Abstract
Long noncoding RNAs (lncRNAs) are an important class of pervasive noncoding RNA involved in a variety of biological functions. Numerous studies have demonstrated their important regulatory role in human disease, especially cancer. However, the mechanism underlying the transcription of lncRNAs is not fully elucidated. Here, a comparison of local chromatin structure of the ROR lncRNA locus revealed a cohesin-complex-mediated intrachromosomal loop that is juxtaposed with an upstream enhancer to the ROR promoter, enabling activation of endogenous ROR lncRNA in tumor cells. This chromosomal interaction was not observed in normal control cells. Knockdown of SMC1 by RNAi or deletion of the enhancer DNA by CRISPR/Cas9 abolished the intrachromosomal interaction, resulting in ROR lncRNA silencing and inhibition of the tumor progression in animals carrying tumor xenografts. Our results reveal a novel mechanism by which the cohesin-orchestrated intrachromosomal looping may serve as a critical epigenetic driver to activate transcription of ROR lncRNA, subsequently inducing tumorigenesis. Our data represent a novel chromosomal folding pattern of lncRNA regulation, thereby providing a novel alternative concept of chromosomal interaction in lncRNA-triggered tumorigenesis.
Keywords: intrachromosomal looping, ROR lncRNA, SMC1, tumorigenesis
Fan and his colleagues demonstrate that a cohesin-orchestrated intrachromosomal looping that formed between the ROR promoter and its newly identified enhancer serve as a critical epigenetic driver that activates transcription of ROR lncRNA in cancer, thereby specifying a new pattern of lncRNA regulation by intrachromosomal interaction.
Introduction
Genomic research revealed that the mammalian transcriptome encodes a large number of long noncoding RNA (lncRNA) transcripts in multiple organisms.1, 2, 3 These lncRNAs show clear evolutionary conservation4, 5 and are involved in epigenetic regulatory networks,6, 7 such as Xist, which is involved in X chromosome inactivation,8 and Kcnq1ot1, which is involved in genomic imprinting.9 Consequently, a greater number of studies were performed to profile the lncRNA expression patterns in different tissues and/or organisms to discover new molecular biomarkers or therapeutic targets for human disease. Indeed, increasing evidence suggests that altered lncRNA expression levels are strongly associated with various human diseases, including cancer,10, 11 and correlate with patient clinicopathological features and prognosis. For example, HOTAIR lncRNA was upregulated in breast tumors and associated with metastases,12 and MALAT1 lncRNA was significantly higher in HCC tissues that correlated with HCC prognosis.13 However, the precise mechanism of the transcription of most lncRNAs remains to be fully elucidated.
The process of transcriptional regulation mainly includes cis-control elements, trans-control elements, and epigenetic modification.14, 15 Intriguingly, emerging studies have unraveled the role of nonlinear interactions, which are also called three-dimensional (3D) chromosomal interactions, in transcriptional regulation.16, 17 The promoters of active β-globin genes interacting with an upstream regulatory sequence serve as the first example of intrachromosomal interactions contributing to transcriptional regulation.18, 19 Similar intrachromosomal interactions have now been described for many other genes that are on average separated by approximately 150 kb, including approximately 30,000 interactions between active promoters and putative enhancers.20 For instance, transcription of HoxD genes in digits integrates the collective activities of several regulatory elements. The multiple regulatory islands that cover approximately 800 kb upstream of the HoxD cluster either have enhancer activity or serve as anchor points.21 CCCTC-binding factor (CTCF), cohesion (SMC1), and mediator (MED12) are the top three factors that have been reported as both essential for cellular functions and correlated with a specific looping interaction using chromosome conformation capture (3C) technology.17, 22 Distinct combinations of CTCF, mediator, and cohesin show widespread enrichment in chromatin interactions at different length scales.23 As Kagey et al.17 previously reported, high-confidence SMC1 binding sites significantly overlapped high-confidence MED12 binding sites. Furthermore, cohesin-mediated bridge-proximal enhancer-promoter interactions are functionally linked to gene expression.23 In addition, we also found that the SMC1-orchestrated intrachromosomal loop is a critical epigenetic barrier to the induction of pluripotency.16 As clearly noted in the examples above, we assume that intrachromosomal interactions are likely to involve transcriptional regulation of ROR lncRNAs, which act as a decoy oncoRNA and play an important regulatory role in tumorigenesis as previously reported.24
In this study, we attempted to identify the potential role of intrachromosomal interactions in the transcriptional regulation of ROR lncRNA. Using 3C approaches, we reveal that the SMC1-orchestrated intrachromosomal looping may serve as a critical epigenetic driver of ROR lncRNA transcription, thereby guiding the induction of tumorigenesis.
Results
A Novel Intrachromosomal Looping Exists at the ROR Locus
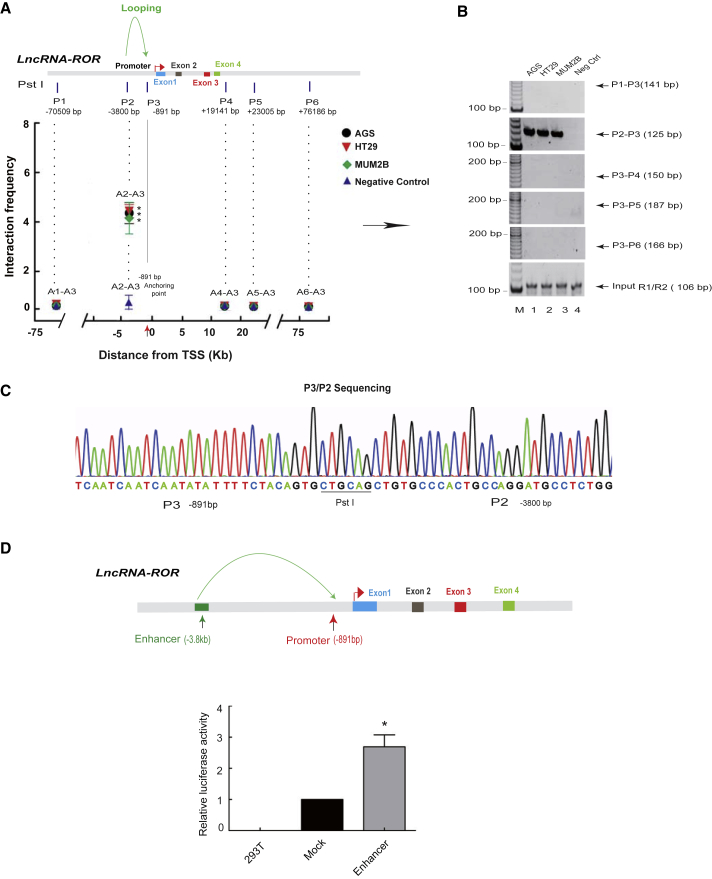
We were interested in exploring the underlying epigenetic modifications, such as local chromatin structure remodeling, that may contribute to ROR transcription. To investigate our hypothesis, we explored whether the remodeling of local chromatin structure contributes to ROR transcription during the progression of tumorigenesis. We used 3C methodology25 to examine the chromatin structure present in human tumor cells compared with normal cells. As expected, in ROR-expressing tumor cells (AGS, HT29, and MUM2B), the ROR promoter DNA interacted frequently with a DNA region that is located approximately 3,800 bp upstream of the promoter (P2-P3 interaction; Figure 1A). This chromatin interaction was not detected in ROR-deficient control cells (Figure 1B, second panel, lane 4). Then, 3C products were confirmed by sequencing, and the identity of the ligated ROR core promoter (P3)/ROR upstream −3,800 bp was revealed (Figure 1C). To address the role of the upstream DNA sequence, we cloned a 962-bp fragment containing the site P2 (Figure 1D, top panel) and tested its activity that regulated gene expression. As noted in Figure 1D, the 962-bp DNA fragment significantly augments promoter activity as measured by luciferase activity. These data indicate that intrachromosomal looping may help bring the upstream enhancer in close proximity to the ROR core promoter, where it activates ROR lncRNA expression in tumor cells. The absence of intrachromosomal looping may be a critical epigenetic barrier that impedes ROR activation.
