Abstract
The Netherlands launched a nationwide implementation study on non-invasive prenatal testing (NIPT) as a first-tier test offered to all pregnant women. This started on April 1, 2017 as the TRIDENT-2 study, licensed by the Dutch Ministry of Health. In the first year, NIPT was performed in 73,239 pregnancies (42% of all pregnancies), 7,239 (4%) chose first-trimester combined testing, and 54% did not participate. The number of trisomies 21 (239, 0.33%), 18 (49, 0.07%), and 13 (55, 0.08%) found in this study is comparable to earlier studies, but the Positive Predictive Values (PPV)—96% for trisomy 21, 98% for trisomy 18, and 53% for trisomy 13—were higher than expected. Findings other than trisomy 21, 18, or 13 were reported on request of the pregnant women; 78% of women chose to have these reported. The number of additional findings was 207 (0.36%); these included other trisomies (101, 0.18%, PPV 6%, many of the remaining 94% of cases are likely confined placental mosaics and possibly clinically significant), structural chromosomal aberrations (95, 0.16%, PPV 32%,) and complex abnormal profiles indicative of maternal malignancies (11, 0.02%, PPV 64%). The implementation of genome-wide NIPT is under debate because the benefits of detecting other fetal chromosomal aberrations must be balanced against the risks of discordant positives, parental anxiety, and a potential increase in (invasive) diagnostic procedures. Our first-year data, including clinical data and laboratory follow-up data, will fuel this debate. Furthermore, we describe how NIPT can successfully be embedded into a national screening program with a single chain for prenatal care including counseling, testing, and follow-up.
Keywords: NIPT, NIPS, fetal trisomy, prenatal screening, cfDNA, genome-wide, implementation study, first tier test, common trisomies, rare autosomal trisomies
Introduction
In recent years, non-invasive prenatal testing (NIPT) for fetal aneuploidy has rapidly transformed the global prenatal screening landscape. Following the discovery of cell-free fetal DNA (cf.f-DNA) by Lo et al.1 in 1997, NIPT was introduced in clinical practice in 2011.2 Since then, the test has been disseminated fast and has become increasingly available to pregnant women worldwide, either in a commercial or in a state-regulated setting.3,4 Although technically many NIPT tests are based on the analysis of the whole genome, reporting is mostly limited to the common trisomies (21, 18, and 13). We present and evaluate the results of implementing genome-wide NIPT as a first-tier screening test in the Netherlands, including reporting and follow-up of findings other than trisomies 21, 18, and 13.
In the Netherlands, screening for untreatable disorders is subject to a governmental license under the Population Screening Act, which has the objective of protecting citizens against the drawbacks of screening.5 In 2007, a nationwide prenatal screening program was established offering first-trimester combined testing (FCT) to all pregnant women to calculate the risk for trisomy 21 (Down syndrome). In 2011, this screening was extended to trisomies 13 (Patau syndrome) and 18 (Edwards syndrome). In the following years, uptake increased from 25% in 2013 to 34% in 2016.6 In 2014, the Dutch NIPT Consortium, a multidisciplinary collaborative partnership among different stakeholders involved in public prenatal care, was granted a governmental license to introduce NIPT in the Netherlands by means of the implementation study Trial by Dutch Laboratories for Evaluation of Non-Invasive Prenatal Testing (TRIDENT). In TRIDENT-1, women at increased risk for the common trisomies (risk ≥ 1/200) based on FCT or medical history, but not advanced maternal age alone, could choose between NIPT or invasive diagnostic testing.7 This study demonstrated a considerable reduction of the number of invasive procedures performed.8
In April 2017, a second governmental license was granted to evaluate the implementation of NIPT as a first-tier screening test for trisomies 21, 18, and 13; this evaluation is embedded in the government-supported national prenatal screening program (TRIDENT-2 study). Implementation aspects studied include both test performance and characteristics, as well as women’s perspectives. All women in the general obstetric population can elect either NIPT (TRIDENT-2) or FCT as a first-tier test. Women with an increased risk, based on the criteria described above, are excluded from TRIDENT-2 but are still eligible to have NIPT and participate in the TRIDENT-1 study, or they can directly choose invasive testing. A unique feature of TRIDENT-2 is that women who elect NIPT can choose a report on chromosomes 21, 18, and 13 either with or without chromosomal aberrations on the other autosomes (size resolution of 10–20Mb). Sex chromosomes are not analyzed.
Previous studies have demonstrated the high sensitivity and specificity of NIPT for the common trisomies in both high- and general-risk populations.9,10 However, few studies describe the nature and clinical relevance of additional findings from whole-genome sequencing (WGS)-based NIPT, as well as the preferences of patients. Moreover, clinical and laboratory follow-up is often absent or very limited, and most of these studies are based on small cohorts or restricted to the increased cytogenetic risk populations.11, 12, 13, 14 Data from large studies within a general obstetric population are needed because the clinical utility of reporting these additional findings is currently the subject of strong debate.11, 12, 13,15, 16, 17, 18
The TRIDENT-1 study, conducted in a high-risk obstetric population, highlighted the potential clinical utility of genome-wide NIPT by showing that 80% of the additional findings originated from the fetus or the placenta (the rest being maternal or unresolved due to lack of follow-up material), and the majority were of clinical relevance for pregnancy management.12 Other research groups have reported similar results.15,19 In contrast, expanding NIPT beyond trisomies 21, 18, and 13 by including screening for sex chromosomal aneuploidy and microdeletion syndromes may lead to unnecessary invasive diagnostic testing and potentially to pregnancy terminations for relatively mild or uncertain phenotypic conditions.17 Here, we present the uptake and performance of genome-wide NIPT for trisomies 21, 18, and 13 and the additional findings, and we evaluate the first-year results of NIPT implementation as a first-tier screening test in the Netherlands.
Subjects and Methods
Study Design
This is an implementation study; the objective is to determine how NIPT could best be introduced as a first-tier test in the national prenatal screening program for Down, Edwards, and Patau syndromes. During the study, NIPT is offered to all pregnant women in the Netherlands, and they are given a choice between FCT, NIPT, or no prenatal screening. Inclusion and exclusion criteria were defined before the start of the study, and these criteria are described in the governmental license or in the application. The study will continue until 2023; it was decided that the first extensive data analysis would be performed after one year. The TRIDENT-2 study consists of two parts. Part one is described here and contains information on implementation aspects (e.g., uptake, test characteristics, technical performance, and logistics). Part two concerns the evaluation of pregnant women’s perspectives (e.g., decision making, psychological well-being, and satisfaction). This part has not been completed yet and will be published later.
Information and Counseling
In general, pregnant women in the Netherlands consult an obstetric professional (most often a primary care midwife) in the first trimester. As part of standard prenatal care and covered by basic health insurance, a viability and dating scan is performed between 8 and 12 weeks of gestation. In case of a twin pregnancy, chorionicity is determined. The next scan offered is the 20-week anomaly scan. During the first-trimester consultation, women are asked if they have an interest in further information on prenatal screening for fetal anomalies.
All pregnant women who express an interest in first-trimester prenatal screening are subsequently offered a thirty-minute counseling session with a certified obstetric counselor, who may be a midwife, trained nurse, or obstetrician, to discuss the different available screening options. Training and certification of the approximately 3,000 counselors nationwide is organized by the Center for Population Screening of the Dutch Institute for Public Health and the Environment (RIVM/CvB) together with the Regional Centers for Prenatal Screening. Counselors were additionally trained before the start of TRIDENT-2. Certified counselors should perform a minimum of 50 counseling sessions each year, pass an e-learning (refresher) course, and attend continuing education courses every two years. For the public, information leaflets about prenatal screening for fetal trisomy, explaining the differences between NIPT and FCT, are available in 11 different languages. Additionally, two websites were launched to provide women with further information regarding prenatal screening and the TRIDENT studies.
