Abstract
The development of hindlimbs in tetrapod species relies specifically on the transcription factor TBX4. In humans, heterozygous loss-of-function TBX4 mutations cause dominant small patella syndrome (SPS) due to haploinsufficiency. Here, we characterize a striking clinical entity in four fetuses with complete posterior amelia with pelvis and pulmonary hypoplasia (PAPPA). Through exome sequencing, we find that PAPPA syndrome is caused by homozygous TBX4 inactivating mutations during embryogenesis in humans. In two consanguineous couples, we uncover distinct germline TBX4 coding mutations, p.Tyr113∗ and p.Tyr127Asn, that segregated with SPS in heterozygous parents and with posterior amelia with pelvis and pulmonary hypoplasia syndrome (PAPPAS) in one available homozygous fetus. A complete absence of TBX4 transcripts in this proband with biallelic p.Tyr113∗ stop-gain mutations revealed nonsense-mediated decay of the endogenous mRNA. CRISPR/Cas9-mediated TBX4 deletion in Xenopus embryos confirmed its restricted role during leg development. We conclude that SPS and PAPPAS are allelic diseases of TBX4 deficiency and that TBX4 is an essential transcription factor for organogenesis of the lungs, pelvis, and hindlimbs in humans.
Keywords: TBX4, small patella syndrome, semi-dominant, lung and pelvis hypoplasia, hindlimb amelia, Xenopus, PAPPAS, allelic diseases, animal models, SPS
Main Text
Small patella syndrome (SPS, MIM: 147891), also known as ischio-pubic-patellar syndrome, ischiocoxopodopatellar syndrome with or without pulmonary arterial hypertension, coxo-podo patellar syndrome, ischiopatellar dysplasia, or Scott-Taor syndrome, is a rare autosomal dominant syndrome characterized by patellar aplasia or hypoplasia and absent, delayed, or irregular ossification of the ischiopubic junctions and/or the infra-acetabular axe-cut notches.1, 2, 3, 4, 5 Additional features include pediatric-onset pulmonary arterial hypertension, sandal gap, short fourth and fifth rays of the feet, and occasionally pes planus.6, 7, 8, 9, 10, 11, 12 In 2004, Bongers et al. identified a critical region of 5.6 cM on chromosome 17q11 via haplotype analysis in six families with SPS, and found heterozygous loss-of-function (LoF) mutations in the T-box transcription factor 4 (TBX4) gene (MIM: 601719) to be responsible for this syndrome.13 Recently, the occurrence of rare coding TBX4 variants with putative hypomorphic non-coding mutations in one of its lung enhancers has been recognized as a cause of pediatric pulmonary hypertension and lethal lung developmental disease, which is associated with congenital heart disease, foot anomalies, and developmental disability.14, 15, 16, 17
T-box genes are a family of evolutionary-conserved transcription factors characterized by a shared DNA-binding domain. They have restricted spatio-temporal patterns of expression during embryogenesis, and play distinct roles during organogenesis as documented by genetic studies performed in animal models and humans.18, 19, 20, 21, 22, 23 Studies in animal models have revealed the importance of Tbx4 in hindlimb development.24, 25, 26 The misexpression of a dominant-negative Tbx4 construct causes legless chickens with deformed and hypoplastic pelvis and absent pubis.26,27 In mice, a homozygous Tbx4-null allele entirely abrogates hindlimb development while forelimbs remain unaffected.24 TBX4 is a transcriptional activator of the short stature homeodomain transcription factor 2 (SHOX2) during murine fore- and hindlimb development, and is itself regulated by SHOX2 specifically in the forelimb bud, possibly via a positive feedback loop.28
Interestingly, autosomal dominant mutations in the paralogous gene TBX3 are responsible for Ulnar-Mammary syndrome (UMS, MIM: 181450), which is predominantly characterized by posterior forelimb deficiencies or duplications, with rare involvement of the hindlimbs, and apocrine and mammary gland hypoplasia or dysfunction, with absent or abnormal nipples.29 In mouse, Tbx3 expression is observed in both forelimbs and hindlimbs,30 and homozygous mutant embryos exhibit both forelimb and hindlimb abnormalities.31 These embryos also present with a deficiency of mammary gland induction.31 In contrast, autosomal dominant mutations in TBX5 lead to Holt-Oram syndrome (HOS, MIM: 142900), which is characterized by anterior upper limb amelia and cardiac abnormalities.32,33 Tbx5 is specifically expressed in the heart and developing forelimb buds, and its inactivation in mice phenocopies HOS.34,35 Tbx4 and Tbx5 are thus crucial in the specification of hindlimbs and forelimbs, respectively,25,36 and are both upstream regulators of FGF10 and RSPO2, which drive the outgrowth of all four limb buds.26,27,37
In this study, we identify two consanguineous families from Iran segregating a recessively inherited lethal embryonic syndrome. In four affected fetuses displaying absent hindlimbs with pelvis and pulmonary hypoplasia, we identified via exome sequencing two distinct germline homozygous LoF mutations in TBX4 as the cause of this hitherto undescribed disease. We propose to name this clinical entity posterior amelia with pelvis and pulmonary hypoplasia syndrome (PAPPAS). Functional tests in Xenopus tropicalis confirmed that TBX4 is needed for posterior limb and pelvis development but not for forelimb development. Because the heterozygous parents of these two families also presented with skeletal features consistent with SPS, we conclude that these two Mendelian diseases are allelic. We propose that the heterozygous loss of TBX4 is incompletely dominant while its recessive form in PAPPAS demonstrates its essentiality for hindlimb, pelvis, and lung development in humans.
