Abstract
Recent work has demonstrated that two archaic human groups (Neanderthals and Denisovans) interbred with modern humans and contributed to the contemporary human gene pool. These findings relied on the availability of high-coverage genomes from both Neanderthals and Denisovans. Here we search for evidence of archaic admixture from a worldwide panel of 1,667 individuals using an approach that does not require the presence of an archaic human reference genome. We find no evidence for archaic admixture in the Andaman Islands, as previously claimed, or on the island of Flores, where Homo floresiensis fossils have been found. However, we do find evidence for at least one archaic admixture event in sub-Saharan Africa, with the strongest signal in Khoesan and Pygmy individuals from Southern and Central Africa. The locations of these putative archaic admixture tracts are weighted against functional regions of the genome, consistent with the long-term effects of purifying selection against introgressed genetic material.
Keywords: ghost admixture, archaic humans, GenomeAsia project, Neanderthal, Denisovan
Introduction
Anatomically modern humans are thought to have evolved in one or more locations in Africa more than 100 Kya.1, 2, 3 From there they expanded throughout Africa, into Eurasia, and throughout the rest of the world, with remote islands being colonized only within the past 1,000 years.4,5 As modern humans migrated to new areas, they came into contact with and interbred with various “archaic” human groups. It is now well documented that non-African individuals inherited ∼2% of their DNA from Neanderthals,6, 7, 8 while Melanesian and aboriginal Australian individuals inherited ∼4%–5% of their DNA from an enigmatic archaic human group called Denisovans.9, 10, 11 The identification and quantification of these admixture events was greatly aided by the availability of high-quality draft genome sequences from a single Neanderthal7 and a single Denisovan11 individual. Since only sparse, fragmented DNA has been recovered from any other (i.e., non-Neanderthal, non-Denisovan) archaic human source,12,13 other methods must be used to detect any additional archaic admixture events in human history.
Recently, there have been several studies claiming evidence of past archaic admixture events in sub-Saharan Africa.14, 15, 16, 17, 18 There are two main reasons why these claims are plausible. First, the archeological record shows that modern and archaic humans, as well as groups with a mixture of modern and archaic skeletal features, are likely to have co-existed in the same place and at the same time in Africa for tens of thousands of years,3,19, 20, 21 so there was likely more opportunity for admixture inside Africa compared with outside of Africa. Second, genetic studies have consistently shown that sub-Saharan African genomes contain more long, diverged haplotypes than expected under “null” models without archaic admixture,14, 15, 16 and these diverged haplotypes are exactly what we would expect to observe if archaic admixture did in fact occur.22 However, these previous studies have been limited in scope, because they either considered only a small fraction of the genome,14,15,23 or included a very limited collection of samples/populations,16,17 or used only low-coverage genome sequences.18 Further, none of these other studies explicitly excluded potential copy number variants, which can mimic the appearance of archaic admixture tracts.
In this study, we revisit the question of archaic admixture by analyzing 1,667 diverse, high-coverage human genomes representative of the full spectrum of human genetic diversity. We focus on non-Neanderthal, non-Denisovan admixture and use a linkage-disequilibrium (LD) based approach similar to previous studies. The data come from the GenomeAsia 100K Pilot Project (GAsP),24 and the broad sampling of the GAsP allows us to more fully explore the strength of evidence for archaic admixture into populations from around the world. The in-depth sampling also allows us to test the efficacy of our approach by seeing whether we could detect evidence of Denisovan admixture purely from patterns of LD and without access to the Denisovan genome. (See also Skov et al.25 for an unrelated approach to the same question.)
While quantifying the extent of admixture between modern and archaic humans is clearly of historical and evolutionary interest, it also has direct implications for human disease genetics. There is a growing body of evidence suggesting that introgressed Neanderthal regions are generally deleterious (i.e., harmful) in modern humans.8,26, 27, 28 There are large stretches of the genome completely devoid of Neanderthal (or Denisovan) admixture,8,26,29 which suggests there may be genetic incompatibilities between archaic human and modern human DNA in these regions. Further, regions of the genome subject to stronger purifying selection have on average less Neanderthal and Denisovan introgression,8,29 as expected if most archaic ancestry tracts are being selected against. In addition, introgressed Neanderthal regions are associated with several common diseases, including depression, obesity, coronary atherosclerosis, and myocardial infarction.27 Finally, a temporal analysis of modern human samples claimed that the amount of Neanderthal DNA in Europeans has been decreasing over time,30 as expected due to purifying selection acting against Neanderthal admixture. However, this last observation is unproven and may be an artifact of the methodology used for estimating admixture proportions.31 Overall, we believe that the identification of additional (non-Neanderthal non-Denisovan) introgression tracts will help identify regions that are more likely to contain harmful variants and point to populations that have an elevated disease burden due to ancient admixture events that happened tens of thousands of years ago.
