Abstract
Huntington disease (HD) is a fatal neurodegenerative disorder caused by a gain-of-function mutation in HTT. Suppression of mutant HTT has emerged as a leading therapeutic strategy for HD, with allele-selective approaches targeting HTT SNPs now in clinical trials. Haplotypes associated with the HD mutation (A1, A2, A3a) represent panels of allele-specific gene silencing targets for efficient treatment of individuals with HD of Northern European and indigenous South American ancestry. Here we extend comprehensive haplotype analysis of the HD mutation to key populations of Southern European, South Asian, Middle Eastern, and admixed African ancestry. In each of these populations, the HD mutation occurs predominantly on the A2 HTT haplotype. Analysis of HD haplotypes across all affected population groups enables rational selection of candidate target SNPs for development of allele-selective gene silencing therapeutics worldwide. Targeting SNPs on the A1 and A2 haplotypes in parallel is essential to achieve treatment of the most HD-affected subjects in populations where HD is most prevalent. Current allele-specific approaches will leave a majority of individuals with HD untreated in populations where the HD mutation occurs most frequently on the A2 haplotype. We further demonstrate preclinical development of potent and selective ASOs targeting SNPs on the A2 HTT haplotype, representing an allele-specific treatment strategy for these individuals. On the basis of comprehensive haplotype analysis, we show the maximum proportion of HD-affected subjects that may be treated with three or four allele targets in different populations worldwide, informing current allele-specific HTT silencing strategies.
Keywords: Huntington disease, antisense, gene silencing, antisense oligonucleotides, haplotypes, haplogroups, population genetics, neurogenetics, rare diseases, allele-specific, preclinical development, therapeutics
Introduction
Gene silencing and gene editing approaches for the treatment of monogenic disorders are moving rapidly from preclinical studies into human clinical trials.1,2 Huntington disease (HD) (MIM: 143100), a fatal neurodegenerative disorder caused by a CAG repeat expansion in HTT, is the most common genetic condition to be addressed with gene-silencing drugs to date.3,4 A recent phase I/IIa clinical trial with an intrathecally delivered antisense oligonucleotide (ASO), engaging the HTT transcript for degradation by RNaseH, achieved safety endpoints allowing open-label extension and larger efficacy trials.5 Wave Life Sciences is conducting parallel phase I/IIa trials of two stereopure ASOs complementary to single-nucleotide polymorphisms (SNPs) associated with the HD mutation, each designed to selectively silence mutant HTT over wild-type HTT.3
Selective suppression of mutant HTT may lead to superior therapeutic outcomes versus non-selective suppression of both mutant and wild-type HTT. Non-selective suppression of HTT has been shown to improve motor and cognitive deficits in the BACHD, YAC128, and Hu97/18 mouse models of HD.6,7 However, mice without murine analog Hdh are embryonic lethal, and postnatal ablation of Hdh to ∼10% of endogenous expression by tamoxifen-induced Cre-Lox recombination results in reduced lifespan, progressive motor impairments, and neuropathology, cautioning against prolonged non-selective HTT suppression in humans.8,9 In contrast, heterozygous Hdh(+/−) mice do not manifest Hdh-deficient phenotypes, suggesting that prolonged, selective mutant HTT suppression may be comparatively safe.10 Selective suppression of mutant HTT has been demonstrated to halt and reverse motor and behavioral phenotypes of HD mice with similar efficacy to non-selective HTT suppression, but with improved protection against brain volume loss in humanized HD mice.4,7 Selective suppression of mutant HTT may therefore offer improved tolerability and efficacy over extended, possibly lifelong durations of human HTT suppressive treatment.
Selective targeting of mutant HTT for gene silencing or editing crucially relies on discrimination of mutant HTT from wild-type HTT, either at the expanded CAG repeat itself or associated SNPs found preferentially on the mutant HTT transcript.4 CAG-targeted methods have shown efficacy in mouse models of HD,11 but off-target silencing of other CAG repeats in the transcriptome, including polymorphic wild-type HTT, remains a major concern for the safety and design of clinical trials of CAG-targeted compounds.12 In contrast, SNP-targeted silencing offers a specific RNA target sequence of mutant HTT amenable to antisense drug design and clear inclusion criteria for genetically eligible individuals in clinical trials.4
Thousands of polymorphic sequences within HTT offer potential targets for silencing mutant HTT, but identifying alleles in strong association with the HD mutation, representing the most useful targets for treating the greatest number of HD subjects, requires detailed population-specific genetic studies in large clinical cohorts. The HD mutation occurs on specific haplotypes in populations of Northern European ancestry, and while expanded CAG repeat length and somatic CAG repeat instability do not appear to vary in relation to the most common haplotypes of the mutation,13,14 these haplotypes represent attractive targets for allele-specific HTT silencing.15,16 We have previously shown through high-density HTT haplotype investigations that the A1 and A2 haplotypes represent panels of target alleles for treating the most HD-affected subjects of Northern European ancestry.16 Clinical reports suggest that these target haplotypes also occur in non-European populations which contribute to the global clinical burden of HD. For example, in a clinical investigation of Indian HD subjects, 4/25 subjects of Northern Indian ancestry and 4/10 subjects of Southern Indian ancestry had the Δ2642 codon deletion indicative of the A1 haplotype, suggesting that a proportion of these subjects may be amenable to allele-specific silencing of mutant HTT with A1 haplotype targets.17 The prevalence of HD is unknown in South Asian populations, but the A1 HTT haplotype is known to be enriched in populations where HD is more prevalent and absent in populations where HD is rare.18 Detailed characterization of haplotypes of the HD mutation in non-European populations is therefore necessary to enable planning of allele-specific therapies for the greatest number of HD-affected individuals worldwide. Additionally, population-specific approaches to allele-specific silencing of HTT may be necessary to extend treatment to a majority of HD-affected subjects in all affected population groups. Advances in long-read sequencing technology in clinical diagnostic settings may further accelerate the identification of alleles offering personalized gene silencing or editing approaches.
In addition to understanding the frequency of target alleles and haplotypes on mutant HTT, the frequency of target haplotypes and their constitutive alleles on wild-type HTT is also important for determining the most useful targets for allele-specific treatment. For a HD-affected subject to be treatable by allele-specific HTT silencing approaches, a target allele must be present on the same chromosome as the mutant CAG expansion but not on the corresponding wild-type HTT copy. For example, an allele found on all mutant HTT haplotypes in a population but more than half of control HTT haplotypes will be less heterozygous in HD-affected subjects, and therefore less useful, than a target allele found on only half of mutant HTT haplotypes but absent on the wild-type copy. Owing to the large number of potential allele targets in HTT, comprehensive heterozygosity estimates of HTT haplotypes in different HD-affected populations are necessary to understand the utility of specific targets for allele-specific drug development. Of note, ASO drugs targeting specific HTT alleles are already in human trials despite incomplete knowledge of how many individuals with HD stand to benefit in different affected populations around the world.
In this study, we describe priority targets for development of HD antisense therapies in 13 distinct clinical populations of HD-affected subjects worldwide. Using haplotype heterozygosity to determine the maximum number of HD-affected subjects treatable with different target alleles, we show that selective targeting of the A2 HTT haplotype is essential for maximum benefit of allele-specific therapy in diverse HD populations around the world. Building on our prior work targeting the A1 HTT haplotype, we demonstrate preclinical development of ASOs targeting the A2 HTT haplotype for potent and selective silencing of mutant HTT in patient-derived cells. In the context of allele-specific HTT suppression strategies already in clinical trials, we provide a map toward the development of additional allele-specific HTT silencing and editing compounds with the greatest cumulative impact toward treatment of individuals with HD worldwide.