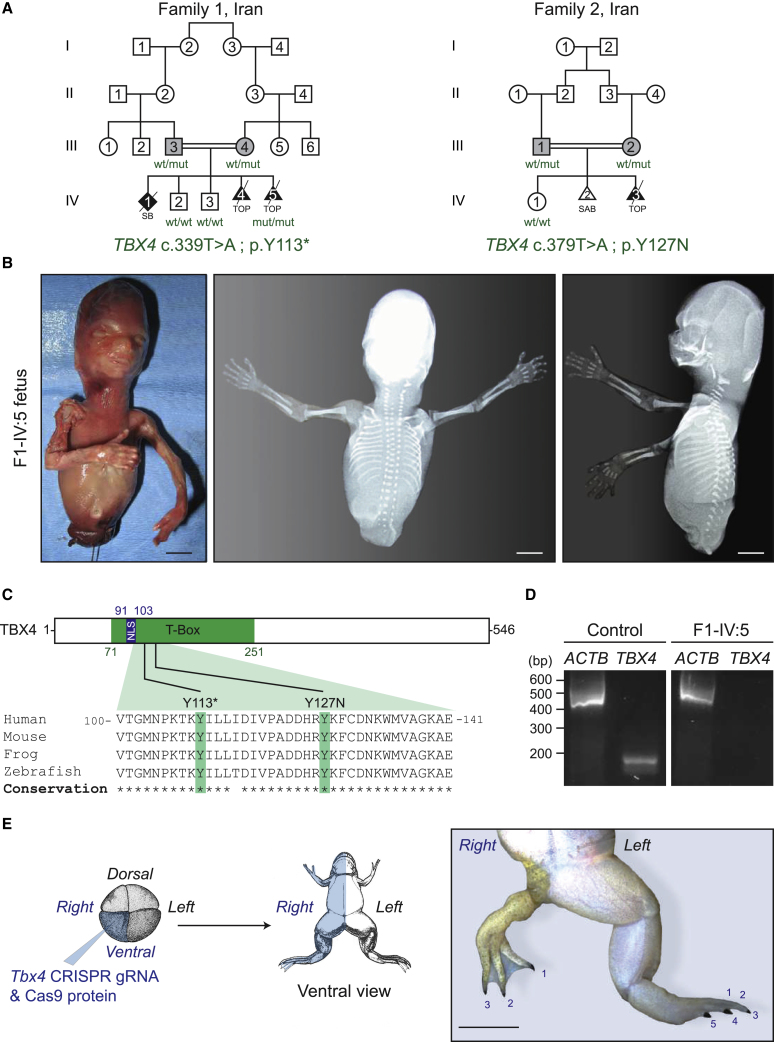
The fetus F1-IV:5 is the result of a fifth pregnancy of related Iranian parents (Figure 1A). The first pregnancy resulted in a stillborn fetus born at term. The genitalia were ambiguous and lower limbs were not formed. Ultrasound examination, X-rays, or pictures were not taken from this first fetus. The second and third pregnancies resulted in healthy boys now aged 14 and 10 years. The fourth pregnancy was followed by ultrasound examination, and the absence of lower limbs was noted at 15 weeks of gestation, and elective abortion was performed at 16 weeks of pregnancy. Autopsy examination was not performed. The fifth pregnancy was controlled with ultrasound, and the absence of lower limbs and severe hypoplasia of pelvic bones was detected at 15 weeks of pregnancy, elective abortion was performed at 16 weeks of pregnancy, and the fetus was sent for autopsy examination. In this and previous pregnancies, the mother did not have any history of drug ingestion or exposure to known teratogens. Gross findings of F1-IV:5 included completely absent lower limbs, anal atresia, ambiguous genitalia, rectoperineal fistula, muscular hypoplasia of the upper limbs, atresia of external auditory canals, hypoplastic lungs, and pulmonary segmentation defect (Figure 1B). Measurements were as follows: crown rump (120 mm, -0.09 SD), head circumference (110 mm, -1.08 SD), and weight (77 grams, -1.08 SD). The heart, liver, gallbladder, stomach, duodenum, jejunum, ileum, and colon were normal. The rectum ended in a blind pouch which was connected to the perineum with a narrow fistula tract. Microscopic findings included severe hypoplasia of the lungs with no apparent differentiation, and mild pyelocaliceal dilatation. X-ray examination showed complete absence of lower limbs, absent pelvic bones, and hypoplastic sacrum (Figure 1B).
Figure 1.
PAPPAS Is Caused by TBX4 Recessive Loss-of-Function Mutations
(A) Pedigrees of two consanguineous Iranian families segregating small patella syndrome in heterozygous TBX4 individuals (gray) and posterior amelia with pelvis and pulmonary hypoplasia syndrome (PAPPAS) in homozygous fetuses (black). The identified mutation and the genotype of available family members are indicated in green. Squares, circles, diamonds, and triangles denote males, females, unknown gender individuals, and fetuses, respectively. Open symbols are used for unaffected family members, and deceased individuals are indicated by a diagonal slash through the symbol. Wt, wildtype; mut, mutant; SB, stillbirth; TOP, termination of pregnancy; SAB, spontaneous abortion.
(B) Photographs and X-rays of affected fetus IV:5 in Family 1 with a germline homozygous p.Y113X TBX4 mutation. Scale bars: 1 cm.
(C) Phylogenetic alignment performed with Clustal O, and position of germline recessive TBX4 mutations in the conserved T-box domain of the transcription factor TBX4. NLS, nuclear localization signal.