Figure 1.
Novel Intra-chromosomal Looping Exists at the ROR Locus
(A and B) Intrachromosomal interaction between the ROR promoter (P3) and enhancer (P2) regions in AGS, HT29, and MUM2B. (B) The intrachromosomal interactions between P1–P3, P2–P3, P3–P4, P3–P5, and P3–P6 as assessed by PCR. R1/R2 PCR was used as the positive control. (A) Gray intensity analysis of the brands (P3 bait, vertical arrows) to each Pst I site and the off-target site (P1–P3, P3–P4, P3–P5, and P3–P6). Numbers under P1–P6: distance from the translation start site (TSS). The interaction frequency was determined by normalizing the 3C PCR signal over that of the positive control (R1/R2 PCR). *p < 0.05 compared with negative control fibroblasts. (C) 3C products are confirmed by DNA sequencing. The 3C products derived from the ROR promoter P2 and P3 interaction (P2/P3) were cloned and sequenced. The 3C products contain the Pst I site (5′-CTGCAG-3′) that was flanked on both sides by the ROR promoter −891 and −3,800 bp. (D) Identification of the ROR upstream interacting region as an ROR enhancer. Enhancer activity was measured as the relative luciferase units in 293T cells. ROR enhancer was inserted upstream of pGL2-promoter-Luc. For comparison, the luciferase expression of the mock insert at 48 h was arbitrarily set as 1 in the calculation. *p < 0.05 compared with mock luciferase expression. 293T, wild-type 293T cells; M, marker; Mock, empty pGL2-promoter-Luc vector.
SMC1 Forms Intrachromosomal Looping at the ROR Locus
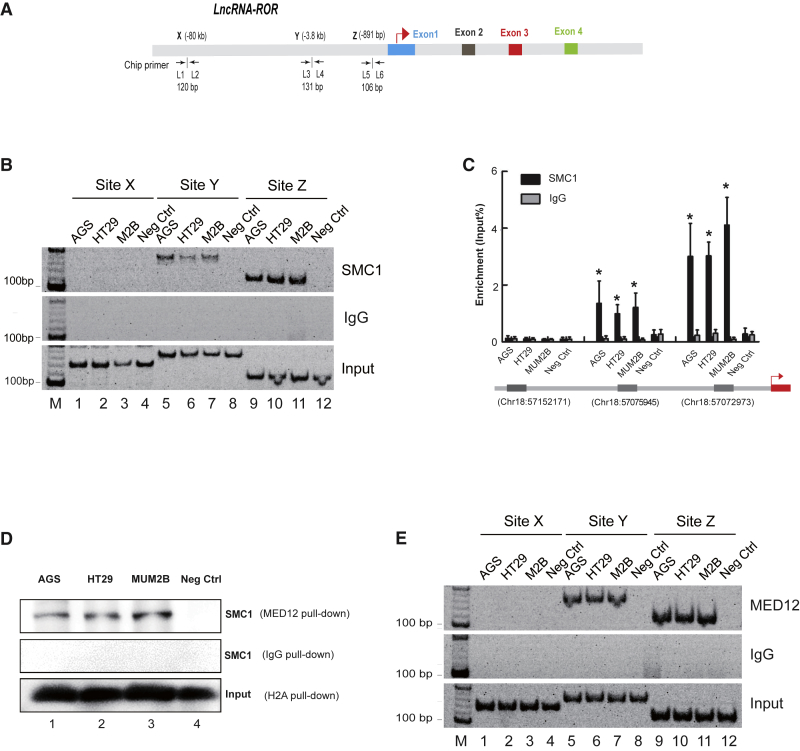
Next, we determined which factor coordinated this chromosomal looping. Because cohesin-complex and mediator-complex were classic elements that orchestrated intrachromosomal loops between active promoters and putative enhancers,16, 17 we were interested in whether a cohesin-complex was involved in the chromosomal loop at the ROR locus (Figure 2A). As expected, we found the ROR promoter (Figure 2B, first panel, lanes 9–11) and its upstream enhancer fragment (−3.8 kb) (Figure 2B, first panel, lanes 5–7) interact with the cohesin-complex protein SMC1 in AGS, HT29, and MUM2B cells, but not in control cells (Figure 2B, first panel, lanes 4, 8, and 12). Similarly, we also detected consistent results in quantitative chromatin immunoprecipitation (ChIP) assays (Figure 2C). Because cohesin and mediator work together to mediate gene expression and chromatin architecture,16, 17 we then further investigated the ability of interaction between mediator and cohesion complexes in AGS, HT29, and MUM2B. As expected, co-immunoprecipitation (coIP) showed that SMC1 protein interacted with MED12 protein (Figure 2D). We next determined the binding of mediator-complex protein MED12 with the ROR promoter and enhancer in AGS, HT29, and MUM2B. We found that MED12 could interact with the ROR promoter (Figure 2E, first panel, lanes 9–11) and its enhancer (Figure 2E, first panel, lanes 5–7). These data indicate that chromatin-binding complex cohesion is likely to recruit mediator complex to form SMC1-mediated intrachromosomal looping between the ROR promoter and enhancer in tumor cells.
Figure 2.
