Abstract
Limb girdle muscular dystrophy type 2A (LGMD2A), caused by mutations in the Calpain 3 (CAPN3) gene, is an incurable autosomal recessive disorder that results in muscle wasting and loss of ambulation. To test the feasibility of an autologous induced pluripotent stem cell (iPSC)-based therapy for LGMD2A, here we applied CRISPR-Cas9-mediated genome editing to iPSCs from three LGMD2A patients to enable correction of mutations in the CAPN3 gene. Using a gene knockin approach, we genome edited iPSCs carrying three different CAPN3 mutations, and we demonstrated the rescue of CAPN3 protein in myotube derivatives in vitro. Transplantation of gene-corrected LGMD2A myogenic progenitors in a novel mouse model combining immunodeficiency and a lack of CAPN3 resulted in muscle engraftment and rescue of the CAPN3 mRNA. Thus, we provide here proof of concept for the integration of genome editing and iPSC technologies to develop a novel autologous cell therapy for LGMD2A.
Keywords: limb girdle muscular dystrophy type 2A, Calpain 3, CAPN3, gene correction, genome editing, induced pluripotent stem cells, iPSCs, myogenic progenitors, autologous cell therapy
Selvaraj et al. show that three different CAPN3 mutations can be corrected in LGMD2A patient-specific iPSCs using a single CRISPR-Cas9-based genome-editing strategy. This gene correction rescues CAPN3 expression in the gene-corrected iPSC derivatives both in vitro and in vivo.
Introduction
Limb girdle muscular dystrophy type 2A (LGMD2A) is an autosomal recessive inherited disorder and the most common form of LGMD.1, 2, 3 Major symptoms are symmetrical and progressive muscle wasting of pelvic and scapular musculature and joint contractures.4 Clinical presentation for LGMD2A varies from severe forms with early onset (as early as 2 years of age) and rapid progression to milder forms with late onset and slower development.1, 5, 6 Different from most muscular dystrophies (MDs) caused by mutations in structural genes, LGMD2A occurs due to loss of functional Calpain 3 (CAPN3), a skeletal muscle-specific isoform of the calcium-sensitive Calpain cysteine protease family.7
CAPN3 was the first non-structural protein linked to MD pathogenesis.7 To date, more than 400 different mutations across the full-length of the CAPN3 gene have been described to be associated with LGMD2A.8 However, the mechanism of pathogenicity underlining LGMD2A is currently not well understood, as the role of CAPN3 remains unclear. One of the existing hypotheses is that CAPN3 may be involved in the assembly and maintenance of sarcomere,9, 10, 11 where CAPN3 directly binds to the structural and scaffold protein titin, changing its localization from the M-lines to the N2 regions as the sarcomere extends.12 Recent studies using CAPN3 mutant and knockout (KO) mouse models provided evidence that CAPN3 is involved in Ca2+ release from the sarcoplasmic reticulum in response to muscle loading,13, 14 which may be the basis for muscle wasting in the context of mutated CAPN3.
Similar to other MD disorders, there is no cure for LGMD2A. Most of current research focuses on the use of adeno-associated virus (AAV) vectors to deliver the CAPN3 gene. Studies using AAV vector-based CAPN3 gene delivery have demonstrated improvement in CAPN3-KO mice, but concerns remain over long-term safety and efficacy.15, 16 One caveat for the use of AAV for this disease is that CAPN3 gene expression is strictly regulated, and, accordingly, leakage of CAPN3 expression in the heart was reported to cause severe toxicity.15, 16 Therefore, current efforts in LGMD2A gene therapy are focused at delivering and maintaining CAPN3 gene expression restricted to skeletal muscle.
Stem cell-based therapies are particularly attractive for MDs since diseased muscle tissue would be replaced with stem cells that can give rise to healthy muscle as well as self-renew. Using conditional expression of PAX7 in human pluripotent stem cells, we have documented long-term engraftment of human skeletal myogenic progenitor cells upon their transplantation into dystrophic mice.17, 18 Whereas either allogeneic or autologous cell transplantation has the potential to lead to an effective treatment in LGMD2A, the autologous approach, which requires in vitro genetic correction of induced pluripotent stem cells (iPSCs) prior to transplantation, is more desirable, as it would circumvent issues associated with immune rejection. Here we show the rescue of CAPN3 expression upon gene editing of LGMD2A iPSCs, providing proof of principle for the potential future therapeutic application of an autologous cell transplantation for LGMD2A patients.
Results
Characterization of LGMD2A iPSCs and Myotube Derivatives
We reprogrammed fibroblast samples from three LGMD2A patients (9015, 0826, and 0989), with three different CAPN3 mutations (Figure 1A), into iPSCs using an integration-free approach.19 Generated iPSC lines express pluripotency markers, display normal karyotype, and in vivo develop teratomas containing cell types from all three germ layers (Figures S1A–S1C). We have previously documented the pluripotency characterization of LGMD2A 9015 iPSCs.18
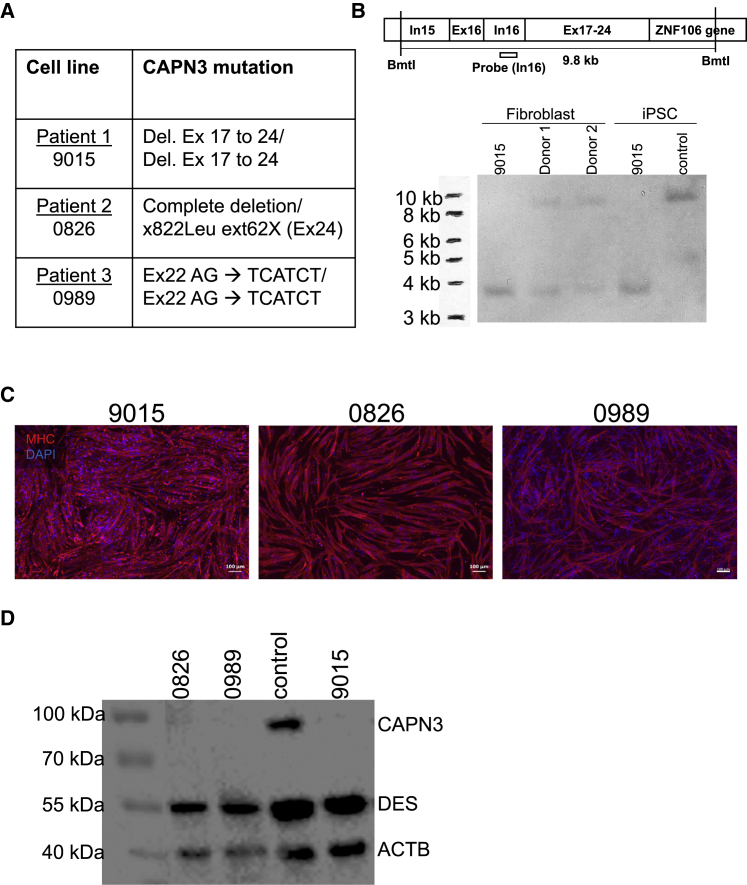
Figure 1.
Characterization of LGMD2A iPSCs and Respective Muscle Derivatives
(A) List of LGMD2A patient-specific fibroblast samples and their respective CAPN3 mutations. (B) Southern blot for detection of genomic DNA deletion in 9015 LGMD2A iPSCs. This individual’s parent fibroblasts (donors 1 and 2) and control unaffected iPSCs were used as reference. Upper panel shows scheme of the genomic DNA region analyzed and the location of the probe (intron 16). (C) Representative images show staining for MHC (in red) in LGMD2A iPSC-derived myotubes. DAPI stains nuclei (blue). Scale bars: 100 μm. (D) Western blot for CAPN3 in myotubes from patient-specific LGMD2A and control iPSCs. DES (Desmin) and ACTB were used as the differentiation and loading controls, respectively.
Because patient 1 (9015) had been identified homozygous for deletion at exons 17–24, we confirmed the extent of deletion by Southern blot in both fibroblasts and respective iPSCs (Figure 1B). Fibroblasts from this individual’s parents (donors 1 and 2) carried the same deletion in heterozygous state (Figure 1B). Based on these data, we designed a gene-editing approach able to correct mutations in all three iPSC lines, as they all have mutations in exons downstream of exon 15 (Figure 1A).