(D) TBX4 and ACTB RT-PCR analysis on cDNA extracted from forelimb tissues of a control and the F1-IV:5 fetuses indicating complete absence of the endogenous TBX4 transcript in the mutant fetus. bp, base pairs.
(E) Xenopus tropicalis crispants show severe hindlimb dysplasia after injection of tbx4_gRNA1 pre-complexed with Cas9 protein in one ventral cell of a four-cell-stage embryo (blue), thereby targeting the ventral lineage unilaterally, i.e. primarily the lateral plate mesoderm and the epidermis. A representative post-metamorphic animal (NF stage 66) injected on the right side is shown; the number of toes is indicated. Scale bar: 0.5 cm, ventral view.
The fetus F2-IV:3 in Family 2 is the result of a third pregnancy of first-cousin Iranian parents (Figure 1A). The first pregnancy resulted in a healthy girl, and the second pregnancy resulted in a miscarriage. For the third pregnancy, sonography at gestational week 16 + 3 showed a living fetus with reduced movements, who demonstrated bilateral absence of the posterior extremities (femur, tibia, and fibula). The parents decided to abort the pregnancy and no photography or detailed autopsy of the fetus could be performed.
Whole-exome sequencing (WES) in fetus VI:5 of Family 1 uncovered a private homozygous nonsense variatiant (c.339T>A) in exon 4 of TBX4 (RefSeq accession number NM_001321120.1) that is predicted to result in a stop codon and premature truncation at codon 113 (p.Tyr113∗) (Figure 1C). Co-segregation study in available family members revealed that both parents were heterozygous and that the two unaffected siblings were non-carriers for this rare TBX4 variant (Figure S1A). Because the c.339T>A variant was predicted to cause a premature stop codon probably leading to nonsense-mediated decay (NMD) of the endogenous mRNA, an RT-PCR analysis was carried out on cDNA extracted from forelimb tissue. This analysis showed no detectable expression levels of endogenous TBX4 transcripts in the F1-IV:5 fetus, while expression was found in forelimb tissue from a control fetus of the same age (Figure 1D). These data confirm that the c.339T>A TBX4 variant leads to complete destabilization of the endogenous transcript by NMD. This may therefore constitute the first protein-null or knockout TBX4 allele in humans.
Exome capture in the parents of Family 2 revealed a single-nucleotide-variant (c.379T>A) in exon 4 of TBX4 (RefSeq accession number NM_001321120.1) that results in a heterozygous missense mutation p.Tyr127Asn in the evolutionary conserved DNA-binding domain of TBX4 (Figure 1C). This rare TBX4 c.379T>A (p.Tyr127Asn) variant was absent from all available population databases including ExAc, and also absent from our in-house database of 10,000 exomes. It has a combined annotation-dependent depletion (CADD,38) score of 29.8, which predicts it to be highly pathogenic, as is also suggested by other in silico predictors such as MutationTaster and SIFT. The heterozygosity of both parents for this variant is consistent with their SPS phenotype, and co-segregation study confirmed that the unaffected sister was wild-type (Figure S1B). Unfortunately, DNA from the affected fetus was not available.
In order to further document the causal link between TBX4 deficiency and hindlimb development defects, we injected tbx4 gRNA1 with Cas9 protein unilaterally in one ventral blastomere of four-cell-stage Xenopus tropicalis embryos (Figure 1E). Successful genome editing was validated via targeted deep sequencing (Table S1). Across two experimental repeats, 10% (n = 31) and 6.66% (n = 30), respectively, of the resulting F0 mosaic tadpoles (crispants) manifested striking unilateral hindlimb defects (Figure 1E), but forelimb anomalies were never observed. The most strongly affected animals had a severely underdeveloped hindlimb with a reduced number of toes that still carried claws, a typical characteristic of hindlimbs in Xenopus species. Strikingly, the affected limbs were completely covered with pigmented skin, which is normally present only on the dorsal side, suggesting that Tbx4 inactivation may lead to dorsalization of the affected hindlimb. The less severely affected animals presented with normal forelimbs on the injected side, while the hindlimbs were smaller and not used for swimming. Alizarin red and alcian blue staining of these animals showed a shorter femur and dislocated joints, both at the hip and the knee (Figure S2). In order to confirm the hindlimb phenotype with a second gRNA and to completely recapitulate hindlimb amelia as seen in patients, we also injected tbx4 gRNA2 pre-complexed with Cas9 protein unilaterally in two-cell-stage Xenopus tropicalis embryos (data not shown). With this gRNA, almost universal genome editing was seen on the injected side (Table S1), and all animals manifested a very apparent growth delay and died before limb formation around 35 days post-injection, potentially due to lung defects. Unfortunately, this precluded the identification of potential hindlimb amelia. Together, these in vivo functional tests confirm the importance of TBX4 during embryonic development in vertebrates, including a role for proper hindlimb morphogenesis.
