Abstract
Our goal is the development of in vivo hematopoietic stem cell (HSC) transduction technology with targeted integration. To achieve this, we modified helper-dependent HDAd5/35++ vectors to express a CRISPR/Cas9 specific to the “safe harbor” adeno-associated virus integration site 1 (AAVS1) locus and to provide a donor template for targeted integration through homology-dependent repair. We tested the HDAd-CRISPR + HDAd-donor vector system in AAVS1 transgenic mice using a standard ex vivo HSC gene therapy approach as well as a new in vivo HSC transduction approach that involves HSC mobilization and intravenous HDAd5/35++ injections. In both settings, the majority of treated mice had transgenes (GFP or human γ-globin) integrated into the AAVS1 locus. On average, >60% of peripheral blood cells expressed the transgene after in vivo selection with low-dose O6BG/bis-chloroethylnitrosourea (BCNU). Ex vivo and in vivo HSC transduction and selection studies with HDAd-CRISPR + HDAd-globin-donor resulted in stable γ-globin expression at levels that were significantly higher (>20% γ-globin of adult mouse globin) than those achieved in previous studies with a SB100x-transposase-based HDAd5/35++ system that mediates random integration. The ability to achieve therapeutically relevant transgene expression levels after in vivo HSC transduction and selection and targeted integration make our HDAd5/35++-based vector system a new tool in HSC gene therapy.
Keywords: helper-dependent adenovirus, hematopoietic stem cell, targeted integration, AAVS1, CRISPR/Cas9, transgenic mice
Graphical Abstract
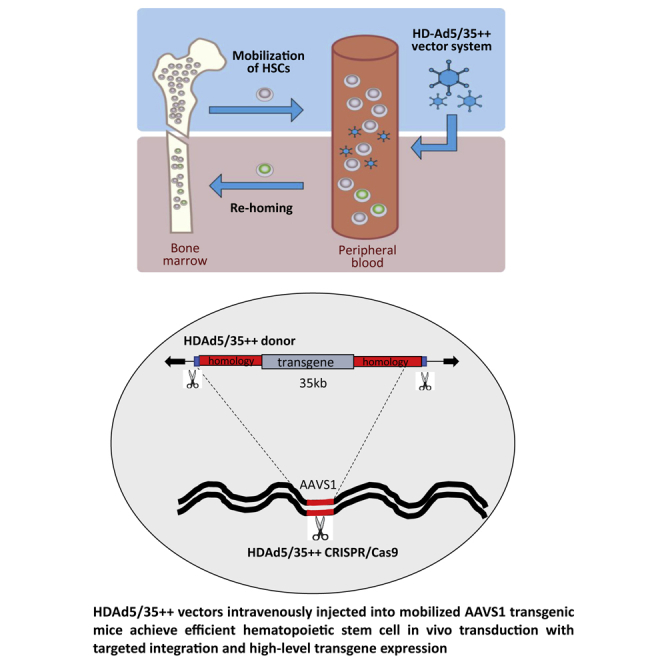
We developed a simple in vivo hematopoietic stem and progenitor cell (HSPC) gene therapy approach that involves GSCF/AMD3100-mediated mobilization of HSPCs from the bone marrow. Although HSPCs circulate at high numbers in the periphery, we transduce them with intravenously injected helper-dependent HDAd5/35++ adenovirus vectors. Transduced cells return to the bone marrow, where they persist long term. Using a HDAd5/35++-CRISPR/Cas9 plus HDAd5/35++-γ-globin donor vector system, we demonstrate efficient targeted integration in AAVS1 transgenic mice. γ-Globin expression levels were significantly higher than those achieved with a Sleeping Beauty (SB100x) transposase-based HDAd5/35++ system that mediated random integration.
Introduction
Current hematopoietic stem cell gene therapies in patients use lentivirus vectors for gene delivery.1, 2 Lentivirus vectors efficiently integrate in the human genome with a strong bias toward actively transcribed genes. This semi-random integration pattern entails a risk of perturbing the expression of neighboring genes, including cancer-related genes. Therefore, a major goal in the field is to target transgene integration to a preselected site. A number of “safe harbors” for targeted integration into the human genome have been suggested (e.g., adeno-associated virus integration site 1 [AAVS1]).3 Among the criteria for a safe harbor site are (1) a distance of more than 50 kb from the 5′ end of any gene, (2) a distance of >300 kb from cancer-related genes, (3) a distance of more than 300 kb from any microRNA, (4) a location outside of a gene transcription unit, and (5) a location outside of ultra-conserved regions. The AAVS1 locus on chromosome (chr)19 is used by wild-type AAV for integration mediated by the virus-encoded protein Rep78, which recognizes a specific motif (Rep-binding site [RBS]) within the AAVS1 site.4, 5 Because a large proportion of the human population has encountered AAV, as evidenced by detectable antibodies against some AAV serotypes, but without any discernable pathology, it was concluded that integration into AAVS1 may be safe.6 Furthermore, this locus contains a DNase I hypersensitive site and an insulator that maintain an open chromatin conformation in CD34+ and induced pluripotent stem cells (iPSCs).7, 8, 9 This allows better access of genome editing tools and, on the other hand, should support high-level transgene expression.7, 10
Targeted transgene integration can be achieved via homology-directed repair (HDR).11 Following cleavage by an engineered site-specific nuclease, DNA double-strand breaks are resolved through non-homologous end joining (NHEJ), an error-prone DNA repair pathway that typically leads to variable insertions or deletions (indels), or HDR, which repairs DNA by copying a homologous donor template. Delivery of exogenous DNA flanked by DNA homologous to the genomic sequence around the break site can lead to incorporation of the exogenous sequence in a site-specific manner.
Current approaches to achieve targeted integration are based on electroporation of hematopoietic stem cells (HSCs) in vitro with endonuclease-encoding mRNA/ribonucleoproteins (RNPs) and donor plasmid DNA,12, 13, 14, 15 integration-deficient lentivirus vectors (IDLVs),11, 16 or rAAV6 vectors.17, 18, 19 We developed helper-dependent adenovirus (HDAd5/35++) vectors to deliver designer integrases20, 21 and, in this study, donor templates. HDAd5/35++ vectors target human CD46, a receptor that is expressed on primitive HSCs.22
The ability of HDAd5/35++ vectors to efficiently deliver their genomes into the nucleus of non-dividing cells allows high amounts of donor DNA, a prerequisite for efficient targeted integration. Because HDAd5/35++ vectors can carry up to 30 kb of foreign DNA, they can accommodate long stretches of donor sequences that are homologous to the given target site. This should increase the efficacy of gene targeting by homologous recombination, which directly correlates with the length of the homology region.23, 24, 25, 26, 27 Because these vectors are easy to produce at high yields and have strong HSC tropism, we employed them for in vivo HSC transduction.22 The central idea of our approach is to mobilize HSCs from the bone marrow using granulocyte colony-stimulating factor (G-CSF)/AMD3100, and while they circulate at high numbers in the periphery, transduce them with an intravenously injected HDAd5/35++ vector. Transduced cells return to the bone marrow, where they persist long term. We have demonstrated the safety and efficacy of the approach in CD46 transgenic (tg) mouse models for hemoglobinopathies either by CRISPR/Cas9-mediated reactivation of endogenous fetal globin20 or by fetal globin gene addition using a hyperactive Sleeping Beauty transposase (SB100x) that mediates efficient random transgene integration.28 Although SB100x-mediated transgene integration is theoretically safer than quasi-random integration of lentivirus vectors, it still raises concerns regarding transgene silencing, undesired effects on neighboring genes, and genomic rearrangements. The goal of this study was therefore to modify our HDAd5/35++-based in vivo HSC transduction approach for targeted integration into AAVS1.
A sequence homologous to the human AAVS1 locus is absent in rodents.29 Two tg rodent models have been reported previously that contain either a 3.5-kb fragment of the AAVS1 locus in the rat (7 head-to-tail copies) or mouse genome (X chromosome).30 A study showed that the open chromatin structure of AAVS1 is maintained in tg mice.31 The Jackson Laboratory distributes AAVS1 tg mice.32 The Jackson Laboratory’s website states that these mice contain five copies of an 8.2-kb human AAVS1 locus fragment inserted into a single genomic site. To make AAVS1 tg mice suitable for transduction with HDAd5/35++ vectors, we crossed them with mice that were tg for the human CD46 locus.33 All animal studies were performed with AAVS1/CD46+/+ mice.
Here we report a novel HDAd5/35++ vector system for targeted integration. We achieved efficient and high-level transgene expression in AAVS1 tg mice after ex vivo and in vivo HSCs transduction.
Results
Design of HDAd-CRISPR and HDAd-Donor Vectors
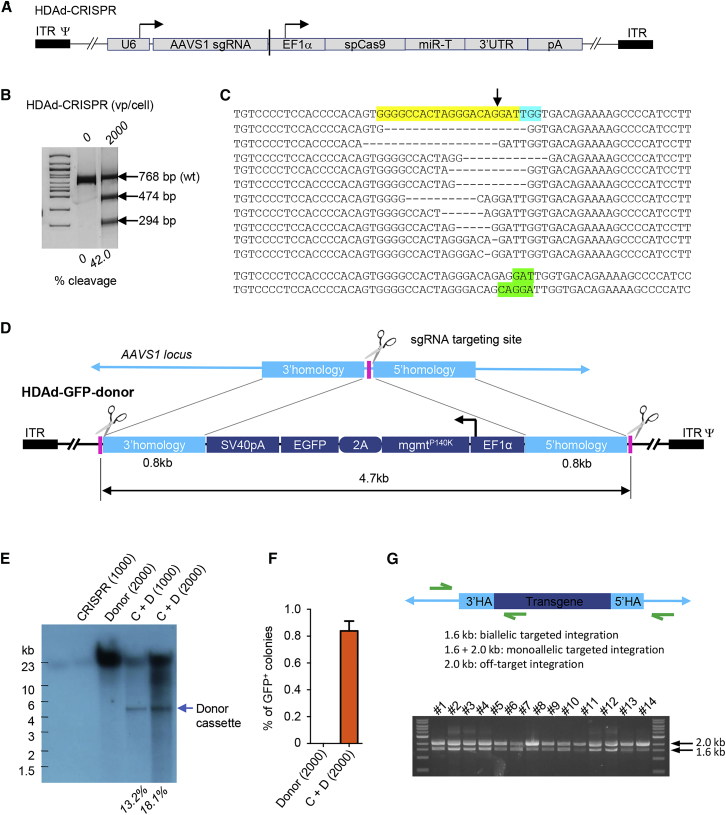
We constructed a HDAd5/35++ vector expressing a CRISPR/Cas9 capable of creating double-stranded DNA (dsDNA) breaks within the AAVS1 locus (Figure 1A). Previous studies demonstrated that site-specific integration into this locus allowed robust transgene expression without side effects in primary human cells.8 To test the activity of the corresponding HDAd-CRISPR vector, we transduced human CD34+ cells, a cell fraction that is enriched for HSCs. AAVS1 site-specific cleavage on day 3 after infection with a frequency of ∼42% was demonstrated by mismatch-sensitive nuclease T7E1 assay (Figure 1B). For deep sequencing of indels, we PCR-amplified an ∼250-bp region surrounding the predicted AAVS1 cleavage site and sequenced the products using an Illumina system (Figure 1C). 80% of indels were deletions ranging from 1–20 bp, and only 10% were 1- to 2-bp micro-insertions.
Figure 1.