Material and Methods
GA100KP Sequence Data
The GenomeAsia 100K project analyzed a total of 1,739 individuals (1,236 newly sequenced genomes and 503 genomes from previous studies), representing 64 countries and more than 200 ethnic groups. All individuals were sequenced to high coverage (∼30×) using standard Illumina paired-end sequencing. A uniform pipeline was used for read mapping in both the public and newly sequenced genomes, followed by joint variant calling in all of the individuals using an approach similar to what was used by the ExAC consortium.32 Originally, 1,863 genomes were analyzed, but after application of strict QC filters this number was reduced to 1,739. Proper informed consent and IRB approval was obtained for all of the new genomes generated by the project. Further details on the samples, sequencing, variant calling, and quality control can be found in the GAsP paper.24
We excluded one individual from every first-degree relative pair to obtain a set of 1,667 putatively unrelated individuals from the GAsP. We accessed vcf files for these individuals from the GenomeAsia website24 and restricted our analyses to biallelic SNPs. To reduce the confounding effects of erroneous genotype calls in repetitive regions, we excluded all genomic regions that were contained within the 1000 Genomes Project strict mask file (see Web Resources). We also excluded all regions containing copy number variants (CNVs) in these samples (see below).
Identifying CNVs
We applied BIC-seq233 to identify the copy number variant regions in each sample separately. For each sample, we used SAMtools34 to determine the leftmost mapping location for all primary alignments that were assigned a mapping quality greater than zero. We then used NBICseq-norm to normalize the counts in non-overlapping bins of 100 bps, using a mappability map35 for reads of 75 bps. NBICseq-norm models the number of reads mapped to a position in the mappability map dependent on local features and calculates the expected number of mapped reads for every position in the mappability map. The ratio between the observed number and the expected number of mapped reads in a region is then a reflection of the copy number of the region. We identified a subset of 206 individuals with GC-associated biases that could not be accounted for by the regression model used in BIC-seq, and those were collated into a separate list. We then used NBICseq-seg to segment the bin counts into segments that have the same copy number state. We used “--bootstrap” to assign confidence values to the CNV calls. All segments that had a pvalue < 0.01 and a log2.copyRatio > 0.2 were subsequently called as duplications, and segments that had a pvalue < 0.01 and a log2.copyRatio < −0.2 were called as deletions.
Sample Selection
While all 1,667 unrelated individuals from the GAsP dataset were analyzed jointly, we subsampled the GAsP genomes into 9 regional groupings based primarily on geographic origin and secondarily on ethnicity for some comparisons with previous results or simulations. These included 12 Khoesan from Southern Africa, 7 Pygmies from Central Africa, 42 West Africans, 24 East Africans, 4 North Africans, 19 Middle Easterners, 75 Europeans, 315 East Asians, and 66 Melanesians (Table 1). These comparisons excluded Bantu language-speaking groups from Eastern and Southern Africa due to potential admixture in the last several thousand years. While any division of the Eurasian samples is somewhat arbitrary, we chose to focus on Europeans and East Asians (and exclude Central Asians, South Asians, and Caucasians) to help align our results with the simulations described below. Finally, we colloquially use “Melanesians” to refer to individuals indigenous to Australia, New Guinea, or the islands directly to the east of New Guinea. The estimated numbers of PGHs for all 1,667 samples is provided in Table S2.
Table 1.
The Average Number of PGHs per Individual across Different Geographical Regions
| Group | Sample Size | PGH Mean | p Valuea | Simulation Mean |
|---|---|---|---|---|
| Khoesan | 12 | 498.9 | 0.0031 | – |
| Pygmy | 7 | 428 | <10−7 | – |
| West Afr. | 42 | 334 | <10−7 | 0.6–33.1 |
| East Afr. | 24 | 266.3 | 9.77 × 10−5 | – |
| North Afr. | 4 | 118.5 | 2.26 × 10−4 | – |
| Middle East | 19 | 68.2 | <10−6 | – |
| European | 75 | 53.6 | <10−7 | 0–26.9 |
| Melanesian | 66 | 42.6 | <10−7 | 0–26.5 |
| East Asian | 315 | 35.4 | – | 0–27.6 |
“PGH mean” shows the observed averages while “Simulation mean” shows the simulated averages under a demographic model proposed by Malaspinas et al.30 with a range of different recombination assumptions. See Material and Methods for details.
p value of a Mann-Whitney U test of whether the PGH values in the specified row are significantly larger than the PGH values in the row directly below.
Identifying Archaic Admixture (“Ghost” Admixture)
We use the term “ghost” admixture to refer to potential archaic-modern human admixture involving unknown groups. We searched for long, diverged haplotypes as candidate regions for archaic human introgression, similar to our previous work.36,37 (We call these regions putative ghost haplotypes or PGHs.) Since we do not know a priori where ghost admixture may have occurred, we did not assume that African genomes had no admixture, as previous studies of Neanderthal and Denisovan admixture have done.8,29,36 (This assumption is relaxed in the next section.) We searched across the autosomes for regions with
-
1
Ten or more SNPs in complete LD (i.e., all pairwise r2 values of 1)
-
2
A minimum distance of 10 bp between consecutive SNPs identified in (1)
-
3
A minimum total length of 5 kb
-
4
A density of diagnostic SNPs of a least 2 per kb
-
5
An average frequency in the Denisovan and Neanderthal genomes of <0.1 for the derived alleles in the SNPs identified in (1)
-
6
A frequency of >0.1% and <20% for the putative haplotype
For step 4, to remove edge effects we calculated (S − 1)/(L − 1), where S is the number of SNPs identified in step 1 and L is the number of assayed (i.e., non-masked) bases between the first and last SNPs in the haplotype. While the density cutoff is arbitrary, we chose it so that the number of PGHs under null demographic models is relatively low (see below). Also, for step 5, we only used the Altai Neanderthal and Denisovan genomes. Allele frequencies are thus 0, 0.5, or 1 for each site, and we average these frequencies over all of the sites in complete LD identified in step 1. Note that our definition of PGHs uses unphased genotypes, but we colloquially refer to them as diverged haplotypes.
