Abstract
In humans, structural or functional defects of the sperm flagellum induce asthenozoospermia, which accounts for the main sperm defect encountered in infertile men. Herein we focused on morphological abnormalities of the sperm flagellum (MMAF), a phenotype also termed “short tails,” which constitutes one of the most severe sperm morphological defects resulting in asthenozoospermia. In previous work based on whole-exome sequencing of a cohort of 167 MMAF-affected individuals, we identified bi-allelic loss-of-function mutations in more than 30% of the tested subjects. In this study, we further analyzed this cohort and identified five individuals with homozygous truncating variants in TTC29, a gene preferentially and highly expressed in the testis, and encoding a tetratricopeptide repeat-containing protein related to the intraflagellar transport (IFT). One individual carried a frameshift variant, another one carried a homozygous stop-gain variant, and three carried the same splicing variant affecting a consensus donor site. The deleterious effect of this last variant was confirmed on the corresponding transcript and protein product. In addition, we produced and analyzed TTC29 loss-of-function models in the flagellated protist T. brucei and in M. musculus. Both models confirmed the importance of TTC29 for flagellar beating. We showed that in T. brucei the TPR structural motifs, highly conserved between the studied orthologs, are critical for TTC29 axonemal localization and flagellar beating. Overall our work demonstrates that TTC29 is a conserved axonemal protein required for flagellar structure and beating and that TTC29 mutations are a cause of male sterility due to MMAF.
Keywords: sperm, asthenozoospermia, MMAF, infertility, TTC29, TPR, flagella, mouse, trypanosome
Introduction
The mammalian sperm flagellum is an evolutionarily conserved organelle shaped on a microtubule-based cytoskeleton, called the axoneme, which also serves as the backbone of motile cilia.1 The core of the axoneme consists of nine peripheral doublets of microtubules surrounding a central pair of microtubules (CP), which confer the canonical (9+2) pattern. The peripheral doublets are connected to each another by the nexin-dynein regulatory complex; in addition, multiprotein T-shaped structures, called the radial spokes (RSs), connect each peripheral doublet to the central pair. Beating of motile cilia and flagella is governed by multiprotein ATPases complexes, called the outer and inner dynein arms (ODAs and IDAs, respectively), which are both harbored by the peripheral microtubules and drive the sliding of the peripheral microtubules responsible for flagellar movement.2 In contrast to motile cilia, the mammalian sperm harbors some peri-axonemal structures, such as the outer dense fibers (ODFs) and the longitudinal columns (LC), which associate to the axoneme nearly all along the flagellum.3 In addition, a mitochondrial helix specifically surrounds the sperm axoneme in the midpiece region and is replaced by the fibrous sheath (FS) in the principal piece of the flagellum. These two accessory structures are, in particular, required for energy production.4,5
Structural and/or functional defects of the sperm flagellum induce asthenozoospermia, which in humans is defined by reduced number or absence of motile spermatozoa in the ejaculate (<32% of progressive sperm), according to the World Health Organization reference values.6 Asthenozoospermia may be associated with ciliary defects, as in primary ciliary dyskinesia (PCD [MIM: 244400]), an autosomal-recessive disease principally characterized by chronic airway infections7,8 but is also evidenced in many infertile men with no other symptomatology (i.e., isolated asthenozoospermia). Overall asthenozoospermia is found with variable degrees of severity in more than 80% of infertile men.9 Herein we focused on isolated asthenozoospermia due to multiple morphological abnormalities of the sperm flagellum (MMAF), a phenotype also termed “short tails” or “stump tails,”10,11 which constitutes one of the most severe sperm morphological defects leading to male sterility.12 MMAF is defined by the presence of a mosaic of sperm cells with absent, short, irregular, and coiled flagellum, associated with a severe disorganization of the peri-axonemal structures such as dysplasia of the fibrous sheath.10,11 The proportion of these anomalies is variable between MMAF-affected individuals but all are constantly present at frequencies largely exceeding those found in fertile men.13
In the last 5 years, high-throughput genetic investigations of MMAF-affected individuals from various ethnical origins allowed the rapid identification of a dozen genes, whose loss of function caused by biallelic variants account for more than one third of the MMAF-affected case subjects. Hence frequent mutations were identified in DNAH1 (MIM: 603332),14, 15, 16 DNAH2 (MIM: 603333),17 CFAP43/WDR96 (MIM: 617558),18,19 CFAP44/WDR52 (MIM: 617559),18, 19, 20 CFAP69 (MIM: 617949),21 CFAP251/WDR66 (MIM: 618146),22,23 FSIP2 (MIM: 615796),24 ARMC2 (MIM: 618424),25 QRICH2 (MIM: 618304),26 TTC21A (MIM: 611430),27 and SPEF2 (MIM: 610172)28 in unrelated MMAF-affected subjects. In addition, mutations in CFAP65 (MIM: 614270),19 CEP135 (MIM: 611423),29 and AK7 (MIM: 615364)30 were reported in single familial MMAF-affected case subjects. With the aim to identify additional genetic causes of human asthenozoospermia related to MMAF, we further analyzed whole exome sequencing data from a cohort of 167 MMAF individuals previously established by our team25 and report the identification and characterization of TTC29 bi-allelic truncating mutations in five unrelated individuals. In addition, by performing in silico, in vitro, and in vivo studies, using T. brucei and M. musculus mutant models, we demonstrate that TTC29 is a conserved axonemal protein required for correct flagellar beating and motility in three evolutionary distant species.
Material and Methods
Study Participants and Whole-Exome Sequencing (WES)
We analyzed data obtained by WES performed for a total of 167 men affected by primary infertility associated with a MMAF phenotype.25 WES and bioinformatics analyses were performed according to our previously described protocol using the human genome assembly GRCh38 as a reference sequence.18 All the recruited individuals displayed isolated infertility with no other clinical features; in particular, primary ciliary dyskinesia (PCD) syndrome was excluded. In this cohort, 83 individuals originated from North Africa (mainly from Algeria, Libya, and Tunisia) and sought consultation for primary infertility at the “Clinique des Jasmins” in Tunis, 52 individuals originated from the Middle East (Iran) and were treated in Tehran at the Royan Institute (Reproductive Biomedicine Research Center) for primary infertility, and 32 individuals were recruited in France, mainly at the Reproductive Department at Cochin Hospital in Paris. All individuals presented with a typical MMAF phenotype, which is characterized by severe asthenozoospermia (total sperm motility below 10%; normal value over 40% according to the World Health Organization reference values,6 in association with increased level of the following sperm flagellar abnormalities—short, absent, coiled, bent, or irregular flagella—in comparison with the normal ranges observed in control fertile individuals13).
Informed consent was obtained from all the individuals participating in the study according to local protocols and the principles of the Declaration of Helsinki. The study was approved by local ethics committees, and samples were then stored in the CRB Germethèque (certification under ISO-9001 and NF-S 96-900) according to a standardized procedure or were part of the Fertithèque collection declared to the French Ministry of Health (DC-2015-2580) and the French Data Protection Authority (DR-2016-392).
Sanger Sequencing
The selected mutations in TTC29 were validated by Sanger sequencing performed on ABI 3130XL (Applied Biosystems); analyses were performed using SeqScape software (Applied Biosystems). Sequences of primers used and expected product sizes are summarized in Table S2.
Semen Analysis
Semen samples were obtained by masturbation after a period of 2 to 7 days of sexual abstinence. Semen samples were incubated at 37°C for 30 min for liquefaction; ejaculate volume and pH, sperm concentration, vitality, morphology, and motility were evaluated according to World Health Organization (WHO) guidelines.6 Sperm vitality was assessed by eosin staining, and sperm morphology was analyzed on Schorr stained semen smears according to David’s classification.31
Transmission Electron Microscopy Analysis of Sperm Cells
Human or mouse sperm cells (10 millions) were fixed by incubation in 0.1 M phosphate buffer (pH 7) supplemented with 3% glutaraldehyde (Grade I; Sigma-Aldrich) for 2 h at room temperature. The samples were washed twice in PBS and resuspended in 0.2 M sodium cacodylate buffer. The samples were then post-fixed by incubation with 1% osmium tetroxide (Electron Microscopy Sciences), after which they were dehydrated by immersion in a graded series of alcohol solutions and embedded in Epon resin (Polysciences Inc.). Semi-thin sections were cut and stained with toluidine blue-Azur II. Ultra-thin sections (90 nm) were cut with a Reichert Ultracut S ultramicrotome (Reichert-Jung AG) and were then stained with uranyl acetate and lead citrate. Sections were analyzed with a JEOL 1011 microscope and digital images were acquired with a Gatan Erlangshen CCD camera and Digital Micrograph software.
RT-PCR Analysis of Human Sperm Cells
200–800 ng of total RNA were extracted from 5–10 million human spermatozoa using NucleoSpin RNA kit (Macherey Nagel) and subjected to reverse transcription with High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Fisher Scientific) following the manufacturer’s instructions. PCR reactions were performed with GoTaq DNA polymerase (Promega) using TTC29 specific primers. Amplicons were gel purified and sequenced (Eurofins Genomics). Sequences of primers used and expected product sizes are summarized in Table S3.
Western Blot Analysis on Sperm Cells or Testis Extracts
Denaturized protein samples corresponding to equal amounts of spermatozoa (from human or mouse) or mouse testis extracts were loaded on SDS-PAGE (12% acrylamide/bisacrylamide [40% 37.5:1]) and transferred onto nitrocellulose membranes. The membranes were blocked in 5% milk in PBS-Tween 0.1% or 3% BSA in TBS-Tween 0.1%, and immunoblot analysis was performed using the indicated primary antibodies. Details of antibodies and dilutions used for western blot assays are provided in Table S4.
Immunofluorescence Analysis of Sperm Cells
10 μL of semen samples were spread onto a Superfrost Plus slide (Menzel Glasbearbeitungswerk, GmbH & Co. KG). Sperm was fixed by incubation with PBS/4% paraformaldehyde for 10 min. The slides were incubated 20 min at 95°C in citrate buffer (H-3300, VectorLabs). The slides were next treated with 0.2% Triton in PBS for permeabilization and then blocked by incubation in 1% BSA for 1 h. They were then incubated with primary antibodies for 2 h at room temperature and then secondary antibodies for 1 h at room temperature. The slides were mounted in Vectashield medium (Vector Laboratories) supplemented with 0.5 μg/mL DAPI. Slides were analyzed with a Zeiss Axiophot epifluorescence microscope. Digital images were acquired with a cooled charge-coupled device (CCD) camera (Hamamatsu Co.), under identical instrument settings, with MetaMorph software (Molecular Devices). Details of antibodies and dilutions used for immunofluorescence assays are provided in Table S4.
CRISPR Mutant Mouse Engineering
Handling of mice and experimental procedures were performed in accordance with institutional and national guidelines for the care and use of laboratory animals. Authorizations were obtained from local and governmental ethical review committees: Authorization APAFIS #14124-2017072510448522 v26, Touré (2018-2025). TTC29 mutant mice were generated by the “Transgenesis and Homologous Recombination” core facility of the Institut Cochin (INSERM U1016, Paris, France), using CRISPR/Cas9 technique. The RNA guide targeting Ttc29 exon 5, 5′-CAAAGGGCTGTCGAAAGAAG-3′, was designed using CRISPOR selection tool. gRNA was pre-incubated with Cas9 protein (RT, 10 min) to obtain functional ribonucleoprotein (RNP) complexes. The final injection mix containing 0.6 μM of gRNA and Cas9 protein (1.5 μM) in TE-0.1 buffer (10 mM Tris-HCl, 0.1 mM EDTA) has been injected into 210 fertilized oocytes of superovulated C57BL/6JRj females. 92 typical 2-cell stage embryos were subsequently implanted into the oviduct of 5 pseudo-pregnant B6CBAF1 females. Subsequent genotyping of CRISPR edited founders was performed by PCR amplification (GoTaq DNA Polymerase, Promega) on DNA extracted from tail biopsies (NucleoSpin Tissue, Macherey-Nagel) and PCR-product sequencing (Eurofins Genomics). Sequences of primers used and expected product sizes are summarized in Table S5. Mice carrying Ttc29 mutational events were bred with C57BL6/JRj mice to ensure germline transmission and eliminate any possible mosaicism.