Characterization of the AAVS1-Specific CRISPR/Cas9 Vector and Donor Vector for HDR-Mediated Integration
(A) HDAd-CRISPR vector structure. The AAVS1-specific sgRNA is transcribed by PolIII from the U6 promoter, and the spCas9 gene is under control of the EF1α promoter. Cas9 expression is controlled by miR-183-5p and miR-218-5p, which suppress Cas9 expression in HDAd producer 116 cells but do not negatively affect Cas9 expression in CD34+ cells.21 The corresponding microRNA target sites (miR-Ts) were embedded into a 3′ UTR of the β-globin gene. (B) Target site cleavage frequency in human CD34+ cells measured by T7E1 assay 3 days after HDAd-CRISPR transduction at a MOI of 2,000vp/cell. The specific cleavage products are 474 bp and 294 bp. The cleavage efficacy is shown below the gel. (C) The top 13 most frequent indels found in HDAd-CRISPR-transduced CD34+ cells. The yellow sequence shows the target of the guide RNA, with the protospacer adjacent motif (PAM) sequence marked in blue. The CRISPR/Cas9 cleavage site is marked by a vertical arrow. Shown in green are insertions caused by NHEJ. (D) Structure of the donor vector for integration into the AAVS1 site (HDAd-GFP-donor). The mgmtP140K gene is linked to the GFP gene through a self-cleaving picornavirus 2A peptide. The genes are under control of the EF1α promoter. pA, SV40 poly-adenylation signal. The transgene cassette is flanked by 0.8-kb regions of homology to the AAVS1 locus and are similar to a study published previously.8 Upstream and downstream of the homology region are recognition sites for the AAVS1-specific CRISPR/Cas9 to release the donor cassette. (E) Release of the donor cassette. CD34+ cells were infected with the HDAd-GFP-donor (D; at MOIs of 1,000 or 2,000 vp/cell) alone or in combination with HDAd-CRISPR (C; MOI of 1,000 vp/cell). Three days later, genomic DNA was subjected to Southern blot with a GFP-specific probe. The (linear) full-length HDAd-donor-GFP genome runs at ∼31 kb. The released cassette runs at 4.7 kb. The cleavage frequency is shown below the gel. (F) Percentage of GFP-positive colonies. Two days after transduction with HDAd-GFP-donor alone (MOI = 2,000 vp/cell) or together with HDAd-CRISPR (MOI = 1,000 vp/cell for each virus), cells were plated for the CFU assay. Fourteen days after plating, GFP-positive colonies were counted and analyzed for GFP expression. (G) In and out PCR analysis of the GFP-positive colonies. The top diagram shows the location of primers. Expected product sizes for various integration patterns are listed. The bottom gel pictures demonstrate that all clones had monoallelic targeted integration. C + D, CRISPR + donor.
We used, for the first time, a HDAd5/35++ vector as donor vector. The first HDAd-donor vector contained an expression cassette for GFP and O-6-methylguanine-DNA methyltransferase (mgmt)P140K flanked on both sides by 0.8-kb-long regions that are homologous to areas immediately adjacent to the CRISPR/Cas9 target site (Figure 1D). Linear double-stranded adenoviral genomes are covalently linked with the virus-produced “terminal protein” (TP) when they enter cells and are translocated to the nucleus.34 The same is the case for HDAd5/35++ genomes, where the TP is helper virus derived. A previous study has shown that cleavage of transgene donors in the context of plasmids promotes nuclease-mediated targeted integration.35 We therefore incorporated the single guide RNA (sgRNA) target sites for the AAVS1 CRISPR into the donor vector flanking the donor transgene cassette (Figure 1D). Co-infection of HDAd-CRISPR and HDAd-GFP-donor should therefore simultaneously create dsDNA breaks in the chromosomal AAVS1 target site and release the donor cassette from the incoming HDAd-donor genome inside the nucleus. We demonstrated HDAd-CRISPR-mediated release of the donor cassette from a co-infected HDAd-GFP-donor vector with an efficacy of 13.2% and 18.1% in CD34+ cells on day 2 after infection at a total MOI of 1,000 and 2,000 viral particles (vp)/cell, respectively (Figure 1E). This finding also indicates that CRISPR/Cas9 is capable of cleaving double-stranded linear adenoviral DNA, which has implications for anti-viral therapies. Notably, because our donor vectors contained CRISPR/Cas9 cleavage sites, it was impossible to generate a vector that contained both the donor cassette and the CRISPR/Cas9 expression cassette.
Targeted Integration In Vitro
We transduced human CD34+ cells with HDAd-GFP-donor + HDAd-CRISPR or HDAd-GFP-donor alone and plated transduced cells for a colony-forming unit (CFU) assay. On day 14 after plating, ∼0.9% of total colonies exhibited bright and uniform GFP expression (Figure 1F). Cells transduced with HDAd-GFP-donor alone did not give rise to colonies stably expressing GFP. We then performed a vector integration analysis in single GFP-positive colonies. Because of the long homology regions flanking the transgene cassette, it was not possible to employ commonly used tools for vector integration site analysis (e.g., linear-amplification mediated PCR [LAM-PCR]). To demonstrate the presence of vector-cellular DNA junctions, we used an inverse PCR (iPCR) method that involves the endonuclease cleavage of genomic DNA (gDNA) into ∼4-kb fragments, their circularization, and subsequent PCR with transgene-specific primers.6 The results showed that all 14 tested colonies had monoallelic targeted integration (Figure 1G). No off-target integration was detected. These data demonstrate the functionality of our vector system design in human cells.
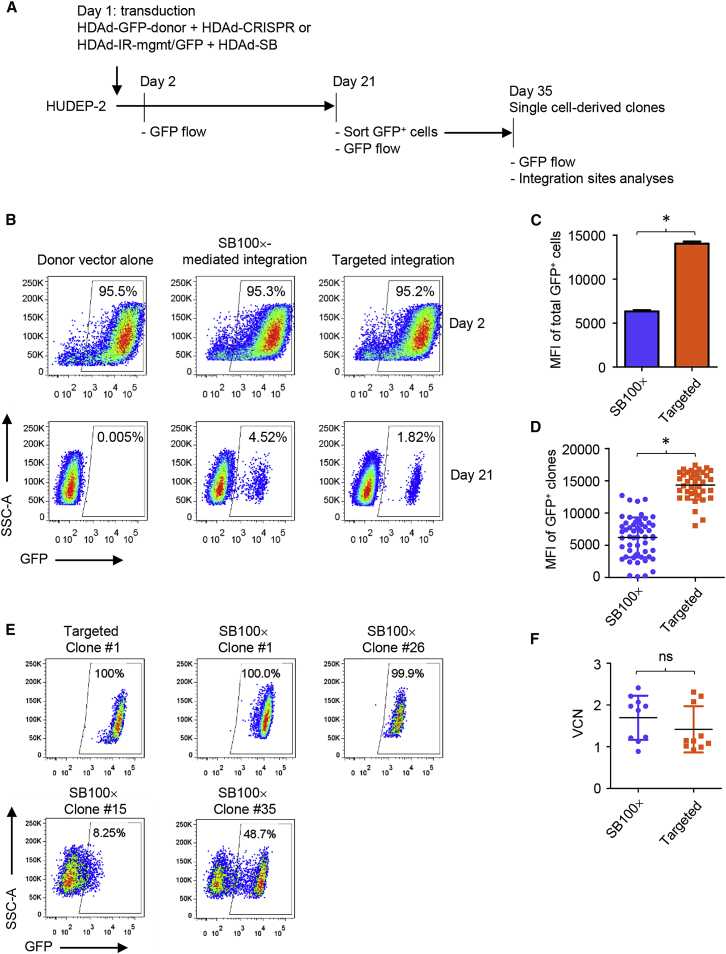
We then tested our HDAd-CRISPR + HDAd-donor vector system for targeted integration in vitro in direct comparison with the SB100x vector system that mediates random integration (Figure 2A). We used HUDEP-2 cells, an immortalized human erythroid precursor cell line.36 This cell line allows the expansion of single colonies, features that facilitate integration site analysis. GFP flow cytometry performed on day 2 after transduction of HUDEP-2 cells demonstrated similar percentages of GFP-positive cells for the SB100x-mediated and targeted integration systems, indicating similar transduction rates (Figure 2B, top panel). GFP expression on day 2 is likely to originate from episomal vector genomes because transduction with HDAd-GFP-donor alone resulted in similar GFP marking. After culturing transduced cells for 21 days, because of cell proliferation, episomal vector genomes nearly disappeared, as indicated by the absence of GFP expression in the HDAd-GFP-donor-alone setting. On day 21, 4.52% and 1.82% of cells were GFP-positive for the SB100x-mediated and targeted integration system, respectively (Figure 2B, bottom panel). This suggests that the SB100x system confers higher stable transduction rates. However, the level of GFP expression, reflected by the mean fluorescence intensity (MFI), was higher in cells transduced with HDAd-CRISPR + HDAd-GFP-donor both in the cell population on day 21 (Figure 2C) and at the single-clone level (Figure 2D). Integration analysis by in and out PCR showed that all tested 36 colonies derived from HDAd-CRISPR + HDAd-GFP-donor-transduced HUDEP-2 cells had transgenes integrated into the AAVS1 site (Figure S1A). Sequencing of amplicons showed that, in all clones except clone 20 Figures S1A and S1C, integration occurred through HDR. This is in line with the homogeneous high level of transgene expression in clones with targeted integration. Figure S1B shows the integration pattern of clone #20. In and out PCR with AAVS1 and transgene-specific primers revealed that integration in 3 of 36 colonies occurred in both alleles; 31 of 36 had monoallelic integrations (Figure S1C). The additional smaller bands in clones 17 and 36 resulted from deletions within the multiple-copy AAVS1 locus (Figures S1D and S1E). In contrast, SB100x-mediated random integration with no preferential targeting of a specific locus28, 37 resulted in varying levels of gene silencing (Figure 2E). Similar levels of vector copy numbers were detected in clones with SB100x and targeted integration (Figure 2F).
Figure 2.
AAVS1-Targeted Integration versus Random SB100x-Mediated Integration in HUDEP-2 Cells
(A) Experimental scheme. HUDEP-2 cells were transduced with the indicated HDAd vectors at a MOI of 1,000 vp/cell for each virus. After expansion for 21 days, GFP-positive cells were sorted into a 96-well plate. Single cell-derived clones were obtained by further expansion for 2 weeks. GFP expression was measured on days 2 and 21 post-transduction in the cell population or on day 35 in cell clones. (B) GFP flow cytometry in cells treated with the donor vector alone or vectors with targeted versus SB100x integration mechanisms on days 2 and 21. (C) Mean fluorescence intensity of GFP in total GFP-positive cells with targeted versus SB100x integration (day 21). Data shown (mean ± SD) represent three independent experiments. *p < 0.05. (D) Mean fluorescence intensity of GFP in single clones. Each symbol represents an individual cell clone. Data shown (mean ± SD) are representative of two independent experiments. *p < 0.05. (E) Flow cytometry showing GFP expression in representative cell clones with targeted or SB100x-mediated integration. (F) Vector copy number in cell clones measured by qPCR using GFP primers. ns, not significant.
In summary, the in vitro studies showed that the HDAd-CRISPR + HDAd-GFP-donor system conferred targeted integration with high efficiency and resulted in higher GFP expression levels than the SB100x-mediated system. The efficacy of stable integration was ∼40% lower for the targeted system.