SMC1 Forms Intrachromosomal Looping at the ROR Locus
(A) Schematic diagram of the ROR lncRNA and the position of primers used for ChIP analyses. L1 through L6, primer names; arrows, transcriptional direction. (B) The interaction between SMC1 and ROR lncRNA. ChIP assay demonstrating that SMC1 binds the ROR promoter (−891 bp) and the upstream fragment (−3.8 kb). IgG is used as a negative control. Sites X, Y, and Z are ChIP detection sites. (C) The qPCR-ChIP assay showing the interaction between SMC1 and the ROR lncRNA. ChIP sites on the chromosome are listed at the bottom. Immunoglobulin G (IgG) is the negative control. All of the data are presented as the mean ± SD. *p < 0.05 compared with the negative control (fibroblast). (D) SMC1 interacted directly with MED12 as detected by co-immunoprecipitation. Cohesin (SMC1) is detected by western blot after AGS, HT29, and MUM2B immunoprecipitation. (E) ChIP assay detecting the interaction between MED12 and the ROR lncRNA. Sites X, Y, and Z are ChIP detection sites. M, marker.
The SMC1-Orchestrated Intrachromosomal Loop Was a Critical Epigenetic Barrier to Activation of ROR Transcription In Vitro
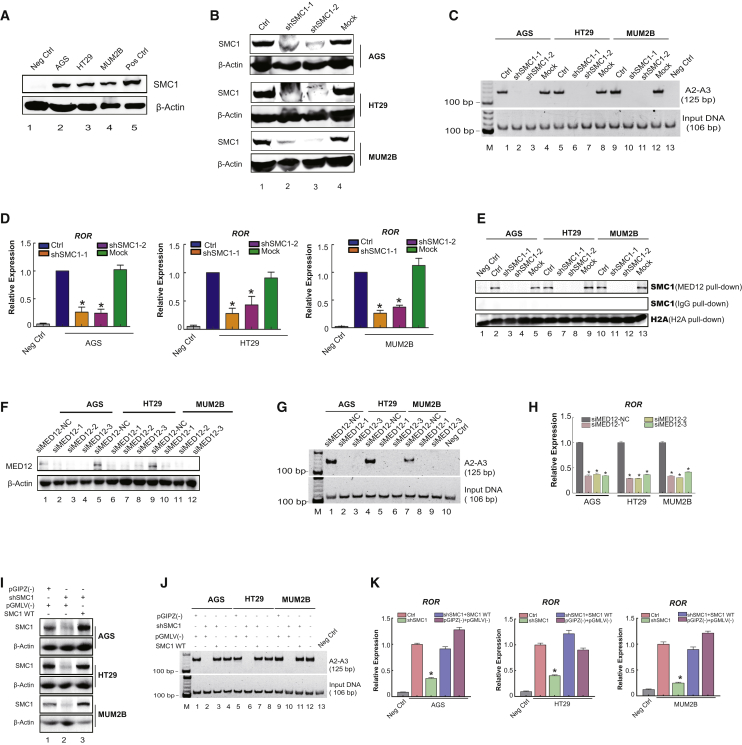
Next, we investigate whether SMC1-mediated intrachromosomal looping controls the expression of endogenous ROR. We first analyzed the expression level of SMC1 in AGS, HT29, and MUM2B. As shown in Figure 3A, SMC1 exhibited significantly higher expression in AGS, HT29, and MUM2B tumor cells (Figure 3A, lanes 2–4). We then used conventional RNAi to successfully knock down SMC1 expression in AGS, HT29, and MUM2B. Real-time PCR (Figure S1) and western blot (Figure 3B) showed that SMC1 protein expression was significantly inhibited in tumor cells. We then examined whether intrachromosomal looping was abolished after SMC1 silencing. As expected, SMC1 knockdown abolished the intrachromosomal looping (Figure 3C, lanes 2, 3, 6, 7, 10, and 11), suggesting a critical role of SMC1 in the formation of intrachromosomal looping of the ROR promoter and enhancer. Notably, the weak expression of ROR in SMC1-silenced cells (Figure 3D) further confirmed the importance of SMC1 in activation of endogenous ROR expression in tumor cells. Moreover, coIP assays showed that the interaction between SMC1 and MED12 was abolished when SMC1 was knocked down in three tumor cells (Figure 3E, lanes 3, 4, 7, 8, 11, and 12). These data support our hypothesis that transcriptional activation of endogenous ROR requires the formation of SMC1-dependent intrachromosomal looping.
Figure 3.
SMC1-Mediated Intrachromosomal Looping Determines Endogenous ROR Transcription
(A) SMC1 expression in a variety of tumor cell lines was detected by western blot. In brief, 293T cells and fibroblasts were used as positive and negative controls, respectively. (B) Western blot detecting SMC1 expression in shRNA-SMC1-treated AGS, HT29, and MUM2B cells. (C) Chromosome conformation capture (3C) was used to examine the intrachromosomal looping between the ROR promoter (P3) and enhancer (P2) regions in SMC1 knockdown AGS, HT29, and MUM2B. Once SMC1 was depleted, intrachromosomal looping was lost. (D) Real-time PCR measurement of ROR expression in SMC1-knockdown tumor cells. ROR expression levels were remarkably decreased in SMC1-silenced tumor cells that lack intrachromosomal looping. (E) Interaction of MED12 and SMC1 detected by co-immunoprecipitation. The interaction of MED12 and SMC1 was abolished after SMC1 knockdown. (F) Silenced expression of MED12 using siRNAs in AGS and HT29 cells. Western blot demonstrated that siMED12 provided the optimal deletion of MED12. Experiments were performed 48 h following siMED12 (50 nM) and control transfection. (G) Loss of the intrachromosomal loop between the ROR promoter and enhancer regions in MED12-knockdown tumor cells as quantified by PCR. (H) qPCR analysis of ROR expression in MED12-knockdown tumor cells. The expression levels of ROR were remarkably decreased following loss of the intrachromosomal loop in MED12-downregulated tumor cells. (I) SMC1 protein levels in cells treated with wild-type SMC1, shSMC1, or empty vector were measured with western blotting. SMC1 was restored in the presence of the overexpressed wild-type SMC1. (J) 3C detected the intrachromosomal loop in the rescue study of SMC1-knockdown cells. Cohesin-mediated intrachromosomal looping was restored in SMC1-knockdown cells by overexpression of wild-type SMC1. (K) qPCR analysis of ROR expression in the rescue study of SMC1 knockdown. ROR expression levels were restored following overexpression of wild-type SMC1 in SMC1-silenced tumor cells. Mock, empty pGIPZ vector; NC, nonsilencing control; Neg Ctrl, fibroblast; pGIZ(−), empty pGIPZ vector; pGMLV(−), empty pGIPZ vector; SMC1 WT (wild-type), SMC1 ORF was cloned into pGMLV plasmids for stable expression.