In TRIDENT-2, women who choose screening can opt to have NIPT or FCT. To create equal access, both tests are offered at comparable costs (€175 for NIPT, €168 for FCT in 2017). The government subsidizes the remaining costs for NIPT. Furthermore, women who elect NIPT can choose reporting on chromosomes 21, 18, and 13 with or without additional findings (here referred to as “open cohort” and “targeted cohort”). Findings indicative of a maternal malignancy and structural chromosomal aberrations (SAs) of chromosomes 21, 18, and 13 are always reported.
Participation and Inclusion
The study population consisted of the general obstetric population who elected to have NIPT performed as a first-tier test between April 1, 2017 and April 1, 2018. Written informed consent was obtained from all participating women. Approval for the study was granted by the Dutch Ministry of Health, Welfare, and Sport (license 1017420-153371-PG) and the Medical Ethical Committee of VU University Medical Center Amsterdam (No. 2017.165).
Exclusion criteria for NIPT were pregnancies with a vanishing or dichorionic twin, fetal ultrasound anomalies including a nuchal translucency of ≥3.5 mm, or gestational age < 11 + 0 weeks. Participants under the age of 18 years or couples known to carry a (balanced) chromosomal abnormality were excluded. Also, women who had a current malignancy; who, in the past three months, had received blood transfusions, stem cell therapy, or immunotherapy to treat a malignancy; or who had an organ transplantation, were excluded. In addition, women had to have a Dutch social security number and Dutch health insurance (mandatory for all Dutch residents) and needed to be able to provide informed consent. Women who elected NIPT and were at high risk for the common trisomies, based on FCT ≥ 1/200 or medical history, but not on advanced maternal age alone, were enrolled in the TRIDENT-1 study and excluded from this paper.
Registration, Ordering, Reporting and Post-Test Counseling
NIPT ordering and reporting was performed using Peridos, the online national digital registration system for prenatal screening. According to recent guidelines,20 results were reported as either low-risk (no aberrations found with NIPT) or high-risk (aberration found). Women with a low-risk result were informed by their obstetric care professional. Counseling after a high-risk result depended on the type of aberration found. If the result indicated high risk for a common trisomy (21, 18, or 13), women were referred to a regional center for prenatal diagnostics for further counseling by a consultant obstetrician, followed by invasive prenatal diagnostics if desired. All women at high risk for an additional findings were referred to and counseled by a clinical geneticist.
Sample Collection
Blood draw for NIPT is scheduled at or after 11 + 0 weeks of gestation, and sampling is performed at 173 different service locations across the Netherlands. Blood is drawn in two 10 mL Cell-Free DNA BCT CE tubes (Streck) and shipped at room temperature by courier or regular mail in specific transport containers. Time between blood collection and plasma isolation was five days at the most.
Laboratory Analysis and Bioinformatics
Clinical Genetic laboratories from three university medical centers (Amsterdam UMC location VUMC, Rotterdam Erasmus MC, and Maastricht UMC+) performed NIPT, including DNA isolation, library preparation, next-generation sequencing (NGS), data analysis, interpretation, and reporting. Cell-free DNA (cf.DNA) was isolated from plasma through the use of QIAsymphony Circulating DNA Kits (QIAgen). DNA libraries were prepared for genome-wide shallow sequencing (0.2×; 51bp single-end), which was performed with either the Illumina HiSeq4000 or the NextSeq500 sequencer (Illumina). Bioinformatic analysis was performed using the WISECONDOR (v2.0.1) algorithm under standard settings to call aneuploidy and other unbalanced chromosomal aberrations.21 For the targeted cohort, a filter was applied to reveal only the results of WISECONDOR analysis of chromosomes 21, 18, and 13, and to mask the other autosomes. In the open cohort, the filter was not applied. Two laboratories routinely used DEFRAG to measure fetal fraction in male fetuses, but only one requested a blood redraw when fetal fraction was lower than 4% or when DEFRAG indicated a “bad cluster.”22
Diagnostic Follow-up Testing
Follow-up diagnostic testing was done in all eight Dutch University Medical Centers. Clinical genetic laboratory follow-up of high-risk NIPT results was performed as described previously.12 The type of follow-up test (amniocentesis [AC] or chorionic villus sampling [CVS])23 was decided based on the type of chromosomal aberration, gestational age, and patient preferences. Postnatal cytogenetic confirmation (including cases of fetal demise) was mostly done on cord blood or skin muscle fascia biopsies. When a maternal origin of the chromosomal aberration was suspected, testing the mother first was recommended. Results of invasive testing and obstetric outcomes were collected at the eight university medical centers, the midwifery practices, and the referral hospitals.
Outcome Categories
The primary outcome variables included in this study were test uptake; choice for reporting additional findings; failure rate; turn-around time (TAT, number of days between blood arrival at the NIPT laboratory and reporting in Peridos); test performance of NIPT for trisomies 21, 18, and 13; and additional findings (rare autosomal trisomies [RATs] and SAs). Test uptake was defined as the percentage of Dutch pregnant women having NIPT. Here we explain the definitions we use for the common trisomies and additional findings.
Definitions for the common trisomies
-
•
True positive (TP): a high-risk NIPT result cytogenetically confirmed in the fetus by invasive testing or in the child after birth. In rare high-risk cases where laboratory follow-up was declined during and after pregnancy, a clinical diagnosis after birth could also confirm the NIPT result.
-
•
Discordant positive (DP): a high-risk NIPT result cytogenetically not confirmed in the fetus by invasive testing or clinically in the child after birth.
-
•
Discordant negative (DN): a low-risk NIPT result but a common trisomy cytogenetically confirmed in the fetus by invasive testing or in the child after birth.
-
•
True negative (TN): a low-risk NIPT result with no abnormal cytogenetic or clinical diagnosis.
As part of the TRIDENT-2 protocol, all discordant negatives had to be reported to the project leader. Because all prenatal cytogenetic testing in the Netherlands is performed in one of the eight university medical centers involved in this study, the chance of missing a discordant negative is low.
Definitions for additional findings (according to Van Opstal et al.12)
-
•
Fetal: a high-risk NIPT result cytogenetically confirmed in the fetus by invasive testing or in the child after birth.
-
•
Placental: a high-risk NIPT result not confirmed in the fetus but confirmed by CVS or placental testing or involving a chromosomal aberration typically involved in confined placental mosaicism (CPM).
-
•
Maternal: a high-risk NIPT result confirmed in the mother as a constitutional chromosomal aberration (e.g., [mosaic] copy number variants [CNVs]) or as an acquired chromosomal aberration (e.g., originating from a malignant or benign tumor).
-
•
Unknown: a high-risk NIPT result of unknown origin (in most cases due to lack of follow-up).