Ex Vivo Transduction of AAVS1/CD46 HSCs with HDAd-CRISPR + HDAd-GFP-Donor and Subsequent Transplantation into Lethally Irradiated Recipients
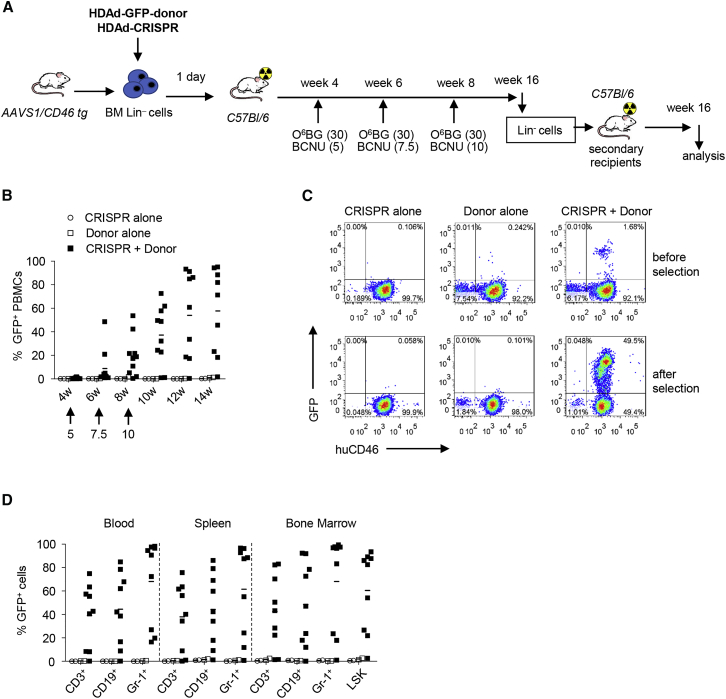
We next tested our targeted integration system in HSCs AAVS1/CD46-tg mice. The target site cleavage frequency after ex vivo transduction of lineage-negative (Lin−) cells, a bone marrow cell fraction enriched for HSCs, was ∼25% after transduction with the HDAd-CRISPR vector at a MOI of 1,000 vp/cell (Figure S2). We chose a total MOI of 500 vp/cell for ex vivo transduction experiments to minimize potential adverse effects associated with high-level CRISPR/Cas9 expression38 while maintaining efficient cleavage activity. AAVS1/CD46 Lin− cells transduced ex vivo with HDAd-CRISPR alone (MOI = 500 vp/cell), HDAd-GFP-donor alone (MOI = 500 vp/cell), and a combination of both (MOI = 250 vp/cell for each vector) were transplanted into lethally irradiated C57BL/6 mice, which were then followed for 16 weeks (Figure 3A). Donor cell engraftment rates were comparable for all three settings (Figure S3), suggesting that the genomic modification introduced in HSCs by the HDAd-CRISPR and HDAd-CRISPR + HDAd-GFP-donor vectors had no detrimental effects on HSC biology, specifically on the multilineage repopulation of lethally irradiated recipients. GFP marking rates reaching up to 100% appeared in peripheral blood mononuclear cells (PBMCs) after three rounds of O6BG/bis-chloroethylnitrosourea (BCNU) selection of HSCs and progenitors that stably expressed transgenes (Figures 3B and 3C). Before selection (4 weeks after transplantation), the percentage of GFP-positive PBMCs was, on average, 1.1%, indicating that targeted integration is a rare event. GFP-positive PBMCs were less than 0.2% on average in mice that were transplanted with Lin− cells transduced with HDAd-GFP-donor only. This points toward the necessity of CRISPR/Cas9-mediated dsDNA breaks to achieve stable transgene expression. Mice analyzed at week 16 after transplantation showed GFP marking in all lineages analyzed in bone marrow, spleen, and PBMCs (Figure 3D). GFP marking rates were maintained for 16 weeks in secondary transplant recipients, demonstrating that primitive HSCs were genetically modified with the HDAd-CRISPR + HDAd-GFP-donor vector system (Figure S4).
Figure 3.
Ex Vivo Transduction of AAVS1/CD46 Lin− Cells with HDAd-CRISPR and HDAd-GFP-Donor and Subsequent Transplantation into Lethally Irradiated Recipients
(A) Schematic of the experiment. Bone marrow was harvested from AAVS1/CD46-tg mice and lineage-negative (Lin−) cells were isolated by MACS. Lin− cells were transduced with HDAd-CRISPR and HDAd-GFP-donor alone or in combination at a total MOI of 500 vp/cell. After 1 day in culture, 1 × 106 transduced cells/mouse were transplanted into lethally irradiated C57BL/6 mice. At week 4, O6BG/BCNU treatment was started and repeated two times every 2 weeks. With each cycle, the BCNU concentration was increased, from 5 mg/kg to 7.5 mg/kg to 10 mg/kg. At week 16, mice were sacrificed, and bone marrow Lin− cells were used for transplantation into lethally irradiated secondary C57BL/6 recipients, which were then followed for 16 weeks. (B) Percentage of GFP-positive cells in peripheral blood mononuclear cells (PBMCs), measured by flow cytometry. Shown are groups that were transplanted with Lin− cells transduced with HDAd-CRISPR only, HDAd-GFP-donor only, and HDAd-CRISPR + HDAd-GFP-donor. Each symbol represents an individual animal. N = 3 for CRISPR only, N = 3 for donor only, N = 10 for CRISPR + donor. p < 0.01 for CRISPR + donor versus CRISPR only and donor only for all time points after week 6. In mice that were transplanted with mock-transduced Lin− cells, less than 0.1% of cells were GFP-positive. (C) Percentage of GFP-positive cells in PBMCs from representative mice transplanted with Lin− cells. Data from week 4 (before selection) and week 12 (after selection) are shown. (D) Percentage of GFP-positive cells in lineage-positive CD3+ cells (T cells), CD19+ cells (B cells), Gr-1+ cells (myeloid cells), and HSCs (LSK cells).
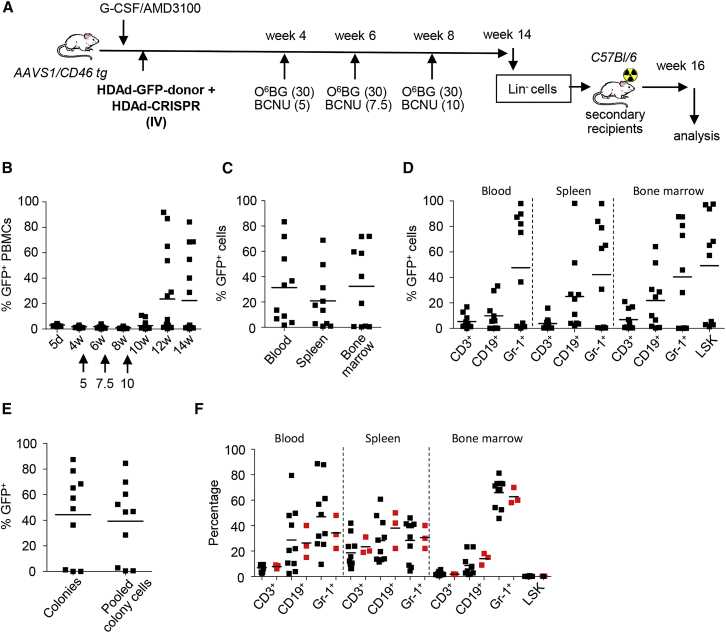
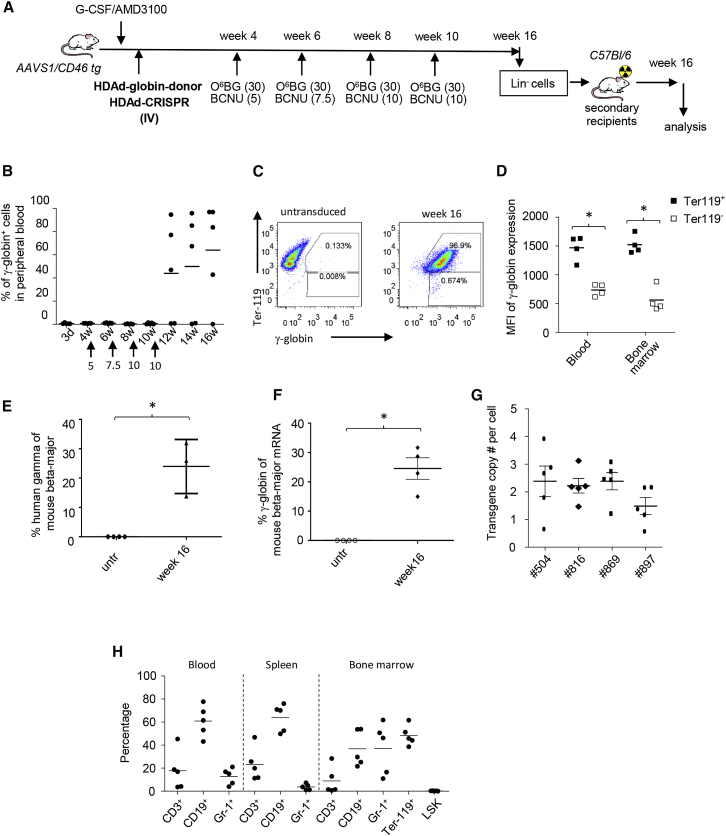
In Vivo HSC Transduction of AAVS1/CD46-tg Mice with HDAd-CRISPR + HDAd-GFP-Donor
Our main focus is the development of in vivo HSC transduction technology with targeted integration. For in vivo HSC transduction of AAVS1/CD46-tg mice, HSCs were mobilized from the bone marrow into the peripheral blood stream by subcutaneous injections of G-CSF/AMD3100 and transduced in vivo by intravenously delivered HDAd-CRISPR + HDAd-GFP-donor vectors (Figure 4A). After in vivo selection with three cycles of O6BG/BCNU, ∼60% of mice showed GFP expression in PBMCs ranging from 35% to 95% GFP-positive PBMCs in individual animals (Figure 4B). At week 14 after in vivo transduction, similar marking was seen in mononuclear cells in the blood, spleen, and bone marrow (Figure 4C). GFP marking was seen in CD3+, CD19+, and Gr-1+ lineage cells in the blood, spleen, and bone marrow (Figure 4D). In the bone marrow of “responders,” more than 50% of Lin−/Sca1+/cKit+ (LSK) cells (a fraction that is enriched for HSCs) were GFP positive (Figure 4D, last group). This was also reflected by a functional assay for HSCs and the ability to form progenitor colonies (Figure 4E). Furthermore, transduction of primitive, long-term repopulating HSCs was shown in secondary recipients (Figure S5). The in vivo HSC transduction and selection procedure had no negative influence on bone marrow cell composition and hematopoiesis (Figure 4F).
Figure 4.
In Vivo Transduction of AAVS1/CD46-tg Mice with HDAd-AAVS1-CRISPR + HDAd-GFP-Donor
(A) Treatment regimen. AAVS1/hCD46tg mice were mobilized and injected intravenously (i.v.) with HDAd-CRISPR + HDAd-GFP-donor (twice, each 4 × 1010 vp of a 1:1 mixture of both viruses). Four weeks later, O6BG/BCNU treatment was started. With each cycle, the BCNU concentration was increased from 5 mg/kg to 7.5 mg/kg and 10 mg/kg. The O6BG concentration was 30 mg/kg for all three treatments. Mice were followed until week 14, when animals were sacrificed for analysis and Lin− cell transplantation into secondary recipients. Secondary recipients were then followed for 16 weeks. (B) Percentage of GFP-positive cells in PBMCs, measured by flow cytometry. N = 13. The percentage of GFP-positive cells in mice that were mock-injected was less than 0.1%. (C) Percentage of GFP-positive cells in PBMCs, spleen, and bone marrow at week 14. (D) Percentage of GFP-positive cells in lineage-positive CD3+ cells (T cells), CD19+ cells (B cells), Gr-1+ cells (myeloid cells), and HSCs (LSK cells). (E) Analysis of GFP-positive colonies. Total bone marrow Lin− cells from week 14 mice were plated, and GFP expression in colonies was analyzed 12 days later. Each symbol is the average GFP-positive colony number for an individual mouse (left panels). Cells from all colonies were pooled and analyzed by flow cytometry (right panels). (F) Percentage of lineage-positive and LSK cells at week 14. Data from mock in vivo-transduced mice are shown as red symbols. The difference between mock and treatment groups was not significant.
