Abstract
While genes with an excess of de novo mutations (DNMs) have been identified in children with neurodevelopmental disorders (NDDs), few studies focus on DNM patterns where the sex of affected children is examined separately. We considered ∼8,825 sequenced parent-child trios (n ∼26,475 individuals) and identify 54 genes with a DNM enrichment in males (n = 18), females (n = 17), or overlapping in both the male and female subsets (n = 19). A replication cohort of 18,778 sequenced parent-child trios (n = 56,334 individuals) confirms 25 genes (n = 3 in males, n = 7 in females, n = 15 in both male and female subsets). As expected, we observe significant enrichment on the X chromosome for females but also find autosomal genes with potential sex bias (females, CDK13, ITPR1; males, CHD8, MBD5, SYNGAP1); 6.5% of females harbor a DNM in a female-enriched gene, whereas 2.7% of males have a DNM in a male-enriched gene. Sex-biased genes are enriched in transcriptional processes and chromatin binding, primarily reside in the nucleus of cells, and have brain expression. By downsampling, we find that DNM gene discovery is greatest when studying affected females. Finally, directly comparing de novo allele counts in NDD-affected males and females identifies one replicated genome-wide significant gene (DDX3X) with locus-specific enrichment in females. Our sex-based DNM enrichment analysis identifies candidate NDD genes differentially affecting males and females and indicates that the study of females with NDDs leads to greater gene discovery consistent with the female-protective effect.
Keywords: sex bias, neurodevelopmental disorder, autism, intellectual disability, female protective effect, X chromosome
Introduction
Sex biases have been observed in a number of disorders1,2 (Figure S1A) with reported estimates in autism (MIM: 209850) of ∼4 males:1 female2,3 and in developmental disorders of ∼1.3 males:1 female.4 Among neurodevelopmental disorders (NDDs), sex biases become more pronounced with less severe symptom manifestations. For example, male to female biases ranging from 4 to 15 have been reported among individuals with high-functioning autism.5,6 The reasons for these differences are not fully understood but potential biological factors include involvement of the sex chromosomes,7 hormonal regulation of genes,8 sex-specific imprinting, or a protective effect in the rarer class (in NDDs the female-protective effect [FPE]).9, 10, 11 The FPE is rooted in the liability (genetic and environment) distribution9 for complex disorders. In a disorder with no sex bias, there is a fixed threshold at which individuals with liability greater than the threshold have the disease. In a sex-biased disorder, there are two thresholds whereby individuals of the rarer class (females in NDDs) require a higher liability to develop the NDD. It has been proposed that the study of individuals of the rare class can identify relevant genes by assessing severe genetic variants11 and potentially pinpoint key gene regulatory network components.12 Since previous NDD studies identified females as having higher mutational burden of deleterious CNVs10,13 and private deleterious SNVs as compared to males,10 we hypothesize that study of females will lead to greater gene discovery than in males. Nonbiological factors include ascertainment bias in the clinical setting and differential presentation of autism in males and females.14
Recently, family-based whole-exome and whole-genome sequencing have been applied to identify de novo mutations (DNMs) and novel risk factors for patients with NDDs. This work has identified a number of genes but has primarily focused on aggregating all individuals (males and females) together as one group15, 16, 17 to increase power for detection of genes with a significant excess of DNM (Figure S1B). While the approach has been powerful, it is also possible that there are sex-specific risk factors for NDDs that have been missed but may have been detected by testing male and female patients with NDDs as two distinct groups (Figure S1B). One important consideration for this analysis is the sex chromosomes. There are numerous examples of syndromes that differentially affect males and females due to dosage differences of the X chromosome (e.g., Rett syndrome [MIM: 312750] and MECP2 [MIM: 300005]18 or Hunter syndrome [MIM: 309900] and IDS [MIM: 300823]19).
In this study, taking advantage of the thousands of individuals with NDD for which we now have DNM,15, 16, 17,20, 21, 22, 23, 24, 25 we ask three main questions. The first is whether there are any genes that are significant in only one sex and not the other. The second is whether gene discovery rates differ by analyzing males and females separately (i.e., are gene discovery analyses based on severe variants more fruitful in females than males as the FPE would suggest?). The third is whether there are genes with differential counts of de novo variants in a direct comparison of males and females with NDDs. Using published available data, in ∼8,825 sequenced trios as a discovery cohort, we sought to address these questions by assessing the frequency and pattern of DNM in a sex-specific manner. We further replicated these findings in a cohort of an additional 18,778 sequenced trios where DNM counts were available for individual genes.
Materials and Methods
Discovery Cohort
We obtained DNM data from denovo-db26 v.1.5 (Table 1) where information on sex (male, female) could be assessed. We excluded individuals from targeted sequencing studies and any duplicate variant sites that were seen in two or more members of a single family. The dataset included probands with autism15,16,20, 21, 22, 23, 24, 25 and developmental delay (DD).17 The combined sex ratio in our NDD discovery cohort was 2.2 with sex ratios of 5.5 and 1.2 in autism and DD, respectively. Sample denominators for de novo enrichment calculation are estimated based on unaffected sibling data from the Simons Simplex Collection (SSC).15,21,22,24 This is because we did not have access to the total number of males and females across all studies—only those with DNMs—and because we wanted to be as conservative as possible with our statistical analyses. In denovo-db we identified 1,467 of the SSC unaffected siblings with one or more variants out of the total of 1,911 siblings (76.8%). By applying this estimate to the female and male sets, we estimated a total 2,779 females (2,133 observed females, Table S1) and 6,046 males (4,641 observed males, Table S1) in our discovery cohort. In total, there were 4,308 DNMs in females and 8,517 DNMs in males. There was a significant difference in the DNM discovery rate in females than males (Poisson p = 3.65 × 10−7) even when considering autosomes only (Poisson p = 3.4 × 10−3). After applying the high-coverage region filter (described later), there were 3,297 DNMs in females and 6,726 in males (Table S2). There was still a significant difference in the DNM discovery rate in females than males (Poisson p = 2.61 × 10−3), but when focusing only on the autosomes there was no longer a significant difference (Poisson p = 0.29).
Table 1.