Cases without diagnostic verification of the NIPT results (“lost to follow-up”) were excluded from the calculation of test performance; these also included early intrauterine fetal demise (IUFD) and termination of pregnancy (TOP) in abortion clinics. We did not report 10q26 terminal deletions because we have proven that these are caused by maternal FRA10B expansion.24
Statistical Analysis
Descriptive statistics were applied to describe mean, standard deviation (SD), and range for uptake, failure rate, TAT, maternal age, gestational age, and weight. Positive predictive value (PPV), sensitivity, specificity, and confidence intervals were calculated using 2 × 2 tables. Mean age of the various NIPT result categories (low-risk, trisomy 21, trisomy 18, trisomy 13, RAT, and SA) were compared with an ANOVA analysis using IBM SPSS statistics 22. A p value of <0.05 was considered significant.
Results
Uptake of NIPT
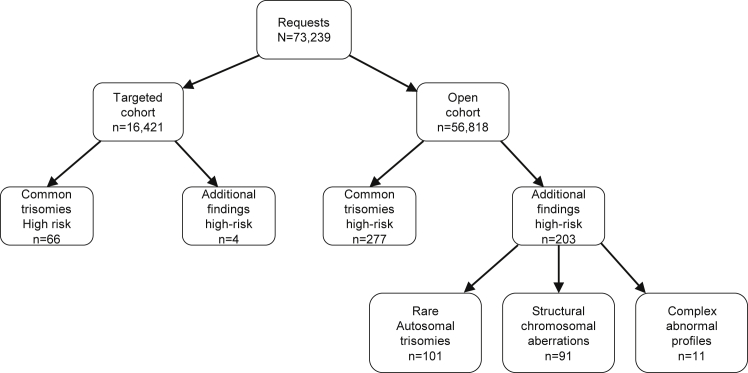
Between April 1, 2017 and April 1, 2018, a total of 73,239 pregnancies were included in the TRIDENT-2 study for NIPT, resulting in a nationwide NIPT uptake of 42%, based on 173,244 pregnancies at 12 weeks of gestation in 2017.25 The open cohort (with additional findings) consisted of 56,818 women (78%), whereas the targeted cohort (without additional findings) consisted of 16,421 women (22%).
Study Population Characteristics
The women included in this study had a mean maternal age of 31.7 years (range 18–52, median 32) which is comparable to the Dutch average age of pregnant women of 31.3 years (source: Statistics Netherlands). The mean gestational age was 11.9 weeks (range 11–41, median 12) at the time of blood draw. Women with high-risk NIPT results for trisomy 21, 18, or 13 (mean 35.2 years, range 22–48, median 36) or high-risk results indicative of an SA (mean 33.4 years, range 20–43, median 33) were significantly older than women with low-risk results (p < 0.001 for both groups). The mean age for women with RATs (mean 32.4 years, range 20–43, median 32) was not significantly different (p = 0.273).
Failure Rate and TAT
In 361 (0.5%) cases, due to protocol violations concerning the collection and transportation instructions, for example mislabeling of blood tubes, no result was issued. In 766 (1%) additional cases, no result could be reported either due to low fetal fraction as defined in the subjects and methods section (605) or due to technical issues (161), of which a low number of reads is most common. In 1,127 (1.5%) cases, no result could be issued after the first blood draw. In 1,009 out of 1,127 women, the test was repeated on a new sample, and for 11 of 1,127 (0.02%) women, the test was repeated twice. For 880 (86%) out of 1,020 repeated tests, a conclusive result could be issued. In total, including repeated testing, for 99.7% of cases, a conclusive result was reported. Mean TAT for all reports was 6.5 business days (range 1–19) and 97.4% were reported within 10 business days. This TAT does not include the time needed for redraw.
Test Performance for Trisomies 21, 18, and 13
A total of 343 high-risk cases were identified by NIPT (Table 1 and Figure 1): 239 (0.33%) cases of trisomy 21 (Down syndrome), 49 (0.07%) cases of trisomy 18 (Edwards syndrome), and 55 (0.08%) cases of trisomy 13 (Patau syndrome). Of the 239 cases of trisomy 21, 16 remained without confirmation of the NIPT result by diagnostic testing. Fourteen of these cases ended in an IUFD, while two cases underwent TOP in a private clinic. In 223 cases, follow-up investigations were performed (cytogenetically or clinically). Cytogenetic confirmatory studies were performed in 221 cases. In 212 out of the 221 cases (96%), trisomy 21 was confirmed by either invasive prenatal testing or postnatal blood analysis, while nine cases (4%) were discordant positives (Table S1). Furthermore, in two other cases, parents had declined karyotyping, but the children had a trisomy 21 phenotype at birth. Five cases of discordant negatives for trisomy 21 were reported (Table S2). Three of these were detected after the birth of a child with Down syndrome, and two cases were identified after IUFD. In summary, the test cohort contained 219 cases of trisomy 21 of which 214 were identified by NIPT and five had low-risk NIPT results. Therefore, the sensitivity of NIPT for trisomy 21 is 98%. Apart from 214 true positive results, there were nine discordant positives, resulting in a PPV of 96% (Table 1).
Table 1.
NIPT Performance for Detecting Trisomies 21, 18, and 13
| - | - | Cases without Confirmatory Testing | Cases with Confirmatory Diagnostic Testing | |||||
|---|---|---|---|---|---|---|---|---|
| NIPT result | n | IUFD (n) | TOP (n) | True Positive (n) | Discordant Positive (n) | Discordant Negative (n) | Sensitivity, % (95% CI) | PPV, % (95% CI) |
| T21 | 239 | 14 | 2 | 214a | 9 | 5 | 98 (95–99) | 96 (93–98) |
| T18 | 49 | 0 | 0 | 48 | 1 | 5 | 91 (79–97) | 98 (87–100) |
| T13 | 55 | 3 | 1 | 27 | 24 | 0 | 100 (87–100) | 53 (43–63) |
CI, confidence interval; IUFD, intrauterine fetal demise; n, number; NIPT, non-invasive prenatal test; PPV, positive predictive value; TOP, termination of pregnancy; T, trisomy
Includes two cases without cytogenetic confirmation, but with a clinical diagnosis of Down syndrome.
Figure 1.
NIPT Flow and Numbers for the Open and Targeted Cohorts
n, number.
For trisomy 18, 49 cases were identified by NIPT as high-risk cases. In 48 cases (98%), trisomy 18 was confirmed by diagnostic testing, whereas in the remaining case, postnatal cord blood analysis indicated a chromosomally normal child (Table S1). Furthermore, five discordant negative cases were reported that were all identified by ultrasound anomalies suggestive of a trisomy 18 phenotype (Table S2). In total, the test cohort contained 53 cases of trisomy 18 of which 48 were identified by NIPT and five received low-risk NIPT results. The sensitivity of NIPT for trisomy 18 is 91%. As one case was discordant positive, the PPV is 98% (Table 1).
Finally, 55 cases of trisomy 13 were identified by NIPT as high-risk cases. Of these, three cases resulted in IUFD and one case in TOP due to ultrasound abnormalities, without diagnostic testing. For 27 (53%) of the remaining 51 cases, the NIPT result was confirmed, but in 24 (47%), the result was not confirmed by either cytogenetic or clinical follow-up (Table S1). No cases of discordant negative trisomy 13 were identified. In total, all 27 cases of trisomy 13 present in the test cohort were identified by NIPT, resulting in a sensitivity of NIPT for trisomy 13 of 100%. However, an additional 24 cases were discordant positives; therefore the PPV is 53% (Table 1).
Additional Findings
NIPT indicated a high risk for 101 (0.18%) RATs and 95 (0.16%) SAs, adding up to a total of 196 additional findings with possible fetal implications. Moreover, in 11 (0.02%) cases, a complex abnormal NIPT profile suggested a possible acquired chromosomal aberration such as a maternal malignancy (Table 2 and Figure 1). Almost all additional findings were reported in the open cohort. In only four cases, additional findings such as a deletion or duplication on chromosome 21, 18, or 13 were reported in the targeted cohort.