Material and Methods
Subject Recruitment and Consenting
Subjects of Swedish, French, Korean, Japanese, Chinese, and European-Canadian origin were identified and consented through the UBC HD Biobank as previously described.16 Subjects of Italian origin were recruited and consented through the CSS-Mendel Institute and Lega Italiana Ricerca Huntington (LIRH) in Rome, Italy. Peruvian and Latin American HD-affected families were recruited and consented for HD research as previously described.19 Brazilian HD-affected subjects were recruited and consented for research through outreach efforts at the Universidade Federal do Estado do Rio de Janeiro (UNIRIO). Our Brazilian study was approved by the Ethics in Research Committees of Gaffree and Guinle University Hospital/UNIRIO (CAAE 26387113.1.0000.5258) and by the Ethics in Research Committees CEP-UNIFAMINAS Muriae (CAAE 71244117.0.0000.5105). HD-affected families of exclusive South Asian ancestry were identified from participants in the UBC HD Biobank or sourced from subjects belonging to the Sinhalese ethnic group seeking clinical testing with research consent through the University of Colombo, Sri Lanka. Subjects of Middle Eastern ancestry were identified from participants in the UBC HD Biobank or from the Mendel Institute in Rome, Italy. Black, white, and mixed ancestry South Africans were obtained through the University of Cape Town and the University of the Witwatersrand as previously described.20
In total, 13 distinct ethnic populations were characterized by examination of more than 1,900 subjects in nearly 800 HD-positive pedigrees (Table S14) to identify 1,288 total HD HTT alleles and 2,646 control HTT alleles.
HTT Haplotype Analysis
DNA samples from all participants in this study were genotyped with variable 96-SNP panels on the Illumina GoldenGate genotyping array, all sharing a core panel of 63 SNPs curated to capture ethnic variation at the HTT locus. Genotypes were called using Illumina GenomeStudio software and reconstructed using PHASE v.2.1 as previously described.16 Haplotypes were manually annotated and phased to CAG repeat length by familial segregation or by CCG repeat length associations.18,20 Haplotype heterozygosity was evaluated for all gene-spanning haplotypes in each clinical population, with heterozygosity of a given haplotype in phase with the HD mutation denoting a subject targetable at defining alleles of that haplotype. For population estimates of haplotype heterozygosity, only probands were used in order to limit the disproportionate effect of counting multiple related subjects with the same HD haplotype in large pedigrees.
In silico determination of SNP alleles in linkage disequilibrium across HTT haplotypes in different populations from the 1000 Genomes project were determined using the LDproxy tool within LDlink. SNP alleles with values of D’ > 0.9 and r2 > 0.8 versus rs362307 (A1), rs2798235 (A2), and rs113407847 (A3a) were identified as proxy SNPs along each respective haplotype.
ASO Synthesis and Preparation
Custom LNA ASO gapmer molecules for site validation at the five unique A2 sites (Figure 1B) and for primary and secondary screening at rs362313 were synthesized by QIAGEN (formerly Exiqon) at 200 nmol scale and purified by standard desalting. Lyophilized ASOs were resuspended in Dulbecco’s phosphate buffered saline (no calcium, no magnesium; GIBCO) and incubated at 37°C for 1 h. ASO concentrations were calculated based on: Concentration (M) = OD260 × dilution factor/ ε260, where ε260 = extinction coefficient of oligo at 260 nm.
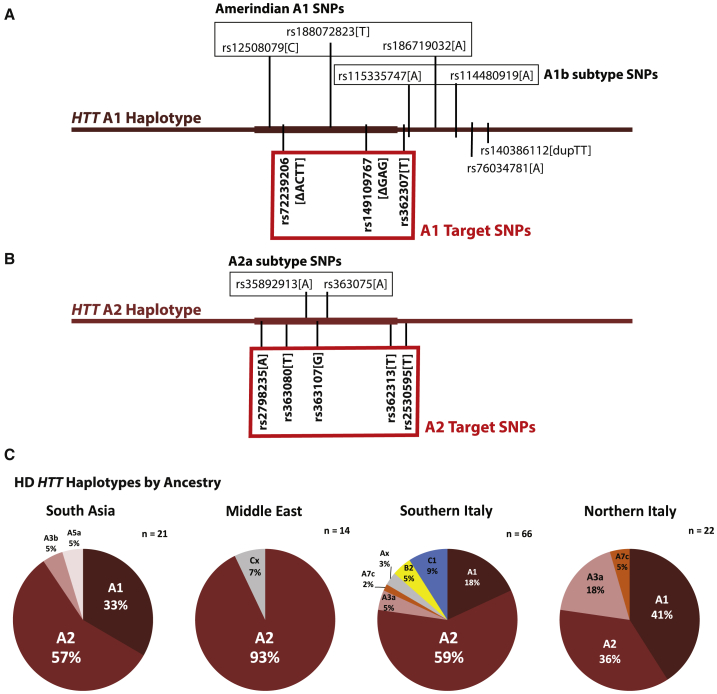
Figure 1.
The HD Mutation Occurs Most Frequently on the A2 HTT Haplotype in HD-Affected Subjects of South Asian, Middle Eastern, and Southern European Ancestry
(A) The extended A1 HTT haplotype is defined by five alleles, three of which occur within transcribed HTT targetable by ASOs. Subtypes A1b and indigenous Amerindian A1 are each defined by two and three additional alleles, respectively.
(B) The extended A2 haplotype is defined by five alleles, all of which occur within transcribed HTT targetable by ASOs. Subtype A2a is defined by two additional alleles.
(C) The A2 HTT haplotype is the most frequent HD haplotype in subjects of South Asian, Middle Eastern, and Southern European ancestry.
In Vitro Screening of A2 HTT Haplotype Allele Targets
The GM02150 cell line was identified as having the HD mutation in phase with the A2 HTT haplotype. For in vitro screening studies, human GM02150 lymphoblasts were grown in RPMI 1640 media (GIBCO) containing 10% fetal bovine serum (GIBCO) and seeded in a 12-well dish at a density of 1.25 × 105 cells/well the day prior to treatment. The following day, a serial dilution was performed with ASOs in growth medium and were added to cells to give final concentrations ranging from 312 nM to 5,000 nM. Cells were incubated for either 120 h for site validation experiments or 240 h for primary and secondary screening experiments then harvested. HTT protein was quantified by allelic separation immunoblotting as in Carroll et al.21 Briefly, 40 μg of total protein was resolved on 10% low-BIS acrylamide gels, transferred to 0.45 μm nitrocellulose, and blotted for total HTT (MAB2166, Millipore) and non-muscle myosin (Ab138498, Abcam) as a loading control. Primary antibodies were detected with IR dye 800CW goat anti-mouse (Rockland 610-131-007) and AlexaFluor 680 goat anti-rabbit (Molecular Probes A21076)-labeled secondary antibodies and scanned using the Li-Cor Odyssey Infrared Imaging system. Densitometry was performed on mHTT, wtHTT, and non-muscle myosin bands using the Li-Cor Image Studio Lite software, and HTT intensities were normalized to non-muscle myosin loading control. n values in text and figure legends represent the number of independent biological replicates performed for each molecule. Statistical analyses of (1) site validation using ASOs at a single dose at each of the A2 haplotype defining SNPs (Figure 4C) and (2) screening with ascending doses of lead ASO molecules at rs362313 (Figures 4D and 4E) were performed by two-way ANOVA with posthoc analysis (Bonferroni multiple comparison test) using GraphPad Prism v.8. Symbols or bars in Figure 4 represent mean of all biological replicates and error bars represent standard error of the mean.
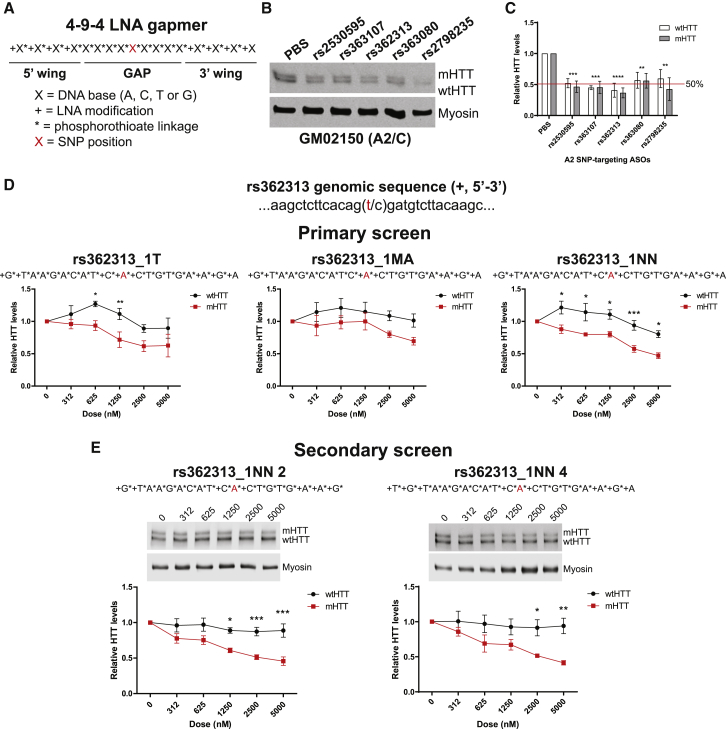
Figure 4.