Testing the PGH Methodology
We tested the approach described above to see whether we could detect the signs of Denisovan admixture if we did not have access to the Denisovan genome. Similar to before, we identified PGHs fulfilling the following criteria:
-
1
Ten or more SNPs in complete LD (i.e., all pairwise r2 values of 1)
-
2
A minimum distance of 10 bp between consecutive SNPs identified in (1)
-
3
A minimum total length of 5 kb
-
4
A density of diagnostic SNPs of a least 2 per kb
-
5
An average frequency in the Neanderthal genome of <0.1 for the derived alleles in the SNPs identified in (1)
-
6
A frequency of >0.1% and <50% for the putative haplotype
-
7
A frequency of 0% among 49 West and Central Africans for the putative haplotype (because our focus for this test case is on archaic admixture outside of Africa)
Null Simulations
To estimate how many PGHs would be expected under a null model with no archaic admixture, we ran coalescent simulations38 using the demographic model proposed by Malaspinas and colleagues36 (see their Figure S07.3). We simulated 1 Neanderthal, 1 Denisovan, 50 West African, 50 European, 50 East Asian, and 50 Melanesian diploid samples in multiple 10 Mb regions and tabulated the average number of PGHs for each population using the same methodology as used for our analyses of the GAsP data.
We made two separate assumptions regarding repetitive regions for our whole-genome simulations. First, we simulated a total of 3 Gb of sequence and assumed that the scaled mutation rate was constant at θ = 0.7 / kb, to match the observed average levels of heterozygosity for the filtered data. This is equivalent to assuming that the proportion of sequence excluded using the 1000 Genomes Project strict mask is constant across the genome. Our second assumption simulated a total of 2.07 Gb of data, with a scaled mutation rate of θ = 1 / kb (which matches the observed levels of heterozygosity for assayed bases in the filtered data). This is equivalent to assuming that all of the sequence excluded using the mask is contiguous. The first assumption will underestimate the number of PGHs expected under a given evolutionary model, while the second will lead to an overestimate. We ran both to obtain a range of expectations.
Since assumptions about the recombination rate have a direct effect on levels of linkage disequilibrium and thus the expected numbers of PGHs, we reran our simulations using a range of different underlying recombination rates. Some of these assumed a constant value for the scaled recombination parameter across the genome, ranging from ρ = 0.05 to 0.4 / kb. (For comparison, the true genome-wide average is estimated to be ρ ≈1 / kb for West Africans.) Note that lower recombination rates for the null (i.e., no ancient admixture) simulations are conservative for the goal of testing whether ancient admixture occurred, since they will tend to overestimate the number of PGHs.
We also ran simulations using a distribution of recombination rates to better mimic the variation in patterns of linkage disequilibrium found in real data. We used the scaled recombination rates estimated from the HapMap YRI data39 and sorted the recombination rate values for non-overlapping 200 kb windows. Then, we divided the genome up into 10 equally sized groups (which we call “deciles”) and calculated the average recombination rate for each decile. Our simulations then involved 10 sets of 21 (or 30, see above) different 10 Mb regions, with each set having a recombination rate equal to the average value over a specific decile, or a recombination rate exactly half of the average for each decile. The results for each recombination scenario were averaged over 5 whole-genome replicates.
PGH Desert Simulations
To estimate the number of large PGH deserts expected by chance, we randomly chose 2,319 PGH locations from a single contiguous 2.068 Gb stretch of assayed sequence. (This is conservative in that modeling each autosome separately will lead to fewer large PGH deserts.) We performed 1,000 replicates and tabulated for each replicate the number of large (>7 Mb) regions that did not contain a single simulated PGH. Results are similar for other size cutoffs.
Overlap of PGHs with Coding Regions and Protein-Altering SNPs
In total, we identified 2,319 PGHs which included 58,313 putative archaic variants (i.e., identified in step 1 in the definition above) spanning 21.3 Mb. Of the putative archaic variants, 123 were non-synonymous. The proportion, 123/58,313, is 47% smaller than the total fraction of variants (with frequency > 0.1% and < 50%) in the GenomeAsia dataset that were non-synonymous (55,563/13,903,374).