Mouse Sperm Morphological Analysis
Spermatozoa were retrieved from cauda epididymes in PBS buffer and spread onto a Superfrost Plus slide (Menzel Glasbearbeitungswerk, GmbH & Co. KG). Sperm cells were fixed by incubation with PBS/4% paraformaldehyde for 10 min and stained following Papanicolaou protocol (Hematoxylin, OG6, EA50).
Mouse Sperm Motility Analysis
Sperm motility was assessed by Computer Aided sperm Analysis (CASA) using CEROS II apparatus (Hamilton Thorne). Briefly, mouse sperm cells expelled from the cauda epididymis were recovered into M2 medium (Sigma-Aldrich). The movements of at least 500 sperm cells per sample were analyzed in 20 μm chambers (Leja Products B.V.) with Zeiss AX10 Lab. A1 microscope (10× objective), using HT CASAII software.
The settings were as follows: acquisition rate, 60 Hz; number of frames, 45; minimum head brightness, 175; minimum tail brightness, 80; minimum head size, 10 μm2; minimum elongation gate, 1%; maximum elongation gate, 100%; objective magnification factor, 1.2.
The principal motility parameters measured were: curvilinear velocity (VCL), average path velocity (VAP), straight-line velocity (VSL), beat/cross frequency (BCF), amplitude of lateral head displacement (ALH). Progressive sperm cells were characterized by average path velocity (VAP) > 45 μm/s and straightness (STR = VSL/VAP) > 45%, respectively.
Gamete Preparation and In Vitro Fertilization
Oocyte preparation: C57BL6/J female mice of 6–8 weeks old (JANVIER LABS, France) were superovulated with 5 IU of pregnant mare serum gonadotropin (PMSG) and 5 IU human chorionic gonadotropin (hCG) (Intervet) 48 h apart. About 14 h after hCG injection, the animals were sacrificed by cervical dislocation. Cumulus oophorus were collected by tearing the ampulla wall of the oviduct, placed in Ferticult medium (FertiPro N.V) supplemented with 3% BSA (Sigma-Aldrich), and maintained at 37°C under 5% CO2 in air under mineral oil (Sigma-Aldrich). When experiments were performed with zona pellucida (ZP)-free oocytes, cumulus cells were removed by a brief exposure to hyaluronidase IV-S (15 mg/mL, Sigma-Aldrich). The ZP was then dissolved with acidic Tyrode’s (AT) solution (pH 2.5) (Sigma-Aldrich) under visual monitoring. The zona-free eggs were rapidly washed five times and kept at 37°C under 5% CO2 in air for 2 to 3 h to recover their fertilization ability.
Capacitated sperm preparation: mouse spermatozoa were obtained from the cauda epididymides of C57BL6/J male mice (8- to 10-week-old) and capacitated at 37°C under 5% CO2 for 90 min in a 500 μL drop Ferticult medium supplemented with 3% BSA, under mineral oil.
In vitro fertilization: cumulus-intact and zona-free eggs were inseminated with capacitated spermatozoa for 3 h in a 100 μL drop of Ferticult medium 3% BSA at a final concentration of 106 or 105 per mL, respectively. Then, they were washed and directly mounted in Vectashield/DAPI (Vector Laboratories) for observation under UV light (Nikon Eclipse E600 microscope). Only oocytes showing at least one fluorescent decondensed sperm head within their cytoplasm were considered fertilized and according to this, the fertilization rate per cumulus-intact (FR) was evaluated. To assess the fertilization index (FI), the number of decondensed sperm heads per zona-free oocyte was recorded.
Mice Breeding Assay
To test the fertility, three pubescent Ttc29−/− (L5 or L3 lines) and 5 wild-type littermates, aged of 8 weeks, were mated each with two C57BL/6 J female mice, during a 4-month period. The number of pups per litter and the number of litters per female were recorded throughout the breeding assay period.
Trypanosoma brucei Culture and Transfection
The trypanosome cell lines used in this study derived from the procyclic parental form T. brucei SmOxP427 strain, co-expressing the T7 RNA polymerase and the tetracycline repressor.32 Cells were cultured at 27°C in SDM79 medium containing 10% (v/v) heat-inactivated fetal calf serum, 10 μg/mL hemin, supplemented with puromycin (1 μg/mL), and transfected as previously described33 using specific transfection buffer,34 and cloned by serial dilution. The culture medium was supplemented, when required, with blasticidin (10 μg/mL), neomycin (10 μg/mL), and phleomycin (5 μg/mL). RNA interference (RNAi) was induced with tetracycline (10 μg/mL).
Trypanosoma brucei Cell Lines Generated for This Study
For RNAi, a fragment of the TbTTC29 ORF was amplified by PCR (bp 352–806, corresponding to amino acids 118–268) and cloned between the XhoI and XbaI sites of p2T7tiB. SmOxP427 cells were transfected with the NotI-linearized plasmid. After selection, several clones were analyzed and one clone was chosen for further studies (cell line RNAiTbTTC29). To produce an endogenous TbTTC29 with a carboxy-terminal 10TY1 epitope-tag,35 RNAiTbTTC29 cells were transfected with a tagging cassette that was obtained by PCR using the pPOTv7-blast-10xTY1 as a template (cell line TbTTC29::TY1/RNAiTbTTC29). A similar approach was used to generate a cell line expressing TbTTC29-Nter::myc (aa 1–120 of TbTTC29) in the TbTTC29::TY1/RNAiTbTTC29 background, using the pPOTv7-10myc-neomycin vector as a template (cell line TbTTC29::TY1/TbTTC29-Nter::myc/RNAiTbTTC29). Sequences of primers used and expected product sizes are summarized in Table S6.
Immunofluorescence Analysis of Trypanosoma brucei
Cells were collected, washed, and processed for immunolabelling on methanol-fixed detergent-extracted cells (cytoskeleton, CSK) as previously described.36 Cytoskeletons were incubated 1 h at RT with primary antibodies and then with secondary antibodies. Nuclei and kinetoplasts were stained with DAPI (10 μg/mL). Images were acquired on a Zeiss Imager Z1 microscope, using a Photometrics Coolsnap HQ2 camera, with a 100× Zeiss objective (NA 1.4) using Metamorph software (Molecular Devices), and processed with ImageJ.
Trypanosoma Sedimentation Assays and Video-microscopy
Sedimentation assays were performed as previously described.37 Briefly, the cultures of parental (WT), non-induced and RNAi-induced cells were placed in cuvettes at a density of 1.107 cells/mL and incubated another 24 h without shaking. The optical density at 600 nm (OD) was then measured before mixing (ODb, the cell density reflecting the “swimming” cells) and after mixing (ODa, the cell density reflecting “swimming” and “sedimenting” cells). The graphs represent the percentage of sedimentation calculated as 100 − (ODb/ODa) × 100 and normalized with the parental cells. Video-microscopy was carried out as previously described.38 Briefly, parental cells and 8-days-RNAi-induced cells were washed in PBS. Cellular mobility was recorded by phase contrast on a Zeiss AxioImager Z1 with a 40× lens (NA 1.3). Twenty-five seconds of digital video from separate regions were captured and analyzed using MetamorphH software (Molecular Devices).
Western Blotting Analysis on Trypanosoma brucei Cells
Proteins from whole cell extracts (5.106 cells per well) or from subcellular fractions (cytoskeleton or flagellum) were separated on 12% SDS-PAGE and semi-dry transferred (BioRad) for 45 min at 25V on PVDF membrane. After a 1 h-blocking step in 5% skimmed milk in TBS-0.2% Tween-20 (blocking solution, BS), the membranes were incubated overnight at 4°C with the primary antibodies in BS. After 10-min washes in BS, then 1 M NaCl, then BS, the membranes were incubated with the HRP-conjugated secondary antibodies, washed twice 10 min in blocking buffer and twice 5 min in TBS. Blots were revealed using the Clarity Western ECL Substrate kit (Bio Rad) on a ImageQuant LAS4000 apparatus and quantified using ImageJ. Stripping of the membranes was performed when required by incubation in glycine 100 mM (pH 3), SDS 1%, NP40 0.1% (2× 10 min), followed by 3 washes in TBS; the membranes were then blocked in BS and incubated as described above with another primary antibody.
Statistical Analysis
Results are expressed as mean ± SEM of at least three independent experiments. The following statistical tests were performed, when appropriate: one-tailed unpaired t tests, one-way ANOVA followed by Tukey’s test, and chi-square t tests. Results were analyzed in GraphPad Prism v.7.00 for Windows, GraphPad Software, www.graphpad.com. Differences were considered as statistically significant when p value < 0.05.
Results
Identification of TTC29 Bi-allelic Truncating Variants in MMAF-Affected Infertile Individuals
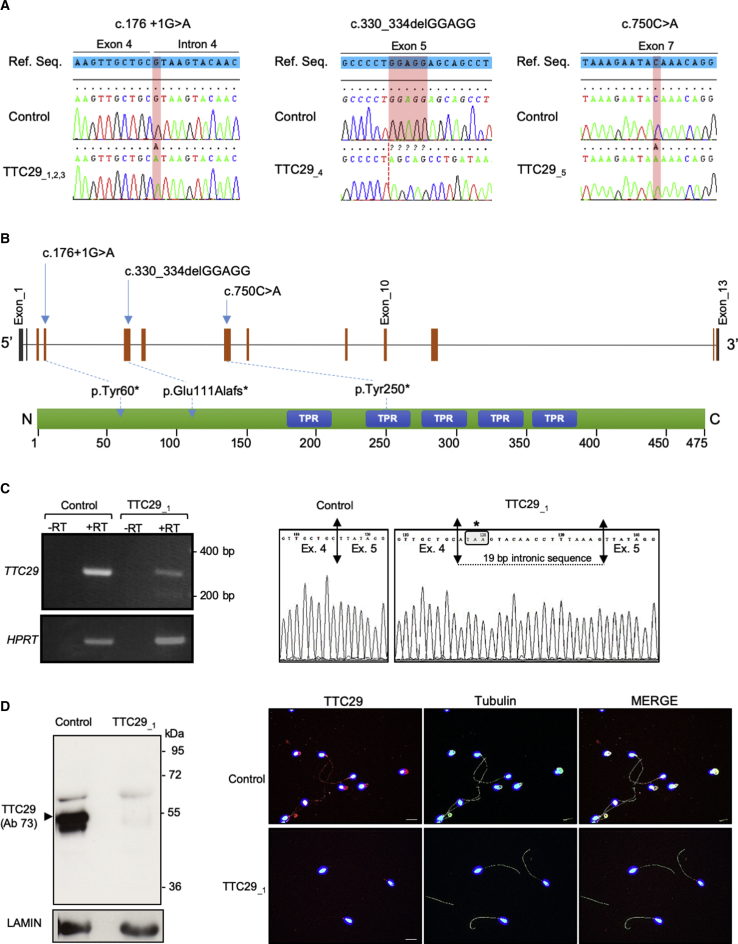
Whole-exome sequencing (WES) data from a cohort of 167 MMAF-affected individuals were analyzed in order to identify new candidate genes for severe asthenozoospermia. Previous analyses of this cohort permitted to identify bi-allelic variants in a total of 58 men (35%) in confirmed MMAF-associated genes (DNAH1, CFAP43, CFAP44, CFAP69, WDR66, FSIP2, ARMC2, TTC21, and SPEF2).21,25,27,28 We therefore consider that the cause of the MMAF phenotype is established for these 58 men, leaving 109 undiagnosed individuals. In these remaining undiagnosed individuals, we identified TTC29 (tetratricopeptide repeat domain 29; Gene ID: 83894) as a very good candidate since five unrelated individuals presented a homozygous truncating variant in this gene. TTC29 is located on chromosome 4 and contains 13 exons (ENST00000325106; GenBank: NM_031956.3), predicting a 475-amino-acid protein (UniProtKB: Q8NA56). The TTC29 protein comprises five tetratricopeptide repeats (TPR), which are 34 amino acid repeats present in a wide variety of proteins, forming alpha helixes that behave as scaffolds for protein-protein interactions and assembly of multiprotein complexes.39, 40, 41 Based on public tissue expression databases (EMBL-EBI Expression Atlas, NCBI, and GTEx), TTC29 is found to be highly expressed in the testis and to a much lesser extent in the lung. Quantitative single-cell RNA sequencing datasets from human adult testis (ReproGenomics Viewer)42,43 indicate an expression in the germ cells from zygotene spermatocyte to late spermatid stage but not in the testicular somatic cells (i.e., Leydig and Sertoli cells), strongly suggesting a role in sperm cells differentiation and/or function. In addition, the TTC29 protein was positively detected in human sperm proteome44 whereas it was near absent in human airway cilia.45 Among the five TTC29 mutated individuals, two originated from North Africa, one from central Africa (Niger), and two from Iran. The three African subjects (TTC29_1, TTC29_2, and TTC29_3) were homozygous for the same variant c.176+1G>A (p.Tyr60∗). This variant alters the canonical donor splice site of TTC29 exon 4 and was identified in gnomAD at a frequency of 3.42e−4. The two remaining Iranian individuals carried two different truncating variants. Individual TTC29_4 was homozygous for a five nucleotide deletion, c.330_334delGGAGG (p.Glu111Alafs∗), which was found in gnomAD at a frequency of 1.21e−5, and individual TTC29_5 was homozygous for the c.750C>A (p.Tyr250∗) variant, absent from the gnomAD database. These two variants are deleterious as they induce a premature stop codon (positions 113 and 250, respectively, in the 475-amino acid TTC29 protein sequence). The presence of all TTC29 homozygous variants was confirmed by Sanger sequencing on DNA samples from the five individuals, as illustrated in Figure 1A, and the consequences of the variants on protein translation are shown in Figure 1B.
