Abstract
Background
Despite numerous publications on mitochondrial DNA (mtDNA) in the last decade it remains to be seen whether mtDNA can be used clinically. We conducted a systematic review to assess circulating cell-free mtDNA as a biomarker of mortality in critically ill patients.
Methods
This systematic review was registered with PROSPERO (CRD42016046670). PubMed, CINAHL, the Cochrane Library, Embase, Scopus, and Web of Science, and reference lists of retrieved articles were searched. Studies measuring circulating cell-free mtDNA and reporting on all-cause mortality in critically ill adult and pediatric patients were included. The primary and secondary outcomes were mortality and morbidity, respectively.
Results
Of the 1,566 initially retrieved publications, 40 studies were included, accounting for 3,450 critically ill patients. Substantial differences between studies were noted in how mtDNA was isolated and measured. Sixteen of the 40 included studies (40%) explored the association between mtDNA levels and mortality; of those 16 studies, 11 (68.8%) reported a statistically significant association. The area under the receiver operating characteristic (AUROC) curve for mtDNA and mortality was calculated for 10 studies and ranged from 0.61 to 0.95.
Conclusions
There is growing interest in mtDNA as a predictor of mortality in critically ill patients. Most studies are small, lack validation cohorts, and utilize different protocols to measure mtDNA. When reported, AUROC analysis usually suggests a statistically significant association between mtDNA and mortality. Standardization of mtDNA protocols and the completion of a large, prospective, multicenter trial may be warranted to firmly establish the clinical usefulness of mtDNA.
Key Words: biomarkers, circulating cell-free DNA, critical illness, mitochondrial DNA, mortality
Abbreviations: APACHE, Acute Physiology and Chronic Health Evaluation; AUC, area under the curve; CSF, cerebrospinal fluid; DAMP, damage-associated molecular pattern; MELD, Model for End-Stage Liver Disease; mtDNA, mitochondrial DNA; QUIPS, Quality in Prognosis Studies; TLR-9, Toll-like receptor 9
The complex mechanisms by which mitochondria regulate innate immunity and whether extracellular mitochondrial DNA (mtDNA) influences the pathogenesis of disease are currently the subject of intense research. Thus far, murine models have linked mtDNA to the development of posttraumatic acute lung injury, ventilator-associated pneumonia, inflammatory arthritis, acute kidney injury, and nonalcoholic steatohepatitis.1, 2, 3, 4, 5 There is also mounting evidence that extracellular mtDNA might be useful as a biomarker of human disease severity. One study of critically ill patients found that mtDNA was significantly associated with mortality, even after adjustment for procalcitonin and lactate.6 Investigators have also found that mtDNA performed similarly to Model for End-Stage Liver Disease (MELD) scores in patients with acetaminophen-induced acute liver failure and outperformed troponin as a biomarker of mortality in patients with pulmonary embolism.7, 8 If mtDNA can be successfully developed as a biomarker, it could provide clinicians with timely insight into disease severity affecting treatment decisions.
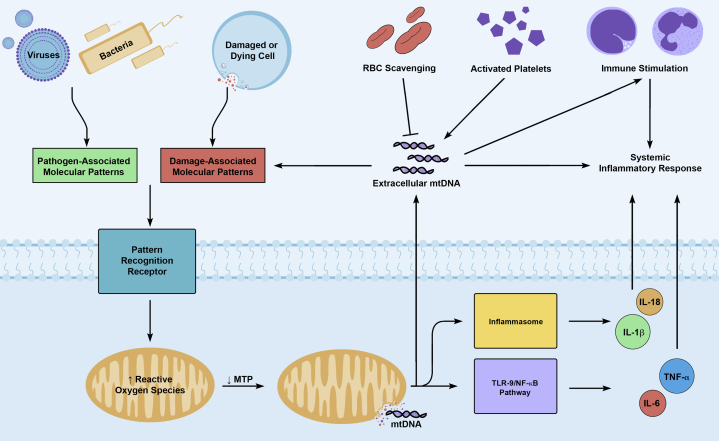
The molecular biology accounting for these observations is deeply rooted in our understanding of mtDNA as a potent damage-associated molecular pattern (DAMP). When cells are exposed to pathogen-associated molecular patterns or DAMPs, mitochondria respond by increasing their production of reactive oxygen species (Fig 1).9, 10 The resulting oxidative stress causes fragmentation of the mitochondrial genome and disruption of the mitochondrial membrane, allowing mitochondrial DAMPs, such as fragmented mtDNA, to leak into the cytosol.9, 11, 12, 13 Because of the bacterial ancestry of mitochondria, mtDNA contains unmethylated CpG repeats that are recognized by pattern recognition receptors.14, 15 This allows cytosolic mtDNA to promote expression of IL-6 and tumor necrosis factor-α or IL-1β and IL-18 via activation of the Toll-like receptor 9 (TLR-9)/nuclear factor-κB pathway or the NALP3 inflammasome, respectively.9, 16 Cytosolic mtDNA may also enter, via necroptosis or NETosis (neutrophil extracellular trap [NET] formation), the extracellular environment, where it is believed to propagate a systemic inflammatory response through recognition as a DAMP.17, 18
Figure 1.
Production and release of extracellular mitochondrial DNA. Pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) are released in the setting of infection or cellular injury, respectively. Both PAMPs and DAMPs can stimulate pattern recognition receptors, leading to increased production of mitochondrial reactive oxygen species. This causes fragmentation of the mitochondrial genome and decreased mitochondrial membrane transition permeability, allowing mitochondrial DNA (mtDNA) to enter the cytosol. mtDNA is a potent DAMP; it can stimulate inflammasomes or the Toll-like receptor 9 (TLR-9)/nuclear factor-κB pathway, leading to the production of proinflammatory cytokines (IL-1β, IL-18, tumor necrosis factor-α, IL-6). Cytosolic mtDNA can also enter the extracellular environment via necroptosis, NETosis, or platelet activation, where it can propagate the initial inflammatory response through further recognition as a DAMP by immune and nonimmune cells expressing TLR-9. RBCs have been shown to scavenge extracellular mtDNA through binding to TLR-9. MTP = membrane transition permeability; NF-κB = nuclear factor-κB; NETosis = formation of neutrophil extracellular traps (NETs); TNF-α = tumor necrosis factor-α.
Despite the strong biologic basis for the potential usefulness of mtDNA, its role as a biomarker of critical illness has yet to be firmly established. To address this, we set out to perform a systematic review to determine whether circulating mtDNA could be clinically useful as a predictor of mortality in critically ill patients.