Description of Cohorts Assessed in This Study
| Cohort | Cohort Type | Number of Males | Number of Females | Total | Sex Ratio | Data Type |
|---|---|---|---|---|---|---|
| denovo-db v.1.5 | discovery | 6,046 | 2,779 | 8,825 | 2.2 | de novo variants |
| GeneDx | replication | 10,379 | 8,399 | 18,778 | 1.2 | de novo variant counts per gene |
| total | 16,425 | 11,178 | 27,603 | 1.5 |
Replication Cohort
The GeneDx replication cohort consisted of 18,778 trios of 8,399 females and 10,379 males diagnosed with an NDD and collected as part of routine clinical exome sequencing (GeneDx, Table 1). Eleven duplicate trios were removed prior to these cohort totals based on screening of a 47-variant panel between the discovery and replication sets. We obtained de novo variant counts per gene only where DNM variants were classified as likely gene-disrupting (LGD) (frameshift, canonical splice site, stop-gained) or missense mutations. Exome capture, sequencing, and alignment was performed as previously described.27 Short reads from whole-exome sequencing were aligned using the Burrows-Wheeler Aligner (BWA; v.0.7.5a)28 to the hg19 reference genome. Alignment BAM files were then converted to CRAM format with SAMtools v.1.3.129 and indexed. Individual GVCF files were called with GATK (v.3.7)30 HaplotypeCaller in GVCF mode by restricting output regions to be plus/minus 50 bp of the refGene31 primary coding regions. Single-sample GVCF files were then combined into multi-sample GVCF files with each combined file containing 200 samples. These multi-sample GVCF files were then joint-genotyped using the GATK tool GenotypeGVCFs chromosome by chromosome. Our entire batch (total 18,789 trios), prior to duplicate individual removal, were joint-genotyped in two separate batches: one with 10,138 trios and the other 8,651 trios. The chromosome-level output VCF files from the previous step then went through variant quality recalibration with the GATK tool VariantRecalibrator (VQSR) for both single-nucleotide polymorphisms (SNPs) and insertions/deletions (indels), with known SNPs from 1000 Genomes Project32 phase 1 high-confidence set and known indels from Mills et al.33 and the 1000 Genomes Project Gold set. Variants in VQSR VCF files were annotated with Ensembl Variant Effect Predictor (VEP).34 The transcript with the most severe consequence was selected and all associated VEP annotations were based on the predicted effect of the variant on that particular transcript. Variants that were called in the proband and not in the parents were selected as potential de novo variants. All de novo calls were restricted to loci where the coverage depth in every sample of the trio was at least 10×. The variant called in the proband also had to have a frequency above 15% if it was an SNV or above 25% if it was an indel, with at least four reads supporting the alternative allele. The potential de novo calls were further filtered by requiring the Genotyping Quality score above 40, while the Phred-scaled p value using Fisher’s exact test to detect strand bias below 30 and the log odds of being a true variant versus being false from VQSR(VQSLOD) above −10. Any variant with a general population frequency above 1% was excluded based on 1000 Genomes Project and Exome Aggregation Consortium (ExAC)35 variant population frequency. Any remaining de novo variant was further filtered if the same variant was called more than four times in the parental samples in the cohort for SNVs, and that cutoff was set to one for insertions and two for deletions. To further polish the DNM call set, we also filtered out any calls with VQSLOD below seven and variant percentage below 30%. Any indels greater than 100 bp were excluded. DNM calls from individuals with more than 10 DNM calls were filtered and any non-X chromosome DNM calls with percentage greater than 90% were also removed. DNM calls overlapping with segmental duplication and simple repeat regions were filtered out as well. Shared DNMs between siblings were included only once. There were 7,318 DNMs in females and 8,727 DNMs in males. When assessing all DNMs, there were significantly more in females than males (Poisson p = 0.03), but this was no longer significant when assessing autosomes only (Poisson p = 0.39).
Sequencing Coverage Analysis
We accounted for sequence read-depth differences across platforms by assessing discovery cohort data from whole-exome sequencing in 9,014 individuals in the SSC22 and 3,317 in the Deciphering Developmental Disorders (DDD) study.36 Using the underlying BAM files, we computed the average depth per sample across each capture region and then averaged that across all individuals in the SSC and DDD, respectively. We retained only those regions with at least 20-fold sequence coverage. We cross-referenced these high-coverage regions with all of the regions annotated as coding sequence (CDS) in RefSeq genes (∼35 Mbp, downloaded from UCSC Genome Browser in May 2017). We then applied a 90% reciprocal overlap, using BEDTools,37 to identify CDS regions with sufficient coverage in both the SSC and DDD (∼26 Mbp). Similarly, the GeneDx replication cohort was assessed with a file of average depth per exon, allowing us to identify all regions with greater than 20-fold sequence coverage. We utilized a 90% reciprocal overlap, using BEDTools,37 to identify CDS regions with sufficient coverage in the SSC, DDD, and GeneDx cohort (∼26 Mbp) including 4 bp to the end of each capture region to recover splice sites. These high-coverage regions are provided in Table S3.
Statistical Analyses
We assessed de novo significance for missense, LGD, and the combination of missense and LGD variants using two statistical methods. One method identifies genes with an excess of DNM based on chimpanzee-human (CH) divergence model20,38,39 while the second, denovolyzeR v.0.2.0, leverages mutation bias based on triplet nucleotide content.40 Genes were considered significant for excess DNM based on a Bonferroni correction (6 tests [LGD, missense, combined; in the two models] and 19,295 genes [p < 4.3 × 10−7]). Significant genes were identified from the union of the CH and the denovolyzeR models.
We generated two categories of downsampled individual-level data from the discovery cohort: one consisting of males only and one consisting of a combination of males and females. In each category, 10,000 different downsampling sets consisting of variant data from 2,133 individuals (representing 2,779 individuals and equal to the females in our study) were tested in both the CH and denovolyzeR models. The number of significant genes, from each downsampling test, was based on a union of the two models.
We applied a Fisher’s exact test on allele counts for combined de novo LGD and missense variants to directly compare males and females. For the discovery set, autosomal variants were assessed from 12,092 chromosomes in males and 5,558 in females. X chromosome variants were assessed from 6,046 chromosomes in males and 5,558 in females. Since we combined the two functional categories, genes were considered significant if they met Bonferroni correction for the 19,295 genes tested (p < 2.6 × 10−6). The same testing strategy was applied to the replication set. Since the replication set contained more individuals, the number of chromosomes assessed on the autosomes was 20,758 in males and 16,798 in females. X chromosome variants were assessed from 10,379 chromosomes in males and 16,798 in females.
We performed power calculations to assess the number of additional parent-child sequenced trios that would be required to identify sex-biased genes from the direct male-to-female combined LGD plus missense count difference comparisons. For this analysis, we utilized the power.fisher.test function from the statmod package41 in R v.3.4.3 (see Web Resources). The number of additional parent-child sequenced trios we assessed ranged from ten thousand to ten million and are shown in Table 2. For each test, the settings included a sex ratio of 4 males:1 female, alpha = 2.6 × 10−6, and the nsim = 1,000, and the proportions were based on the empirical discovery data. Finally, we counted the number of genes reaching significance at 80%, 90%, and 100% power.
Table 2.