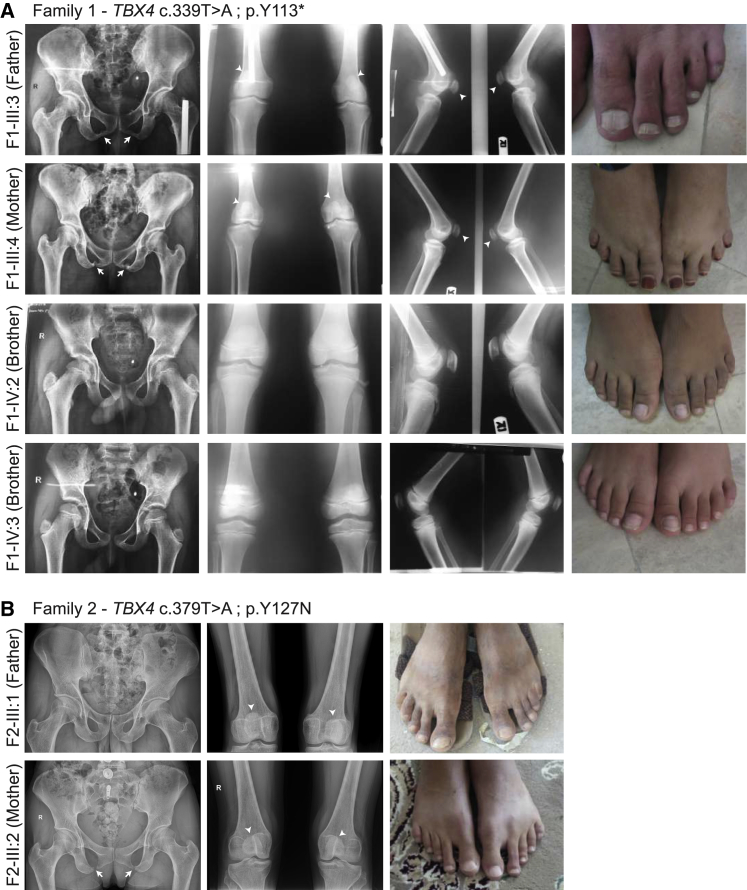
Heterozygous TBX4 mutations are known to lead to dominant SPS.13 We thus examined the parents of the PAPPAS fetuses, who are obligate heterozygous carriers of the identified germline TBX4 mutations. X-ray examination showed the classical findings of SPS in the parents of Family 1 (III:3 and III:4), including patellar hypoplasia, absent or delayed or irregular ossification of the ischiopubic junctions and/or the infra-acetabular axe-cut, and short fourth and fifth rays of the feet (Figure 2A). The two healthy sons (IV:2 and IV:3), who are wild-type for the variant, do not show any of the X-ray findings associated with SPS. For Family 2, X-ray examination of the parents (III:1 and III:2) also revealed phenotypes consistent with SPS, which included patellar hypoplasia, thin ischial bones, and sandal gap between the first and second toes (Figure 2B). Abnormal ischio-pubic junction and short fourth and fifth rays of the feet were also observed in the mother (Figure 2B). Altogether, these findings confirm the pathogenicity of these two germline TBX4 LoF variants. We therefore conclude that while heterozygous TBX4 mutations are responsible for SPS, when homozygously inherited, the same alleles lead to lethal PAPPAS characterized by caudal regression with complete posterior amelia and pelvic aplasia.
Figure 2.
The Heterozygous TBX4 Parents Present with SPS
X-ray and photographs of the indicated family members in Family 1 (A) and Family 2 (B). White arrows point to abnormal ischio-pubic junction, and arrowheads point to hypoplastic patellae in both heterozygous parents of the two families. Note the short fourth and fifth rays in the feet of both parents in Family 1 and the mother in Family 2, and the wide gap between the first and second toes in both parents of Family 2.
Inactivation of Tbx4 has different effects depending on the timing of limb development. When it is deleted before limb bud formation, the hindlimbs fail to develop altogether, but if it is inactivated during limb outgrowth, a dramatic loss of proximal skeletal elements and a mild loss of distal skeletal elements are seen.39 Our results in Xenopus support this as the least affected frogs showed more severe proximal than distal hindlimb anomalies. Our observation that pigmented skin, which is normally restricted to the dorsal side of the animal, was covering the affected limbs of TBX4 crispant froglets could indicate that TBX4 plays a role in specifying, or maintaining, ventral identity in developing hindlimbs. Alternatively, this may be interpreted as a failure of hindlimbs to properly articulate at the levels of the pelvis so that they instead point backwards, a phenomenon seen in conditional Tbx4 mutant mice.39
All of the affected fetuses with homozygous Tbx4 mutations present with complete absence of lower limbs, absent pelvic bones, and hypoplastic sacrum. These phenotypes were to be anticipated based on animal models of TBX4 inactivation. Loss of TBX4 in mice cancels hindlimb development while forelimbs remain unaffected,24 and dominant-negative Tbx4 overexpression leads to chickens bereft of legs, with deformed and hypoplastic pelvis and absent pubis.26,27 Of note, TBX4 in humans does not seem to be essential for umbilical cord formation since TBX4 knockout fetuses can reach full term (F1-IV:1), while mice lacking Tbx4 die at mid-gestation (E10.5) due to defects in the vascularization and fusion of the allantois.24 However, the fetus F1-IV:5 showed additional features including muscular hypoplasia of upper limbs, atresia of external auditory canals, and hypoplastic lungs and pulmonary segmentation defect. The short stature homeodomain transcription factor SHOX2 is a key player during limb development; its loss in mouse is associated with altered muscular development and innervation defects of the proximal forelimbs.40 Interestingly, Glaser et al. showed that Tbx4-/- mouse embryos present with a reduced expression of Shox2 in forelimbs and hindlimbs, and suggested that regulation of Tbx4 by SHOX2 may contribute to the development of muscular and skeletal elements in the developing mammalian forelimb.28 Therefore, the hypoplasia of upper limbs observed in the F1-IV:5 fetus is in line with the suggested role of TBX4 in forelimb muscular development. This fetus also had hypoplastic lungs and pulmonary segmentation defects. The role of TBX4 on lung development has recently been studied in humans. Complex non-coding variants in the lung enhancer of TBX4 in conjunction with coding mutations in TBX4 cause severe lung abnormalities such as acinar dysplasia, congenital alveolar dysplasia, pulmonary hypoplasia, and alveolar growth abnormality.14, 15, 16, 17,41 Of note, the F1-IV:5 fetus also showed atresia of external auditory canal. We are not aware of previous reports of abnormalities in external ear development in association with TBX4 variants or CNVs in humans or other organisms, but there are reports of deletions and/or duplications of the region containing the TBX4 gene associated with hearing loss and abnormal auricles.42, 43, 44
From an evolutionary perspective, TBX4 is not essential in lower vertebrate species. The naturally occurring Tbx4 mutant zebrafish known as “pelvic fin-loss” is fertile and shows a viable phenotype with absent pelvic fins, which are analogous to posterior limbs in tetrapod species.45 Notably, Tbx4 is entirely lost in the seahorse Hippocampus comes who has only retained Tbx5 for the specification of pectoral fins, which are analogous to forelimbs in tetrapods.46
Taken together, our clinical, genetic, molecular, and embryological studies define a heretofore unknown monogenic syndrome caused by homozygous null TBX4 mutations. This embryonic lethal condition, which we propose to name PAPPAS, is allelic to SPS. Together, these Mendelian conditions delineate a spectrum of severity driven by either monoallelic or biallelic TBX4 deficiency in humans.