Figure 1.
Identification of TTC29 Mutations in Five Unrelated Infertile Men and Functional Consequences of the c.176+1G>A TTC29 Variant
(A) Illustration of Sanger sequencing data for the TTC29 mutations identified in MMAF-affected infertile men: individuals TTC29_1, TTC29_2, and TTC29_3 carried the c.176+1G>A homozygous mutation; individual TTC29_4 carried the c.330_334delGGAGG homozygous mutation; individual TTC29_5 carried the c.750C>A homozygous mutation.
(B) Schematic representation of TTC29 exons structure (top) and predicted protein domains (bottom), according to SMART webtool and Uniprot, with position of the three different mutations identified in the gene. TTC29 encodes a protein of 475 amino acids that contains five tetratricopeptide domains (blue boxes).
(C) At the left, RT-PCR analysis on sperm sample from individual TTC29_1, carrying the c.176+1G>A mutation, which indicates a reduced amount of TTC29 transcript compared to control individual while HPRT transcript level was unaffected. At the right, electrophoregram of Sanger sequencing of the amplified transcript in sperm from individual TTC29_1, which shows an abnormal exon 4–5 boundary resulting in a premature stop codon, in comparison with control individual.
(D) Western blot and immunofluorescence analyses of sperm sample from individual TTC29_1, carrying the c.176+1G>A mutation, which both indicates the absence of TTC29 protein compared to control individual.
Lack of TTC29 Protein in Sperm from Individual TTC29_1, Carrying the c.176+1G>A Mutation
All three TTC29 identified variants are predicted to introduce a premature stop codon in the first half of the protein sequence, thereby potentially inducing mRNA decay and if not, a truncated protein, which would lack most of the TPR functional domains. In order to provide further arguments for the pathogenicity of the identified variants, we analyzed the transcript and levels of protein in semen samples available from individual TTC29_1 carrying the c.176+1G>A mutation, which affects exon 4 consensus splice donor site. We first amplified TTC29 transcripts in sperm cells from control individual and individual TTC29_1, using primers located in exons 3 and 5. The levels of TTC29 transcripts were reduced in sperm cells from individual TTC29_1, as compared to what was observed in a control individual while the expression levels of HPRT (an ubiquitous housekeeping gene) was similar in both individuals (Figure 1C), indicating that the mutated transcript was subject to incomplete mRNA decay. Sanger sequencing of the amplified transcript showed an abnormal exon 4/5 boundary with an additional 19 nucleotides retained from intron 4, inducing a premature stop codon one nucleotide after the end of exon 4 (Figure 1C). We then performed western blot experiments using two different antibodies raised against TTC29: antibody Ab73, binding amino acids 9–92 (N terminus) and antibody Ab06, binding amino acids 174–260 (middle region). When using antibody Ab73, the TTC29 protein was detected at the predicted molecular weight of 55 kDa in sperm protein extracts from the control individual but no protein was observable in sample from individual TTC29_1, whereas Lamin, a component of the nuclear membrane, was equally detected in samples from both individuals (Figure 1D). Similar results were observed when using the antibody Ab06, although the signal intensity was much lower (Figure S1A). To confirm these results, we performed immunodetection assays using antibody Ab73; we observed that the TTC29 protein was detected along the sperm flagellum of the control individual but absent in sperm cells from individual TTC29_1, which were able to assemble a flagellum as visualized by α-Tubulin staining (Figures 1D and S1B). Taken together, the results confirm the deleterious effect of the c.176+1G>A splice mutation, which results in the total absence of TTC29 protein in sperm from individual TTC29_1.
Phenotypical Characterization of MMAF-Affected Individuals Harboring TTC29 Truncating Variants
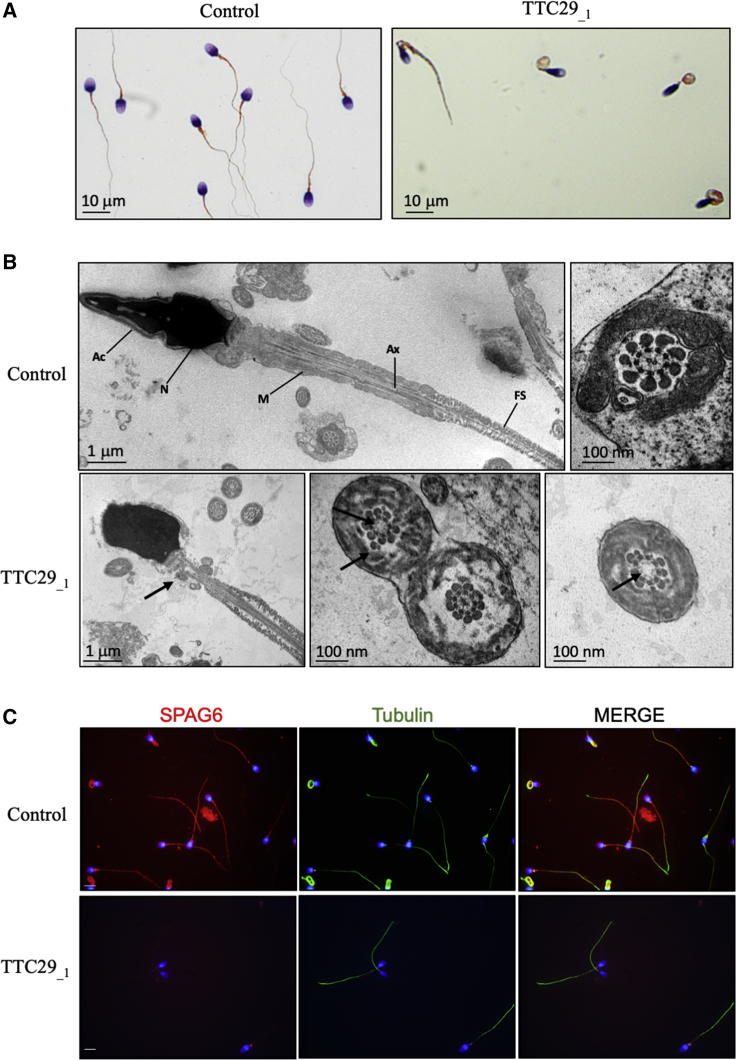
All five individuals harboring TTC29 truncating mutations had a normal somatic karyotype (46, XY) with normal bilateral testicular size, hormone levels, and secondary sexual characteristics. Semen analysis revealed normal vitality and normal sperm number, except for individual TTC29_1 who showed a moderate decrease of sperm number (16.8 million; normal value: 39 million) (Table 1). All five individuals presented with severe asthenozoospermia, with nearly zero progressive spermatozoa present in the ejaculate (between 0% and 2%; normal value > 32%) and less than 10% of total motile spermatozoa (normal value > 40%) (Table 1). In addition, no spermatozoa with normal morphology were recorded (0% typical forms; normal value > 23%) for any of the five individuals. As illustrated for individual TTC29_1 (Figure 2A), quantitative sperm morphological analysis confirmed a typical MMAF phenotype defined by sperm with absent, short, and irregular flagella in rates largely exceeding the distribution ranges observed in control fertile men13 (Table 1). In addition, the global average of semen parameters from the five individuals carrying mutations in TTC29 was found similar to that of the other MMAF-affected individuals from the cohort with known or unknown genetic causes (Table 2). Ultrastructure analysis of the sperm cells from individual TTC29_1 was performed by transmission electron microscopy (TEM) and showed abnormal midpiece and fibrous sheath together with severe axonemal disorganization (Figure 2B), as previously described for MMAF-affected individuals. Quantification on a total number of 23 transversal sections of sperm cells from individual TTC29_1 indicated 36% of abnormal axonemal structure including 27% of sections lacking the central pair (9+0 pattern) and a few sections displaying global microtubule doublets disorganization (Figure 2B). The percentage of sections with axonemal defects observed in sperm from individual TTC29_1 (36%) was, however, much lower than what was previously reported for MMAF-affected individuals carrying mutations in CFAP43 and CFAP44, also encoding axonemal proteins (95% in average including 81.8% and 66.7% of sections lacking the central pair, respectively).18 Despite this observation, we could confirm the extent of the axonemal anomalies, as immuno-marking of SPAG6, a component of the central pair complex, gave nearly no signal in sperm from individual TTC29_1 (Figure 2C). In addition, although positive, immunodetection with a set of antibodies specifically marking other functional protein complexes of the axoneme, such as the ODAs (DNAI1), IDAs (DNALI1), and RSs (RSPH1), appeared weaker than the patterns obtained in sperm from control individuals (Figures S2 and S3) while FS staining with AKAP4 antibody was unaffected (Figures S2 and S3). Overall, this indicated that the absence of TTC29 had a strong impact on the positioning of other axonemal proteins.
Table 1.
Semen Parameters and Sperm Morphological Defects (Flagellum, Head, and Acrosome) of MMAF-Affected Individuals Carrying Mutations in TTC29
|
General Semen Characteristics |
Flagellum Defects |
Head Defects |
Acrosome Defects |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Volume (mL) | Total Sperm Count (106) | Total Motility | Progressive Motility | Vitality | Typical Forms | Absent | Short | Irregular | Coiled | Bent | Tapered | Thin | Micro-cephalic | Macro-cephalic | Post-acrosomal | Acrosomal | |
| TTC29_1c.1761+G>A | 2.8 | 16.8 | 4 | 1 | 57 | 0 | 15 | 11 | 23 | 10 | 18 | 4 | 55 | 8 | 0 | 31 | 95 |
| TTC29_2c.1761+G>A | 4 | 168 | 10 | 2 | 64 | 0 | 12 | 42 | 60 | 8 | ND | 28 | 12 | 12 | 0 | 34 | 60 |
| TTC29_3c.1761+G>A | 4,8 | 91.2 | 7 | 2 | 65 | 0 | 30 | 28 | 46 | 14 | 0 | 4 | 4 | 6 | 2 | 15 | 98 |
| TTC29_4c.330_334delGGAGG | 3 | 36 | 0 | 0 | 88 | 0 | 8 | 92 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND |
| TTC29_5c.750C>A | 6.5 | 38 | 0 | 0 | 80 | 0 | 0 | 90 | 0 | 0 | ND | 7 | 0 | 0 | 0 | 0 | 0 |
| Reference limitsa | 1.5 (1.4–1.7) | 39 (33–46) | 40 (38–42) | 32 (31–34) | 58 (55–63) | 23 (20–26) | 5 (4–6) | 1 (0–2) | 2 (1–3) | 17 (15–19) | 13 (11–15) | 3 (2–4) | 14 (12–16) | 7 (5–9) | 1 (0–2) | 42 (39–45) | 60 (57–63) |
Values are expressed in percent, unless specified otherwise. Bold indicates abnormal values. ND, not determined.