Power Calculations
| Additional Parent-Child Sequenced Trios | Genes at 80% Power | Genes at 90% Power | Genes at 100% Power |
|---|---|---|---|
| 10,000 | 1 | 1 | 1 |
| 20,000 | 4 | 4 | 1 |
| 30,000 | 7 | 4 | 1 |
| 40,000 | 17 | 8 | 2 |
| 50,000 | 19 | 18 | 4 |
| 100,000 | 148 | 126 | 18 |
| 500,000 | 4,208 | 1,918 | 1,886 |
| 1,000,000 | 4,253 | 4,245 | 4,208 |
| 5,000,000 | 4,660 | 4,623 | 4,615 |
| 10,000,000 | 4,660 | 4,660 | 4,657 |
Single-Cell Expression of Enriched Genes
A total of 15,928 single nuclei were isolated from the middle temporal gyrus of adult post-mortem brains of three human donors and profiled with RNA-seq.38,42 Unsupervised clustering identified 75 distinct transcriptomic clusters, including 45 GABAergic (inhibitory) neuronal, 24 glutamatergic (excitatory) neuronal, and 6 non-neuronal cell types. For each gene, the expression pattern was characterized as the number of cell types with appreciable expression (median counts per million > 1) in three broad classes: inhibitory and excitatory neurons and non-neuronal cells. Heatmaps were constructed of median log-normalized expression, log2(CPM + 1), of NDD sex-biased risk genes across cell types. Genes were ordered by hierarchical clustering using Ward’s linkage. The number of inhibitory and excitatory neuronal and glial types that expressed NDD risk genes and control genes were quantified and visualized as empirical cumulative distributions. Shapiro-Wilk tests rejected (p < 0.05) the null hypothesis that distributions were normally distributed of cell-type counts for each broad class, maximum average expression, and marker scores. Therefore, distributions were compared with two-sided Wilcoxon rank sum tests, and p values were Bonferroni corrected for multiple testing.
Protein Network Analysis
We performed a protein network analysis using the STRING database43 v.11 with the multiple proteins input option (June 21, 2019). Network statistics were derived from the “Network Stats” section of the “Analysis” category. GO enrichments were derived from the functional enrichments in the “Your Network” section of the “Analysis” category. Basic settings used in this analysis included: “confidence for meaning of network edges,” “textmining,” “experiments,” “databases,” “co-expression,” “neighborhood for active interaction sources,” and “minimum required interaction score” of 0.400. Subcellular localization data, for assessing nuclear localization, was downloaded from The Human Protein Atlas44 (June 19, 2019). We removed genes with the “Reliability” label of uncertain.
Inheritance Analysis of Private Mutations
Whole-exome sequencing data from 2,377 families from the SSC and 1,055 families from the DDD15,17,22 were assessed for heterozygous variants and observed only once in the parental cohort, which we refer to as private variants. Variants from the SSC were previously called using GATK45 and FreeBayes,22 and variants from the DDD were generated with GATK and FreeBayes using the same parameters. For both study cohorts, the FreeBayes and GATK call sets were merged together using GATK CombineVariants. We defined private variants separately for each cohort by counting the number of parents carrying a variant in the union call set of GATK and FreeBayes, which met all of the following criteria: (1) quality score (QUAL) > 50, (2) total read depth (INFO/DP) ≥ 20, (3) heterozygous in only one parent, (4) followed Mendelian transmission patterns in the carrier family (other families with a child that may carry the same variant as potential DNMs are ignored), and (5) in a high-coverage region (Table S3). These variants were then mapped for transmission and required sites to be called by both GATK and FreeBayes in the final analysis set. The merged SSC and DDD call sets were annotated with gene name, variant function, and CADD score using SnpEff v4.3t (build 2017-11-24) for hg19.22,46 Allele frequencies from ExAC’s nonpsychiatric subset were also annotated on the variant calls. We further refined the private variant call set by removing variants that were reported in the nonpsychiatric subset of ExAC. For the meta-analysis, we intersected the variant counts from the SSC and DDD cohorts. Variants unique to only one family in the combined set and not observed in the nonpsychiatric subset of the ExAC database were retained as the meta-analysis private variant set. For each of the 54 genes showing sex-biased enrichment for DNMs, we calculated the total number of private, inherited SNVs and indels for male and female probands, mothers, and fathers in the SSC, DDD, and combination of SSC and DDD. We tested male and female probands separately for deviations from 50% Mendelian transmission on a per-gene basis using a binomial test. In addition, we also directly compared the number of private mutations observed in males versus females in both probands and parents using a binomial test where the expected proportion of variants is equal to the proportion of female chromosomes over the total number of chromosomes (e.g., 1,680/6,864 for autosomal genes and 1,680/4,272 for X chromosome genes in probands). A Bonferroni-corrected significance threshold of 1.54 × 10−4 was used for all male-versus-female comparisons (54 genes × 3 variant classes × 2 disease states), and a Bonferroni-corrected significance threshold of 7.72 × 10−5 was used for single-sex enrichment analyses (54 genes × 3 variant classes × 2 sexes × 2 disease states [probands, parents]). The binomial test was performed in R v.3.4.1 using the function binom.test with an expected probability of 0.5.
Results
De Novo Enrichment in Males and Females
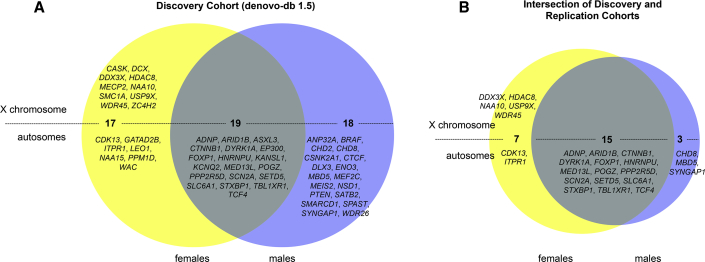
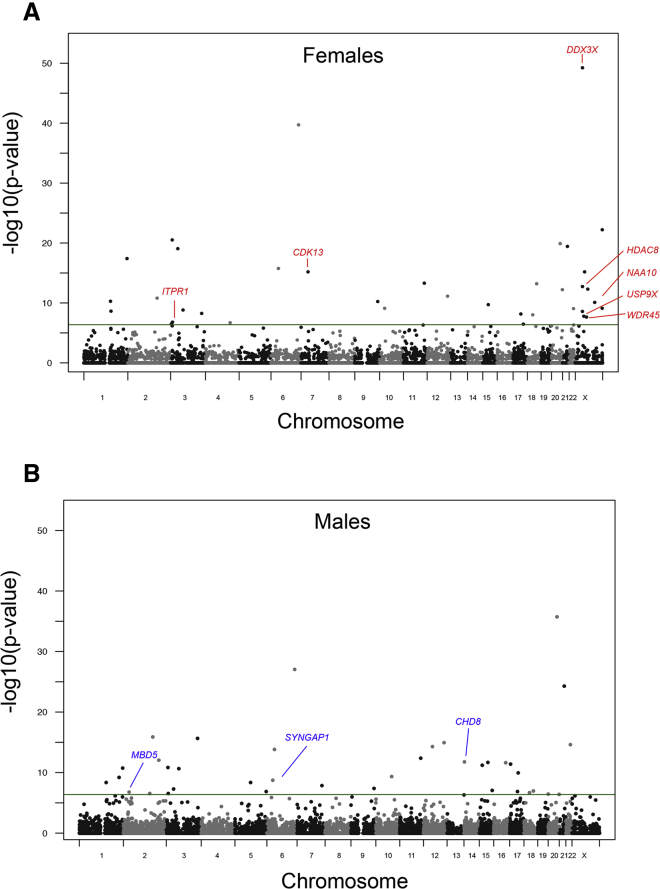
We analyzed DNM data from ∼8,825 parent-child trios and assessed male and female probands with NDDs separately for enrichment of de novo LGD, missense, or the combination of LGD and missense variants for 19,295 genes. We considered two DNM enrichment models: one based on CH divergence20,38 and another based on triplet repeat context (i.e., denovolyzeR40). In total, 54 distinct genes reached genome-wide significance in the union of the CH and denovolyzeR models when stratified by sex (17 in females, 18 in males, and 19 that were enriched for DNM in both sexes; Figures 1A and 2, Table S4). We tested replication of this sex-based classification by assessing counts of DNM from a second cohort of 18,778 parent-child trios subjected to exome sequencing by GeneDx. While 46 of the original 54 discovery set genes remained significant for DNM (Table S4), only 25 are significant based on their original classification (7 female-only, 3 male-only, and 15 in both sexes; Figures 1B, 2, and 3A, Table S4). Of the replicated female-only genes, 71.4% map to the X chromosome. Two autosomal genes (CDK13 [MIM: 603309], ITPR1 [MIM: 147265]) replicate as female enriched and, similarly, three genes (CHD8 [MIM: 610528], MBD5 [MIM: 611472], SYNGAP1 [MIM: 603384]) replicate as male biased.