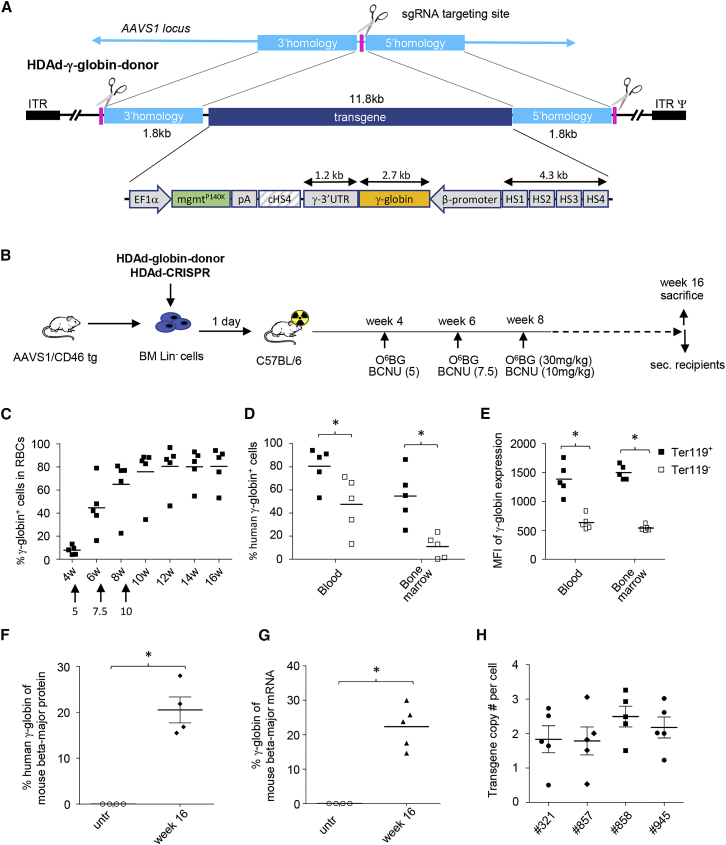
Ex Vivo and In Vivo HSC Transduction with the HDAd-CRISPR and HDAd-Globin-Donor Vector
Although studies with the HDAd-GFP-donor vector suggest stable HSC transduction in the majority of animals, a higher rate of responders would be desirable. This would require increasing the efficacy of HDR-mediated integration, which can be achieved by increasing the length of the homology arms.23, 24, 25 We therefore generated a new HDAd-donor vector with 1.8-kb regions that were homologous to AAVS1 genomic sequences surrounding the CRISPR/Cas9 cleavage site (Figure 5A). For potential application in gene therapy of hemoglobinopathies, we used the human γ-globin gene (HBG1) under control of a mini-β-globin locus control region (LCR). We tested this HDAd-globin-donor vector both in the ex vivo and in vivo HSC transduction protocols. In the ex vivo transduction setting (Figure 5B), we observed that all mice responded, with 4 of 5 mice expressing γ-globin in 80% of peripheral red blood cells (RBCs) (Figure 5C). The percentage of γ-globin-positive erythroid (Ter119+) cells in the blood and bone marrow was significantly higher than that of non-erythroid (Ter119−) cells (Figure 5D). The same was the case for the γ-globin MFI (Figure 5E). This suggests that the mini-LCR confers preferential expression in erythroid cells. At week 16, the level of γ-globin was 20.52% (±5.66%) of that of adult mouse β-globin, as measured by high-performance liquid chromatography (HPLC) (Figure 5F), and 22.33% (±6.21%) by qRT-PCR (Figure 5G). In a previous study, performed under the same regimen with the SB100x system, γ-globin expression levels were 15.74% (+±2.69%) by HPLC and 15.40% (±9.21%) by qRT-PCR.39 This implies that the level of γ-globin expression is higher for the targeted integration system compared with the SB100x system. In fact, for the targeted integration system, it would be in the range of curative levels, which is thought to be 20% γ-globin of adult globin for patients with β0/β0-thalassemia or sickle cell disease.28 In agreement with previous studies,28 we measured, in single Lin− cell-derived colonies, on average, two integrated vector copies per genome at week 16 (Figure 5H). Ex vivo HSC transduction of Lin− cells did not affect their ability for multilineage engraftment and complete hematopoietic reconstitution in lethally irradiated recipients (Figure S6). Analysis of secondary HSC transplant recipients showed that ex vivo transduction with the HDAd-CRISPR + HDAd-globin-donor vector followed by in vivo selection did not affect the pool of HSCs capable of long-term repopulation (Figure S7).
Figure 5.
Ex Vivo Transduction of AAVS1/CD46 Lin− Cells with HDAd-AAVS1 and HDAd-γ-Globin-Donor Vectors and Subsequent Transplantation into Lethally Irradiated Recipients
(A) Structure of the donor. The overall structure is the same as for the HDAd-GFP-donor vector (Figure 1D). The regions of homology are longer (1.8 kb versus 0.8 kb) in the new HDAd-globin-donor vector. The γ-globin expression cassette contains a 4.3-kb version of the β-globin LCR consisting of four DNase hypersensitivity (HS) regions and the β-globin promoter.59 The full-length γ-globin cDNA, including the 3′ UTR (for mRNA stabilization in erythrocytes), was used. The mgmtP140K gene is under control of the ubiquitously active EF1α promoter. The bi-directional SV40 poly-adenylation signal is used to terminate transcription. To avoid interference between the LCR/β-promoter and EF1α promoter, a 1.2-kb chicken HS4 chromatin insulator60 was inserted between the cassettes. (B) The treatment regimen is the same as shown in Figure 3A. (C) Percentage of human γ-globin-positive cells in peripheral red blood cells (RBCs), measured by flow cytometry. In mice that were transplanted with mock-transduced Lin− cells, less than 0.1% of cells were γ-globin positive. N = 5. (D and E) Percentage (D) and mean fluorescence intensity (E) of human γ-globin-positive cells in erythroid (Ter119+) and non-erythroid (Ter119−) cells in blood and bone marrow at week 16 after in vivo transduction. *p < 0.05. (F) Percentage of γ-globin chains relative to mouse β-major chains measured in RBCs at week 16 by HPLC. *p < 0.05. (G) Percentage of γ-globin mRNA relative to mouse β-major RNA measured in RBCs at week 16 by qRT-PCR. *p < 0.05. (H) Vector copy number per cell in colonies derived from Lin− cells. Each symbol represents one colony. Differences between animals are not significant.
In the in vivo HSC transduction studies with the HDAd-CRISPR + HDAd-globin-donor vector (Figure 6A), after in vivo selection, 4 of 5 mice showed stable γ-globin expression in RBCs, ranging from 40% to 97% γ-globin+ RBCs in individual mice (Figure 6B). γ-Globin expression was found preferentially in erythroid cells (Figures 6C and 6D). The γ-globin expression levels in RBCs were 23.97% (±7.22%) by HPLC (Figure 6E) and 24.53% (±7.34%) by qRT-PCR (Figure 6F) of that of adult mouse β-globin. The vector copy number per cell ranged from 1.5 to 2.5 in individual mice. In the same in vivo HSC transduction and selection setting, using the SB100x-based γ-globin vectors, γ-globin levels were 10.5% (±3.1%) by HPLC and 12.17% (±3.38%) by qRT-PCR, with an average of 2 integrated vector copies per genome (Figure 6G).28 The composition of lineage-positive cells in the blood, spleen, and bone marrow and LSK cells in bone marrow at week 16 after in vivo transduction was normal (Figure 6H). Transplantation of bone marrow Lin− cells harvested at week 16 after in vivo transduction with HDAd-CRISPR + HDAd-globin-donor into lethally irradiated recipients showed 100% engraftment and stable γ-globin expression in RBCs over 16 weeks, with an average γ-globin level of 24% of that of adult β-globin (Figure S8).
Figure 6.
In Vivo Transduction of AAVS1/CD46-tg Mice with HDAd-CRISPR + HDAd-γ-Globin-Donor
A) Treatment regimen. (B) Percentage of γ-globin-positive RBCs. The percentage of γ-globin-positive cells in mice that were mock-injected was less than 0.1%. N = 5. (C) Representative flow cytometry analysis showing the percentage of γ-globin-positive cells in peripheral RBCs from untransduced control mice or mice at week 16 after in vivo transduction. (D) Mean fluorescence intensity of γ-globin in erythroid (Ter119+) and non-erythroid (Ter119−) cells in blood and bone marrow. *p < 0.05. (E) Percentage of γ-globin chains relative to mouse β-major chains measured in RBCs at week 16 by HPLC. *p < 0.05. (F) Percentage of γ-globin mRNA relative to mouse β-major RNA measured in RBCs at week 16 by qRT-PCR. *p < 0.05. (G) Vector copy number per cell in colonies derived from Lin− cells from four responder mice. Each symbol represents one colony. Differences between animals are not significant. (H) Composition of lineage-positive cells in blood, spleen, and bone marrow and LSK cells in bone marrow at week 16 after in vivo transduction.
In summary, our HSC transduction studies with HDAd-CRISPR + HDAd-globin-donor resulted in stable γ-globin expression at levels that are significantly higher than those achieved in previous studies with the SB100x-based system. Clearly, to conclude that this is the result of targeted integration into AAVS1 (which has an open chromatin profile) requires integration site analysis in bone marrow cells of transduced AAVS1/CD46-tg mice.
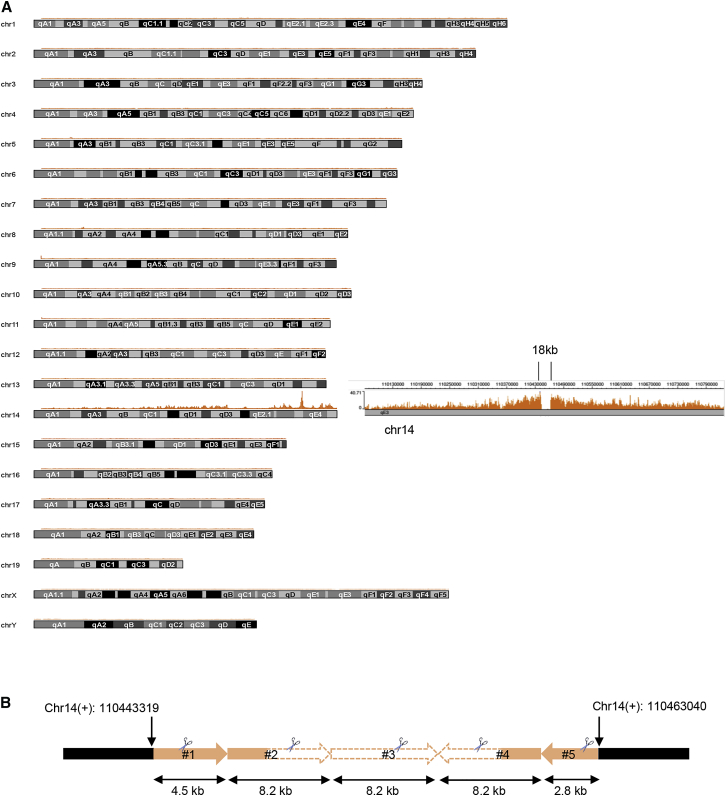
Localization of the AAVS1 Locus in AAVS tg Mice
iPCR for integration site analysis requires knowledge of AAVS1 locus localization in the genome of AAVS1/CD46-tg mice. To determine this, we used targeted locus amplification (TLA)-PCR technology that involves crosslinking of physically proximal sequences40 (Materials and Methods). The TLA data obtained from bone marrow cells from AAVS1/CD46-tg mice were then aligned with a reference mouse genome. The TLA results indicate that the AAVS1 transgene is integrated into chr14 at the location (chr14:110443871–110461834), accompanied by an 18-kb deletion of the mouse genome sequence (Figure 7A). Using this information, we then used primers to amplify and sequence into the locus (Figure 7B). We found repeats of the AAVS1 locus facing left to right and right to left. Both terminal repeats (1 and 5) were truncated and 4.5 and 2.8 kb long, respectively. Repeat 5 lacked a complete 5′ homology region. This constellation of target sites complicated the integration site analysis. Some of the theoretical outcomes for integration by the HDAd-CRISPR + HDAd-donor system are outlined in Figure S9.