Figure 2.
Characterization of the Sperm Morphological and Ultra-structural Phenotype of Individual TTC29_1 Carrying the c.176+1G>A Mutation
(A) Schorr staining of semen smears from individual TTC29_1, carrying the c.176+1G>A mutation, showing sperm without normally constituted flagella such as coiled flagella and flagella of irregular caliber. Scale bars represent 10 μm.
(B) TEM analysis of spermatozoa from individual TTC29_1, carrying the c.176+1G>A mutation, showing abnormal flagellum assembly with a reduced mitochondrial sheath and abnormal axonemal cross sections with a lack of the central pair. Ac, acrosome; N, nucleus; M, mitochondria; Ax, axoneme; FS, fibrous sheath. Black arrows indicate abnormal structures. Scale bars represent 1 μm and 100 nm.
(C) Immunofluorescence staining of spermatozoa from control and individual TTC29_1, carrying the c.176+1G>A mutation, with SPAG6 antibody (in red) and Tubulin (in green). Cells were counterstained with DAPI (blue) as nuclei marker. Scale bars represent 5 μm.
Table 2.
Comparison of Semen Parameters and Sperm Morphological Defects of Individuals Carrying Mutations in TTC29 (n = 5) versus Other MMAF-Affected Subjects of the Cohort (n = 162)
|
General Semen Characteristics |
Flagellum Defects |
Head Defects |
Acrosome Defects |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Volume (mL) | Total Sperm Count (×106) | Total Motility | Progressive Motility | Vitality | Typical Forms | Absent | Short | Irregular | Coiled | Bent | Tapered | Thin | Micro-cephalic | Macro-cephalic | Acrosomal Region∗ | Post-acrosomal Region | |
| TTC29 mutated individuals | 40.5 ± 3.1 (n’ = 5) | 70 ± 61.3 (n’ = 5) | 4.2 ± 4.3 (n’ = 5) | 1.0 ± 1.0 (n’ = 5) | 70.8 ± 12.7 (n’ = 5) | 0.0 ± 0.0 (n’ = 5) | 11.4 ± 12.4 (n’ = 5) | 52.6 ± 36.7 (n’ = 5) | 25.8 ± 27.0 (n’ = 5) | 6.4 ± 6.2 (n’ = 5) | 4.5 ± 9.0 (n’ = 4) | 10.2 ± 10.1 (n’ = 5) | 14.2 ± 23.3 (n’ = 5) | 5.2 ± 5.2 (n’ = 5) | 0.4 ± 0.8 (n’ = 5) | 63.2 ± 45.5 (n’ = 4) | 20.0 ± 15.7 (n’ = 4) |
| Other MMAF individuals | 40.3 ± 7.4 (n’ = 116) | 62.2 ± 69.4 (n’ = 148) | 4.6 ± 6.9 (n’ = 153) | 2.0 ± 4.5 (n’ = 153) | 56.2 ± 23.0 (n’ = 150) | 1.2 ± 3.2 (n’ = 150) | 14.9 ± 15.2 (n’ = 141) | 50.8 ± 29.0 (n’ = 147) | 26.4 ± 26.9 (n’ = 142) | 11.1 ± 9.9 (n’ = 144) | 3.7 ± 7.6 (n’ = 96) | 16.0 ± 17.6 (n’ = 139) | 6.3 ± 10.0 (n’ = 136) | 4.7 ± 6.6 (n’ = 138) | 0.6 ± 1.3 (n’ = 137) | 50.2 ± 37.8 (n’ = 140) | 20.9 ± 22.66 (n’ = 136) |
| p Value | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 |
Values are expressed in percent, unless specified otherwise. Values are mean ± SD; n = total number of individuals in each group; n′ = number of individuals used to calculate the average based on available data. We compared statistical differences between MMAF due to TTC29 mutations versus MMAF (multiple morphological abnormalities of the flagella) due to other known or unknown causes. Statistical analysis: unpaired t test was used to compare the two groups.
Phenotypical Characterization of CRISPR-Engineered Mice Lacking TTC29 Protein
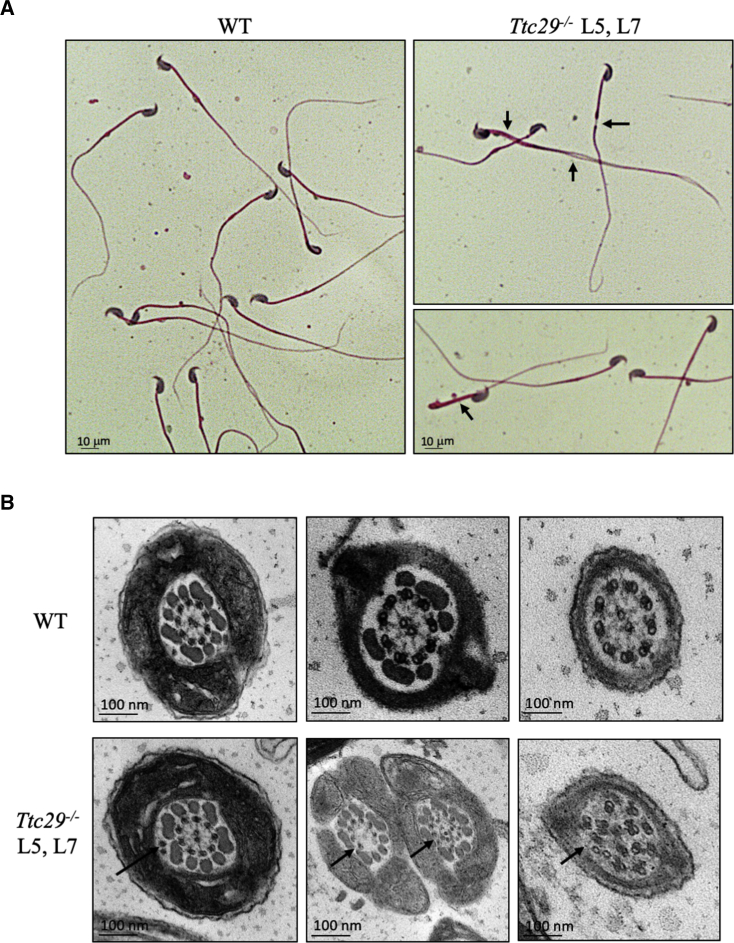
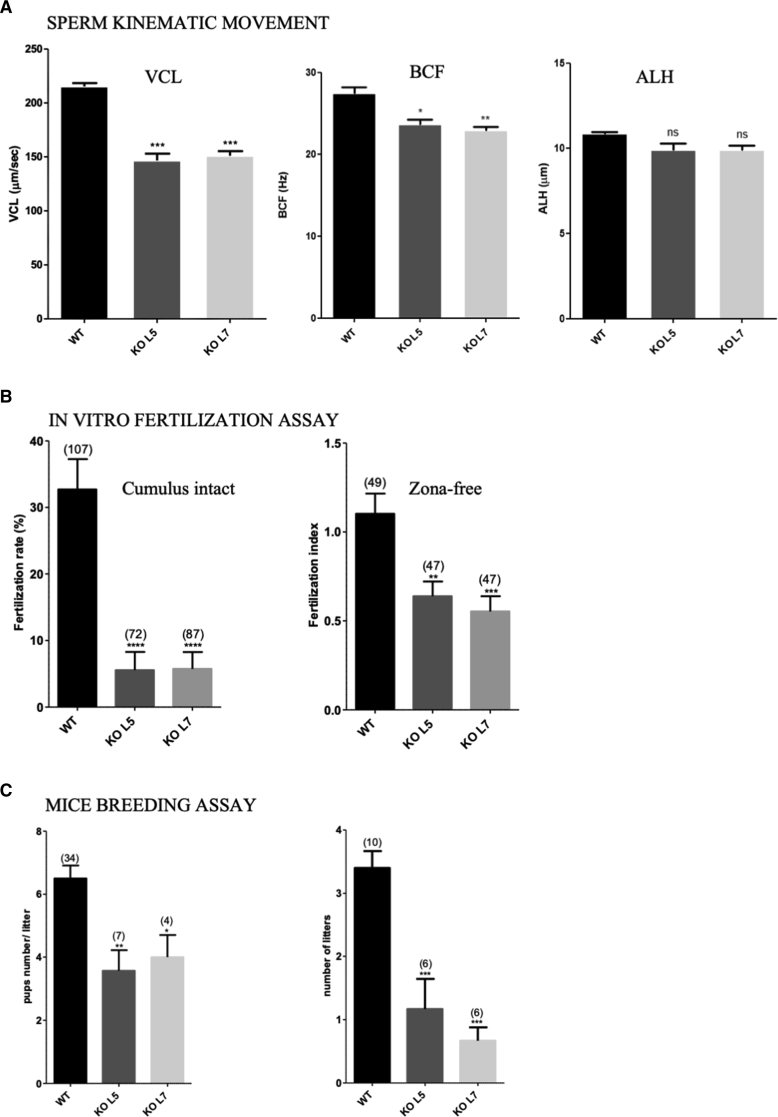
In mouse, Ttc29 is located on chromosome 8 and contains 14 exons predicting a 471-amino-acid protein (UniProtKB: Q80VM3; GenBank: NP_898919.3), which share 75% of identity with human TTC29 protein. Similar to humans, information retrieved from public expression databases and from the literature46 indicate a strong expression of Ttc29 in rodent testis and germ cells, which prompted us to further investigate the mouse ortholog. Using the CRISPR-Cas9 technique, we generated and characterized two different mouse lines, Ttc29 L5 and L7, with mutational events targeting Ttc29 exon 5, comparable to the region mutated in individual TTC29_1. The mutations generated in Ttc29 L5 and L7 mouse lines corresponded to a 1 nucleotide insertion and 17 nucleotide deletion, respectively, and were both expected to induce a translational frameshift and the production of a truncated protein, if any (Figures S4A and S4C). Heterozygous mutant animals of L5 and L7 mutant lines were mated to generate homozygous offspring, which were obtained at the expected Mendelian frequencies of 26.3% (n = 81) and 22.7% (n = 68) for L5 and L7, respectively, thus excluding embryonic or perinatal lethality due to Ttc29 mutations. We first validate the mutant mouse models by performing western blot experiments on testes protein extracts from Ttc29 L5 and L7 homozygous mutant mice, which indicated the absence of TTC29 protein and of any truncated protein (Figure S4B). To confirm this result, we prepared purified sperm flagella preparations from wild-type versus mutant mice, following previously described protocols,47 and performed tandem mass spectrometry (MS/MS) analysis. TTC29 peptides were readily identified in wild-type sperm preparations (Table S6), confirming that in the mouse TTC29 protein also locate to the flagellum. However, no peptides from TTC29 protein could be recovered from the mutant samples (n = 4), thereby formally excluding the production of a truncated protein, similar to what was observed in individual TTC29_1 carrying the c.176+1G>A mutation (Table S1). We next analyzed the phenotype and the male reproductive functions of Ttc29 L5 and L7 homozygous mutant males. Both mutant males were viable and indistinguishable from their wild-type littermates in survival rate and general appearance. Normal body, testes, and epididymides weights were also reported (Table 3). Histological analysis indicated normal cytoarchitecture of the testes and epididymides. In addition, all stages of germ cells differentiation were observed within the seminiferous tubules of the testes, indicating that spermatogenesis occurred normally in those mutant animals (Figures S5 and S6). Consistent with this, no increase in germ cells apoptosis was detected in seminiferous tubules of Ttc29 mutant testes (Figure S7) and epididymal sperm counts were normal (Table 3). Importantly, Ttc29 mutant sperm did not display a typical MMAF phenotype and appeared overall normal in length (Figure 3A). A significant increase in more subtle morphological defects of the flagellum was, however, observed in comparison with wild-type mice (Table 3). In particular, Ttc29 L5 and Ttc29 L7 sperm flagella sometime displayed a disjunction at midpiece-principal piece junction, a folding concerning this same region and an increase of flagella with irregular caliber (L5: 7.52% and L7: 6.00% compared to wild type: 3.48%; p values < 0.01 and 0.05, respectively) (Figure 3A). Ultra-structural analyses performed by TEM indicated an increased number of transversal sections with an abnormal axonemal organization such as supernumerary or missing outer microtubule doublets and global microtubule disorganization (Figure 3B). Similar to sperm from individual TTC29_1, we performed immunofluorescence detection of DNAI1, DNALI1, RSPH1, and SPAG6 axonemal proteins in sperm from Ttc29 mutant mice but we did not observe obvious differences compared to wild-type (data not shown). Furthermore, we analyzed the label-free quantification intensity of DNAI1, DNALI1, RSPH1, and SPAG6 peptides, which we identified by MS/MS analyses on flagellum fractions from wild-type and mutant Ttc29 mice and did not observe statistically significant changes between the two genotypes (Table S7). Overall, this confirmed that in contrast to sperm from individual TTC29_1, the DNAI1, DNALI1, RSPH1, and SPAG6 proteins are present in sperm from ttc29 mutant mice, which is somewhat consistent with their minor axonemal defects. Importantly, while the percentages of viable and total motile sperm in Ttc29 mutant mice were normal, the fraction of progressive sperm was reduced (Table 3; Figure S8) and detailed assessment of the sperm kinematic parameters by means of computer-assisted sperm analysis (CASA) confirmed a severe alteration of sperm velocity and flagellar beating. The curvilinear velocity (VCL), the straight-line velocity (VSL), and the average-path velocity (VAP), which are three kinematics assessing sperm velocity, were strongly diminished in Ttc29 mutant sperm (Figures 4A and S8). The beat/cross frequency (BCF), which is a measure of flagellar beating, was also found slightly decreased in Ttc29 mutant sperm while the head movements assessed by the amplitude of lateral head displacement (ALH) were unaffected (Figure 4A). Notably, when sperm from Ttc29 L5 and L7 mutant mice were capacitated in vitro, they displayed the normal associated-tyrosine protein phosphorylation (Figure S9) but were not efficient in fertilizing intact or ZP-free oocytes collected from wild-type females (Figure 4B). Hence the fertilization rates (FR; percentage of fertilized oocytes) of Ttc29−/− L5 and L7 sperm were of 5.56 ± 0.03 and 5.74 ± 0.03, respectively, compared to 32.71 ± 0.05 for control sperm (mean ± SEM) (p values < 0.01 and 0.001). Similarly, the fertilization index (FI; mean number of male nuclei per oocyte) of Ttc29 L5 and L7 mutant sperm were severely diminished and of 0.64 ± 0.08 and 0.55 ± 0.09, respectively, compared to 1.10 ± 0.11 for control sperm (mean ± SEM) (p values < 0.01 and 0.001). In line with these results, when Ttc29 L5 and L7 mutant males were mated with wild-type females, they produced litters reduced in size and number, in comparison with their control littermates (Figure 4C). The number of pups per litter during a breeding period of 124 days was 3.57 ± 0.65 and 4.00 ± 0.71 for Ttc29−/− L5 and L7 males as compared to 6.50 ± 0.41 for control males (Figure 4C). In addition, over the same breeding period, the number of litters per female breeder crossed with Ttc29−/− L5 and L7 males was 1.17 ± 0.48 and 0.67 ± 0.21, in comparison with 3.40 ± 0.27 for control males (Figure 4C). Overall, we demonstrated that in the mouse, Ttc29 homozygous loss of function is associated with minor morphological and ultra-structural defects of the sperm flagella while it results in a profound alteration of sperm flagellar velocity and beating frequency, strongly impairing fertilization, both in vitro and in vivo.