ASOs Targeting the A2 Haplotype Potently and Selectively Silence Mutant HTT in HD Patient-Derived Cells
(A) Graphical representation of 4-9-4 ASO gapmer configuration for site validation studies.
(B and C) Representative western blot (B) and quantification (C) of HTT suppression after 120 h treatment with 4-9-4 ASOs at each of the defining A2 SNPs (two-way ANOVA HTT p = 0.4571, treatment p < 0.0001, interaction p = 0.9579; Sidak’s multiple comparison test ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; n = 3).
(D) Primary screen quantification of HTT levels after 240 h treatment with 3 ASO molecules with novel LNA configurations against the A2 defining rs362313 SNP (rs362313_1T: two-way ANOVA HTT p < 0.0001, dose p = 0.0006, interaction p = 0.2144, n = 4; rs362313_1MA: two-way ANOVA HTT p = 0.0029, dose p = 0.2867, interaction p = 0.7207, n = 4; rs362313_1NN: two-way ANOVA HTT p < 0.0001, dose p < 0.0001, interaction p = 0.0684, n = 5; Bonferroni posttests ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(E) Secondary screen western blots and quantifications of HTT levels after 240 h treatment with 2 lead ASOs based on the 1NN parent molecule (rs362313_1NN 2: two-way ANOVA HTT p < 0.0001, dose p < 0.0001, interaction p = 0.0209, n = 6; rs362313_1NN 4: two-way ANOVA HTT p < 0.0001, dose p = 0.0211, interaction p = 0.1708, n = 5; Bonferroni posttests ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
All error bars represent standard error of the mean among biological replicates.
Results
Target A1, A2, and A3a HTT Haplotypes Are Conserved in Ethnically Distinct Populations Worldwide
The targetable A1 HTT haplotype is the most common haplotype of the HD mutation in subjects of Northern European ancestry.16,22 The defining intragenic alleles of the A1 HTT haplotype—rs72239206 (a 4 bp intron deletion), rs149109767 (a 3 bp codon deletion), and rs362307 (a 3′ UTR SNP)—are found in European, admixed European, and South Asian reference populations of the 1000 Genomes Project, but not in populations of East Asian or black African ancestry (Table S1).16,23 SNPs in lower pairwise linkage disequilibrium with the three intragenic A1 markers, but exclusive to the A1 HTT haplotype, define subtypes of A1 worldwide, including the indigenous American A1 HTT haplotype in Latin America and the A1b subtype found only in Europeans (Figure 1A).19 The A2 HTT haplotype is the second most common haplotype of the HD mutation in subjects of Northern European ancestry and is defined by five intragenic SNPs in high pairwise linkage disequilibrium (Table S2).16 Two additional intragenic SNPs, rs35892913 and rs363075, define the A2a subtype in roughly half of all A2 HTT haplotypes in European ancestry populations (Figure 1B). The A3a haplotype, the third most common haplotype of the HD mutation in subjects of Northern European ancestry, is found in European, admixed European, and South Asian reference populations of the 1000 Genomes Project, but not in populations of East Asian or black African ancestry (Table S3).
Haplotypes are disrupted distally and proximally by recombination. More than 95% of intragenic HTT haplotypes in European ancestry populations show no evidence of recombination within the ∼170 kb transcribed gene sequence.16 However, association signals with the HD mutation have been shown to span a 1 Mb region of chromosome 4 from rs11248108 (chr4:2479763) to rs1730768 (chr4:3407632), implying that the A1 and A2 haplotypes may extend beyond HTT, with potential application to selective gene editing or cis-modifier studies.13,24 To assess conservation and length of the A1 and A2 HTT haplotypes in different global populations, we examined haplotypes of HTT across a 1.1 Mb window spanning these variants (chr4:2400000–3500000) in the 1000 Genomes Phase III reference populations. Stringent pairwise LD analysis in this region identified only two additional defining SNPs of the A1 haplotype, rs76034781 and rs140386112, both located less than 30 kb proximal of the HTT 3′ UTR (Figure 1A). A1 markers rs362307, rs149109767, rs76034781, and rs140386112 occur in high pairwise LD in all European, Latin American, and South Asian reference populations (Tables S4–S6). A fifth A1-defining allele, the 4 bp deletion rs72239206, occurs in high pairwise LD with the other four A1 markers in all European and Latin American populations, but is additionally found in a subset of South Asian haplotypes lacking the other A1 markers (Table S1).
No extragenic SNPs in the 1.1 Mb window occur in exclusive pairwise LD with the five defining markers of the intragenic A2 HTT haplotype in populations where A2 is present, although numerous extragenic SNPs with lower pairwise association are observed (Figure 1B, Tables S7–S9). Variant rs362313 occurs in high pairwise LD with the other four A2 markers in European and South Asian ancestry populations but is also found on an African ancestry haplotype lacking the other A2 markers entirely (Table S2). A small number of haplotypes with rs362313 but not the other A2 markers are observed in admixed American populations, likely of African origin. These data suggest that rs362313 is ancestral to subsequent A2 markers but that all five A2 markers including rs362313 occur in close conjunction in populations outside Africa.
As we previously observed in Northern European populations,16 there are no SNP alleles in high pairwise LD with A3a-defining rs113407847 in the European, admixed European, or South Asian populations where this allele is found (Tables S10–S12).
The A1 and A2 HTT haplotypes, each defined by five HTT variants, are therefore highly conserved in populations of European, South Asian, and indigenous American ancestry, with no specific markers upstream of the 5′ UTR or further downstream than 33 kb of the 3′ UTR. Notable exceptions are the HD-associated indel rs72239206, found in a small number of non-A1 haplotypes in South Asians, and HD-associated rs362313, found in some non-A2 African ancestry haplotypes. Haplotypes with rs113407847 remain uniquely tagged by this variant in all European, admixed European, and South Asian populations in which it occurs.
A2 Is the Most Common HD Haplotype in Subjects from South Asia, the Middle East, and Southern Europe
The UBC HD Biobank includes more than 8,000 samples from HD-affected subjects and family members representing a wide range of ethnic ancestry groups across North America, Europe, South America, East Asia, and Africa.16,19,20,22,25 In North American and Northern European clinical cohorts, approximately half of all HD mutations occur on the A1 haplotype, also referred to as the major European HD haplotype.13,16,24 In HD-affected subjects from Latin America, the HD mutation occurs predominantly on the indigenous American A1 subtype, which shares defining markers of European A1 in addition to variants of indigenous ancestry.19
To investigate dense HTT haplotypes in HD-affected subjects from additional populations where the A1 and A2 haplotypes are shown to occur (Tables S1 and S2), we selected HD-affected subjects and relatives from 17 families of exclusive South Asian ancestry and 13 families with exclusive ancestry from the Middle East. Family members were genotyped with a custom panel of 96 SNPs across the HTT gene region, designed to reflect haplotype diversity of the locus in reference populations. In South Asian HD-affected families, 57% (12/21) of HD mutations occur on the A2 haplotype while only 33% (7/21) occur on the A1 haplotype (Figure 1C). Strikingly, 93% (13/14) of unrelated HD mutations from our Middle Eastern families occur on the A2 haplotype. The proportion of HD chromosomes occurring on the A2 haplotype in our Middle Eastern subjects (93%) is higher than in any other HD clinical cohort previously characterized.
