to the polymorphisms found among these isolates.
The differences in susceptibility in these isolates could be also related to an increased fitness of tolerant parasites as described as a MF resistance mechanism in L. (L.) donovani (Deep et al., 2017). Such changes would be apparent when parasites were submitted to other challenges and that was tested determining ampB activity against these isolates. A small degree of variability was observed in the susceptibility to ampB in these isolates but without significance or correlation with the differences in MF susceptibility. Similar findings were described by Mondelaers et al. (2018) on MF resistant L. infantum selected in vitro and/or recovered from patients that retained ampB susceptibility.
Drug efflux is a well-known drug-resistance mechanism in Leishmania described mainly in association with pentavalent antimonials (Hefnawy et al., 2017) but that has also been correlated with MF resistance (Perez-Victoria et al., 2006b). The analysis of fluorescence decay in parasites loaded with MF indicated that both sensitive and tolerant isolates presented the same reduction in fluorescence units, suggesting that efflux was not different in these parasites. It should be stressed that, as we were not able to measure MF externalized from the parasite, we cannot ensure whether the reduction in MF inside the parasite over time was a result of efflux or of drug metabolism. Nevertheless, the decay in MF resulting from efflux and/or drug metabolism does not seem to be the cause of the differential susceptibility in these isolates.
Confocal microscopy of MT-EtBDP labeled LTCP 16012 and LTCP 19446 isolates revealed that MF localizes partially to the mitochondrion. In addition, the MF localization did not differ among these isolates. MF partial co-localization with the mitochondrion is an expected finding since the mechanism of MF's action involves cytochrome C oxidase inhibition resulting in mitochondrial depolarization (Luque-Ortega and Rivas, 2007; Santa-Rita et al., 2004).
The analysis of the transcriptome of the study isolates was then employed as a tool to identify the reasons accounting for the unequivocal differences in drug accumulation observed. Among the DEG found in the comparisons of tolerant and sensitive parasites, Ros3 stood out. The Ros3 gene encodes the beta-subunit of the MF-Ros3 complex, and although a higher abundance of Ros3-mRNA was found in the more sensitive strain, the same was not observed for MT transcripts. This is somewhat surprising since the complex seems to act as a whole with mutually dependent components. However, similar results were observed by Sanchez-Canete et al. (2009) in L. (V.) braziliensis Peruvian isolates, in which Ros3 was shown to act as a limiting factor in L. (V.) braziliensis MT-Ros3 transport rate. Ros3 gene overexpression was enough to increase both, MT and Ros3 levels in the parasite membrane (Sanchez-Canete et al., 2009) and could explain why MT transcripts were not found upregulated in these isolates.
This study was inspired by the urgency in making available new agents for the treatment of Brazilian tegumentary leishmaniasis caused by L. (V.) braziliensis. Two clinical trials conducted in Brazil found approximately 70% cure rates in CL patients treated with MF. It is not yet known if parasite susceptibility was a playing factor in failure and that will be an important investigation to be pursued. Further studies are necessary in order to evaluate whether or not these differences in in vitro susceptibility reflect different cure rates. Furthermore, our results again point to the need of new molecules for Leishmania parasites and molecule modifications in MF in order to overcome the MT-Ros3 complex essentiality for MF entrance in Leishmania parasites. We should however stress that one of the limitations of this study is that only three natural isolates were evaluated, and from only one region in Brazil. There is therefore a clear need for a functional evaluation of the pathways studied here in a larger number of isolates.
In conclusion, reduced MF susceptibility in Brazilian L. (V.) braziliensis was found to be related to decreased drug accumulation. Decreased transport seems to be caused by reduced Ros3 mRNA expression rather than polymorphisms in MT-Ros3 complex coding genes. The data described here illustrates the need for parasite susceptibility studies as a recommendation before drug implementation. New molecules and modifications in MF that could overcome MT-Ros3 dependence are highly encouraged by our findings.
Declaration of competing interests
No competing interests.
Funding
This work was supported by research grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2011/20484-7 and 2015/09080-2) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, 473343/2012-6), Brazil. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. SRBU is the recipient of a senior researcher scholarship from CNPq. ACC and CRE were fellows supported by FAPESP (2012/14629-5 and 2016/23405-4).