Before performing gene correction of LGMD2A iPSCs, we differentiated these cells into myotubes to confirm their absence of CAPN3. Of relevance to these studies, Fougerousse and colleagues have reported that CAPN3 protein expression becomes detectable only at later stages of human development (from 8 weeks).20 Consistent with this, we were unable to detect CAPN3 in control iPSC-derived myotubes using our standard differentiation conditions,17, 18 as immature myotubes do not express CAPN3 (Figures S2A and S2B). Of note, CAPN3 is easily detectable in human myotubes from adult myoblasts.21 To address this, we performed an unbiased screening using a small molecule library, which identified four small molecule compounds that enhance the differentiation and maturation of iPSC-derived myotubes (S.S. and R.M.-G., unpublished data). Exposure to this cocktail of inhibitors resulted in thicker myotubes that displayed enhanced fusion (Figure S2A, lower panel), and, most importantly, expressed CAPN3 protein (Figure S2B), demonstrating that this protocol generates more mature myotubes. This was validated in samples from multiple PSC lines (Figure S2B) using an antibody that recognizes the N-terminal epitope of CAPN3 (NCL-CALP-2C4).22 Subsequently, we generated LGMD2A iPSC-derived myotubes (Figure 1C) using conditional expression of PAX7,17, 18 followed by the optimized differentiation protocol discussed above. Western blot analysis confirmed that all three LGMD2A samples lacked expression of the CAPN3 protein (Figure 1D). This is in accordance with a previous study showing complete lack of CAPN3 protein expression in LGMD2A muscle biopsies for various CAPN3 mutations, including point mutations.23
To further validate our western blot results, we generated a CAPN3-KO isogenic iPSC line using CRISPR-Cas9-based deletion of exon 1 (Figure S2C) in control (PLZ) iPSCs. Because PCR of genomic DNA showed heterogeneity in the prospective CAPN3-KO clone (KO-C11) (Figure S2D), we sub-cloned KO-C11 cells to obtain two homogeneous sub-clones (KO-C11-7 and KO-C11-17) (Figure S2D). Staining for Myosin-Heavy-Chain (MHC) showed that both CAPN3-KO clones (C11-7 and C11-17) differentiated efficiently into myotubes in vitro (Figure S2E), and, as expected, they lacked CAPN3 at both RNA and protein levels (Figures S2F and S2G, respectively), whereas parental counterparts expressed CAPN3 abundantly. Taken together, these data confirm our ability to reliably detect CAPN3 in iPSC-derived myotubes.
Gene Correction Rescues CAPN3 Protein Expression in LGMD2A iPSC-Derived Myotubes
Having validated the absence of CAPN3 in LGMDA iPSC-derived myotubes, we proceeded to correct CAPN3 mutations in these LGMD2A iPSC lines using a knockin approach. Since all three iPSC lines have mutations downstream of exon 15 in the CAPN3 gene, we targeted exon 14 for the knocking in of cDNA encoding exons 15–24 to circumvent the mutations downstream of exon 14. As shown in Figure 2A, our approach consisted of creating a double-stranded break in exon 14 using CRISPR-Cas9, followed by homology-directed repair (HDR) from an exogenous donor vector to enable gene knockin. Donor vector contained exons 15–24, an SV40 poly(A) signal sequence, and a selection cassette. Following transfection with CRISPR-Cas9 and HDR donor plasmids, 9015 iPSCs were cultured with G418 to select for neomycin-resistant iPSC clones, which represent clones that are positive for insertion. To select against non-specific integration, we included the thymidine kinase gene under a constitutive promoter in the backbone of the donor vector, and we selected cells with ganciclovir. iPSC colonies that survived this double selection were expanded into cell lines.
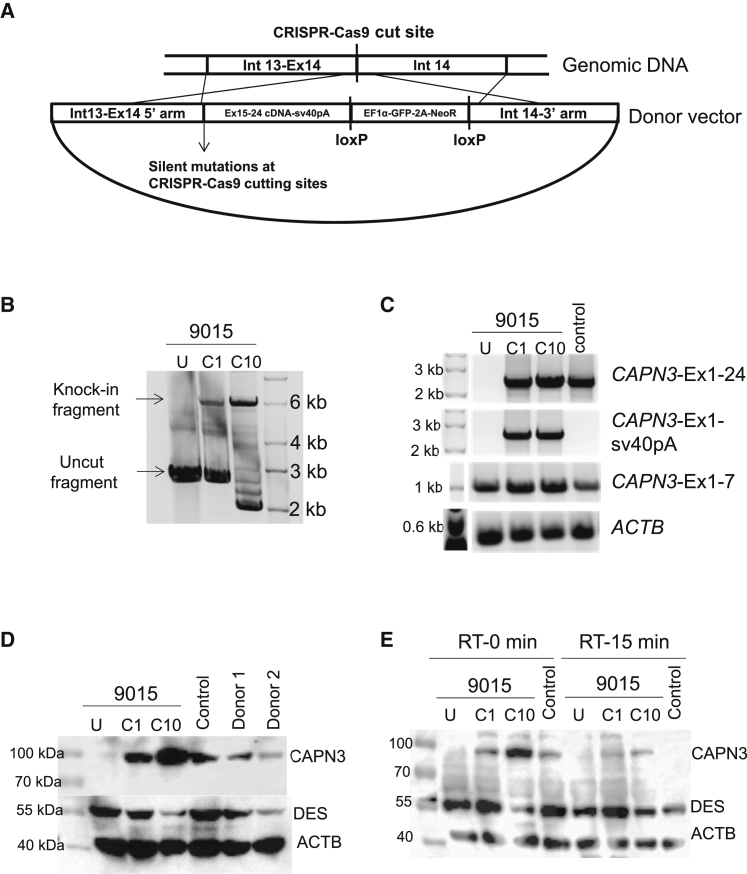
Figure 2.
Gene Correction of CAPN3 Mutation in 9015 LGMD2A iPSCs
(A) Schematic of the homology-directed repair strategy utilized for gene knockin-based correction of CAPN3 mutations. (B) PCR shows amplification of the region spanning the knockin in genomic DNA from uncorrected (U) and gene-corrected (C1 and C10) LGMD2A iPSCs. (C) RT-PCR analysis of gene-corrected and uncorrected 9015 iPSC-derived myotubes as well as of unaffected myotubes (control). Upper panel shows amplification of full-length CAPN3 (Ex1-24). The second panel shows amplification of CAPN3 exon 1 to sv40pA sequence, which is specific to the insert. The third panel denotes amplification of CAPN3 exons 1–7, which is present in all samples. The lower panel indicates ACTB used as loading control. (D) Western blot shows rescue of CAPN3 protein expression in gene-corrected (C1 and C10) LGMD2A iPSC-derived myotubes, whereas uncorrected counterparts are absent. The patient’s parents and unrelated control iPSC-derived myotubes were used as reference. (E) Autocatalytic activity of CAPN3 is shown by western blot analysis on lysates obtained from myotubes incubated for 0 or 15 min at room temperature. DES and ACTB were used as the differentiation and loading controls respectively.
PCR amplification of the region spanning the knockin on two presumptive corrected iPSC clones (C1 and C10) showed the amplification of an approximately 6-kb fragment, indicating the gene knockin (Figure 2B). We then differentiated these two clones into MHC+ myotubes (Figure S3A). Gene expression analysis of gene-corrected and uncorrected myotubes showed that only gene-corrected myotubes (C1 and C10) expressed full-length CAPN3 mRNA (exons 1–24), indicating the rescue of CAPN3 expression (Figure 2C). In addition, we confirmed that rescued CAPN3 mRNA expression was specifically due to the gene knockin, as shown by the amplification of full-length CAPN3 with knockin-specific primers (exon 1 to sv40 pA; Figure 2C). As expected, 9015 uncorrected iPSC-derived myotubes did express correctly spliced CAPN3 exons 1–7, which were unaffected by this mutation (Figure 2C). Most importantly, western blot confirmed the rescue of CAPN3 protein expression in corrected clones, corroborating the efficacy of this genome-editing approach (Figure 2D). Because genome editing with CRISPR-Cas9 ribonucleoprotein (RNP) has been reported to be highly specific and efficient,24, 25 we also assessed whether CRISPR-Cas9 RNP could be used as an alternative to plasmid. As shown in Figure S3B, CRISPR-Cas9 RNP also resulted in the gene knockin of 9015 iPSCs, and this led to the rescue of CAPN3 protein in the resulting myotubes (Figure S3C).