In contrast to Northern European HD-affected populations, A2 is the most common haplotype of the HD mutation in Italian HD-affected families.16 For this study we haplotyped a large number of Italian families (n = 88) allowing analysis of HD haplotypes by geographic origin within Italy. When divided into Northern Italian and Southern Italian cohorts, HD mutations in Central/Southern Italians occur predominantly on the A2 haplotype (59%), whereas HD mutations in Northern Italians occur at nearly equal frequency on the A1 and A2 haplotypes (41% and 36%, respectively) (Figure 1C). Only 18% of HD mutations in the Central/Southern Italian cohort occurred A1 haplotypes. These data support a geographic gradient of HD haplotypes across Europe, from higher frequency of A1 in the North to higher frequency of A2 in the South, further extending to populations from the Middle East and South Asia where the A2 HD haplotype predominates.16
The HD Mutation Occurs on A1 and A2 Haplotypes in Admixed Populations
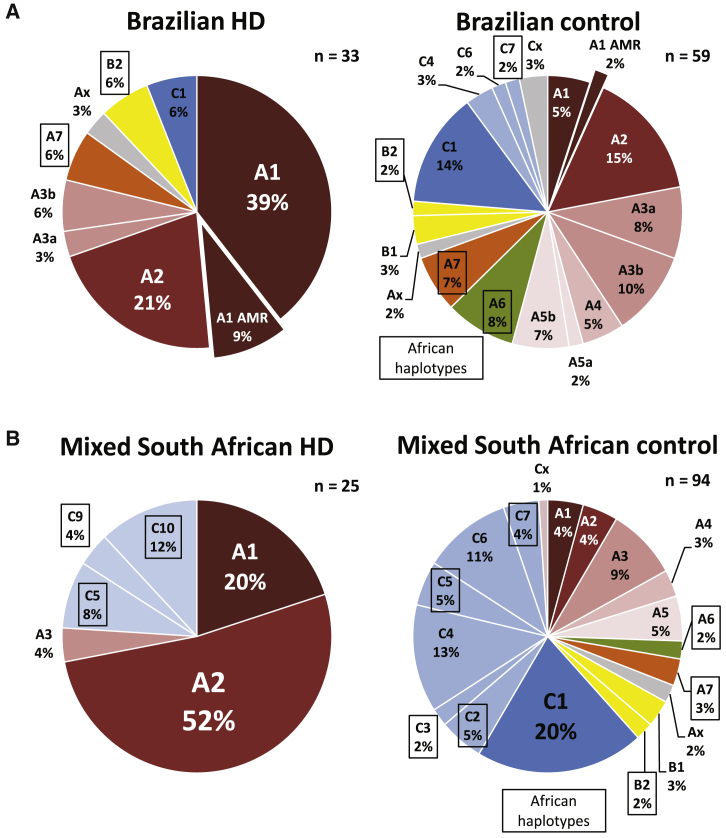
Haplotypes of the HD mutation remain poorly characterized in numerous admixed populations around the world. Our recent investigations of the HD mutation in subjects from Peru and elsewhere in Latin America suggest that a majority of HD mutations in Latin America occur on the indigenous American A1 haplotype, which shares common targetable SNP alleles with the parent A1 haplotype found in Europe and North America. In collaboration with UNIRIO, we recruited and haplotyped 33 additional HD-affected families from Brazil to characterize targetable haplotypes of the HD mutation in a distinct and highly admixed population (Figure 2A). Similar to our previously described cohorts from Peru and the UBC HD Biobank, the HD mutation in Brazil occurs most frequently on the A1 haplotype (49%). However, in contrast to Peruvian subjects, where HD occurs overwhelmingly on the indigenous American A1 variant, HD mutations in Brazil occur most frequently on the parent A1 haplotype of European origin (39% European A1 versus 9% Amerindian A1). African haplotypes (A6, A7, B2, C7) are also represented among Brazilian HD and control haplotypes, reflecting diverse admixture of European, African, and indigenous American ancestry in the Brazilian population.
Figure 2.
The HD Mutation Occurs on a Diversity of Ancestry-Specific HTT Haplotypes in Admixed Populations
(A) The HD mutation in Brazil occurs most frequently on the A1 haplotype, typically of European origin. Control haplotypes in Brazil are highly diverse, representing European, African, and indigenous American origins.
(B) The HD mutation in the mixed South African population occurs most frequently on the A2 haplotype. A minority of HD mutations are found on European A1 and various African haplotypes. Control haplotypes in the mixed South African population are diverse, reflecting the highly admixed origins of this population from Europe, Africa, and South Asia.
The mixed ancestry subpopulation of South Africa, concentrated in the province of Western Cape, is a unique admixed group with origins in African, European, and South Asian populations. Haplotype analysis of 25 independent HD-affected families, roughly double our previous report in this population, shows that more than half of HD mutations in mixed ancestry South African HD-affected families occur on the A2 haplotype (52%), while only a minority occur on the A1 haplotype (20%) (Figure 2B). African haplotypes (C5, C9, C10) are also observed in 24% of mixed South African HD chromosomes, supporting local black African ancestry in a proportion of mutations in this population. As in Brazil, control haplotypes in the mixed South African population are highly diverse, with haplotypes of African, European, and possibly South Asian ancestry represented.
Targeting the A2 HTT Haplotype Allows Allele-Specific Treatment of the Most HD-Affected Subjects in Diverse Population Groups Worldwide
The A1 and A2 haplotypes are constituent haplotypes of the AB haplogroup, which includes other A and B haplotypes not associated with the HD mutation. All other HTT haplotypes belong to the C haplogroup. Targets currently under development for allele-specific suppression of mutant HTT by Wave Life Sciences are specific to the A1 haplotype or to the AB haplogroup, with the latter targeting HD-associated A1 and A2 haplotypes only when these haplotypes are heterozygous with the C haplogroup. We analyzed haplotype heterozygosity in phase with the HD mutation for each panel of priority targets—AB haplogroup, A1 haplotype, or A2 haplotype—in HD-affected subjects from a range of ancestry groups, to measure which targets allow treatment of the most individuals with HD in each population (Table 1).
Table 1.
Targeting the A1 and A2 HD Haplotypes Allows Maximal Treatment of HD across Diverse Population Groups
| Population | n |
One Target |
Two Targets |
Three Targets |
Four Targets |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatable AB | Treatable A1 | Treatable A2 | Treatable A1+AB | Treatable A1+A2 | Treatable A1+A2+AB | Treatable A1+A2+A3a | Treatable A1+A2+AB+C | Treatable A1+A2+A3a+C | Treatable A1+A2+A3a+AB | ||
| Brazil | 33 | 27% | 45% | 21% | 61% | 67% | 76% | 73% | 82% | 76% | 79% |
| Italy | 88 | 27% | 20% | 42% | 36% | 63% | 65% | 69% | 72% | 76% | 72% |
| White S. African | 30 | 47% | 27% | 57% | 57% | 83% | 87% | 90% | 90% | 93% | 90% |
| Canada | 283 | 44% | 44% | 24% | 64% | 68% | 73% | 78% | 77% | 82% | 79% |
| France | 53 | 51% | 43% | 21% | 74% | 64% | 79% | 87% | 83% | 91% | 89% |
| Sweden | 51 | 47% | 47% | 18% | 69% | 65% | 76% | 92% | 80% | 96% | 92% |
| Peru | 62 | 34% | 56% | 11% | 61% | 68% | 68% | 68% | 76% | 76% | 68% |
| Latin America | 17 | 41% | 53% | 18% | 65% | 71% | 82% | 82% | 82% | 82% | 88% |
| Middle East | 14 | 57% | 0% | 86% | 57% | 86% | 86% | 86% | 93% | 93% | 86% |
| South Asia | 21 | 24% | 33% | 52% | 52% | 86% | 86% | 86% | 86% | 86% | 86% |
| Mixed S. African | 25 | 44% | 16% | 52% | 52% | 68% | 68% | 72% | 76% | 80% | 72% |
| East Asian | 55 | 7% | 0% | 0% | 7% | 0% | 7% | 0% | 40% | 33% | 7% |
| Black African | 41 | 51% | 0% | 0% | 51% | 0% | 51% | 0% | 66% | 15% | 51% |
Bold values represent the target combination allowing treatment of the highest proportion of the individuals in a population.
In contrast to our previous findings from HD-affected subjects of Northern European ancestry, where A1 and AB haplogroup targets are the most heterozygous and therefore the most useful, A2 haplotype targets allow treatment of more individuals with HD (42%–86%) than either A1 haplotype targets or AB haplogroup targets in a range of global populations from Italy, South Asia, the Middle East, and non-black populations of South Africa. Targeting the A1 haplotype would allow allele-specific treatment of the most individuals with HD across Latin American populations (45%–56%), whereas defining alleles of the AB haplogroup targets would allow treatment of the most individuals with HD in populations of Northern European ancestry (44%–51%) and black African ancestry (51%).