Table 2.
Additional Findings (Findings Other than Trisomy 21, 18, or 13)
| - | - | Cases without Confirmatory Testing | Cases with Confirmatory Diagnostic Testing | ||||
|---|---|---|---|---|---|---|---|
| NIPT result | n | IUFD (n) | TOP (n) | Missing (n) | Confirmed (n) | Discordant (n) | % Confirmed (PPV) |
| RATs | 101 | 0 | 1 | 3 | 6 (fetus) | 91a | 6 |
| SAs | 95 | 1 | 0 | 3 | 29 (fetus) | 62a | 32 |
| Complex abnormal profiles | 11 | 0 | 0 | 0 | 7b (mother) | 4 | 64b |
IUFD, intrauterine fetal demise; n, number; NIPT, non-invasive prenatal test; PPV, positive predictive value; RAT, rare autosomal trisomy; SA, structural aberration; TOP, termination of pregnancy
Further clinical details are provided in Tables S3–S5.
Also including cases in which the NIPT findings were confirmed in the placenta or chorion villi or in the mother but not in the fetus.
Confirmed is defined as cases in which a maternal malignancy was present.
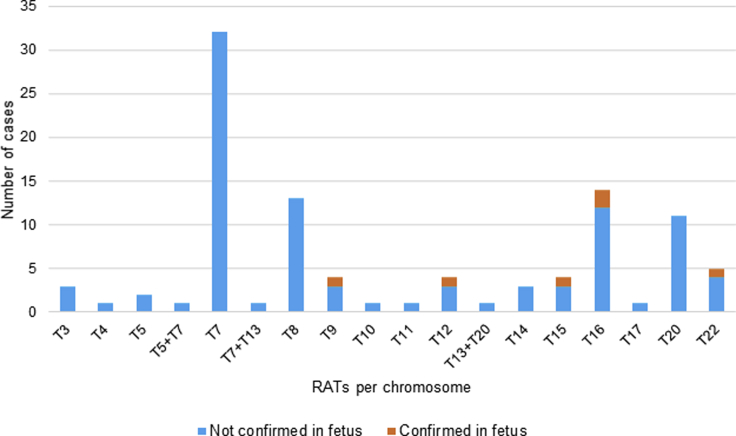
An overview of the RATs is presented in Figure 2. Trisomy 7 was most frequently detected (n = 32), followed by trisomy 16 (n = 14), trisomy 8 (n = 13), and trisomy 20 (n = 11). In one case, the pregnancy was terminated before confirmatory diagnostic testing, and three cases were lost to follow-up. Diagnostic follow-up was available for 97 of 101 cases. For six of 97 cases (6%), the trisomy or a resulting uniparental disomy (UPD) was confirmed in the fetus (Table 2 and Table S3). This included two cases of mosaic trisomy 16 and one case of mosaic trisomy 22. Furthermore, three cases of fetal UPD were confirmed of chromosomes 9, 12, and 15 after NIPT indicated a trisomy for these chromosomes. Only one of these UPDs is pathogenic by itself: the maternal UPD 15, which causes Prader-Willi syndrome. In four (4%) cases, the trisomy was confirmed with CVS (three trisomy 8 and one trisomy 13 + 20), but follow-up fetal investigations in amniotic fluid (AF) or cord blood were normal, confirming the diagnosis of CPM. In 87 of 97 cases (90%), diagnostic follow-up, mostly in AF during pregnancy or in cord blood after birth, showed no trisomy in the fetus. However, it should be mentioned that these tests do not exclude the presence of a CPM. The overall sensitivity for RATs cannot be calculated because we have no data on discordant negative cases in the open cohort. The overall PPV is 6%, but it should be noted that this percentage differs per chromosome.
Figure 2.
Distribution of Rare Autosomal Trisomies
NIPT, non-invasive prenatal test; RAT, rare autosomal trisomy; T, trisomy
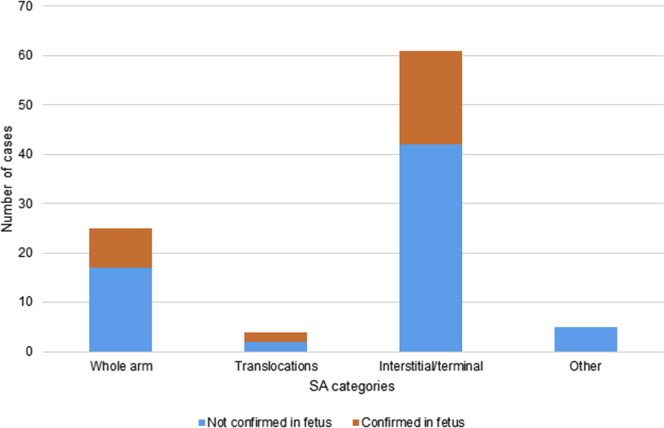
In 95 cases, NIPT indicated an SA (Table 2, Table S4, Figure 3). In this group, one case of IUFD without diagnostic testing occurred, and three were lost to follow-up. In 91 cases, follow-up was available, and in 29 cases (32%), the finding was confirmed in the fetus through diagnostic testing. We categorized and counted the detected unbalanced SAs into four categories: (i) whole-arm aberrations (n = 25), (ii) potential translocations (n = 4), (iii) interstitial/terminal aberrations (n = 61), and (iv) other aberrations (n = 5) including three combined events. An overview of each category can be found in the supplemental data (Table S4).
Figure 3.
Distribution of Structural Chromosomal Aberrations
NIPT, non-invasive prenatal test; SA, structural chromosomal aberration
Of the 25 (i) whole-arm aberrations, one case resulted in IUFD, one apparently healthy child was born, and one child was born with multiple congenital abnormalities not fitting the expected clinical picture; all cases were not karyotyped. Follow-up information was available for the remaining 22 cases. Eight of these (36%) aberrations were confirmed by AC or a fetal skin muscle fascia biopsy. Two (9%) were confirmed to be placental, and the remaining 12 cases (59%) were not confirmed in AF (11) or CVS (1) and are classified as having an unknown origin. Furthermore, four (ii) potential unbalanced translocations were detected, of which two were confirmed in AF, one was confirmed in CVS while AF was normal (CPM), and one case had no diagnostic follow-up and resulted in an apparently healthy live born child, so this case was classified as unknown. In addition, 61 (iii) interstitial/terminal aberrations were identified. For 58 out of 61, follow-up was complete. The NIPT results were confirmed in the fetus for 19 out of 58 cases (33%). Furthermore, seven cases (12%) were confirmed to be of constitutional or acquired maternal origin. For 32 of 58 cases (55%), the diagnostic results in the fetus or the mother were discordant from the NIPT result. Within the group of interstitial/terminal aberrations, we identified eight 5q deletions and five 20q deletions (total 22%) of the critical region associated with myeloid neoplasms.26 For all 13 cases, maternal blood was tested; only in one case was the deletion (20q) constitutionally present as a mosaic in maternal blood. In two cases, cytogenetic analysis of the bone marrow was also performed and revealed the presence of the deletion (one 5q and one 20q deletion) without signs of myeloid malignancies. The hemato-oncological follow-up for several other 5q- and 20q-deletion cases is still ongoing. Finally, five (iv) other aberrations were detected, of which three were combined events of RAT and SA, one was a deletion and duplication of chromosome 6q combined with a 10q duplication, and one was a monosomy 10. None of these were confirmed in the fetus, but one combined event was found back in the placenta. Again, the overall sensitivity for SAs cannot be calculated because we have no data on any missed cases. The overall PPV for SAs is 32%, and 37% if the subgroup of 5q- and 20q-deletions is not included.