Table 3.
General and Sperm Parameters of TTC29−/− L5 and L7 Mouse Lines Compared to Control Mice
| Mouse Lines | Body Weight (g) | Testis Weight (mg) | Apoptosis | Sperm Count (M) | Sperm Vitality (%) | Morphological Defects (%) | Axonemal Defects (%) |
|---|---|---|---|---|---|---|---|
| Control | 27.38 ± 0.95 | 111.3 ± 10.8 | 0.34 ± 0.15 | 18.41 ± 1.73 | 48.70 ± 3.89 | head: 7.88 ± 1.79 tail: 3.48 ± 0.59 |
– 2.2 ± 1.3 |
| KO L5 | 27.28 ± 0.96 | 93.1 ± 2.6 | 0.09 ± 0.02 | 19.33 ± 2.56 | 49.00 ± 2.63 | head: 6.96 ± 1.32 tail: 7.52 ± 0.87∗∗ |
– 21.6 ± 11.0∗∗∗ |
| KO L7 | 27.38 ± 1.12 | 113.3 ± 14.7 | 0.48 ± 0.17 | 16.89 ± 3.72 | 45.45 ± 2.75 | head: 7.13 ± 1.74 tail: 6.00 ± 0.96∗ |
– 13.0 ± 4.9∗∗ |
The mean age of the animals is 4 months. Wild-type (WT mice): n = 15, Ttc29−/− L5 and L7 mice: n = 10. Testis weight of each animal is pondered by mean body weight of its group; WT: n = 7, Ttc29−/− L5: n = 4, and Ttc29−/− L7: n = 5. Apoptosis rate is indicated as mean number of apoptotic cells/seminiferous tubule; WT: n = 5, Ttc29−/− L5: n = 5, and Ttc29−/− L7: n = 6. Sperm counts are indicated in millions (M) as mean number of sperm cells obtained from dissection of two cauda epididymes; WT: n = 15, Ttc29−/− L5 and L7: n = 10. The percentage of morphological defects of the sperm head and tail is obtained by analysis of at least 100 cells for each animal on Papanicolaou-stained smears; WT: n = 6, Ttc29−/− L5: n = 5, and Ttc29−/− L7: n = 4.The percentage of axonemal defects is obtained by TEM analysis of 20 to 50 axonemal cross-sections; WT: n = 4, Ttc29−/− L5: n = 4, and Ttc29−/− L7: n = 3. The percentage of sperm vitality was assessed by eosine-nigrosin staining and counting of at least 200 cells per animal; WT: n = 8, Ttc29−/− L5: n = 6, and Ttc29−/− L7: n = 7. Statistical analysis: values are indicated as mean ± SEM. One-way ANOVA was used to compare the three groups, except for the analysis of the morphological defects, which was performed with a chi-square test (2 df). p > 0.05 was considered statistically not significant. Significant p values are indicated as (∗): p value < 0.05, (∗∗): p value < 0.01, and (∗∗∗): p value < 0.001.
Figure 3.
Characterization of the Sperm Morphological and Ultra-structural Phenotype of Ttc29−/− L5 and L7 Mutant Mice
(A) Papanicolaou staining of epididymal sperm from Ttc29−/− L5 and L7 mutant mice, showing the presence of morphological abnormalities (black arrows) such as flagella bending, flagella of irregular caliber, and midpiece-principal piece disjunction as compared with control sperm (WT). Scale bars represent 10 μm.
(B) TEM analysis of epididymal sperm from Ttc29−/− L5 and L7 mutant mice, showing abnormal cross sections of the axoneme with ectopic peripheral doublets, lack of the central pair, and global disorganization of the structure as compared to control sperm with (9+2) canonical structure (WT). Scale bars represent 100 nm.
Figure 4.
Characterization of Sperm Motility and Fertilization Potential of Ttc29−/− L5 and L7 Mutant Mice
(A) Assessment of sperm kinematic movement of Ttc29−/− L5 and L7 mutant mice by CASA, indicating reduced curvilinear velocity (VCL) and beat/cross frequency (BCF) as compared with controls (WT) while the amplitude of lateral head displacement (ALH) was found unaffected. Data are represented as the mean ± SEM p value < 0.05 (∗); p value < 0.01 (∗∗); p value < 0.001 (∗∗∗); non-significant (ns).
(B) In vitro fertilization assays with capacitated sperm from Ttc29−/− L5 and L7 epididymes, showing reduced fertilization rates and fertilization index when inseminated with cumulus-intact oocytes and Zona-free oocytes, respectively, as compared with control sperm (WT). The total number of analyzed oocytes is indicated above the histogram of each mouse genotype. Data are represented as the mean ± SEM p value < 0.01 (∗∗); p value < 0.001 (∗∗∗); p value < 0.0001 (∗∗∗∗). Each experiment was repeated three times.
(C) In vivo breeding assays of Ttc29−/− L5 and L7 mutant mice with wild-type females, showing a reduced number of pups per litter (left) and of litters per female (right) over a breeding period of 124 days, as compared to control males (WT). The total numbers of litters analyzed and the number of female breeders used for each genotype is indicated at the top of the histograms (left and right, respectively). Data are represented as the mean ± SEM p value < 0.05 (∗); p value < 0.01 (∗∗); p value < 0.001 (∗∗∗).
Localization of TTC29 Ortholog Protein in T. brucei Flagella
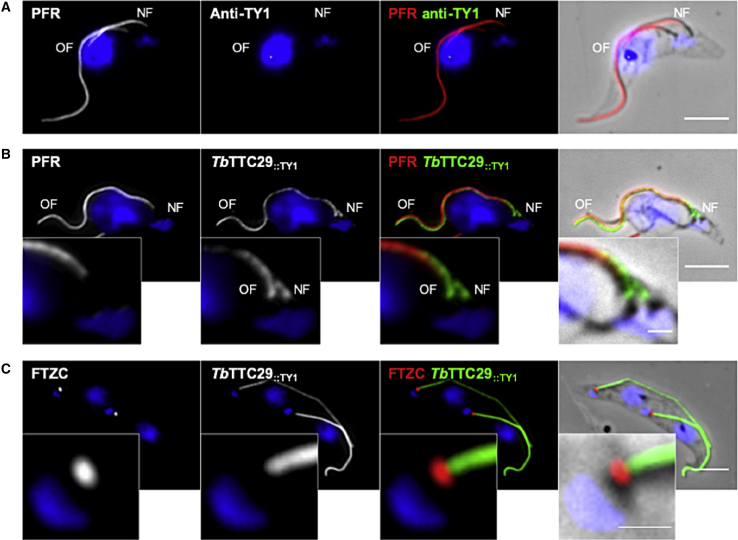
In order to further assess the importance of the TTC29 protein in flagellar beating, we chose to investigate the localization and function of its ortholog in Trypanosoma brucei (T. brucei), a flagellated protozoan, which has largely contributed to elucidating the molecular pathogeny of human ciliopathies.48 The T. brucei axoneme is similar to that of mammalian sperm flagella but its para-axonemal structure, although comparable in function, is different. In T. brucei, the sperm FS and ODFs present in mammalian flagella are replaced by the paraflagellar rod (PFR), a complex structure connecting with axonemal doublets (4–7).49 The PFR plays a role in flagellum motility50,51 and serves as a platform for metabolic and signaling enzymes.52, 53, 54 By performing BlastP analysis on the T. brucei genome database55 using the human TTC29 protein sequence, we identified the T. brucei ortholog Tb927.3.1990 (XP_843793.1), namely TbTTC29 in this study, which shares 22% of identity (40% of similarity) with the human ortholog. TbTTC29 is a 481 amino acids protein and annotated as a Tetratricopeptide repeat putative protein. We generated a trypanosome cell line expressing a tagged version of the endogenous TbTTC29 protein, TbTTC29::TY1, and investigated the protein localization by immunofluorescence on detergent-extracted cytoskeleton preparations (CSK) (Figures 5 and S10). TbTTC29::TY1 was found to localize to the flagellum with staining adjoined to that of the paraflagellar rod protein PFR2. In addition, TbTTC29::TY1 was labeled at both the old flagellum (OF) and the new flagellum (NF), even when the latter has not assembled a PFR structure yet. Our data therefore indicate that TbTTC29::TY1 is an axonemal-associated protein, which localizes along the axoneme from the transition zone, specifically labeled with anti-FTZC56 to the distal tip (Figure 5).
Figure 5.
Localization of TTC29 Orthologous Protein, TbTTC29, in T. brucei Flagellar Axoneme
(A) Representative picture of detergent-extracted parental cells immunolabelling, with anti-PFR (red) that labels the para-axonemal paraflagellar rod structure and anti-TY1 (green) antibodies, as a negative experimental control.
