Abstract
Background
Lymphangioleiomyomatosis (LAM), a destructive lung disease that affects primarily women, is caused by loss-of-function mutations in TSC1 or TSC2, leading to hyperactivation of mechanistic/mammalian target of rapamycin complex 1 (mTORC1). Rapamycin (sirolimus) treatment suppresses mTORC1 but also induces autophagy, which promotes the survival of TSC2-deficient cells. Based on the hypothesis that simultaneous inhibition of mTORC1 and autophagy would limit the availability of critical nutrients and inhibit LAM cells, we conducted a phase 1 clinical trial of sirolimus and hydroxychloroquine for LAM. Here, we report the analyses of plasma metabolomic profiles from the clinical trial.
Methods
We analyzed the plasma metabolome in samples obtained before, during, and after 6 months of treatment with sirolimus and hydroxychloroquine, using univariate statistical models and machine learning approaches. Metabolites and metabolic pathways were validated in TSC2-deficient cells derived from patients with LAM. Single-cell RNA-Seq was employed to assess metabolic enzymes in an early-passage culture from an LAM lung.
Results
Metabolomic profiling revealed changes in polyamine metabolism during treatment, with 5′-methylthioadenosine and arginine among the most highly upregulated metabolites. Similar findings were observed in TSC2-deficient cells derived from patients with LAM. Single-cell transcriptomic profiling of primary LAM cultured cells revealed that mTORC1 inhibition upregulated key enzymes in the polyamine metabolism pathway, including adenosylmethionine decarboxylase 1.
Conclusions
Our data demonstrate that polyamine metabolic pathways are targeted by the combination of rapamycin and hydroxychloroquine, leading to upregulation of 5′-methylthioadenosine and arginine in the plasma of patients with LAM and in TSC2-deficient cells derived from a patient with LAM upon treatment with this drug combination.
Trial Registry
ClinicalTrials.gov; No.: NCT01687179; URL: www.clinicaltrials.gov. Partners Human Research Committee, protocol No. 2012P000669.
Key Words: 5′-methylthioadenosine, angiomyolipoma, arginine, autophagy, hydroxychloroquine, lymphangioleiomyomatosis, mechanistic/mammalian target of rapamycin complex 1, metabolomics, polyamines, rapamycin, single-cell transcriptomics, sirolimus, tuberous sclerosis
Abbreviations: AMD1, adenosylmethionine decarboxylase 1; DMEM, Dulbecco’s modified Eagle’s medium; DMSO, dimethyl sulfoxide; EBAM, empirical Bayes analysis of microarrays; FBS, fetal bovine serum; FDR, false discovery rate; LAM, lymphangioleiomyomatosis; LPC, lysophosphosphatidylcholine; MTA, 5′-methylthioadenosine; mTORC1, mechanistic/mammalian target of rapamycin complex 1; ODC1, ornithine decarboxylase 1; PRMT5, protein arginine methyltransferase 5; SAIL, Sirolimus and Autophagy Inhibition in Lymphangioleiomyomatosis; SMS, spermine synthase; SOM, self-organizing map; SRM, spermidine synthase; TAG, triacylglycerol; TSC, tuberous sclerosis complex
Lymphangioleiomyomatosis (LAM) is characterized by cystic lung destruction and diffuse proliferation of smooth muscle-like “LAM cells,” which carry inactivating mutations in the TSC1 or TSC2 tumor suppressor gene,1, 2, 3 leading to hyperactivation of mTORC1.4, 5 Aberrant activation of mTORC1 in LAM cells suppresses autophagy via the kinase ULK1 and the transcription factor TFEB.6 In tumor cells and in TSC2-deficient cells, upregulation of autophagy can promote cell survival by providing nutrients and metabolic precursors.7 TSC2-deficient cells have distinctive autophagy-dependent pentose phosphate pathway alterations, increases in glucose utilization, decreases in mitochondrial oxygen consumption, and increases in mitochondrial reactive oxygen species production.8 Rapamycin (sirolimus) is approved by the Food and Drug Administration for the treatment of LAM, but continuous treatment is required because lung function declines when the therapy is discontinued.9
We have previously found in mouse models, using both genetic and pharmacologic approaches, that the combination of mTORC1 and autophagy inhibition was more effective than either treatment alone in inhibiting the survival of TSC2-deficient cells.10 To test this in humans, we conducted a phase 1 clinical trial (Sirolimus and Autophagy Inhibition in Lymphangioleiomyomatosis [SAIL])11 of combinatorial treatment with sirolimus and hydroxychloroquine for 24 weeks, followed by 24 weeks of observation off treatment.12 The SAIL trial showed potential efficacy in preventing lung function decline when the therapy was discontinued. Because both mTORC1 and autophagy are critical regulators of cellular metabolism, we hypothesized that the impact of the therapy could be detected in the plasma metabolome. We performed metabolomics analyses on plasma samples collected before, during, and after treatment with sirolimus and hydroxychloroquine. We found an unexpected upregulation of many intermediate metabolites in polyamine metabolism, with arginine and 5′-methylthioadenosine (MTA) being the most significantly elevated. Alterations of selected metabolites and pathways were validated using a TSC2-deficient cell line (621-101) derived from a patient with LAM and using single-cell transcriptomic profiling of a primary culture derived from a lung specimen obtained from a patient with LAM at the time of lung transplantation.
Taken together, these data indicate a major metabolic alteration induced by treatment with sirolimus and hydroxychloroquine in LAM that is detectable in patient plasma as well as in a cellular model of LAM. These findings provide novel candidate biomarkers of treatment efficacy in LAM and reveal, to our knowledge for the first time, that polyamine metabolism is regulated by the combination of sirolimus and hydroxychloroquine.
Materials and Methods
Plasma Sample Preparation
Samples were prepared using the Microlab STAR system (Hamilton Company). Recovery standards were added for quality control purposes. Proteins were removed by methanol precipitation. Two fractions of metabolite extract were analyzed by two separate reversed-phase ultraperformance liquid chromatography-tandem mass spectrometry methods with positive ion mode electrospray ionization.
Clinical data of patients enrolled in the clinical trial (previously reported12) can be found in e-Table 1.