We did not report 10q26 terminal deletions because we have proven that these are caused by maternal FRA10B expansion.24 However, we have registered them, and a total of 37 (0.05%) were found.
Complex Abnormal Profiles Suggesting an Acquired Chromosomal Aberration
We encountered 11 (0.015%) complex abnormal NIPT profiles indicative of possible acquired maternal malignancies (Table S5). One woman declined further testing except for ultrasound analysis of the breasts and abdomen. All other cases were subjected to extensive clinical and laboratory follow-up. In nine cases (81%), the potential sources of the complex abnormal profiles could be found. Hematological malignancies were discovered in five cases: three Hodgkin lymphomas and two non-Hodgkin lymphomas. Furthermore, two solid tumors were identified: one pre-malignant breast ductal carcinoma-in situ (DCIS gravida) and one breast carcinoma. We also detected two benign uterine leiomyomas. For the remaining case, no explanation could be found, despite the exclusion of other known causes such as vitamin B12 deficiency and systemic lupus erythematosus.27,28 When cf.DNA sequencing of maternal blood was repeated after childbirth, it showed a low-risk result, therefore this complex NIPT profile was most likely caused by a placental mosaicism.
Invasive Tests
During the first year of TRIDENT-2, a total of 455 invasive tests were performed, 294 (65%) to confirm common trisomies and 161 (35%) to confirm additional findings. In order to confirm high-risk NIPT results for a common trisomies, 163 ACs (55%) and 131 CVSs (45%) were performed. To confirm high-risk NIPT results for additional findings, 141 ACs (88%) and 20 CVSs (12%) were conducted. In five cases, both ACs and CVSs were performed; in four of these five cases, both were performed in order to examine additional findings. Overall, invasive testing beyond the common trisomies revealed that in 35 cases (22% of invasive tests), the aberration was fetal (Table 2).
Discussion
The Dutch TRIDENT-2 study concerns the national implementation of NIPT offered as a first-tier screening test for the detection of trisomies 21, 18, and 13 in the general obstetric population. The nationwide set-up and the fact that all relevant partners joined the Dutch NIPT Consortium allowed us to acquire follow-up data for most of the pregnancies (100% for trisomies 21, 18, and 13, >97% overall). The results from diagnostic follow-up testing confirm a high sensitivity for screening for trisomies 21, 18, and 13 of 98%, 91%, and 100% respectively. The frequency of the common trisomies for this general obstetric population in TRIDENT-2 is, as expected, much lower than that observed in the high-risk population of the TRIDENT-1 study (Table 3). This table also shows that the PPV for the common trisomies in TRIDENT-2 is similar to that in TRIDENT-1 (high-risk women) and thus higher than expected considering the fact that this is a general population risk group. This observation is confirmed when we compare our data to the meta-analysis of 35 studies in both general-risk and high-risk populations by Gil et al.10 which reported sensitivity rates for trisomy 21 between 94% and 100%, for trisomy 18 between 87% and 100%, and for trisomy 13 between 40% and 100%.
Table 3.
Comparison of TRIDENT-1 and TRIDENT-2
| - |
TRIDENT-1 (High-Risk Population) |
TRIDENT-2 (General-Risk Population) |
||
|---|---|---|---|---|
| NIPT Result Frequency (%) | PPV (%) | NIPT Result Frequency (%) | PPV (%) | |
| T21 | 2.24a | 94a | 0.33 | 96 |
| T18 | 0.36a | 80a | 0.07 | 98 |
| T13 | 0.43a | 67a | 0.08 | 53 |
| RATs | 1.11b | 15b | 0.18 | 6 |
| SAs | 0.47b | 50b | 0.16 | 32 |
| Complex abnormal profiles | 0b | 0b | 0.02 | 64 |
Table 3 compares TRIDENT-1 and TRIDENT-2. In the first year of TRIDENT-2, nationwide uptake for NIPT was 42% and for FCT was 4%,25 resulting in a national uptake for first-trimester prenatal screening of 46%. In 2016, before the introduction of NIPT as first-tier test in the Netherlands, FCT uptake was 34%, meaning that the introduction of NIPT resulted in a drastic reduction in the uptake of FCT.6 In addition to the 34% of women who chose FCT, an unknown but significant number of women chose commercial NIPT testing abroad. Based on public records, we estimate this number to be approximately 5,000–8,000 women every year, or 3%–5% of Dutch pregnancies. Therefore, the introduction of NIPT as a first-tier test in 2017 resulted in a small increase in the participation of first-trimester prenatal screening. Less than half of the pregnant women in the Netherlands participate, which is relatively low compared to other European countries.29, 30, 31 Studies have suggested several factors that may explain the low uptake rate in the Netherlands; these factors include the way the offer of screening is framed, by focusing on the “right not to know,” the costs of the test, a positive attitude toward Down syndrome, and a negative attitude toward TOP.32, 33, 34
In 2016, before the start of TRIDENT-2, 3,250 pregnancies received high-risk FCT results (≥1:200) for trisomies 21, 18, and 13 in the Netherlands (screen positive rate 5.4%).6 This is six times higher than the 549 high-risk results after NIPT reported here (screen positive rate 0.75%), a reduction of 83%. If we correct for the increase in participation, the reduction is 86%.
A unique aspect of our study using genome-wide NIPT is that after pre-test counseling, women could choose to have analysis of chromosomes 21, 18, and 13 with (open cohort) or without (targeted cohort) a report of additional findings. The vast majority (78%) of women chose to have additional findings reported, suggesting that pregnant women in general want to know more about the fetus’ health than just the presence of the common trisomies. This is in line with what was expected based on earlier questionnaire studies.35,36
Important differences are observed when comparing RATs between the high-risk group included in the TRIDENT-1 study and the general obstetric risk groups included in TRIDENT-2 (Table 3). Both the frequency (1.1%) and the PPV (15%) in the high-risk group are higher compared to the 0.14% and 6% in TRIDENT-2. This not only confirms the well-known association of abnormal FCT with CPM,37,38 but also suggests that FCT aimed at the common trisomies enriches for the NIPT-detection of fetal (mosaic) RATs, as the PPV of RATs in TRIDENT-1 is higher. The vast majority (94%) of the RATs we identified were discordant positives, most likely explained by CPM. It is known that CPM can be associated with a broad range of both adverse fetal and adverse maternal outcomes, such as multiple congenital anomalies, fetal growth restriction, preterm birth, and stillbirth.12,38,39 Expert ultrasound scans and ultrasound monitoring of fetal growth was recommended for all cases of RATs.
A similar observation was made for the SAs, of which 32% were confirmed in the fetus after invasive testing, which is lower than the PPV of 50% found for SAs in the high-risk population (Table 3).12 However, similar to the RATs, the number of SAs found in this general-risk population (0.13%) is much lower than in the increased-risk group of TRIDENT-1 (0.47%, Table 3). Therefore, FCT does also enrich for SAs and for the fact that they can be retraced to the fetus, although the difference in PPV between increased risk and general risk is smaller than it is for RATs. Our data confirm earlier findings of enrichment for RATs and SAs by FCT. This has implications when FCT is used as a first-tier test, and follow-up testing after high-risk FCT does not reveal a common trisomy in the fetus.40 Women with high-risk NIPT results for SAs were significantly older than women with low-risk results, suggesting that, similar to the risk of common trisomies, the risk of a high-risk SA result increases with maternal age. This unexpected relationship was not found for the RATs, and although it was a statistically significant difference, we have no biological explanation for this finding, which therefore needs confirmation by other studies.