(B) Representative picture of detergent-extracted TbTTC29::TY1 cell line immunolabelling with anti-PFR (red) and anti-TY1 (green) antibodies, showing axonemal localization of the endogenous recombinant TbTTC29::TY1 at both the old flagellum (OF) and the new flagellum (NF).
(C) Representative picture detergent-extracted TbTTC29::TY1 cell line immunolabelling with anti-FTZC labeling the transition zone (red) and anti-TY1 (green) antibodies, showing localization of TbTTC29::TY1 from the transition zone to the distal tip of the axoneme. The mitochondrial genome and the nuclei are stained with DAPI. Scale bars represent 5 μm in main figures and 1 μm in zoom insets. OF and NF indicated the old flagellum and the new flagellum, respectively.
Phenotypical Characterization of TbTTC29 RNAi-Knockdown
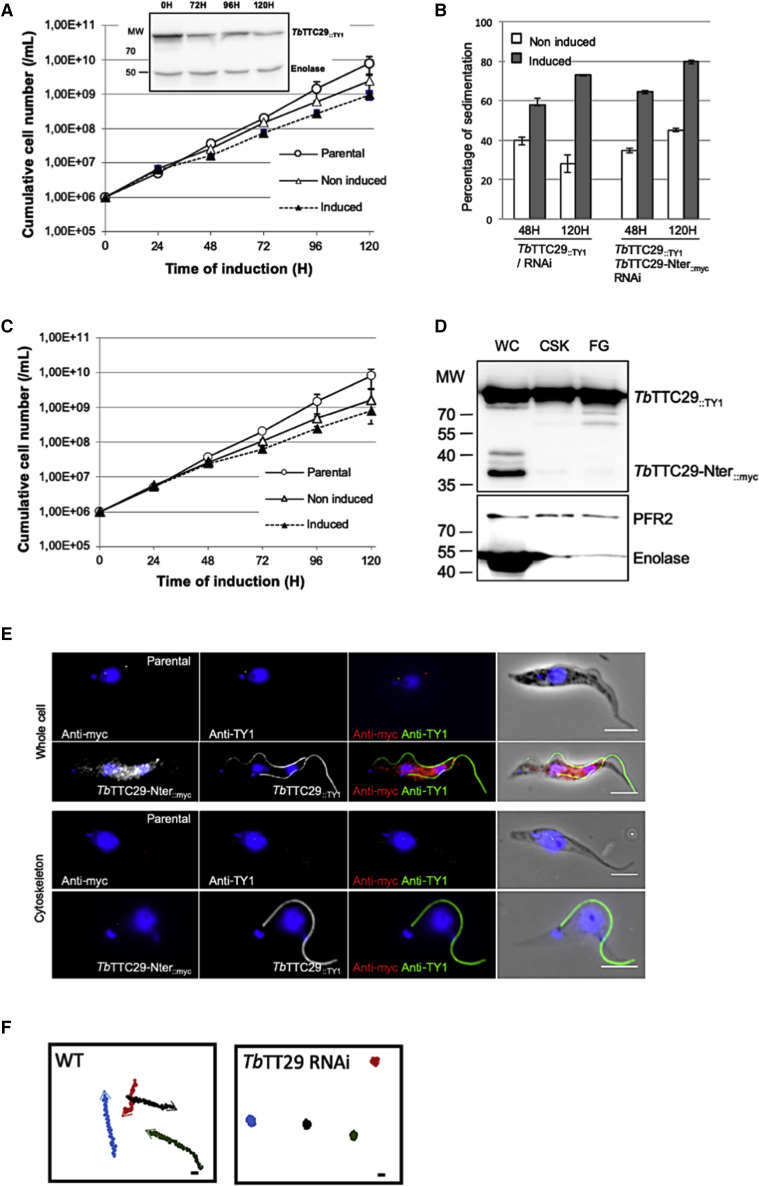
To assess the functional role of TbTTC29 in the parasite, we used the tetracycline-inducible RNA interference (RNAi) system. We generated an RNAi inducible TbTTC29::TY1/RNAiTbTTC29 cell line by targeting nucleotides 352 to 806 of the transcript sequence. Induction of the RNAi led to a substantial reduction of TbTTC29::TY1 expression, as demonstrated by western blotting analysis (Figure 6A insert). Functional analysis of RNAi induced cells showed that TbTTC29 knockdown did not dramatically affect the growth rate nor the morphology of the cells, in comparison with non-induced or parental cells (Figure 6A). In addition, when performing TEM analysis, the flagella appeared normal in number and length and their overall structure was preserved (data not shown). However, we observed that RNAi induced cells were more prone to sediment in comparison to non-induced cells (Figure 6B). Hence, quantitative sedimentation assays indicated that after 120 h of tetracycline treatment, 70% of the RNAi induced cells had sedimented compared to 30% for the non-induced cells. Such an increase in sedimentation is directly correlated to defects in flagellar motility and cell mobility, which we confirmed by video microscopy (Videos S1 and S2) and mobility tracking (Figure 6F). Overall, this functional study demonstrated that in T. brucei, similar to what we observed in the mice and in humans, the TTC29 protein is involved in flagellar beating and motility.
Figure 6.
Characterization of the Cell Motility Phenotype Induced by TbTTC29 RNAi-Induced Knockdown in T. brucei
(A) Comparative growth curve of parental, non-induced, and RNAi-induced TbTTC29::TY1/RNAiTbTTC29 cells. Inset: Western blot analysis of RNAi knockdown by immunolabelling of TbTTC29::TY1 at 0, 72, 96, and 120 h of induction with tetracycline. Anti-enolase was used for loading normalization.
(B) Sedimentation assays at 48 and 120 h post-RNAiTbTTC29 induction. Percentages of sedimentation were normalized to basal levels of sedimentation in the parental cells.
(C) Comparative growth curves for parental, non-induced, and RNAi-induced TbTTC29::TY1/TbTTC29-Nter::myc/RNAiTbTTC29 cells.
(D) Western blot analysis of TbTTC29::TY1 and TbTTC29-Nter::myc in protein extracts from whole cells (WC), cytoskeleton (CSK), and flagella fractions (FG), showing the presence of TbTTC29::TY1 in all preparations while TbTTC29-Nter::myc is detected only in the whole cell fraction. Anti-PFR2 was used as a control for CSK and FG fractions, anti-enolase was used as a control for the cytosolic fraction.
(E) Representative picture of of TbTTC29-Nter::myc (anti-myc) and of TbTTC29::TY1 (anti-TY1) immunolabelling on whole cells or detergent-extracted cytoskeleton preparations, showing the absence of axonemal localization for the truncated N terminus protein. The kinetoplasts and the nuclei are stained with DAPI. Scale bars represent 5 μm.
(F) Mobility graphs obtained from movies of non-induced (Video S1) and RNAiTbTTC29 (Video S2). The positions of individual cells are plotted at 0.2 s intervals. Open circle: starting position of each cell. Arrowhead: ending position.
Bars represent 10 μm. In (A), (B), and (C), data are represented as the mean ± SEM.
Video microscopy indicating flagellum and cell motility defects of the RNAi-induced mutant cells, which remained primarily at one location while the WT cells traveled long distances.
Determination of the Protein Region Required for Localization and Function of TTC29 Protein in T. brucei
Based on the functional similarity observed between HsTTC29, MmusTTC29 and TbTTC29, we decided to further utilize the T. brucei model in order to investigate the role of the TPR domains in TTC29 protein localization and function. We generated a construct encoding the N terminus domain of TbTTC29 protein (amino acids 1–120) and lacking the TPR repeats, TbTTC29-Nter::myc, which was expressed in the TbTTC29::TY1/RNAiTbTTC29 background cell line; thus generating the cell line TbTTC29::TY1/TbTTC29-Nter::myc/RNAiTbTTC29. In this cell line the RNAi was designed to specifically target the full-length TbTTC29 protein with the N terminus construct, therefore, being not affected. The endogenous expression level of TbTTC29-Nter::myc did not further affect the cell growth (Figure 6C); the slight growth delay observed during induction of RNAiTbTTC29 was found to persist. However, in contrast to TbTTC29::TY1 that was detected in the whole cell (WC), the detergent-extracted cell (cytoskeleton, CSK), and the flagellum (FG) fractions by means of western blotting, the TbTTC29-Nter::myc protein was detected only in the WC fraction (Figure 6D), therefore indicating that the N terminus domain is not sufficient to position the protein within the axoneme. This was further confirmed by immunodetection of TbTTC29-Nter::myc, which showed its localization within the cytoplasm of non-extracted cells but not in the flagellum from detergent-extracted cytoskeleton preparations (Figures 6E and S10). Importantly, expression of TbTTC29-Nter::myc could not rescue the sedimentation phenotype induced by TbTTC29 RNAi knockdown, as shown in Figure 6B. Taken together, these experiments demonstrate that the TPR domains located in the second half of the TTC29 protein are required for proper localization of the protein into the axoneme and proper function in flagella beating and motility.
Structural Analysis of HsTTC29, MmusTTC29, and TbTTC29 Proteins
High preference for α-helicity in HsTTC29 protein can be predicted on the basis of its primary sequence, with only four strong helix-forming residues (alanine, glutamic acid, histidine, and leucine) accounting for a third of all residues.57,58 In fact, five tandem helical repeats were initially identified as TPR domains using SMART prediction software (aa 182–215, 234–267, 274–307, 314–347, 354–387) (Figure 1). As SMART analysis is only based on primary sequence and may provide incomplete information on structural motifs, we performed a secondary structure prediction59 followed by manual alignment of suspect peptides.41 By doing that, we identified seven pairs of helices (36–40 amino acids) compactly disposed throughout the central part of the protein sequence, flanked by unpaired N-terminal and C-terminal helices. On the basis of significant conservation both within and outside of the consensus TPR sites (4, 7, 8, 11, 20, 24, 27, and 32),41 we hypothesize that HsTTC29 contains up to seven TPRs, which are stabilized by additional helices that have been referred to as “capping” or “solubilization” helices in the context of TPR proteins60 (Figure S11A). The N-terminal repeat 1 and 2, featuring non-canonical residues at the key 8 and 20 positions (e.g., F104 instead of G/A/S, K117 and R159 instead of A/S/E), were not recognized by SMART as TPRs. However, the peculiar helical pairing and significant conservation outside of the consensus sites make us hypothesize that these regions fold into TPR-like structures. Furthermore, an attempt to identify a suitable template for creating a homology model of HsTTC29, yields an adaptor LGN protein involved in spindle orientation (e.g., PDB: 6HC2) at 18% sequence identity, which features 8 compactly disposed TPRs with similar poor consensus retention at the N-terminal repeat,61 thus confirming the identified TPR domains in HsTTC29. In MsTTC29, which shares 75% of identity (85% of similarity) with HsTTC29, 7 TPR domains were similarly identified. In TbTTC29, the N-terminal TPR was found severely truncated in the middle of the sequence, although some sequence conservation with the corresponding segments in human and murine orthologs persists (Figure S11B). Overall, as illustrated in Figure S9B, HsTTC29, MmusTTC29, and TbTTC29 proteins display a significant level of conservation, particularly within the TPR regions internal to the protein, suggesting that in mammals and T. brucei, the TPR domains are likely to be critical for localization and function of TTC29 orthologs. The compact and extended nature of the TPR repeats in these proteins also indicates their ability to participate in protein-protein interactions via concave amphipathic super-helical structures; such repeats are known to self-assemble,62 which is likely responsible for functional roles displayed by these proteins.
Discussion
We report here the identification of bi-allelic mutations in TTC29 inducing a MMAF phenotype, asthenozoospermia, and sterility in men. TTC29_2 and TTC29_3 individuals were born from related parents (2nd and 1st degree cousins, respectively) but familial information was not available for the other three individuals. The presence of the described TTC29 homozygous truncating variants in five seemingly unrelated individuals is highly suggestive of recessive inheritance. Unfortunately, we could not test any other family members to obtain additional genotype-phenotype correlation. However, all affected men were conceived spontaneously from very likely heterozygous fathers (and mothers) and the five mutated subjects had at least one sibling. This constitutes strong indirect evidence that men carrying a heterozygous TTC29 truncating variant are fertile and that TTC29-associated MMAF is transmitted under a recessive mode of inheritance. Importantly, by analyzing TTC29 orthologs in the mouse and in the flagellated parasite T. brucei, we provide further demonstration of the importance of TTC29 proteins in flagellar beating and motility. We show that TTC29 mutations identified in the five infertile individuals result in the total absence of the protein or at best to truncated proteins lacking the critical TPR region. We demonstrated that this TPR region is important for TTC29 localization and function in T. brucei, by using a truncated TbTTC29 protein, which failed to locate to the axoneme and to rescue the cell mobility defects induced by silencing the endogenous TbTTC29 expression.