To further confirm that the band detected by western blot was specific to expression from the knockin alleles, we performed small hairpin RNA (shRNA) knockdown using an shRNA construct that targets the wild-type (WT) 3′ UTR (Figures S3D–S3F). As anticipated, we found that the shRNA targeting the 3′ UTR of CAPN3 (C3-95) could only knock down the CAPN3 protein expression of control iPSC-derived myotubes, but not of gene-corrected LGMD2A counterparts (Figures S3D–S3F), indicating that the detected CAPN3 protein was specific to the knockin sequence since the 3′ UTR was supplanted by the sv40 poly(A) signal sequence. Meanwhile, the other two shRNA tested (C3-94 and C3-96), targeting exons 1 and 22, respectively, led to CAPN3 knockdown in all three samples, further confirming that the detected band is indeed that of CAPN3 protein (Figures S3D–S3F). Taking advantage of the distinctive autocatalytic activity of CAPN3 protein26 upon protein extraction and in vitro incubation under non-denaturing conditions, we assessed autocatalytic activity in gene-edited iPSC-derived myotubes, along with uncorrected and unaffected control counterparts. As shown in Figure 2E, our data clearly showed reduced detection of CAPN3 protein in both control and gene-edited iPSC-derived myotubes upon 15-min exposure to room temperature, indicating that the detected protein was autocatalytically active.
Next, we assessed whether this same gene-editing approach would effectively correct 0826 and 0989 LGMD2A iPSCs, which have mutations in exons 24 and 22, respectively. Following this approach, we obtained two presumptive gene-corrected clones for 0826 (C5 and C10) and 0989 (C1 and C3) LGMD2A iPSCs, which were positive for the gene knockin, as indicated by PCR amplification of the region spanning the knockin (Figures 3A and 3B). When subjected to terminal differentiation, gene-corrected iPSC clones gave rise to MHC+ myotubes (Figure 3C). Consistently, RT-PCR analysis of gene-corrected myotubes showed the expression of CAPN3 mRNA specifically from the gene knockin sequence (Figure 3D), and western blot confirmed the rescue of CAPN3 protein expression (Figure 3E). Similar to the gene-edited 9015 sample (Figure 2E), CAPN3 protein in gene-corrected 0826 and 0989 iPSC-derived myotubes was shown to be autocatalytically active (Figures S4A and S4B).
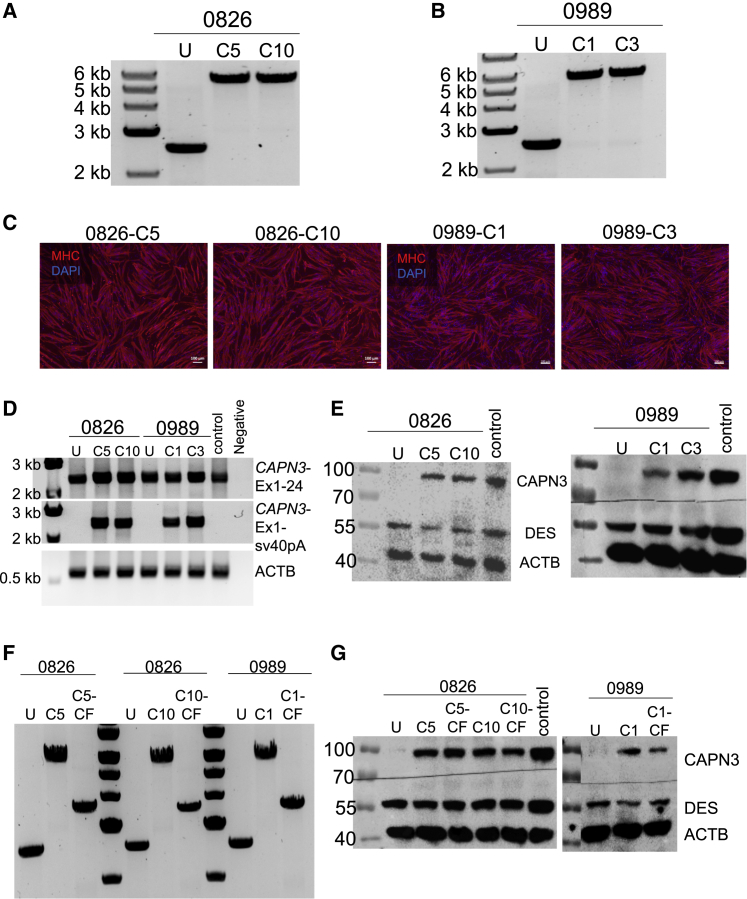
Figure 3.
Gene Correction of 0826 and 0989 iPSCs
(A and B) PCR shows amplification of the region spanning the knockin in genomic DNA from uncorrected (U) and corrected clones for samples 0826 (C5, C10) (A) and 0989 (C1, C3) (B). (C) Representative images show MHC (in red) staining in gene-corrected 0826 (C5, C10) and 0989 (C1, C3) iPSC-derived myotubes. DAPI stains nuclei (in blue). Scale bars: 100 μm. (D) RT-PCR analysis of gene-corrected and uncorrected 0826 and 0989 iPSC-derived myotubes. Upper panel shows amplification of full-length CAPN3 (Ex1-24). The second panel shows amplification of CAPN3 exon 1 to sv40pA sequence, which is specific to the insert. The lower panel indicates ACTB used as loading control. (E) Western blot shows CAPN3 protein expression in gene-corrected 0826 (left) and 0989 (right) iPSC-derived myotubes. (F) PCR shows amplification of the region spanning the knockin in genomic DNA from gene-corrected iPSCs post-excision of the selection cassette (cassette-free, CF). (G) Western blot shows CAPN3 protein expression in cassette-free gene-corrected 0826 and 0989 iPSC-derived myotubes.
When considering future potential therapeutic applications for this approach, excision of the selection cassette is important for the elimination of GFP and Neomycin resistance transgenes. Using Cre recombinase, we successfully excised the selection cassette in gene-corrected 0826 and 0989 iPSCs, as indicated by the PCR amplification of the region spanning the knockin (Figure 3F). Importantly, western blot for CAPN3 in the cassette-free gene-edited LGMD2A iPSC-derived myotubes confirmed that the expression of CAPN3 protein remained unaffected upon excision of the cassette (Figure 3G).
Minimal Off-Target Effects
If one considers utilizing iPSC derivatives combined with CRISPR-Cas9-based genome editing for therapeutic applications, it is important to evaluate the karyotypic stability and potential off-target effects. We confirmed that genome editing did not affect the karyotype of gene-edited 9015 iPSCs (C10) (Figure S5). We then used the Off-Spotter software27 to identify the top 5 predicted off-target sites for the guide RNA that was used to target exon 14 of the CAPN3 gene. We analyzed the off-target effects in the gene-corrected iPSC clones from 9015, 0826, and 0989, as well as in the bulk population from 0989. The genomic regions of these predicted off-targets were amplified by PCR in the corrected iPSCs, in addition to uncorrected counterparts. PCR amplicons were then sequenced for evaluation of the off-target effects. Sequencing chromatograms were analyzed by tracking of indels by decomposition (TIDE)28 and inference of CRISPR edits (ICE)29 programs to determine the percentage of off-target mutations. These analyses showed that the predicted off-target percentage was quite low (less than 6%) in each of the five analyzed sites in all cell lines. These results are summarized in Figures S6A and S6B. Taken together, these results suggest that the CRISPR-Cas9-based genome-editing approach applied to LGMD2A iPSCs in this study is safe and potentially suitable for therapeutic applications.
Generation of an Immunodeficient CAPN3-KO Mouse Model
To assess CAPN3 correction in vivo, we generated an immunodeficient CAPN3-KO (C3KO) mouse model by recombining C3KO30 with NOD/SCID and interleukin-2 (IL-2) receptor gamma mutations.31, 32, 33 This model provides an ideal environment for the transplantation of human cells (xenogeneic). Upon obtaining homozygous C3KO-NSG mice, we performed fluorescence-activated cell sorting (FACS) analysis on the peripheral blood of these mice, using specific antibodies, to assess the presence of B (CD19), T (CD3ε), and natural killer (NK) (double positive for CD49b and NK1.1) cells.34 This analysis confirmed the lack of T, B, and NK cells in C3KO-NSG mice (Figure S7A). Moreover, using three different monoclonal antibodies to CAPN3 (E6, 12A2, and IS2), we confirmed the lack of CAPN3 protein expression in the muscles of these mice (Figure S7B). H&E and Masson’s trichrome staining of muscle sections from both C3KO and C3KO-NSG mice showed only minor signs of fibrosis (Figures S7C and S7D), as previously documented for this mouse model.30 Quantification of collagen deposition using Masson’s trichrome staining showed no differences between C3KO and C3KO-NSG mice (Figure S7E).