Because SMC1 and MED12 combined at the ROR locus, we then determined whether downregulated MED12 could influence intrachromosomal looping. Thus, to decipher the potential role of MED12 in the chromatin architecture, we aimed to knock down MED12 expression using conventional RNAi methodology (Figure 3F). 3C was then performed to detect the effect on intrachromosomal loop in MED12-knockdown cells. Intriguingly, we detected that intrachromosomal looping was abolished after MED12 silencing (Figure 3G, lanes 2, 3, 5, 6, 8, and 9). Similarly, ROR expression was remarkably decreased once the intrachromosomal loop was lost (Figure 3H).
To obtain further insights into the specific role of SMC1 in this intrachromosomal looping, we performed a rescue study of SMC1 knockdown. The results showed the remarkably increased expression levels of SMC1 after SMC1-knockdown tumor cells were transfected with wild-type SMC1 plasmid (Figure 3I, first, third, and fifth panels, lane 3). Interestingly, the cohesin-orchestrated intrachromosomal looping was successfully restored (Figure 3J, first panel, lanes 3, 7, and 11). ROR expression was dependent on the rescued intrachromosomal looping that was effectively restored by overexpression of the wild-type SMC1 (Figure 3K). Moreover, no direct positive correlation was noted between ROR and epigenetic regulator (SMC1 and MED12) expression in colon cancer (Figure S2A), gastric cancer (Figure S2B), and uveal melanoma (UVM) (Figure S2C) based on the GEPIA Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn).26 These data indicated that SMC1 itself was not able to activate ROR lncRNA transcription and likely helped to bring the upstream enhancer in close proximity to the ROR core promoter, where it activates ROR lncRNA expression in tumor cells. As suggested, we transfected normal cells (NCM60, GES-1, PIG1) with low ROR and SMC1 expression with wild-type SMC1 (Figures S3A and S3B). As shown in Figure S3C, SMC1 was overexpressed in NCM60, GES-1, and PIG1. However, cohesin-orchestrated intrachromosomal looping does not occur (Figure S3D) despite SMC1 overexpression.
Enhancer Deletion Disrupts the Intrachromosomal Looping and Downregulated ROR Expression
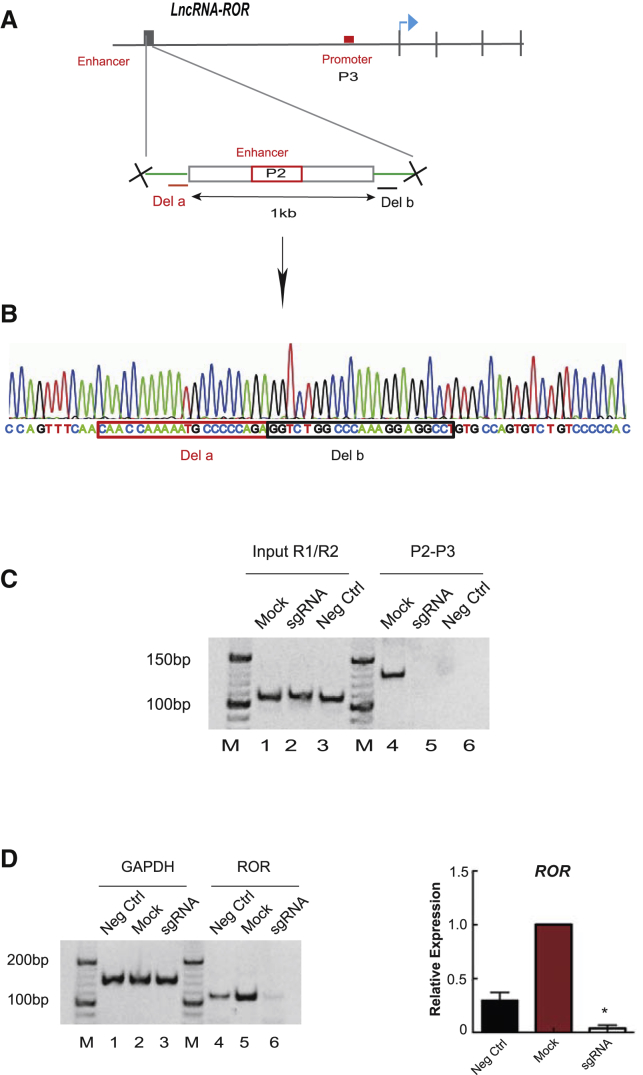
To further investigate the role of intrachromosomal looping in ROR regulation, we next deleted the ∼1-kb enhancer DNA using the CRISPR/Cas9 method. As shown in Figures 4A and 4B, the 1-kb enhancer was deleted from the genome. Next, we found that the intrachromosomal looping between ROR promoter and the upstream enhancer DNA was abolished (Figure 4C, lane 5). To determine whether the deletion would control ROR transcription, we assessed ROR lncRNA expression. As expected, weak ROR expression was detected in enhancer-deleted tumor cells (Figure 4D, lane 6). Similarly, qPCR data were consistent with this finding (Figure 4D, right). Taken together, these data indicate that the intrachromosomal looping between the ROR promoter and the enhancer controls endogenous ROR expression.
Figure 4.
Enhancer Deletion Disrupts Intrachromosomal Looping and ROR Expression
(A) Schematic of lncRNA-ROR fragment deletion using CRISPR-Cas9. (B) Deletion of 1-kb P2-containing region from the genome by CRISPR-Cas9 as confirmed by sequence. (C) Loss of intrachromosomal looping in MUM2B-sgRNA using CRISPR-Cas9. (D) Weak expression of lncRNA-ROR in MUM2B-CRISPR-Cas9. *p < 0.05 compared with the mock. Mock, empty CRISPR-Cas9 vector.