Single-Cell Experiment and Analysis
Primary cells were derived from an LAM end-stage lung on transplantation. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), with a doubling time of approximately 5 days. The culture (passage 2) was treated with 20 nM rapamycin or dimethylsulfoxide (DMSO) control for 24 hours, and then immediately subjected to single-cell transcriptomic profiling. Single-cell transcriptomics profiling was performed according to the 10× Genomics protocol (https://www.10xgenomics.com). All analysis was done with R package Seurat.13
Cell Metabolomics Experiment
621-101 cells (passage 24), 621-103 (passage 31), or normal human fibroblasts (Medical Research Council cell strain 5) were cultured in DMEM with 10% FBS to 80% confluency. Cells were treated for 24 hours with DMSO (1 μL/10 mL of medium), 20 nM rapamycin, 5 μM hydroxychloroquine, or a combination of 20 nM rapamycin and 5 μM hydroxychloroquine. Metabolomic profiling was done as previously described.14
Univariate Analysis
The R Limma package was used to determine fold change, and R base was used for t-testing and for generating volcano plots. For multigroup analysis, MetaboAnalyst was used for one-way analysis of variance with associated post hoc analyses and correlation analyses to identify significant metabolite biomarkers of treatment.
Detailed methods are provided in the online supplement.
Results
Study Design and Analysis Pipeline
Seventy-eight plasma samples were collected from 13 subjects with LAM over seven visits: before treatment (n = 13, one visit), during treatment (n = 47, four visits) and after coadministration (n = 18, two visits) of sirolimus and hydroxychloroquine (Figs 1A, 1B, e-Table 1). All samples were obtained after overnight fasting and processed according to standardized methodology.15 None of the patients enrolled in the SAIL trial had tuberous sclerosis complex (TSC). Clinical data from the patients can be found in e-Table 1.
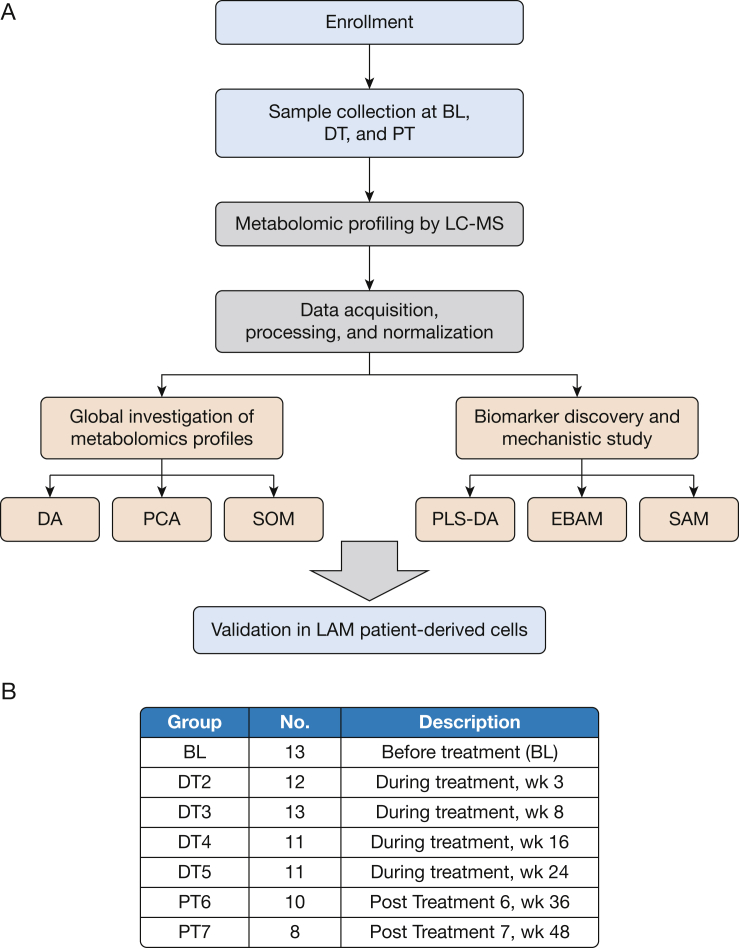
Figure 1.
Overview of data collection and analysis. A, Sample collection and analysis pipeline. Plasma samples were collected before treatment as baseline reference, during a 24-week treatment period consisting of four visits, and from the posttreatment observation period consisting of two visits. Metabolomic profiling was performed by liquid chromatography-mass spectrometry. Principal component analysis and a self-organizing map were used for global metabolomic profiles analysis. Differential analysis was used for identification of changes of metabolite levels caused by treatment. Three statistical models were employed for biomarker identification: partial least squares-discriminant analysis, empirical Bayes analysis of microarrays, and significance analysis of microarrays. Identified biomarkers and metabolic pathways were validated in a TSC2-deficient cell line (621-101) derived from a patient with lymphangioleiomyomatosis. B, Summary of sample collections. Thirteen subjects were enrolled in the study and provided baseline plasma samples. During the period of treatment, plasma samples were collected at up to four visits, resulting in a total of 47 samples. In total, 18 samples were collected at two posttreatment visits. BL = baseline; DA = differential analysis; DT = during treatment; EBAM = empirical Bayes analysis of microarrays; LAM = lymphangioleiomyomatosis; LC-MS = liquid chromatography-mass spectrometry; PCA = principal component analysis; PLS-DA = partial least squares-discriminant analysis; PT = posttreatment; SAM = significance analysis of microarrays; SOM = self-organizing map.
Metabolomic profiling was performed according to established protocols.15 Two platforms were used for metabolomic and lipidomic analysis: the HD4 platform and complex lipids panel (Metabolon), respectively, with 1,975 unique chemicals detected (e-Table 2).
Principal component analysis showed that the primary driver of clustering was intersubject variance, but within each individual a treatment effect was evident (e-Fig 1A), with during-treatment samples (circles) migrating away from the baseline (squares) and the posttreatment samples (triangles). To capture this treatment-dependent variance, we employed the supervised normalization of microarray algorithm,16 which is widely used in high-throughput data analyses to remove confounding factors. After normalization, the variance attributed to treatment effect became a more prominent driver of clustering (e-Figs 1B, 1C).