Purifying Selection
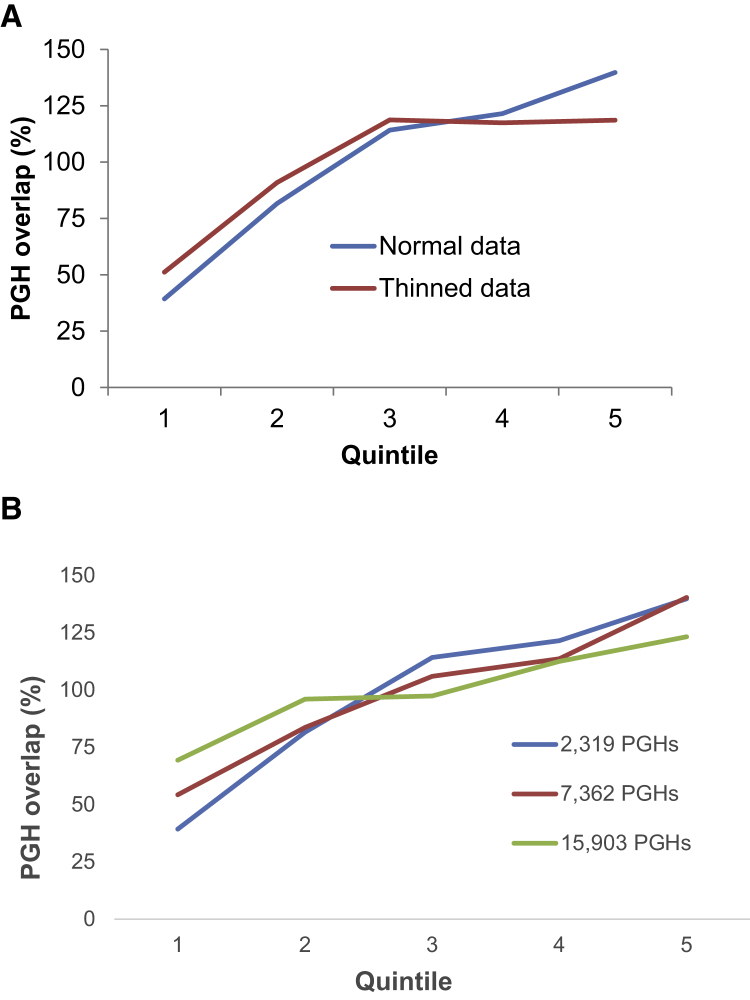
We partitioned the genome into five roughly equally sized quintiles based on the estimated strength of background selection (B, cf. McVicker et al.40). We then tabulated the total sequence overlap between the 2,319 identified PGHs and each quintile (blue line, Figure 3A). As expected, the density of SNPs varies across the quintiles. Since our PGH definition implicitly is affected by SNP density, it is possible that the quintile-PGH overlap will vary even in the absence of purifying selection on PGHs. To correct for this possibility, we randomly removed SNPs from quintiles until the density of SNPs was the same, and recalculated quintile-PGH overlaps (red line, Figure 3A).
Figure 3.
Percent Overlap of PGHs Relative to Expectations, Stratified by the Strength of Purifying Selection
Quintile 1 is the fifth of the genome with the strongest purifying selection, while quintile 5 has the weakest.
(A) Results for 2,319 PGHs in blue and results for a highly conservative thinned version of the data in red.
(B) Results using increasingly less stringent definitions for PGHs. See Material and Methods for details.
Alternate PGH Definitions
The initial PGH definition was formulated in part to minimize the number of false positives. So, it is likely to be highly conservative, with many true ghost haplotypes being missed. We explored two alternative (less stringent) criteria for PGHs and examined the evidence for purifying selection in these additional PGHs. These criteria were the same as before, except as noted below:
-
1
Eight or more SNPs in complete LD, a minimum total length of 4 kb, a density of diagnostic SNPs of at least 1.6 per Kb, and a frequency of <50% for the putative haplotype.
-
2
Same as (1), but with six or more SNPs in complete LD and a density of diagnostic SNPs of at least 1.2 per kb
Under these criteria, we identified 7,362 and 15,903 PGHs, respectively. This means there were 5,043 PGHs that satisfied (1) but not the original criteria, and 8,541 PGHs that satisfied (2) but not (1). The relative overlap between these additional classes of PGHs and the background selection quintiles is shown in Figure 3B.
Results
We looked for evidence of admixture with unknown archaic hominins by searching for long, diverged haplotypes unlikely to be inherited from Neanderthals or Denisovans, using a method similar to our previous work.36,37 We call these regions “putative ghost haplotypes,” or PGHs.