To date, the precise function of TTC29 in humans and the details of its molecular interactions are unknown, but it is very likely that, similar to T. brucei, the TPRs are important for TTC29 function in human sperm. TPR motifs are usually found in arrays of 2 to 20 repeats, which provide coiled and super-helical structures for protein interactions. The TPR-containing (TTC) proteins are widely present from bacteria to humans and are involved in various important biological processes, such as intracellular transport, vesicle fusion, protein folding, cell cycle, and transcriptional regulation.40 By performing in silico structural analyses and alignment of human, mouse, and trypanosome TTC29 protein, we provide a better characterization of TPR structural domains in human TTC29 protein and confirmed the high conservation of such domains in mouse and trypanosome. Most importantly, we identify TTC29 homology with LGN, which was described to associate with cytoplasmic dynein/dynactin that positions the spindle microtubules during cell division.63 Such similarity is in favor of a possible function of TTC29 in the transport and/or positioning of microtubules and axonemal proteins.
Interestingly, TTC proteins were described to be enriched in the intraflagellar transport (IFT) and the BBSome complexes, both being critical for cilia and flagella assembly.64, 65, 66, 67 Accordingly, in humans, some TTC loss-of-function mutations were described to induce defects in ciliary structure and function.68,69 In addition, recent work from Liu et al. reported the identification of MMAF-affected infertile individuals with truncating mutations in TTC21A (MIM: 611430), also known as IFT139A, encoding for an IFT-A subcomplex protein.27 So far, no experimental evidences indicating that TTC29 is part of IFT complexes have been reported in humans, but in silico prediction software (String) identified TTC29 in the functional association network including TTC26 and TTC30A/B, two known components of the anterograde IFT-B complex. In line with these elements, work from Cheung et al. identified Ttc29 in Xenopus as a direct target of RFX2, a transcription factor coordinating ciliogenesis and demonstrated that Ttc29 knockdown in multi-ciliated epithelial cells of Xenopus, strongly impacted the size and number of cilia, indicating that TTC29 was part of the IFT-B complex.70 Taken together, these results suggest that in humans, the function of TTC29 is likely to be related to IFT, in line with the MMAF phenotype observed in individuals carrying TTC29 truncating mutations. The situation in the mouse and in T. brucei seems to be slightly different as the observed morphological and ultra-structural defects were less severe than in human although flagellar beating and motility were profoundly impaired. Interestingly, in Leishmania, deletion of TTC29 induced both shortening of the flagellum and motility defect.71 In keeping with TTC29 possible function in IFT, these observations would suggest a partial functional compensation of TTC29 deficiency in mouse and in the parasite. In this regard, our observation that the sperm parameters and axonemal defects of individuals carrying mutations in TTC29 were less severe than those observed in individuals carrying mutations in two other MMAF-related genes, CFP43 and CFPA44, is stimulating as it could relate to similar redundancy in human TTC protein function.
Another essential point concerns the motility and flagellar beating defects, which constitute a shared phenotype in the three studied models. All TTC29 mutated individuals presented zero progressive motility, and the flagellar beating was profoundly impacted in the mouse and in T. brucei, precluding cell progression. This could be due to a direct consequence of the IFT defect, impacting the composition and functionality of protein complexes involved in beating activity and/or regulation. It also raises the hypothesis that in addition to structural function mediated through IFT, the TTC29 protein could also fulfill some specific functions required for flagellar beating itself. In line with this later hypothesis, studies performed in Chlamydomonas, previously reported that the TTC29 ortholog, called p44, is a component of IDAs.31 TTC29 may therefore be directly involved in the dynein complexes, which are well established as critical protein complexes orchestrating the beating of cilia and flagella.
Overall, the combination of our data obtained in human, mouse, and T. brucei are consistent with a function of TTC29 in IFT and flagellar beating, in line with previously published data regarding TTC29 orthologs localization and function in Xenopus and Chlamydomonas, respectively. Further studies using those evolutionarily distant and complementary study models should help in precisely defining the molecular mechanisms underlying TTC29 function in flagella assembly and beating.
Declaration of Interests
The authors have no conflict of interest to declare.
Acknowledgments
We thank all the individuals and their families for their cooperation, as well as all the referring physicians. We thank all the technicians from the Service de Biologie de la Reproduction at the Hôpital Cochin (Paris) for routine semen sample evaluation (Jacques Bras, Nathalie Chériau, Véronique Hernandorena, Jean-Claude Cambronne, and Caroline Villalon). We thank the core facilities of the Institut Cochin: Transgenesis-Homologous recombination-Cryosconservation, Cellular Imaging Facility for electron microscopy procedures, histology facility HistIM (Rachel Onifarasoaniaina) together with the 3P5 proteomic facility of the University Paris Descartes (Virginie Salnot, Cédric Broussard, and Evangeline Bennana). We thank Keith Gull for the L8C4 antibody (Oxford University), Klaus Ersfeld (Bayreuth University) for the anti-myc antibody (9E10), Nicolas Biteau (Bordeaux University) for the anti-PFR2 antibody, Frédéric Bringaud (Bordeaux University) for the anti-enolase and anti-FTZC, Philippe Bastin (Institut Pasteur, Paris) for the BB2 antibody, and Samuel Dean (Oxford University) for the pPOT vectors. This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National de la Recherche Scientifique (CNRS), the Université Paris Descartes, the University of Bordeaux, the French National Research Agency (grant MUCOFERTIL ANR-12-BSV1-0011-01 to A.T. and M.B., grant MASFLAGELLA ANR-14-CE15-0002-03 to P.F.R., A.T., and M.B.), the LabEx ParaFrap (ANR-11-LABX-0024 to D.R.R.), the FEDER (2007-2013 Operational Programme for Competitiveness Factors and employment to F.G.), and the Canceropole Ile de France (funding to F.G.).
Published: November 14, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.10.007.
Web Resources
CRISPOR, http://crispor.tefor.net/
EMBL EBI Expression Atlas, https://www.ebi.ac.uk/gxa/home
gnomAD Browser, http://gnomad.broadinstitute.org
GTEx Portal, https://gtexportal.org/home/
MaxEntScan, http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html
OMIM, https://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
ReproGenomics Viewer, https://rgv.genouest.org/app/#/
String, https://string-db.org/
Uniprot, https://www.uniprot.org/
Accession Numbers
The TTC29 variants reported in this manuscript are accessible in ClinVar with the submission number SUB6408157.
Supplemental Data
References
- 1.Satir P., Christensen S.T. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa T. Axoneme Structure from Motile Cilia. Cold Spring Harb. Perspect. Biol. 2017;9:9. doi: 10.1101/cshperspect.a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holstein A.F.C., Roosen Runge E.C. Grosse Verlag; Berlin: 1981. Atlas of Human Spermatogenesis. [Google Scholar]
- 4.Eddy E.M., Toshimori K., O’Brien D.A. Fibrous sheath of mammalian spermatozoa. Microsc. Res. Tech. 2003;61:103–115. doi: 10.1002/jemt.10320. [DOI] [PubMed] [Google Scholar]
- 5.Ferramosca A., Zara V. Bioenergetics of mammalian sperm capacitation. BioMed Res. Int. 2014;2014:902953. doi: 10.1155/2014/902953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 7.Bisgrove B.W., Yost H.J. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- 8.Mitchison H.M., Valente E.M. Motile and non-motile cilia in human pathology: from function to phenotypes. J. Pathol. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- 9.Curi S.M., Ariagno J.I., Chenlo P.H., Mendeluk G.R., Pugliese M.N., Sardi Segovia L.M., Repetto H.E., Blanco A.M. Asthenozoospermia: analysis of a large population. Arch. Androl. 2003;49:343–349. doi: 10.1080/01485010390219656. [DOI] [PubMed] [Google Scholar]
- 10.Chemes H.E., Brugo S., Zanchetti F., Carrere C., Lavieri J.C. Dysplasia of the fibrous sheath: an ultrastructural defect of human spermatozoa associated with sperm immotility and primary sterility. Fertil. Steril. 1987;48:664–669. doi: 10.1016/s0015-0282(16)59482-5. [DOI] [PubMed] [Google Scholar]
- 11.Escalier D. Arrest of flagellum morphogenesis with fibrous sheath immaturity of human spermatozoa. Andrologia. 2006;38:54–60. doi: 10.1111/j.1439-0272.2006.00711.x. [DOI] [PubMed] [Google Scholar]
- 12.Ray P.F., Toure A., Metzler-Guillemain C., Mitchell M.J., Arnoult C., Coutton C. Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin. Genet. 2017;91:217–232. doi: 10.1111/cge.12905. [DOI] [PubMed] [Google Scholar]
- 13.Auger J., Jouannet P., Eustache F. Another look at human sperm morphology. Hum. Reprod. 2016;31:10–23. doi: 10.1093/humrep/dev251. [DOI] [PubMed] [Google Scholar]
- 14.Ben Khelifa M., Coutton C., Zouari R., Karaouzène T., Rendu J., Bidart M., Yassine S., Pierre V., Delaroche J., Hennebicq S. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sha Y., Yang X., Mei L., Ji Z., Wang X., Ding L., Li P., Yang S. DNAH1 gene mutations and their potential association with dysplasia of the sperm fibrous sheath and infertility in the Han Chinese population. Fertil. Steril. 2017;107:1312–1318.e2. doi: 10.1016/j.fertnstert.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Wang X., Jin H., Han F., Cui Y., Chen J., Yang C., Zhu P., Wang W., Jiao G., Wang W. Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin. Genet. 2017;91:313–321. doi: 10.1111/cge.12857. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Sha Y., Wang X., Ding L., Liu W., Ji Z., Mei L., Huang X., Lin S., Kong S. DNAH2 is a novel candidate gene associated with multiple morphological abnormalities of the sperm flagella. Clin. Genet. 2019;95:590–600. doi: 10.1111/cge.13525. [DOI] [PubMed] [Google Scholar]
- 18.Coutton C., Vargas A.S., Amiri-Yekta A., Kherraf Z.E., Ben Mustapha S.F., Le Tanno P., Wambergue-Legrand C., Karaouzène T., Martinez G., Crouzy S. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat. Commun. 2018;9:686. doi: 10.1038/s41467-017-02792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang S., Wang X., Li W., Yang X., Li Z., Liu W., Li C., Zhu Z., Wang L., Wang J. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H., Li W., He X., Liu C., Fang Y., Zhu F., Jiang H., Liu W., Song B., Wang X. NovelCFAP43 andCFAP44 mutations cause male infertility with multiple morphological abnormalities of the sperm flagella (MMAF) Reprod. Biomed. Online. 2019;38:769–778. doi: 10.1016/j.rbmo.2018.12.037. [DOI] [PubMed] [Google Scholar]
- 21.Dong F.N., Amiri-Yekta A., Martinez G., Saut A., Tek J., Stouvenel L., Lorès P., Karaouzène T., Thierry-Mieg N., Satre V. Absence of CFAP69 Causes Male Infertility due to Multiple Morphological Abnormalities of the Flagella in Human and Mouse. Am. J. Hum. Genet. 2018;102:636–648. doi: 10.1016/j.ajhg.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kherraf Z.E., Amiri-Yekta A., Dacheux D., Karaouzène T., Coutton C., Christou-Kent M., Martinez G., Landrein N., Le Tanno P., Fourati Ben Mustapha S. A Homozygous Ancestral SVA-Insertion-Mediated Deletion in WDR66 Induces Multiple Morphological Abnormalities of the Sperm Flagellum and Male Infertility. Am. J. Hum. Genet. 2018;103:400–412. doi: 10.1016/j.ajhg.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auguste Y., Delague V., Desvignes J.P., Longepied G., Gnisci A., Besnier P., Levy N., Beroud C., Megarbane A., Metzler-Guillemain C., Mitchell M.J. Loss of Calmodulin- and Radial-Spoke-Associated Complex Protein CFAP251 Leads to Immotile Spermatozoa Lacking Mitochondria and Infertility in Men. Am. J. Hum. Genet. 2018;103:413–420. doi: 10.1016/j.ajhg.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez G., Kherraf Z.E., Zouari R., Fourati Ben Mustapha S., Saut A., Pernet-Gallay K., Bertrand A., Bidart M., Hograindleur J.P., Amiri-Yekta A. Whole-exome sequencing identifies mutations in FSIP2 as a recurrent cause of multiple morphological abnormalities of the sperm flagella. Hum. Reprod. 2018;33:1973–1984. doi: 10.1093/humrep/dey264. [DOI] [PubMed] [Google Scholar]