Upregulation of Polyamine Metabolism and MTA Levels by Combined Sirolimus and Hydroxychloroquine Treatment
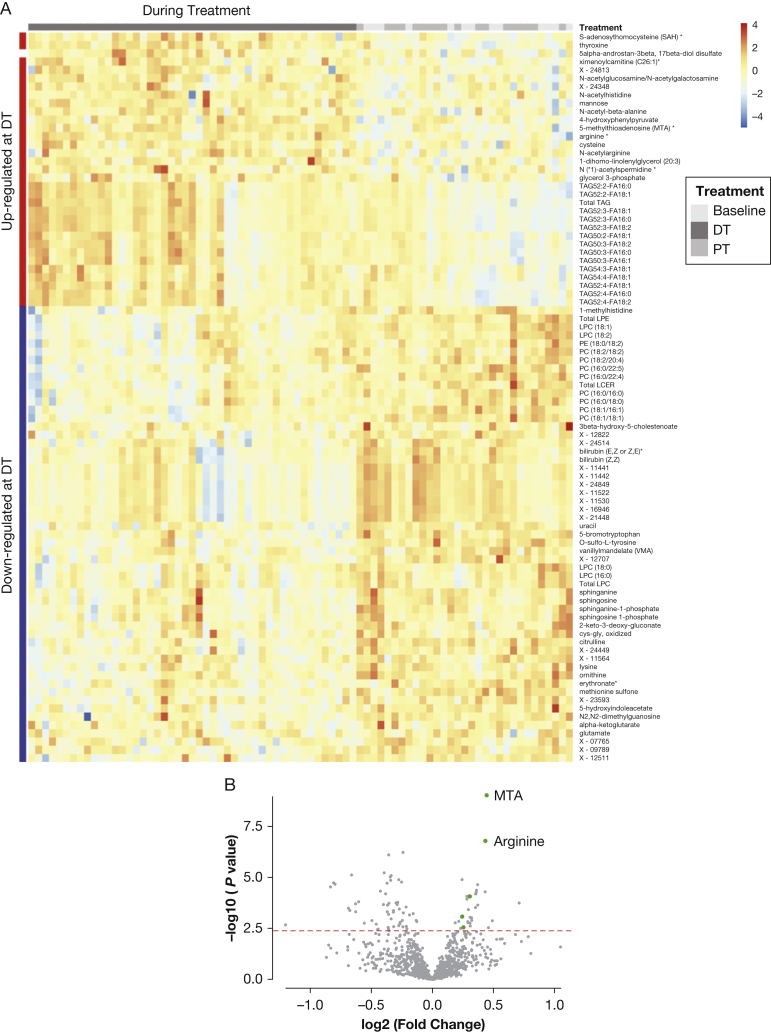
Clustering analysis showed that samples collected during treatment separated from the baseline and posttreatment samples (Fig 2A). Differential analysis was performed with the R Limma package.17 After false discovery rate (FDR) correction, 87 metabolites were significantly changed during treatment compared with baseline (Fig 2A). Only one metabolite was altered posttreatment compared with baseline (Fig 2B, e-Table 3).
Figure 2.
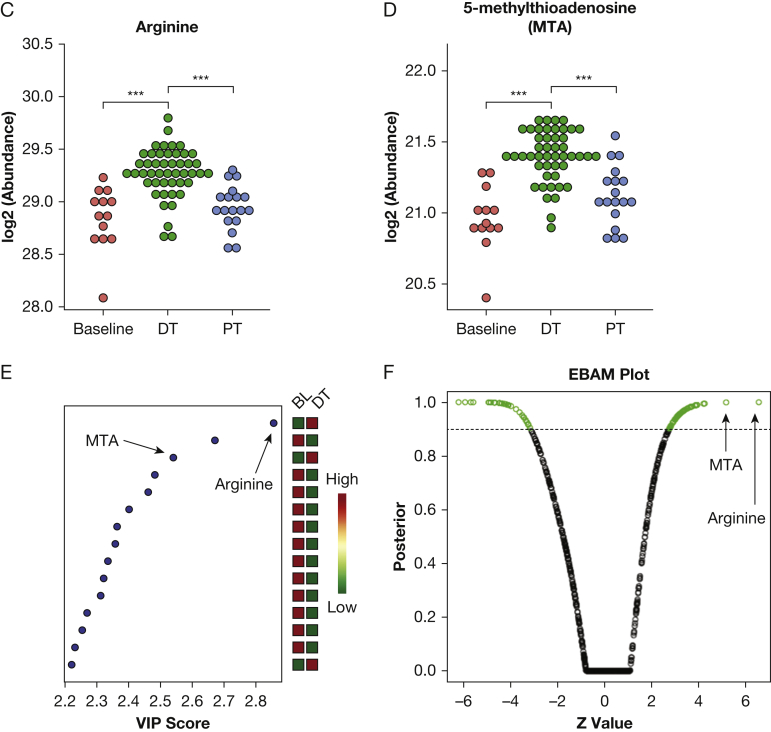
Differential analysis of metabolomic profiles across baseline (BL), during treatment (DT), and posttreatment (PT) identified upregulation of 5′-methylthioadenosine (MTA)-related polyamine metabolism by combinatorial treatment of rapamycin and hydroxychloroquine. A, Heatmap of significantly upregulated and downregulated metabolites (rows) DT and PT (columns). Total triacylglycerol (TAG), specific TAG species, and many metabolites in polyamine metabolism were upregulated DT (indicated as red bar on left side), including MTA, arginine, S-adenosyl-l-homocysteine, and acetylspermidine. B, MTA and arginine in polyamine metabolism showed the most significant upregulation DT compared with baseline. Five of 32 upregulated metabolites are intermediate metabolites in polyamine metabolism (green dots). Dots on the right sides represent upregulated metabolites during treatment, whereas dots on the left side represent downregulated metabolites during treatment. The x axis represents fold change; the y axis represents negative log10-transformed P values. The dashed red line indicates a false discovery rate (FDR) cutoff of 0.05. C, Arginine was significantly upregulated DT compared with BL and PT (***P < .001). The y axis represents normalized log2-transformed values. The DT samples from four visits (n = 47) were grouped for analysis, and PT samples from two visits (n = 18) were grouped for analysis. D, MTA was significantly upregulated DT compared with BL and PT (***P < .001). The y axis represents normalized log2-transformed values. E, MTA and arginine are among the most significant metabolites identified by partial least squares-discriminate analysis. The colored boxes on the right indicate the relative concentrations of the corresponding metabolite in BL and DT samples. The variable importance in projection (VIP) score is a weighted sum of squares of the partial least squares loadings. A full list of significant metabolites with VIP scores is provided in e-Table 4. F, MTA and arginine are the most significant biomarkers discriminating treatment group from BL identified by empirical Bayes analysis. The green circles represent metabolites that exceed the significance threshold (dotted line). MTA and arginine are labeled. All metabolites identified as biomarkers and their FDR values can be found in e-Table 5. EBAM = empirical Bayes analysis of microarrays; FA = fatty acid; LCER = lactosylceramide; LPC = lysophosphosphatidylcholine; LPE = lysophosphosphatidylethanolamine; PC = phosphatidylcholine; PE = phosphatidylethanolamine. See Figure 1 legend for expansion of other abbreviations.