Methods
Protocol and Registration
This systematic review was registered with PROSPERO (the International Prospective Register of Systematic Reviews) (www.crd.york.ac.uk/prospero; record No. CRD42016046670)19 and conducted in accordance with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement.20
Data Sources
A systematic review of the literature was performed by using the key phrase, “mitochondrial DNA AND (critically ill OR emergency department OR intensive care),” to search PubMed, CINAHL (the Cumulative Index to Nursing and Allied Health Literature), the Cochrane Library, Embase, Scopus, and the Web of Science from database inception to June 1, 2018. The reference lists of included studies were manually reviewed for additional publications.
Selection of Studies
Two authors (J. S. H. and I. I. S.) independently searched the databases and performed study selection, with disagreements resolved by consensus. Observational studies measuring circulating cell-free mtDNA in critically ill patients (adult or pediatric) while reporting all-cause mortality were considered eligible for inclusion. We defined “critically ill” as an admitting diagnosis that could result in admission to an ICU (further clarification provided in e-Appendix 1). No limitations were applied to the publications on the basis of time or language.
Data Extraction
Two authors (J. S. H. and I. I. S.) independently read and extracted data from the included studies according to a standard data extraction form (e-Table 1). Any disagreement between the authors was resolved by consensus. Corresponding authors of the selected studies were contacted by e-mail to request any missing data, which was then incorporated into our analysis.
Outcomes of This Systematic Review
All-cause mortality, assessed at any time point, was the primary outcome for this systematic review. Morbidity, as suggested by ICU length of stay, hospital length of stay, need for mechanical ventilation, and need for vasopressors, served as secondary outcomes.
Assessment of Risk of Bias of Included Studies
Studies in this review were assessed for bias according to a modified version of the Quality in Prognosis Studies (QUIPS) tool.21 QUIPS assesses studies of prognostic factors by prompting reviewers to classify a study as having low, moderate, or high risk of bias according to the following domains: study participation, study attrition, prognostic factor measurement, outcome measurement, study confounding, and statistical analysis. Details are provided in e-Appendix 1.
Results
Characteristics of Selected Studies
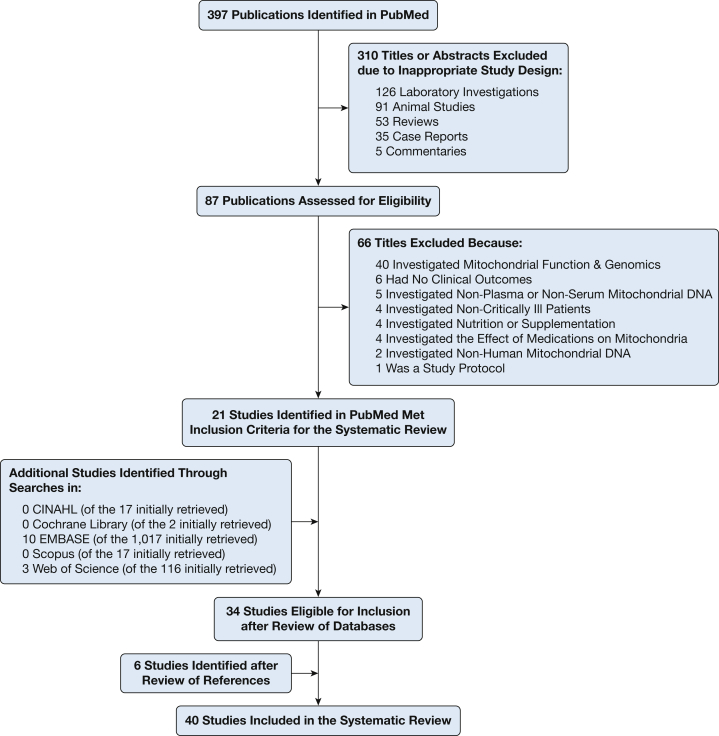
The flow diagram illustrating the study selection process can be found in Figure 2. Our search of databases yielded 1,566 publications. Thirty-four of them met our inclusion criteria.2, 6, 7, 8, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51 On reviewing the references of these publications, we found six additional studies that were deemed relevant.52, 53, 54, 55, 56, 57 Thus, 40 studies,2, 6, 7, 8, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57 involving 41 cohorts and a total of 3,450 critically ill patients (range, 12 to 753 patients per study), were included in our systematic review.
Figure 2.
Flow diagram illustrating study selection process. CINAHL = Cumulative Index to Nursing and Allied Health Literature.
Characteristics of the included studies can be found in Table 1. All of the studies were published in 2004 and later, with the majority (75%) published after 2012. Ten studies were conducted in North America, 15 studies in Europe, 13 studies in Asia, one study in Africa, and one study in Australia. All of the included studies were observational and five (12.5%) were multicenter.6, 7, 22, 28, 46 Only one study had a validation cohort.6 Patients were admitted to medicine in 17 studies (42.5%),6, 7, 8, 28, 29, 30, 32, 33, 36, 39, 46, 47, 48, 49, 52, 53, 55 to surgery in 13 studies (32.5%),2, 23, 25, 26, 27, 31, 34, 37, 40, 43, 51, 54, 57 and to neurology in five studies (12.5%).41, 44, 45, 50, 56 Four studies22, 24, 38, 42 enrolled medical and surgical patients. Only one study enrolled pediatric patients35; all of these patients had sepsis. Severity of illness scores for the individual studies are presented in Table 1.
Table 1.