Figure 1.
Genes Significant by DNM by Sex
(A) Discovery cohort results show genes reaching DNM genome-wide significance in females only (n = 17, yellow), males only (n = 18, blue), or both sexes (n = 19, gray).
(B) Replication cohort: while 46 genes remained significant post-replication, only 25 are significant by their original classification. The Venn diagrams are split to show the genes on the X chromosome (above the line) and those on the autosomes (below). The genes significant for de novo variants were from the union of the CH and denovolyzeR methods.
Figure 2.
Sex-Biased DNM Enrichment
Manhattan plots for de novo testing in (A) females and (B) males. Shown are the minimum p values from the discovery cohort analyses. Labeled are all genes with replicated significance in females only or males only, respectively. Unlabeled significant enrichments represent those shared between sexes and those that were not significant in the replication cohort.
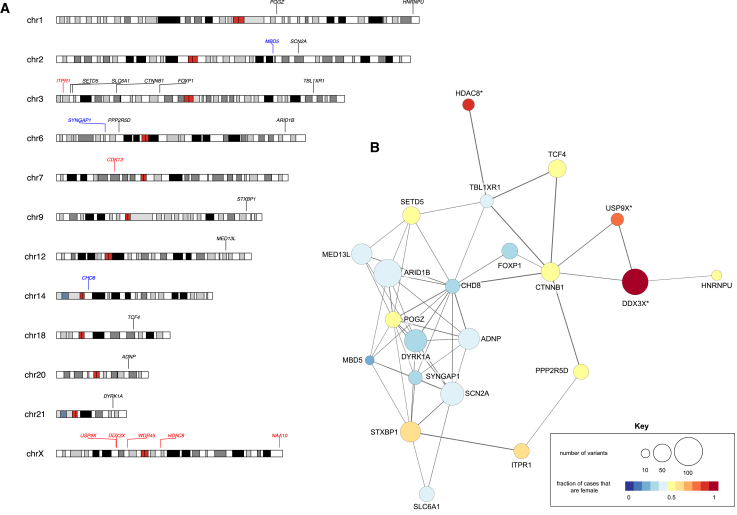
Figure 3.
Genomic Location and Protein-Protein Interaction Analysis of Significant Genes
(A) Replicated, significant genes are labeled at their genomic coordinate with genes in black as significant in both males and females, in blue significant in males only, and in red significant in females only.
(B) STRING protein-protein interaction network highlighting genes enriched for DNMs in females and/or males. The size of the circle corresponds to the number of mutations and the shading corresponds to the number of variants that are female (100% = red) or male (100% = blue) with shades in between based on the amount that are male or female. This network is shown to emphasize the high amount of interaction, at the protein level, between the genes surviving replication; 22 of the 25 genes surviving replication are within this same network. It also provides a visualization of proportionally how many of the variants are coming from females or males. Asterisk (∗) next to the protein name indicates the gene is on the X chromosome. Note: (B) includes data from both the discovery and replication sets.
We compared the number of affected individuals carrying a DNM in the sex-biased genes. Among female probands, 6.5% (180/2,779) carried a de novo LGD or missense DNM in one of the 22 female-enriched genes. In contrast, for the 6,046 males there were only 165 males (2.7%) carrying an LGD or missense de novo variant in one of the 18 male-enriched genes. Focusing on the seven female-biased genes, for example, identifies 69 female probands with DNMs (2.5%), whereas assessing the three male-biased genes identifies only 20 male probands with DNMs (0.3%). We should note that in the processing of these data, it was critical to QC for coverage and differences due to the number of X chromosomes. For example, we initially observed a significant difference in DNM rate discovery in males and females (Poisson p = 3.65 × 10−7) even when considering autosomes only (Poisson p = 3.4 × 10−3). However, after applying a high-coverage region filter (see Material and Methods) the higher DNM discovery rate in females was only observed when including the X chromosome (Poisson p = 2.61 × 10−3) and was no longer found among the autosomes (Poisson p = 0.29).
Since there are 124 genes known to reach genome-wide significance for de novo variants in NDDs,38 we decided to assess these genes for enrichment in males or females using the combined discovery and replication cohorts. Of the 124 genes, the number of genome-wide significant genes for de novo variants were 58 in both males and females, 13 in females only, and 15 in males only. There were 38 genes that were not significant in either males or females. The female significant genes included DDX3X (MIM: 300160), EEF1A2 (MIM: 602959), HDAC8 (MIM: 300269), ITPR1, KCNQ3 (MIM: 602232), LEO1 (MIM: 610507), NAA10 (MIM: 300013), QRICH1 (MIM: 617387), SMARCA4 (MIM: 603254), SMC1A (MIM: 300040), SOX5 (MIM: 604975), USP9X (MIM: 300072), and WDR45 (MIM: 300526) and the male significant genes included ANP32A (MIM: 600832), ASH1L (MIM: 607999), CAPRIN1 (MIM: 601178), CUL3 (MIM: 603136), DLG4 (MIM: 602887), DLX3 (MIM: 600525), ENO3 (MIM: 131370), MBD5, NONO (MIM: 300084), PIK3CA (MIM: 171834), PPP2R1A (MIM: 605983), SETBP1 (MIM: 611060), SMARCD1 (MIM: 601735), SYNGAP1, and ZBTB18 (MIM: 608433).
Single-Cell Expression and Protein Network Properties of Genes with DNM Enrichment
Overall, the 25 replicated genes enriched for DNM are significant for protein interactions (STRING; 6 expected versus 53 observed edges; p < 1.0 × 10−16, Figure 3B), especially proteins associated with regulation of transcriptional processes (p = 5.4 × 10−4) and chromatin binding (p = 4.7 × 10−3). Gene ontology analyses also reveal nuclear subcellular localization of many of these genes (nuclear lamin p = 1.2 × 10−6, nucleoplasm p = 2.92 × 10−5). Using Human Protein Atlas data (see Web Resources), we assess subcellular localization data for 22 of the 25 proteins where localization data are available. The majority (90.9%) localize to the nucleus—a significant enrichment (Fisher’s exact test p = 9.3 × 10−4, OR = 7.4) when compared to the full set of genes with subcellular localization data (57% or 6,537/11,377).