Testing Our Approach
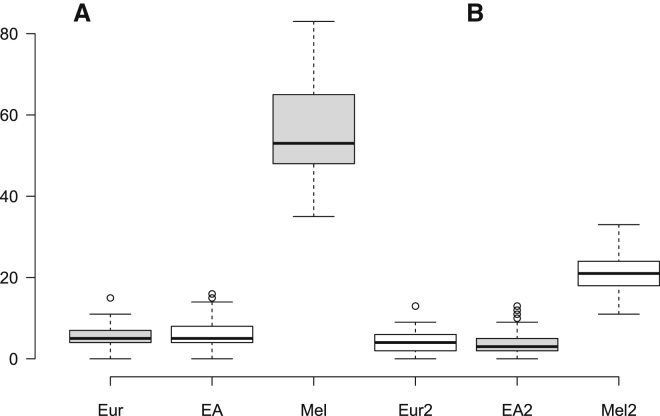
As proof-of-principle, we first analyzed the non-African samples from the GAsP assuming that the Neanderthal genome7 was known but the Denisovan genome11 was unknown. We wanted to see whether our approach could identify Melanesian genomes (thought to have experienced ∼4%–5% Denisovan admixture9,11) as likely candidates for archaic introgression. (Here we colloquially use the term “Melanesian” to refer to individuals in the GAsP indigenous to Australia or Papua New Guinea.) We tabulated the number of non-African, non-Neanderthal PGHs across 66 Melanesian, 75 European, and 315 East Asian genomes (Figure 1A). The Melanesian genomes contained 10–20 times more PGHs than did the European and East Asian genomes, and this difference is highly significant (Mann-Whitney U test, p < 10−8). We then recalculated the number of PGHs for each genome excluding haplotypes sharing similarity with the Altai Denisovan genome (Figure 1B). The relative differences in average numbers of PGHs across groups was substantially smaller but still significant (p < 10−5). These results show that archaic admixture events (such as the previously documented Denisovan admixture) can be detected purely from patterns of linkage disequilibrium (see also Skov et al.25). However, the differences between continental groups that remain in Figure 1B also reflect the difficulty in identifying Denisovan admixture tracts given the large estimated divergence between the Altai Denisovan genome and the Denisovans that admixed with the ancestors of present-day Melanesians11 and perhaps also the effects of additional admixture events with an unknown hominin group36 (see below for further discussion).
Figure 1.
Boxplot Showing the Distribution of the Number of Putative Ghost Haplotypes (PGHs) per Individual for Different Continental Groups
Eur refers to 75 European samples, EA to 315 East Asian samples, and Mel to 66 Melanesian samples. The whiskers are defined as 1.5 times the interquartile range.
(A) PGHs identified assuming the Denisovan genome was unknown.
(B) PGHs identified assuming the Denisovan genome was known.
Numbers of PGHs Vary across Populations
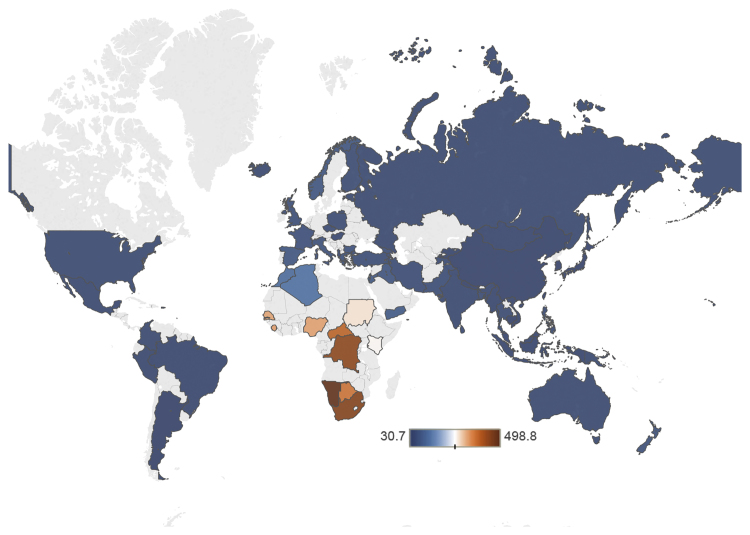
We next searched for PGHs across the full GAsP dataset. We found a total of 2,319 autosomal PGHs across all individuals. We also observed a striking gradient in the number of PGHs, with sub-Saharan African individuals containing 5–15 times more PGHs than non-African individuals (Figure 2; Table S2). The average number of PGHs was highest in Khoesan genomes, followed by Central African Pygmy genomes, West African genomes, East African genomes, North African genomes, Middle Eastern genomes, and other Eurasian genomes (Table 1). This is consistent with a primary admixture event occurring in the ancestors of present-day Khoesan populations, with subsequent migration between modern human populations leading to the observed gradient in PGH density across populations.
Figure 2.
Heatmap Showing the Average Number of PGHs Stratified by Country of Origin
Countries with no samples are shaded light gray. The raw PGH counts on which this figure is based are provided in Table S2.
Since our PGH definition depends indirectly on SNP density, it is likely that the expected number of PGHs varies across populations even under a null model with no archaic admixture. To estimate the magnitude of this effect and to determine whether the number of PGHs across populations is consistent with the expectation of null models not containing archaic admixture, we simulated whole-genome sequence data using the demographic model proposed by Malaspinas and colleagues,36 and a range of different assumptions about recombination (see Material and Methods). We found that the number of PGHs was 10–500 times smaller in the simulated datasets than what was observed in the actual West African data, while the simulations and results were roughly consistent for the three non-African groups (Tables 1 and S3). We conclude that (1) the differences in numbers of PGHs across populations is much larger than expected under simple models of modern human demography (which incorporate factors such as the presumed population bottleneck in non-African populations) and (2) the excess of PGHs across all sub-Saharan African populations provides evidence for at least one major archaic admixture event, likely subsequent to the initial divergence between African and non-African populations.
