Abstract
As a type of severe asthenoteratospermia, multiple morphological abnormalities of the flagella (MMAF) are characterized by the presence of immotile spermatozoa with severe flagellar malformations. MMAF is a genetically heterogeneous disorder, and the known MMAF-associated genes can only account for approximately 60% of human MMAF cases. Here we conducted whole-exome sequencing and identified bi-allelic truncating mutations of the TTC29 (tetratricopeptide repeat domain 29) gene in three (3.8%) unrelated cases from a cohort of 80 MMAF-affected Han Chinese men. TTC29 is preferentially expressed in the testis, and TTC29 protein contains the tetratricopeptide repeat domains that play an important role in cilia- and flagella-associated functions. All of the men harboring TTC29 mutations presented a typical MMAF phenotype and dramatic disorganization in axonemal and/or other peri-axonemal structures. Immunofluorescence assays of spermatozoa from men harboring TTC29 mutations showed deficiency of TTC29 and remarkably reduced staining of intraflagellar-transport-complex-B-associated proteins (TTC30A and IFT52). We also generated a Ttc29-mutated mouse model through the use of CRISPR-Cas9 technology. Remarkably, Ttc29-mutated male mice also presented reduced sperm motility, abnormal flagellar ultrastructure, and male subfertility. Furthermore, intracytoplasmic sperm injections performed for Ttc29-mutated mice and men harboring TTC29 mutations consistently acquired satisfactory outcomes. Collectively, our experimental observations in humans and mice suggest that bi-allelic mutations in TTC29, as an important genetic pathogeny, can induce MMAF-related asthenoteratospermia. Our study also provided effective guidance for clinical diagnosis and assisted reproduction treatments.
Keywords: male subfertility, mouse model, sequencing, sperm flagella, TTC29, asthenoteratospermia, CRISPR, intraflagellar transport, MMAF, WES
Introduction
Male infertility has been reported as a complex multifactorial pathological condition with highly heterogeneous phenotypic presentations ranging from the total absence of spermatozoa in the testis to obvious alterations of sperm quality.1, 2, 3 Suboptimal semen qualities, including poorly motile sperm and abnormal sperm morphology, have become important factors leading to the inability to have offspring in 30%–50% of sub-fertile couples.3,4
Multiple morphological abnormalities of the flagella (MMAF) is one of the most severe sperm malformations. MMAF is characterized as asthenoteratospermia involving absent, short, bent, coiled, and/or irregular-caliber flagella without any other symptoms of primary ciliary dyskinesia (PCD).5 MMAF is considered to be a disorder of genetic origin, and several genes have been reported to be responsible for sperm flagellar defects; these genes include AK7 (MIM: 615364), AKAP4 (MIM: 300185), ARMC2 (MIM: 618424), CFAP43 (MIM: 617558), CFAP44 (MIM: 617559), CFAP65 (MIM: 614270), CFAP69 (MIM: 617949), DNAH1 (MIM: 603332), FSIP2 (MIM: 618153), QRICH2 (MIM: 618304), SPEF2 (MIM: 610172), TTC21A (MIM: 611430), and WDR66 (also known as CFAP251, MIM: 618146).6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 However, previous findings only account for approximately 60% of human MMAF cases. These findings highlight the genetic heterogeneity of MMAF and the potential involvement of new genetic factors in asthenoteratospermia.
Cilia and flagella are microtubule-based organelles whose assembly requires a motile process, known as intraflagellar transport (IFT). IFT, first discovered in the green alga Chlamydomonas reinhardtii, was shown to rely on a bi-directional transport system directed by IFT protein complexes and motors.20 IFT protein complexes have been distinguished into complexes A and B, and they contain approximately 20 IFT proteins in Chlamydomonas.21 Previous studies have revealed that mutations in IFT protein complexes cause several ciliopathies and male infertility.22, 23, 24 For example, IFT140 (a component of IFT complex A) is a key regulator for male fertility, and Ift140-mutated mice were infertile and affected by significantly reduced sperm counts and multiple morphological abnormalities.22 Furthermore, IFT74, a component of IFT complex B, is also essential for spermiogenesis in mice.24 Therefore, these animal studies support the association of IFT protein complexes with male fertility and suggest that more IFT transport genes could be involved in spermiogenesis or related ciliopathies.
In this study, we conducted genetic analyses in a large cohort of 80 MMAF-affected Chinese men through the use of whole-exome sequencing (WES). Interestingly, bi-allelic truncating mutations of TTC29 (tetratricopeptide repeat domain 29) were identified in two unrelated consanguineous families and in one simplex individual. Furthermore, our generated Ttc29-mutated male mice also showed reduced sperm motility, abnormal flagellar ultrastructure, and damaged fertility. Despite poor upshots of in vitro fertilization (IVF) performed for Ttc29-mutated male mice, the intracytoplasmic sperm injection (ICSI) treatment showed that both Ttc29-mutated mice and men harboring TTC29 mutations acquired satisfactory outcomes. These findings strongly suggest that bi-allelic mutations of TTC29 can induce asthenoteratospermia and male subfertility in humans and mice.
Material and Methods
Study Participants
The cohort of 80 MMAF-affected Chinese men was enrolled from the First Affiliated Hospital of Anhui Medical University and the Affiliated Suzhou Hospital of Nanjing Medical University in China. All individuals presented with a typical MMAF phenotype characterized by severe asthenoteratospermia due to a combination of sperm flagellar defects as follows: absent, short, bent, coiled, and/or irregular-caliber flagella. Moreover, PCD-associated symptoms (such as sinusitis, bronchitis, pneumonia, and otitis media)25 were reviewed very carefully and excluded from the MMAF cohort. Clinical investigation revealed that development of male external genitalia, bilateral testicular sizes, hormone levels, and secondary sexual characteristics were normal in all of the MMAF-affected men in this study. Peripheral whole blood samples were collected for genetic analyses. The chromosomal karyotypes are also normal (46; XY), and no large-scale deletions were found in the Y chromosome. This study was approved by the institutional review boards at Fudan University, the First Affiliated Hospital of Anhui Medical University, and the Affiliated Suzhou Hospital of Nanjing Medical University. Signed informed consent was obtained from all of the subjects participating in the study.
Whole-Exome Sequencing and Bioinformatic Analysis
Genomic DNA was isolated from peripheral blood samples from human subjects through the use of the DNeasy Blood and Tissue Kit (QIAGEN). 1 μg of genomic DNA was utilized to enrich the human exome through the use of the SureSelect XT Human All Exon Kit (Agilent). Next-generation sequencing was conducted with the Illumina HiSeq X-TEN platform at Cloud Health Genomics. Reads were mapped to the human genome reference assembly (GRCh37/hg19) by using Burrows Wheeler Aligner (BWA) software to put the original mapping result into BAM format.26 Picard software was employed to remove PCR duplicates and evaluate the quality of variants. Then ANNOVAR software was used for functional annotation with information from OMIM, Gene Ontology, KEGG Pathway, SIFT, PolyPhen-2, MutationTaster, 1000 Genomes Project, ExAC, and gnomAD.27, 28, 29, 30, 31, 32, 33 Finally, the variants with read depths less than 4 × were filtered out according to the Genome Analysis Toolkit.34
According to previous pedigree analyses, MMAF has been assumed to follow an autosomal recessive inheritance.35,36 Therefore, we mainly focused on bi-allelic rare variants identified through WES. Considering the fact that MMAF leads to male infertility, the genetic variants with allele frequencies ≥0.01 in the human population genome datasets (e.g., the ExAC Browser and 1000 Genomes Project) were filtered out. Nonsense, frameshift, and essential splice-site variants were preferred. Missense variants predicted to be deleterious simultaneously by the bioinformatic tools of SIFT, PolyPhen-2, and/or MutationTaster were also included for further evaluation.
Semen Characteristics Analysis
Semen analysis was conducted in the source laboratories during routine biological examination of the individuals according to the World Health Organization (WHO) guidelines. Semen samples from the men harboring TTC29 mutations (A002 IV-1, A038 IV-1, and S003 II-1) were collected through masturbation after 2–7 days of sexual abstinence and evaluated after liquefaction for 30 min at 37°C. Analyses of semen volume, sperm concentration, and motility were carried out and replicated in the source hospitals during routine examination. The morphology of the sperm cells was assessed with hematoxylin and eosin (H&E) staining and scanning electron microscopy (SEM) assay. For each subject, at least 200 spermatozoa were examined to evaluate the percentages of morphologically abnormal spermatozoa.
Semen samples from mice were collected from the cauda epididymides obtained through dissection of adult male mice and diluted in 1 mL solution of capacitation for 15 min at 37°C. Semen characteristics were further analyzed using a computer-assisted analysis system. At least three 8-week-old male C57BL/6 mice were analyzed for each group.
Electron Microscopy Evaluation
For SEM assay, the sperm specimens were deposited on poly-L-lysine-coated coverslips and immersed in 2.5% glutaraldehyde, rinsed in 0.1 mol/L phosphate buffer for 30 min, and post-fixed in osmic acid. After being rinsed thoroughly in the same buffer for 30 min, the specimens were progressively dehydrated with an ethanol and isoamyl acetate gradient and dried through the use of a CO2 critical-point dryer (Eiko HCP-2, Hitachi). Afterward, the specimens were mounted on aluminum stubs, sputter coated through the use of an ionic sprayer meter (Eiko E-1020, Hitachi), and analyzed via SEM (Stereoscan 260) under an accelerating voltage of 20 kV.