Detection of CAPN3 Rescue upon Transplantation
To test the rescue of CAPN3 expression in vivo, we transplanted gene-corrected iPSC-derived myogenic progenitors into cardiotoxin-pre-injured tibialis anterior (TA) muscles of C3KO-NSG mice. A cohort of NSG mice was used as control recipients. Then, 6 weeks later, we assessed engraftment and CAPN3 expression. Immunofluorescence staining for human DYSTROPHIN and human LAMIN A/C showed engraftment of corrected LGMD2A 9015 iPSC-derived myogenic progenitors as well as of uncorrected counterparts and control unaffected myogenic progenitors (Figures 4A and S8). In spite of significant engraftment, we were not able to detect CAPN3 protein expression in the transplanted muscles by western blot (Figure S9A). Immunofluorescence staining with available CAPN3 antibodies does not provide reliable staining.
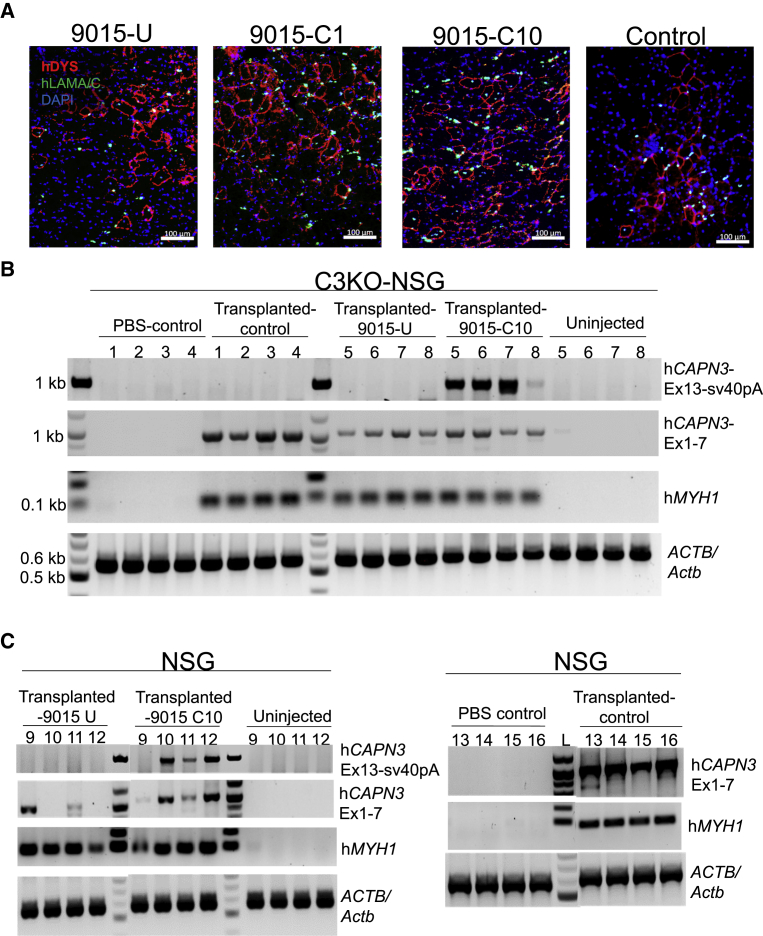
Figure 4.
Transplantation of Gene-Corrected iPSC-Derived Myogenic Progenitors into C3KO-NSG Mice
(A) Representative images show immunofluorescence staining for human DYSTROPHIN (DYS, in red) and human LAMIN A/C (LAM A/C, in green) in TA muscles that had been transplanted with myogenic progenitors from corrected (C1, C10), uncorrected (U) 9015 iPSCs, or control iPSCs. DAPI stains nuclei (in blue). Scale bars: 100 μm. Images depict a close up of the area of engraftment in transplanted muscle. (B) RT-PCR analysis of TA muscles from C3KO-NSG mice that had been transplanted with control (PLZ), 9015 uncorrected (U-right TA), and corrected (C10-left TA) iPSC-derived myogenic progenitors for human CAPN3 insert (Exon 13 to sv40 pA), human CAPN3 (exons 1–7), and human MYH1. ACTB/Actb was used as loading control. 1, 2, 3, and 4 are TA muscles that had been injected with control (PLZ) iPSC-derived myogenic progenitors or PBS (contra-lateral), whereas 5, 6, 7 and 8 are TA muscles that had been transplanted with 9015 uncorrected or corrected (contra-lateral) iPSC-derived myogenic progenitors. Non-injected control denotes the gastrocnemius muscle of corresponding mice. (C) RT-PCR analysis for human CAPN3 (Ex 13-sv40pA), CAPN3 (Ex 1-7), human MYH1 and ACTB/Actb in TA muscles from NSG mice that had been transplanted with 9015 uncorrected (9015-U, lanes 9 to 12, left), corrected (9015-C10, lanes 9 to 12, middle), and control (PLZ, lanes 13 to 16, left) iPSC-derived myogenic progenitors (left TA). Negative controls included non-injected gastrocnemius muscles of the corresponding mice (lanes 9 to 12, right) and PBS injected contra-lateral TA muscle (lanes 13 to 16, right).
To understand whether lack of protein detection by western blot was due to sensitivity limit, we titrated NSG (WT) and C3KO-NSG muscle lysates at different percentages to mimic the different levels of engraftment. This analysis revealed that the reliable detection limit for CAPN3 by western blot is approximately 20%–25% of NSG in C3KO-NSG lysates, a result that was consistent across three different CAPN3 antibodies (Figure S9B). Quantification of engraftment in recipients that had been transplanted with gene-corrected (9015-C1-10) iPSC-derived myogenic progenitors showed up to 200 fibers co-expressing human DYSTROPHIN and human LAMIN A/C (Figure S9C), which was superior than control (PLZ) and uncorrected counterparts (Figure S9C) but below the threshold detection limit of 20%–25% (∼10% if we consider the TA muscle has about 2,000 total fibers).
Nevertheless, RT-PCR for human CAPN3 in engrafted muscles clearly showed that muscles that had been transplanted with LGMD2A 9015 iPSC-derived myogenic progenitors displayed the rescued expression of CAPN3 mRNA specifically from the knockin sequence (Figure 4B), as indicated by PCR amplification of exon 13 to sv40pA. As expected, muscles transplanted with uncorrected counterparts were negative for the knockin sequence, but they did express CAPN3 exons 1–7 (Figure 4B), similar to in vitro counterparts (Figure 2C). Similarly, muscles transplanted with unaffected control iPSC-derived myogenic progenitors also showed efficient RT-PCR amplification, indicating the rescue of CAPN3 mRNA expression, whereas PBS-injected muscles showed none (Figure 4B). Transplantation of gene-corrected and control unaffected cells into NSG mice corroborated the rescue of CAPN3 mRNA (Figure 4C). These results confirm that in vitro gene editing of LGMD2A iPSCs leads to the rescue of CAPN3 mRNA expression in vivo. Thus, we show proof of principle for the integration of genome editing and iPSC technologies to develop a novel autologous cell therapy for LGMD2A.
Discussion
Because pluripotent stem cells have essentially unlimited expansion potential and can differentiate into virtually any somatic cell type, they represent an attractive option for regenerative therapies. The derivation of iPSCs from somatic cells through reprogramming technology35, 36, 37 brings this type of therapy much closer to clinical application, since it eliminates the ethical and immunological issues associated with embryonic stem cells, allowing for the derivation of patient-specific cell types. Accordingly, significant progress has been achieved in the last decade, and clinical trials are in progress for macular degeneration38 and Parkinson’s disease.39, 40
When considering the treatment of genetic diseases, such as MDs, correction of specific mutations is required prior to developing an autologous iPSC-based therapy. We have previously reported proof of principle for this strategy in mouse-to-mouse studies using the severe dystrophin and utrophin double-KO (dKO) mouse model for Duchenne MD (DMD), in which corrected engrafted cells showed stable expression of the transgene and improved muscle force.41 Gene correction of patient-specific iPSCs has been documented for DMD42, 43, 44, 45 and LGMD types 2B and 2D,46, 47 diseases associated with structural proteins linked to the dystrophin-glycoprotein complex, including dystrophin, dysferlin, and α-sarcoglycan, respectively. From these six studies, only two reported transplantation and engraftment of gene-edited iPSC derivatives.42, 47
Contrary to most MDs, which are linked to mutations in DGC or other structural proteins, LGMD2A is due to mutations in the CAPN3 gene, which encodes a protein with enzymatic activity. We designed a gene-editing strategy able to correct several mutations, as demonstrated here by the in vitro rescue of CAPN3 expression at both RNA and protein levels in three patient-specific iPSC lines bearing different mutations (Figures 2 and 3). This genome-editing strategy is theoretically applicable to correction of mutations in the 3′ end of any gene.