Article Characteristics
| Study/Year | Country | Study Design | Discipline | Population | Setting | No. of Patients | Severity of Illnessa | Validation Cohort |
|---|---|---|---|---|---|---|---|---|
| Aslami et al22/2018 | The Netherlands | Post hoc analysis of RCT with healthy control subjects | Medicine and surgery | Post-cardiac arrest | ICU | 20 | 36°C group APACHE II: 23.8 ± 7.0 | No |
| 33°C group APACHE II: 19.3 ± 8.9 | ||||||||
| Leijte et al23/2018 | The Netherlands | Pro cohort with healthy control subjects | Surgery | Post CRS-HIPEC | OR → ICU | 20 | ECOG performance score: 1 (1-1) | No |
| Jansen et al24/2018 | The Netherlands | Pro case-control plus in vivo and in vitro | Medicine and surgery | Noninfectious SIRS | ICU | 37 | NR | No |
| Paunel-Görgülü et al25/2017 | Germany | Pro cohort with healthy control subjects plus in vitro | Surgery | Cardiopulmonary bypass | OR → ICU | 48 | CPB < 100 SAPS II: 28.5 ± 8.9 | No |
| CPB > 100 SAPS II: 29.8 ± 7.4 | ||||||||
| Hampson et al26/2017 | United Kingdom | Pro cohort with healthy control plus ex vivo | Surgery | Burn | ICU vs floor | 63 | APACHE II: 26 (12-31) | No |
| Simmons et al27/2017 | United States | Pro case control | Surgery | Trauma | ICU | 14 | No ARDS ISS: 24.5 ± 4.3 | No |
| ARDS ISS: 23.0 ± 6.3 | ||||||||
| Donnino et al28/2017 | United States | Pro cohort with healthy control subjects | Medicine | Post-cardiac arrest | ED → ICU | 102 | NR | No |
| Qin et al29/2017 | China | Pro cohort with control subjects | Medicine | Myocardial infarction | NR | 38 | NR | No |
| Simmons et al2/2017 | United States | Pro cohort with in vivo | Surgery | Ventilator-associated pneumonia | ICU | 31 | Non-VAP ISS: 27 ± 8 | No |
| VAP ISS 23 ± 8 | ||||||||
| Marenzi et al30/2016 | Italy | Pro cohort | Medicine | Myocardial infarction | NR | 753 | Nondetectable cyt c TIMI score: 4.2 ± 2.2 | No |
| Detectable cyt c TIMI score: 4.3 ± 2.3 | ||||||||
| Mohamed et al31/2016 | Egypt | Pro cohort with healthy control subjects | Surgery | Trauma | ED → ICU | 61 | NR | No |
| Timmermans et al32/2016 | The Netherlands | Pro cohort with healthy control subjects plus in vivo | Medicine | Sepsis | ICU | 121 | APACHE II: 23 (11-45) | No |
| Omura et al33/2016 | Japan | Pro cohort | Medicine | Post-cardiac arrest | ED → ICU | 21 | “Favorable” APACHE II: 24 (15-29) | No |
| “Unfavorable” APACHE II: 30 (26-37) | ||||||||
| Qin et al34/2016 | China | Pro cohort | Surgery | Cardiopulmonary bypass | OR → ICU | 46 | NR | No |
| Di Caro et al35/2016 | United States | Pro cohort with healthy control subjects plus in vitro | Pediatrics | Sepsis | ICU | 28 | PRISM score: 9.5 (1-32) | No |
| Schäfer et al36/2016 | Germany | Pro cohort with healthy control subjects plus in vivo and in vitro | Medicine | Sepsis | ICU | 165 | SAPS II: 32 (23-42) | No |
| Timmermans et al37/2016 | The Netherlands | Pro cohort with healthy control subjects plus ex vivo | Surgery | Trauma | Field → ED → ICU | 166 | ISS: 26 (17-37) | No |
| Krychtiuk et al38/2015b | Austria | Pro cohort with healthy control subjects | Medicine and surgery | Mixed | ICU | 228 | APACHE II: 20 (13-25) | No |
| Timmermans et al52/2015 | The Netherlands | Pro cohort with healthy control subjects plus ex vivo | Medicine | Post-cardiac arrest | ICU | 14 | APACHE: 24 (15-39) | No |
| Bhagirath et al39/2015 | Canada | Pro cohort with healthy control subjects plus in vitro | Medicine | Sepsis | ICU | 12 | APACHE II: 24 (11-41) | No |
| McIlroy et a l40/2015 | Australia | Pro cohort with healthy control subjects | Surgery | Trauma | ICU vs floor | 35 | ISS: 14 (9-22) | No |
| McGill et al7/2014 | United States | Pro cohort with healthy control subjects | Medicine | Acetaminophen-induced acute liver failure | NR | 69 | NR | No |
| Wang et al41/2014 | Taiwan | Pro cohort with healthy control subjects | Neurology | Traumatic brain injury | ED → ICU | 88 | GCS: 15 (13-15) ISS: 16 (11-20) |
No |
| Fernández-Ruiz et al53/2014 | Spain | Pro cohort with healthy control subjects plus in vitro | Medicine | Myocardial infarction | NR | 75 | NR | No |
| Nakahira et al6/2013c | United States | Pro cohort | Medicine | MICU | ICU | 200 | APACHE II: 24 (18-30) | Yes |
| ARDS | 243 | APACHE II: 22 (18-26) | ||||||
| Yamanouchi et al42/2013d,e | Japan | Pro cohort with healthy control subjects | Medicine and surgery | Sepsis | ED | 23d | APACHE II: 14 (11-19)d | No |
| Trauma | 37e | APACHE II: 11 (6-15)e | ||||||
| ISS: 25 (16-34)e | ||||||||
| Simmons et al43/2013 | United States | Pro cohort | Surgery | Trauma | ICU | 13 | ISS: 21.1 ± 1.7 | No |
| Gu et al54/2013 | China | Pro cohort with healthy control subjects | Surgery | Trauma | ICU | 86 | ISS: 18 (13-22) | No |
| Arnalich et al8/2013f | Spain | Pro cohort with healthy control subjects | Medicine | Pulmonary embolism | ED → ICU | 74 | PESI class III: 11, 0f | No |
| PESI class IV: 20, 16f | ||||||||
| PESI class V: 6, 21f | ||||||||
| Wang et al44/2013 | Taiwan | Pro cohort with healthy control subjects | Neurology | Subarachnoid hemorrhage | ED → ICU | 21 | “Good outcome” GCS: 15 (14.25-15) | No |
| “Poor outcome” GCS 11 (6-13) | ||||||||
| Wang et al45/2012 | Taiwan | Pro cohort with healthy control subjects | Neurology | Intracerebral hemorrhage | ICU | 60 | “Good outcome” GCS: 15 (15-15) | No |
| “Poor outcome” GCS 14 (9-15) | ||||||||
| Arnalich et al55/2012 | Spain | Retro cohort | Medicine | Post-cardiac arrest | ED | 85 | Survivors APACHE II: 35 (32-38) | No |
| Nonsurvivors APACHE II: 38 (33-41) | ||||||||
| Puskarich et al46/2012 | United States | Pro cohort with ED control subjects | Medicine | Sepsis | ED | 69 | Sepsis SOFA: 1 (0-3) | No |
| Septic shock SOFA: 4 (1-8) | ||||||||
| Kung et al47/2012 | Taiwan | Pro cohort with healthy control subjects | Medicine | Sepsis | ED → ICU vs floor | 67 | Survivors APACHE II: 18.9 ± 6.9 | No |
| Nonsurvivors APACHE II: 20.9 ± 3.8 | ||||||||
| McGill et al48/2012 | United States | Pro cohort with healthy control subjects plus in vivo | Medicine | Acetaminophen-induced acute liver failure | ED → ICU | 40 | NR | No |
| Garrabou et al49/2012 | Spain | Pro cohort with healthy control subjects plus ex vivo | Medicine | Sepsis | NR | 19 | SAPS II 45.5 ± 13.3 | No |
| Tsai et al56/2011 | Taiwan | Pro cohort with at-risk control subjects | Neurology | Acute ischemic stroke | NR | 50 | NIHSS (day 1): 7 (1-24) | No |
| Lu et al50/2010 | Taiwan | Pro cohort with healthy control subjects | Neurology | Bacterial meningitis | NR | 22 | GCS: 11 ± 4.1 | No |
| Chou et al51/2008 | Taiwan | Pro cohort | Surgery | Corrosive ingestion | ED → NR | 48 | NR | No |
| Lam et al57/2004 | Hong Kong | Pro cohort with healthy control subjects | Surgery | Trauma | ED | 38 | ISS < 16: 28 patients | No |
| ISS > 16: 10 patients |
APACHE II = Acute Physiology and Chronic Health Evaluation II; CPB < 100 = cardiopulmonary bypass less than 100 minutes; CPB > 100 = cardiopulmonary bypass greater than 100 minutes; CRS-HIPEC = cytoreductive surgery with hyperthermic intraperitoneal chemotherapy; cyt c = cytochrome c; ECOG = Eastern Cooperative Oncology Group; GCS = Glasgow Coma Scale; ISS = Injury Severity Score; MICU = medical ICU; NIHSS = National Institutes of Health Stroke Scale; NR = not reported; OR = operating room; PESI = Pulmonary Embolism Severity Index; PRISM = Pediatric Risk of Mortality; Pro = prospective; RCT = randomized control trial; Retro = retrospective; SAPS II = Simplified Acute Physiology Score II; SIRS = systemic inflammatory response syndrome; SOFA = Sepsis-Related Organ Failure Assessment; TIMI = thrombolysis in myocardial infarction; VAP = ventilator-associated pneumonia.