Weak Evidence for Non-Neanderthal, Non-Denisovan Archaic Admixture outside of Africa
One recent study claimed to find a signal of unknown archaic admixture in South Asians (and shared with Melanesians).41 Despite our dense sampling of caste and language groups throughout South Asia (which included Jarawa and Onge from the Andaman Islands), we find no increase in the estimated number of PGHs in any South Asian or Melanesian group, when compared to all other non-African populations (Table S2). Similarly, we studied samples from three different locations on the island of Flores, including individuals of small stature (adult height < 150 cm) from Rampasasa, close to the Liang Bua cave where H. floresiensis samples have been found. These groups do not show any increase in the number of PGHs compared with other populations in Southeast Asia. Given the magnitude of difference between observed and expected numbers of PGHs across populations (Table 1), we conclude that the evidence for any non-Neanderthal, non-Denisovan admixture outside of Africa is weak, considering the uncertainties in demographic model and recombination rate. In contrast, the gap between simulated and actual numbers of PGHs is much larger in sub-Saharan African populations. Given the size of this gap across a wide range of assumptions about the recombination rate, we believe our results are most easily explained by long-term isolation and population structure within sub-Saharan Africa (see Discussion).
Purifying Selection on PGHs
There is a growing body of evidence suggesting that introgressed Neanderthal regions are generally deleterious (i.e., harmful) in modern humans.8,26,28 If the PGHs we identified are truly the result of introgression from unknown archaic human sources, we would expect them to display the same indirect signs of past and current purifying selection, since at the time of admixture the archaic human group probably had a small effective population size (and thus a large genetic load). Consistent with these expectations, we find that PGH locations are preferentially located away from coding regions (61% reduction over the expectation if PGH locations were randomly and uniformly distributed across the genome), and that amino-acid-altering mutations are underrepresented in SNPs contained in PGHs (47% reduction; see Material and Methods). Further, the genome contains large regions bereft of PGHs (Table 2), which is expected if there are allelic incompatibilities between archaic and modern human DNA. The observed number of large (>7 Mb of assayed sequence) PGH deserts is much higher than expected if PGH locations were randomly chosen (13 observed versus 0.89 expected, p < 10−7).
Table 2.
Locations of Large (>7 Mbp of Assayed Bases) Genomic Regions without Any PGHs, in hg19 Coordinates
| Chromosome | Approximate Position (Mbp) |
|---|---|
| 2 | 22.47–34.46 |
| 2 | 51.49–72.34 |
| 2 | 140.79–150.87 |
| 3 | 45.69–56.19 |
| 3 | 74.15–83.93 |
| 5 | 85.87–94.42 |
| 5 | 135.26–145.38 |
| 7 | 126.70–139.28 |
| 9 | 12.12–21.40 |
| 9 | 92.23–105.18 |
| 14 | 58.13–68.22 |
| 15 | 66.80–79.64 |
| 16 | 65.01–75.93 |
To explore the issue of purifying selection against PGHs further, we subdivided the genome into quintiles, based on the estimated strength of purifying selection at each genomic location.40 Among these quintiles, we find a strong negative correlation between the strength of purifying selection and the amount of overlap with PGH regions (Figure 3). For example, the quintile of the genome subject to the strongest purifying selection contains only 1.3 Mb of PGH regions, compared with 5.0 Mb in the quintile with the weakest purifying selection. The same qualitative pattern still holds true even if we conservatively thin the data to have a uniform density of SNPs across the quintiles (Figure 3A; see Material and Methods for details).
Alternate PGH Definitions
Our PGH definition was purposely conservative, in order to minimize the number of false positives expected. We also considered two less stringent definitions of PGHs (which identified 7,362 and 15,903 total haplotypes) and explored whether these expanded list of PGHs also showed evidence of purifying selection. As before, we found strong negative correlations between the strength of purifying selection and the amount of overlap with PGH regions (Figure 3B). This provides indirect evidence that we are capturing mostly archaic human introgressed regions even with the least stringent PGH definition (which includes 15,903 PGHs spanning a total of 113.5 Mb of sequence) and suggests that there are many more real ghost haplotypes besides the ones analyzed in this study.
Discussion
In summary, our analyses documented a striking surplus of long, diverged haplotypes (PGHs) in sub-Saharan African genomes, with a strong gradient from south to north within Africa. There are several complementary lines of evidence suggesting that these haplotypes are the result of recent introgression between modern and archaic humans. First, the excess of PGHs in sub-Saharan African groups is unexpected under standard models of modern human demography (Table 1). Second, our stringent filtering minimizes any confounding effects due to mapping and alignment errors. Third, our PGH approach would have easily identified Denisovan admixture into Melanesian genomes even without the presence of the Denisovan genome. In addition, there is strong indirect evidence that the identified PGHs have been subject to purifying selection, as expected if they really originated from introgression with archaic humans.
We note that the extent of admixture between modern and archaic humans, while clearly of historical and evolutionary interest, also has direct implications for human disease genetics. If selection against introgressed DNA is ongoing, then our results suggest that individuals with sub-Saharan African ancestry are likely to have an elevated disease burden due to the presence in their genomes of maladapted archaic human DNA. Given the much larger difference in the number of PGHs between African and non-African populations (Figure 2) compared with the differences between non-African populations shown in Figure 1, we speculate that the magnitude of this admixture (in terms of the divergence of the introgressing archaic human population, the amount of admixture, or both) is substantially greater than the previously documented Neanderthal and Denisovan admixture events. The results shown in Figure 3B are consistent with high (e.g., ≥5%) rates of admixture into some sub-Saharan African populations. This is consistent with the admixture rate estimates obtained in another recent study.18
One caveat though is that our approach cannot directly distinguish between models of long-term isolation and population structure within sub-Saharan African modern humans and the archaic admixture scenario explored above. We believe that archaic admixture is more likely due to the evidence that PGHs have been subject to purifying selection, but more work needs to be done to rigorously compare different historical scenarios. In addition, further sequencing and analysis of genomes from indigenous Central and Southern African groups will be required to obtain a better null model for African modern human demographic history than the Malaspinas model36 used here.