A major subgroup (n = 13, 22%) of the interstitial SAs concerned 5q and 20q deletions that may be associated with myeloid neoplasms.26 As far as we know now, none of these women showed any clinical signs of leukemia, including the two cases where we could retrace the deletion to bone marrow cells. Therefore, currently we cannot predict whether these findings are medically relevant, i.e., whether these women are prone to develop a hematological malignancy later in life. We will monitor these women at regular intervals in the following years.
For eleven samples, NIPT indicated a possible acquired maternal malignancy, which equals approximately one in 6,500 in the studied population. Only seven of these proved to be malignant or pre-malignant, the others being benign (uterine leiomyoma) or unknown because of lack of follow-up. This leads to a final frequency of malignant or pre-malignant tumors of one in 10,500 women tested via NIPT. According to the Surveillance, Epidemiology, and End Results Program of the National Institutes of Health, the overall incidence of cancer in this population in the United States is estimated to be approximately one in 1,300,41 meaning that only one out of nine cancer cases will be identified by NIPT, presumably because the fraction of cell-free tumor DNA is too low, or because the tumor does not have an unbalanced chromosome profile that can be detected by NIPT. The majority of detected malignancies (five out of seven) were Hodgkin’s and non-Hodgkin’s lymphomas, a result similar to what has previously been reported.42
One of the original benefits of introducing NIPT was a reduction of the number of invasive tests. This was demonstrated during the introduction of NIPT as a second-tier test in TRIDENT-1, which resulted in a reduction of at least 62%.8 A possible argument against genome-wide NIPT in contrast to targeted NIPT is that this would increase the number of invasive tests. Although this is confirmed by our data, the number of extra invasive tests to confirm additional findings is limited to 161, comprising 35% of all invasive tests performed to confirm NIPT results. Testing beyond the common trisomies revealed 35 fetal aberrations, most of them expected to have severe clinical consequences, that would not have been detected by targeted NIPT. Further studies on the clinical relevance of these findings, including those with CPM, are ongoing.
It should be noted that reporting back results of genome-wide NIPT may result in an increase of parental concern and anxiety in cases of discordant-positive results or when the clinical relevance of the additional finding is not immediately clear and/or affects the mother rather than the fetus.43 Studies on this subject highlight the importance of adequate pre-test counseling to establish patient knowledge and expectations.44,45 Such pre-test counseling is currently limited because no PPVs for additional findings are available. Studies like the one presented here will fill this gap. Within the context of TRIDENT-2, we will also study women’s perspectives and experiences through the use of questionnaires and in-depth interviews.
This study also had limitations. We did not analyze the sex chromosomes, because this was not part of our protocol and governmental license. Therefore, we cannot present data on the sex chromosomal aneuploidies. Another limitation is that we cannot be sure that all missed common trisomies were reported to us. We do not expect that we have missed many, because it was part of our study to report missed trisomies to the project leader, and because all cytogenetic laboratory follow-up in the Netherlands is done in one of the eight academic centers, which are all part of the Consortium. The final limitation is that the test was performed in three different laboratories. Although we aimed at using the same laboratory set-up in all three, there were some differences. For instance, one of three laboratories used a NextSeq for sequencing, whereas the others used HiSeq. Furthermore, only one of the labs rejected samples because of low fetal fraction. As a result, this lab had a higher failure rate. Although we do not expect that these differences had a big impact on our results, we cannot exclude it completely. Most of these issues have now been solved as all laboratories changed to a uniform laboratory protocol solution in July 2018.
In conclusion, this study has confirmed that genome-wide NIPT is a reliable and robust screening test for the detection of fetal trisomies 21, 18, and 13. As expected, the PPV for the additional findings, and for RATs in particular, is lower than for a single common trisomy, but still higher than the PPV of FCT for trisomy 21, 18, or 13 (combined: 4.4%).46 For SAs, the overall PPV of 32% is relatively high for a screening test.47 The number of findings indicative of maternal cancer is very low (one in 6,500 reported, one in 10,500 malignancies or pre-malignancies). The debate on the usefulness of genome-wide compared to targeted NIPT revolves to a great extent around the lack of information on the test’s characteristics, its scope, and its limitations. Although this study is a significant contribution towards resolving the debate, further research is needed on the clinical relevance of the additional findings, as well as on the emotional impact on women, to support implementation of genome-wide NIPT for screening of relevant fetal pathology and adverse pregnancy outcomes. As TRIDENT-2 will continue until April 2023, we will collect and publish more relevant data on additional findings (including clinical and molecular follow-up performed both during and after pregnancy in confirmed fetal cases and cases with CPM). This will allow us to set specific guidelines for pre- and post-test counseling, and for clinical and laboratory follow-up for the most common additional findings. In our opinion, this study derived great benefit from the incorporation of NIPT in a government-supported prenatal screening program, which ensured a prompt chain of counseling, testing, and, in case of high-risk NIPT results, effective clinical follow-up and diagnostic testing.
Consortia
Members of the Dutch NIPT Consortium (excluding authors): A. Polstra, E. Voorhoeve, M.P.R. Lombardi, I.M.C. Bakker, E.J. Bradley, C. Louwerens-Zintel, M. Smit, S. van ‘t Padje, E. Pajkrt, L. Martin, J.T. Gitsels-van der Wal, F. Sleutels, W. de Valk, W.H. Deelen, L.C.P. Govaerts, A.M.S. Joosten, M.E. Redeker, A.T.J.I Go, S. Galjaard, A.K.E. Prinsen, A.P.G. Braat, A. van der Wijngaard, H. Scheffer, L.H. Houben, M.A.A. van Esch-Lennarts, H. Brunner, A.B.C. Coumans, D.E.M. Oldeweghuis, T. Hofste, I. Derks-Prinsen, K. Bouman, E. van Vliet-Lachotzki, and K. Dolsma.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors would like to thank all participating women for their contribution to the TRIDENT-2 study. Moreover, we thank all who have contributed to the set-up and execution of the study, including health professionals, laboratory staff, and other supporting staff. Staff members of RIVM/CvB, the Regional Centers for Prenatal Screening, and Peridos are acknowledged for their collaboration. Sandra van ‘t Padje is acknowledged for all her work as functional manager of the Consortium. This work was supported by a grant from the Netherlands Organization for Health Research and Development (ZonMw, No. 543002001).
Published: November 7, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.10.005.
Data and Material Availability
All data associated with this study are present in the paper or Supplemental Data or are available under a data use agreement and subject to the limitations of the informed consent document.
For affiliations see Supplemental Appendix A in Supplemental Data.