Severity of illness is preferentially reported for the entire cohort. However, some studies described severity of illness only according to subpopulations or “good vs bad outcome.” When this occurred, severity of illness was reported for each individual group. Please see e-Appendix 1 for further clarification of the subpopulations or on how “good vs bad outcome” was defined.
This study had a “mixed” population composed of medicine and post-cardiothoracic surgery patients.
This study was composed of a derivation cohort and a validation cohort. The data presented at the top of the row represent characteristics of the derivation cohort, whereas the data at the bottom of the row represent characteristics of the validation cohort.
This was a single study in which patients were analyzed within a sepsis or a trauma subgroup. Data denoted by a d represent characteristics of the sepsis subgroup whereas data denoted by an e represent characteristics of the trauma subgroup.
Severity of illness was reported according to PESI classification. The first and second numbers, for a given class, correspond to the number of patients with submassive and massive pulmonary embolism, respectively.
Measurement of mtDNA
Characteristics of the protocols used by investigators to measure mtDNA can be found in Table 2. Twenty-four studies (60%)2, 22, 23, 25, 26, 27, 28, 29, 32, 33, 34, 37, 40, 41, 42, 43, 44, 45, 47, 48, 50, 51, 52, 56 performed serial mtDNA measurements. The specific time points, including when mtDNA peaked for those studies, can be found in e-Table 2.
Table 2.
Description of mtDNA Assay
| Study/Year | Blood Draw(s)a | Sample Processingb | Serum or Plasma | Primer | Standards | Units of Measurement | Cutoffc |
|---|---|---|---|---|---|---|---|
| Aslami et al22/2018 | Serial, arterial, NR | WB 600g 10 4°C | Plasma | MT-ND1, MT-ND2, MT-CO3, MT-CYB | cDNA | AU | NR |
| Leijte et al23/2018 | Serial, arterial, EDTA | WB 1,600g 10 4°C → 16,000g 10 4°C | Plasma | MT-ND1 | Healthy volunteers | Fold change | — |
| Jansen et al24/2018 | Single, arterial, heparin | WB 1,550g 20 RT → 1,550g 20 RT | Plasma | MT-ND1, MT-ND2, MT-CO3 | NR | AU | NR |
| Paunel-Görgülü et al25/2017 | Serial, NR, heparin | WB 3,000g 10 | Plasma | MT-ATP8 | Nuclear DNA | mtDNA:nDNA | NR |
| Hampson et al26/2017 | Serial, NR, variable | WB 2,000g 20 RT → 13,000g 20 | Plasma | MT-CYB | K562 mtDNA | ng/mL | NR |
| Simmons et al27/2017d | Serial, venous, citrate | WB 1,200g 25 21°C | Serum | MT-ND6 | NR | ng/mL | NR |
| Donnino et al28/2017 | Serial, NR, variable | WB NR → 16,000g 10 | Plasma | D-loop, MT-tRNALeu | Whole blood DNA | Fold change | NR |
| Qin et al29/2017 | Serial, NR, EDTA | WB 1,000 rpm 15 4°C → 16,000g 15 4°C | Plasma | MT-ND1 | Plasmid DNA | Copies/μL | — |
| Simmons et al2/2017e | Serial, NR, NR | WB 1,200g 25 21°C | Serum | MT-ND6 | cDNA | ng/mL | — |
| Marenzi et al30/2016 | Single, venous, NR | P 700g 5 4°C → 18,000g 15 4°C | Plasma | MT-ND1 | Plasmid DNA | Copies/μL | NR |
| Mohamed et al31/2016 | Single, venous, EDTA | P 16,000g 5 4°C → 18,000g 15 4°C | Plasma | MT-ND1 | Plasmid DNA | Copies/μL | NR |
| Timmermans et al32/2016f | Serial, arterial, EDTA | WB 1,600g 10 4°C → 16,000g 10 4°C | Plasma | MT-ND1 | Healthy volunteers | Fold change | NR |
| Omura et al33/2016 | Serial, NR, NR | WB 1,600 rpm 10 → 0.22-μm filter | Plasma | MT-ND1, MT-CO3, MT-CYB | A549 mtDNA | μg/mL | NR |
| Qin et al34/2016 | Serial, NR, NR | NR | NR | For: 5′-CGAGCAGTAGCCCAAACAAT-3′ Rev: 5′-TGTGATAAGGGTGGAGAGGTT-3′ |
NR | Copies/μL | — |
| Di Caro et al35/2016 | Single, NR, heparin | WB 1,914g 10 | Plasma | MT-CO1 | Plasmid DNA | Copies/mL | NR |
| Schäfer et al36/2016 | Single, NR, NR | NR | Serum | MT-ATP6, D-loop | NR | fg/μL | NR |
| Timmermans et al37/2016g | Serial, venous, EDTA | WB 1,600g 10 4°C → 16,000g 10 4°C | Plasma | MT-ND1 | Healthy volunteers | Fold change | NR |
| Krychtiuk et al38/2015 | Single, arterial or venous, EDTA | WB 2,500g 30 4°C | Plasma | MT-ND2, MT-CO3, MT-CYB | Human smooth muscle cells | ng/mL | 38.2 ng/mL |
| Timmermans et al52/2015 | Serial, arterial, EDTA | WB 1,600g 10 4°C → 16,000g 10 4°C | Plasma | For: 5′-GCCCCAACGTTGTAGGCCCC-3′ Rev: 5′-AGCTAAGGTCGGGGCGGTGA-3′ |
Healthy volunteers | Fold change | NR |
| Bhagirath et al39/2015 | Single, NR, citrate | WB 2,000g 10 → 3,000g 10 | Plasma | MT-CYB, Mit 3130F, Mit 3301R | Purified mtDNA | μg/mL | NR |
| McIlroy et al40/2015 | Serial, NR, NR | P 12,000g 10 | Plasma | MT-ND3, MT-CO3 | Purified mtDNA | ng/mL | — |