Multiple metabolites in the polyamine metabolism pathway were significantly upregulated by the treatment (Fig 2B). This was evident at individual time points during treatment (Fig 2A) and in the combined group (e-Fig 2A). MTA and arginine, components of the polyamine metabolic pathway, were the two most significantly upregulated metabolites during treatment, with FDR < 0.05 (Fig 2B). Multiple other polyamine pathway metabolites were also increased during treatment, including S-adenosyl-l-homocysteine and acetylspermidine. Arginine and MTA were upregulated at all time points during the treatment, returning to baseline after treatment (Figs 2C, 2D).
Arginine and MTA were identified as the biomarkers that best separate treatment from baseline, using three distinct approaches: partial least squares-discriminant analysis18 (Fig 2E, e-Table 4), empirical Bayes analysis of microarrays (EBAM)17 (Fig 2F, e-Table 5), and significance analysis of microarrays (e-Table 6). EBAM is an empirical Bayesian method based on moderated t-statistics, which uses a two-group mixture model for null and significant features.18
Combined Sirolimus and Hydroxychloroquine Treatment Alters Lipid Profiles
Multiple lipid species were significantly impacted by combined sirolimus and hydroxychloroquine treatment, including a significant increase in plasma triacylglycerol (TAG) levels, evident in the combined and individual TAG level (Fig 2A, e-Fig 2A). TAGs remained elevated throughout treatment and dropped to baseline levels after treatment. Interestingly, within the TAGs, shorter chain-length compounds (total carbons, 36 to 46 in the three chains) were not elevated by the treatment. In comparison, 14 plasma TAGs, with chain lengths of 56 to 60 carbons, were elevated during treatment (Fig 2A, e-Fig 2A). Sphinganine, sphinganine 1-phosphate, sphingosine, sphingosine 1-phosphate, and specifically the total lactosylceramide were decreased during treatment (Fig 2A, e-Fig 2A) and returned to baseline after treatment.
Interestingly, while TAGs increased during treatment, some lysophosphosphatidylcholine (LPC) and lysophosphosphatidylethanolamine molecules normally derived from TAGs decreased (Fig 3A), including LPC (16:0), LPC (18:0), LPC (18:1), and LPC (18:2). This is of particular interest because elevated LPC levels have been previously found in plasma from patients with LAM compared with healthy control subjects, including LPC (16:0), LPC (18:0), and LPC (18:1).14
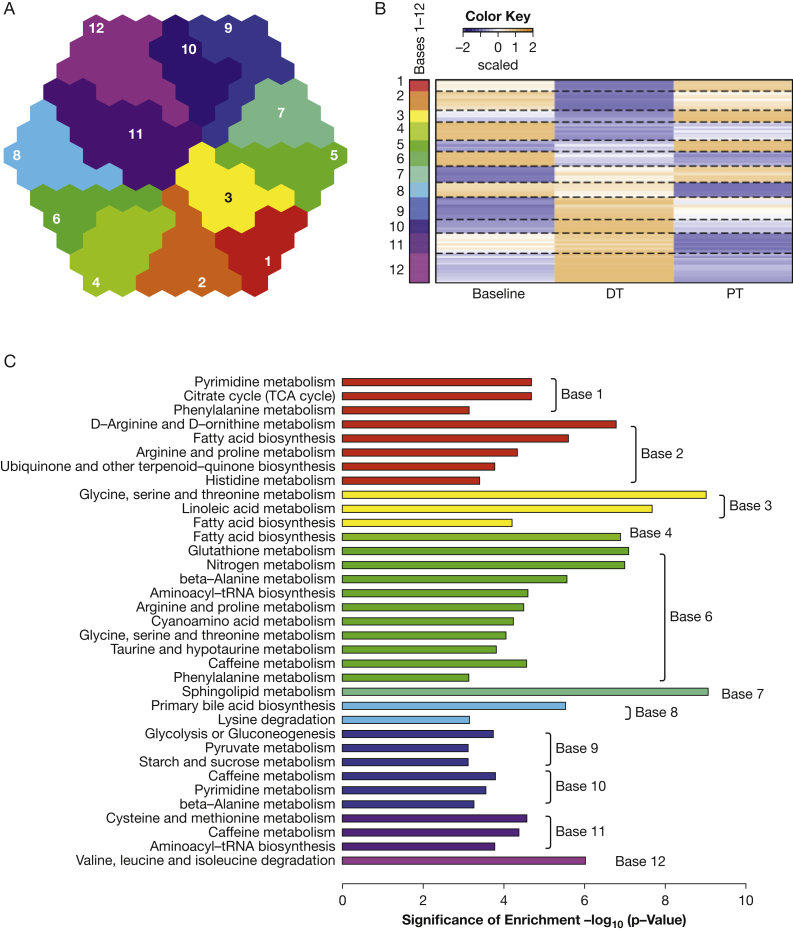
Figure 3.