For transmission electron microscopy (TEM), the prepared spermatozoa were washed and immersed in 2.5% phosphate buffered glutaraldehyde, washed with 0.1 mol/L phosphate buffer (PB, pH7.2) three times, and post fixed with 1% osmium tetroxide in 0.1 mol/L PB for 1–1.5 h at 4°C. Dehydration was performed using graded ethanol (50%, 70%, 90%, and 100%) and 100% acetone followed by infiltration with 1:1 acetone and SPI-Chem resin overnight at 37°C. After infiltration and embedding in Epon 812, the specimens were sliced with ultra-microtome and stained with uranyl acetate and lead citrate, and then they were observed and photographed via TEM (TECNAI-10, Philips) with an accelerating voltage of 80 kV. For quantification of axonemal anomalies by TEM, at least 100 flagella with cross sections and several longitudinal sections were counted.
Real-Time Quantitative PCR and Reverse Transcription PCR
Total RNA of sperm was extracted using the RNeasy Mini Kit (QIAGEN) and converted into cDNAs using SuperScript III Reverse Transcriptase (Invitrogen) and oligo (dT) primers (TaKaRa). The obtained cDNAs were individually diluted 5-fold to be used as templates for the subsequent real-time fluorescence quantitative PCR with AceQ qPCR SYBR Green Master Mix (Vazyme). GAPDH/Gapdh was used as an internal control. The expression of mRNA was quantified according to the 2-ΔΔCt method.
Immunoblot Analysis
The proteins of mouse sperm cells and testis tissues were extracted using Minute™ Total Protein Extraction Kit for Animal Cultured Cells and Tissues (Invent), then denatured at 95°C for 10 min. The denatured proteins were separated on 10% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) for the immunoblot analysis. After a 1 h blocking step in 5% milk diluted with TBST (TBS-0.1% Tween-20), the membranes were incubated overnight at 4°C using the anti-TTC29 (PA5-70409, Invitrogen, 1:1000) directed toward the N-terminal region (14 amino acid immunogen in ARQKLPCSSRKIPRSQLIKEKDDIDHYLEVNFKEEVAYRNSYKKN) of the TTC29 protein and HRP-conjugated beta actin (HRP-60008, Proteintech, 1:2000) diluted in blocking buffer. Then the membrane was washed in TBST and incubated with HRP-conjugated secondary antibody (Abmart, M21002) at a 1:2500 dilution in blocking solution for 1 h at room temperature. After washing three times in TBST, blots were revealed using the Chemistar™ High-sig ECL Western Blotting Substrate (Tanon) with the Tanon 5200.
Immunofluorescence Analysis
Sperm cells were washed in phosphate buffer saline (PBS) and fixed in 4% paraformaldehyde for 30 min at room temperature and coated on slides which were pre-coated with 0.1% poly L-lysine (Thermo Scientific). Then the slides of sperm cells and cryosections of mouse testes were washed in 1 mL PBS and blocked in 10% donkey serum before being incubated overnight at 4°C with the following primary antibodies: rabbit polyclonal anti-TTC29 (PA5-70409, Invitrogen, 1:100), anti-TTC30A (sc-393206, Santa Cruz Biotechnology, 1:100), anti-IFT52 (17534-1-AP, Proteintech, 1:100), anti-α-tubulin antibody (EP1332Y, abcam, 1:500), and monoclonal mouse anti-α-tubulin (T9026, Sigma, 1:500). Washes were performed using PBS with 0.1% (v/v) Tween20, followed by 1 h incubation at room temperature with highly cross-absorbed secondary antibodies Alexa Fluor 488 anti-Mouse IgG (34106ES60, Yeasen, 1:1000) and Cy3-conjugated AffiniPure Goat Anti-Rabbit IgG (111-165-003, Jackson, 1:4000). Images were captured with a confocal microscope (Zeiss LSM 880).
Mouse Model Generation Using CRISPR-Cas9
The frameshift mutations in Ttc29 were generated in mice by using CRISPR-Cas9 technology according to previous studies.37, 38, 39 The single-guide RNA (sgRNA) was specifically designed against mouse chr8:78333554–78333613 (GRCm38/mm10) at Ttc29 exon 12 according to the position of TTC29 stop-gain variant p.Tyr369∗, which was shared by two human subjects, A002 IV-1 and S003 II-1, of this study. In brief, the sgRNAs were synthesized, annealed, and ligated to the pX458 plasmid, which had been digested with BbsI. Then the pX458 plasmid harboring corresponding sgRNA was transfected into embryonic stem cells which were separated with flow cytometry and plated on dishes 24 h later. Single colonies were picked out and expanded for genotyping. Then the embryonic stem cells carrying genetic modifications were injected into blastocysts, which were generated by mating superovulated female mice with wild-type (WT) C57BL/6 male mice. After a short in vitro culture, the injected blastocysts were transferred into pseudopregnant female mice. The frameshift mutation in Ttc29 was identified in founder mice and their offspring through the use of PCR and Sanger sequencing. All animal experiments were carried out in accordance with the recommendations of the US National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. This study was approved by the animal ethics committee at the School of Life Science, Fudan University. Adult mice (aged 6 weeks or older) were used in this study.
In Vitro Fertilization and Intracytoplasmic Sperm Injection
Two-month-old B6D2F1 (C57BL/6 × DBA2) female mice were superovulated as previously described.40 In brief, the female mice were superovulated by injecting 5–7.5 IU of pregnant mare serum gonadotropin (PMSG), followed by 5–7.5 IU of human chorionic gonadotropin (hCG) 48 h later. For IVF, sperm samples were collected from epididymis and added into the human tubal fluid (HTF; Millipore, Cat. # MR-070-D) drop. Then cumulus-intact oocytes, collected from superovulated females, were transferred into the sperm-containing HTF drop. After 5–6 h of incubation, the embryos were washed in HTF and transferred into KSOM medium (Millipore, Cat. # MR-106-D) to further culture at 37°C under 5% CO2. Fertilization rates were evaluated by recording the numbers of two-cell embryos and late-stage blastocysts at 20 h and 91 h later, respectively.
For ICSI, oocytes were obtained from superovulated females, and sperm heads were injected into oocytes through the use of a Piezo-driven pipette as previously described.41 Then the injected oocytes were cultured in KSOM medium at 37°C under 5% CO2. Two-cell embryos and blastocysts were counted 20 h and 96 h later, respectively.
Results
Identification of Bi-allelic Truncating Mutations of TTC29 in MMAF-Affected Men
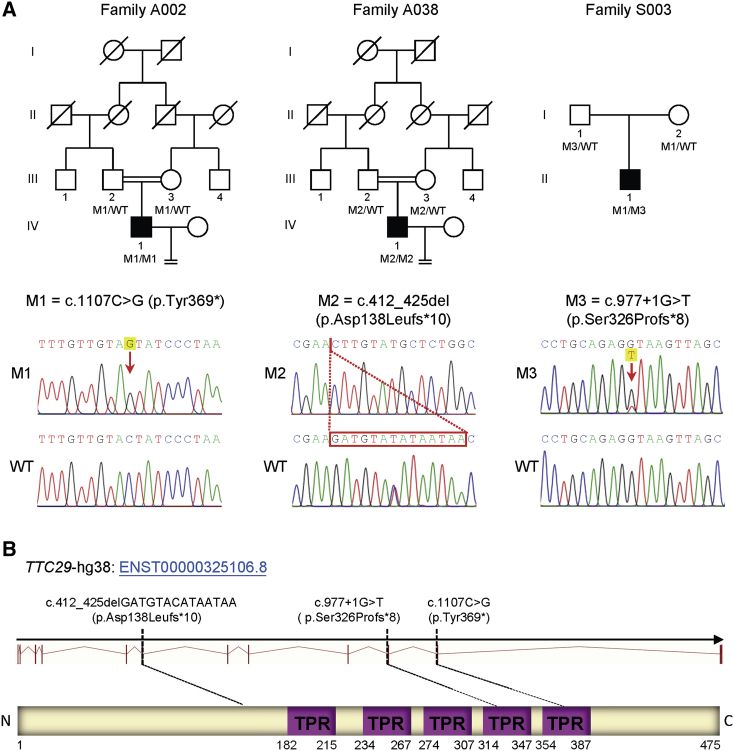
In this study, high-throughput exome sequencing and bioinformatic analyses were performed on 80 MMAF-affected men according to our previously described protocol.14 We identified bi-allelic truncating mutations of TTC29 (GenBank: NM_031956.3) in three (3.8%) unrelated cases of 80 MMAF-affected men (Figure 1A, Table S1). These TTC29 truncating mutations were further verified through Sanger sequencing (Table S2) and illustrated in Figure 1A. TTC29 is located on the human chromosome 4, and it contains 13 exons which encode a predicted 475-amino-acid protein (NCBI: NP_114162.2; UniProt: Q8NA56). TTC29 protein contains tetratricopeptide repeat (TPR) domains, and TTC29 protein is preferentially expressed in the human testis according to the data from the Encyclopedia of DNA Elements (ENCODE), the Functional Annotation of the Mammalian Genome (FANTOM), and the Genotype Tissue Expression (GTEx) project.
Figure 1.
Identification of Bi-allelic Mutations in TTC29
(A) Pedigrees of three families carrying TTC29 truncating mutations. All three affected individuals have bi-allelic mutations with a recessive inheritance mode. The probands from both consanguineous families A002 and A038 have homozygous truncating mutations in TTC29. The proband from family S003 carries compound heterozygous variants of TTC29 derived from his parental, heterozygous carriers. Sanger sequencing results are shown below the pedigrees. The position of each variant is indicated by a red arrow or box.
(B) Schematic representation of the functional domains of TTC29 and locations of TTC29 mutations identified in this study. The purple boxes indicate tetratricopeptide repeat domains as described by the Uniprot server. TTC29 mutations are annotated in accordance to recommendations of the Human Genome Variation Society. Abbreviations: M1, mutation 1; M2, mutation 2; M3, mutation 3; WT, wild type.