Online Materials and Methods
Editorial Policies and Ethical Considerations
This study was approved in Iran by the ethics committees of the Kariminejad-Najmabadi Pathology and Genetics Center for Family 1, and of Mashhad University of Medical Sciences and Pardis Genetic Laboratory for Family 2. The parents of both families provided written informed consent to participate in this study and to publish their family pedigrees and clinical data. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Genetic Analysis
WES was performed on gDNA of aborted fetus IV:5 of Family 1 after obtaining written informed consent. Exome capture was performed using the Agilent SureSelect Human All Exon V6 Kit with 150-bp paired-end read sequences generated on a HiSeq4000; 2× 100bp. Sequences were aligned to hg19, and variants were identified through the GATK pipeline. Variations were annotated with ANNOVAR. Identified variants were checked against public databases dbSNP ver.149, Exome Variant Server, ExAC, and Iranome. Co-segregation of the variant was examined in the available family members via bidirectional Sanger sequencing (Figure S1).
Exome capture in the parents of Family 2 was conducted following the Illumina Nextera Rapid Capture Enrichment library preparation protocol using 50 ng of genomic DNA. Paired-end sequencing of the library was performed using a NextSeq500 desktop sequencer (Illumina) with a v2 reagent kit (Illumina). The generated sequences were demultiplexed and mapped to the human genome reference (NCBI build37/hg19 version) with Burrows-Wheeler Aligner. Variant calling and analysis was conducted using GensearchNGS software (PhenoSystems SA). Variants with a coverage of ≤20, a Phred-scaled quality of ≤15, a frequency of ≤20, and a MAF of ≥1% were filtered out. Alamut® Visual (Interactive Biosoftware), with its incorporated prediction tools like SIFT, MutationTaster, and PolyPhen-2, was used for variant prioritization. Co-segregation of the variant was examined in the available family members via bidirectional Sanger sequencing.
Transcript Analysis
Total RNA was extracted from forelimb tissue of affected fetus of Family 1 (IV:5) through the use of the Total RNA Isolation System (Promega). Synthesis of first-strand complementary DNA (cDNA) was performed using the iScript cDNA Synthesis kit (Bio-Rad Laboratories), according to the manufacturer's instructions using 0.4 μg total RNA. To analyze the impact of the c.339T>A variant in transcript expression of TBX4 in the affected fetus of Family 1, reverse-transcriptase (RT)-PCR was performed on cDNA extracted from forelimb tissue with primer pairs covering part of exon 1 through exon 4 generating an amplicon of 181 bp (forward primer 5′-CAG ACC ATC GAG AAC ATC AAG-3′ and reverse primer 5′-GCA GGG ACA ATG TCA ATC AG-3′). The 496 bp amplification of the cDNA encoding beta-actin (ACTB) served as an endogenous quality control (forward primer 5′-GGC ATC CTC ACC CTG AAG TA-3′ and reverse primer 5′-CCA TCT CTT GCT CGA AGT CC-3′).
Generation of Xenopus Tropicalis Mosaic Mutants by CRISPR/Cas9
All experiments on Xenopus tropicalis were executed in accordance with the guidelines and regulations of Ghent University, faculty of Sciences, Belgium. Approval was obtained by the ethical committee of Ghent University, faculty of Sciences (permit number EC2017-093). In brief, two guide RNA targeting Tbx4 were designed with CRISPRScan, generated as previously described,47,48 and yield was quantified by Qubit BR RNA assay (ThermoFischer Scientific). Embryos were injected either in one ventral blastomere at the four-cell stage (tbx4_gRNA1) or unilaterally in a two-cell-stage embryo (tbx4_gRNA2) with 47 pg of tbx4 gRNA and 900 pg of NLS-Cas9-NLS (VIB/UGent Protein Service Facility), and adult animals were analyzed at Nieuwkoop-Faber stage 66. The gRNA and PCR primer sequences used are as follows: tbx4_gRNA1, gRNA target 5′-GGT GTT GAG TGG CAA GTC CT-3′, forward primer 5′-AGG CAA AGA CTG GTA TCA A-3′, reverse primer 5′-ACC TGG GTA GAA TGA GAA-3′ ; tbx4_gRNA2, gRNA target 5′-TGA TGA CAG TGA CCT CCG TGT GG-3′, forward primer 5′-ATA TTT ACC CAC ACC AGC C-3′, reverse primer 5′-TTT CCA ATG CCA GTA TCC AC-3′. For quantitative analysis of genome editing, nine embryos per injected clutch were pooled and lysed overnight in lysis buffer (50 mM Tris pH 8.8, 1 mM EDTA, 0.5% Tween-20, 200 μg/ml proteinase K) at 55°C.