Figure 7.
Localization and Structure of the AAVS1 Locus in AAVS1/CD46-tg Mice
(A) TLA data showing mismatches on chromosome 14. The right panel shows an enlarged section of chromosome 14 with the 18-kb gap visible. The gap corresponds to the added human AAVS1 loci. (B) Detailed structure of the AAVS1 loci, indicating the genomic localization. The shaded AAVS1 areas were confirmed by Sanger sequencing. The empty areas were deduced from restriction analysis (see below) and information from The Jackson Laboratory. The CRISPR/Cas9 cleavage sites are indicated by scissors. Repeats 2–4 are complete 8.2-kb human AAVS1 EcoRI fragments, whereas repeats 1 and 5 only contain a fraction of the EcoRI fragment. Notably, repeat 5 lacks a complete 5′ homology arm.
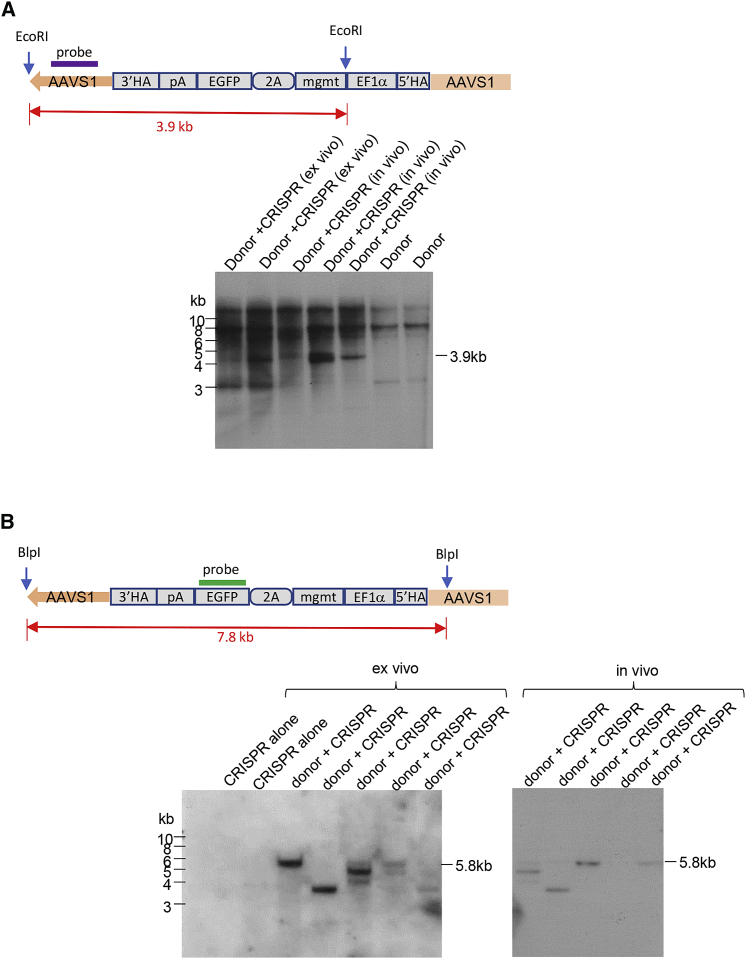
Chromosomal Integration after Ex Vivo and In Vivo HSC Transduction with HDAd-CRISPR+HDAd-Donor
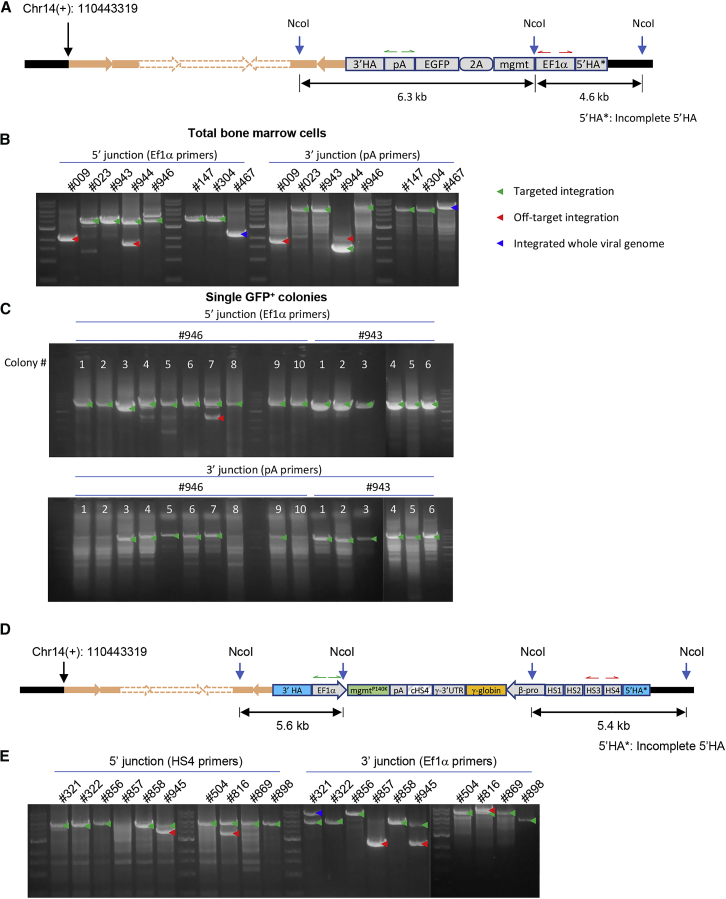
First we performed a genomic Southern blot on DNA from bone marrow cells harvested at week 16 (Figure 8). Hybridization of EcoRI-digested gDNA with an AAVS1-specific probe showed, in all analyzed mice, a 3.9-kb-specific band indicative of integration of the donor cassette into one (or more) repeats of the AAVS1 locus (Figure 8A). Hybridization of BlpI-digested DNA with the GFP probe resulted in 5.8-kb signals in 5 of 10 mice, representative of integration into the repeat 5-containing-only partial 5′ homology region (Figure 8B). The signals of other size could be the result of integration into repeats 1–4. Two of ten mice appeared to have integrations into several AAVS1 motif repeats. To demonstrate the presence of transgene-chromosome junctions, we performed iPCR on gDNA from mice (Figures 9A and 9B). Six of eight mice analyzed displayed PCR products consistent with HDR-mediated integration into the AAVS1 site (Figure 9B, green arrows). Several of these mice had additional bands that resulted from integration into one of the CRISPR/Cas9 off-target sites on ch5 (Figure 9B, red arrows). We also found bands that originated from integration of the full-length HDAd genome involving the inverted terminal repeats (ITRs) as junctions. Interestingly, these integrated full-length HDAd vector genomes were on chr14, the chromosome containing the CRISPR AAVS1 target site (Figure 9B, blue arrows). In an attempt to de-complex these results derived from a pool of bone marrow cells, we plated GFP-positive bone marrow Lin− cells to generate progenitor colonies derived from single cells (Figure 9C). Analysis of colonies from mice with only one band specific for HDR integration into AAVS1 (e.g., mouse 943) showed homogeneous signals in all colonies, whereas colonies from mice with additional off-target integration (e.g. 946) showed a chimeric pattern: nine of ten colonies with only on-target integration and one colony containing both on-target and off-target integrations, which is possible because the average number of integrated transgenes per genome is ∼2. Integration site analysis of bone marrow cells in ex vivo and in vivo transduction studies with HDAd-CRISPR and HDAd-globin-donor vector revealed a similar outcome (Figures 9D and 9E). Off-targeted integration into off-target sites in chr2 or chr5 was also observed (Figures S10 and S11). In the ex vivo HSC transduction setting with HDAd-CRISPR + HDAd-globin-donor, a higher rate of animals with targeted integrations was found compared with the in vivo HSC transduction study with the HDAd-GFP-donor vector. Based on previous reports,23, 24, 25 we speculate that this could be due to higher HDR efficiency based on longer homology regions. A summary of integration site analyses by iPCR can be found in Figure S12.
Figure 8.
Integration Site Analysis by Southern Blotting of Genomic DNA Isolated at Week 16 after Ex Vivo or In Vivo HSC Transduction with HDAd-CRISPR + HDAd-GFP-Donor
(A) Hybridization with an AAVS1-specific probe. The top panel shows the expected EcoRI fragment size and the localization of the probe. The bottom panel shows the analysis of individual mice from the ex vivo and in vivo transduction setting. The larger bands represent non-targeted AAVS1 locus repeats. (B) Hybridization of BlpI-digested DNA with a GFP-specific probe. The 7.8-kb predicted band is based on integration into a complete AAVS1 EcoRI fragment. The observed 5.8-kb band resulted from integration into repeat 5, which lacks a complete 5′ homology region. The band pattern is discussed in more detail in the text.
Figure 9.
Integration Site Analysis by Inverse PCR (iPCR) of Genomic DNA Isolated at Week 16 after Ex Vivo or In Vivo HSC Transduction with HDAd-CRISPR + HDAd-GFP-Donor and HDAd-CRISPR + HDAd-γ-Globin-Donor
(A) The locations of NcoI sites and primers (half arrows; red, EF1α primers for 5′ junctions; green, pA primers for 3′ junctions). The expected amplicon size at each site for targeted integration in repeat 5 is indicated. (B) iPCR results using genomic DNA from total bone marrow cells. Each lane represents an individual mouse. 009, 023, 943, 944, and 946 are mice after ex vivo HSC transduction. 147, 304, and 467 are in vivo-transduced animals. (C) iPCR analysis of GFP-positive colonies. Bone marrow Lin− cells from week 14 mice were plated, and genomic DNA was isolated from GFP-positive colonies 20 days later and used for iPCR. Mice 943 and 946 were analyzed. Each lane represents one colony. Green arrows, targeted integration; red arrows, off-target integration; blue arrows, integrated whole HDAd viral genome. (D) The diagram shows the locations of NcoI sites and primers (half arrows; red, EF1α primers for 5′ junctions; green, pA primers for 3′ junctions). The expected amplicon size at each side for targeted integration in repeat 5 is shown. (E) iPCR results using genomic DNA from total bone marrow cells. Each lane represents one mouse. 321, 322, 856, 857, 858, and 945 are mice after ex vivo transduction. 504, 816 869, and 898 are in vivo-transduced animals.
Overall, our integration studies indicate a high frequency of targeted integrations into AAVS1 loci.