Interruption of SMC1-Mediated Intrachromosomal Looping Inhibits Tumorigenesis
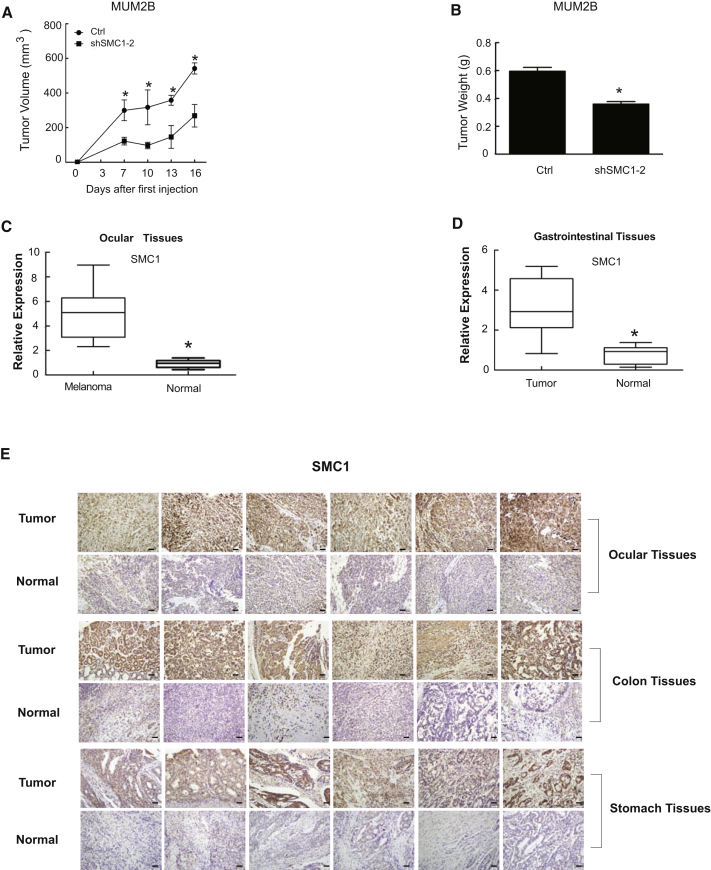
To apply the in vitro findings to the in vivo situation, we generated a xenograft tumor model of MUM2B cells in nude mice. As expected, we noticed that tumor growth was significantly reduced in those animals that received the SMC1-silencing MUM2B cells (n = 6; *p < 0.05) compared with wild-type (Ctrl) MUM2B cells (Figure 5A). In addition, SMC1 knockdown resulted in approximately 40% reduction in tumor weight (Figure 5B). To verify the clinical significance of SMC1, we collected a set of tumor tissues paired with adjacent normal tissue from diagnosed patients, including gastric tumors (n = 8) and ocular melanomas (n = 11). Notably, we detected a prominent increase in SMC1 expression in all of the tumor tissues compared with adjacent healthy specimens (Figures 5C and 5D). We then performed immunohistochemistry-staining assays to detect SMC1 protein expression in tumor tissues. The results clearly showed that SMC1 protein expression was remarkably increased in melanoma tumors (Figure 5E, first row), colon cancer (Figure 5E, first row), and gastric cancer (Figure 5E, third row) tissues compared with normal tissues (Figure 5E, second, fourth, and sixth). These data further highlight the clinical importance of SMC1 in gastrointestinal cancer and ocular melanoma.
Figure 5.
SMC1 Knockdown Inhibits Tumorigenesis
(A) Tumor volume following SMC1 knockdown in MUM2B cells (n = 6). Suppressive effects on tumor growth in mice treated with knockdown SMC1. *p < 0.05 compared with control. (B) Tumor weight on day 20 (n = 6). Knockdown of SMC1 expression results in tumor weight reduction. *p < 0.05 compared with control. (C and D) SMC1 expression levels in normal, colon cancer, and melanoma tissues. SMC1 was highly expressed in colon cancer and melanoma tissues compared with normal tissues. *p < 0.05. Scale bars, 50 μm. (E) Immunohistochemical staining of SMC1 in tumor and normal tissues. SMC1 expression in tumor sections from six melanoma patients, six colon cancer patients, and six gastric cancer patients were higher than normal tissues (original magnification ×200).
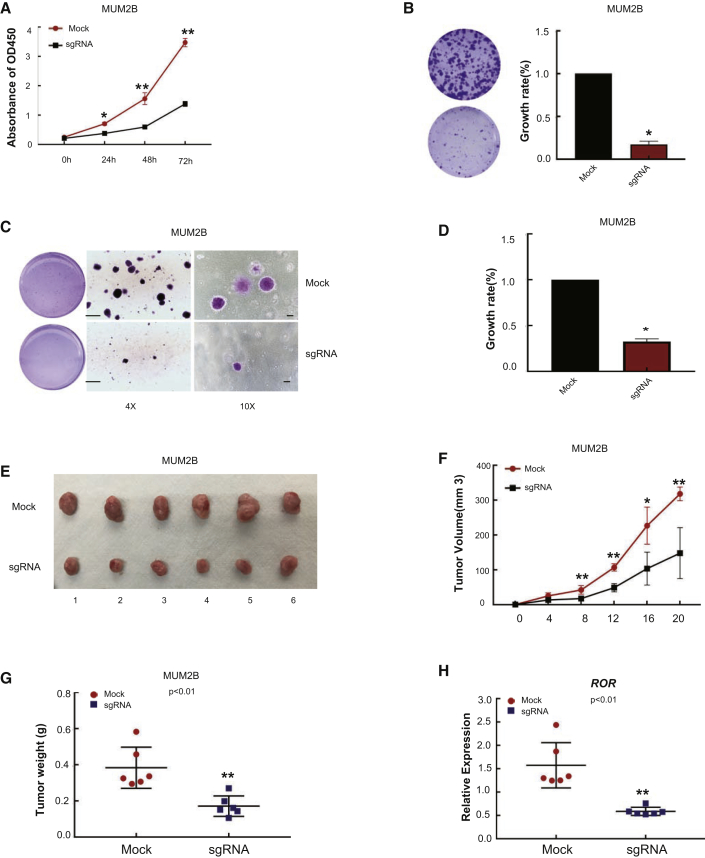
We next tested whether intrachromosomal looping deficiency also inhibits tumor progression in another enhancer-deletion model. We then examined cell phenotype changes. CCK8 assays showed that cell growth was significantly reduced in enhancer-deleted tumor cells (Figure 6A). Moreover, two colony formation methods were used to evaluate the capacity of tumor colony formation, and the data showed that the number of macrocolonies and microcolonies was significantly decreased in enhancer-deleted tumor cells using plate (Figure 6B) and soft agar (Figures 6C and 6D) methods. To examine the role of intrachromosomal looping in vivo in animal experiments, we established a xenograft model in nude mice using enhancer-deleted MUM2B cells. We then evaluated the size of the resultant tumors every 2 days for 20 days. As expected, both tumor volume (Figure 6F) and weight (Figure 6G) in the enhancer-deleted group were significantly reduced compared with the empty vector group. Tumor size was also reduced in the enhancer-deleted group (Figure 6E). In addition, a remarkable decrease in ROR expression was observed in the enhancer-deleted group (Figure 6H). These data suggested that the cohesin-complex-mediated intrachromosomal loop is a key regulator of ROR-mediated tumorigenesis both in vitro and in vivo.
Figure 6.