Plasma metabolomic profiling reveals metabolic pathways targeted by combinatorial treatment of sirolimus and hydroxychloroquine. A, Base representation of self-organizing maps. Clusters of metabolites of similar patterns were merged into bases, which resulted in 12 bases representing clusters of metabolites differentially regulated by the treatments. Each color represents metabolites that behave similarly among subjects and across treatment points. B, Heatmap of metabolites upregulated or downregulated during treatment and posttreatment in each base, color-coded to match (A). The color indicates the direction of change. For example, metabolites in base 1 (red, top) were downregulated during treatment. C, Enriched metabolism pathways representative of each base shown in (B). Metabolites in polyamine metabolism are included in arginine and proline metabolism in base 6. TCA = tricarboxylic acid. See Figure 1 legend for expansion of other abbreviations.
Treatment-Dependent Features of Plasma Metabolomic Profiles
To further examine the association between treatment and metabolic features, we used the self-organizing map (SOM) algorithm and clustering analyses.19 After normalization and log2 transformation, longitudinal feature values were subjected to SOM analysis. The resulting SOMs of significantly variable features across baseline, during-treatment, and posttreatment samples were projected onto suprahexagonal landscapes (e-Fig 2B), using the R package supraHex19 (e-Fig 2C). Compounds within a cluster (hexagon) have similar metaprint patterns and are predicted to function in related biochemical pathways. On the basis of these similarities, hexagonal clusters were further merged into larger modules, or “bases,” for pathway analysis (Fig 3A). Features in each base display distinct upregulation or downregulation during treatment compared with baseline (Fig 3B). Pathway analysis of each base identified metabolomic pathways altered during treatment (Fig 3C). Some pathways (eg, fatty acid metabolism and pyrimidine metabolism) were enriched in multiple bases, presumably reflecting interconnected network regulation. Many of these metabolic pathways were previously shown to be associated with TSC, including prior work showing that TSC2-deficient cells have increased lipid metabolism14 and nucleotide metabolism.20, 21 Polyamine metabolism was also enriched in multiple bases.
MTA Is Elevated in TSC2-Deficient Cells Treated With Rapamycin and Chloroquine
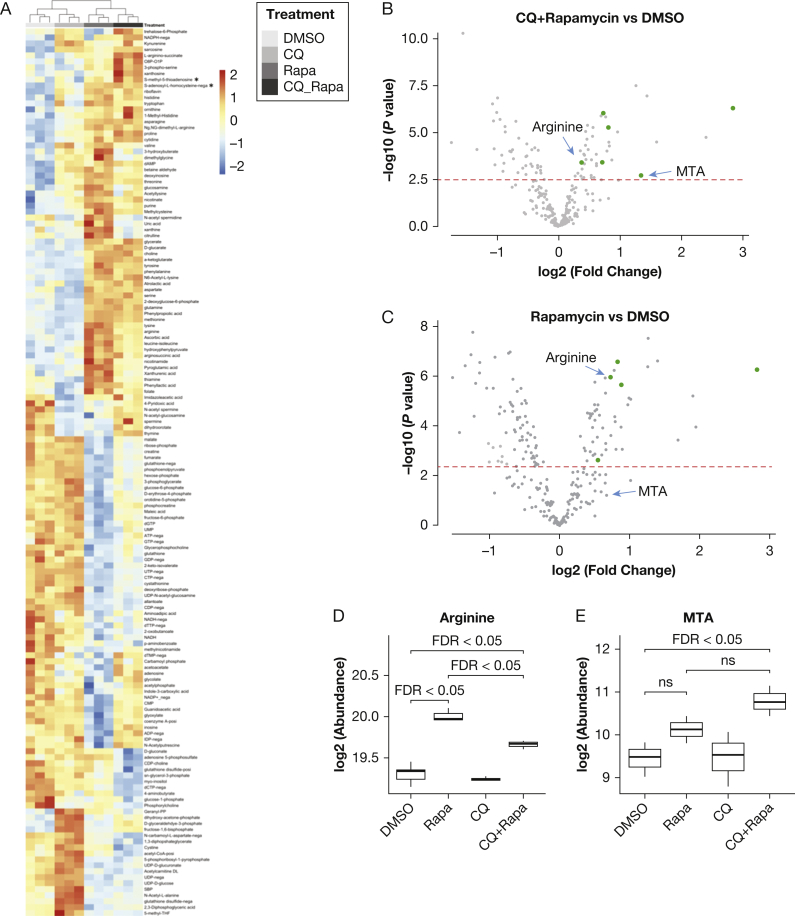
We next performed metabolomic profiling in 621-101 cells, which are derived from a sporadic LAM-associated renal angiomyolipoma and carry biallelic inactivation of TSC2. 621-101 cells were treated in triplicate for 24 hours with 20 nM rapamycin, 5 μM chloroquine, the combination of rapamycin and chloroquine, or DMSO (1 μL/10 mL of medium) as a control. Over 100 metabolites were significantly changed (FDR < 0.05) on rapamycin treatment alone compared with control, and over 100 metabolites were altered by combinatorial treatment with rapamycin and chloroquine, with many metabolites showing similar up/down patterns between these two groups (Fig 4A, e-Table 7). Samples were clustered by triplicates of treatment conditions, demonstrating the reproducibility of the biologic replicates.
Figure 4.
5′-Methylthioadenosine (MTA)-related polyamine metabolism was upregulated by combinatorial treatment of rapamycin and chloroquine in TSC2-deficient 621-101 cells derived from a patient with lymphangioleiomyomatosis. A, Heatmap of significantly upregulated or downregulated metabolites (rows) across treatment conditions (columns). 621-101 cells (passage 24) were treated for 24 hours with 20 nM rapamycin, 5 μM chloroquine, a combination of 20 nM rapamycin and 5 μM chloroquine, or dimethylsulfoxide (DMSO) control. Many intermediate metabolites in polyamine metabolism were upregulated by the combinatorial treatment with rapamycin and chloroquine, including MTA, arginine, and S-adenosyl-l-homocysteine. B, Volcano plot shows that polyamine metabolites were upregulated by treatment with rapamycin and chloroquine (CQ) compared with DMSO control, including MTA and arginine (labeled). Dots on the right sides represent metabolites upregulated by the treatment, and dots on the left side represent metabolites downregulated by the treatment. Metabolites in the polyamine metabolism pathway are depicted as green dots. The x axis represents fold change; the y axis represents negative log10-transformed P values. The dashed red line indicates a false discovery rate (FDR) cutoff of 0.05. Same experiment as described in (A). C, Arginine, but not MTA, was significantly upregulated by rapamycin. Metabolites in the polyamine pathway are represented as green dots. The horizontal dashed red line indicates an FDR cutoff of 0.05. Same experiment as described in (A). D, Arginine was significantly upregulated by the treatment with rapamycin and by the combinatorial treatment with rapamycin and CQ (FDR <0.05). Combinatorial treatment of rapamycin and chloroquine significantly decreased arginine compared with rapamycin treatment alone. E, MTA was significantly upregulated by the combinatorial treatment with rapamycin and CQ, but not by either rapamycin or chloroquine alone. No significant difference is observed between rapamycin treatment alone and combinatorial treatment of rapamycin and chloroquine (denoted as ns). ns = not significant; Rapa = rapamycin.