Discussion
Self-inactivating lentivirus vectors, in contrast to gamma-retrovirus vectors, have not been associated with insertion site-associated malignant clonal expansions in clinical HSC gene therapy trials. However, this risk cannot be completely excluded, as indicated by a recent study in non-human primates.41 Theoretically, the random integration pattern mediated by SB100x transposase and the lack of a preference for integration into activate genes and promoters should be safer, but concerns about genotoxicity remain. Therefore, a major effort in the field is aimed toward targeted transgene integration into preselected sites such as the AAVS1 site. Zinc-finger nuclease mRNA and AAV6-mediated donor template delivery in human HSCs resulted in more than 50% targeted integration into the AAVS1 locus.17 In other studies that employed an AAVS1-specific CRISPR/Cas9 RNP and AAV6 to deliver the donor template, the frequency of site-specific integration was up to 90%.19, 42 Similar rates were achieved for targeted integration into CCR5.18
Our approach for targeted integration into AAVS1 has a number of new aspects. (1) We used a helper-dependent, capsid-modified HDAd5/35++ vector to deliver the donor template. Corresponding vector genomes are double-stranded, linear DNA covalently linked on both ends to the viral TP. It is thought that, in contrast to single-stranded AAV6 donor vectors, double-stranded, linear adenoviral DNA is not an optimal template for HDR. To compensate for this potential disadvantage, we incorporated AAVS1 CRISPR/Cas9 cleavage sites into the HDAd-donor vectors to create free “recombinogenic” DNA ends. (2) Because the insert capacity of HDAd5/35++ vectors is ∼30 kb, we were able to incorporate homology arms that would exceed the packaging capacity of rAAV6 or IDLVs. Previous studies suggest that increasing the homology can improve HDR.23, 24, 25. (3) The large HDAd5/35++ insert capacity also allowed us to include the mgmtP140-based in vivo selection cassette into the donor template, mediating selective survival and expansion of progeny cells without affecting the pool of transduced primitive HSCs by short-term treatment with low-dose O6BG/BCNU.43 Considering the low efficacy of HDR and, consequently, targeted integration in HSCs,44 in vivo HSC selection appears to be crucial to achieve high transgene marking levels in peripheral blood cells. (4) Finally, because of the ease to produce high yields of HDAd5/35++ vectors and their tropism for primitive HSCs, they can be used for in vivo HSC transduction via intravenous injection into mobilized animals. Notably, although CD46 is expressed on all nucleated cells in humans, transgene expression after intravenous injection of HDAd5/35++ vectors was only detected in peripheral blood cells and in a rare subset of splenic cells.22 This could be due to low CD46 receptor levels and/or poor receptor accessibility in other tissues.
To achieve stable transgene (GFP or γ-globin) expression, coinfection of HDAd-donor and HDAd-CRISPR was essential, suggesting that CRISPR-mediated gDNA breaks and, most likely, the release of the donor template from the HDAd-donor vector greatly stimulated integration. An indicator of transgene integration into HSCs after in vivo transduction with HDAd-donor + HDAd-CRISPR was the fraction of mice that displayed stable high-level transgene expression after completion of in vivo selection (i.e., responders). It was 6 of 16 (37.5%) for HDAd-GFP-donor + HDAd-CRISPR and 4 of 5 (80%) for HDAd-globin-donor + HDAd-CRISPR. Notably, the responder rate with a high frequency of targeted integration was 100% for both vectors in the ex vivo transduction setting. This indicates that a limiting factor for our targeted in vivo HSC transduction approach is the efficacy of HSC infection. The initial infection step could theoretically be improved by an optimized HSC mobilization regimen45 and two rounds of HDAd injection 1 day apart.
Although we did not compare HDAd-donor and AAV6 donor vectors side by side, our data indicate that our vector system is an efficient tool to achieve targeted integrations in HSCs in ex vivo and in vivo transduction settings. We speculate that key for this is the high efficacy of HDAd-donor vector delivery to the nucleus of non-dividing cells and the HDAd vectors’ capacity to incorporate large homology regions.
An important finding in our study was that the targeted integration system conferred higher transgene expression levels than the SB100x-based system in the in vitro transduction setting, with the same number of integrated transgene copies per cell (Figures 2C–2F). Compared with previous studies with the SB100x system done with the same HSC transduction and selection regimen,28, 39 γ-globin levels were also higher after targeted integration (10%–12% γ-globin of mouse β-major globin for SB100x versus 20%–25% γ-globin of mouse β−major globin). Notably, the vector copy number (VCN) in the studies cited above using the SB100x integration system was ∼2 copies/cell and comparable with the copy number we found in mice after targeted integration (Figures 5H and 6G).
This is particularly relevant for gene therapy of hemoglobinopathies (β0/β0-thalassemia and sickle cell disease), which requires γ-globin at levels that are more than 20% of adult globin levels. In responder mice that were transduced ex vivo or in vivo with HDAd-CRISPR + HDAd-globin-donor, we achieved these theoretically curative levels. This is an important improvement over our previous study in a thalassemia mouse model, where we used the SB100x transposase system for γ-globin gene addition.28 We speculate that epigenomic effects on transgene expression are less pronounced after integration into the AAVS1 locus, which is known to maintain an open chromatin configuration in HSCs2, 5, 7 and in AAVS1 tg mice.31 On the other hand, it cannot be excluded that random SB100x-mediated integration places transgenes into regions that are subjected to silencing.
Our integration site analyses suggest a nearly 100% targeted integration efficacy after in vitro transduction of HUDEP-2 cells. In ex vivo and in vivo HSC transduction studies, both Southern blot and iPCR on genomic bone marrow DNA suggested efficient targeted integration in bone marrow HSCs. For example, iPCR of integration junctions documented targeted integration in 75% of mice, with most of these mice having no off-target integration. This was further confirmed by analysis of colonies derived from single CFUs. At a low frequency, integrations were also found in two of the in silico predicted CRISPR Cas9 off-target sites. Furthermore, we found full-length HDAd-donor genomes integrated in chr14, the chromosome that carries the AAVS1 loci. We found previously that HDAd ITRs are prone to DNA breaks and that this can result in inefficient integration into genomic sites in which DNA breaks occur.46, 47 Considering recent studies on CRISPR/Cas9-induced undesired large deletions and/or translocations (7–8 kb) around the target site,48 we speculate that CRISPR-Cas9 DNA breaks far away from the target site could be implicated in the integration of complete HDAd genomes. Overall, reports of large deletions and/or translocations question the safety of CRISPR/Cas9. On the other hand, because no developmental effects associated with CRISPR/Cas9-mediated germline editing in animals have been reported so far, it is likely that cells with such deleterious chromosomal changes are selected out during development. Support for this hypothesis comes from a recent non-human primate (NHP) study in which CRISPR/Cas9-edited HSCs were transplanted, and a 4.9-kb deletion in the HBG1/2 region disappeared in PBMCs over time (O. Humbert et al., 2019, ASGCT, conference). To prove that our HDAd-CRISPR + HDAd-donor in vivo HSC transduction approach is safe, similar long-term studies (optimally employing high-accuracy CRISPR/Cas9 versions) in NHPs are required.
From our studies, we also concluded that the AAVS1tg mouse model is suboptimal for targeted integration studies involving CRIRPR/Cas9 because of the presence of multiple AAVS1 target loci, some of which were truncated to a degree where they lost areas of homology with the HDAd-donor vector. The presence of truncated AAVS1 loci also suggests that rearrangement can occur in AAVS1 tg mice, as reported previously.49 Future studies should be done with a model that contains only mono- or bi-allelic copies of the target site. For example, the human βs-globin gene or one of the human α-globin genes present in the Townes mouse model for sickle cell disease50 could be considered as targets, provided that these loci display an open chromatin structure and are accessible to CRISPR/Cas9 in HSCs.
In our studies with AAVS1/CD46-tg mice, we provided proof of targeted integration in HSCs after ex vivo or in vivo transduction with HDAd-CRISPR + HDAd-donor vector. Although the initial in vivo HSC transduction and transgene integration frequency requires improvement, the fact that AAVS1-targeted integration yields higher transgene expression levels and a theoretically safer integration pattern is a critical advantage of our HDAd-based vector system over other currently used approaches.
Materials and Methods
Cells
CD34+ cells from G-CSF-mobilized adult donors were provided by Fred Hutch Hematopoietic Cell Procurement and Processing Services. Cells were recovered from frozen stocks and incubated overnight in StemSpan H3000 (STEMCELL Technologies, Vancouver, Canada) with penicillin/streptomycin, Flt3 ligand (Flt3L, 25 ng/mL), interleukin 3 (10 ng/mL), thrombopoietin (TPO; 2 ng/mL), and stem cell factor (SCF; 25 ng/mL). Cells were transduced with HDAd vectors at a MOI of 1,000 or 2,000 vp/cell and analyzed as indicated. HUDEP-2 cells36 were provided by Ryo Kurita (Central Blood Institute, Japanese Red Cross Society, Department of Research and Development, Tokyo 135-8521, Japan) and Yukio Nakamura (RIKEN BioSource Center, Cell Engineering Division, Ibaraki 305-007, Japan). HUDEP-2 cells were cultured in the presence of SCF, erythropoietin (EPO), doxycycline, and dexamethasone, as described previously.51 A total of 2 × 105 cells were plated in 24-well ultra-low attachment surface plates (Corning) with 1 mL of medium/well. HDAd vectors at a MOI of 500–1,000 vp/cell were added. The medium was changed 24 h later.
HDAd5/35++ Vectors
HDAd-SB, HDAd-inverted repeat (IR)-GFP/mgmt, and HDAd-IR-γ-globin/mgmt have been described before.39, 43 Details regarding the generation of HDAd-CRISPR, HDAd-GFP-donor, and HDAd-globin-donor are described in the Supplemental Materials and Methods.
For production of HDAd5/35++ vectors, corresponding plasmids were linearized with PmeI and rescued in 116 cells52 with the Ad5/35++-Acr helper vector20 as described in detail elsewhere.52 Helper virus contamination levels were found to be <0.05%. Titers were 6–12 × 1012 vp/mL.
All HDAd vectors used in this study contain chimeric fibers composed of the Ad5 fiber tail, the Ad35 fiber shaft, and the affinity-enhanced Ad35++ fiber knob.53
Mismatch-Sensitive Nuclease T7E1 Assay
gDNA was isolated as described previously.54 Genomic segments encompassing the AAVS1 target site were amplified by polymerase from Thermococcus kodakaraensis (KOD) Hot Start DNA Polymerase (Millipore Sigma, Burlington, MA) using the following primers: AAVS1 forward, 5′-ATCTCACAGGTAAAACTGACGCACGGAGGAACA-3′; reverse, 5′- CGGGTCACCTCTCACTCCTTTCATTTGGGC-3′. PCR products were hybridized and treated with 2.5 units of T7EI (New England Biolabs) for 20 min at 37°C. Digested PCR products were resolved by 6% Tris-borate-EDTA (TBE) PAGE (Bio-Rad) and stained with ethidium bromide. Band intensity was analyzed using ImageJ software. Percent cleavage = (1-sqrt(parental band/(parental band + cleaved bands))) × 100%.
Flow cytometry is described in the Supplemental Materials and Methods.
Lin− Magnetic-Activated Cell Sorting (MACS) and Ex Vivo Transduction
For depletion of lineage-committed cells, a mouse lineage cell depletion kit (Miltenyi Biotec, San Diego, CA) was used according to the manufacturer’s instructions. For ex vivo transduction, a total of 1 × 107 cells were plated in 10-cm ultra-low attachment surface dishes (Corning Life Sciences) with 7 mL of medium. HDAd vectors were added. Twenty-four hours later, cells were harvested, washed with PBS, and resuspended in PBS at 1 × 106 cells per 200 μL for transplantation.