| McGill et al7/2014 | Single, NR, NR | S 20,000g 10 | Serum | MT-ND, MT-CO3 | HepaRG mtDNA | ng/mL | 14 ng/mL |
| Wang et al41/2014 | Serial, venous, citrate | WB 3,000 rpm 10 | Plasma | MT-ND2 | Human gDNA | ng/mL | — |
| Fernández-Ruiz et al53/2014 | Single, venous, NR | NR | Plasma | For: 5′-CCACGGGAAACAGCAGTGAT-3′ Rev: 5′-CTATTGACTTGGGTTAATCGTGTGA-3′ |
NR | Copies/μL | NR |
| Nakahira et al6/2013 | Single, NR, EDTA | P 700g 5 4°C → 18,000g 15 4°C | Plasma | MT-ND1 | Plasmid DNA | Copies/μL | 3,200 copies/μL |
| Yamanouchi et al42/2013 | Serial, NR, NR | WB 1,600 rpm 10 → 0.22-μm filter | Plasma | MT-ND1, MT-CO3, MT-CYB | A549 mtDNA | μg/mL | NR |
| Simmons et al43/2013 | Serial, venous, citrate | WB 1,200g 25 21°C | Plasma | MT-ND1, MT-ND6, MT-CO1, D-loop | NR | Fold increase | NR |
| Gu et al54/2013 | Single, venous, EDTA | WB 900g 10 → 9,600g 10 | Plasma | MT-ND2 | THP-1 mtDNA | pg/mL | 1.3185 μg/mL |
| Arnalich et al8/2013 | Single, NR, NR | WB 1,800g 10 4°C → 16,000g 10 4°C | Plasma | MT-ND2 | Human gDNA | GE/mL | 3,380 GE/mL or 22.308 ng/mL |
| Wang et al44/2013 | Serial, venous, EDTA | WB 3,000 rpm 10 → 10,000 rpm 10 | Plasma | MT-ND2 | Human gDNA | ng/mL | NR |
| Wang et al45/2012 | Serial, venous, citrate | WB 3,000g 10 4°C | Plasma | MT-ND2 | Human gDNA | ng/mL | — |
| Arnalich et al55/2012 | Single, venous, NR | WB 1,600g 10 4°C → 16,000g 10 | Plasma | MT-ATP8 | Human gDNA | GE/mL | 3,495 GE/mL |
| Puskarich et al46/2012 | Single, NR, NR | NR | Plasma | MT-ND1, MT-CO3, MT-CYB | Purified PCR products | μg/mL | NR |
| Kung et al47/2012 | Serial, venous, EDTA | WB 3,000 rpm 10 → 10,000 rpm 10 | Plasma | MT-ND2 | Human gDNA | ng/mL | 198 ng/mL |
| McGill et al48/2012 | Serial, NR, NR | NR | Plasma | MT-ND, MT-CO3 | HepaRG mtDNA | ng/mL | NR |
| Garrabou et al49/2012 | Single, NR, EDTA | WB 1,500g 15 → 0.22-μm filter | Plasma | MT-ND2 | NR | Gene/mL | NR |
| Tsai et al56/2011 | Serial, venous, EDTA | WB 3,000 rpm 10 → 10,000 rpm 10 | Plasma | MT-ND2 | Human gDNA | Kilogenome equivalents/L | — |
| Lu et al50/2010 | Serial, venous, EDTA | WB 3,000 rpm 10 → 10,000 rpm 10 | Plasma | MT-ND2 | gDNA | ng/mL | 58.9 ng/mL |
| Chou et al51/2008 | Serial, venous, EDTA | WB 1,500g 5 | Plasma | Mit 3130F, Mit 3301R | Plasmid DNA | Kilogenome equivalents/L | NR |
| Lam et al57/2004 | Single, venous, EDTA | WB 1,600g 10 | Plasma | Mit 3130F, Mit 3301R | Plasmid DNA | Copies/mL | NR |
AU = arbitrary unit; cDNA = complementary DNA; For = forward primer; g = force of gravity; gDNA = genomic DNA; GE = genome equivalents; MT-ATP8 = mitochondrially encoded ATP synthase 8; MT-CO1 = mitochondrially encoded cytochrome c oxidase I; MT-CO2 = mitochondrially encoded cytochrome c oxidase II; MT-CO3 = mitochondrially encoded cytochrome c oxidase III; MT-CYB = mitochondrially encoded cytochrome b; mtDNA = mitochondrial DNA; MT-ND = mitochondrially encoded NADH dehydrogenase; MT-ND1 = mitochondrially encoded NADH dehydrogenase I; MT-ND2 = mitochondrially encoded NADH dehydrogenase II; MT-ND6 = mitochondrially encoded NADH dehydrogenase VI; nDNA = nuclear DNA; P = plasma; Rev = reverse primer; RT = room temperature; S = serum; WB = whole blood. See Table 1 legend for expansion of other abbreviation.
“Blood draw(s)” consists of three distinct elements: the first (single or serial) corresponds to whether the study consisted of a single or serial mtDNA measurements; the second (venous or arterial) defines the source of the whole blood specimen; and the third element (EDTA, heparin, citrate, or variable) describes the type of vacutainer used for whole blood collection. The term “variable” means a combination of tubes were used; these details can be found in e-Appendix 1.
The notation for “sample processing” is as follows: WB, P, and S represent the initial specimen; these abbreviations correspond to whole blood, plasma, and serum, respectively. After the specimen designation there will be two to three numbers. The first number corresponds to the centrifugation speed, the second the centrifugation time in minutes, and the third the temperature in Celsius. If a third number is not present it means the study did not report this information. An “→” supplements the use of the word “then;” it means an additional step was performed. As an example, “WB 1,600g 10 4°C → 16,000g 10 4°C” reads as “whole blood was initially centrifuged at 1,600g for 10 min at 4°C and then the sample was centrifuged at 16,000g for 10 min at 4°C.”