Targeting the A1, A2, and A3a HTT Haplotypes Allows Allele-Specific Treatment of the Highest Cumulative Proportion of HD-Affected Subjects
Combinatorial targeting of HD-associated alleles is necessary to extend allele-specific therapies to the most people with HD and to treat a majority of people with HD across ethnically distinct populations where different haplotype targets have different heterozygosity rates. For example, in our Brazilian clinical cohort, 45% of HD-affected subjects are heterozygous for the A1 HD haplotype but only 27% are heterozygous for the AB HD haplogroup, due to the high frequency of the AB haplogroup among control chromosomes (Figure 2A). When both panels of targets are analyzed jointly, 61% of Brazilian HD-affected subjects are heterozygous for either the A1 haplotype or AB haplogroup in phase with the HD mutation.
Allele targets on the AB haplogroup, the A1 haplotype, and the A2 haplotype have high heterozygosity rates in distinct clinical populations (Table 1). We therefore calculated the cumulative on-target heterozygosity (i.e., phased to the HD mutation) when using more than one panel of targets in each of 13 HD-affected cohorts representing different ancestry groups around the world. In 9 of 13 HD-affected clinical cohorts, targeting the A1 and A2 haplotypes in parallel would enable treatment of more individuals with HD than targeting the A1 haplotype and AB haplogroup in parallel (Figure 3). Notably, targeting the A1 haplotype and AB haplogroup, but not the A2 haplotype, excludes from treatment the majority of individuals with HD in the Italian population, whereas targeting the A1 and A2 haplotypes would treat 63%. Variants rs362307 and rs362331, targeted by stereopure ASOs in clinical trials by Wave Life Sciences, represent A1 haplotype and AB haplogroup targets and therefore may treat fewer people with HD worldwide relative to two targets representing the A1 and A2 haplotypes instead.
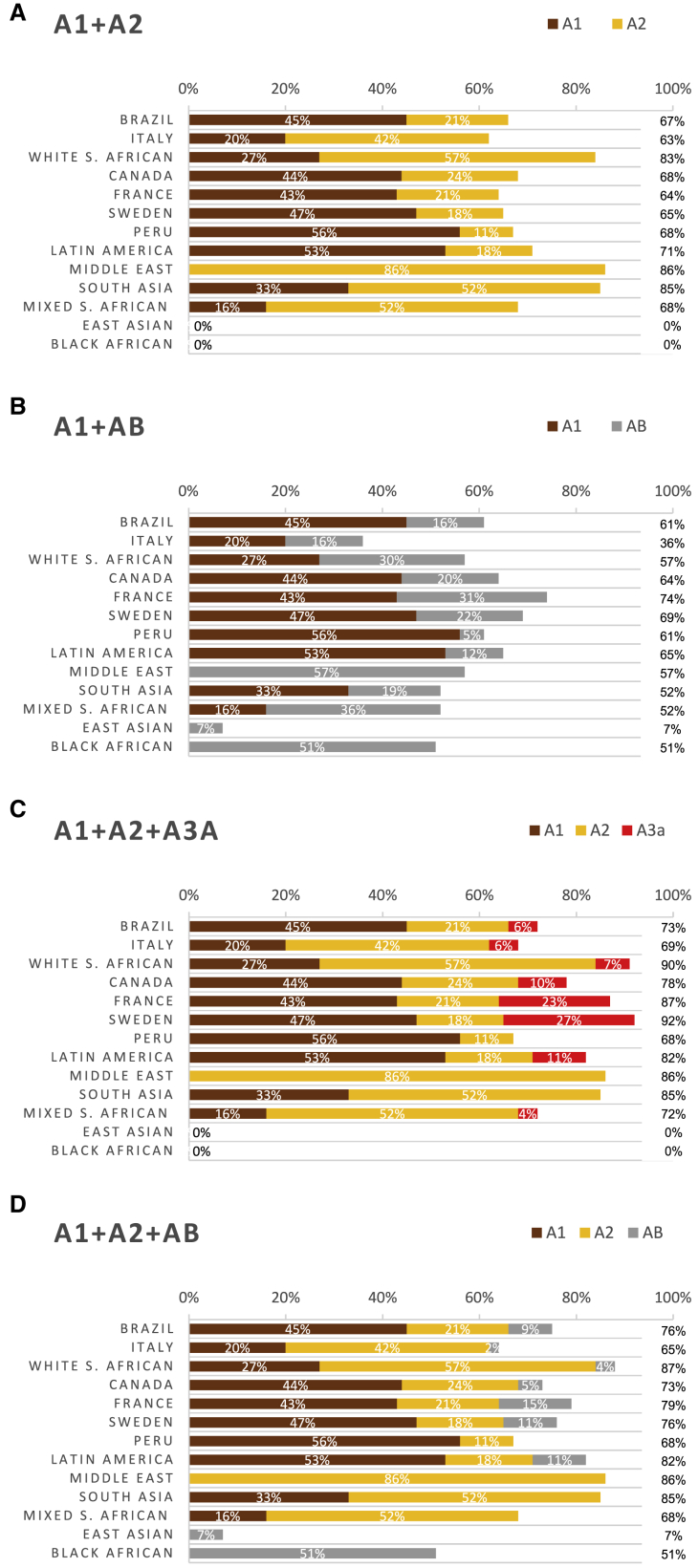
Figure 3.
Targeting the A1 and A2 Haplotypes Allows Treatment of the Most HD-Affected Subjects across Diverse Population Groups when Two or Three Allele Targets Are Considered Cumulatively
(A) Percentage of HD-affected subjects from each study population who are heterozygous for the HTT A1 or A2 haplotype in phase with the HD mutation and therefore treatable with two defining allele targets of A1 and A2, respectively.
(B) Percentage of HD-affected subjects from each study population who are heterozygous for the HTT A1 haplotype or AB haplogroup in phase with the HD mutation and therefore treatable with two defining allele targets of A1 and AB, respectively.
(C) Percentage of HD-affected subjects from each study population who are heterozygous for the HTT A1 or A2 or A3a haplotypes in phase with the HD mutation and therefore treatable with three defining allele targets of A1, A2, and A3a, respectively.
(D) Percentage of HD-affected subjects from each study population who are heterozygous for the HTT A1 or A2 haplotypes or AB haplogroup in phase with the HD mutation and therefore treatable with three defining allele targets of A1, A2, or AB, respectively.
When a third allele target is considered in populations of Northern European ancestry, the maximum number of treatable individuals with HD is achieved by targeting the A3a haplotype.16 We evaluated the utility of a third target panel—either the AB haplogroup or the A3a haplotype, when A1 and A2 haplotypes are chosen as primary and secondary targets—in all 13 HD-affected populations in our study (Table 1 and Figure 3). In 10 of these 13 HD-affected clinical cohorts, tertiary targeting of the A3a haplotype allows treatment of the same or more individuals with HD than tertiary targeting of the AB haplogroup, or an equivalent number.
In HD-affected subjects of East Asian ancestry, in contrast to other populations, the HD mutation most commonly occurs on the C haplogroup. On this basis, targeting the C haplogroup as a complementary strategy to targeting the AB haplogroup has been previously proposed.26 We evaluated the utility of targeting a fourth allele target cumulative to the three-target strategies described above, employing either a C haplogroup target (cumulative to A1+A2+A3a or A1+A2+AB) or an AB haplogroup target (cumulative to A1+A2+A3a) (Figure S1). When four targets are considered, the A1+A2+A3a+C target combination enables treatment of 33% of East Asians with HD and incrementally more in other populations, whereas the A1+A2+AB+C target combination enables treatment of the most individuals with HD from black African and East Asian ancestry groups (66% and 40%, respectively) at the expense of treating fewer individuals with HD in European populations by omission of A3a.
The A1 and A2 haplotypes are entirely absent from both African and East Asian ancestry groups, and HD instead occurs on an assortment of local haplotypes without specific association.20,25 These population-specific differences in HTT allele frequency and heterozygosity crucially inform strategies for worldwide development of HTT silencing compounds, revealing a potential role for AB haplogroup and C haplogroup targets in treating genetically distinct HD-affected populations from Africa and East Asia that do not share the A1 and A2 HTT haplotypes which predominate in HD mutations from major affected populations worldwide.
Targets of the A2 HTT Haplotype Are Not Represented in Current Preclinical and Clinical Development Programs
To evaluate current leads for haplotype-specific HTT suppression, we performed a literature search for preclinical investigations of allele-specific HTT suppressive approaches and analyzed the haplotype relationship of reported targets. Seventeen studies were identified using allele-specific methods for suppressing or editing mutant HTT with target human variants (Table 2).7,16,21,26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 Two HD-associated alleles, rs362307 and rs362331, are targets of active clinical investigation by Wave Life Sciences using stereopure ASOs targeting the A1 haplotype and AB haplogroup, respectively (Table 3). Among all 17 preclinical studies and 2 current clinical trials, our previous investigation of HTT haplotypes in European ancestry subjects was the only study to identify variants on the A2 haplotype for allele-specific therapy in the maximum number of people with HD.16
Table 2.