Web Resources
English information leaflet about prenatal screening for fetal trisomy, https://www.rivm.nl/documenten/folder-informatie-over-screening-op-down-edwards-en-patausyndroom-engels-english
Further information regarding prenatal screening, https://onderzoekvanmijnongeborenkind.nl/
About NIPT and the TRIDENT studies, https://www.meerovernipt.nl
Online national digital registration system for prenatal screening Peridos, https://www.peridos.nl
Supplemental Data
References
- 1.Lo Y.M., Corbetta N., Chamberlain P.F., Rai V., Sargent I.L., Redman C.W., Wainscoat J.S. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekharan S., Minear M.A., Hung A., Allyse M. Noninvasive prenatal testing goes global. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008704. 231fs15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi D.W., Chiu R.W.K. Sequencing of Circulating Cell-free DNA during Pregnancy. N. Engl. J. Med. 2018;379:464–473. doi: 10.1056/NEJMra1705345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green E.D., Rubin E.M., Olson M.V. The future of DNA sequencing. Nature. 2017;550:179–181. doi: 10.1038/550179a. [DOI] [PubMed] [Google Scholar]
- 5.van El C.G., Pieters T., Cornel M. Genetic screening and democracy: lessons from debating genetic screening criteria in the Netherlands. J. Community Genet. 2012;3:79–89. doi: 10.1007/s12687-011-0063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liefers J., Cruijsberg J., Harmsen M., Atsma F. Nijmegen: Scientific Center for Quality of Healthcare (IQ healthcare) 2017. Monitor 2016 Prenatale screening op downsyndroom en het Structureel Echoscopisch Onderzoek. [Google Scholar]
- 7.van Schendel R.V., van El C.G., Pajkrt E., Henneman L., Cornel M.C. Implementing non-invasive prenatal testing for aneuploidy in a national healthcare system: global challenges and national solutions. BMC Health Serv. Res. 2017;17:670. doi: 10.1186/s12913-017-2618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oepkes D., Page-Christiaens G.C., Bax C.J., Bekker M.N., Bilardo C.M., Boon E.M., Schuring-Blom G.H., Coumans A.B., Faas B.H., Galjaard R.H., and for the Dutch NIPT Consortium Trial by Dutch laboratories for evaluation of non-invasive prenatal testing. Part I-clinical impact. Prenat. Diagn. 2016;36:1083–1090. doi: 10.1002/pd.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor-Phillips S., Freeman K., Geppert J., Agbebiyi A., Uthman O.A., Madan J., Clarke A., Quenby S., Clarke A. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: a systematic review and meta-analysis. BMJ Open. 2016;6:e010002. doi: 10.1136/bmjopen-2015-010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil M.M., Accurti V., Santacruz B., Plana M.N., Nicolaides K.H. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet. Gynecol. 2017;50:302–314. doi: 10.1002/uog.17484. [DOI] [PubMed] [Google Scholar]
- 11.Ehrich M., Tynan J., Mazloom A., Almasri E., McCullough R., Boomer T., Grosu D., Chibuk J. Genome-wide cfDNA screening: clinical laboratory experience with the first 10,000 cases. Genet. Med. 2017;19:1332–1337. doi: 10.1038/gim.2017.56. [DOI] [PubMed] [Google Scholar]
- 12.Van Opstal D., van Maarle M.C., Lichtenbelt K., Weiss M.M., Schuring-Blom H., Bhola S.L., Hoffer M.J.V., Huijsdens-van Amsterdam K., Macville M.V., Kooper A.J.A. Origin and clinical relevance of chromosomal aberrations other than the common trisomies detected by genome-wide NIPS: results of the TRIDENT study. Genet. Med. 2018;20:480–485. doi: 10.1038/gim.2017.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pertile M.D., Halks-Miller M., Flowers N., Barbacioru C., Kinnings S.L., Vavrek D., Seltzer W.K., Bianchi D.W. Rare autosomal trisomies, revealed by maternal plasma DNA sequencing, suggest increased risk of feto-placental disease. Sci Transl Med. 2017;9:eaan1240. doi: 10.1126/scitranslmed.aan1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pescia G., Guex N., Iseli C., Brennan L., Osteras M., Xenarios I., Farinelli L., Conrad B. Cell-free DNA testing of an extended range of chromosomal anomalies: clinical experience with 6,388 consecutive cases. Genet. Med. 2017;19:169–175. doi: 10.1038/gim.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiorentino F., Bono S., Pizzuti F., Duca S., Polverari A., Faieta M., Baldi M., Diano L., Spinella F. The clinical utility of genome-wide non invasive prenatal screening. Prenat. Diagn. 2017;37:593–601. doi: 10.1002/pd.5053. [DOI] [PubMed] [Google Scholar]
- 16.Chitty L.S., Hudgins L., Norton M.E. Current controversies in prenatal diagnosis 2: Cell-free DNA prenatal screening should be used to identify all chromosome abnormalities. Prenat. Diagn. 2018;38:160–165. doi: 10.1002/pd.5216. [DOI] [PubMed] [Google Scholar]
- 17.Gregg A.R., Skotko B.G., Benkendorf J.L., Monaghan K.G., Bajaj K., Best R.G., Klugman S., Watson M.S. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet. Med. 2016;18:1056–1065. doi: 10.1038/gim.2016.97. [DOI] [PubMed] [Google Scholar]
- 18.Benn P., Grati F.R. Genome-wide non-invasive prenatal screening for all cytogenetically visible imbalances. Ultrasound Obstet. Gynecol. 2018;51:429–433. doi: 10.1002/uog.19014. [DOI] [PubMed] [Google Scholar]
- 19.Bayindir B., Dehaspe L., Brison N., Brady P., Ardui S., Kammoun M., Van der Veken L., Lichtenbelt K., Van den Bogaert K., Van Houdt J. Noninvasive prenatal testing using a novel analysis pipeline to screen for all autosomal fetal aneuploidies improves pregnancy management. Eur. J. Hum. Genet. 2015;23:1286–1293. doi: 10.1038/ejhg.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deans Z.C., Allen S., Jenkins F., Khawaja F., Hastings R., Mann K., Patton S., Sistermans E.A., Chitty L.S. Recommended practice for laboratory reporting of non-invasive prenatal testing (NIPT) of trisomies 13, 18 and 21: a consensus opinion. Prenat. Diagn. 2017;37:699–704. doi: 10.1002/pd.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straver R., Sistermans E.A., Holstege H., Visser A., Oudejans C.B.M., Reinders M.J.T. WISECONDOR: detection of fetal aberrations from shallow sequencing maternal plasma based on a within-sample comparison scheme. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gkt992. e31–e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Beek D.M., Straver R., Weiss M.M., Boon E.M.J., Huijsdens-van Amsterdam K., Oudejans C.B.M., Reinders M.J.T., Sistermans E.A. Comparing methods for fetal fraction determination and quality control of NIPT samples. Prenat. Diagn. 2017;37:769–773. doi: 10.1002/pd.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Opstal D., Srebniak M.I. Cytogenetic confirmation of a positive NIPT result: evidence-based choice between chorionic villus sampling and amniocentesis depending on chromosome aberration. Expert Rev. Mol. Diagn. 2016;16:513–520. doi: 10.1586/14737159.2016.1152890. [DOI] [PubMed] [Google Scholar]