The cutoffs presented correspond to different types of all-cause mortality; consequently, direct comparisons cannot be made. The two exceptions are Reference 54 (Gu et al, 2013) and Reference 50 (Lu et al, 2010), where the cutoffs correspond to the development of posttraumatic SIRS and “poor outcome” (including mortality), respectively. In addition, this field does not apply to studies where the mortality was zero, which is represented by a dash (—).
There are two separate Simmons et al (2017) studies: References 2 and 27. Data represented by d correspond to their study on trauma patients receiving blood transfusions, while data represented by e correspond to their study on ventilator-associated pneumonia.
There are two separate Timmermans et al (2016) studies: References 32 and 37. Data represented by f correspond to their study on sepsis while data represented by g correspond to their study in patients with trauma.
As shown in Table 2, heterogeneity was observed amongst studies regarding the preparation of cell-free mtDNA, specifically in terms of centrifugation settings, the type of specimen (plasma or serum), and expression of mtDNA levels (concentration, a copy number, a fold change, genome equivalents, arbitrary units, or as a ratio to nuclear DNA). Heterogeneity was also observed regarding the primers used to recognize a single portion of the mitochondrial genome, with NADH dehydrogenase (or one of its subunits) being the most commonly used followed by cytochrome c oxidase III, cytochrome b, the D-loop, ATPase 8, cytochrome c oxidase I, cytochrome c oxidase II, ATPase 6, and tRNALeu.
Risk of Bias Within Studies
The results of the modified QUIPS for each individual study can be found in e-Table 3. A moderate risk of bias was common.
Mortality
All of the studies in this systematic review reported on mortality.2, 6, 7, 8, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57 Mortality was assessed at variable time points, such as at ICU22, 31, 35 or hospital discharge2, 25, 28, 30, 34, 39, 40, 45, 46, 47, 51, 52, 54; at 3,55 15,8 21,7 28,6, 23, 24, 32, 37, 42 and 3033, 36, 38 days; and at 3,24, 50, 56 6,44 and 1230, 53 months. Eight studies (20%)26, 27, 28, 41, 43, 48, 49, 57 did not provide details on the timing of their mortality assessment. Eleven studies (27.5%)6, 7, 8, 31, 36, 38, 43, 47, 51, 55, 57 reported cell-free mtDNA levels were higher in nonsurvivors than survivors; while four studies (10%)26, 28, 30, 46 showed there was no association between mtDNA and mortality. Interestingly, one study42 showed mtDNA was significantly elevated in nonsurvivors of trauma, but not significantly elevated in nonsurvivors of sepsis. Two studies44, 50 found cell-free mtDNA to be significantly higher at one or more time points in patients with poor outcome (including mortality) than in patients with a good outcome from subarachnoid hemorrhage and bacterial meningitis. Another study found cell-free mtDNA to be significantly elevated on admission in post-cardiac arrest patients, but mtDNA did not correlate with unfavorable neurological outcome (including mortality).33 For the remaining studies, the association between mortality and levels of mtDNA could not be assessed, either because the mortality was zero2, 23, 29, 34, 40, 41, 45, 54, 56 or it was not discussed in the source material.22, 24, 25, 27, 32, 35, 37, 39, 48, 49, 52, 53 Of these 23 studies without a formal assessment between mtDNA and mortality, only two studies23, 45 did not find mtDNA to be significantly elevated in their population of interest. Mortality data for studies, stratified by patient population, can be found in e-Table 2.
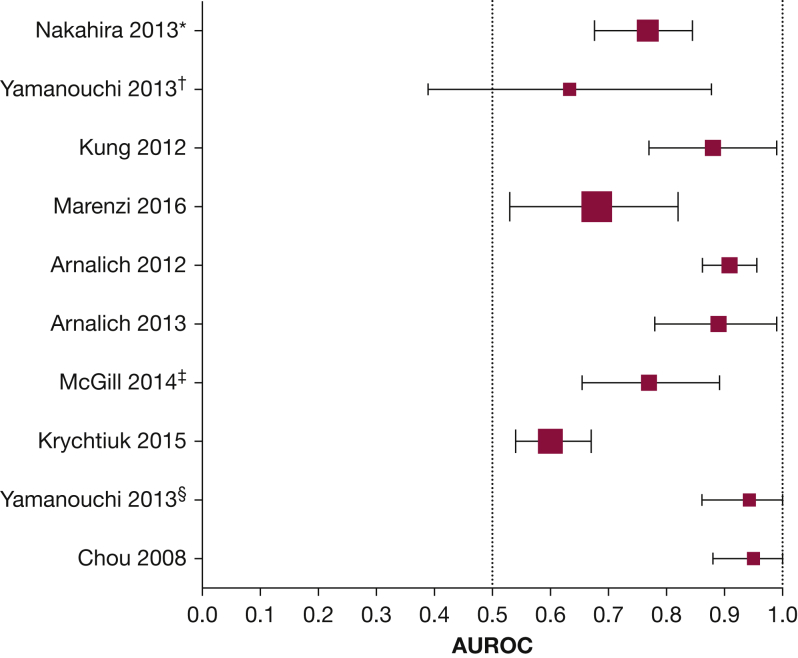
Data on the receiver operating characteristic curves for mtDNA and mortality were reported in eight studies (20%).6, 7, 8, 31, 38, 47, 51, 55 Five investigators7, 30, 35, 38, 42 furnished additional information on receiver operating characteristic curves from their studies on request. The data are presented in Figure 3. The area under the curve (AUC) ranged from 0.61 (95% CI, 0.52-0.70)38 to 0.95 (95% CI, 0.88-1.01).51 In three studies, the AUC for mtDNA and mortality was compared with a currently accepted biomarker/clinical model (namely, lactate, Acute Physiology and Chronic Health Evaluation [APACHE], troponin, and MELD).6, 7, 8 These details can be found within the “Observation” column of e-Table 2.
Figure 3.
Forest plot illustrating the area under the receiver operating characteristic curve with CIs for mtDNA and all-cause mortality in critically ill patients. The data presented here were either readily available within the articles or made available by the authors on request. The studies are organized according to discipline, with medicine patients near the top and surgery patients near the bottom of the plot. The size of each square denotes the proportional size of each cohort. *Data derived from the Brigham and Women’s Hospital cohort. †Data derived from the sepsis subgroup. ‡Data collected from use of NADH as a primer. §Data derived from the trauma subgroup.