Summary of SNP-Specific HTT Suppression Studies
|
Primary |
Secondary |
||||||
|---|---|---|---|---|---|---|---|
| Reference | Modality | SNP Target | Target Allele(s) | Haplotype/Haplogroup | SNP Target | Target Allele(s) | Haplotype/Haplogroup |
| Schwarz et al., 200627 | siRNA | rs362331 | unspecified | AB | – | – | – |
| van Bilsen et al., 200828 | siRNA | rs363125 | C | C2, C4, C6 | – | – | – |
| Zhang et al., 200929 | siRNA | rs149109767 | delGAG | A1 | – | – | – |
| Pfister et al., 200930 | siRNA | rs362307 | U | A1 | – | – | – |
| Lombardi et al., 200931 | siRNA | rs362331 | U | AB | – | – | – |
| Carroll et al., 201121 | ASO | rs7685686 | A | AB | – | – | – |
| Ostergaard et al., 201332 | ASO | rs7685686 | A | AB | – | – | – |
| Skotte et al., 201426 | ASO | rs7685686 | A | AB | – | – | – |
| Southwell et al., 201433 | ASO | rs7685686 | A | AB | – | – | – |
| Ostergaard et al., 201534 | ASO | rs7685686 | A | AB | – | – | – |
| Kay et al., 201516 | ASO | rs72239206, rs149109767, rs362307 | delACTT, delGAG, U | A1 | rs2798235, rs363080, rs363107, rs362313, rs2530595 | A, U, G, U, U | A2 |
| Miniarikova et al., 201635 | miRNA | rs362307 | U | A1 | rs362331 | U | AB |
| Shin et al., 201636 | CRISPR/Cas9 | rs1313774, rs16843804 | C | B1, C1 | – | – | – |
| Ostergaard et al., 201737 | ASO | rs7685686 | A | AB | – | – | – |
| Miller et al., 201738 | siRNA | rs362331 | U | AB | – | – | – |
| Monteys et al., 201739 | CRISPR/Cas9 | rs2857935 | unspecified | C1 | – | – | – |
| Southwell et al., 20187 | ASO | rs7685686 | A | AB | rs6446723 | G | C |
Table 3.
SNP-Specific HTT Suppression Clinical Trials
| Trial Name | Compound | Year Initiated | Phase | Modality | SNP Target | Allele Target | Haplotype/Haplogroup | ClinicalTrials.gov Identifier |
|---|---|---|---|---|---|---|---|---|
| PRECISION-HD1 | WVE-120101 | 2017 | Phase 1b/2a | Stereopure ASO | rs362307 | U | A1 | NCT03225833 |
| PRECISION-HD2 | WVE-120102 | 2017 | Phase 1b/2a | Stereopure ASO | rs362331 | U | AB | NCT03225846 |
Five preclinical studies and one clinical trial—WVE-120101 by Wave Life Sciences—target defining alleles of the A1 HTT haplotype. Eight preclinical studies and one clinical trial—WVE-120102 by Wave Life Sciences—target defining alleles of the AB haplogroup. Development of A1 haplotype and AB haplogroup targets are therefore well represented in the allele-specific HTT suppression literature, whereas focus on the A2 haplotype has been absent despite its high association with the HD mutation in numerous populations around the world, including Southern Europe, South Asia, and the Middle East.
The A2 Haplotype Can Be Selectively Silenced with ASOs in Patient-Derived Cells
To evaluate whether defining alleles of the A2 haplotype can be targeted to achieve selective silencing of mutant HTT, we first needed to identify a patient-derived cell line heterozygous for the A2 haplotype on the CAG expanded allele. We chose to genotype Epstein-Barr virus (EBV)-immortalized B-lymphocyte continuous lines (lymphoblasts) derived from peripheral blood mononuclear cells over fibroblasts from the Coriell NIGMS Human Genetic Cell Repository because levels of HTT have been shown to be low in fibroblasts and these cells become senescent after a limited number of passages. We genotyped 11 HD patient-derived lymphoblast lines and identified GM02150 as heterozygous for the A2 haplotype, in phase with the mutant allele, and a C haplotype on the wild-type allele (CAG = 45/18).
We next sought to validate all five A2 defining SNPs (Figure 1B) to determine whether these sites were active and amenable to silencing using ASOs. SNPs found in rapidly spliced intronic regions or within RNA secondary structure can be inaccessible to antisense oligonucleotide (ASO) binding making site validation an important first step prior to expanding screening efforts.26,33 To validate the A2 defining SNPs, we designed ASO molecules complementary to each target sequence that would be predicted to be potent at the expense of selectivity and other pharmacological properties. Locked nucleic acids (LNA) are base modifications that can be incorporated into oligonucleotides to confer improved hybridization affinity and specificity for a complementary target sequence.40 Several LNA residues can be incorporated at the 5′ and/or 3′ termini or “wings” of an oligonucleotide to increase resistance to nucleases, activate RNase H, and promote antisense activity to inhibit gene expression.40,41 Therefore, we designed ASO molecules with 4 locked nucleic acid (LNA) modified base wings flanking a 9 nucleotide “gap” where the entire molecule was modified with phosphorothioate linkages between nucleotides (4-9-4 gapmer configuration) and the SNP was centered in the gap (Figure 4A). For site validation experiments, A2 lymphoblasts were treated with 5 μM ASO at each site for a duration of 120 h and cell lysates were run by western blot to quantify the level of both mutant HTT (mHTT) and wild-type HTT (wtHTT) suppression (Figures 4B and 4C). We observed that each site was indeed active and that each molecule induced significant reduction of HTT, but none of the molecules tested showed selectivity preference for the target mutant allele over the non-target wild-type allele (n = 3; two-way ANOVA HTT p = 0.4571, SNP p < 0.0001, interaction p = 0.9579). We also tested a number of other molecule designs with configurations predicted to improve allelic discrimination but we did not observe improved selectivity for the target mutant allele with these molecules (data not shown).
In an effort to improve selectivity for the mutant HTT allele, we used a rational design approach based on previously defined base mismatch discrimination values for LNA probes to design a novel iteration of LNA ASOs.42,43 We chose to narrow our screening targets to a single A2 defining SNP, rs362313, based on the increased activity observed from the site validation experiments and because this variant is ancestral to all other A2 variants and may therefore offer the broadest coverage of individuals with HD. We designed three ASO gapmers with different configurations of LNA bases at or surrounding the rs362313 SNP binding position and screened them at five increasing doses (312–5,000 nM) in the GM02150 lymphoblasts for a treatment duration of 240 h (Figure 4D). The rs362313 triplet molecule (rs362313_1T) with LNAs positioned at the SNP binding base and flanking bases showed selectivity for the mutant allele but also lowered wtHTT at the two highest doses (n = 4; two-way ANOVA HTT p < 0.0001, dose p = 0.0006, interaction p = 0.2144; Bonferroni posttest ∗p < 0.05, ∗∗p < 0.01). The rs362313 modified-after SNP molecule (rs362313_1MA), with LNAs positioned at the SNP binding site and the 3′ neighboring base, showed only modest selectivity for the mutant allele but no dose-dependent silencing effect (n = 4; two-way ANOVA HTT p = 0.0029, dose p = 0.2867, interaction p = 0.7207). Lastly, the rs362313 nearest neighbor molecule (rs362313_1NN) with LNAs positioned flanking the SNP binding base showed selectivity for the mutant allele at all doses tested but led to reduction of wtHTT at the two highest doses (n = 4; two-way ANOVA HTT p < 0.0001, dose p < 0.0001, interaction p = 0.0684; Bonferroni posttest ∗p < 0.05, ∗∗∗p < 0.001).