Genotyping PCRs were performed with the respective primer pairs. Targeted deep sequencing of amplicons was performed as previously described and analyzed by the BATCH-GE software package.49 Indel frequency data and sequence variants for all targeted deep sequencing are shown in Table S1. Note that the maximum expected efficiency is 25% for gRNA1 and 50% for gRNA2 because the injections were performed in one cell out of four or two cells, respectively. For staining of the skeletons of the mutant animals, premetamorphic tadpoles and postmetamorphic froglets were euthanized using a 0.05% benzocaine solution. Animals were fully eviscerated and skinned and eyes were removed. Whole mount alcian blue/alizarin red stainings were performed as follows: 95% EtOH (4 days, change after 24h), 100% acetone (48 h), 0.15% alcian blue 8G× (Sigma-Aldrich A3157) in 76% EtOH/20% glacial acetic acid/4% H2O (24 h), 70% EtOH (24 h), 95% EtOH (12 h), 1% KOH (6 h), 0.05% alizarin red S (Sigma-Aldrich A5533) in 1% KOH (48 h), 1% KOH (48 h). Imaging was performed with a Leica MZ FLIII stereomicroscope/Leica DC300F camera.
Accession Numbers
The WES data for Family 1 and Family 2 have been deposited in the NCBI Sequence Read Archive (SRA) database under accession numbers PRJNA578898 and PRJNA578086, respectively.
Declaration of Interests
The authors declare no competing interests.
Acknowledgements
We thank members of both families who provided samples and clinical information for this study. H.T. was supported by grants from the European Union’s Seventh Framework Programme for research, technological development, and demonstration under grant agreement no. 608473 and the Swedish Research Council. E.S-R. is supported by a National Medical Research Council Open Fund—Young Individual Research Grant (OF-YIRG, #OFYIRG18May-0053). T.N. is funded by Kom op tegen Kanker (Stand up to Cancer), the Flemish cancer society. B.R. is a fellow of the Branco Weiss Foundation, an A∗STAR Investigator, a National Research Foundation and Amsterdam Academic Alliance (AAA) fellow, and an European Molecular Biology Organization (EMBO) Young Investigator. This work was partly funded by a Strategic Positioning Fund on Genetic Orphan Diseases from A∗STAR, Singapore. The funders had no role in the design of the study and collection, analysis, decision to publish, interpretation of data, or preparation of the manuscript.
Published: November 21, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.10.013.
Contributor Information
Pooneh Nikuei, Email: poonehnikuei@gmail.com.
Bruno Reversade, Email: bruno@reversade.com.
Web Resources
1000 Genomes Project Database, http://browser.1000genomes.org/index.html
CRISPRScan, https://www.crisprscan.org
Exome Aggregation Consortium (ExAC), http://exac.broadinstitute.org
Exome Variant Server from NHLBI Exome Sequencing Project (ESP), https://evs.gs.washington.edu/EVS/
Genome Aggregation Database (GnomAD), http://gnomad.broadinstitute.org/
Greater Middle East (GME) Variome web, http://igm.ucsd.edu/gme/index.php
NCBI dbSNP, https://www.ncbi.nlm.nih.gov/SNP/
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org
Iranome database, http://www.iranome.com
Supplemental Data
References
- 1.Vanĕk J. [Ischiopatellar dysplasia (Scott and Taor’s syndrome of the small patella)] RoFo Fortschr. Geb. Rontgenstr. Nuklearmed. 1981;135:354–356. [PubMed] [Google Scholar]
- 2.Morin P., Vielpeau C., Fournier L., Denizet D. [The coxopodopatellar syndrome] J. Radiol. 1985;66:441–446. [PubMed] [Google Scholar]
- 3.Scott J.E., Taor W.S. The “small patella” syndrome. J. Bone Joint Surg. Br. 1979;61-B:172–175. doi: 10.1302/0301-620X.61B2.438269. [DOI] [PubMed] [Google Scholar]
- 4.Dellestable F., Péré P., Blum A., Régent D., Gaucher A. The ‘small-patella’ syndrome. Hereditary osteodysplasia of the knee, pelvis and foot. J. Bone Joint Surg. Br. 1996;78:63–65. [PubMed] [Google Scholar]
- 5.Poznanski A.K. Comments on the ischio-pubic-patellar syndrome. Pediatr. Radiol. 1997;27:428–429. doi: 10.1007/s002470050161. [DOI] [PubMed] [Google Scholar]
- 6.Sandhaus Y.S., Ben-Ami T., Chechick A., Goodman R.M. A new patella syndrome. Clin. Genet. 1987;31:143–147. doi: 10.1111/j.1399-0004.1987.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 7.Kozlowski K., Nelson J. Small patella syndrome. Am. J. Med. Genet. 1995;57:558–561. doi: 10.1002/ajmg.1320570408. [DOI] [PubMed] [Google Scholar]
- 8.Azouz E.M., Kozlowski K. Small patella syndrome: a bone dysplasia to recognize and differentiate from the nail-patella syndrome. Pediatr. Radiol. 1997;27:432–435. doi: 10.1007/s002470050163. [DOI] [PubMed] [Google Scholar]
- 9.Habboub H.K., Thneibat W.A. Ischio-pubic-patellar hypoplasia: is it a new syndrome? Pediatr. Radiol. 1997;27:430–431. doi: 10.1007/s002470050162. [DOI] [PubMed] [Google Scholar]