Morbidity
The association between indicators of morbidity (ie, ICU length of stay) or specific organ failures (ie, acute kidney injury or acute respiratory distress syndrome) and mtDNA was not consistently reported. The available details can be found in e-Table 2.
Discussion
By systematically reviewing data from almost 3,500 subjects included in 40 clinical studies, we found that there is a growing international and multidisciplinary interest in mtDNA as a biomarker in critically ill patients. Over one-quarter of the studies in this systematic review found mtDNA to be significantly elevated in nonsurvivors relative to survivors. When reported, AUC analysis usually suggested a statistically significant association between the levels of mtDNA and mortality of critically ill patients.
An important clinical question is whether mtDNA outperforms established biomarkers (eg, lactate) or prognostic tools (eg, APACHE). Nakahira et al6 found mtDNA, procalcitonin, and lactate to be significantly associated with 28-day mortality in critically ill patients. They further found that mtDNA remained associated with mortality after adjustment for procalcitonin and lactate, but the odds ratio for lactate was significantly attenuated when adjusted for mtDNA. Although the above results from a single study imply that mtDNA might outperform lactate,6 it should be emphasized that this is not a robust way to compare biomarkers and that further studies are warranted before drawing any conclusions. Most studies did not provide a detailed comparison between mtDNA and current prognostic tools. Of what was reported, three of five studies found no correlation between mtDNA and APACHE II6, 33, 38, 42, 54 and two studies found that mtDNA did not correlate with Sepsis-Related Organ Failure Assessment (SOFA), while a third study actually found a slight negative association between mtDNA and SOFA in sepsis.38, 42, 46 On the other hand, mtDNA was found to correlate with Injury Severity Score in trauma42, 54 and to have an AUC similar to MELD for 21-day mortality in acetaminophen-induced acute liver failure.7
Even though most studies reporting on the association between mtDNA and mortality found a positive association, a handful of studies found no association. Such discrepant results may be due to unreported differences in patient characteristics or management. Recently, Hotz et al58 provided compelling data that RBCs scavenge circulating mtDNA through TLR-9. They theorized that mtDNA levels rise during critical illness because the inflammatory milieu either saturates or impairs RBC scavenging. Interestingly, TLR-9 is not ubiquitously expressed on the surface of RBCs, and while the factors influencing expression have not been elucidated there may be differences in RBC scavenging based on blood type. In contrast, other investigators have shown negative correlations between plasma mtDNA levels and the administration of intraoperative IV fluids or antibiotics.40, 47 We acknowledge that differences in ABO blood type, blood transfusion, resuscitation, and timing of antibiotics are unlikely to impact mtDNA measurements on a large scale, but such differences may become significant in small cohorts, a prominent feature of the studies in this review.
Apart from differences in patient characteristics, another explanation for discrepant results could be related to a significant amount of variation in the isolation and measurement of mtDNA. Care must be taken in the preparation of plasma to ensure adequate removal of mitochondria-containing cells, with two-step centrifugation, higher speeds, and filtration yielding platelet-poor plasma of higher quality.59 Furthermore, while most investigators selected primers to subunits of NADH dehydrogenase, the actual sequence or size of the amplicon is seldom reported—thus, it should not be assumed that different investigators are measuring identical regions of the reported genes. Primer differences may lead to variation in the number of CpG repeats within measured mtDNA fragments, with CpG-dense fragments more likely to yield significant associations due to a stronger interaction with TLR-9.16, 17
Another consideration is whether the prognostic ability of plasma mtDNA is affected by the timing of specimen retrieval. An interesting observation lies in the kinetics provided by Wang and colleagues44 during their study of patients with aneurysmal subarachnoid hemorrhage. Cerebrospinal fluid (CSF) mtDNA, not plasma mtDNA, was significantly elevated in patients with poor outcome on day 1. As CSF mtDNA trended down, plasma mtDNA rose until it became significantly elevated on day 8, corresponding to when CSF mtDNA began to nadir. Similarly, in a study of patients with ventilator-associated pneumonia, Simmons et al2 found mtDNA to be significantly elevated in bronchoalveolar lavage in patients on day 1, but 24 hours needed to transpire before mtDNA was significantly elevated in serum. Together, these findings suggest it may be prudent to trend mtDNA like troponin.60
This systematic review has limitations. Although our key phrase was broad, permitting a rigorous review of the literature, we recognize our search results may nonetheless be subject to publication bias. Also, while the generalizability of this study is increased by the diversity of the patient populations, we recognize that the small size and lack of external validation cohorts is limiting, especially when attempting to assess the prognostic value of a candidate biomarker.61 The small size of included studies may also explain the considerable imprecision regarding the association between mtDNA levels and mortality, as indicated by the wide 95% CI of Figure 3. In addition, a meta-analysis could not be performed because of considerable clinical heterogeneity among the included studies, mainly in terms of measurement of mtDNA without risk of providing a summary treatment effect that could be inaccurate or misleading.62 Plus, six different units of measurement were used to report mtDNA levels, rendering it impossible to synthesize the data in a statistically meaningful manner. Given that clinical heterogeneity precluded carrying out a meta-analysis, we chose not to quantify statistical heterogeneity (using I2) in Figure 3. Also, data were not consistently available to examine the association between mtDNA and specific organ failures, such as acute kidney injury or acute respiratory distress syndrome. Last, while there are encouraging data about the use of mtDNA as a biomarker for a variety of conditions, we remain hesitant to draw any firm conclusions since almost half of the studies had either a mortality of zero or the investigators did not comment on the relationship between mtDNA and mortality. With respect to the latter, we reached out to corresponding authors for additional mortality data and included it when available.
Conclusion
There is a growing international interest in evaluating the prognostic value of circulating cell-free mtDNA for predicting mortality of critically ill patients. Most of the published studies are relatively small, without validation cohorts, and vary considerably in terms of how mtDNA is measured. The results of most studies that performed AUC analysis suggest there is a statistically significant association between mtDNA levels and the mortality of critically ill patients. To move forward, the field needs to take steps to standardize how mtDNA is measured to facilitate large, prospective, multicenter trials to further define the ability of mtDNA to predict outcomes in critically ill patients.
Acknowledgments
Author contributions: J. S. H.: Study conception, data collection and analysis, and manuscript drafting. J. W. H., E. J. S., and K. N.: Data analysis and critical manuscript revision. I. I. S.: Study conception, data collection and analysis, and critical manuscript revision. A. M. K. C.: Data analysis and critical manuscript revision. All authors provided final manuscript approval.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: A. M. K. C. is a cofounder, stock holder, and serves on the scientific advisory board for Proterris, which develops therapeutic uses for carbon monoxide. A. M. K. C. also has a use patent on CO. A. M. K. C. served as a consultant for an advisory board meeting of Teva Pharmaceutical Industries, July 2018. None declared (J. S. H., J.-W. H., E. J. S., K. N., I. I. S.)