- 25.Coutton C., Martinez G., Kherraf Z.E., Amiri-Yekta A., Boguenet M., Saut A., He X., Zhang F., Cristou-Kent M., Escoffier J. Bi-allelic Mutations in ARMC2 Lead to Severe Astheno-Teratozoospermia Due to Sperm Flagellum Malformations in Humans and Mice. Am. J. Hum. Genet. 2019;104:331–340. doi: 10.1016/j.ajhg.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y., Zhang F., Li F., Jiang X., Yang Y., Li X., Li W., Wang X., Cheng J., Liu M. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat. Commun. 2019;10:433. doi: 10.1038/s41467-018-08182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W., He X., Yang S., Zouari R., Wang J., Wu H., Kherraf Z.E., Liu C., Coutton C., Zhao R. Bi-allelic Mutations in TTC21A Induce Asthenoteratospermia in Humans and Mice. Am. J. Hum. Genet. 2019;104:738–748. doi: 10.1016/j.ajhg.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C., Lv M., He X., Zhu Y., Amiri-Yekta A., Li W., Wu H., Kherraf Z.E., Liu W., Zhang J. Homozygous mutations in SPEF2 induce multiple morphological abnormalities of the sperm flagella and male infertility. J. Med. Genet. 2019 doi: 10.1136/jmedgenet-2019-106011. jmedgenet-2019-106011. [DOI] [PubMed] [Google Scholar]
- 29.Sha Y.W., Xu X., Mei L.B., Li P., Su Z.Y., He X.Q., Li L. A homozygous CEP135 mutation is associated with multiple morphological abnormalities of the sperm flagella (MMAF) Gene. 2017;633:48–53. doi: 10.1016/j.gene.2017.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Lorès P., Coutton C., El Khouri E., Stouvenel L., Givelet M., Thomas L., Rode B., Schmitt A., Louis B., Sakheli Z. Homozygous missense mutation L673P in adenylate kinase 7 (AK7) leads to primary male infertility and multiple morphological anomalies of the flagella but not to primary ciliary dyskinesia. Hum. Mol. Genet. 2018;27:1196–1211. doi: 10.1093/hmg/ddy034. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto R., Yanagisawa H.A., Yagi T., Kamiya R. Novel 44-kilodalton subunit of axonemal Dynein conserved from chlamydomonas to mammals. Eukaryot. Cell. 2008;7:154–161. doi: 10.1128/EC.00341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poon S.K., Peacock L., Gibson W., Gull K., Kelly S. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biol. 2012;2:110037. doi: 10.1098/rsob.110037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wirtz E., Leal S., Ochatt C., Cross G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 34.Baker N., Alsford S., Horn D. Genome-wide RNAi screens in African trypanosomes identify the nifurtimox activator NTR and the eflornithine transporter AAT6. Mol. Biochem. Parasitol. 2011;176:55–57. doi: 10.1016/j.molbiopara.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastin P., Bagherzadeh Z., Matthews K.R., Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- 36.Florimond C., Sahin A., Vidilaseris K., Dong G., Landrein N., Dacheux D., Albisetti A., Byard E.H., Bonhivers M., Robinson D.R. BILBO1 is a scaffold protein of the flagellar pocket collar in the pathogen Trypanosoma brucei. PLoS Pathog. 2015;11:e1004654. doi: 10.1371/journal.ppat.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bastin P., Pullen T.J., Sherwin T., Gull K. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci. 1999;112:3769–3777. doi: 10.1242/jcs.112.21.3769. [DOI] [PubMed] [Google Scholar]
- 38.Oberholzer M., Lopez M.A., Ralston K.S., Hill K.L. Approaches for functional analysis of flagellar proteins in African trypanosomes. Methods Cell Biol. 2009;93:21–57. doi: 10.1016/S0091-679X(08)93002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Riba A., Itzhaki L.S. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr. Opin. Struct. Biol. 2019;54:43–49. doi: 10.1016/j.sbi.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 40.Zeytuni N., Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Blatch G.L., Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 42.Darde T.A., Lecluze E., Lardenois A., Stévant I., Alary N., Tüttelmann F., Collin O., Nef S., Jégou B., Rolland A.D., Chalmel F. The ReproGenomics Viewer: a multi-omics and cross-species resource compatible with single-cell studies for the reproductive science community. Bioinformatics. 2019;35:3133–3139. doi: 10.1093/bioinformatics/btz047. [DOI] [PubMed] [Google Scholar]
- 43.Darde T.A., Sallou O., Becker E., Evrard B., Monjeaud C., Le Bras Y., Jégou B., Collin O., Rolland A.D., Chalmel F. The ReproGenomics Viewer: an integrative cross-species toolbox for the reproductive science community. Nucleic Acids Res. 2015;43(W1):W109-16. doi: 10.1093/nar/gkv345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G., Guo Y., Zhou T., Shi X., Yu J., Yang Y., Wu Y., Wang J., Liu M., Chen X. In-depth proteomic analysis of the human sperm reveals complex protein compositions. J. Proteomics. 2013;79:114–122. doi: 10.1016/j.jprot.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Blackburn K., Bustamante-Marin X., Yin W., Goshe M.B., Ostrowski L.E. Quantitative Proteomic Analysis of Human Airway Cilia Identifies Previously Uncharacterized Proteins of High Abundance. J. Proteome Res. 2017;16:1579–1592. doi: 10.1021/acs.jproteome.6b00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohta M., Ohyama K., Asano A., Yokota S., Khalid A.M., Yamano Y. Regulation of rat tetratricopeptide repeat domain 29 gene expression by follicle-stimulating hormone. Biosci. Biotechnol. Biochem. 2012;76:1540–1543. doi: 10.1271/bbb.120293. [DOI] [PubMed] [Google Scholar]
- 47.Hashemitabar M., Sabbagh S., Orazizadeh M., Ghadiri A., Bahmanzadeh M. A proteomic analysis on human sperm tail: comparison between normozoospermia and asthenozoospermia. J. Assist. Reprod. Genet. 2015;32:853–863. doi: 10.1007/s10815-015-0465-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vincensini L., Blisnick T., Bastin P. 1001 model organisms to study cilia and flagella. Biol. Cell. 2011;103:109–130. doi: 10.1042/BC20100104. [DOI] [PubMed] [Google Scholar]
- 49.de Souza W., Souto-Padrón T. The paraxial structure of the flagellum of trypanosomatidae. J. Parasitol. 1980;66:229–236. [PubMed] [Google Scholar]
- 50.Bastin P., Sherwin T., Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- 51.Santrich C., Moore L., Sherwin T., Bastin P., Brokaw C., Gull K., LeBowitz J.H. A motility function for the paraflagellar rod of Leishmania parasites revealed by PFR-2 gene knockouts. Mol. Biochem. Parasitol. 1997;90:95–109. doi: 10.1016/s0166-6851(97)00149-7. [DOI] [PubMed] [Google Scholar]
- 52.Pullen T.J., Ginger M.L., Gaskell S.J., Gull K. Protein targeting of an unusual, evolutionarily conserved adenylate kinase to a eukaryotic flagellum. Mol. Biol. Cell. 2004;15:3257–3265. doi: 10.1091/mbc.E04-03-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridgley E., Webster P., Patton C., Ruben L. Calmodulin-binding properties of the paraflagellar rod complex from Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;109:195–201. doi: 10.1016/s0166-6851(00)00246-2. [DOI] [PubMed] [Google Scholar]
- 54.Oberholzer M., Marti G., Baresic M., Kunz S., Hemphill A., Seebeck T. The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. FASEB J. 2007;21:720–731. doi: 10.1096/fj.06-6818com. [DOI] [PubMed] [Google Scholar]
- 55.Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M., Depledge D.P., Fischer S., Gajria B., Gao X. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bringaud F., Robinson D.R., Barradeau S., Biteau N., Baltz D., Baltz T. Characterization and disruption of a new Trypanosoma brucei repetitive flagellum protein, using double-stranded RNA inhibition. Mol. Biochem. Parasitol. 2000;111:283–297. doi: 10.1016/s0166-6851(00)00319-4. [DOI] [PubMed] [Google Scholar]
- 57.Chou P.Y., Fasman G.D. Empirical predictions of protein conformation. Annu. Rev. Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 58.Costantini S., Colonna G., Facchiano A.M. Amino acid propensities for secondary structures are influenced by the protein structural class. Biochem. Biophys. Res. Commun. 2006;342:441–451. doi: 10.1016/j.bbrc.2006.01.159. [DOI] [PubMed] [Google Scholar]
- 59.Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43(W1):W389-94. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D’Andrea L.D., Regan L. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 61.Takayanagi H., Yuzawa S., Sumimoto H. Structural basis for the recognition of the scaffold protein Frmpd4/Preso1 by the TPR domain of the adaptor protein LGN. Acta Crystallogr. F Struct. Biol. Commun. 2015;71:175–183. doi: 10.1107/S2053230X14028143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuzawa S., Kamakura S., Iwakiri Y., Hayase J., Sumimoto H. Structural basis for interaction between the conserved cell polarity proteins Inscuteable and Leu-Gly-Asn repeat-enriched protein (LGN) Proc. Natl. Acad. Sci. USA. 2011;108:19210–19215. doi: 10.1073/pnas.1110951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pirovano L., Culurgioni S., Carminati M., Alfieri A., Monzani S., Cecatiello V., Gaddoni C., Rizzelli F., Foadi J., Pasqualato S., Mapelli M. Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions. Nat. Commun. 2019;10:2208. doi: 10.1038/s41467-019-09999-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Y., Cao J., Huang S., Feng D., Zhang W., Zhu X., Yan X. Characterization of tetratricopeptide repeat-containing proteins critical for cilia formation and function. PLoS ONE. 2015;10:e0124378. doi: 10.1371/journal.pone.0124378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., Cole D.G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taschner M., Bhogaraju S., Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83:S12–S22. doi: 10.1016/j.diff.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Dam T.J., Townsend M.J., Turk M., Schlessinger A., Sali A., Field M.C., Huynen M.A. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc. Natl. Acad. Sci. USA. 2013;110:6943–6948. doi: 10.1073/pnas.1221011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tadenev A.L., Kulaga H.M., May-Simera H.L., Kelley M.W., Katsanis N., Reed R.R. Loss of Bardet-Biedl syndrome protein-8 (BBS8) perturbs olfactory function, protein localization, and axon targeting. Proc. Natl. Acad. Sci. USA. 2011;108:10320–10325. doi: 10.1073/pnas.1016531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis E.E., Zhang Q., Liu Q., Diplas B.H., Davey L.M., Hartley J., Stoetzel C., Szymanska K., Ramaswami G., Logan C.V., NISC Comparative Sequencing Program TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chung M.I., Kwon T., Tu F., Brooks E.R., Gupta R., Meyer M., Baker J.C., Marcotte E.M., Wallingford J.B. Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife. 2014;3:e01439. doi: 10.7554/eLife.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beneke T., Demay F., Hookway E., Ashman N., Jeffery H., Smith J., Valli J., Becvar T., Myskova J., Lestinova T. Genetic dissection of a Leishmania flagellar proteome demonstrates requirement for directional motility in sand fly infections. PLoS Pathog. 2019;15:e1007828. doi: 10.1371/journal.ppat.1007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video microscopy indicating flagellum and cell motility defects of the RNAi-induced mutant cells, which remained primarily at one location while the WT cells traveled long distances.