- 10.Bongers E.M., Van Bokhoven H., Van Thienen M.N., Kooyman M.A., Van Beersum S.E., Boetes C., Knoers N.V., Hamel B.C. The small patella syndrome: description of five cases from three families and examination of possible allelism with familial patella aplasia-hypoplasia and nail-patella syndrome. J. Med. Genet. 2001;38:209–214. doi: 10.1136/jmg.38.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy M., Eyries M., Szezepanski I., Ladouceur M., Nadaud S., Bonnet D., Soubrier F. Genetic analyses in a cohort of children with pulmonary hypertension. Eur. Respir. J. 2016;48:1118–1126. doi: 10.1183/13993003.00211-2016. [DOI] [PubMed] [Google Scholar]
- 12.Kerstjens-Frederikse W.S., Bongers E.M.H.F., Roofthooft M.T.R., Leter E.M., Douwes J.M., Van Dijk A., Vonk-Noordegraaf A., Dijk-Bos K.K., Hoefsloot L.H., Hoendermis E.S. TBX4 mutations (small patella syndrome) are associated with childhood-onset pulmonary arterial hypertension. J. Med. Genet. 2013;50:500–506. doi: 10.1136/jmedgenet-2012-101152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bongers E.M.H.F., Duijf P.H.G., van Beersum S.E.M., Schoots J., Van Kampen A., Burckhardt A., Hamel B.C.J., Losan F., Hoefsloot L.H., Yntema H.G. Mutations in the human TBX4 gene cause small patella syndrome. Am. J. Hum. Genet. 2004;74:1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galambos C., Mullen M.P., Shieh J.T., Schwerk N., Kielt M.J., Ullmann N., Boldrini R., Stucin-Gantar I., Haass C., Bansal M. Phenotype characterisation of TBX4 mutation and deletion carriers with neonatal and paediatric pulmonary hypertension. Eur. Respir. J. 2019;54:1801965. doi: 10.1183/13993003.01965-2018. [DOI] [PubMed] [Google Scholar]
- 15.Karolak J.A., Szafranski P., Kilner D., Patel C., Scurry B., Kinning E., Chandler K., Jhangiani S.N., Coban Akdemir Z.H., Lupski J.R. Heterozygous CTNNB1 and TBX4 variants in a patient with abnormal lung growth, pulmonary hypertension, microcephaly, and spasticity. Clin. Genet. 2019;96:366–370. doi: 10.1111/cge.13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suhrie K., Pajor N.M., Ahlfeld S.K., Dawson D.B., Dufendach K.R., Kitzmiller J.A., Leino D., Lombardo R.C., Smolarek T.A., Rathbun P.A. Neonatal lung disease associated with TBX4 mutations. J. Pediatr. 2019;206:286–292.e1. doi: 10.1016/j.jpeds.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szafranski P., Coban-Akdemir Z.H., Rupps R., Grazioli S., Wensley D., Jhangiani S.N., Popek E., Lee A.F., Lupski J.R., Boerkoel C.F., Stankiewicz P. Phenotypic expansion of TBX4 mutations to include acinar dysplasia of the lungs. Am. J. Med. Genet. A. 2016;170:2440–2444. doi: 10.1002/ajmg.a.37822. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann B.G., Kispert A. The T genes in embryogenesis. Trends Genet. 1994;10:280–286. doi: 10.1016/0168-9525(90)90011-t. [DOI] [PubMed] [Google Scholar]
- 19.Smith J. T-box genes: What they do and how they do it. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- 20.Papaioannou V.E., Silver L.M. The T-box gene family. BioEssays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 21.Papaioannou V.E. T-box genes in development: from hydra to humans. Int. Rev. Cytol. 2001;207:1–70. doi: 10.1016/s0074-7696(01)07002-4. [DOI] [PubMed] [Google Scholar]
- 22.Haworth K., Putt W., Cattanach B., Breen M., Binns M., Lingaas F., Edwards Y.H. Canine homolog of the T-box transcription factor T; failure of the protein to bind to its DNA target leads to a short-tail phenotype. Mamm. Genome. 2001;12:212–218. doi: 10.1007/s003350010253. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh T.K., Brook J.D., Wilsdon A. T-box genes in human development and disease. Curr. Top. Dev. Biol. 2017;122:383–415. doi: 10.1016/bs.ctdb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Naiche L.A., Papaioannou V.E. Loss of Tbx4 blocks hind limb development and affects vascularization and fusion of the allantois. Development. 2003;130:2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Esteban C., Tsukui T., Yonei S., Magallon J., Tamura K., Izpisua Belmonte J.C. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature. 1999;398:814–818. doi: 10.1038/19769. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi J.K., Koshiba-Takeuchi K., Suzuki T., Kamimura M., Ogura K., Ogura T. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development. 2003;130:2729–2739. doi: 10.1242/dev.00474. [DOI] [PubMed] [Google Scholar]
- 27.Agarwal P., Wylie J.N., Galceran J., Arkhitko O., Li C., Deng C., Grosschedl R., Bruneau B.G. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–633. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- 28.Glaser A., Arora R., Hoffmann S., Li L., Gretz N., Papaioannou V.E., Rappold G.A. Tbx4 interacts with the short stature homeobox gene Shox2 in limb development. Dev. Dyn. 2014;243:629–639. doi: 10.1002/dvdy.24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bamshad M., Lin R.C., Law D.J., Watkins W.C., Krakowiak P.A., Moore M.E., Franceschini P., Lala R., Holmes L.B., Gebuhr T.C. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat. Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 30.Gibson-Brown J.J., Agulnik S.I., Chapman D.L., Alexiou M., Garvey N., Silver L.M., Papaioannou V.E. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hind limb identity. Mech. Dev. 1996;56:93–101. doi: 10.1016/0925-4773(96)00514-x. [DOI] [PubMed] [Google Scholar]