We searched for signs of ongoing purifying selection using two different approaches. First, we looked for overlap between SNPs used to identify PGHs and homozygous protein truncating variants (PTVs, which are likely to be deleterious) identified in the GAsP dataset.24 Out of the 212,225 total SNPs in the expanded list of 15,903 PGHs, none of them are homozygous PTVs. We also interrogated a list of 725 likely pathogenic autosomal variants identified using ClinVar (Table S4b in GenomeAsia100K Consortium24). None of these were present in the expanded list of PGH sites. So, there is no evidence of overlap between mutations thought to be deleterious and PGHs. We tentatively conclude that if purifying selection against PGHs is ongoing, it is likely acting on weakly deleterious variants with more subtle effects on fitness.
Declaration of Interests
E.S. was previously a full-time employee of Genentech, Inc. and is currently a full-time employee of MedGenome, Inc. J.D.W. is a consultant for Genentech, Inc., and MedGenome, Inc.
Acknowledgments
J.D.W. was supported in part by NIH grant R01 GM115433. A.R. was supported in part by NIH grant P30 CA044579 to the UVA Cancer Center.
Published: December 5, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.11.005.
Contributor Information
Jeffrey D. Wall, Email: jeff.wall@ucsf.edu.
the GenomeAsia 100K Consortium:
Jeffrey D. Wall, Eric Stawiski, Aakrosh Ratan, Hie Lim Kim, Changhoon Kim, Ravi Gupta, Kushal Suryamohan, Elena S. Gusareva, Rikky Wenang Purbojati, Tushar Bhangale, Vadim Stepanov, Vladimir Kharkov, Markus S. Schrӧder, Vedam Ramprasad, Jennifer Tom, Steffen Durinck, Qixin Bei, Jiani Li, Joseph Guillory, Samir Phalke, Analabha Basu, Jeremy Stinson, Sandhya Nair, Sivasankar Malaichamy, Nidhan K. Biswas, John C. Chambers, Keith C. Cheng, Joyner T. George, Seik Soon Khor, Jong-Il Kim, Belong Cho, Ramesh Menon, Thiramsetti Sattibabu, Akshi Bassi, Manjari Deshmukh, Anjali Verma, Vivek Gopalan, Jong-Yeon Shin, Mahesh Pratapneni, Sam Santhosh, Katsushi Tokunaga, Badrul M. Md-Zain, Kok Gan Chan, Madasamy Parani, Purushothaman Natarajan, Michael Hauser, R. Rand Allingham, Cecilia Santiago-Turla, Arkasubhra Ghosh, Santosh Gopi Krishna Gadde, Christian Fuchsberger, Lukas Forer, Sebastian Shoenherr, Herawati Sudoyo, J. Stephen Lansing, Jonathan Friedlaender, George Koki, Murray P. Cox, Michael Hammer, Tatiana Karafet, Khai C. Ang, Syed Q. Mehdi, Venkatesan Radha, Viswanathan Mohan, Partha P. Majumder, Sekar Seshagiri, Jeong-Sun Seo, Stephan Schuster, and Andrew S. Peterson
Web Resources
1000 Genomes Project strict mask file (which contains a list of excluded regions), ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/accessible_genome_masks/20141020.strict_mask.whole_genome.bed
GenomeAsia 100K, https://browser.genomeasia100k.org/
Supplemental Data
References
- 1.White T.D., Asfaw B., DeGusta D., Gilbert H., Richards G.D., Suwa G., Howell F.C. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 2.Clark J.D., Beyene Y., WoldeGabriel G., Hart W.K., Renne P.R., Gilbert H., Defleur A., Suwa G., Katoh S., Ludwig K.R. Stratigraphic, chronological and behavioural contexts of Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:747–752. doi: 10.1038/nature01670. [DOI] [PubMed] [Google Scholar]
- 3.Scerri E.M.L., Thomas M.G., Manica A., Gunz P., Stock J.T., Stringer C., Grove M., Groucutt H.S., Timmermann A., Rightmire G.P. Did our species evolve in subdivided populations across Africa, and why does it matter? Trends Ecol. Evol. 2018;33:582–594. doi: 10.1016/j.tree.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilmshurst J.M., Hunt T.L., Lipo C.P., Anderson A.J. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proc. Natl. Acad. Sci. USA. 2011;108:1815–1820. doi: 10.1073/pnas.1015876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slocum J. New York Century Co.; New York: 1900. Sailing alone around the world; p. 212. [Google Scholar]
- 6.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reich D., Green R.E., Kircher M., Krause J., Patterson N., Durand E.Y., Viola B., Briggs A.W., Stenzel U., Johnson P.L. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reich D., Patterson N., Kircher M., Delfin F., Nandineni M.R., Pugach I., Ko A.M., Ko Y.C., Jinam T.A., Phipps M.E. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer M., Fu Q., Aximu-Petri A., Glocke I., Nickel B., Arsuaga J.L., Martínez I., Gracia A., de Castro J.M., Carbonell E., Pääbo S. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2014;505:403–406. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 13.Meyer M., Arsuaga J.L., de Filippo C., Nagel S., Aximu-Petri A., Nickel B., Martínez I., Gracia A., Bermúdez de Castro J.M., Carbonell E. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature. 2016;531:504–507. doi: 10.1038/nature17405. [DOI] [PubMed] [Google Scholar]