Using one-way analysis of variance, we found that multiple metabolites in the polyamine metabolism pathway were significantly upregulated by combinatorial treatment of rapamycin and chloroquine (Fig 4B) or by rapamycin treatment alone (Fig 4C), similar to the plasma findings. Both methionine and arginine were among the top upregulated metabolites in the cells treated with rapamycin alone or with rapamycin plus chloroquine (Figs 4A-C). Importantly, the combination of rapamycin and chloroquine significantly upregulated MTA (FDR < 0.05) (Figs 4B, 4E), but neither rapamycin treatment nor chloroquine treatment alone elevated MTA (Figs 4C, 4E). Multiple other metabolites in the polyamine pathway, including S-adenosyl-l-homocysteine, ornithine, methionine, and arginine, were upregulated in the combinatorial treatment group (Fig 4B). These results parallel the discovery of upregulation of polyamine and MTA metabolism in the clinical trial plasma samples. Finally, 621-103 cells (TSC2-deficent 621-101 cells with TSC2 re-expression) and MRC5 cells (human lung fibroblasts) were treated with DMSO, chloroquine, rapamycin, or chloroquine and rapamycin for 24 hours. No alteration in MTA levels was seen in these cells (e-Figs 3A, 3B), in contrast to what we observed in the TSC2-deficient cells, indicating that the MTA changes appear to be LAM-specific and TSC-dependent. A statistically significant increase in arginine was observed in the MRC-5 cells treated with rapamycin plus chloroquine, but not in the 621-103 cells, suggesting that the arginine changes may be TSC-dependent.
Critical Enzymes Involved in MTA Metabolism in Polyamine Metabolism Pathway Are Upregulated in LAM-Activated Fibroblasts by Rapamycin Treatment
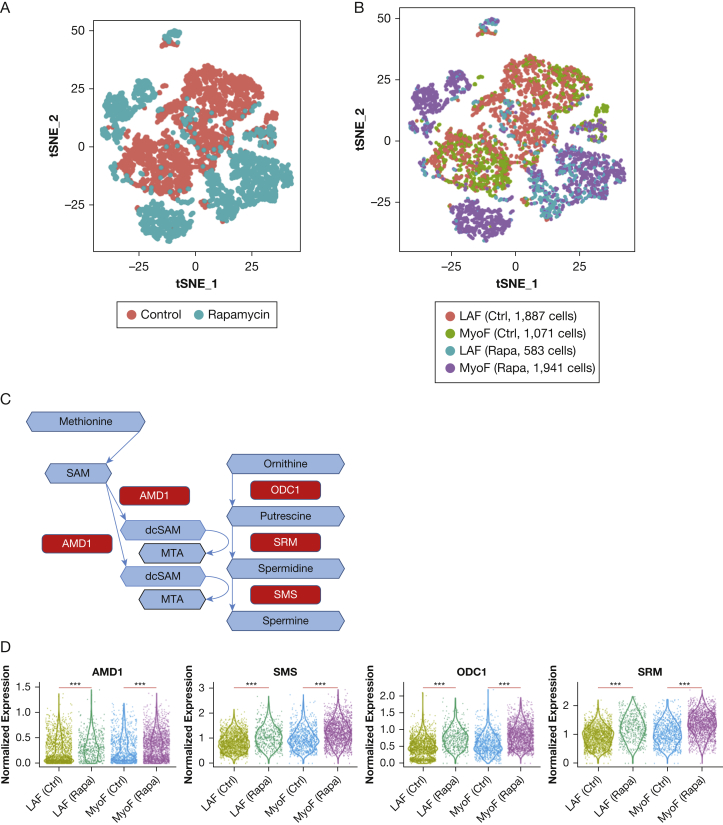
To understand the mechanisms through which MTA is upregulated by rapamycin and hydroxychloroquine in LAM, we performed single-cell transcriptomic profiling (single-cell RNA-Seq) on primary cell cultures derived from the lung of a woman with end-stage LAM who underwent lung transplantation. The patient was not receiving sirolimus or hydroxychloroquine treatment at the time of lung transplantation. Cells at passage 2 were treated with DMSO or 20 nM rapamycin for 24 hours. After initial quality control, 5,600 single-cell transcriptomes were analyzed, with an average of 1,500 genes measured per cell. Cell types were annotated by projection onto the cell-type-specific bulk RNA-Seq database Blueprint-encode, using the R SingleR package.22 On the basis of this annotation, fibroblasts represented over 95% of all cells identified (Fig 5A). Based on our analysis of LAM marker genes, including CTSK (Cathepsin K) and PMEL, we believe that very few LAM cells are in the primary culture.
Figure 5.