- 31.Davenport T.G., Jerome-Majewska L.A., Papaioannou V.E. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- 32.Basson C.T., Bachinsky D.R., Lin R.C., Levi T., Elkins J.A., Soults J., Grayzel D., Kroumpouzou E., Traill T.A., Leblanc-Straceski J. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 33.Li Q.Y., Newbury-Ecob R.A., Terrett J.A., Wilson D.I., Curtis A.R., Yi C.H., Gebuhr T., Bullen P.J., Robson S.C., Strachan T. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat. Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 34.Rallis C., Bruneau B.G., Del Buono J., Seidman C.E., Seidman J.G., Nissim S., Tabin C.J., Logan M.P.O. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130:2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- 35.Bruneau B.G., Nemer G., Schmitt J.P., Charron F., Robitaille L., Caron S., Conner D.A., Gessler M., Nemer M., Seidman C.E., Seidman J.G. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi J.K., Koshiba-Takeuchi K., Matsumoto K., Vogel-Höpker A., Naitoh-Matsuo M., Ogura K., Takahashi N., Yasuda K., Ogura T. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature. 1999;398:810–814. doi: 10.1038/19762. [DOI] [PubMed] [Google Scholar]
- 37.Szenker-Ravi E., Altunoglu U., Leushacke M., Bosso-Lefèvre C., Khatoo M., Thi Tran H., Naert T., Noelanders R., Hajamohideen A., Beneteau C. RSPO2 inhibition of RNF43 and ZNRF3 governs limb development independently of LGR4/5/6. Nature. 2018;557:564–569. doi: 10.1038/s41586-018-0118-y. [DOI] [PubMed] [Google Scholar]
- 38.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naiche L.A., Papaioannou V.E. Tbx4 is not required for hind limb identity or post-bud hind limb outgrowth. Development. 2007;134:93–103. doi: 10.1242/dev.02712. [DOI] [PubMed] [Google Scholar]
- 40.Vickerman L., Neufeld S., Cobb J. Shox2 function couples neural, muscular and skeletal development in the proximal forelimb. Dev. Biol. 2011;350:323–336. doi: 10.1016/j.ydbio.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 41.Arora R., Metzger R.J., Papaioannou V.E. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8:e1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimmakayalu M., Major H., Sheffield V., Solomon D.H., Smith R.J., Patil S.R., Shchelochkov O.A. Microdeletion of 17q22q23.2 encompassing TBX2 and TBX4 in a patient with congenital microcephaly, thyroid duct cyst, sensorineural hearing loss, and pulmonary hypertension. Am. J. Med. Genet. A. 2011;155A:418–423. doi: 10.1002/ajmg.a.33827. [DOI] [PubMed] [Google Scholar]
- 43.Peterson J.F., Ghaloul-Gonzalez L., Madan-Khetarpal S., Hartman J., Surti U., Rajkovic A., Yatsenko S.A. Familial microduplication of 17q23.1–q23.2 involving TBX4 is associated with congenital clubfoot and reduced penetrance in females. Am. J. Med. Genet. A. 2014;164A:364–369. doi: 10.1002/ajmg.a.36238. [DOI] [PubMed] [Google Scholar]
- 44.Schönewolf-Greulich B., Ronan A., Ravn K., Baekgaard P., Lodahl M., Nielsen K., Rendtorff N.D., Tranebjaerg L., Brøndum-Nielsen K., Tümer Z. Two new cases with microdeletion of 17q23.2 suggest presence of a candidate gene for sensorineural hearing loss within this region. Am. J. Med. Genet. A. 2011;155A:2964–2969. doi: 10.1002/ajmg.a.34302. [DOI] [PubMed] [Google Scholar]
- 45.Don E.K., de Jong-Curtain T.A., Doggett K., Hall T.E., Heng B., Badrock A.P., Winnick C., Nicholson G.A., Guillemin G.J., Currie P.D. Genetic basis of hind limb loss in a naturally occurring vertebrate model. Biol. Open. 2016;5:359–366. doi: 10.1242/bio.016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Q., Fan S., Zhang Y., Xu M., Zhang H., Yang Y., Lee A.P., Woltering J.M., Ravi V., Gunter H.M. The seahorse genome and the evolution of its specialized morphology. Nature. 2016;540:395–399. doi: 10.1038/nature20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naert T., Van Nieuwenhuysen T., Vleminckx K. TALENs and CRISPR/Cas9 fuel genetically engineered clinically relevant Xenopus tropicalis tumor models. Genesis. 2017;55:e23005. doi: 10.1002/dvg.23005. [DOI] [PubMed] [Google Scholar]
- 48.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.-D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boel A., Steyaert W., De Rocker N., Menten B., Callewaert B., De Paepe A., Coucke P., Willaert A. BATCH-GE: Batch analysis of Next-Generation Sequencing data for genome editing assessment. Sci. Rep. 2016;6:30330. doi: 10.1038/srep30330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.