Acknowledgments
We acknowledge the contribution of Dr. A.U.Acuña (CSIC, Madrid, Spain) to the design and preparation of MF fluorescent analogues. We are grateful to Dr. Mauro Javier Cortez Veliz for helping with microscopy and Carmen S. A. Takata for performing Sanger sequencing. The authors also thank Jenicer K. U. Yokoyama-Yasunaka for technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2019.02.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Andrews S. 2010. FASTQC. A quality control tool for high throughput sequence data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc Available at. [Google Scholar]
- Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M., Depledge D.P., Fischer S., Gajria B., Gao X., Gardner M.J., Gingle A., Grant G., Harb O.S., Heiges M., Hertz-Fowler C., Houston R., Innamorato F., Iodice J., Kissinger J.C., Kraemer E., Li W., Logan F.J., Miller J.A., Mitra S., Myler P.J., Nayak V., Pennington C., Phan I., Pinney D.F., Ramasamy G., Rogers M.B., Roos D.S., Ross C., Sivam D., Smith D.F., Srinivasamoorthy G., Stoeckert C.J., Jr., Subramanian S., Thibodeau R., Tivey A., Treatman C., Velarde G., Wang H. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuls A.L., Hide M., Prugnolle F. Leishmania and the leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv. Parasitol. 2007;64:1–109. doi: 10.1016/S0065-308X(06)64001-3. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Bhandari V., Kulshrestha A., Deep D.K., Stark O., Prajapati V.K., Ramesh V., Sundar S., Schonian G., Dujardin J.C., Salotra P. Drug susceptibility in Leishmania isolates following miltefosine treatment in cases of visceral leishmaniasis and post kala-azar dermal leishmaniasis. PLoS Neglected Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burza S., Croft S.L., Boelaert M. Leishmaniasis. Lancet. 2018;392:951–970. doi: 10.1016/S0140-6736(18)31204-2. [DOI] [PubMed] [Google Scholar]
- Chrusciak-Talhari A., Dietze R., Chrusciak Talhari C., da Silva R.M., Gadelha Yamashita E.P., de Oliveira Penna G., Lima Machado P.R., Talhari S. Randomized controlled clinical trial to access efficacy and safety of miltefosine in the treatment of cutaneous leishmaniasis Caused by Leishmania (Viannia) guyanensis in Manaus, Brazil. Am. J. Trop. Med. Hyg. 2011;84:255–260. doi: 10.4269/ajtmh.2011.10-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho A.C., Boisvert S., Mukherjee A., Leprohon P., Corbeil J., Ouellette M. Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Neglected Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho A.C., Trinconi C.T., Costa C.H., Uliana S.R. In vitro and in vivo miltefosine susceptibility of a Leishmania amazonensis isolate from a patient with diffuse cutaneous leishmaniasis. PLoS Neglected Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojean S., Houze S., Haouchine D., Huteau F., Lariven S., Hubert V., Michard F., Bories C., Pratlong F., Le Bras J., Loiseau P.M., Matheron S. Leishmania resistance to miltefosine associated with genetic marker. Emerg. Infect. Dis. 2012;18:704–706. doi: 10.3201/eid1804.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deep D.K., Singh R., Bhandari V., Verma A., Sharma V., Wajid S., Sundar S., Ramesh V., Dujardin J.C., Salotra P. Increased miltefosine tolerance in clinical isolates of Leishmania donovani is associated with reduced drug accumulation, increased infectivity and resistance to oxidative stress. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada C.R., Ribeiro-Dias F., Dorta M.L., de Araujo Pereira L.I., de Carvalho E.M., Machado P.R., Yokoyama-Yasunaka J.K., Coelho A.C., Uliana S.R. Susceptibility to miltefosine in Brazilian clinical isolates of Leishmania (Viannia) braziliensis. Am. J. Trop. Med. Hyg. 2017;96:656–659. doi: 10.4269/ajtmh.16-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Prada C., Vincent I.M., Brotherton M.C., Roberts M., Roy G., Rivas L., Leprohon P., Smith T.K., Ouellette M. Different mutations in a P-type ATPase transporter in Leishmania parasites are associated with cross-resistance to two leading drugs by distinct mechanisms. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0005171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Jr., Tesh R.B., McMahon-Pratt D. A review of the geographic distribution and epidemiology of leishmaniasis in the New World. Am. J. Trop. Med. Hyg. 1989;41:687–725. doi: 10.4269/ajtmh.1989.41.687. [DOI] [PubMed] [Google Scholar]