In consanguineous family A002, a homozygous stop-gain mutation of TTC29 (c.1107C>G [p.Tyr369∗]) was identified in proband A002 IV-1, and this variant was inherited from his heterozygous parental carriers (Figure 1A). Furthermore, a homozygous frameshift mutation in TTC29 (c.412_425del [p.Asp138Leufs∗10]) was identified in proband IV-1 from consanguineous family A038 (Figure 1A). Both of these TTC29 truncating mutations in two consanguineous families introduce premature stop codons and are consequently expected to induce nonsense-mediated mRNA decay that can affect protein synthesis.
Notably, the TTC29 c.1107C>G (p.Tyr369∗) variant was recurrently identified in another unrelated family, S003 (Figure 1A), in which the mother transmitted a TTC29 c.1107C>G (p.Tyr369∗) allele to the proband. Furthermore, a TTC29 splice-site variant, c.977+1G>T, was identified at the paternal allele in proband S003 II-1 (Figure 1A). According to the Human Splicing Finder (HSF), the TTC29 c.977+1G>T variant abrogates the consensus donor site, leading to altered splicing and subsequent frameshift. To further investigate the predicted alteration of gene splicing, reverse-transcription PCR (RT-PCR) was performed with the cDNA reversely transcribed from sperm RNA (Table S3). The RT-PCR product obtained from proband S003 II-1 displayed two bands; one had the same length (455 bp) with the control, but the other was short (363 bp), suggesting a splicing alteration (Figure S1A). Sanger sequencing of cDNA indicated the skipping of TTC29 exon 9 at the mutated allele (Figure S1B). Therefore, the TTC29 c.977+1G>T variant in the canonical donor site of intron 9 was predicted to lead to a subsequent frameshift (p.Ser326Profs∗8) secondary to skipping of exon 9 (Figure S1C).
Asthenoteratospermia Phenotypes in Men Harboring TTC29 Mutations
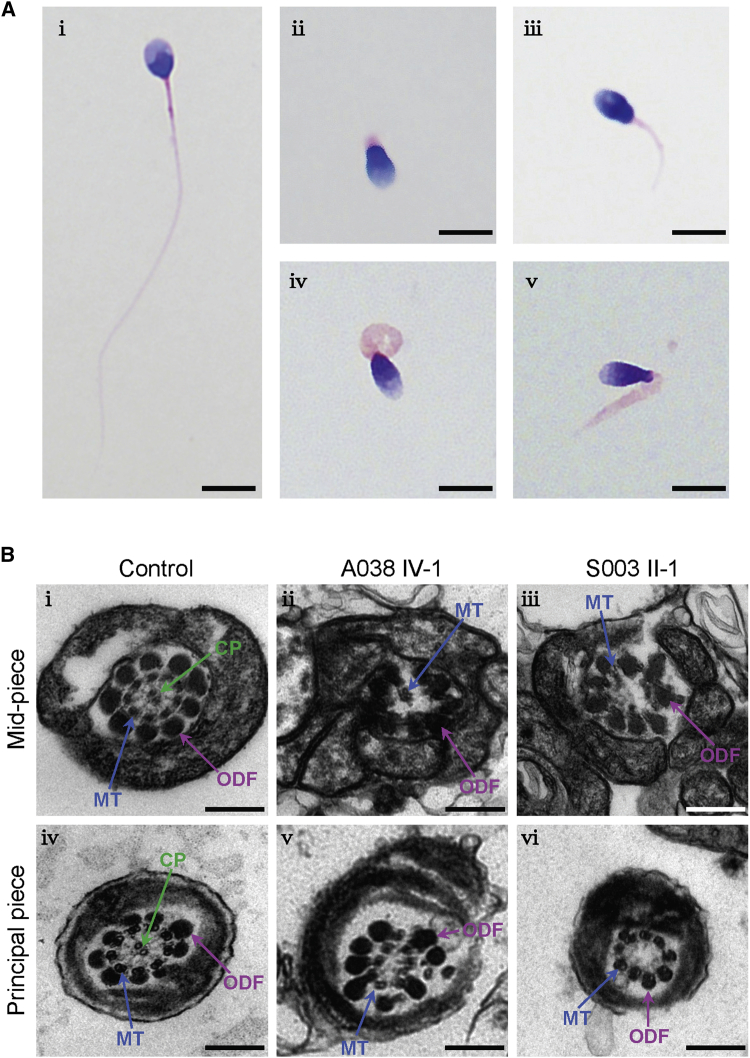
Semen analysis was conducted in the source laboratories during routine examination of the individuals according to WHO guidelines.42 All men harboring TTC29 mutations were shown to have severe-to-complete asthenoteratospermia according to semen characteristics (Table 1). Notably, no spermatozoa with progressive motility were observed in any subjects affected by bi-allelic truncating mutations of TTC29. The morphology of the sperm cells was assessed with H&E staining and SEM. More than 80% of the spermatozoa from men harboring TTC29 mutations displayed abnormal flagella (Table 1). Subject A038 IV-1 with the early truncating mutation of TTC29 (c.412_425del [p.Asp138Leufs∗10]) had a higher malformation rate of sperm flagella than those of the other two subjects with late truncating mutations of TTC29. When compared with the long and thin tails in the normal spermatozoa from a healthy control man, the spermatozoa from men harboring TTC29 mutations displayed obvious MMAF phenotypes, including short, absent, coiled flagella and irregular caliber (Figures 2A and S2).
Table 1.
Semen Characteristics and Sperm Morphology in the Men Carrying Bi-allelic TTC29 Mutations
| - | Human Subject | |||
|---|---|---|---|---|
| - | A002 IV-1 | A038 IV-1 | S003 II-1 | Reference Limits |
| Semen Parameters | ||||
| Semen volume (mL) | 4.5 | 2.5 | 3.0 | 1.5a |
| Sperm concentration (106/mL) | 9.0 | 57.6 | 39.5 | 15.0a |
| Motility (%) | 1.3 | 0.0 | 0.8 | 40.0a |
| Progressive motility (%) | 0.0 | 0.0 | 0.0 | 32.0a |
| Sperm Morphology | ||||
| Normal flagella (%) | 16.0 | 0.0 | 5.0 | 23.0b |
| Absent flagella (%) | 17.5 | 5.5 | 12.0 | 5.0b |
| Short flagella (%) | 39.5 | 63.5 | 36.5 | 1.0b |
| Coiled flagella (%) | 26.0 | 26.5 | 33.0 | 17.0b |
| Angulation (%) | 1.0 | 4.5 | 2.0 | 13.0b |
| Irregular caliber (%) | 0.0 | 0.0 | 11.5 | 2.0b |
| Flagellar Ultrastructural Abnormalityc | ||||
| Abnormal mid-piece (%) | 80.0 | 86.6 | 81.8 | 11.8 (8.6–15.1)d |
| Abnormal end piece (%) | 92.1 | 87.5 | 75.1 | 8.1 (5.2–11.2)d |
Reference limits according to the WHO standards.62
Reference limits according to the distribution range of morphologically normal spermatozoa observed in 926 fertile individuals.63
More than one hundred cross sections were analyzed.
Values represent the mean (range) and are from the statistics of more than three healthy and fertile individuals in this study.
Figure 2.
Sperm Morphology and Ultrastructure Analyses for Men Harboring TTC29 Mutations
(A) Light microscopy analysis of spermatozoa from the control (i) and men harboring TTC29 mutations (ii–v). Most spermatozoa from men harboring TTC29 mutations have flagella that are absent (ii), short (iii), coiled (iv), or of irregular caliber (v). The spermatozoa from subject A038 IV-1 are given as examples of typical MMAF phenotypes observed in men harboring TTC29 mutations. Scale bars: 5 μm
(B) TEM analyses of sperm cells from a fertile control and two men harboring TTC29 mutations. Cross-sections of the mid-piece (i) and principal piece (iv) of the flagella in a control man show the typical ‘‘9 + 2’’ microtubule structure, including nine peripheral microtubule doublets paired with nine outer dense fibers and the central pair of microtubules, surrounded by the organized mitochondrial sheath or fibrous sheath. The ultrastructure observed in cross sections of the flagella in men harboring TTC29 mutations indicates the misarranged outer dense fibers (ii) and disorganization of the peripheral microtubules with lack of the central pair of microtubules (iii, v, and vi). Scale bars: 200 nm. Abbreviations: CP, central pair of microtubules (green arrows); MT, peripheral microtubule doublet (blue arrows); ODF, outer dense fiber (purple arrows).
TEM was conducted according to a previously described protocol14 to further investigate sperm flagellar ultrastructure in the subjects with TTC29 mutations. Higher rates of abnormal flagellar ultrastructures were detected in men harboring TTC29 mutations when compared with those in fertile individuals (Table 1). The cross sections displayed the typical “9 + 2” microtubule structure (nine peripheral microtubule doublets and a central pair of microtubules) in sperm flagella from a control specimen, but presented a dramatic disorganization in axonemal or other peri-axonemal structures in the spermatozoa from men harboring TTC29 mutations (Figure 2B). For example, the unassembled outer dense fibers (ODFs) were found at the mid-piece of sperm flagella, and the absent (9 + 0 conformation) or misplaced central-pair microtubules were also constantly observed in the sections of the principal piece (Figure 2B).