Real-Time RT-PCR
Total RNA was extracted from 50∼100 μL blood by using TRIzol reagent (Thermo Fisher Scientific) following the manufacture’s phenol-chloroform extraction method and then reverse transcribed to generate cDNA using the Quantitect reverse transcription kit from QIAGEN. Potential gDNA contamination was eliminated by treatment of the RNA samples with gDNA wipe-out reagents provided in the kit. Comparative real-time PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems) and run on a StepOnePlus real-time PCR system (Applied Biosystems). The following primer pairs were used: mouse RPL10 (housekeeping) forward 5′-TGAAGACATGGTTGCTGAGAAG-3′ and reverse 5′-GAACGATTTGGTAGGGTATAGGAG-3′; human γ-globin forward 5′-GGGGCAAGGTGAATGTGGAAGA-3′ and reverse 5′-CATGATGGCAGAGGCAGAGGAC-3′; mouse β-major globin forward 5′-ATGCCAAAGTGAAGGCCCAT-3′ and reverse 5′-CCCAGCACAATCACGATCAT-3′.
Globin HPLC
Individual globin chain levels were quantified on a Shimadzu Prominence instrument with an SPD-10AV diode array detector and an LC-10AT binary pump (Shimadzu, Kyoto, Japan). A 38%–58% gradient mixture of 0.1% trifluoroacetic acid in water-acetonitrile was applied at a rate of 1 mL/min using a Vydac C4 reverse-phase column (Hichrom, UK).
CFU Assay
2,500 Lin− cells were plated in triplicates in ColonyGEL 1202 mouse complete medium (ReachBio, Seattle, WA) and incubated for 12 days at 37°C in 5% CO2 and maximum humidity. Colonies were enumerated using a Leica MS 5 dissection microscope (Leica Microsystems). For colonies derived from HDAd-GFP-donor-transduced mice, GFP-positive colonies were counted, picked, and analyzed. CFU assays with transduced CD34+ cells were performed using ColonyGEL 1102 human complete medium (ReachBio) according to the manufacturer’s protocol. GFP-positive colonies were counted, picked, and analyzed on day 14 after plating.
Measurement of Vector Copy Number
Total DNA from bone marrow cells or single colonies was extracted by PureLink Genomic DNA Mini Kit (Invitrogen). Viral DNA extracted from HDAd-GFP-donor or HDAd-globin-donor was serially diluted and served as a standard curve. qPCR was conducted in duplicate using Power SYBR Green PCR master mix on a StepOnePlus real-time PCR system (Applied Biosystems). 5 ng DNA was used for a 10-μL reaction. The following primer pairs were used: GFP forward 5′- TCGTGACCACCCTGACCTAC-3′ and reverse 5′-GGTCTTGTAGTTGCCGTCGT-3′; mgmt forward 5′-GCTGTCTGGTTGTGAGCAGGGTCT-3′ and reverse 5′-CGGGCTGGTGGAAATAGGCATTC-3′. Human γ-globin primers are described in Real-Time RT-PCR.
Localization of the AAVS1 Locus in AAVS1 tg Mice
TLA libraries were prepared as described previously.40 Briefly, formaldehyde-crosslinked DNA from total bone marrow cells was digested with NlaIII. After ligation and reverse crosslinking, DNA was purified. This product was further digested with NspI and ligated to obtain circular chimeric DNA of approximately 2 kb. Chimeric DNA was PCR amplified using AAVS1-specific TLA primers: forward, 5′-GGTTGTCCAGAAAAACGGTGAT-3′; reverse, 5′-CCTCTCACTCCTTTCATTTGGG-3′. TLA libraries from PCR-amplified products were prepared using the Illumina Nextera XT NGS kit according to the manufacturer’s protocol. Paired-end sequencing was performed on an Illumina NovaSeq system. The TLA protocol leads to reshuffling of DNA; thus, reads were aligned using Split-Read Aware Aligner BWA55 using settings bwasw –b 7 as suggested previously (https://github.com/Cergentis/Cergentis_common).56 The aligned bam files were converted to reads per kilobase of transcript, per million mapped reads (RPKM)-normalized bigwig files using deepTools.57. Genome-wide distribution was visualized using the WashU Epigenome Browser.58
Southern Blot
gDNA from mouse bone marrow was digested with either EcoRI or BlpI and subjected to Southern blotting with either an AAVS1- or GFP-specific probe labeled with 32P using the Prime-It RmT Random Primer labeling kit (Agilent Technologies). Non-incorporated 32P deoxycytidine triphosphate (dCTP) was removed by centrifugation through MicroSpin G25 columns (GE Healthcare). Hybridization was performed in PerfectHyb Plus hybridization buffer (Sigma). Blots were exposed to Amersham Hybond-XL films (GE Healthcare).
Inverse PCR and in and out PCR are described in the Supplemental Materials and Methods.
In Silico Prediction of Off-Target Cleavage Sites
Off-target sites of the AAVS1 guide sequence in the human or mouse genome were predicted using an online tool (https://www.sanger.ac.uk/htgt/wge/find_off_targets_by_seq).
Animal Studies
All experiments involving animals were conducted in accordance with the institutional guidelines set forth by the University of Washington. The University of Washington is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AALAC), and all live animal work conducted at this university is in accordance with the Office of Laboratory Animal Welfare (OLAW) Public Health Assurance (PHS) policy, United States Department of Agriculture (USDA) Animal Welfare Act and Regulations, the Guide for the Care and Use of Laboratory Animals, and University of Washington’s Institutional Animal Care and Use Committee (IACUC) policies. The studies were approved by the University of Washington IACUC (protocol 3108-01). Mice were housed in specific pathogen-free facilities. AAVS1 tg mice (C3;B6-Tg(AAVS1)A1Xob/J) (The Jackson Laboratory) were recovered from cryopreserved embryos of mice developed by Xandra O. Breakefield.32 Mice were hemizygous for the human AAVS1 locus. AAVS1 tg mice were crossed with human CD46+/+ mice to obtain AAVS1+/−/CD46+/− mice for ex vivo studies and AAVS1+/−/CD46+/+ mice for in vivo HSC transduction studies. The following primers were used for genotyping of CD46 mice: forward, 5′-GCCAGTTCATCTTTTGACTCTATTAA-3′; reverse, 5′-AATCACAGCAATGACCCAAA-3′. Mice homozygous or heterozygous for CD46 were identified by different intensities of CD46 expression on PBMCs, detected by flow cytometry. Genotyping of the AAVS1 transgene was performed by PCR according to The Jackson Laboratory’s recommended protocol.
Bone Marrow Lin− Cell Transplantation
Recipients were female C57BL/6 mice, 6–8 weeks old. On the day of transplantation, recipient mice were irradiated with 1,000 rad. Four hours after irradiation, 1 × 106 Lin− cells were injected intravenously through the tail vein. This protocol was used for transplantation of ex vivo-transduced Lin− cells and for transplantation into secondary recipients.
HSC Mobilization and In Vivo Transduction
This procedure has been described previously.22 Briefly, HSCs were mobilized in mice by subcutaneous (s.c.) injections of human recombinant G-CSF (5 μg/mouse/day, 4 days) (Amgen, Thousand Oaks, CA), followed by an s.c. injection of AMD3100 (5 mg/kg) (Sigma-Aldrich) on day 5. In addition, animals received dexamethasone (10 mg/kg) intraperitoneally (i.p.) 16 h and 2 h before virus injection. Thirty and 60 min after AMD3100, animals were injected intravenously with HDAd-CRISPR and HDAd-GFP-donor or HDAd-globin-donor through the retro-orbital plexus with a dose of 4 × 1010 vp for each virus per injection. Four weeks later, mice were injected with O6-BG (15 mg/kg, i.p.) twice, 30 min apart. One hour after the second injection of O6-BG, mice were injected with BCNU (5 mg/kg, i.p.). The BCNU dose was increased to 7.5 and 10 mg/kg in the second and third cycle, respectively. Both BCNU and O6-BG were from Sigma-Aldrich.
Statistical Analyses
For comparisons of multiple groups, one-way and two-way ANOVA with Bonferroni post-testing for multiple comparisons was employed. Statistical analysis was performed using GraphPad Prism version 6.01 (GraphPad, La Jolla, CA).
Author Contributions
A.L. provided the conceptual framework for the study. C.L. and A.L. designed the experiments. C.L., A.S.M., S.G., M.W., and A.G. performed the experiments. T.P. and R.D.H. provided critical comments on the manuscript. A.L. wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Nikoletta Psatha for helpful advice and technical help. We thank Pavel Sova for support with next-generation sequencing. The study was supported by grants R21CA193077, R01HL128288, and R01HL141781; a grant from the Bill and Melinda Gates Foundation (to A.L.); and grant R01 DK 101328 (to T.P.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.08.006.