Expression upregulation of key enzymes in 5′-methylthioadenosine (MTA) metabolism in primary lymphangioleiomyomatosis (LAM)-activated lung fibroblasts on rapamycin treatment. A, Primary culture (passage 2) derived from an LAM lung was treated for 24 hours with rapamycin (20 nM) or dimethylsulfoxide (DMSO) control. Single-cell RNA-Seq was performed. The t-distributed stochastic neighbor embedding (t-SNE) plot shows 5,482 fibroblasts with 2,524 cells from the rapamycin treatment group (cyan) and 2,958 cells from the DMSO control group (blue). B, t-SNE plot showing LAM-activated fibroblasts (LAFs) and myofibroblasts in control and rapamycin groups. LAFs were defined by higher average expression of a panel of activated fibroblasts marker genes (FAP, PDPN, MMP2, MMP11, PDGFRA, PDGFRL, CTSK) over the mean value of all cells. C, Schematic showing selected enzymes involved in MTA metabolism. Metabolites are represented by blue hexagons. Enzymes are represented by red rectangles. D, Single-cell gene expression of key enzymes measured in LAM lung primary cultures treated with 20 nM rapamycin and DMSO (control) for 24 hours. ***FDR < 0.001. All enzymes showed upregulation by rapamycin treatment in both myofibroblasts and LAFs. The y axis represents normalized expression values. AMD1 = adenosylmethionine decarboxylase 1; Ctrl = control; dcSAM = decarboxylated S-adenosylmethionine; MyoF = myofibroblast; ODC1 = ornithine decarboxylase 1; Rapa = rapamycin; SAM = S-adenosylmethionine; SMS = spermine synthase; SRM = spermidine synthase.
A subpopulation of cells, which we named LAM-activated fibroblasts, highly expressed genes associated with activated fibroblasts,23 including fibroblast activation protein and podoplanin, shown in Figures 5B and 5C. The remaining fibroblasts were named myofibroblasts as these cells expressed high levels of myofibroblast marker genes, including smooth muscle actin (ACTA2) and myosin light-chain protein.
To determine whether the transcription of genes involved in MTA metabolism was altered in LAM-activated fibroblasts treated with rapamycin, we focused on four enzymes (Fig 5D): adenosylmethionine decarboxylase 1 (AMD1), ornithine decarboxylase 1 (ODC1), spermidine synthase (SRM), and spermine synthase (SMS). S-Adenosylmethionine is converted to decarboxylated S-adenosylmethionine by AMD1. ODC1 catabolizes ornithine to putrescine. In two consecutive reactions, the aminopropyl group of decarboxylated S-adenosylmethionine is transferred to putrescine by SRM and to spermidine by SMS to yield two molecules of MTA.24 On rapamycin treatment, expression of all four genes encoding enzymes that generate MTA was upregulated in both myofibroblasts and LAM-activated fibroblasts (Fig 5D).
Discussion and Conclusions
This study investigated the metabolomic profiles of LAM patients before, during, and after combined sirolimus and hydroxychloroquine treatment in the SAIL clinical trial. A major shift of metabolic profile was observed during treatment. Multiple significant metabolite alterations were observed during treatment, including changes in LPC, TAG, and sphingolipid levels, and metabolites in the polyamine pathway. Importantly, several LPC molecules previously found to be elevated in plasma of patients with LAM compared with healthy control subjects,14 including LPC (16:0), LPC (18:0), LPC (18:1), were decreased during treatment in the present study. This suggests that the phospholipases responsible for their production are among the targets of constitutively active mTORC1.
In our clinical specimens, MTA and multiple polyamine metabolism intermediates increased during treatment, followed by a return to baseline after the end of treatment. Furthermore, in TSC2-deficient cells derived from a patient with LAM, combinatorial treatment of rapamycin and chloroquine increased MTA production. These data suggest that chloroquine enhance rapamycin’s upregulation of MTA and polyamine metabolism in a TSC2-deficient context. These results are particularly interesting in light of a study demonstrating that aberrant activation of mTORC1 upregulates MTA-related polyamine metabolism in a prostate cancer model by activating AMD1 which converts S-adenosylmethionine to decarboxylated S-adenosylmethionine.25 Based on these data, it would be predicted that mTORC1 inhibition could lead to decreased MTA levels, in contrast to the increased MTA levels observed in our patient plasma samples and cellular model. This suggests that the mechanisms of polyamine metabolism in TSC2-deficient, patient-derived cells are distinct from other settings with hyperactive mTORC1.
We and others have previously shown that the majority of patients with sporadic LAM have somatic inactivating TSC2 mutations in their LAM cells.3, 26 We believe, therefore, that these results from TSC2-deficient cells and the clinical trial have implications for the majority of patients with sporadic LAM.
Using single-cell RNA sequencing, we observed upregulation of the four key metabolic enzymes involved in MTA production (AMD1, SMS, SRM, ODC1) on rapamycin treatment of early-passage (P2) LAM lung-derived primary cultures. Upregulation of these enzymes is consistent with the elevated MTA in the plasma samples. Interestingly, 95% of the cells in the primary culture were annotated as fibroblasts, using cell-type-specific marker gene algorithms. Using established marker genes of tumor-associated fibroblasts, we designated one-half of them as LAM-activated fibroblasts and one-half as myofibroblasts. We hypothesize that these “LAM-activated fibroblasts” and “myofibroblasts” are wild-type fibroblasts that are stimulated by LAM cells in the LAM microenvironment. So-called LAM-activated fibroblasts with wild-type TSC1 and TSC2 are an emerging concept in this field.27 Increases in MTA pathway enzymes were evident in both LAM-activated fibroblasts and myofibroblasts. This may have implications from a therapeutic perspective: MTA is a potent inhibitor of protein arginine methyltransferase (PRMT), and MTA accumulation may create a hypomorphic PRMT5 state, potentially sensitizing LAM cells to PRMT5 inhibition.28
In summary, we have found striking parallels between the effects of rapamycin and hydroxychloroquine on polyamine metabolism in patient plasma, a cell line derived from a patient with LAM, and primary LAM-derived cultures. These results reveal a previously unknown impact of mTORC1 inhibition on this critical pathway. MTA and other polyamine metabolites represent novel candidate metabolic biomarkers of treatment effect in LAM. These metabolic data support the hypothesis that the combination of rapamycin and chloroquine may have therapeutic efficacy in LAM. The SAIL trail was a phase 1 safety trial with only 13 enrolled patients and was not powered to assess whether the combinatorial treatment led to a better therapeutic response compared to sirolimus treatment alone. These metabolic biomarkers could potentially be combined with other metabolic, genetic, and protein biomarkers14, 29, 30, 31, 32 to predict disease activity and response to therapeutic interventions.
Acknowledgments
Author contributions: E. P. H. and C. P. conceived and designed the study. A. T.-D., H. J. G., S. B., I. O. R., S. E.-C., and J. M. participated in the design and conduct of the clinical trial and the acquisition and processing of specimens for the study. Y. T. performed the cellular metabolomic and single-cell RNA-Seq experiments. Y. T. performed the data analyses. Y. T., C. P., S. E.-C., and E. P. H. interpreted the data. Y. T. and E. P. H. wrote the manuscript.