Enhancer Deletion Inhibits Tumor Growth In Vitro and In Vivo
(A) CCK8 assays show tumor cell growth after enhancer deletion. Cell growth was obviously restrained at day 3 in enhancer-deleted MUM2B cells. The absorbance values were detected at 24, 48, and 72 h, and the control was arbitrarily set at 100% on day 1. *p < 0.05, **p < 0.01 compared with mock. Scale bars, 500 μm (left; original magnification ×4), 100 μm (right; original magnification ×10). (B–D) Plate clone formation assays (B) and soft agar assays (C and D) demonstrate that the tumor formation ability was significantly reduced in enhancer-deleted MUM2B cells. *p < 0.05 compared with the mock. (E and F) Suppressive effects on tumor volume (F) and weight (G) in mice. *p < 0.05, **p < 0.01 compared with mock. (G) Mice were sacrificed, and tumors were isolated 20 days after inoculation. The growth of enhancer-deleted MUM2B cells was compared with empty vector-transfected cells. **p < 0.01 compared with mock. (H) Real-time PCR showing that ROR lncRNA expression was significantly decreased in the enhancer-deleted group. **p < 0.01 compared with mock.
Discussion
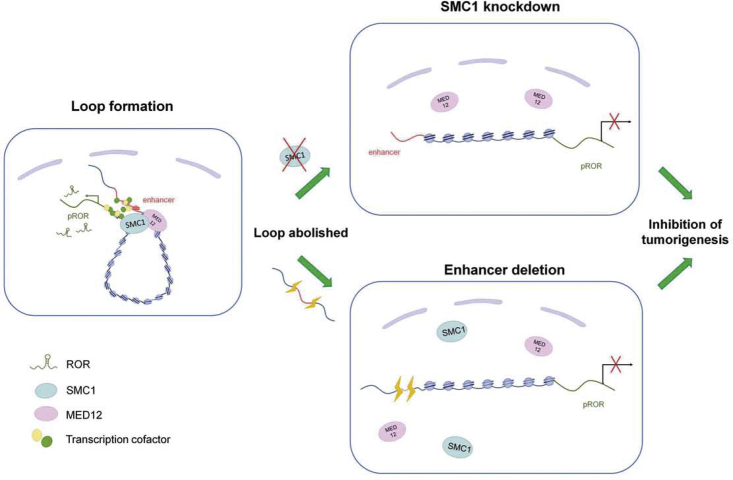
To date, numerous studies have focused on lncRNA functions in a variety of diseases, especially cancers.27, 28, 29, 30 A great number of cancer-associated lncRNAs have been reported in various tumors, such as CCAT1,31 MALAT1,13 HOTAIR,12 and CANT1;32 however, the transcriptional regulation of most lncRNAs remains unclear. We thus began to reveal the precise mechanism underlying the transcription of lncRNAs. In the past decades, a large number of cis-regulatory linear sequences have been reported to control gene transcription.15 Given that the genome exhibits 3D organization, long-range chromatin interactions between these elements are easily formed to control their target genes. For example, a long-range intrachromosomal looping between KvDMR1 and the Kcnq1 promoter maintained mono-allelic expression of the Kcnq1 genes,9 and EVI1 oncogene activation is required for the formation of chromosomal looping between the EVI1 promoter and GATA2 enhancer in leukemia.33 Given these facts, it raises the possibility that the long-range 3D chromosomal architecture may also be involved in the transcription of ROR lncRNAs. In support of this hypothesis, here, we for the first time demonstrate that an intrachromosomal looping formed between the ROR promoter and its newly identified enhancer to subsequently activate the expression of ROR and drivers of tumorigenesis, thereby representing new insight into our understanding of lncRNA transcription in cancer (Figure 7).
Figure 7.
A Schematic Model of the Intrachromosomal Looping in ROR Transcription and ROR-Mediated Tumorigenesis
A novel oncogenetic mechanism in which the intrachromosomal interaction bound by cohesin mediator serves as an epigenetic driver to activate the expression of oncogenetic lncRNA and subsequently trigger lncRNA-mediating tumorigenesis. However, when SMC1 was silenced or enhancer DNA was deleted, the intrachromosomal looping between ROR promoter and enhancer was abolished, subsequently inhibiting ROR expression and disrupting ROR-triggered tumorigenesis.
The cohesin complex is a major player in transcriptional regulation during interphase.34 The complex acts together with the mediator complex to maintain the physical interaction between promoters and enhancers of target genes.16, 17 Here, we suggest that cohesin-orchestrated intrachromosomal looping is a critical epigenetic activator of transcription initiation of ROR lncRNA. This concept is supported by the fact that the ROR lncRNA was silenced after interruption of the intrachromosomal interaction by knockdown of SMC1 or enhancer deletion. To our knowledge, this is the first example whereby intrachromosomal interaction mediated by cohesin contributes to transcriptional activation of lncRNA.
SMC1 is one of the components of cohesin that was recently shown to be involved in chromatin loop organizer function in the interphase nucleus, thereby regulating gene expression;17, 35, 36, 37 evidence indicates that SMC1 is closely correlated to tumorigenesis and development.38 We detected a prominent increase in SMC1 expression in tumors that was further highlighted in clinical ocular melanoma. Knockdown of SMC1 expression leads to significantly repressed tumor progression in vivo. Notably, targeted suppression of SMC1 expression in tumors silences ROR expression, and intrachromosomal looping between the ROR promoter and upstream DNA was abolished. Thus, we suggest that high SMC1 expression enhances its role as a stabilizer of the interaction between the ROR promoter and the upstream DNA, thereby contributing to ROR lncRNA expression to promote tumorigenesis.
It should also be noted that rescue study of SMC1 knockdown successfully restored the cohesin-orchestrated intrachromosomal looping in SMC1 knockdown cells and ultimately effectively restored ROR expression. However, this successful “bail-out” was confined to SMC1-silenced tumor cells. Thus, the role of SMC1 in tumors may not involve the induction of tumorigenesis directly. Rather, SMC1 holds the ROR promoter and the upstream DNA together to activate endogenous ROR lncRNA, leading to tumorigenesis. We cannot theoretically eliminate the involvement of other genetic or epigenetic factors in ROR expression, especially in this intrachromosomal looping. It would be of great interest to focus on the identification of other factors to better understand ROR lncRNA transcriptional regulation. Further studies should be focused on the identification of other factors that can organize or occupy chromatin loops to activate ROR lncRNA expression. Nevertheless, to our knowledge, this is the first study to imply that ROR lncRNA expression is highly dependent on intrachromosomal interactions that juxtapose the enhancer and promoter regions of ROR.
In summary, our results reveal a completely novel mechanism in which the intrachromosomal interaction bound by the cohesin mediator serves as an epigenetic driver to activate the expression of oncogenetic lncRNA, subsequently triggering lncRNA-mediated tumorigenesis. Most importantly, because many lncRNAs exhibit variable expression levels in different organs and play an important role in variant diseases, it raises the possibility that the chromosomal interaction might be a critical cause in the pathology of diseases, thereby suggesting new inspiration for exploring the importance of chromosomal interactions in lncRNA transcription and lncRNA-mediated diseases.