- 24.Huijsdens-van Amsterdam K., Straver R., van Maarle M.C., Knegt A.C., Van Opstal D., Sleutels F., Smeets D., Sistermans E.A. Mosaic maternal 10qter deletions are associated with FRA10B expansions and may cause false-positive noninvasive prenatal screening results. Genet. Med. 2018;20:1472–1476. doi: 10.1038/gim.2018.32. [DOI] [PubMed] [Google Scholar]
- 25.Liefers J., Cruijsberg J., Atsma F. (Nijmegen: Scientific Center for Quality of Healthcare (IQ healthcare) 2018. Monitor 2017 Prenatale screening op down-,edwards en patausyndroom en het Structureel Echoscopisch Onderzoek. [Google Scholar]
- 26.Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 27.Schuring-Blom H., Lichtenbelt K., van Galen K., Elferink M., Weiss M., Vermeesch J.R., Page-Christiaens L. Maternal vitamin B12 deficiency and abnormal cell-free DNA results in pregnancy. Prenat. Diagn. 2016;36:790–793. doi: 10.1002/pd.4863. [DOI] [PubMed] [Google Scholar]
- 28.Chan R.W., Jiang P., Peng X., Tam L.S., Liao G.J., Li E.K., Wong P.C., Sun H., Chan K.C., Chiu R.W., Lo Y.M. Plasma DNA aberrations in systemic lupus erythematosus revealed by genomic and methylomic sequencing. Proc. Natl. Acad. Sci. USA. 2014;111:E5302–E5311. doi: 10.1073/pnas.1421126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hui L., Hutchinson B., Poulton A., Halliday J. Population-based impact of noninvasive prenatal screening on screening and diagnostic testing for fetal aneuploidy. Genet. Med. 2017;19:1338–1345. doi: 10.1038/gim.2017.55. [DOI] [PubMed] [Google Scholar]
- 30.Ekelund C.K., Petersen O.B., Jørgensen F.S., Kjaergaard S., Larsen T., Olesen A.W., Skibsted L., Skovbo P., Sommer S., Sperling L., Danish Fetal Medicine Research Group The Danish Fetal Medicine Database: establishment, organization and quality assessment of the first trimester screening program for trisomy 21 in Denmark 2008-2012. Acta Obstet. Gynecol. Scand. 2015;94:577–583. doi: 10.1111/aogs.12581. [DOI] [PubMed] [Google Scholar]
- 31.Blondel B., Coulm B., Bonnet C., Goffinet F., Le Ray C., National Coordination Group of the National Perinatal Surveys Trends in perinatal health in metropolitan France from 1995 to 2016: Results from the French National Perinatal Surveys. J. Gynecol. Obstet. Hum. Reprod. 2017;46:701–713. doi: 10.1016/j.jogoh.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Bakker M., Birnie E., Pajkrt E., Bilardo C.M., Snijders R.J.M. Low uptake of the combined test in The Netherlands--which factors contribute? Prenat. Diagn. 2012;32:1305–1312. doi: 10.1002/pd.4001. [DOI] [PubMed] [Google Scholar]
- 33.Crombag N.M., Vellinga Y.E., Kluijfhout S.A., Bryant L.D., Ward P.A., Iedema-Kuiper R., Schielen P.C., Bensing J.M., Visser G.H., Tabor A., Hirst J. Explaining variation in Down’s syndrome screening uptake: comparing the Netherlands with England and Denmark using documentary analysis and expert stakeholder interviews. BMC Health Serv. Res. 2014;14:437. doi: 10.1186/1472-6963-14-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill M., Johnson J.-A., Langlois S., Lee H., Winsor S., Dineley B., Horniachek M., Lalatta F., Ronzoni L., Barrett A.N. Preferences for prenatal tests for Down syndrome: an international comparison of the views of pregnant women and health professionals. Eur. J. Hum. Genet. 2016;24:968–975. doi: 10.1038/ejhg.2015.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Schendel R.V., Dondorp W.J., Timmermans D.R., van Hugte E.J., de Boer A., Pajkrt E., Lachmeijer A.M., Henneman L. NIPT-based screening for Down syndrome and beyond: what do pregnant women think? Prenat. Diagn. 2015;35:598–604. doi: 10.1002/pd.4579. [DOI] [PubMed] [Google Scholar]
- 36.van der Steen S.L., Diderich K.E.M., Riedijk S.R., Verhagen-Visser J., Govaerts L.C.P., Joosten M., Knapen M.F.C.M., Van Opstal D., Srebniak M.I., Tibben A., Galjaard R.J. Pregnant couples at increased risk for common aneuploidies choose maximal information from invasive genetic testing. Clin. Genet. 2015;88:25–31. doi: 10.1111/cge.12479. [DOI] [PubMed] [Google Scholar]
- 37.Eckmann-Scholz C., Mallek J., von Kaisenberg C.S., Arnold N.K., Jonat W., Reiner S., Caliebe A., Heidemann S. Chromosomal mosaicisms in prenatal diagnosis: correlation with first trimester screening and clinical outcome. J. Perinat. Med. 2012;40:215–223. doi: 10.1515/jpm.2011.130. [DOI] [PubMed] [Google Scholar]
- 38.Toutain J., Goutte-Gattat D., Horovitz J., Saura R. Confined placental mosaicism revisited: Impact on pregnancy characteristics and outcome. PLoS ONE. 2018;13:e0195905. doi: 10.1371/journal.pone.0195905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodfellow L.R., Batra G., Hall V., McHale E., Heazell A.E.P. A case of confined placental mosaicism with double trisomy associated with stillbirth. Placenta. 2011;32:699–703. doi: 10.1016/j.placenta.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 40.Srebniak M.I., Knapen M.F.C.M., Polak M., Joosten M., Diderich K.E.M., Govaerts L.C.P., Boter M., Kromosoeto J.N.R., van Hassel D.A.C.M., Huijbregts G. The influence of SNP-based chromosomal microarray and NIPT on the diagnostic yield in 10,000 fetuses with and without fetal ultrasound anomalies. Hum. Mutat. 2017;38:880–888. doi: 10.1002/humu.23232. [DOI] [PubMed] [Google Scholar]
- 41.National Cancer Institute, S., Epidemiology, and End Results Program (SEER). SEER Cancer Statistics Fact Sheets: Hodgkin and Non-Hodgkin Lymphoma. In. (Bethesda, MD: National Cancer Institute.)
- 42.Lenaerts L., Vandenberghe P., Brison N., Che H., Neofytou M., Verheecke M., Leemans L., Maggen C., Dewaele B., Dehaspe L. Genomewide copy number alteration screening of circulating plasma DNA: potential for the detection of incipient tumors. Ann. Oncol. 2019;30:85–95. doi: 10.1093/annonc/mdy476. [DOI] [PubMed] [Google Scholar]
- 43.Horn R., Parker M. Opening Pandora’s box?: ethical issues in prenatal whole genome and exome sequencing. Prenat. Diagn. 2018;38:20–25. doi: 10.1002/pd.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gammon B.L., Jaramillo C., Riggan K.A., Allyse M. Decisional regret in women receiving high risk or inconclusive prenatal cell-free DNA screening results. J. Matern. Fetal Neonatal Med. 2018 doi: 10.1080/14767058.2018.1519541. Published online October 1, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis C., Hill M., Chitty L.S. Women’s Experiences and Preferences for Service Delivery of Non-Invasive Prenatal Testing for Aneuploidy in a Public Health Setting: A Mixed Methods Study. PLoS ONE. 2016;11:e0153147. doi: 10.1371/journal.pone.0153147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gezondheidsraad . Wet op het bevolkingsonderzoek: NIPT als eerste test voor de syndromen van Down, Patau en Edwards. Den Haag.; 2016. [Google Scholar]
- 47.Nederend J., Duijm L.E.M., Louwman M.W., Groenewoud J.H., Donkers-van Rossum A.B., Voogd A.C. Impact of transition from analog screening mammography to digital screening mammography on screening outcome in The Netherlands: a population-based study. Ann. Oncol. 2012;23:3098–3103. doi: 10.1093/annonc/mds146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.