Financial/nonfinancial disclosures: None declared.
Role of sponsors: The sponsors had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Other contributions: The authors thank Julie Nijmeh, PhD, for help in manuscript preparation.
Footnotes
FUNDING/SUPPORT: This study was supported by a Department of Defense Grant [W81XWH-12-1-0578] to E. P. H. and in part by the Division of Intramural Research, National Institutes of Health/National Heart, Lung, and Blood Institute (J. M.) and the Engles Fund for TSC and LAM Research.
Supplementary Data
References
- 1.Henske E.P., McCormack F.X. Lymphangioleiomyomatosis—a wolf in sheep’s clothing. J Clin Invest. 2012;122(11):3807–3816. doi: 10.1172/JCI58709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henske E.P., Jozwiak S., Kingswood J.C., Sampson J.R., Thiele E.A. Tuberous sclerosis complex. Nat Rev Dis Primers. 2016;2:16035. doi: 10.1038/nrdp.2016.35. [DOI] [PubMed] [Google Scholar]
- 3.Carsillo T., Astrinidis A., Henske E.P. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc Natl Acad Sci U S A. 2000;97(11):6085–6090. doi: 10.1073/pnas.97.11.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimobayashi M., Hall M.N. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15(3):155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 6.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkhitko A.A., Priolo C., Coloff J.L. Autophagy-dependent metabolic reprogramming sensitizes TSC2-deficient cells to the antimetabolite 6-aminonicotinamide. Mol Cancer Res. 2014;12(1):48–57. doi: 10.1158/1541-7786.MCR-13-0258-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormack F.X., Inoue Y., Moss J. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N Engl J Med. 2011;364(17):1595–1606. doi: 10.1056/NEJMoa1100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkhitko A., Myachina F., Morrison T.A. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc Natl Acad Sci U S A. 2011;108(30):12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Institutes of Health Clinical Center . National Institutes of Health; Bethesda, MD: 2012. Safety study of sirolimus and hydroxychloroquine in women with lymphangioleiomyomatosis (SAIL). NCT01687179. ClinicalTrials.gov.https://clinicaltrials.gov/ct2/show/NCT01687179 Updated October 11, 2018. [Google Scholar]
- 12.El-Chemaly S., Taveira-Dasilva A., Goldberg H.J. Sirolimus and autophagy inhibition in lymphangioleiomyomatosis: results of a phase I clinical trial. Chest. 2017;151(6):1302–1310. doi: 10.1016/j.chest.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Priolo C., Ricoult S.J., Khabibullin D. Tuberous sclerosis complex 2 loss increases lysophosphatidylcholine synthesis in lymphangioleiomyomatosis. Am J Respir Cell Mol Biol. 2015;53(1):33–41. doi: 10.1165/rcmb.2014-0379RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans A.M., Bridgewater B.R., Liu Q. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics Open Access. 2014;4(2):1–7. [Google Scholar]
- 16.Mecham B.H., Nelson P.S., Storey J.D. Supervised normalization of microarrays. Bioinformatics. 2010;26(10):1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie M.E., Phipson B., Wu D. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chong J., Soufan O., Li C. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang H., Gough J. supraHex: an R/Bioconductor package for tabular omics data analysis using a supra-hexagonal map. Biochem Biophys Res Commun. 2014;443(1):285–289. doi: 10.1016/j.bbrc.2013.11.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Sahra I., Manning B.D. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valvezan A.J., Turner M., Belaid A. mTORC1 couples nucleotide synthesis to nucleotide demand resulting in a targetable metabolic vulnerability. Cancer Cell. 2017;32(5):624–638.e5. doi: 10.1016/j.ccell.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aran D., Looney A.P., Liu L. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madar S., Goldstein I., Rotter V. “Cancer associated fibroblasts”—more than meets the eye. Trends Mol Med. 2013;19(8):447–453. doi: 10.1016/j.molmed.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Williams-Ashman H.G., Seidenfeld J., Galletti P. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem Pharmacol. 1982;31(3):277–288. doi: 10.1016/0006-2952(82)90171-x. [DOI] [PubMed] [Google Scholar]
- 25.Zabala-Letona A., Arruabarrena-Aristorena A., Martin-Martin N. mTORC1-dependent AMD1 regulation sustains polyamine metabolism in prostate cancer. Nature. 2017;547(7661):109–113. doi: 10.1038/nature22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badri K.R., Gao L., Hyjek E. Exonic mutations of TSC2/TSC1 are common but not seen in all sporadic pulmonary lymphangioleiomyomatosis. Am J Respir Crit Care Med. 2013;187(6):663–665. doi: 10.1164/ajrccm.187.6.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dongre A., Clements D., Fisher A.J., Johnson S.R. Cathepsin K in lymphangioleiomyomatosis: LAM cell-fibroblast interactions enhance protease activity by extracellular acidification. Am J Pathol. 2017;187(8):1750–1762. doi: 10.1016/j.ajpath.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Mavrakis K.J., McDonald E.R., III, Schlabach M.R. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351(6278):1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 29.Li C., Lee P.S., Sun Y. Estradiol and mTORC2 cooperate to enhance prostaglandin biosynthesis and tumorigenesis in TSC2-deficient LAM cells. J Exp Med. 2014;211(1):15–28. doi: 10.1084/jem.20131080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller S., Coveney C., Johnson J. The vitamin D binding protein axis modifies disease severity in lymphangioleiomyomatosis. Eur Respir J. 2018;52(5):1800951. doi: 10.1183/13993003.00951-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamattina A.M., Taveira-Dasilva A., Goldberg H.J. Circulating biomarkers from the phase 1 trial of sirolimus and autophagy inhibition for patients with lymphangioleiomyomatosis. Chest. 2018;154(5):1070–1082. doi: 10.1016/j.chest.2018.08.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nijmeh J., El-Chemaly S., Henske E.P. Emerging biomarkers of lymphangioleiomyomatosis. Expert Rev Respir Med. 2018;12(2):95–102. doi: 10.1080/17476348.2018.1409622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.