Deficiency of TTC29 and Abnormal Intraflagellar Transport
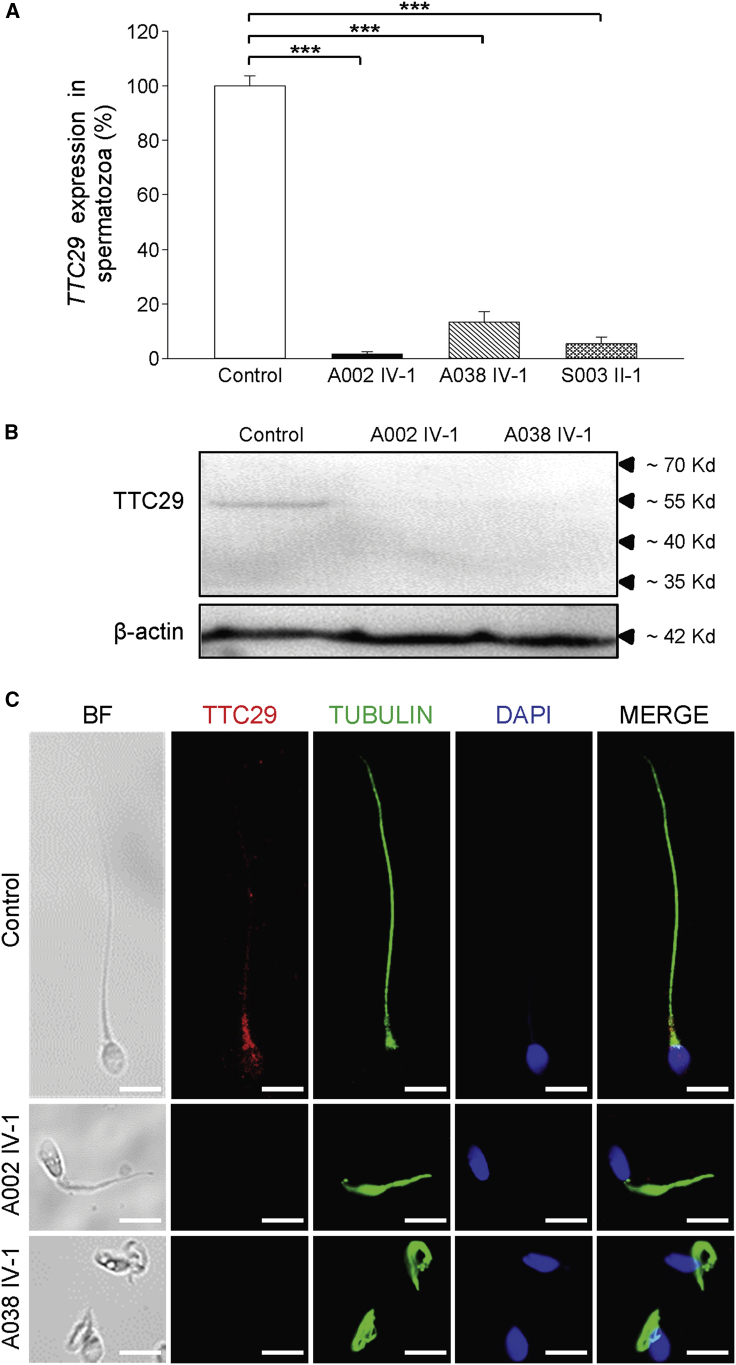
To investigate pathogenicity of the MMAF-associated TTC29 mutations, we newly obtained the sperm samples from a fertile control man and from men harboring TTC29 mutations. The expressions of TTC29 mRNA and protein were investigated using real-time quantitative PCR (RT-qPCR) and immunoblot assays, respectively. It was shown that the expression of TTC29 mRNA in sperm was reduced dramatically in men harboring TTC29 mutations when compared to the control (Figure 3A). Immunoblotting analysis of control sperm lysates through the use of a commercial antibody against TTC29 detected a band of ∼55 Kd (Figure 3B), which is consistent with the predicted molecular weight of the protein encoded by the longest TTC29 transcript (Ensembl: ENST00000513335.5). The corresponding band was absent when we investigated the sperm from men harboring TTC29 mutations (Figure 3B). Our experimental observations on TTC29 expression revealed the possibility of nonsense-mediated mRNA decay triggered by premature translation termination.
Figure 3.
Expression Analysis of TTC29 mRNA and Location of TTC29 Protein in Sperm Flagella
(A) The expression of TTC29 mRNA in the spermatozoa from a normal control and men harboring TTC29 mutations. RT-qPCR assays suggested that the level of TTC29 mRNA was reduced significantly in the sperm from men harboring TTC29 mutations when compared to the level in sperm from a normal control man. Data represent the means ± standard error of the means (SEM) of three independent experiments. Two-tailed Student’s paired or unpaired t tests were used as appropriate. ∗∗∗p < 0.001.
(B) Immunoblotting analysis of TTC29 protein in sperm lysates from a normal control and from men harboring TTC29 mutations (A002 IV-1 and A038 IV-1). β-actin was used as a loading control.
(C) TTC29 immunostaining in human spermatozoa from a normal control and from men harboring TTC29 mutations. Sperm cells were stained with anti-TTC29 (red) and anti-α-tubulin (green) antibodies. DNA was counterstained with DAPI as a nuclei marker. TTC29 staining is concentrated at the base of sperm flagella and faintly along flagella in the fertile control but is almost absent in the sperm flagella of men harboring TTC29 mutations. Scale bars: 5 μm.
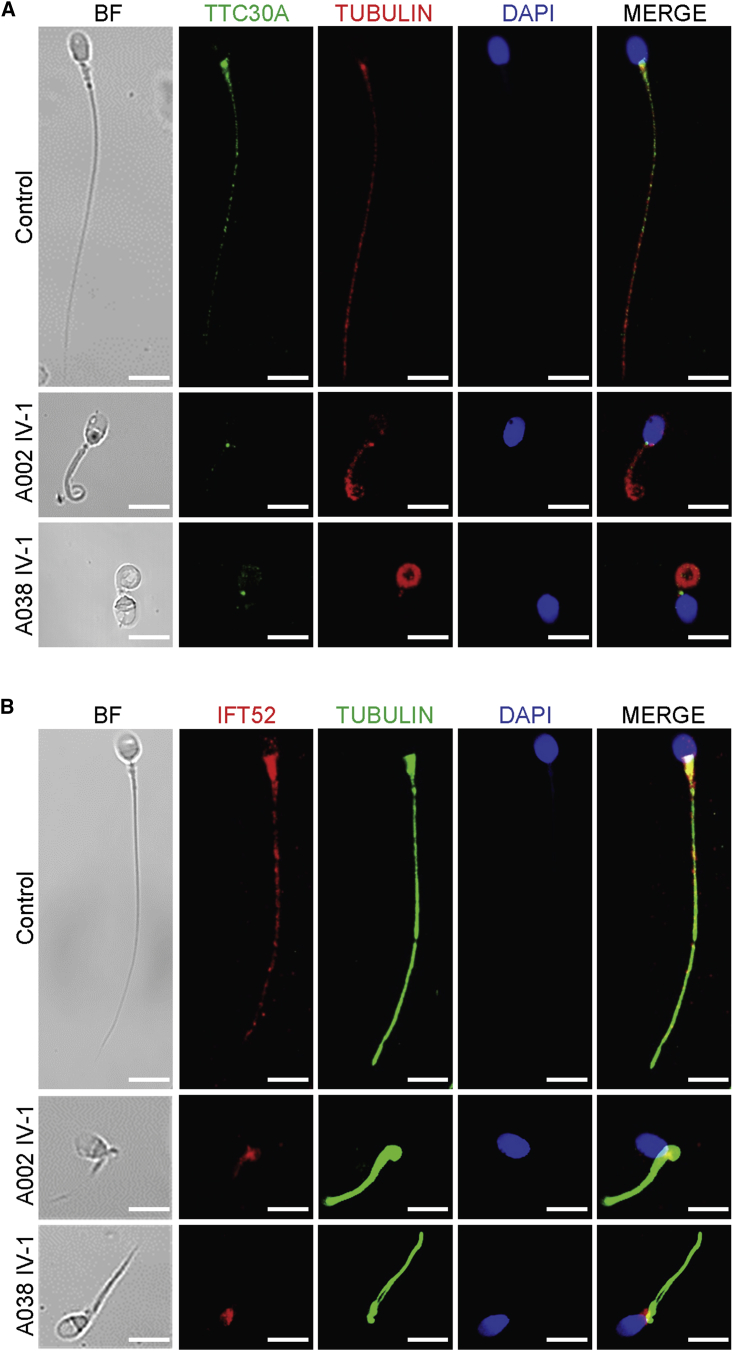
To determine the localization of TTC29 in normal spermatozoa and the effect of TTC29 deficiency on the assembly and/or anchorage of axonemal protein complexes, immunofluorescence (IF) analysis was performed with anti-TTC29 and other antibodies targeting proteins located to various axonemal and peri-axonemal structures of the sperm flagella. We observed that TTC29 immunostaining was localized mainly at the base of sperm flagella and faintly along flagella in normal spermatozoa, but it was almost absent in the spermatozoa from men harboring TTC29 mutations (Figures 3C and S3). Additionally, the analysis on TTC30A (also known as IFT70A, a component of IFT complex B), which is essential for anterograde IFT and ciliogenesis,43,44 revealed a significantly reduced TTC30A staining in the spermatozoa from men harboring TTC29 mutations (Figures 4A and S4). Furthermore, IFT52 (a key component of IFT complex B), which plays a crucial role in microtubule anchorage and centrosome cohesion,45,46 showed an atypical localization on the sperm flagella of men harboring TTC29 mutations in comparison with the control. IFT52 staining was found mainly at the flagellar base and faintly along flagella in the control, but it was weak and only aggregated at the base of sperm flagella in men harboring TTC29 mutations (Figures 4B and S5). These findings indicated the TTC29-associated abnormal assembly of anterograde complex in IFT.
Figure 4.
Affected TTC30A and IFT52 Immunostaining in the Spermatozoa from Men Harboring TTC29 Mutations
(A) Sperm cells from a normal control and from men harboring TTC29 mutations were stained with anti-TTC30A (green, a component of IFT complex B) and anti-α-tubulin (red) antibodies. DNA was counterstained with DAPI as a nuclei marker. In spermatozoa from the fertile control, TTC30A staining appears to be located mainly in the basal body and lightly decorates the sperm flagellum. In the sperm cells from men harboring TTC29 mutations, TTC30A staining is strongly reduced or totally absent. Scale bars: 5 μm.