- Hefnawy A., Berg M., Dujardin J.C., De Muylder G. Exploiting knowledge on Leishmania drug resistance to support the quest for new drugs. Trends Parasitol. 2017;33:162–174. doi: 10.1016/j.pt.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Hendrickx S., Eberhardt E., Mondelaers A., Rijal S., Bhattarai N.R., Dujardin J.C., Delputte P., Cos P., Maes L. Lack of correlation between the promastigote back-transformation assay and miltefosine treatment outcome. J. Antimicrob. Chemother. 2015;70:3023–3026. doi: 10.1093/jac/dkv237. [DOI] [PubMed] [Google Scholar]
- Hornillos V., Carrillo E., Rivas L., Amat-Guerri F., Acuna A.U. Synthesis of BODIPY-labeled alkylphosphocholines with leishmanicidal activity, as fluorescent analogues of miltefosine. Bioorg. Med. Chem. Lett. 2008;18:6336–6339. doi: 10.1016/j.bmcl.2008.10.089. [DOI] [PubMed] [Google Scholar]
- Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kumar D., Kulshrestha A., Singh R., Salotra P. In vitro susceptibility of field isolates of Leishmania donovani to Miltefosine and amphotericin B: correlation with sodium antimony gluconate susceptibility and implications for treatment in areas of endemicity. Antimicrob. Agents Chemother. 2009;53:835–838. doi: 10.1128/AAC.01233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte M.C., Leprohon P., Legare D., Ouellette M. Deep-sequencing revealing mutation dynamics in the miltefosine transporter gene in Leishmania infantum selected for miltefosine resistance. Parasitol. Res. 2016;115:3699–3703. doi: 10.1007/s00436-016-5195-y. [DOI] [PubMed] [Google Scholar]
- Laffitte M.N., Leprohon P., Papadopoulou B., Ouellette M. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Research. 2016;5:2350. doi: 10.12688/f1000research.9218.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C., Alhamdoosh M., Su S., Smyth G., Ritchie M. 2016. RNA-seq Analysis Is Easy as 1-2-3 with Limma, Glimma and edgeR [version 2; Referees: 3 Approved]. F1000Research; p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luque-Ortega J.R., Rivas L. Miltefosine (hexadecylphosphocholine) inhibits cytochrome c oxidase in Leishmania donovani promastigotes. Antimicrob. Agents Chemother. 2007;51:1327–1332. doi: 10.1128/AAC.01415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado P.R., Ampuero J., Guimaraes L.H., Villasboas L., Rocha A.T., Schriefer A., Sousa R.S., Talhari A., Penna G., Carvalho E.M. Miltefosine in the treatment of cutaneous leishmaniasis caused by Leishmania braziliensis in Brazil: a randomized and controlled trial. PLoS Neglected Trop. Dis. 2010;4 doi: 10.1371/journal.pntd.0000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. [Google Scholar]
- Mondelaers A., Hendrickx S., Van Bockstal L., Maes L., Caljon G. Miltefosine-resistant Leishmania infantum strains with an impaired MT/ROS3 transporter complex retain amphotericin B susceptibility. J. Antimicrob. Chemother. 2018;73:392–394. doi: 10.1093/jac/dkx407. [DOI] [PubMed] [Google Scholar]
- Mondelaers A., Sanchez-Canete M.P., Hendrickx S., Eberhardt E., Garcia-Hernandez R., Lachaud L., Cotton J., Sanders M., Cuypers B., Imamura H., Dujardin J.C., Delputte P., Cos P., Caljon G., Gamarro F., Castanys S., Maes L. Genomic and molecular characterization of miltefosine resistance in Leishmania infantum strains with either natural or acquired resistance through experimental selection of intracellular amastigotes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obonaga R., Fernandez O.L., Valderrama L., Rubiano L.C., Castro Mdel M., Barrera M.C., Gomez M.A., Gore Saravia N. Treatment failure and miltefosine susceptibility in dermal leishmaniasis caused by Leishmania subgenus Viannia species. Antimicrob. Agents Chemother. 2014;58:144–152. doi: 10.1128/AAC.01023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria F.J., Castanys S., Gamarro F. Leishmania donovani resistance to miltefosine involves a defective inward translocation od the drug. Antimicrob. Agents Chemother. 2003;47:2397–2403. doi: 10.1128/AAC.47.8.2397-2403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria F.J., Gamarro F., Ouellette M., Castanys S. Functional cloning of the miltefosine transporter. A novel P-type phospholipid translocase from Leishmania involved in drug resistance. J. Biol. Chem. 2003;278:49965–49971. doi: 10.1074/jbc.M308352200. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria F.J., Sanchez-Canete M.P., Castanys S., Gamarro F. Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J. Biol. Chem. 2006;281:23766–23775. doi: 10.1074/jbc.M605214200. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria J.M., Cortes-Selva F., Parodi-Talice A., Bavchvarov B.I., Perez-Victoria F.J., Munoz-Martinez F., Maitrejean M., Costi M.P., Barron D., Di Pietro A., Castanys S., Gamarro F. Combination of suboptimal doses of inhibitors targeting different domains of LtrMDR1 efficiently overcomes resistance of Leishmania spp. to Miltefosine by inhibiting drug efflux. Antimicrob. Agents Chemother. 