Although it is well-recognized that the C3KO mouse model has only a mild phenotype,30, 48, 49 it nevertheless represents a valuable model when combined with immunodeficiency to demonstrate engraftment of gene-corrected human myogenic cells. Transplantation of gene-edited LGMD2A iPSC-derived myogenic progenitors into C3KO-NSG mice resulted in reasonable numbers of donor-derived myofibers (∼200), but these levels are lower than the detection limit for CAPN3 on western blot. Despite research advances, efficiency of xenotransplantation of human myogenic cells in mouse models remains low.17, 18, 50, 51, 52 Nevertheless, we validated that the corrected cells rescue CAPN3 mRNA expression upon transplantation, whereas this was absent following the engraftment of uncorrected counterparts (Figure 4). In addition, the C3KO mouse model does not recapitulate the severity of the human disease, and, therefore, better animal models are needed to reliably validate the therapeutic efficacy of cells or any other treatment approach. Interestingly, studies in mice and primary samples have suggested impaired muscle regeneration in LGMD2A,8 thus supporting the potential benefit for the use of cell-based therapy to treat this disease. Although there are several clinical trials underway for MDs, there is none for LGMD2A. In light of the findings we present here, there is strong evidence for the therapeutic application of an autologous gene-corrected iPSC-based therapy to target LGMD2A.
Materials and Methods
Cell Culture and Myogenic Differentiation of iPSCs
Studies involved de-identified fibroblast samples according to procedures approved by the Institutional Review Board of the University of Minnesota. To reprogram fibroblasts into iPSCs, we utilized the Cytotune 2.0 Sendai virus-based reprogramming kit (Thermo Fisher Scientific), as per the manufacturer’s instructions. This kit includes three different sendai viral vectors consisting of cMYC, KLF4, and polycistronic KLF4-OCT3/4-SOX2. Fibroblasts were transduced with these sendai viral vectors and cultured as per the manufacturer’s recommendation to derive iPSC clones. The clones that maintained typical iPSC morphology and pluripotency marker expression were utilized for further studies.
iPSCs were maintained on matrigel-coated flasks in mTeSR1 medium (STEMCELL Technologies). To generate myogenic progenitors, iPSCs were transduced with pSAM2-iPAX7-ires-mCherry and FUGW-rtTA, as previously described.17 To induce myogenic differentiation, transduced iPAX7 iPSCs were dissociated into single cells using accutase (Innovative Cell Technologies), and 1 million cells were seeded onto non-adherent 60-mm Petri dishes, in the presence of mTeSR1 supplemented with 10 μM ROCK inhibitor Y-27632 (APExBIO), and incubated at 37°C and 5% CO2 on a shaker at 60 rpm to derive embryoid bodies (EBs). Then 2 days later, medium was switched to 10 μM GSK3β inhibitor- (CHIR 990217; Tocris) supplemented EB myogenic medium.17 After 48 h, EBs were switched to EB myogenic medium supplemented with 200 nM LDN-193189 (Cayman Chemical) and 10 μM SB-431542 (Cayman Chemical) and incubated for 24 h. At this point, medium was supplemented with 1 μg/mL Doxycycline (Dox; Sigma-Aldrich) to induce PAX7 expression and incubated for an additional 24 h. Then, cells were switched to EB myogenic medium supplemented with Dox for 48 h.
Next, one-fifth of the EBs were plated on gelatin-coated T75 flasks in EB myogenic medium supplemented with 5 ng/mL basic fibroblast growth factor (bFGF; PeproTech) and Dox. Then 4 days later, cells were sorted via FACS for mCherry to purify the PAX7-positive myogenic progenitor cells, which were plated in the medium supplemented with Dox, bFGF. These myogenic progenitors were expanded every 3 days at a ratio of 1:6 for up to 4–5 passages. To induce terminal differentiation, myogenic progenitors were plated at a density of 37,500 cells/cm2 and allowed to grow confluent for 3 days. Once they were confluent, terminal differentiation into myotubes was induced by switching to low nutrient differentiation medium made up of KnockOut DMEM supplemented with 20% KnockOut Serum Replacement (KOSR; Gibco), 1% Non-essential amino acids, 1% Glutamax (Gibco), and 1% penicillin-streptomycin (Invitrogen). To induce CAPN3 protein expression in iPSC-derived myotubes, the differentiation medium was supplemented with 10 μM each of SB-431542, DAPT, Forskolin, and Dexamethasone (Cayman Chemical and Selleckchem). For the terminal differentiation of 0989 iPSCs, we also included 10 μM PD0325901 (Cayman Chemical) in the differentiation medium, in addition to the other four compounds, which improved differentiation and, thereby CAPN3 protein expression.
Karyotype Analysis
Karyotype analysis was performed at the Cytogenomics shared resource at the Masonic Cancer Center in the University of Minnesota. iPSCs were treated with colcemid for 3 h to arrest cells in metaphase. A total of 20 different metaphases were then analyzed by Giemsa banding at a resolution of 400–450 band level.
Mice
Animal experiments were carried out according to protocols approved by the University of Minnesota Institutional Animal Care and Use Committee. For the generation of immunodeficient CAPN3-KO mice, we crossed the CAPN3-KO mouse model (C3KO),30 kindly provided by Melissa Spencer (UCLA), with NSG mice (Jackson ImmunoResearch Laboratories), and the C3KO heterozygous litters were backcrossed with NSG until the derivation of mice homozygous for Prkdcscid and IL2rg mutations (NSG). These immunodeficient C3KO heterozygous litters were then crossed to obtain the C3KO-NSG mouse model.
Peripheral Blood FACS Analysis
FACS analysis on the peripheral blood for B, T, and NK cells was carried out upon staining with the following antibodies: CD19 (Phycoerythrin-Cy7, eBio1D3), CD3ε (Phycoerythrin, 145-2C11), CD49b (Phycoerythrin, DX5), and NK1.1 (Allophycocyanin, PK136) (all from eBioscience), as previously described.34 FACS analysis was performed using a FACSAria (BD Biosciences). Data were analyzed using FlowJo software.
Transplantation
For transplantation studies, TA muscles of 12- to 16-week-old C3KO-NSG mice or 6- to 8-week-old NSG mice were pre-injured with cardiotoxin (15 μL of 10 μM stock; Latoxan) and then injected with 1 × 106 myogenic progenitors. 6 weeks later, muscles were collected for engraftment and rescue assessment.
Teratoma Assay
For teratoma experiments, 1.5 × 106 human iPSCs resuspended in 50 μL 1:1 DMEM/F12:Matrigel were injected into the quadriceps of 8-week-old male immunodeficient mice (NSG, Jackson ImmunoResearch Laboratories). Teratomas were collected, fixed, sectioned, and processed for H&E staining (SelecTech H+E, Leica Biosystems).
CRISPR-Cas9-Based Gene Correction
CRISPR-Cas9 construct was generated by cloning the gRNA sequence into the pX458 plasmid (Addgene 48138; kindly provided by Feng Zhang). The guide RNA sequence targeting CAPN3-exon 14 (5′-CATCTCCGTGGATCGGCCAG-3′) was cloned using the BbsI restriction enzyme sites in the pX458 vector. HDR donor vector was constructed in the pBluescript plasmid backbone. The selection cassette of loxP-flanked GFP-2A-neoR driven by HEF1-eIF4g for positive selection was cloned into the pBluescript backbone. Herpes simplex virus thymidine kinase (HSV-tk) driven by MC1 promoter was cloned into the backbone for negative selection. The 5′ homology arm consisting of intron 13 (∼950 bp), the knockin insert consisting of exons 15–24 cDNA, and sv40 poly(A) signal sequence were cloned upstream of GFP-2A-neoR cassette by PCR amplification. The 3′ homology arm consisting of the In14 (∼950 bp) was cloned downstream of the GFP-2A-neoR cassette by PCR amplification.