- 14.Hammer M.F., Woerner A.E., Mendez F.L., Watkins J.C., Wall J.D. Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. USA. 2011;108:15123–15128. doi: 10.1073/pnas.1109300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wall J.D., Lohmueller K.E., Plagnol V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol. Biol. Evol. 2009;26:1823–1827. doi: 10.1093/molbev/msp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh P., Woerner A.E., Wall J.D., Lachance J., Tishkoff S.A., Gutenkunst R.N., Hammer M.F. Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies. Genome Res. 2016;26:291–300. doi: 10.1101/gr.196634.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lachance J., Vernot B., Elbers C.C., Ferwerda B., Froment A., Bodo J.M., Lema G., Fu W., Nyambo T.B., Rebbeck T.R. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durvasula A., Sankararaman S. Recovering signals of ghost archaic admixture in the genomes of present-day Africans. bioRxiv. 2018 [Google Scholar]
- 19.Brauer G. The origin of modern anatomy: By speciation or intraspecific evolution? Evol. Anthropol. 2008;17:22–37. [Google Scholar]
- 20.Rightmire G.P. Out of Africa: modern human origins special feature: middle and later Pleistocene hominins in Africa and Southwest Asia. Proc. Natl. Acad. Sci. USA. 2009;106:16046–16050. doi: 10.1073/pnas.0903930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvati K., Stringer C., Grün R., Aubert M., Allsworth-Jones P., Folorunso C.A. The Later Stone Age calvaria from Iwo Eleru, Nigeria: morphology and chronology. PLoS ONE. 2011;6:e24024. doi: 10.1371/journal.pone.0024024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wall J.D. Detecting ancient admixture in humans using sequence polymorphism data. Genetics. 2000;154:1271–1279. doi: 10.1093/genetics/154.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plagnol V., Wall J.D. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GenomeAsia100K Consortium The GenomeAsia 100K project: enabling genetic discoveries across Asia. Nature. 2019 doi: 10.1038/s41586-019-1793-z. Published online November 27, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skov L., Hui R., Hobolth A., Scally A., Schierup M.H., Durbin R. Detecting archaic introgression without archaic reference genomes. bioRxiv. 2018 doi: 10.1371/journal.pgen.1007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vernot B., Akey J.M. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 27.Simonti C.N., Vernot B., Bastarache L., Bottinger E., Carrell D.S., Chisholm R.L., Crosslin D.R., Hebbring S.J., Jarvik G.P., Kullo I.J. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 2016;351:737–741. doi: 10.1126/science.aad2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris K., Nielsen R. The Genetic Cost of Neanderthal Introgression. Genetics. 2016;203:881–891. doi: 10.1534/genetics.116.186890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sankararaman S., Mallick S., Patterson N., Reich D. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 2016;26:1241–1247. doi: 10.1016/j.cub.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Q., Posth C., Hajdinjak M., Petr M., Mallick S., Fernandes D., Furtwängler A., Haak W., Meyer M., Mittnik A. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petr M., Pӓӓbo S., Kelso J., Vernot B. The limits of long-term selection against Neandertal introgression. bioRxiv. 2018 doi: 10.1073/pnas.1814338116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xi R., Lee S., Xia Y., Kim T.M., Park P.J. Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants. Nucleic Acids Res. 2016;44:6274–6286. doi: 10.1093/nar/gkw491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derrien T., Estellé J., Marco Sola S., Knowles D.G., Raineri E., Guigó R., Ribeca P. Fast computation and applications of genome mappability. PLoS ONE. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malaspinas A.S., Westaway M.C., Muller C., Sousa V.C., Lao O., Alves I., Bergström A., Athanasiadis G., Cheng J.Y., Crawford J.E. A genomic history of Aboriginal Australia. Nature. 2016;538:207–214. doi: 10.1038/nature18299. [DOI] [PubMed] [Google Scholar]
- 37.Wall J.D., Yang M.A., Jay F., Kim S.K., Durand E.Y., Stevison L.S., Gignoux C., Woerner A., Hammer M.F., Slatkin M. Higher levels of neanderthal ancestry in East Asians than in Europeans. Genetics. 2013;194:199–209. doi: 10.1534/genetics.112.148213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudson R.R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 39.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McVicker G., Gordon D., Davis C., Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mondal M., Casals F., Xu T., Dall’Olio G.M., Pybus M., Netea M.G., Comas D., Laayouni H., Li Q., Majumder P.P., Bertranpetit J. Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat. Genet. 2016;48:1066–1070. doi: 10.1038/ng.3621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.