2006;50:3102–3110. doi: 10.1128/AAC.00423-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati V.K., Sharma S., Rai M., Ostyn B., Salotra P., Vanaerschot M., Dujardin J.C., Sundar S. In vitro susceptibility of Leishmania donovani to miltefosine in Indian visceral leishmaniasis. Am. J. Trop. Med. Hyg. 2013;89:750–754. doi: 10.4269/ajtmh.13-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotomanga M., Blanc S., Gaudin K., Chaminade P., Loiseau P.M. Miltefosine affects lipid metabolism in Leishmania donovani promastigotes. Antimicrob. Agents Chemother. 2007;51:1425–1430. doi: 10.1128/AAC.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sanchez-Canete M.P., Carvalho L., Perez-Victoria F.J., Gamarro F., Castanys S. Low plasma membrane expression of the miltefosine transport complex renders Leishmania braziliensis refractory to the drug. Antimicrob. Agents Chemother. 2009;53:1305–1313. doi: 10.1128/AAC.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Rita R.M., Henriques-Pons A., Barbosa H.S., de Castro S.L. Effect of the lysophospholipid analogues edelfosine, ilmofosine and miltefosine against Leishmania amazonensis. J. Antimicrob. Chemother. 2004;54:704–710. doi: 10.1093/jac/dkh380. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K., Perez-Victoria F.J., Stettler M., Sanchez-Canete M.P., Castanys S., Gamarro F., Croft S.L. Inactivation of the miltefosine transporter, LdMT, causes miltefosine resistance that is conferred to the amastigote stage of Leishmania donovani and persists in vivo. Int. J. Antimicrob. Agents. 2007;30:229–235. doi: 10.1016/j.ijantimicag.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shaw C.D., Lonchamp J., Downing T., Imamura H., Freeman T.M., Cotton J.A., Sanders M., Blackburn G., Dujardin J.C., Rijal S., Khanal B., Illingworth C.J., Coombs G.H., Carter K.C. In vitro selection of miltefosine resistance in promastigotes of Leishmania donovani from Nepal: genomic and metabolomic characterization. Mol. Microbiol. 2016;99:1134–1148. doi: 10.1111/mmi.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Mishra J., Gupta A.K., Singh A., Shankar P., Singh S. Laboratory confirmed miltefosine resistant cases of visceral leishmaniasis from India. Parasites Vectors. 2017;10:49. doi: 10.1186/s13071-017-1969-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyoto T., Potet J., Boelaert M. Why miltefosine-a life-saving drug for leishmaniasis-is unavailable to people who need it the most. BMJ global health. 2018;3 doi: 10.1136/bmjgh-2018-000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utaile M., Kassahun A., Abebe T., Hailu A. Susceptibility of clinical isolates of Leishmania aethiopica to miltefosine, paromomycin, amphotericin B and sodium stibogluconate using amastigote-macrophage in vitro model. Exp. Parasitol. 2013;134:68–75. doi: 10.1016/j.exppara.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Vacchina P., Norris-Mullins B., Abengozar M.A., Viamontes C.G., Sarro J., Stephens M.T., Pfrender M.E., Rivas L., Morales M.A. Genomic appraisal of the multifactorial basis for in vitro acquisition of miltefosine resistance in Leishmania donovani. Antimicrob. Agents Chemother. 2016;60:4089–4100. doi: 10.1128/AAC.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckx S., Del-Favero J., Rademakers R., Claes L., Cruts M., De Jonghe P., Van Broeckhoven C., De Rijk P. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005;15:436–442. doi: 10.1101/gr.2754005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner A., Drobot B., Herrmann A., Sanchez-Canete M.P., Gamarro F., Castanys S., Gunther Pomorski T. Disruption of the lipid-transporting LdMT-LdRos3 complex in Leishmania donovani affects membrane lipid asymmetry but not host cell invasion. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2018. Leishmaniases: Epidemiological Report of the Americas, Report Leishmaniases Nº 6–2018.https://www.who.int/leishmaniasis/resources/who_paho_era6/en/ Available at. [Google Scholar]
- Yardley V., Croft S.L., De Doncker S., Dujardin J.C., Koirala S., Rijal S., Miranda C., Llanos-Cuentas A., Chappuis F. The sensitivity of clinical isolates of Leishmania from Peru and Nepal to miltefosine. Am. J. Trop. Med. Hyg. 2005;73:272–275. [PubMed] [Google Scholar]
- Zauli-Nascimento R.C., Miguel D.C., Yokoyama-Yasunaka J.K., Pereira L.I., Pelli de Oliveira M.A., Ribeiro-Dias F., Dorta M.L., Uliana S.R. In vitro sensitivity of Leishmania (Viannia) braziliensis and Leishmania (Leishmania) amazonensis Brazilian isolates to meglumine antimoniate and amphotericin B. Trop. Med. Int. Health. 2010;15:68–76. doi: 10.1111/j.1365-3156.2009.02414.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.