The CRISPR-Cas9 and the HDR donor plasmids were transfected in iPSCs using the Amaxa Nucleofector II (Lonza), as per the manufacturer’s instructions. Then 3 days later, cells were selected with 50 μg/mL Geneticin (G418, Thermo Fisher Scientific) for 8 days and, subsequently, with 4 μM Ganciclovir (Invivogen) for 5 days. iPSCs that survived this double selection were plated sparsely in medium supplemented with cloneR (STEMCELL Technologies) to obtain single-cell clones. For each iPSC line, we isolated approximately 20–25 single-cell clones. These were analyzed by PCR to amplify the region spanning the knockin. PCR was performed by using the primeSTAR GXL DNA polymerase (Takara) with primers that bind to the genomic DNA upstream and downstream of the homology arms (forward primer [In13], 5′-ttccagcctgaggggcttcg-3′ and reverse primer [In14], 5′-cccacgccccaacattcctg-3′). Clones that were positive for this PCR were selected for further analysis. In general, 3–5 clones, from the total of 20–25 single-cell clones, were found to be positive for the knockin based on PCR.
For CRISPR-Cas9 RNP-based correction, guide RNA and Cas9 protein were obtained from Synthego and IDT, respectively. iPSCs were seeded at a density of 75,000 cells/well of a 24-well plate a day before transfection. RNP complex and the donor plasmid were transfected using Lipofectamine CRISPRMAX Cas9 Transfection Reagent (Thermo Fisher Scientific), as per the manufacturer’s instructions.
Generation of CAPN3-KO iPSC Line
To generate a CAPN3-KO iPSC line, we designed a guide RNA pair spanning exon 1 to intron 1. Sequences of the guide RNAs used were 5′-ATGACGGTCGGCATGGCAAG-3′ (exon 1) and 5′-GCTGGGTTTTCCCCCCACGG-3′ (intron 1). PLZ iPSCs were seeded at a density of 75,000 cells/well in a 24-well plate in mTESR1 media, transfected after 24 h with CRISPR Cas9 RNP, and maintained for 48–72 h. Single-cell colonies were picked as described in the previous section. PCR was performed on genomic DNA to amplify the region that spans exon 1 to intron 1 (650 bp), which gets disrupted in an event of deletion. Clones that showed genomic deletion based on PCR were differentiated into myotubes and further analyzed for mRNA and protein expression of CAPN3.
CRISPR-Cas9 Off-Target Analysis
To evaluate the off-target effects, we analyzed the top 5 predicted off-target regions for guide RNA targeting exon 14, as predicted by the Off-Spotter software.27 The genomic regions were PCR amplified in uncorrected and corrected 9015, 0826, and 0989 iPSCs, and the sequencing chromatograms were compared between the two using the TIDE28 and ICE29 software to analyze for the percentage of off-target editing, as per the manufacturer instructions. These analyses give an estimate of the frequency of mutations generated due to non-homologous end-joining upon CRISPR-Cas9-induced double-stranded break in a specific genomic site. TIDE and ICE software predict the off-target frequency based on the level of alterations in sequencing chromatograms around the CRISPR-Cas9 cut site in the genome-edited sample comparatively to that of the unedited control.
Excision of Selection Cassette
To excise the loxP-flanked selection cassette consisting of GFP-2A-NeoR, gene-corrected 0826-C5, -C10, and 0989-C1 iPSC clones were transfected with Cre recombinase protein (EG-1001, Excellgen) using Lipofectamine Stem reagent (Thermo Fisher Scientific), as per the manufacturer’s instructions. Next, iPSC clones were checked for excision of selection cassette using PCR amplification of the region spanning knockin. The Cre transfection protocol was repeated 4–5 times to make sure the selection cassette was excised in every cell.
RNA Isolation and RT-PCR Analysis
Total RNA was isolated from cells using Trizol reagent (Thermo Fisher Scientific). For RNA isolation from skeletal muscle tissue, the tissue was pulverized in liquid nitrogen-cooled mortar and pestle to obtain a powder, which is dissolved in Trizol reagent. RNA was isolated using the PureLink RNA mini kit (Thermo Fisher Scientific) with on-column DNase treatment, as per the manufacturer’s instructions. Reverse transcription of the RNA was performed using SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific), as per the manufacturer’s instructions. PCR was performed using cDNA corresponding to 20–50 μg RNA using the GoTaq Flexi DNA Polymerase, as per the manufacturer’s instructions. Following are the RT-PCR amplicons and their corresponding primer sequences: hCAPN3 exons 1–24 (forward 5′-gcgcatctgtggctccaagg-3′, reverse 5′-gagctgcagccactccagaacg-3′), hCAPN3 exon 1-sv40 pA (forward 5′-gcgcatctgtggctccaagg-3′, reverse 5′-cactgcattctagttgtggtttgtcca-3′), hCAPN3 exons 1–7 (forward 5′-gcgcatctgtggctccaagg-3′, reverse 5′-cctcatccagccccgtgaca-3′), hCAPN3 exon 13-sv40 pA (forward 5′-tgcacgggaacaagcagcac-3′, reverse 5′-cactgcattctagttgtggtttgtcca-3′), hMYH1 (forward 5′-gcggctgatcaatgacctcaca-3′, reverse 5′-tgtccttttcatctagctggcgt-3′), and ACTB/Actb (human and mouse) (forward 5′-gcgacgaggcccagagcaag-3′, reverse 5′-tggccgtcaggcagctcgta-3′).
Western Blot Analysis
Cells were lysed in lysis buffer consisting of 20 mM Tris HCl, 0.1 mM EDTA, 1 mM DTT, 20 μg/mL soybean trypsin inhibitor, 28 μM E64, and 2 mM phenylmethylsulfonyl fluoride (PMSF) plus 1× Laemmli sample buffer (diluted from 6×). The lysate was collected using cell scraper, boiled at 95°C for 10 min, and centrifuged for 5 min at 15,000 rpm to remove cell debris. Total protein concentration was measured using a Bradford assay. A total of 50 μg protein was electrophoresed in 7.5% SDS-PAGE gels. After electrophoresis, proteins were transferred to immobilon-FL polyvinylidene fluoride (PVDF) membrane (Millipore). After transfer, the blot was blocked with 5% dry milk in TBST (Tris-buffered saline and 0.1% Tween 20) for 1 h at room temperature (RT). After the blocking, the blot was incubated with primary antibody diluted in 3% BSA in TBST and incubated at 4°C overnight.
The next day, the blot was washed thrice in TBST and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at RT. The blot was then washed three times with TBST, and the protein detection was performed using the Supersignal West chemiluminescent substrate (Thermo Fisher Scientific). The chemiluminescence signal was captured in the chemidoc imager (Bio-Rad) or the X-ray film. The following antibodies were used: CAPN3-2C4 (CALP-2C4, Leica Biosystems; 1:50), CAPN3-12A2 (CALP-12A2, Leica Biosystems; 1:30), CAPN3-IS2 (COP-COP-080048, Cosmo Bio; 1:1,000), DES (RD301, Santa Cruz Biotechnology; 1:500), and ACTB (C4, Santa Cruz Biotechnology; 1:1,000).
For titration experiments between NSG and C3KO-NSG muscle lysates, the two samples were mixed such that the NSG percentage varied from 100% to 0% (100, 75, 50, 25, 20, 15, 10, 5, and 0). 200 μg total protein was loaded in the SDS-PAGE gel, and the western blot was performed with three different CAPN3 antibodies (E6, 12A2, and IS2).
Immunofluorescence Staining
Following fixation with 4% PFA (paraformaldehyde) for 20 min at RT, cells were permeabilized with 0.3% Triton X-100 in PBS for 20 min at RT. Cells were then blocked with 3% BSA for 1 h at RT and then incubated with primary antibody diluted in 3% BSA overnight at 4°C. The next day, cells were washed three times with PBS and then incubated with secondary antibody and DAPI for 45 min at RT. After secondary antibody incubation, the cells were washed three times with PBS and stored in the dark at 4°C until imaging. For tissues, muscles were cryosectioned at 10-μm thickness. Sections were allowed to dry at RT for 15 min, rehydrated in PBS, and then fixed with 4% PFA for 10 min. The rest of the staining protocol was similar to that of the cells. After final washing, the slides were mounted with coverslips using ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific). The stained sections were imaged in the Zeiss upright microscope. Following are the antibodies used for immunofluorescence staining: human DYSTROPHIN (MANDYS106, DSHB; 1:50), human LAMIN A/C (ab108595, Abcam; 1:500), MHC (MF20, DSHB; 1:100), OCT3/4 (C-10; 1:50), SOX2 (Y-17; 1:50), NANOG (H-2; 1:50), SSEA-4 Antibody (813-70; 1:50) (all from Santa Cruz Biotechnology), Alexa Fluor 488 anti-rabbit immunoglobulin G (IgG) (A-11008; 1:500), Alexa Fluor 555 anti-mouse IgG (A21424; 1:500), and Alexa Fluor 555 anti-goat IgG (A21424; 1:500) (all from Thermo Fisher Scientific).