Conclusions
In this study, we reveal for the first time a novel mechanism in which the intrachromosomal interaction bound by cohesin mediator serves as an epigenetic driver to activate oncogenetic ROR lncRNA expression, subsequently triggering lncRNA-mediating tumorigenesis. Our data provide a potential therapeutic biomarker and reveal the novel chromosomal folding pattern of lncRNA regulation, thereby providing a novel alternative concept of chromosomal interaction in lncRNA-mediated tumorigenesis.
Materials and Methods
3C
The 3C assay was performed as described previously.16 In brief, 1.0 × 107 cells were cross-linked with 2% formaldehyde and quenched with 0.125 M glycine. Cells were lysed with cell lysis buffer (10 mM Tris [pH 8.0], 10 mM NaCl, 0.2% Nonidet P-40 [NP-40], and protease inhibitors), and nuclei were collected. Nuclei were resuspended in 1× restriction enzyme buffer in the presence of 0.3% SDS and incubated at 37°C for 1 h. Triton X-100 was then added to a final concentration of 1.8% to sequester the SDS. An aliquot of nuclei (2 × 106) was digested with 800 U of restriction enzyme PstI at 37°C overnight. Then, 1.6% SDS was added, and the mixture was incubated at 65°C for 20 min to stop the reaction. Chromatin DNA was diluted with T4 ligation reaction buffer, and 2 μg DNA was ligated with 4,000 U of T4 DNA ligase (Takara, Japan) at 16°C for 4 h (final DNA concentration, 2.5 μg/mL). After treatment with 10 mg/mL Proteinase K at 65°C overnight to reverse cross-links and 0.4 μg/mL RNase A for 30 min at 37°C, DNA was extracted with phenol-chloroform, ethanol precipitated, and used for PCR amplification of the ligated DNA products. PCR primers used in this study are listed in Table S1.
ROR Enhancer-Luciferase Assays
ROR enhancer was amplified from genomic DNA using primers (Table S3) incorporating restriction enzyme sites (Kpn I-Xho I) and cloned into the Kpn I-Xho I sites upstream of the promoter-Luc transcriptional unit. Luciferase assays were performed in 24-well white plates using the Luciferase Assay System (Promega, USA) according to the manufacturer’s protocol.
ChIP
ChIP assays were performed as previously described.16 One hundred million cells were fixed with 1% formaldehyde and sonicated for 8 min (10 s on and 15 s off) on ice with a 2-mm microtip at 40% output control and 90% duty cycle settings. The sonicated chromatin (1 mL) was clarified by centrifugation and snap-frozen in liquid nitrogen. To perform ChIP, sonicated chromatin (150 μL) was diluted 10-fold, and protein G-agarose (60 μL) (Millipore, USA) was added with shaking at 4°C for 2 h. Samples were then briefly centrifuged at 1,000 rpm for 5 min at 4°C, and the supernatant was collected into a new tube. SMC1 and MED12 antibodies were obtained from Abcam (USA) and added to the supernatant overnight at 4°C. PureProteome Protein A and Protein G Magnetic Beads (60 μL) (Millipore, USA) were used to pull down the protein at 4°C for 6 h. DNA released from the bound chromatin after cross-linking reversal and Proteinase K treatment was precipitated and diluted in 100 μL of 0.2 M glycine.
PCRs (3 μL under liquid wax) contained 2 μL ChIP (or input) DNA, 0.5 mM appropriate primer pairs, 50 μM deoxynucleotide triphosphate, and 0.2 U Klen-TaqI (Ab Peptides, USA). The PCR conditions were 95°C for 5 min followed by 34 cycles of 95°C for 30 s, 30 s of optimal annealing temperature, and 72°C for 30 s of extension (Table S2). The PCR products were separated on 8% polyacrylamide-urea gels.
coIP
coIP was performed according to the Nuclear Complex coIP kit protocol (Active motif, USA). In brief, nuclear extracts were prepared by suspending cells in 1× hypotonic buffer for 15 min on ice. The cells were homogenized and centrifuged for 30 s at 14,000 × g. Immunoprecipitation (IP) was performed with 300 μg nuclear protein and 5 μg anti-MED12 antibody (Abcam, CA, USA) in 500 μL IP incubation buffer at 4°C overnight. The reaction mixtures were incubated with PureProteome Protein A and Protein G Magnetic Beads (50 μL) (Millipore, USA) at 4°C for 1 h on a rotator. The immunoprecipitated complexes were washed twice with IP wash buffer supplemented with 1 mg/mL BSA and without BSA. The washed beads were incubated with 2× reduction loading buffer and boiled at 100°C for 5 min. The protein released from components of the complexes was examined by SDS-PAGE and western blotting with anti-SMC1.
RNA Extraction and RT-PCR Analysis
Total RNA was extracted using TRI-REAGENT (Invitrogen, USA) according to the manufacturer’s instructions, and cDNA was synthesized using the PrimeScript RT reagent kit (Takara, Japan).
Western Blot Analysis
Cells were harvested at the indicated time and washed twice with cold PBS. Cell extracts were prepared with lysis buffer and centrifuged at 13,000 × g for 30 min at 4°C. Protein samples were separated by SDS-PAGE in 7.5% (w/v) polyacrylamide gels and transferred to polyvinylidene fluoride membranes. Membranes were immunoblotted with anti-SMC1 (Proteintech, USA). Membranes were incubated with a secondary antibody conjugated to a fluorescent tag. The bands were visualized using the Odyssey infrared Imaging System (LI-COR, Lincoln, NE, USA).
Plasmid, Retrovirus, and Selection
SMC1 short hairpin RNA (shRNA)-mir constructs and verified nonsilencing shRNA were purchased from Open Biosystems (Lafayette, CO, USA) (Table S3). Selection was performed by incubating with 4 μg/mL puromycin for 2 weeks; the clones were screened for GFP expression. The cDNA encoding the SMC1 open reading frame (ORF) was amplified by PCR using primers (Table S3) incorporating restriction enzyme sites (XhoI-BamHI). The PCR fragment was cloned into the digested plasmids of the pGMLV vector.
Lentivirus Packaging and Generation of Stable Cell Lines
The Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was incubated with Opti-MEM I Reduced Serum Medium (GIBCO, USA) and used to transfect 239T cells with 3 μg SMC1 shRNAs or pGMLV-SMC1 plasmids, 3 μg pMD2.D plasmids, and 6 μg PsPax plasmid. The medium was replaced with 10 mL of fresh medium 5 h after transfection. The supernatant containing the viruses was collected at 48 and 72 h, filtered, and concentrated. Twenty-four hours prior to transfection, cells were seeded in a 60-mm dish at 3.0 × 105 cells/dish. The medium was replaced with 25 μL/mL virus-containing supernatant supplemented with 10 ng/mL polybrene (Sigma-Aldrich, USA). After 48 h of transduction, selection of stable SMC1-knockdown cell lines was performed by incubation with 4 μg/mL puromycin (InvivoGen, USA) for 2 weeks; the clones were screened for GFP. For the SMC1 rescue experiment, 4 μg/mL puromycin and 6 μg/mL hygromycin were applied for selection.