Supplemental Information
References
- 1.Naldini L. Genetic engineering of hematopoiesis: current stage of clinical translation and future perspectives. EMBO Mol. Med. 2019;11:e9958. doi: 10.15252/emmm.201809958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang G.P., Ciuffi A., Leipzig J., Berry C.C., Bushman F.D. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papapetrou E.P., Lee G., Malani N., Setty M., Riviere I., Tirunagari L.M., Kadota K., Roth S.L., Giardina P., Viale A. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 5.Hüser D., Gogol-Döring A., Lutter T., Weger S., Winter K., Hammer E.M., Cathomen T., Reinert K., Heilbronn R. Integration preferences of wildtype AAV-2 for consensus rep-binding sites at numerous loci in the human genome. PLoS Pathog. 2010;6:e1000985. doi: 10.1371/journal.ppat.1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henckaerts E., Linden R.M. Adeno-associated virus: a key to the human genome? Future Virol. 2010;5:555–574. doi: 10.2217/fvl.10.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rensburg R., Beyer I., Yao X.Y., Wang H., Denisenko O., Li Z.Y., Russell D.W., Miller D.G., Gregory P., Holmes M. Chromatin structure of two genomic sites for targeted transgene integration in induced pluripotent stem cells and hematopoietic stem cells. Gene Ther. 2013;20:201–214. doi: 10.1038/gt.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombardo A., Cesana D., Genovese P., Di Stefano B., Provasi E., Colombo D.F., Neri M., Magnani Z., Cantore A., Lo Riso P. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 9.Ogata T., Kozuka T., Kanda T. Identification of an insulator in AAVS1, a preferred region for integration of adeno-associated virus DNA. J. Virol. 2003;77:9000–9007. doi: 10.1128/JVI.77.16.9000-9007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voigt K., Izsvák Z., Ivics Z. Targeted gene insertion for molecular medicine. J. Mol. Med. (Berl.) 2008;86:1205–1219. doi: 10.1007/s00109-008-0381-8. [DOI] [PubMed] [Google Scholar]
- 11.Lombardo A., Naldini L. Genome editing: a tool for research and therapy: targeted genome editing hits the clinic. Nat. Med. 2014;20:1101–1103. doi: 10.1038/nm.3721. [DOI] [PubMed] [Google Scholar]
- 12.Blair J.D., Bateup H.S., Hockemeyer D.F. Establishment of Genome-edited Human Pluripotent Stem Cell Lines: From Targeting to Isolation. J. Vis. Exp. 2016;108:e53583. doi: 10.3791/53583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyer A.K., Hoffmann D., Lachmann N., Ackermann M., Steinemann D., Timm B., Siler U., Reichenbach J., Grez M., Moritz T. TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells. Biomaterials. 2015;69:191–200. doi: 10.1016/j.biomaterials.2015.07.057. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn A., Ackermann M., Mussolino C., Cathomen T., Lachmann N., Moritz T. TALEN-mediated functional correction of human iPSC-derived macrophages in context of hereditary pulmonary alveolar proteinosis. Sci. Rep. 2017;7:15195. doi: 10.1038/s41598-017-14566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S.J., Luo Y., Zhang L.M., Yang W., Zhang G.G. Targeted introduction and effective expression of hFIX at the AAVS1 locus in mesenchymal stem cells. Mol. Med. Rep. 2017;15:1313–1318. doi: 10.3892/mmr.2017.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rio P., Baños R., Lombardo A., Quintana-Bustamante O., Alvarez L., Garate Z., Genovese P., Almarza E., Valeri A., Díez B. Targeted gene therapy and cell reprogramming in Fanconi anemia. EMBO Mol. Med. 2014;6:835–848. doi: 10.15252/emmm.201303374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Ravin S.S., Reik A., Liu P.Q., Li L., Wu X., Su L., Raley C., Theobald N., Choi U., Song A.H. Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 2016;34:424–429. doi: 10.1038/nbt.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung K.L., Meitlis I., Hale M., Chen C.Y., Singh S., Jackson S.W., Miao C.H., Khan I.F., Rawlings D.J., James R.G. Engineering Protein-Secreting Plasma Cells by Homology-Directed Repair in Primary Human B Cells. Mol. Ther. 2018;26:456–467. doi: 10.1016/j.ymthe.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson M.J., Laoharawee K., Lahr W.S., Webber B.R., Moriarity B.S. Engineering of Primary Human B cells with CRISPR/Cas9 Targeted Nuclease. Sci. Rep. 2018;8:12144. doi: 10.1038/s41598-018-30358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C., Psatha N., Sova P., Gil S., Wang H., Kim J., Kulkarni C., Valensisi C., Hawkins R.D., Stamatoyannopoulos G., Lieber A. Reactivation of γ-globin in adult β-YAC mice after ex vivo and in vivo hematopoietic stem cell genome editing. Blood. 2018;131:2915–2928. doi: 10.1182/blood-2018-03-838540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saydaminova K., Ye X., Wang H., Richter M., Ho M., Chen H., Xu N., Kim J.S., Papapetrou E., Holmes M.C. Efficient genome editing in hematopoietic stem cells with helper-dependent Ad5/35 vectors expressing site-specific endonucleases under microRNA regulation. Mol. Ther. Methods Clin. Dev. 2015;1:14057. doi: 10.1038/mtm.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter M., Saydaminova K., Yumul R., Krishnan R., Liu J., Nagy E.E., Singh M., Izsvák Z., Cattaneo R., Uckert W. In vivo transduction of primitive mobilized hematopoietic stem cells after intravenous injection of integrating adenovirus vectors. Blood. 2016;128:2206–2217. doi: 10.1182/blood-2016-04-711580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balamotis M.A., Huang K., Mitani K. Efficient delivery and stable gene expression in a hematopoietic cell line using a chimeric serotype 35 fiber pseudotyped helper-dependent adenoviral vector. Virology. 2004;324:229–237. doi: 10.1016/j.virol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 24.Ohbayashi F., Balamotis M.A., Kishimoto A., Aizawa E., Diaz A., Hasty P., Graham F.L., Caskey C.T., Mitani K. Correction of chromosomal mutation and random integration in embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl. Acad. Sci. USA. 2005;102:13628–13633. doi: 10.1073/pnas.0506598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki K., Mitsui K., Aizawa E., Hasegawa K., Kawase E., Yamagishi T., Shimizu Y., Suemori H., Nakatsuji N., Mitani K. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl. Acad. Sci. USA. 2008;105:13781–13786. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 27.Li K., Wang G., Andersen T., Zhou P., Pu W.T. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS ONE. 2014;9:e105779. doi: 10.1371/journal.pone.0105779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Georgakopoulou A., Psatha N., Li C., Capsali C., Samal H.B., Anagnostopoulos A., Ehrhardt A., Izsvák Z., Papayannopoulou T. In vivo hematopoietic stem cell gene therapy ameliorates murine thalassemia intermedia. J. Clin. Invest. 2019;129:598–615. doi: 10.1172/JCI122836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samulski R.J., Zhu X., Xiao X., Brook J.D., Housman D.E., Epstein N., Hunter L.A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzuto G., Gorgoni B., Cappelletti M., Lazzaro D., Gloaguen I., Poli V., Sgura A., Cimini D., Ciliberto G., Cortese R. Development of animal models for adeno-associated virus site-specific integration. J. Virol. 1999;73:2517–2526. doi: 10.1128/jvi.73.3.2517-2526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young S.M., Jr., McCarty D.M., Degtyareva N., Samulski R.J. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 2000;74:3953–3966. doi: 10.1128/jvi.74.9.3953-3966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakowska J.C., Di Maria M.V., Camp S.M., Wang Y., Allen P.D., Breakefield X.O. Targeted transgene integration into transgenic mouse fibroblasts carrying the full-length human AAVS1 locus mediated by HSV/AAV rep(+) hybrid amplicon vector. Gene Ther. 2003;10:1691–1702. doi: 10.1038/sj.gt.3302061. [DOI] [PubMed] [Google Scholar]
- 33.Kemper C., Leung M., Stephensen C.B., Pinkert C.A., Liszewski M.K., Cattaneo R., Atkinson J.P. Membrane cofactor protein (MCP; CD46) expression in transgenic mice. Clin. Exp. Immunol. 2001;124:180–189. doi: 10.1046/j.1365-2249.2001.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shenk T. Adenoviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 35.Cristea S., Freyvert Y., Santiago Y., Holmes M.C., Urnov F.D., Gregory P.D., Cost G.J. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol. Bioeng. 2013;110:871–880. doi: 10.1002/bit.24733. [DOI] [PubMed] [Google Scholar]
- 36.Kurita R., Suda N., Sudo K., Miharada K., Hiroyama T., Miyoshi H., Tani K., Nakamura Y. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS ONE. 2013;8:e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boehme P., Zhang W., Solanki M., Ehrke-Schulz E., Ehrhardt A. A High-Capacity Adenoviral Hybrid Vector System Utilizing the Hyperactive Sleeping Beauty Transposase SB100X for Enhanced Integration. Mol. Ther. Nucleic Acids. 2016;5:e337. doi: 10.1038/mtna.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C., Psatha N., Gil S., Wang H., Papayannopoulou T., Lieber A. HDAd5/35++ Adenovirus Vector Expressing Anti-CRISPR Peptides Decreases CRISPR/Cas9 Toxicity in Human Hematopoietic Stem Cells. Mol. Ther. Methods Clin. Dev. 2018;9:390–401. doi: 10.1016/j.omtm.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C., Psatha N., Wang H., Singh M., Samal H.B., Zhang W., Ehrhardt A., Izsvák Z., Papayannopoulou T., Lieber A. Integrating HDAd5/35++ Vectors as a New Platform for HSC Gene Therapy of Hemoglobinopathies. Mol. Ther. Methods Clin. Dev. 2018;9:142–152. doi: 10.1016/j.omtm.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vree P.J., de Wit E., Yilmaz M., van de Heijning M., Klous P., Verstegen M.J., Wan Y., Teunissen H., Krijger P.H., Geeven G. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat. Biotechnol. 2014;32:1019–1025. doi: 10.1038/nbt.2959. [DOI] [PubMed] [Google Scholar]
- 41.Espinoza D.A., Fan X., Yang D., Cordes S.F., Truitt L.L., Calvo K.R., Yabe I.M., Demirci S., Hope K.J., Hong S.G. Aberrant Clonal Hematopoiesis following Lentiviral Vector Transduction of HSPCs in a Rhesus Macaque. Mol. Ther. 2019;27:1074–1086. doi: 10.1016/j.ymthe.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin R.M., Ikeda K., Cromer M.K., Uchida N., Nishimura T., Romano R., Tong A.J., Lemgart V.T., Camarena J., Pavel-Dinu M. Highly Efficient and Marker-free Genome Editing of Human Pluripotent Stem Cells by CRISPR-Cas9 RNP and AAV6 Donor-Mediated Homologous Recombination. Cell Stem Cell. 2019;24:821–828.5. doi: 10.1016/j.stem.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Wang H., Richter M., Psatha N., Li C., Kim J., Liu J., Ehrhardt A., Nilsson S.K., Cao B., Palmer D. A Combined In Vivo HSC Transduction/Selection Approach Results in Efficient and Stable Gene Expression in Peripheral Blood Cells in Mice. Mol. Ther. Methods Clin. Dev. 2017;8:52–64. doi: 10.1016/j.omtm.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Psatha N., Sgouramali E., Gkountis A., Siametis A., Baliakas P., Constantinou V., Athanasiou E., Arsenakis M., Anagnostopoulos A., Papayannopoulou T. Superior long-term repopulating capacity of G-CSF+plerixafor-mobilized blood: implications for stem cell gene therapy by studies in the Hbb(th-3) mouse model. Hum. Gene Ther. Methods. 2014;25:317–327. doi: 10.1089/hgtb.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H., Shayakhmetov D.M., Leege T., Harkey M., Li Q., Papayannopoulou T., Stamatoyannopolous G., Lieber A. A capsid-modified helper-dependent adenovirus vector containing the beta-globin locus control region displays a nonrandom integration pattern and allows stable, erythroid-specific gene expression. J. Virol. 2005;79:10999–11013. doi: 10.1128/JVI.79.17.10999-11013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Lieber A. A helper-dependent capsid-modified adenovirus vector expressing adeno-associated virus rep78 mediates site-specific integration of a 27-kilobase transgene cassette. J. Virol. 2006;80:11699–11709. doi: 10.1128/JVI.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linden R.M., Winocour E., Berns K.I. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu L.C., Sun C.W., Ryan T.M., Pawlik K.M., Ren J., Townes T.M. Correction of sickle cell disease by homologous recombination in embryonic stem cells. Blood. 2006;108:1183–1188. doi: 10.1182/blood-2006-02-004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Canver M.C., Smith E.C., Sher F., Pinello L., Sanjana N.E., Shalem O., Chen D.D., Schupp P.G., Vinjamur D.S., Garcia S.P. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmer D., Ng P. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 53.Wang H., Liu Y., Li Z., Tuve S., Stone D., Kalyushniy O., Shayakhmetov D., Verlinde C.L., Stehle T., McVey J. In vitro and in vivo properties of adenovirus vectors with increased affinity to CD46. J. Virol. 2008;82:10567–10579. doi: 10.1128/JVI.01308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller J.C., Holmes M.C., Wang J., Guschin D.Y., Lee Y.L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 55.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cain-Hom C., Splinter E., van Min M., Simonis M., van de Heijning M., Martinez M., Asghari V., Cox J.C., Warming S. Efficient mapping of transgene integration sites and local structural changes in Cre transgenic mice using targeted locus amplification. Nucleic Acids Res. 2017;45:e62. doi: 10.1093/nar/gkw1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku365. W187-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou X., Maricque B., Xie M., Li D., Sundaram V., Martin E.A., Koebbe B.C., Nielsen C., Hirst M., Farnham P. The Human Epigenome Browser at Washington University. Nat. Methods. 2011;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lisowski L., Sadelain M. Locus control region elements HS1 and HS4 enhance the therapeutic efficacy of globin gene transfer in beta-thalassemic mice. Blood. 2007;110:4175–4178. doi: 10.1182/blood-2007-08-108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emery D.W., Yannaki E., Tubb J., Stamatoyannopoulos G. A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc. Natl. Acad. Sci. USA. 2000;97:9150–9155. doi: 10.1073/pnas.160159597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.