For quantification, the number of fibers double-positive for human DYSTROPHIN and human LAMINA/C were counted in 3 different sections (100 μm apart) for each transplanted muscle.
Histology Analysis
H&E staining was performed using the SelecTech kit (Leica Biosystems). Masson’s trichrome staining was utilized to detect collagen content (Polysciences). Fiji ImageJ software was used to quantify percentage area of collagen staining.
Autocatalytic Assay
For this, cells were collected in saline using a cell scraper, and the collected sample was incubated either at 0 min or 15 min at RT, after which 1× Laemmli sample buffer was added and the samples were boiled at 95°C for 10 min. The resulting samples were subsequently evaluated by western blot as described previously.26
Southern Blot Analysis
Genomic DNA was extracted using the Purelink Genomic DNA mini kit as per the manufacturer’s instructions. Genomic DNA was digested with BmtI restriction enzyme overnight at 37°C, and following electrophoresis in 1% agarose gel, DNA was transferred to positively charge Nylon membrane and probed with digoxigenin (DIG)-labeled probe. PCR DIG probe synthesis kit (Roche) was used for labeling the probe. Detection was performed using DIG luminescent detection kit (Roche).
shRNA-Based Gene Knockdown
We utilized three different shRNA clones targeting various regions of the CAPN3 gene, including exon 1 (TRCN0000003494), 3′ UTR (TRCN0000003495), and exon 22 (TRCN0000003496), respectively. The shRNA lentiviral vectors were packaged into 293T cells to generate the lentiviral particles. Day 2 myotubes were transduced with lentiviral vectors by spin infection at 2,500 rpm for 90 min. shRNA-based gene silencing was analyzed 7 days later by western blot.
Author Contributions
S.S. designed and performed experiments, analyzed the data, and wrote the manuscript. N.R.D. designed and performed experiments and analyzed the data. J.K., S.T., R.M.-G., G.K., and V.K.P.O. performed experiments and analyzed the data. A.J.M.-A. and A.L.d.M. provided patient samples. R.C.R.P. contributed with experimental design and interpretation of the data and wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank the generous support from ADVault, Inc. and MyDirectives.com (R.C.R.P.). This project was also supported by funds from the NIH, grants R01 AR055299 and AR071439 (R.C.R.P.), the Coalition to Cure Calpain 3 (R.C.R.P.), and Instituto Carlos III, Fondo Investigaciones Sanitarias-ALM, PI17/01841 (A.J.M.-A and A.L.d.M). The cytogenetic analyses were performed in the Cytogenomics Shared Resource at the University of Minnesota with support from the comprehensive Masonic Cancer Center NIH grant P30 CA077598-09. The monoclonal antibody to MHC was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. We thank Melissa Spencer (UCLA) for the C3KO mice and Michael Kyba for critical reading of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.08.011.
Supplemental Information
References
- 1.Guglieri M., Straub V., Bushby K., Lochmüller H. Limb-girdle muscular dystrophies. Curr. Opin. Neurol. 2008;21:576–584. doi: 10.1097/WCO.0b013e32830efdc2. [DOI] [PubMed] [Google Scholar]
- 2.Piluso G., Politano L., Aurino S., Fanin M., Ricci E., Ventriglia V.M., Belsito A., Totaro A., Saccone V., Topaloglu H. Extensive scanning of the calpain-3 gene broadens the spectrum of LGMD2A phenotypes. J. Med. Genet. 2005;42:686–693. doi: 10.1136/jmg.2004.028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore S.A., Shilling C.J., Westra S., Wall C., Wicklund M.P., Stolle C., Brown C.A., Michele D.E., Piccolo F., Winder T.L. Limb-girdle muscular dystrophy in the United States. J. Neuropathol. Exp. Neurol. 2006;65:995–1003. doi: 10.1097/01.jnen.0000235854.77716.6c. [DOI] [PubMed] [Google Scholar]
- 4.Mercuri E., Bushby K., Ricci E., Birchall D., Pane M., Kinali M., Allsop J., Nigro V., Sáenz A., Nascimbeni A. Muscle MRI findings in patients with limb girdle muscular dystrophy with calpain 3 deficiency (LGMD2A) and early contractures. Neuromuscul. Disord. 2005;15:164–171. doi: 10.1016/j.nmd.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Fanin M., Nascimbeni A.C., Fulizio L., Trevisan C.P., Meznaric-Petrusa M., Angelini C. Loss of calpain-3 autocatalytic activity in LGMD2A patients with normal protein expression. Am. J. Pathol. 2003;163:1929–1936. doi: 10.1016/S0002-9440(10)63551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelini C., Nardetto L., Borsato C., Padoan R., Fanin M., Nascimbeni A.C., Tasca E. The clinical course of calpainopathy (LGMD2A) and dysferlinopathy (LGMD2B) Neurol. Res. 2010;32:41–46. doi: 10.1179/174313209X380847. [DOI] [PubMed] [Google Scholar]
- 7.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 8.Hauerslev S., Sveen M.L., Duno M., Angelini C., Vissing J., Krag T.O. Calpain 3 is important for muscle regeneration: evidence from patients with limb girdle muscular dystrophies. BMC Musculoskelet. Disord. 2012;13:43. doi: 10.1186/1471-2474-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckmann J.S., Spencer M. Calpain 3, the “gatekeeper” of proper sarcomere assembly, turnover and maintenance. Neuromuscul. Disord. 2008;18:913–921. doi: 10.1016/j.nmd.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramerova I., Beckmann J.S., Spencer M.J. Molecular and cellular basis of calpainopathy (limb girdle muscular dystrophy type 2A) Biochim. Biophys. Acta. 2007;1772:128–144. doi: 10.1016/j.bbadis.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Ojima K., Kawabata Y., Nakao H., Nakao K., Doi N., Kitamura F., Ono Y., Hata S., Suzuki H., Kawahara H. Dynamic distribution of muscle-specific calpain in mice has a key role in physical-stress adaptation and is impaired in muscular dystrophy. J. Clin. Invest. 2010;120:2672–2683. doi: 10.1172/JCI40658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ojima K., Ono Y., Doi N., Yoshioka K., Kawabata Y., Labeit S., Sorimachi H. Myogenic stage, sarcomere length, and protease activity modulate localization of muscle-specific calpain. J. Biol. Chem. 2007;282:14493–14504. doi: 10.1074/jbc.M610806200. [DOI] [PubMed] [Google Scholar]
- 13.Kramerova I., Ermolova N., Eskin A., Hevener A., Quehenberger O., Armando A.M., Haller R., Romain N., Nelson S.F., Spencer M.J. Failure to up-regulate transcription of genes necessary for muscle adaptation underlies limb girdle muscular dystrophy 2A (calpainopathy) Hum. Mol. Genet. 2016;25:2194–2207. doi: 10.1093/hmg/ddw086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ojima K., Ono Y., Ottenheijm C., Hata S., Suzuki H., Granzier H., Sorimachi H. Non-proteolytic functions of calpain-3 in sarcoplasmic reticulum in skeletal muscles. J. Mol. Biol. 2011;407:439–449. doi: 10.1016/j.jmb.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartoli M., Roudaut C., Martin S., Fougerousse F., Suel L., Poupiot J., Gicquel E., Noulet F., Danos O., Richard I. Safety and efficacy of AAV-mediated calpain 3 gene transfer in a mouse model of limb-girdle muscular dystrophy type 2A. Mol. Ther. 2006;13:250–259. doi: 10.1016/j.ymthe.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Roudaut C., Le Roy F., Suel L., Poupiot J., Charton K., Bartoli M., Richard I. Restriction of calpain3 expression to the skeletal muscle prevents cardiac toxicity and corrects pathology in a murine model of limb-girdle muscular dystrophy. Circulation. 2013;128:1094–1104. doi: 10.1161/CIRCULATIONAHA.113.001340. [DOI] [PubMed] [Google Scholar]