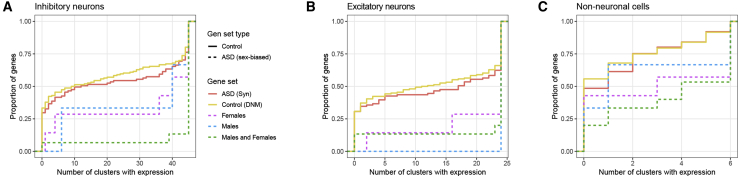
The ten sex-biased genes show broad expression across adult human cortical cell types, including inhibitory and excitatory neurons and, to a lesser extent, non-neuronal cells (Figure 4). Of the sex-shared risk genes, 87% are expressed in all inhibitory neuron types and 80% are expressed in all excitatory neuron types, while only 20% of control genes are as broadly expressed in inhibitory neurons and 34% in excitatory neurons. Shared risk genes are expressed in significantly more cell types than control genes based on a Wilcoxon rank sum test for inhibitory neurons (Bonferroni-corrected p = 9.6 × 10−5), excitatory neurons (p = 0.022), and non-neuronal cells (p = 0.0059). Shared risk genes were enriched in significantly (p = 0.0064) more neuronal cell types (average 89% of types) than non-neuronal cell types (average 61%) based on a Wilcoxon rank sum test. These cell-type enrichments are not driven by sex-specific expression since nuclei isolated from male and female donors showed similar expression patterns across neuronal and non-neuronal cell types (Figure S3).
Figure 4.
Sex-Biased NDD Risk Genes Show Significant Pan-neuronal Enrichment in Human Cortex
Empirical cumulative distributions of the number of cell type clusters recently identified in human cortex42 that express (CPM > 1 in > 50% of nuclei) replicated sex-biased NDD genes versus control genes for three broad classes of cells: (A) inhibitory neurons, (B) excitatory neurons, and (C) non-neuronal cells.
(A) 87% of NDD risk genes shared by males and females are expressed in all 45 types of inhibitory neurons while only 20% of control genes are expressed in all types. This enrichment in inhibitory types is statistically significant based on a Wilcoxon rank sum test (Bonferroni-corrected p = 9.6 × 10−5).
(B) More than two-thirds of male- and female-biased NDD risk genes and 80% of male- and female-shared risk genes are expressed in all 25 excitatory neuron types, which is significantly (Bonferroni-corrected p = 0.022) more than 34% of control genes.
(C) Approximately one-third of sex-biased risk genes are expressed in all six non-neuronal cell types compared to 10% of control genes, and risk genes shared by both males and females show statistically significant (Bonferroni-corrected p = 0.0059) enrichment compared to control genes.
Greater DNM Significant Gene Discovery in Females
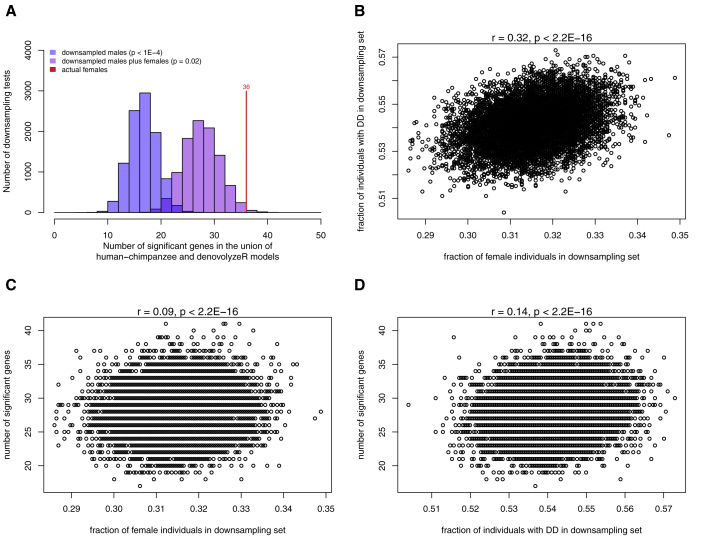
Our initial observations identify proportionally more enriched genes in females than in males. Because male and female sample sizes differed significantly in our discovery cohort, we formally tested whether this was a real effect by performing a downsampling analysis. We generated 10,000 sets of male only and 10,000 sets of male plus female data with the same size as our discovery cohort female dataset. For each permutation, we performed the DNM enrichment analyses with the CH and denovolyzeR models based on the union set of the two statistical models. The observed number of females carrying a DNM mutation in our discovery set (n = 36) is significantly greater than the combined male and female analysis (p = 0.02) and the male-only analysis (p < 1 × 10−4) (Figure 5A). As there are more female individuals with DD than autism we also tested the expected correlation between the fraction of individuals with DD and fraction of female individuals in our downsampling experiments (r = 0.32, p < 2.2 × 10−16, Figure 5B). We also tested whether there is a correlation between the number of significant genes identified in the downsampling and the fraction of females. We saw a positive correlation (r = 0.09, p < 2.2 × 10−16, Figure 5C) in this regard although a similar positive correlation is also observed for DD (r = 0.14, p < 2.2 × 10−16, Figure 5D). Interestingly, we observe significantly lower IQ in females than in males (female mean = 75.0, male mean = 82.1, Mann-Whitney p = 1.8 × 10−6) in individuals with autism from the SSC subset of the discovery cohort.
Figure 5.
Results of Downsampling Experiments
(A) Shown are the number of genes significant in each downsampling test from the discovery cohort. In blue are those from downsamplings consisting of males only and in purple are downsamplings from males and females. The red line indicates the actual result of the female set.
(B) Fraction of individuals with DD versus the fraction of females in each downsampling set.
(C) Number of significant genes versus the fraction of females in each downsampling set.
(D) Number of significant genes versus the fraction of individuals in the discovery cohort with DD.
Above the plots for (B), (C), and (D) are the results of the correlation test.
Direct Comparison of DNM in Males and Females
To determine whether there are genes with a sex bias based on differential counts of DNMs, we directly compared the number of LGD plus missense DNMs between male and female individuals with NDDs and tested for significance using a Fisher’s exact test. In the discovery cohort one gene, DDX3X, reached genome-wide significance with a female bias (26 DNMs in females, 0 DNMs in males, p = 4.73 × 10−9, OR = Inf; Table 3). Utilizing the same test in our replication cohort, we identified DDX3X as reaching genome-wide significance with a female bias in this dataset as well (65 DNMs in female, 2 DNMs in males, p = 1.76 × 10−11, OR = 20.15). Three additional genes reach nominal significance (p < 0.05) in both the discovery and replication cohorts (MECP2, NAA10, HDAC8; Table 3). We performed a power analysis to estimate the number of additional parent-child sequenced trios that would be required beyond the discovery cohort to identify additional genes reaching genome-wide significance (Table 2, Figure S2). Our estimates based on empirical data suggest we will detect additional genes with 90% power at 20,000 additional parent-child sequenced trios and 100% power at 40,000 additional parent-child sequenced trios. While it is notable that many genes will become genome-wide significant at 500,000 additional parent-child sequenced trios, it is important to note that most of these genes are predicted to have relatively small effect sizes. We also tested the 124 known NDD genes38 using the combined discovery and replication data and found that two genes (DDX3X, MECP2) reached genome-wide significance. This result further suggests that more genes will be discovered by this approach with the addition of more exome and/or genome data.