(B) Sperm cells from a normal control and from men harboring TTC29 mutations were stained with anti- IFT52 (red, a key component of the IFT-B complex) and anti-α-tubulin (green) antibodies. DNA was counterstained with DAPI as a nuclei marker. IF52 staining was found mainly at the base of the flagellum and faintly along cilia in control spermatozoa but revealed weak staining and only aggregated at the base of sperm flagella in men harboring TTC29 mutations. Scale bars: 5 μm.
Ttc29-Mutated Male Mice Resembled Asthenoteratospermia Phenotypes
Ttc29-mutations in mice were constructed using the CRISPR-Cas9 system37, 38, 39 to further investigate the roles of TTC29 in sperm flagellar formation. Sanger sequencing of Ttc29 homozygous mutated mice confirmed the presence of a frameshift mutation (c.1128del), which was predicted to cause premature translational termination (p.Phe377Serfs∗11) (Figure S6, Table S4). RT-qPCR assay (Table S5) was performed to compare the level of Ttc29 transcription in the spermatozoa of WT, heterozygous carrier (Ttc29+/mut) and Ttc29-mutated (Ttc29mut/mut) male mice. As expected, the level of Ttc29 mRNA was significantly lower in Ttc29mut/mut male mice than in WT male mice, indicating a partial nonsense-mediated mRNA decay triggered by premature translational termination (Figure S7A). Immunoblotting assay revealed the near absence of Ttc29 protein in the spermatozoa from Ttc29mut/mut male mice compared to the WT male mice, and the expression of Ttc29 protein in epididymal spermatozoa from WT male mice was lower than that in mouse testis (Figures S7B and S8). Consistently, IF analysis was also performed on the spermatozoa and testicular sections from Ttc29mut/mut and WT male mice with TTC29 antibody described above. The TTC29 immunostaining was almost absent in the spermatozoa and testes from Ttc29mut/mut male mice, further suggesting a low level of TTC29 protein (Figures S9 and S10). Furthermore, the expression and location of Ttc30a1 (an ortholog of human TTC30A in mouse) and Ift52 were also analyzed in the spermatozoa and testicular sections from WT and Ttc29mut/mut male mice. Ttc30a1 and Ift52 immunostaining were both located in the mid-piece of sperm flagella or weakly along flagella from both WT and Ttc29-mutated male mice (Figures S11 and S12). For testicular sections, Ttc30a1 and Ift52 immunostaining were mainly concentrated in the elongating spermatids in testicular sections from WT and Ttc29-mutated male mice (Figures S13 and S14).
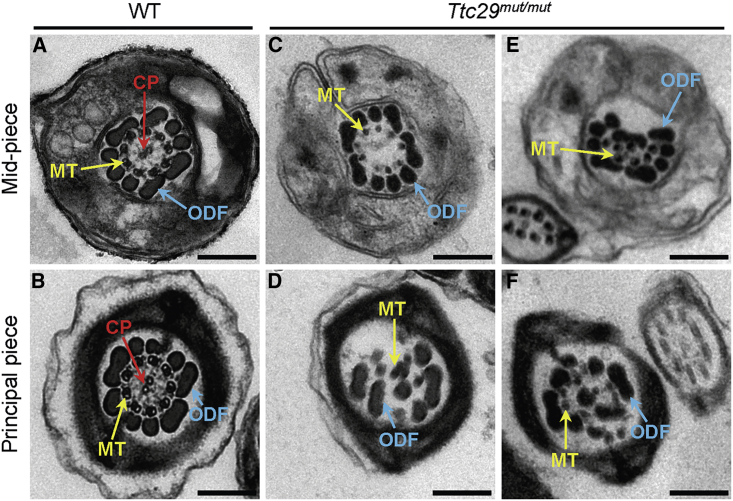
Semen characteristics, sperm morphology, and ultrastructure were investigated in Ttc29-mutated male mice. Notably, sperm motility was significantly reduced in Ttc29mut/mut male mice when compared to that of WT male mice (Table 2). In particular, a significantly lower rate of sperm progressive motility and a significantly higher rate of immotility were observed in Ttc29mut/mut male mice when compared with WT male mice (Figure S15A). Additionally, three kinematic parameters for sperm velocity, including curvilinear velocity (VCL), straight-line velocity (VSL), and average-path velocity (VAP), were significantly reduced in Ttc29mut/mut male mice when compared with WT male mice (Figure S15B). There seems to be no significant difference between WT and Ttc29mut/mut male mice in flagellar beating assessed based on beat/cross frequency (BCF). Also, no significant difference in testis weight was observed between WT and Ttc29mut/mut male mice (Figure S16). Light microscopy revealed a higher rate of bent flagella in Ttc29mut/mut male mice than in wild type (Table 2, Figure S17). Furthermore, TEM observations on flagellar ultrastructure of the spermatozoa from mouse cauda epididymis revealed the lack of central-pair microtubules or the ODF disorganization in Ttc29mut/mut male mice (Figure 5). The rates of spermatozoa with abnormal flagellar ultrastructure were significantly higher in Ttc29mut/mut male mice when compared with that in WT male mice (Table 2).
Table 2.
Semen Characteristics, Ultrastructural Abnormalities, and Sperm Morphology in the Ttc29-Mutated Male Mice
| - | WT Male Micea | Ttc29-Mutated Male Micea |
|---|---|---|
| Semen Parameterb | ||
| Sperm concentration (106/mL) | 19.1 (15.7–21.7) | 12.8 (8.9–18.0) |
| Motility (%) | 81.8 (81.0–82.6) | 51.2 (3.0–71.0)∗ |
| Progressive motility (%) | 65.8 (64.0–67.0) | 39.6 (2.0–57.0)∗ |
| Sperm Morphology | ||
| Absent flagella (%) | 4.3 (2.5–6.1) | 5.7 (2.0–12.1) |
| Short flagella (%) | 3.0 (2.4–3.6) | 4.6 (1.5–9.5) |
| Coiled flagella (%) | 0.8 (0.6–1.1) | 0.9 (0.5–2.5) |
| Irregular caliber (%) | 2.3 (2.0–3.2) | 1.4 (0.6–2.7) |
| Bent flagella (%) | 2.8 (0.5–5.4) | 6.3 (2.5–16.1)∗∗∗ |
| Flagellar Ultrastructural Abnormalityc | ||
| Abnormal mid-piece (%) | 2.7 (2.1–3.3) | 14.0 (8.0–23.5)∗∗ |
| Abnormal end piece (%) | 3.1 (1.8–4.4) | 32.7 (11.4–55.0)∗∗∗ |
∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001. The items with statistical significance are shown in bold.
Values represent the mean (range).
Per single epididymis.
More than one hundred cross sections were analyzed.
Figure 5.
TEM Cross-Sections of Spermatozoa from Ttc29mut/mut Male Mice Reveal Multiple Structural Axonemal Defects
(A) Cross-sections of the mid-piece of sperm flagellum in a WT male mouse. The axoneme is composed of nine peripheral microtubules doublets and a central pair of singlet microtubules, surrounded by nine outer dense fibers and the mitochondrial sheath.
(B) Cross-sections of the principal piece of sperm flagellum from a WT male mouse. Typical ‘‘9 + 2’’ microtubule structure was observed and surrounded by outer dense fibers and the fibrous sheath.
(C–F) Various axonemal anomalies can be observed, including the lack of the central pair of microtubules (C, D) or randomly oriented outer dense fibers (E, F).
Abbreviations: CP, central pair of microtubules (red arrows); MT, peripheral microtubule doublet (yellow arrows); ODF, outer dense fiber (blue arrows); WT, wild type. Scale bars: 200 nm.
Damaged Fertility and Poor Outcomes of IVF in Ttc29-Mutated Male Mice
To assess the fertility and reproductive behavior of Ttc29-mutated mice, the WT and Ttc29-mutated male mice (6 weeks of age or older) were mated to similarly aged WT females, and the numbers of pups per litter were counted. Notably, the average number of pups per litter (0‒2 litters produced per female over 3 months, 2.50 ± 2.81 pups per litter) in Ttc29mut/mut male mice was significantly less than that in WT male mice (3‒4 litters produced per female over 3 months, 7.33 ± 0.13 pups per litter) during 3 months of breeding (Figure S18). No obvious difference in fertility was observed between Ttc29mut/mut female mice and WT females when mated with WT male mice (Figure S18).
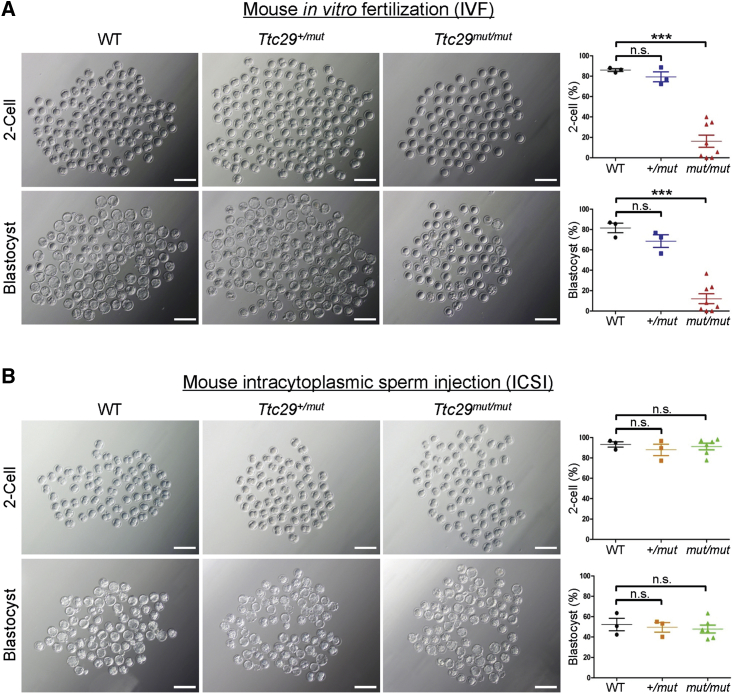
IVF was also performed as described in previous studies47,48 to further investigate the fertility of Ttc29-mutated male mice. Notably, rates of both two-cell embryos and blastocysts were significantly lower in Ttc29mut/mut male mice than in WT male mice (Figure 6A). Our IVF findings strongly suggest that TTC29 plays an important role in sperm quality and male fertility.