Based on the lead nearest neighbor (NN) design from the primary screen, we designed seven additional molecules maintaining the LNA modifications flanking the SNP binding base but altering the 5′ LNA wing length, the 5′ gap length, and the 3′ gap length (Table S13). Once again we screened these molecules at five increasing doses (312–5,000 nM) in the GM02150 lymphoblast line. Of the seven molecules tested, we identified two lead molecules that showed nearly perfect selectivity for the mutant over the wild-type HTT allele (Figure 4E). The 1NN 2 design showed a selectivity for the target mutant HTT allele and potently reduced levels of mHTT in dose-dependent manner up to 55% at the highest dose (n = 6; two-way ANOVA HTT p < 0.0001, dose p < 0.0001, interaction p = 0.0209; Bonferroni posttest ∗p < 0.05, ∗∗∗p < 0.001). Similarly, the 1NN 4 design was both selective and potent for the mutant allele suppressing levels of mHTT by 59% (n = 5; two-way ANOVA HTT p < 0.0001, dose p < 0.0211, interaction p = 0.1708; Bonferroni posttest ∗p < 0.05, ∗∗p < 0.01).
Discussion
The strong association of the HD mutation with A1 and A2 HTT haplotypes in populations with the highest prevalence of the disease suggests that targeting of these haplotypes may enable allele-specific gene silencing treatment of the greatest number of individuals with HD worldwide. HD is found preferentially on A1 and A2 HTT haplotypes in HD-affected subjects from Europe, North America, South America, South Asia, and the Middle East. This study provides comprehensive estimates of the proportion of individuals with HD in each of these major populations that may be treatable with allele-specific HTT silencing or editing strategies.
A crucial principle for allele-specific targeting of mutant HTT is to develop the minimal number of approaches that treat the maximal number of individuals with HD, to ensure that the least number of individuals with HD will be ineligible for this therapeutic approach. Among all populations studied to date, the HD mutation predominates on A1 and A2 haplotypes except in East Asians and black South Africans, in whom the disease is estimated to be ten times less prevalent.23 This suggests a close relationship between the frequency of HD mutations on the A1 and A2 haplotypes in a population and the prevalence of the disease, as we have suggested previously in the context of intermediate alleles.18
To understand the utility of allele-specific suppression of mutant HTT, it is crucial to take into account the frequency of these alleles on wild-type HTT. Frequencies of target alleles on wild-type HTT are necessary for accurate estimation of heterozygosity rates and, by extension, the number of HD-affected subjects who can be treated with a given allele-specific approach to suppressing mutant HTT. We reveal that current HTT allele targets in development, representing the A1 haplotype and AB haplogroup, may exclude a majority of individuals with HD in various populations of European (e.g., Italian) and non-European (e.g., South Asian and East Asian) ancestry.
To estimate the proportion of individuals with HD treatable by targeting the A1 and A2 haplotypes in parallel, we calculated the heterozygosity of these haplotypes in phase with the HD mutation for each clinical population for which we had dense HTT haplotype data. Remarkably, combinatorial targeting of the A1 and A2 HTT haplotypes for allele-specific gene silencing enables treatment of >63% of individuals with HD in all populations where these haplotypes are found. In contrast, targeting of the A1 haplotype and AB haplogroup, exemplified by allele targets under development by Wave Life Sciences, treats comparatively fewer individuals with HD than targeting the A1 and A2 haplotypes specifically. Addition of A3a as a tertiary target, marked by rs113407847, increases overall coverage to nearly 80% of individuals with HD in populations of European and admixed European ancestry, in whom HD is most prevalent. However, AB haplogroup alleles may be preferable as tertiary targets given the advantage of extending treatment to black African and East Asian HD-affected subjects at the expense of treating fewer individuals with HD elsewhere.
Additional HTT haplotyping in populations from Eastern Europe, Central Asia, and different regions of Africa will be necessary to guide a comprehensive, global allele-specific silencing strategy. However, our analysis suggests that targeting of the A1 and A2 HD haplotypes offers superior coverage of individuals with HD in populations where the prevalence of HD is highest. This suggests that defining alleles of A1 and A2 may represent optimal allele-specific HTT silencing targets for treating the most people with HD worldwide. Targeting the AB haplogroup and C haplogroup may nonetheless allow treatment of a major proportion of individuals with HD with ancestry in Africa and East Asia. Notably, the AB haplogroup offers a wide panel of potential targets, whereas the A3a haplotype offers only one specific targetable allele,16 suggesting that development of an antisense drug targeting the AB haplogroup may be more tractable. Indeed, stereopure ASO drugs targeting the AB haplogroup via rs362331 have already entered human trials (Table 3).
To date, no target specific to the A2 HTT haplotype has been the subject of preclinical studies for therapeutic allele-specific silencing or editing of HTT. Our study demonstrates that targeting any of the five defining alleles of the A2 haplotype, all of which are found in HTT pre-mRNA, would enable treatment of >40% of individuals with HD in a range of populations across Europe, South Asia, and the Middle East. Further, targeting the A1 and A2 haplotypes in parallel enables treatment of more individuals with HD in Europe, North America, South America, South Asia, and the Middle East than any other combinatorial approach of two alleles in HTT. We have shown that ASOs targeting one of these alleles, rs362313, are capable of potent and selective silencing of mutant HTT in patient-derived cells, demonstrating direct translation of our genetic targets to preclinical application. Targeting the A2 haplotype should therefore be prioritized in the development of allele-specific HTT silencing therapies to extend these breakthrough approaches to treating HD to the greatest proportion of people living with HD around the world.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are grateful to all the patients and families from the Centre for HD Clinic at UBC, across Canada and the world, who have donated samples and clinical data to the HD BioBank at UBC and continue to make this research possible. The research leading to these results has received funding from the E-Rare Research Programme which is a transnational R&D program jointly funded by national funding organizations within the framework of the ERA-NET E-Rare 3. This work was supported by the Canadian Institutes of Health Research (ERT-155723 and FDN-154278 to M.R.H.). We thank the Fondazione Lega Italiana Ricerca Huntington e malattie correlate (LIRH) for contributing to DNA sample collection in Italy and the Middle East. We wish to thank the patients and families that consented to the use of biological samples for this research project and the Human Genetics Unit Development Fund for funding to recruit the subjects in Colombo, Sri Lanka. We also thank the Brazilian National Council for Scientific and Technological Development (CNPq) for the financial support and postdoctoral fellowship granted to L.A.A. (Science without Borders Program). Postdoctoral support for N.S.C. was provided by the Ripples of Hope Pfizer Trainee Award, Canadian Institutes of Health Research, James Family Fellowship, and the Huntington’s Disease Society of America Berman/Topper HD Career Development Fellowship.
Published: November 7, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.10.011.