Table 3.
Genes Reaching a Minimum of Nominal Significance in Both Discovery and Replication in the Male versus Female Comparison
| Gene | Chrom |
Discovery |
Replication |
Genome-wide Significant in Both | Nominal Significance in Both | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Male Counts | Female Counts | Fisher’s Exact Test p Value | Fisher’s Exact Test Odds Ratio | Male Counts | female Counts | Fisher’s Exact Test p Value | Fisher’s Exact Test Odds Ratio | ||||
| DDX3X | chrX | 0 | 26 | 4.73E−09 | Inf | 2 | 65 | 1.76E−11 | 20.15 | yes | yes |
| MECP2 | chrX | 1 | 12 | 1.26E−03 | 13.08 | 8 | 54 | 1.77E−05 | 4.18 | no | yes |
| NAA10 | chrX | 0 | 5 | 2.52E−02 | Inf | 0 | 11 | 9.12E−03 | Inf | no | yes |
| HDAC8 | chrX | 1 | 7 | 3.23E−02 | 7.62 | 0 | 11 | 9.12E−03 | Inf | no | yes |
Parental Transmission and Significant Sex-Biased Genes
Since sex-biased transmission of private variants in autism has been observed in published studies,10,22 we tested whether there was any evidence for parent-of-origin or transmission bias for private inherited variants in the genes that originally showed sex-biased enrichments for DNMs. We identified private LGD and missense variants in 3,432 families where a child was diagnosed with autism or DD and assessed parental transmission of private mutations. Although we observe sex-biased trends for several of the 54 genes (Table S5), there are too few transmissions of severe mutations to establish significance for either parental carrier or transmission disequilibrium at the individual locus level. Reassuringly, the gene MECP2 shows a trend of under transmission to males (0/7 possible transmissions, p = 0.0156, binomial test) and is completely depleted of private variants in fathers (0/10 variants, p = 0.037, binomial test). In MBD5, which is also DNM enriched in males, we identified a trend of enrichment of private, missense mutations in carrier fathers (p = 8.78 × 10−4, binomial test) but identified no patterns of unusual transmission to either male or female probands. We hypothesize that MBD5 may represent a region of sex-specific hypermutation, where DNMs occur more often than expected by chance. This mechanism would explain the enrichment of DNMs and private mutations in male probands and fathers, respectively, and the pattern of expected Mendelian transmission among children. We observe a similar occurrence in the gene USPX9, where private, missense mutations are overrepresented in mothers (19/21 private mutations, p = 0.0196, binomial test) and DNMs are enriched in females, but there is no pattern of transmission-bias among affected children. However, it should be noted that we were underpowered to detect any statistically significant transmission bias between mothers and affected sons (as compared to daughters) for a single gene or in aggregate for LGD and missense mutations in the five X-linked genes.
Discussion
An outstanding question in the study of complex or multifactorial human phenotypes is “why do some diseases show a preference for one sex and what are the biological factors underlying this preference”? While we acknowledge that there are potential societal, ascertainment, and diagnostic reasons for this difference in NDDs, we hypothesize that some of this relates to the biology of the gene. Our study provides an initial comparison, for the autosomes and the X chromosome, of the two sexes directly for detection of genes with potential excess of variants in one sex or the other. To our knowledge, this is the first global assessment of de novo variants in NDDs focusing separately on enrichments in males and females as well as performing direct comparisons of males to females.
Through assessment of 8,825 sequenced parent-child trios, we initially identified 54 genes with an enrichment of de novo variants in males and/or females with NDDs. By replication analysis in an additional 18,778 sequenced parent-child trios, we find that 25 of these genes remain significant with respect to their original classification. Proportionally, DNMs in these genes were greater in females than in males. The increased prevalence in females may be due to the greater proportion of female patients represented in the intellectual disability/DD cohort in contrast to autism (see also our downsampling analyses). The seven genes replicated as enriched in females only overwhelmingly reside on the X chromosome (5/7), an effect not seen among male-enriched genes. The most likely explanation is male lethality for mutations in the hemizygous state and subtler NDD phenotypic consequence in females. The nature of genic X inactivation is particularly important in this regard. Interestingly, three of the genes showing female significance are either constitutive (DDX3X, USP9X) or facultative (NAA10) escapers of X-inactivation47 and exhibit higher expression in females than males. Constitutive escapers comprise 40% (2/5) of the X chromosome enriched genes in females in contrast to 13% of X chromosome genes escaping inactivation generally (89/683).
The five genes on the X chromosome (DDX3X, HDAC8, NAA10, USP9X, WDR45) and significant in females only are well-established genes in NDDs. In particular, DDX3X has been identified as one of the most common DNM-enriched gene in females with NDDs seen in an estimated 1% of all female case subjects with intellectual disability (MIM: 300958).48 DDX3X genotype-phenotype correlations have been studied in great detail by Blok et al.48 and the gene is a known escaper of X-inactivation involved in Wnt signaling. In particular, males developing intellectual disability have been identified carrying less severe DDX3X mutations than their female counterparts. In such cases the mothers of these males have been shown to be unaffected carriers. Cornelia de Lange syndrome 5 (MIM: 300882) is caused by mutations in HDAC8 and more severe intellectual disability has been observed in males than in females.49 It has also been observed that females harboring mutations in this gene will skew their X chromosome inactivation toward inactivation of the mutant allele resulting in less severe phenotype.49 Ogden syndrome (MIM: 300855) is another X chromosome-related syndrome, which is due to mutations in NAA10.50 Mutations in USP9X are associated with intellectual disability in females.51 The final significant X chromosome gene, WDR45, is associated with a clinical phenotype described as neurodegeneration with brain iron accumulation 5 (MIM: 300894) and nearly exclusively affects females.52
There are two autosomal genes with female-specific enrichment (CDK13, ITPR1). Mutations of CDK13 are involved in both NDDs and congenital heart defects.53 Missense mutations, in particular, have been shown to cluster in specific portions of the protein among reported cases of NDD.54 Mutations in ITPR1 have been identified in Gillespie syndrome (MIM: 206700)55 and spinocerebellar ataxia.56 The protein product is involved in intracellular calcium signaling57 with no previously reported sex biases. There are also three autosomal genes with male-specific enrichment (CHD8, MBD5, SYNGAP1). The first, CHD8, is one of the most well-known DNM genes involved in autism58,59 and is involved in chromatin regulation. Both MBD560 and SYNGAP161 have been shown to be involved in intellectual disability. As a group, we observe an enrichment of nuclear localization and involvement in chromatin and transcriptional regulation for significant genes in this study. This combined with their expression in neurons is consistent with a role of these genes as potential key gene regulatory network components involved in the control of the expression of other genes.