Figure 6.
Impaired Fertilization Capability by Loss of Ttc29 Function Could Be Rescued by Intracytoplasmic Sperm Injection
(A) Representative two-cell embryos and blastocysts from in vitro fertilization. Scale bar, 200 μm. The two-cell rates and blastocyst rates were counted after caudal epididymal sperm fertilized oocytes that were collected from superovulated wild-type (WT) females. Here, the blastocyst rates were shown as the percentage of blastocysts out of total oocytes. The WT, Ttc29+/mut, and Ttc29mut/mut groups consisted of three, three, and eight male mice, respectively. A total of 1,305 embryos were counted. Data are represented as means ± SEM; ∗∗∗p < 0.001. Abbreviations: n.s., not significant; WT, wild type.
(B) Representative two-cell embryos and blastocysts from intracytoplasmic sperm injection. Scale bar, 200 μm. The two-cell rates and blastocyst rates were counted after sperm heads of Ttc29-mutated male mice were injected into oocytes that were collected from superovulated wild-type (WT) females. Here, the blastocyst rates were shown as the percentage of blastocysts out of total injected oocytes. The WT, Ttc29+/mut, and Ttc29mut/mut groups consisted of three, three, and six male mice, respectively. A total of 759 embryos were counted. Data are represented as means ± SEM. Abbreviations: n.s., not significant; WT, wild type.
Good Prognosis of ICSI in Men Harboring TTC29 Mutations and Ttc29-Mutated Male Mice
A previous study revealed that MMAF-affected men with DNAH1 mutations have a good clinical outcome following ICSI.49 To examine whether the male infertility induced by TTC29 mutations could also be overcome by ICSI treatment, we performed ICSI for Ttc29-mutated male mice according to a previously described method.41 As expected, the rates of two-cell embryos and blastocysts in Ttc29mut/mut male mice were almost equivalent to those in WT male mice (Figure 6B). Our experimental observations indicated that the male subfertility in Ttc29-mutated mice could be overcome by ICSI. Therefore, we speculated that ICSI might also be applicable to the patients with TTC29-associated MMAF.
Coincidentally, the three men harboring TTC29 mutations in this study had undergone assisted reproductive therapy by ICSI and acquired good clinical outcomes. As shown in Table 3, the overall rates (median) of fertilization, cleavage, eight-cell formation, and blastocyst formation after ICSI were 74.1%, 100.0%, 40.9%, and 31.8%, respectively. We compared the TTC29-mutated group with the age-matched control group with oligoasthenoteratozoospermia (OAT) and with the DNAH1-mutated MMAF group that had been known to have successful ICSI clinical outcomes.49 There were no statistically significant differences in rates of fertilization, cleavage, 8-cell formation, blastocyst formation, implantation, and clinical pregnancy between the TTC29-mutated group and either the DNAH1-mutated group or the OAT controls (Table 3). Our findings suggest that TTC29-associated MMAF and male subfertility can be treated by ICSI.
Table 3.
Clinical Outcomes of ICSI Cycles Using Spermatozoa of TTC29-Mutated, DNAH1-Mutated Men with MMAF and Controls
| - | Group A | Group B | Group C | Comparisons of Three Groups | A versus B | A versus C | B versus C | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| - | TTC29-Mutated MMAF | DNAH1-Mutated MMAF | OAT as Controls | F/H Value | p | Test Statistic | p | Test Statistic | p | Test Statistic | p |
| No. of couples | 3 | 11 | 301 | NA | NA | NA | NA | NA | NA | NA | NA |
| Mean male age (years) | 31.7 ± 6.7 | 32.7 ± 4.7 | 30.1 ± 4.9 | 1.718 | 0.181 | N/A | 1.000 | N/A | 1.000 | N/A | 0.231 |
| Mean female age (years) | 27.3 ± 3.2 | 28.3 ± 4.2 | 28.4 ± 4.2 | 2.500 | 0.084 | N/A | 0.491 | N/A | 1.000 | N/A | 0.088 |
| No. of ICSI cycles | 3 | 11 | 301 | NA | NA | NA | NA | NA | NA | NA | NA |
| No. of oocytes injected | 11 (7,24) | 10 (6, 20) | 11(1, 34) | 0.362 | 0.834 | −0.472 | 1.000 | −0.413 | 1.000 | −0.435 | 1.000 |
| Fertilization rate (%) | 71.4 (63.6, 91.7) | 63.6 (40.0, 86.0) | 83.3 (12.5, 100.0) (3219/4077) | 8.700 | 0.013 | −0.707 | 1.000 | −0.726 | 1.000 | −2.872 | 0.012∗ |
| Cleavage rate (%) | 100.0 (80.0, 100.0) | 100.0 (70.0, 100.0) | 100.0 (66.7, 100.0) (3188/3219) | 4.455 | 0.108 | −0.091 | 1.000 | −0.966 | 1.000 | −1.909 | 0.168 |
| 8-cell formation rate (%) | 40.9 (0, 85.7) | 57.1(0, 100.0) | 70.6 (10.5, 100.0) | 2.859 | 0.239 | −1.101 | 0.873 | −1.349 | 0.531 | −1.031 | 0.909 |
| Blastocyst formation rate (%) | 31.8 (0, 57.1) | 53.5 (0, 100.0) | 60.0 (0, 100.0) | 4.979 | 0.083 | −1.018 | 1.000 | −1.882 | 0.180 | −1.232 | 0.654 |
| No. of transfer cycles | 2 | 9 | 301 | NA | NA | NA | NA | NA | NA | NA | NA |
| Number of embryos transferred per cycle | 1 (0, 2) | 2 (0,3) | 2 (1, 2) | 1.993 | 0.369 | −0.742 | 1.000 | −1.368 | 0.513 | −0.421 | 1.000 |
| Implantation rate (%) | 66.7 (2/3) | 31.3 (5/16) | 47.9 (229/478) | NA | NA | N/A | 0.592b | 0.005 | 0.945a | 1.723 | 0.190 |
| Clinical pregnancy rate per transfer cycle (%) | 100.0 (2/2) | 55.6 (5/9) | 57.1 (172/301) | NA | NA | 0.136 | 0.712a | 0.254 | 0.614a | 0.061 | 0.805a |
| Miscarriage rate (%) | 0 (0/2) | 20.0 (1/5) | 11.6 (20/172) | NA | NA | NA | NA | N/A | NA | N/A | 0.945b |
Symbols: NA, not available data or not being compared; N/A, not applicable, statistical analysis was performed by Fisher’s exact test and thus without the Chi-square value; ∗, the corrected inspection level, α of 0.0167 was used to analyze pairwise comparisons among three groups for categorical data.
Annotations: A versus B, group A compared with group B; A versus C, group A compared with group C; B versus C, group B compared with group C; OAT, oligoasthenoteratozoospermia; F, statistics of ANOVA; H, statistics of Kruskal-Wallis test.
Statistical analyses were performed by an independent statistician blinded to clinical outcomes. Descriptive statistics were expressed using mean ± standard deviation for normally distributed variables. ANOVA was used for measurement data. The Chi-square test or Fisher’s exact test was performed to analyze categorical data. The non-normal distributed variables were shown with median (min, max) and analyzed with independent-samples Kruskal-Wallis test. All the tests were performed using a two-sided test of difference where the inspection level α of 0.05 and a difference with p < 0.05 were considered statistically significant. Pairwise comparisons among the three groups were corrected by Bonferroni method for measurement data.
Yate’s corrected Chi-square test
Fisher’s exact test
Discussion
As mentioned above, we identified bi-allelic truncating mutations of TTC29 in three (3.8%) unrelated cases of 80 MMAF-affected men, and this result cannot be explained by any known MMAF-associated genes. Notably, all of these TTC29 variants are novel and absent from human population genome datasets archived in the 1000 Genomes Project, Exome Aggregation Consortium (ExAC), and Genome Aggregation Database (gnomAD) (Table S1). Through functional experiments, the absence of TTC29 was observed in the spermatozoa of men harboring TTC29 mutations. Therefore, the MMAF phenotypes in these Chinese men are likely to be explained by the bi-allelic mutations in TTC29. Intriguingly, TTC29 mutations were also newly identified in MMAF-affected individuals of African and Iranian origins (Lores et al., a related manuscript in press at the American Journal of Human Genetics).
TTC29 is an evolutionarily conserved and TPR-domain-containing protein. The TPR domain contains many repeated motifs that form structural domains in proteins, and these structural domains can act as interaction scaffolds in the formation of multi-protein complexes involved in numerous cellular processes.50 Previous studies also indicated that TPR family proteins could play an important role in cilia- and flagella-associated functions. For example, IFT88 is important for primary cilia assembly in mammals, and the defects in mouse Ift88 (also known as TPR repeat protein 10) can lead to polycystic kidney disease.51 Our recent study reported that bi-allelic mutations in TTC21A, which encodes a member of the TPR family, induced asthenoteratospermia in both humans and mice.14 All of these studies remind us of the important role of TPR-domain-containing proteins in the assembly of cilia and flagella. The TPR domains are required for proper localization of TTC29 into the axoneme in Trypanosoma brucei (Lores et al., a related manuscript in press at American Journal of Human Genetics). In our study, all of the MMAF-associated truncating mutations of TTC29 are located at or before the TPR domains, leading to the damage of TPR domains in TTC29 protein (Figure 1B). Consistently, our RT-qPCR, immunoblotting, and IF assays also revealed the significantly reduced expression of TTC29 at mRNA and protein levels in the spermatozoa of men harboring TTC29 mutations (Figure 3). Therefore, it is speculated that deficiency of TPR domains contained in TTC29 protein may be an important factor for MMAF phenotypes.