CCK8 Cell Viability Assay and Soft Agar Tumor Formation Assays
For CCK8 assay, cells were seeded into a flat-bottomed 96-well culture plate at 2,000 cells per well with 100 μL culture medium. In brief, 10 μL of CCK8 (Dojindo, Japan) solution was added to each well. The samples were incubated for 4 h; then the absorbance was measured at 450 nm in a microplate reader (Varioskan Flash; Thermo, USA) for 4 consecutive days. Soft agar colony formation assays were performed in six-well plates as described in our previous study.24 To calculate the colony formation rate after loss of intrachromosomal looping in MUM2B, the mock cell was set to 1.
Small Interfering RNA
MED12 knockdown was achieved using small interfering RNA (siRNA). Cells were seeded at 200,000 cells per well in six-well plates and transfected using Lipofectamine 2000 (Invitrogen) in Opti-MEM I Reduced Serum Medium with 50 nM siRNA (Invitrogen). Forty-eight hours posttransfection, cells were harvested in TRIzol for RNA isolation (Invitrogen) or lysed in radioimmunoprecipitation assay (RIPA) lysis buffer for western blotting.
Plate Colony Formation Assay
A plate colony formation assay was performed in six-well plates. A total of 1,000 cells were suspended in 2.0 mL of complete medium and seeded into each well. The cells were cultured with complete medium for 10 days. For quantification, the colonies grown in plates were stained with 1% crystal violet and then photographed. The number and size of the colonies were determined using ImageJ.
Tumor Xenograft Model in Nude Mice
Animal protocols were approved by the Animal Care and Use Committee at Shanghai Jiao Tong Medical College. Male 3-week-old nude mice were deeply anesthetized. MUM2B, shSMC1 MUM2B, MUM2B-CRISPR/Cas9-P2, or guide RNA (gRNA) empty vector cells in a 0.2-mL sterile saline solution were subcutaneously injected into the right flank. Tumor growth was monitored using a caliper every 3 days. Tumor volume was calculated using the formula: length (mm) × width (mm)2/2. Six mice from each group were sacrificed, and the tumors were weighed.
CRISPR/Cas9-Mediated Deletions
Four small gRNAs (sgRNAs) were cloned separately into lentiGuide-Puro plasmids. To delete the 1-kb P2-containing region from the genome, MUM2B cells were transfected with plasmids containing gRNAs (and Cas9) targeting the left and right side of the region to be deleted. Colonies were derived from single cells and tested for the loss of the targeted region. The control group was transfected with gRNA-empty vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Author Contributions
J.F., Y.X., and X.W. designed and performed the experiments and drafted the manuscript; R.J. and S.G. were responsible for sample collection and data analysis; H.Z. and X.F. wrote and approved the manuscript. All of the authors approved this manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Key Research and Development Plan (grants 2018YFC1106100 and 2017YFE9126300), the National Natural Science Foundation of China (grants 81872339, 31870748, 81772875, 81802739, and 81802702), the ShuGuang Project of Shanghai Municipal Education Commission and Shanghai Education Development Foundation (grant 17SG19), the Outstanding Yong Medical Scholar of Shanghai Municipal Commission of Health and Family Planning (grant 2017YQ067), the Shanghai Ninth People’s Hospital training programs (grant jyyq09201713), the Young doctors’ innovation team (grant QC201805), the Science and Technology Commission of Shanghai (grant 17DZ2260100), the Shanghai Science and Technology Development Funds (grant 19QA1405100), the Shanghai “Rising Stars of Medical Talent” Youth Development Program, and the Youth Medical Talents-Specialist Program.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.07.020.
Contributor Information
Renbing Jia, Email: renbingjia@sjtu.edu.cn.
He Zhang, Email: zhanghe@tongji.edu.cn.
Xianqun Fan, Email: fanxq@sjtu.edu.cn.
Supplemental Information
References
- 1.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 3.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponjavic J., Ponting C.P., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marahrens Y., Panning B., Dausman J., Strauss W., Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H., Zeitz M.J., Wang H., Niu B., Ge S., Li W., Cui J., Wang G., Qian G., Higgins M.J. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J. Cell Biol. 2014;204:61–75. doi: 10.1083/jcb.201304152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Chang H.Y. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo J.H., Ren B., Keryanov S., Tseng G.C., Rao U.N., Monga S.P., Strom S., Demetris A.J., Nalesnik M., Yu Y.P. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Jiao W., Sun L., Fan J., Chen M., Wang H., Xu X., Shen A., Li T., Niu B. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell. 2013;13:30–35. doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter D., Chakalova L., Osborne C.S., Dai Y.F., Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 19.Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 20.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montavon T., Soshnikova N., Mascrez B., Joye E., Thevenet L., Splinter E., de Laat W., Spitz F., Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 22.Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., Lee C.W., Ye C., Ping J.L., Mulawadi F. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S., Ong C.T., Hookway T.A., Guo C., Sun Y. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan J., Xing Y., Wen X., Jia R., Ni H., He J., Ding X., Pan H., Qian G., Ge S. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015;16:139. doi: 10.1186/s13059-015-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 26.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 30.Beltran M., Puig I., Peña C., García J.M., Alvarez A.B., Peña R., Bonilla F., de Herreros A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang F., Xue X., Bi J., Zheng L., Zhi K., Gu Y., Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J. Cancer Res. Clin. Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 32.Xing Y., Wen X., Ding X., Fan J., Chai P., Jia R., Ge S., Qian G., Zhang H., Fan X. CANT1 lncRNA triggers efficient therapeutic efficacy by correcting aberrant lncing cascade in malignant uveal melanoma. Mol. Ther. 2017;25:1209–1221. doi: 10.1016/j.ymthe.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gröschel S., Sanders M.A., Hoogenboezem R., de Wit E., Bouwman B.A.M., Erpelinck C., van der Vel den V.H.J., Havermans M., Avellino R., van Lom K. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 34.Merkenschlager M., Odom D.T. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 35.Hadjur S., Williams L.M., Ryan N.K., Cobb B.S., Sexton T., Fraser P., Fisher A.G., Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishiro T., Ishihara K., Hino S., Tsutsumi S., Aburatani H., Shirahige K., Kinoshita Y., Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seitan V.C., Hao B., Tachibana-Konwalski K., Lavagnolli T., Mira-Bontenbal H., Brown K.E., Teng G., Carroll T., Terry A., Horan K. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barber T.D., McManus K., Yuen K.W., Reis M., Parmigiani G., Shen D., Barrett I., Nouhi Y., Spencer F., Markowitz S. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.