Our study shows that the assessment of females with NDDs enables greater gene discovery with a greater proportion of cases explained by DNM. The differential discovery of significant genes on the X chromosome is an obvious reason. However, we were unable to disentangle this from greater phenotypic severity and possible ascertainment biases. In our study, we show that more females had DD than autism and that females with autism tended to have lower IQ than males with autism. In a previous study, we showed that while females with autism had lower IQ, the female protective effect was stronger than the effects due to lower IQ alone.10 This is an important point because it could mean that females are being differentially ascertained in the clinic or in genetic studies. We note that while in principle, stratifying the analysis of DNMs by sex may increase gene discovery, we cannot verify this with our current sample size.
Beyond assessment of DNMs in males and females as separate populations, we also performed a direct comparison of DNM counts in males and females. This analysis revealed only one gene reaching genome-wide statistical significance (DDX3X) and it is confirmed as specifically enriched in females in the replication cohort. We note three other genes that fall below the level of genome-wide significance but are nominally significant in both discovery and replication cohorts. Our power calculations suggested that at least 20,000 additional parent-child sequenced trios would be required to detect new statistically significant genes at 90% power. Based on the current rate of discovery, we estimate that DNM data from 50,000 parent-child trios would lead to the discovery of 19, 18, and 4 sex-biased genes at 80%, 90%, and 100% power, respectively (Table 2). Given large-scale clinical whole-exome sequencing efforts and research studies (i.e., SPARK62), it is likely that such genes will soon be discovered, enhancing genetic diagnoses in the future.
On a final note, we reflect on the importance of studying NDDs with the sex biases in mind. First, there are genes that are definitely important and different between the two sexes and many reside on the X chromosome (i.e., DDX3X, HDAC8, NAA10, USP9X, WDR45). There are a few replicated sex-biased genes residing on the autosomes (CDK13, ITPR1 in females and CHD8, MBD5, SYNGAP1 in males). Follow-up of these genes will be critical and the potential underlying biological mechanisms may provide insight into the differences between variant effects in these genes on the two sexes. Second, our sex-partitioned analysis indicates that fewer female individuals are required to reach significance than males. Therefore, a study where samples are limiting would be more powerful with a female-only dataset. Third, a major limitation of our study is that we have not considered the Y chromosome. Currently, there are no priors for DNM burden on the Y chromosome and this is especially challenging given the repetitive content of the chromosome.63 Thus, if there are any genes on the Y chromosome with a DNM enrichment, they remain undiscovered as part of this analysis. With advances in long-read sequencing technology and the ability to assemble more complex portions of the genomes,64,65 the patterns of DNM and other potentially disease-associated complex loci may soon be uncovered.
Declaration of Interests
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. Z.Z., R.I.T., and K.R. are employed by GeneDx. K.R. is a shareholder of OPKO.
Acknowledgments
We thank Tonia Brown for assistance in editing this manuscript. This research was supported, in part, by the US National Institutes of Health (NIH R01MH101221) to E.E.E. This work was also supported by a postdoctoral fellowship grant from the Autism Science Foundation (#16-008, T.N.T.) and a grant from the National Institute of Mental Health (1K99MH117165, T.N.T.). E.E.E. is an investigator of the Howard Hughes Medical Institute. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 04/11/2018. Single-nucleus RNA-seq data from human temporal cortex was obtained from the Allen Cell Types Database.
Published: November 27, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.11.003.
Web Resources
Allen Cell Types Database, http://celltypes.brain-map.org/rnaseq
OMIM, https://www.omim.org
R statistical software, https://www.r-project.org/
STRING database, https://string-db.org
The Human Protein Atlas, http://www.proteinatlas.org/
Supplemental Data
Figures S1–S3
Tables S1–S5
References
- 1.Ober C., Loisel D.A., Gilad Y. Sex-specific genetic architecture of human disease. Nat. Rev. Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Psychiatric Association. Kennedy P.J. American Psychiatric Publishing; Washington, DC: 2015. Understanding Mental Disorders: Your Guide to DSM-5. [DOI] [PubMed] [Google Scholar]
- 3.Fombonne E. Epidemiological surveys of autism and other pervasive developmental disorders: an update. J. Autism Dev. Disord. 2003;33:365–382. doi: 10.1023/a:1025054610557. [DOI] [PubMed] [Google Scholar]
- 4.Piton A., Redin C., Mandel J.L. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am. J. Hum. Genet. 2013;93:368–383. doi: 10.1016/j.ajhg.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Werling D.M., Geschwind D.H. Sex differences in autism spectrum disorders. Curr. Opin. Neurol. 2013;26:146–153. doi: 10.1097/WCO.0b013e32835ee548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehnhardt F.G., Falter C.M., Gawronski A., Pfeiffer K., Tepest R., Franklin J., Vogeley K. Sex-related cognitive profile in autism spectrum disorders diagnosed late in life: implications for the female autistic phenotype. J. Autism Dev. Disord. 2016;46:139–154. doi: 10.1007/s10803-015-2558-7. [DOI] [PubMed] [Google Scholar]
- 7.Piton A., Gauthier J., Hamdan F.F., Lafrenière R.G., Yang Y., Henrion E., Laurent S., Noreau A., Thibodeau P., Karemera L. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol. Psychiatry. 2011;16:867–880. doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarachana T., Xu M., Wu R.C., Hu V.W. Sex hormones in autism: androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PLoS ONE. 2011;6:e17116. doi: 10.1371/journal.pone.0017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falconer D.S. The inheritance of liability to certain diseases, estimated from incidence among relatives. Ann. Hum. Genet. 1965;29:51–76. [Google Scholar]
- 10.Jacquemont S., Coe B.P., Hersch M., Duyzend M.H., Krumm N., Bergmann S., Beckmann J.S., Rosenfeld J.A., Eichler E.E. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am. J. Hum. Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner T.N., Sharma K., Oh E.C., Liu Y.P., Collins R.L., Sosa M.X., Auer D.R., Brand H., Sanders S.J., Moreno-De-Luca D. Loss of δ-catenin function in severe autism. Nature. 2015;520:51–56. doi: 10.1038/nature14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravarti A., Turner T.N. Revealing rate-limiting steps in complex disease biology: The crucial importance of studying rare, extreme-phenotype families. BioEssays. 2016;38:578–586. doi: 10.1002/bies.201500203. [DOI] [PubMed] [Google Scholar]
- 13.Polyak A., Rosenfeld J.A., Girirajan S. An assessment of sex bias in neurodevelopmental disorders. Genome Med. 2015;7:94. doi: 10.1186/s13073-015-0216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson E.B., Lichtenstein P., Anckarsäter H., Happé F., Ronald A. Examining and interpreting the female protective effect against autistic behavior. Proc. Natl. Acad. Sci. USA. 2013;110:5258–5262. doi: 10.1073/pnas.1211070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 19.Migeon B. Females Are Mosaics: X Inactivation and Sex Differences in Disease. Gend. Med. 2014;4:97–105. doi: 10.1016/s1550-8579(07)80024-6. [DOI] [PubMed] [Google Scholar]