- 17.Darabi R., Arpke R.W., Irion S., Dimos J.T., Grskovic M., Kyba M., Perlingeiro R.C. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell. 2012;10:610–619. doi: 10.1016/j.stem.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magli A., Incitti T., Kiley J., Swanson S.A., Darabi R., Rinaldi F., Selvaraj S., Yamamoto A., Tolar J., Yuan C. PAX7 Targets, CD54, Integrin α9β1, and SDC2, Allow Isolation of Human ESC/iPSC-Derived Myogenic Progenitors. Cell Rep. 2017;19:2867–2877. doi: 10.1016/j.celrep.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ban H., Nishishita N., Fusaki N., Tabata T., Saeki K., Shikamura M., Takada N., Inoue M., Hasegawa M., Kawamata S., Nishikawa S. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fougerousse F., Durand M., Suel L., Pourquié O., Delezoide A.L., Romero N.B., Abitbol M., Beckmann J.S. Expression of genes (CAPN3, SGCA, SGCB, and TTN) involved in progressive muscular dystrophies during early human development. Genomics. 1998;48:145–156. doi: 10.1006/geno.1997.5160. [DOI] [PubMed] [Google Scholar]
- 21.Toral-Ojeda I., Aldanondo G., Lasa-Elgarresta J., Lasa-Fernández H., Fernández-Torrón R., López de Munain A., Vallejo-Illarramendi A. Calpain 3 deficiency affects SERCA expression and function in the skeletal muscle. Expert Rev. Mol. Med. 2016;18:e7. doi: 10.1017/erm.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson L.V., Davison K., Moss J.A., Richard I., Fardeau M., Tomé F.M., Hübner C., Lasa A., Colomer J., Beckmann J.S. Characterization of monoclonal antibodies to calpain 3 and protein expression in muscle from patients with limb-girdle muscular dystrophy type 2A. Am. J. Pathol. 1998;153:1169–1179. doi: 10.1016/S0002-9440(10)65661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fanin M., Fulizio L., Nascimbeni A.C., Spinazzi M., Piluso G., Ventriglia V.M., Ruzza G., Siciliano G., Trevisan C.P., Politano L. Molecular diagnosis in LGMD2A: mutation analysis or protein testing? Hum. Mutat. 2004;24:52–62. doi: 10.1002/humu.20058. [DOI] [PubMed] [Google Scholar]
- 24.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fanin M., Nascimbeni A.C., Angelini C. Screening of calpain-3 autolytic activity in LGMD muscle: a functional map of CAPN3 gene mutations. J. Med. Genet. 2007;44:38–43. doi: 10.1136/jmg.2006.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pliatsika V., Rigoutsos I. “Off-Spotter”: very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs. Biol. Direct. 2015;10:4. doi: 10.1186/s13062-015-0035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsiau T., Conant D., Rossi N., Maures T., Waite K., Yang J., Joshi S., Kelso R., Holden K., Enzmann B.L., Stoner R. Inference of CRISPR Edits from Sanger Trace Data. bioRxiv. 2019 doi: 10.1089/crispr.2021.0113. [DOI] [PubMed] [Google Scholar]
- 30.Kramerova I., Kudryashova E., Tidball J.G., Spencer M.J. Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet. 2004;13:1373–1388. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 31.Cao X., Shores E.W., Hu-Li J., Anver M.R., Kelsall B.L., Russell S.M., Drago J., Noguchi M., Grinberg A., Bloom E.T. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 32.Bosma G.C., Custer R.P., Bosma M.J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 33.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 34.Arpke R.W., Darabi R., Mader T.L., Zhang Y., Toyama A., Lonetree C.L., Nash N., Lowe D.A., Perlingeiro R.C., Kyba M. A new immuno-, dystrophin-deficient model, the NSG-mdx(4Cv) mouse, provides evidence for functional improvement following allogeneic satellite cell transplantation. Stem Cells. 2013;31:1611–1620. doi: 10.1002/stem.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 38.Mandai M., Kurimoto Y., Takahashi M. Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N. Engl. J. Med. 2017;377:792–793. doi: 10.1056/NEJMc1706274. [DOI] [PubMed] [Google Scholar]
- 39.Studer L. Strategies for bringing stem cell-derived dopamine neurons to the clinic-The NYSTEM trial. Prog. Brain Res. 2017;230:191–212. doi: 10.1016/bs.pbr.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Barker R.A., Parmar M., Studer L., Takahashi J. Human Trials of Stem Cell-Derived Dopamine Neurons for Parkinson’s Disease: Dawn of a New Era. Cell Stem Cell. 2017;21:569–573. doi: 10.1016/j.stem.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Filareto A., Parker S., Darabi R., Borges L., Iacovino M., Schaaf T., Mayerhofer T., Chamberlain J.S., Ervasti J.M., McIvor R.S. An ex vivo gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nat. Commun. 2013;4:1549. doi: 10.1038/ncomms2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young C.S., Hicks M.R., Ermolova N.V., Nakano H., Jan M., Younesi S., Karumbayaram S., Kumagai-Cresse C., Wang D., Zack J.A. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi I.Y., Lim H., Estrellas K., Mula J., Cohen T.V., Zhang Y., Donnelly C.J., Richard J.P., Kim Y.J., Kim H. Concordant but Varied Phenotypes among Duchenne Muscular Dystrophy Patient-Specific Myoblasts Derived using a Human iPSC-Based Model. Cell Rep. 2016;15:2301–2312. doi: 10.1016/j.celrep.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 44.Ifuku M., Iwabuchi K.A., Tanaka M., Lung M.S.Y., Hotta A. Restoration of Dystrophin Protein Expression by Exon Skipping Utilizing CRISPR-Cas9 in Myoblasts Derived from DMD Patient iPS Cells. Methods Mol. Biol. 2018;1828:191–217. doi: 10.1007/978-1-4939-8651-4_12. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Long C., Li H., McAnally J.R., Baskin K.K., Shelton J.M., Bassel-Duby R., Olson E.N. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv. 2017;3:e1602814. doi: 10.1126/sciadv.1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turan S., Farruggio A.P., Srifa W., Day J.W., Calos M.P. Precise Correction of Disease Mutations in Induced Pluripotent Stem Cells Derived From Patients With Limb Girdle Muscular Dystrophy. Mol. Ther. 2016;24:685–696. doi: 10.1038/mt.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tedesco F.S., Gerli M.F., Perani L., Benedetti S., Ungaro F., Cassano M., Antonini S., Tagliafico E., Artusi V., Longa E. Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci. Transl. Med. 2012;4:140ra89. doi: 10.1126/scitranslmed.3003541. [DOI] [PubMed] [Google Scholar]
- 48.Fougerousse F., Gonin P., Durand M., Richard I., Raymackers J.M. Force impairment in calpain 3-deficient mice is not correlated with mechanical disruption. Muscle Nerve. 2003;27:616–623. doi: 10.1002/mus.10368. [DOI] [PubMed] [Google Scholar]
- 49.Richard I., Roudaut C., Marchand S., Baghdiguian S., Herasse M., Stockholm D., Ono Y., Suel L., Bourg N., Sorimachi H. Loss of calpain 3 proteolytic activity leads to muscular dystrophy and to apoptosis-associated IkappaBalpha/nuclear factor kappaB pathway perturbation in mice. J. Cell Biol. 2000;151:1583–1590. doi: 10.1083/jcb.151.7.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J., Matthias N., Lo J., Ortiz-Vitali J.L., Shieh A.W., Wang S.H., Darabi R. A Myogenic Double-Reporter Human Pluripotent Stem Cell Line Allows Prospective Isolation of Skeletal Muscle Progenitors. Cell Rep. 2018;25:1966–1981.e4. doi: 10.1016/j.celrep.2018.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hicks M.R., Hiserodt J., Paras K., Fujiwara W., Eskin A., Jan M., Xi H., Young C.S., Evseenko D., Nelson S.F. ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs. Nat. Cell Biol. 2018;20:46–57. doi: 10.1038/s41556-017-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Wal E., Herrero-Hernandez P., Wan R., Broeders M. In: Large-Scale Expansion of Human iPSC-Derived Skeletal Muscle Cells for Disease Modeling and Cell-Based Therapeutic Strategies. Stem Cell Reports. ‘t Groen S.L.M., van Gestel T.J.M., van IJcken W.F.J., Cheung T.H., van der Ploeg A.T., Schaaf G.J., Pijnappel W.W.M.P., editors. Vol. 10. 2018. pp. 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.