IFT is a conserved mechanism essential for the assembly and maintenance of most eukaryotic cilia and flagella, and it has a bi-directional protein transport system inside cilia. Anterograde transport, which is regulated by IFT complex B, carries ciliary components from the cell body to the tip of cilia, while retrograde transport is regulated by IFT complex A and sends the products of turnover back to the cell body from cilia.20,21,52,53 Previous studies reported that IFT complex B plays a major role in the assembly and maintenance of cilia and flagella.21,54 In most cases, mutations or knockdown of IFT complex B proteins result in absent or very short cilia or flagella.51,55, 56, 57, 58 For example, the mice with a homozygous Ift20 mutation displayed significantly reduced sperm counts, significantly reduced motility, and even abnormally shaped spermatid heads.58 Here we identified TTC29, a potential member of IFT complex B,43 as an asthenoteratospermia-associated protein. TTC29 was involved in functional interaction with the IFT machinery, and partial knockdown of Ttc29 in Xenopus multi-ciliated epithelial cells resulted in a significant decrease in the mean rate of anterograde IFT,43 suggesting the association of Ttc29 with components of the anterograde IFT-B complex. Furthermore, SPAG6, a component of the central pair complex, was almost absent in the sperm flagella from men harboring TTC29 mutations, indicating the impact of TTC29 protein on the positioning of axonemal proteins (Lores et al., a related manuscript in press at American Journal of Human Genetics). Coincidentally, our analyses of sperm morphology and ultrastructure displayed serious defects in external morphology and internal ultrastructure of spermatozoa from men harboring TTC29 mutations and Ttc29-mutated mice, defects which may be caused by failed anterograde IFT.
Furthermore, we also investigated the effect of TTC29 mutations on human TTC30A and IFT52, two members of IFT complex B. A significantly reduced TTC30A staining was observed in the spermatozoa from men harboring TTC29 mutations, and this result further supports the predicted linkage of TTC29 to TTC30A by bioinformatic analysis in a previous study.43 Furthermore, IFT52 staining only aggregated at the base of sperm flagella in men harboring TTC29 mutations, and was weaker than that in the control subject as well. Coincidentally, a recent study demonstrated that a robust interaction of IFT70A (TTC30A) with IFT52–IFT88 dimer in the IFT-B complex is required for ciliogenesis.44 Therefore, all of these previous reports and our findings in this study jointly revealed the participation of TTC29 (as a potential member of IFT-B complex) in anterograde IFT.
TTC30A and IFT52 staining on the sperm and testis samples from Ttc29-mutated male mice seem not to be impacted by the deficiency of mouse Ttc29; this result is different from our experimental observations in humans. This difference may be due to the divergent protein interaction networks between human and mouse as predicted by the in silico tool String (Figure S19). The functional association between TTC29 and TTC30A was identified in human, but no supporting evidence was revealed in mouse. Therefore, TTC29/Ttc29 may participate in the IFT in different ways between human and mouse due to the evolutionarily divergent protein interaction networks. This may also be partially related to the difference in TTC29/Ttc29-associated phenotypic severities between human and mouse. The observed morphological defects of sperm flagella from Ttc29-mutated male mice seem to be mildly impacted when compared with the severe flagella defects of men harboring TTC29 mutations.
Assisted reproduction techniques (ART), such as IVF and ICSI, have become important tools for treating infertile couples.59 For MMAF-associated asthenoteratospermia, no empirical medicinal treatment has been reported to improve the semen parameters; therefore, ICSI could be the only choice for the MMAF-affected cases.60 However, previous studies suggested that MMAF cases caused by different mutations exhibit different prognoses following ICSI. For example, the patients with DNAH1-associated MMAF have good clinical outcomes following ICSI.49 In contrast, a CEP135-associated MMAF subject had a failed pregnancy.61 Therefore, comparative studies on ICSI outcomes between different MMAF-associated genes would be informative to clinicians before ART treatment recommendation. For ICSI treatment in this study, the rates of two-cell embryo and blastocyst in Ttc29mut/mut male mice are similar to those in WT male mice, suggesting that male subfertility of Ttc29-mutated mice could be overcome by ICSI. Coincidentally, all of the three men harboring TTC29 mutations underwent ICSI with their own sperm, and the rates of fertilization, cleavage, eight-cell formation, and blastocyst formation were similar to those of the DNAH1-mutated group and OAT controls. The partners of two affected individuals (A002 IV-1 and S003 II-1) acquired successful clinical pregnancy. Therefore, our findings supported the contention that ICSI can be recommended for TTC29-associated asthenoteratospermia.
In conclusion, our genetic and functional analyses in human subjects and a mouse model strongly suggest that bi-allelic mutations of TTC29 are a crucial genetic cause of MMAF-associated asthenoteratospermia. Furthermore, the effect of TTC29 deficiency on human TTC30A and IFT52 proteins indicated that TTC29 may participate in the anterograde IFT of flagella as a potential member of IFT-B complex. A good pregnancy outcome could be acquired through ICSI using the spermatozoa of men harboring TTC29 mutations. Our study provides new knowledge to clinicians and genetic counselors for understanding the genetic etiology of asthenoteratospermia and male infertility.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank the families for participating and supporting this study. We also thank the Center of Cryo-electron Microscopy at Zhejiang University for technical support. This work was supported by National Natural Science Foundation of China (grant numbers 31625015, 31521003, and 81601340), Special Foundation for Development of Science and Technology of Anhui Province (2017070802D150), Jiangsu Commission of Health (H2018050), Foundation of the Education Department of Anhui Province (KJ2016A370), Shanghai Medical Center of Key Programs for Female Reproductive Diseases (2017ZZ01016), and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01).
Published: November 14, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.10.010.
Contributor Information
Yunxia Cao, Email: caoyunxia6@126.com.
Feng Zhang, Email: zhangfeng@fudan.edu.cn.
Web Resources
1000 Genomes Project, https://www.internationalgenome.org
ENCODE, https://www.encodeproject.org
Ensembl, http://www.ensembl.org
ExAC Browser, http://exac.broadinstitute.org
FANTOM, http://fantom.gsc.riken.jp
HUGO Gene Nomenclature Committee, https://www.genenames.org/
Human Protein Atlas, https://www.proteinatlas.org
Human Splicing Finder, http://www.umd.be/HSF3/
OMIM, https://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
UniProt, https://www.uniprot.org
Supplemental Data
References
- 1.Tournaye H., Krausz C., Oates R.D. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017;5:544–553. doi: 10.1016/S2213-8587(16)30040-7. [DOI] [PubMed] [Google Scholar]
- 2.Robay A., Abbasi S., Akil A., El-Bardisi H., Arafa M., Crystal R.G., Fakhro K.A. A systematic review on the genetics of male infertility in the era of next-generation sequencing. Arab J. Urol. 2018;16:53–64. doi: 10.1016/j.aju.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramalingam M., Kini S., Mahmood T. Male fertility and infertility. Obstet. Gynaecol. Reprod. Med. 2014;24:326–332. [Google Scholar]
- 4.Fujihara Y., Satouh Y., Inoue N., Isotani A., Ikawa M., Okabe M. SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development. 2012;139:3583–3589. doi: 10.1242/dev.081778. [DOI] [PubMed] [Google Scholar]
- 5.Ben Khelifa M., Coutton C., Zouari R., Karaouzène T., Rendu J., Bidart M., Yassine S., Pierre V., Delaroche J., Hennebicq S. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baccetti B., Collodel G., Estenoz M., Manca D., Moretti E., Piomboni P. Gene deletions in an infertile man with sperm fibrous sheath dysplasia. Hum. Reprod. 2005;20:2790–2794. doi: 10.1093/humrep/dei126. [DOI] [PubMed] [Google Scholar]
- 7.Amiri-Yekta A., Coutton C., Kherraf Z.E., Karaouzène T., Le Tanno P., Sanati M.H., Sabbaghian M., Almadani N., Sadighi Gilani M.A., Hosseini S.H. Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum. Reprod. 2016;31:2872–2880. doi: 10.1093/humrep/dew262. [DOI] [PubMed] [Google Scholar]
- 8.Tang S., Wang X., Li W., Yang X., Li Z., Liu W., Li C., Zhu Z., Wang L., Wang J. Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong F.N., Amiri-Yekta A., Martinez G., Saut A., Tek J., Stouvenel L., Lorès P., Karaouzène T., Thierry-Mieg N., Satre V. Absence of CFAP69 causes male infertility due to multiple morphological abnormalities of the flagella in human and mouse. Am. J. Hum. Genet. 2018;102:636–648. doi: 10.1016/j.ajhg.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kherraf Z.E., Amiri-Yekta A., Dacheux D., Karaouzène T., Coutton C., Christou-Kent M., Martinez G., Landrein N., Le Tanno P., Fourati Ben Mustapha S. A homozygous ancestral SVA-insertion-mediated deletion in WDR66 induces multiple morphological abnormalities of the sperm flagellum and male infertility. Am. J. Hum. Genet. 2018;103:400–412. doi: 10.1016/j.ajhg.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auguste Y., Delague V., Desvignes J.P., Longepied G., Gnisci A., Besnier P., Levy N., Beroud C., Megarbane A., Metzler-Guillemain C., Mitchell M.J. Loss of calmodulin- and radial-spoke-associated complex protein CFAP251 leads to immotile spermatozoa lacking mitochondria and infertility in men. Am. J. Hum. Genet. 2018;103:413–420. doi: 10.1016/j.ajhg.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coutton C., Martinez G., Kherraf Z.E., Amiri-Yekta A., Boguenet M., Saut A., He X., Zhang F., Cristou-Kent M., Escoffier J. Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am. J. Hum. Genet. 2019;104:331–340. doi: 10.1016/j.ajhg.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez G., Kherraf Z.E., Zouari R., Fourati Ben Mustapha S., Saut A., Pernet-Gallay K., Bertrand A., Bidart M., Hograindleur J.P., Amiri-Yekta A. Whole-exome sequencing identifies mutations in FSIP2 as a recurrent cause of multiple morphological abnormalities of the sperm flagella. Hum. Reprod. 2018;33:1973–1984. doi: 10.1093/humrep/dey264. [DOI] [PubMed] [Google Scholar]