- 20.O’Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Roak B.J., Stessman H.A., Boyle E.A., Witherspoon K.T., Martin B., Lee C., Vives L., Baker C., Hiatt J.B., Nickerson D.A. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto R., Nakazawa T., Tsurusaki Y., Yasuda Y., Nagayasu K., Matsumura K., Kawashima H., Yamamori H., Fujimoto M., Ohi K. Whole-exome sequencing and neurite outgrowth analysis in autism spectrum disorder. J. Hum. Genet. 2016;61:199–206. doi: 10.1038/jhg.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner T.N., Hormozdiari F., Duyzend M.H., McClymont S.A., Hook P.W., Iossifov I., Raja A., Baker C., Hoekzema K., Stessman H.A. Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am. J. Hum. Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.C Yuen R.K., Merico D., Bookman M., L Howe J., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner T.N., Yi Q., Krumm N., Huddleston J., Hoekzema K.F., Stessman H.A., Doebley A.-L., Bernier R.A., Nickerson D.A., Eichler E.E. denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 2016;45:D804–D811. doi: 10.1093/nar/gkw865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 28.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 1303.3997. [Google Scholar]
- 29.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protocol. Bioinform. 2013;11:11.10.11–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills R.E., Luttig C.T., Larkins C.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coe B.P., Stessman H.A.F., Sulovari A., Geisheker M.R., Bakken T.E., Lake A.M., Dougherty J.D., Lein E.S., Hormozdiari F., Bernier R.A., Eichler E.E. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat. Genet. 2019;51:106–116. doi: 10.1038/s41588-018-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H., Wang T., Wu H., Long M., Coe B.P., Li H., Xun G., Ou J., Chen B., Duan G. Inherited and multiple de novo mutations in autism/developmental delay risk genes suggest a multifactorial model. Mol. Autism. 2018;9:64. doi: 10.1186/s13229-018-0247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ware J.S., Samocha K.E., Homsy J., Daly M.J. Interpreting de novo variation in human disease using denovolyzeR. Curr. Protoc. Hum. Genet. 2015;87 doi: 10.1002/0471142905.hg0725s87. 7.25.21-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smyth G.K. Wiley; London: 1998. Numerical Integration. [Google Scholar]
- 42.Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 45.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tukiainen T., Villani A.C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. [Google Scholar]
- 48.Snijders Blok L., Madsen E., Juusola J., Gilissen C., Baralle D., Reijnders M.R., Venselaar H., Helsmoortel C., Cho M.T., Hoischen A., DDD Study Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am. J. Hum. Genet. 2015;97:343–352. doi: 10.1016/j.ajhg.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaiser F.J., Ansari M., Braunholz D., Concepción Gil-Rodríguez M., Decroos C., Wilde J.J., Fincher C.T., Kaur M., Bando M., Amor D.J., Care4Rare Canada Consortium. University of Washington Center for Mendelian Genomics Loss-of-function HDAC8 mutations cause a phenotypic spectrum of Cornelia de Lange syndrome-like features, ocular hypertelorism, large fontanelle and X-linked inheritance. Hum. Mol. Genet. 2014;23:2888–2900. doi: 10.1093/hmg/ddu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rope A.F., Wang K., Evjenth R., Xing J., Johnston J.J., Swensen J.J., Johnson W.E., Moore B., Huff C.D., Bird L.M. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reijnders M.R., Zachariadis V., Latour B., Jolly L., Mancini G.M., Pfundt R., Wu K.M., van Ravenswaaij-Arts C.M., Veenstra-Knol H.E., Anderlid B.M., DDD Study De novo loss-of-function mutations in USP9X cause a female-specific recognizable syndrome with developmental delay and congenital malformations. Am. J. Hum. Genet. 2016;98:373–381. doi: 10.1016/j.ajhg.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haack T.B., Hogarth P., Kruer M.C., Gregory A., Wieland T., Schwarzmayr T., Graf E., Sanford L., Meyer E., Kara E. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 2012;91:1144–1149. doi: 10.1016/j.ajhg.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamilton M.J., Caswell R.C., Canham N., Cole T., Firth H.V., Foulds N., Heimdal K., Hobson E., Houge G., Joss S. Heterozygous mutations affecting the protein kinase domain of CDK13 cause a syndromic form of developmental delay and intellectual disability. J. Med. Genet. 2018;55:28–38. doi: 10.1136/jmedgenet-2017-104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geisheker M.R., Heymann G., Wang T., Coe B.P., Turner T.N., Stessman H.A.F., Hoekzema K., Kvarnung M., Shaw M., Friend K. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat. Neurosci. 2017;20:1043–1051. doi: 10.1038/nn.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerber S., Alzayady K.J., Burglen L., Brémond-Gignac D., Marchesin V., Roche O., Rio M., Funalot B., Calmon R., Durr A. Recessive and dominant de novo ITPR1 mutations cause Gillespie syndrome. Am. J. Hum. Genet. 2016;98:971–980. doi: 10.1016/j.ajhg.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van de Leemput J., Chandran J., Knight M.A., Holtzclaw L.A., Scholz S., Cookson M.R., Houlden H., Gwinn-Hardy K., Fung H.C., Lin X. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3:e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berridge M.J. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 58.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernier R., Golzio C., Xiong B., Stessman H.A., Coe B.P., Penn O., Witherspoon K., Gerdts J., Baker C., Vulto-van Silfhout A.T. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Talkowski M.E., Mullegama S.V., Rosenfeld J.A., van Bon B.W., Shen Y., Repnikova E.A., Gastier-Foster J., Thrush D.L., Kathiresan S., Ruderfer D.M. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am. J. Hum. Genet. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berryer M.H., Hamdan F.F., Klitten L.L., Møller R.S., Carmant L., Schwartzentruber J., Patry L., Dobrzeniecka S., Rochefort D., Neugnot-Cerioli M. Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency. Hum. Mutat. 2013;34:385–394. doi: 10.1002/humu.22248. [DOI] [PubMed] [Google Scholar]
- 62.SPARK Consortium. Electronic address: pfeliciano@simonsfoundation.org; SPARK Consortium SPARK: A US cohort of 50,000 families to accelerate autism research. Neuron. 2018;97:488–493. doi: 10.1016/j.neuron.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hughes J.F., Page D.C. The biology and evolution of mammalian Y chromosomes. Annu. Rev. Genet. 2015;49:507–527. doi: 10.1146/annurev-genet-112414-055311. [DOI] [PubMed] [Google Scholar]
- 64.Vollger M.R., Dishuck P.C., Sorensen M., Welch A.E., Dang V., Dougherty M.L., Graves-Lindsay T.A., Wilson R.K., Chaisson M.J.P., Eichler E.E. Long-read sequence and assembly of segmental duplications. Nat. Methods. 2019;16:88–94. doi: 10.1038/s41592-018-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vollger M.R., Logsdon G.A., Audano P.A., Sulovari A., Porubsky D., Peluso P., Wenger A.M., Concepcion G.T., Kronenberg Z.N., Munson K.M. Improved assembly and variant detection of a haploid human genome using single-molecule, high-fidelity long reads. Ann. Hum. Genet. 2019 doi: 10.1111/ahg.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S3
Tables S1–S5