Web Resources
ClinicalTrials.gov, https://clinicaltrials.gov
LDlink, https://ldlink.nci.nih.gov/
OMIM, https://www.omim.org/
Supplemental Data
References
- 1.Southwell A.L., Skotte N.H., Bennett C.F., Hayden M.R. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol. Med. 2012;18:634–643. doi: 10.1016/j.molmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F. RNA-Targeted Therapeutics. Cell Metab. 2018;27:714–739. doi: 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Lane R.M., Smith A., Baumann T., Gleichmann M., Norris D., Bennett C.F., Kordasiewicz H. Translating Antisense Technology into a Treatment for Huntington’s Disease. Methods Mol. Biol. 2018;1780:497–523. doi: 10.1007/978-1-4939-7825-0_23. [DOI] [PubMed] [Google Scholar]
- 4.Kay C., Skotte N.H., Southwell A.L., Hayden M.R. Personalized gene silencing therapeutics for Huntington disease. Clin. Genet. 2014;86:29–36. doi: 10.1111/cge.12385. [DOI] [PubMed] [Google Scholar]
- 5.van Roon-Mom W.M.C., Roos R.A.C., de Bot S.T. Dose-Dependent Lowering of Mutant Huntingtin Using Antisense Oligonucleotides in Huntington Disease Patients. Nucleic Acid Ther. 2018;28:59–62. doi: 10.1089/nat.2018.0720. [DOI] [PubMed] [Google Scholar]
- 6.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Southwell A.L., Kordasiewicz H.B., Langbehn D., Skotte N.H., Parsons M.P., Villanueva E.B., Caron N.S., Østergaard M.E., Anderson L.M., Xie Y. Huntingtin suppression restores cognitive function in a mouse model of Huntington’s disease. Sci. Transl. Med. 2018;10:10. doi: 10.1126/scitranslmed.aar3959. [DOI] [PubMed] [Google Scholar]
- 8.Dietrich P., Johnson I.M., Alli S., Dragatsis I. Elimination of huntingtin in the adult mouse leads to progressive behavioral deficits, bilateral thalamic calcification, and altered brain iron homeostasis. PLoS Genet. 2017;13:e1006846. doi: 10.1371/journal.pgen.1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dragatsis I., Dietrich P., Ren H., Deng Y.P., Del Mar N., Wang H.B., Johnson I.M., Jones K.R., Reiner A. Effect of early embryonic deletion of huntingtin from pyramidal neurons on the development and long-term survival of neurons in cerebral cortex and striatum. Neurobiol. Dis. 2018;111:102–117. doi: 10.1016/j.nbd.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeitlin S., Liu J.P., Chapman D.L., Papaioannou V.E., Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat. Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 11.Hu J., Matsui M., Gagnon K.T., Schwartz J.C., Gabillet S., Arar K., Wu J., Bezprozvanny I., Corey D.R. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butland S.L., Devon R.S., Huang Y., Mead C.-L., Meynert A.M., Neal S.J., Lee S.S., Wilkinson A., Yang G.S., Yuen M.M. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J.-M., Gillis T., Mysore J.S., Ramos E.M., Myers R.H., Hayden M.R., Morrison P.J., Nance M., Ross C.A., Margolis R.L. Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am. J. Hum. Genet. 2012;90:434–444. doi: 10.1016/j.ajhg.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos E.M., Gillis T., Mysore J.S., Lee J.M., Gögele M., D’Elia Y., Pichler I., Sequeiros J., Pramstaller P.P., Gusella J.F. Haplotype analysis of the 4p16.3 region in Portuguese families with Huntington’s disease. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2015;168B:135–143. doi: 10.1002/ajmg.b.32289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao M.J., Gillis T., Atwal R.S., Mysore J.S., Arjomand J., Harold D., Holmans P., Jones L., Orth M., Myers R.H. Haplotype-based stratification of Huntington’s disease. Eur. J. Hum. Genet. 2017;25:1202–1209. doi: 10.1038/ejhg.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kay C., Collins J.A., Skotte N.H., Southwell A.L., Warby S.C., Caron N.S., Doty C.N., Nguyen B., Griguoli A., Ross C.J. Huntingtin Haplotypes Provide Prioritized Target Panels for Allele-specific Silencing in Huntington Disease Patients of European Ancestry. Mol. Ther. 2015;23:1759–1771. doi: 10.1038/mt.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saleem Q., Roy S., Murgood U., Saxena R., Verma I.C., Anand A., Muthane U., Jain S., Brahmachari S.K. Molecular analysis of Huntington’s disease and linked polymorphisms in the Indian population. Acta Neurol. Scand. 2003;108:281–286. doi: 10.1034/j.1600-0404.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 18.Kay C., Collins J.A., Wright G.E.B., Baine F., Miedzybrodzka Z., Aminkeng F., Semaka A.J., McDonald C., Davidson M., Madore S.J. The molecular epidemiology of Huntington disease is related to intermediate allele frequency and haplotype in the general population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2018;177:346–357. doi: 10.1002/ajmg.b.32618. [DOI] [PubMed] [Google Scholar]
- 19.Kay C., Tirado-Hurtado I., Cornejo-Olivas M., Collins J.A., Wright G., Inca-Martinez M., Veliz-Otani D., Ketelaar M.E., Slama R.A., Ross C.J. The targetable A1 Huntington disease haplotype has distinct Amerindian and European origins in Latin America. Eur. J. Hum. Genet. 2016;25:332–340. doi: 10.1038/ejhg.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baine F.K., Kay C., Ketelaar M.E., Collins J.A., Semaka A., Doty C.N., Krause A., Greenberg L.J., Hayden M.R. Huntington disease in the South African population occurs on diverse and ethnically distinct genetic haplotypes. Eur. J. Hum. Genet. 2013;21:1120–1127. doi: 10.1038/ejhg.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., Hung G., Bennett C.F., Freier S.M., Hayden M.R. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol. Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warby S.C., Montpetit A., Hayden A.R., Carroll J.B., Butland S.L., Visscher H., Collins J.A., Semaka A., Hudson T.J., Hayden M.R. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kay C., Fisher E.R., Hayden M. Oxford University Press; 2014. Epidemiology (in Huntington’s Disease, 4th ed) [Google Scholar]
- 24.Lee J.M., Kim K.H., Shin A., Chao M.J., Abu Elneel K., Gillis T., Mysore J.S., Kaye J.A., Zahed H., Kratter I.H. Sequence-Level Analysis of the Major European Huntington Disease Haplotype. Am. J. Hum. Genet. 2015;97:435–444. doi: 10.1016/j.ajhg.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warby S.C., Visscher H., Collins J.A., Doty C.N., Carter C., Butland S.L., Hayden A.R., Kanazawa I., Ross C.J., Hayden M.R. HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur. J. Hum. Genet. 2011;19:561–566. doi: 10.1038/ejhg.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skotte N.H., Southwell A.L., Østergaard M.E., Carroll J.B., Warby S.C., Doty C.N., Petoukhov E., Vaid K., Kordasiewicz H., Watt A.T. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS ONE. 2014;9:e107434. doi: 10.1371/journal.pone.0107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz D.S., Ding H., Kennington L., Moore J.T., Schelter J., Burchard J., Linsley P.S., Aronin N., Xu Z., Zamore P.D. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Bilsen P.H., Jaspers L., Lombardi M.S., Odekerken J.C., Burright E.N., Kaemmerer W.F. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington’s disease patient-derived fibroblasts. Hum. Gene Ther. 2008;19:710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Engelman J., Friedlander R.M. Allele-specific silencing of mutant Huntington’s disease gene. J. Neurochem. 2009;108:82–90. doi: 10.1111/j.1471-4159.2008.05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfister E.L., Kennington L., Straubhaar J., Wagh S., Liu W., DiFiglia M., Landwehrmeyer B., Vonsattel J.-P., Zamore P.D., Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombardi M.S., Jaspers L., Spronkmans C., Gellera C., Taroni F., Di Maria E., Donato S.D., Kaemmerer W.F. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Østergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southwell A.L., Skotte N.H., Kordasiewicz H.B., Østergaard M.E., Watt A.T., Carroll J.B., Doty C.N., Villanueva E.B., Petoukhov E., Vaid K. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol. Ther. 2014;22:2093–2106. doi: 10.1038/mt.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Østergaard M.E., Thomas G., Koller E., Southwell A.L., Hayden M.R., Seth P.P. Biophysical and biological characterization of hairpin and molecular beacon RNase H active antisense oligonucleotides. ACS Chem. Biol. 2015;10:1227–1233. doi: 10.1021/cb500880f. [DOI] [PubMed] [Google Scholar]
- 35.Miniarikova J., Zanella I., Huseinovic A., van der Zon T., Hanemaaijer E., Martier R., Koornneef A., Southwell A.L., Hayden M.R., van Deventer S.J. Design, Characterization, and Lead Selection of Therapeutic miRNAs Targeting Huntingtin for Development of Gene Therapy for Huntington’s Disease. Mol. Ther. Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin J.W., Kim K.H., Chao M.J., Atwal R.S., Gillis T., MacDonald M.E., Gusella J.F., Lee J.M. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 2016;25:4566–4576. doi: 10.1093/hmg/ddw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Østergaard M.E., Nichols J., Dwight T.A., Lima W., Jung M.E., Swayze E.E., Seth P.P. Fluorinated Nucleotide Modifications Modulate Allele Selectivity of SNP-Targeting Antisense Oligonucleotides. Mol. Ther. Nucleic Acids. 2017;7:20–30. doi: 10.1016/j.omtn.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller J.R.C., Pfister E.L., Liu W., Andre R., Träger U., Kennington L.A., Lo K., Dijkstra S., Macdonald D., Ostroff G. Allele-Selective Suppression of Mutant Huntingtin in Primary Human Blood Cells. Sci. Rep. 2017;7:46740. doi: 10.1038/srep46740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monteys A.M., Ebanks S.A., Keiser M.S., Davidson B.L. CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo. Mol. Ther. 2017;25:12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahlestedt C., Salmi P., Good L., Kela J., Johnsson T., Hökfelt T., Broberger C., Porreca F., Lai J., Ren K. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braasch D.A., Liu Y., Corey D.R. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 2002;30:5160–5167. doi: 10.1093/nar/gkf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurreck J., Wyszko E., Gillen C., Erdmann V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You Y., Moreira B.G., Behlke M.A., Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.