- 14.Liu W., He X., Yang S., Zouari R., Wang J., Wu H., Kherraf Z.E., Liu C., Coutton C., Zhao R. Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am. J. Hum. Genet. 2019;104:738–748. doi: 10.1016/j.ajhg.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorès P., Coutton C., El Khouri E., Stouvenel L., Givelet M., Thomas L., Rode B., Schmitt A., Louis B., Sakheli Z. Homozygous missense mutation L673P in adenylate kinase 7 (AK7) leads to primary male infertility and multiple morphological anomalies of the flagella but not to primary ciliary dyskinesia. Hum. Mol. Genet. 2018;27:1196–1211. doi: 10.1093/hmg/ddy034. [DOI] [PubMed] [Google Scholar]
- 16.Coutton C., Vargas A.S., Amiri-Yekta A., Kherraf Z.E., Ben Mustapha S.F., Le Tanno P., Wambergue-Legrand C., Karaouzène T., Martinez G., Crouzy S. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat. Commun. 2018;9:686. doi: 10.1038/s41467-017-02792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y., Zhang F., Li F., Jiang X., Yang Y., Li X., Li W., Wang X., Cheng J., Liu M. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat. Commun. 2019;10:433. doi: 10.1038/s41467-018-08182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C., Lv M., He X., Zhu Y., Amiri-Yekta A., Li W., Wu H., Kherraf Z.E., Liu W., Zhang J. Homozygous mutations in SPEF2 induce multiple morphological abnormalities of the sperm flagella and male infertility. J. Med. Genet. 2019 doi: 10.1136/jmedgenet-2019-106011. jmedgenet-2019-106011. [DOI] [PubMed] [Google Scholar]
- 19.Li W., Wu H., Li F., Tian S., Kherraf Z.-E., Zhang J., Ni X., Lv M., Liu C., Tan Q. Biallelic mutations in CFAP65 cause male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. J. Med. Genet. 2019 doi: 10.1136/jmedgenet-2019-106344. jmedgenet-2019-106344. [DOI] [PubMed] [Google Scholar]
- 20.Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa H., Marshall W.F. Intraflagellar transport and ciliary dynamics. Cold Spring Harb. Perspect. Biol. 2017;9:a021998. doi: 10.1101/cshperspect.a021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Liu H., Li W., Zhang Z., Zhang S., Teves M.E., Stevens C., Foster J.A., Campbell G.E., Windle J.J. Intraflagellar transporter protein 140 (IFT140), a component of IFT-A complex, is essential for male fertility and spermiogenesis in mice. Cytoskeleton (Hoboken) 2018;75:70–84. doi: 10.1002/cm.21427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Liu H., Li W., Zhang Z., Shang X., Zhang D., Li Y., Zhang S., Liu J., Hess R.A. Intraflagellar transporter protein (IFT27), an IFT25 binding partner, is essential for male fertility and spermiogenesis in mice. Dev. Biol. 2017;432:125–139. doi: 10.1016/j.ydbio.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi L., Zhou T., Huang Q., Zhang S., Li W., Zhang L., Hess R.A., Pazour G.J., Zhang Z. Intraflagellar transport protein 74 is essential for spermatogenesis and male fertility in micedouble dagger. Biol. Reprod. 2019;101:188–199. doi: 10.1093/biolre/ioz071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles M.R., Zariwala M., Leigh M. Primary ciliary dyskinesia. Clin. Chest Med. 2016;37:449–461. doi: 10.1016/j.ccm.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 31.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 33.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S.M., Li H.B., Wang J.X., Shi Y.C., Cheng H.B., Wang W., Li H., Hou J.Q., Wen D.G. Morphological characteristics and initial genetic study of multiple morphological anomalies of the flagella in China. Asian J. Androl. 2015;17:513–515. doi: 10.4103/1008-682X.146100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coutton C., Escoffier J., Martinez G., Arnoult C., Ray P.F. Teratozoospermia: Spotlight on the main genetic actors in the human. Hum. Reprod. Update. 2015;21:455–485. doi: 10.1093/humupd/dmv020. [DOI] [PubMed] [Google Scholar]
- 37.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y., Oura S., Matsumura T., Oji A., Sakurai N., Fujihara Y., Shimada K., Miyata H., Tobita T., Noda T. CRISPR/Cas9-mediated genome editing reveals 30 testis-enriched genes dispensable for male fertility in mice. Biol. Reprod. 2019;101:501–511. doi: 10.1093/biolre/ioz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L., Li M.Y., Qu C., Miao W.Y., Yin Q., Liao J., Cao H.T., Huang M., Wang K., Zuo E. CRISPR-Cas9-mediated genome editing in one blastomere of two-cell embryos reveals a novel Tet3 function in regulating neocortical development. Cell Res. 2017;27:815–829. doi: 10.1038/cr.2017.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu T.P., Guo F., Yang H., Wu H.P., Xu G.F., Liu W., Xie Z.G., Shi L., He X., Jin S.G. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Yang J., Jia Y., Xiong C., Meng T., Guan H., Xia W., Ding M., Yuchi M. Variability in the morphologic assessment of human sperm: Use of the strict criteria recommended by the World Health Organization in 2010. Fertil. Steril. 2014;101:945–949. doi: 10.1016/j.fertnstert.2013.12.047. [DOI] [PubMed] [Google Scholar]
- 43.Chung M.I., Kwon T., Tu F., Brooks E.R., Gupta R., Meyer M., Baker J.C., Marcotte E.M., Wallingford J.B. Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife. 2014;3:e01439. doi: 10.7554/eLife.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takei R., Katoh Y., Nakayama K. Robust interaction of IFT70 with IFT52-IFT88 in the IFT-B complex is required for ciliogenesis. Biol. Open. 2018;7:bio033241. doi: 10.1242/bio.033241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dupont M.A., Humbert C., Huber C., Siour Q., Guerrera I.C., Jung V., Christensen A., Pouliet A., Garfa-Traoré M., Nitschké P. Human IFT52 mutations uncover a novel role for the protein in microtubule dynamics and centrosome cohesion. Hum. Mol. Genet. 2019;28 doi: 10.1093/hmg/ddz091. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W., Taylor S.P., Nevarez L., Lachman R.S., Nickerson D.A., Bamshad M., Krakow D., Cohn D.H., University of Washington Center for Mendelian Genomics Consortium IFT52 mutations destabilize anterograde complex assembly, disrupt ciliogenesis and result in short rib polydactyly syndrome. Hum. Mol. Genet. 2016;25:4012–4020. doi: 10.1093/hmg/ddw241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin Q., Shen J., Wan X., Liu Q., Zhou Y., Zhang Y. Impaired sperm maturation in conditional Lcn6 knockout mice. Biol. Reprod. 2018;98:28–41. doi: 10.1093/biolre/iox128. [DOI] [PubMed] [Google Scholar]
- 48.Guan M., Bogani D., Marschall S., Raspa M., Takeo T., Nakagata N., Fray M. In vitro fertilization in mice using the MBCD-GSH protocol. Curr. Protoc. Mouse Biol. 2014;4:67–83. doi: 10.1002/9780470942390.mo140059. [DOI] [PubMed] [Google Scholar]
- 49.Wambergue C., Zouari R., Fourati Ben Mustapha S., Martinez G., Devillard F., Hennebicq S., Satre V., Brouillet S., Halouani L., Marrakchi O. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum. Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 50.Allan R.K., Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16:353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., Cole D.G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenbaum J. Intraflagellar transport. Curr. Biol. 2002;12:R125. doi: 10.1016/s0960-9822(02)00703-0. [DOI] [PubMed] [Google Scholar]
- 53.Scholey J.M. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 54.Hou Y., Qin H., Follit J.A., Pazour G.J., Rosenbaum J.L., Witman G.B. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brazelton W.J., Amundsen C.D., Silflow C.D., Lefebvre P.A. The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 2001;11:1591–1594. doi: 10.1016/s0960-9822(01)00485-7. [DOI] [PubMed] [Google Scholar]
- 56.Haycraft C.J., Schafer J.C., Zhang Q., Taulman P.D., Yoder B.K. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 2003;284:251–263. doi: 10.1016/s0014-4827(02)00089-7. [DOI] [PubMed] [Google Scholar]
- 57.Sun Z., Amsterdam A., Pazour G.J., Cole D.G., Miller M.S., Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 58.Follit J.A., Tuft R.A., Fogarty K.E., Pazour G.J. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palermo G., Joris H., Devroey P., Van Steirteghem A.C. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet. 1992;340:17–18. doi: 10.1016/0140-6736(92)92425-f. [DOI] [PubMed] [Google Scholar]
- 60.Chemes H.E., Alvarez Sedo C. Tales of the tail and sperm head aches: Changing concepts on the prognostic significance of sperm pathologies affecting the head, neck and tail. Asian J. Androl. 2012;14:14–23. doi: 10.1038/aja.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sha Y.W., Xu X., Mei L.B., Li P., Su Z.Y., He X.Q., Li L. A homozygous CEP135 mutation is associated with multiple morphological abnormalities of the sperm flagella (MMAF) Gene. 2017;633:48–53. doi: 10.1016/j.gene.2017.08.033. [DOI] [PubMed] [Google Scholar]
- 62.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 63.Auger J., Jouannet P., Eustache F. Another look at human sperm morphology. Hum. Reprod. 2016;31:10–23. doi: 10.1093/humrep/dev251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.