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Other contributions: The authors extend very special thanks to M. R. McGill, H. Jaeschke, S. Yamanouchi, R. K. Aneja, and W. S. Speidl, who provided additional information to assist with the completion of this systematic review.
Additional information: The e-Appendix and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
FUNDING/SUPPORT: This work was supported by the National Institutes of Health to A. M. K. C. [Grants R01 HL055330 and P01 HL108801] and by the National Institutes of Health/National Center for Advancing Translational Sciences to E. J. S. [Grant KL2TR000458-10] and to K. N. [Grant KL2-TR-002385].
Supplementary Data
References
- 1.Gan L., Chen X., Sun T. Significance of serum mtDNA concentration in lung injury induced by hip fracture. Shock. 2015;44(1):52–57. doi: 10.1097/SHK.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 2.Simmons J.D., Freno D.R., Muscat C.A. Mitochondrial DNA damage associated molecular patterns in ventilator-associated pneumonia: prevention and reversal by intratracheal DNase I. J Trauma Acute Care Surg. 2017;82(1):120–125. doi: 10.1097/TA.0000000000001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins L.V., Hajizadeh S., Holme E., Jonsson I.M., Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol. 2004;75(6):995–1000. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- 4.Tsuji N., Tsuji T., Ohashi N., Kato A., Fujigaki Y., Yasuda H. Role of mitochondrial DNA in septic AKI via Toll-like receptor 9. J Am Soc Nephrol. 2016;27(7):2009–2020. doi: 10.1681/ASN.2015040376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Martinez I., Santoro N., Chen Y. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest. 2016;126(3):859–864. doi: 10.1172/JCI83885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakahira K., Kyung S.Y., Rogers A.J. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10(12) doi: 10.1371/journal.pmed.1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGill M.R., Staggs V.S., Sharpe M.R., Lee W.M., Jaeschke H. Acute Liver Failure Study Group. Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology. 2014;60(4):1336–1345. doi: 10.1002/hep.27265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnalich F., Maldifassi M.C., Ciria E. Plasma levels of mitochondrial and nuclear DNA in patients with massive pulmonary embolism in the emergency department: a prospective cohort study. Crit Care. 2013;17(3):R90. doi: 10.1186/cc12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakahira K., Haspel J.A., Rathinam V.A. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West A.P., Brodsky I.E., Rahner C. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472(7344):476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Won J.H., Park S., Hong S., Son S., Yu J.W. Rotenone-induced impairment of mitochondrial electron transport chain confers a selective priming signal for NLRP3 inflammasome activation. J Biol Chem. 2015;290(45):27425–27437. doi: 10.1074/jbc.M115.667063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimada K., Crother T.R., Karlin J. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami T., Ockinger J., Yu J. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci U S A. 2012;109(28):11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington J.S., Choi A.M.K., Nakahira K. Mitochondrial DNA in sepsis. Curr Opin Crit Care. 2017;23(4):284–290. doi: 10.1097/MCC.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groot G.S., Kroon A.M. Mitochondrial DNA from various organisms does not contain internally methylated cytosine in –CCGG– sequences. Biochim Biophys Acta. 1979;564(2):355–357. doi: 10.1016/0005-2787(79)90233-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J.Z., Liu Z., Liu J., Ren J.X., Sun T.S. Mitochondrial DNA induces inflammation and increases TLR9/NF-κB expression in lung tissue. Int J Mol Med. 2014;33(4):817–824. doi: 10.3892/ijmm.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Kaczmarek A., Vandenabeele P., Krysko D.V. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38(2):209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Lood C., Blanco L.P., Purmalek M.M. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016;22(2):146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrington J., Schenck E., Nakahira K., Siempos I., Choi A. Mitochondrial DNA as predictor of mortality in critically ill patients. PROSPERO, August 29, 2016. https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42016046670 [DOI] [PMC free article] [PubMed]
- 20.Moher D., Liberati A., Tetzlaff J., Altman D.G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7) doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden J.A., Côté P., Bombardier C. Evaluation of the quality of prognosis studies in systematic reviews. Ann Intern Med. 2006;144(6):427–437. doi: 10.7326/0003-4819-144-6-200603210-00010. [DOI] [PubMed] [Google Scholar]
- 22.Aslami H., Beurskens C.J.P., Tuip A.M., Horn J., Juffermans N.P. Induced hypothermia is associated with reduced circulating subunits of mitochondrial DNA in cardiac arrest patients. Mitochondrial DNA A DNA Mapp Seq Anal. 2018;29(4):525–528. doi: 10.1080/24701394.2017.1315568. [DOI] [PubMed] [Google Scholar]
- 23.Leijte G.P., Custers H., Gerretsen J. Increased plasma levels of danger-associated molecular patterns are associated with immune suppression and postoperative infections in patients undergoing cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. Front Immunol. 2018;9:663. doi: 10.3389/fimmu.2018.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen M.P.B., Pulskens W.P., Butter L.M. Mitochondrial DNA is released in urine of SIRS patients with acute kidney injury and correlates with severity of renal dysfunction. Shock. 2018;49(3):301–310. doi: 10.1097/SHK.0000000000000967. [DOI] [PubMed] [Google Scholar]
- 25.Paunel-Görgülü A., Wacker M., El Aita M. cfDNA correlates with endothelial damage after cardiac surgery with prolonged cardiopulmonary bypass and amplifies NETosis in an intracellular TLR9-independent manner. Sci Rep. 2017;7(1):17421. doi: 10.1038/s41598-017-17561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hampson P., Dinsdale R.J., Wearn C.M. Neutrophil dysfunction, immature granulocytes, and cell-free DNA are early biomarkers of sepsis in burn-injured patients: a prospective observational cohort study. Ann Surg. 2017;265(6):1241–1249. doi: 10.1097/SLA.0000000000001807. [DOI] [PubMed] [Google Scholar]
- 27.Simmons J.D., Lee Y.L., Pastukh V.M. Potential contribution of mitochondrial DNA damage associated molecular patterns in transfusion products to the development of acute respiratory distress syndrome after multiple transfusions. J Trauma Acute Care Surg. 2017;82(6):1023–1029. doi: 10.1097/TA.0000000000001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donnino M.W., Liu X., Andersen L.W. National Post Arrest Research Consortium (NPARC) Investigators. Characterization of Mitochondrial Injury after Cardiac Arrest (COMICA) Resuscitation. 2017;113:56–62. doi: 10.1016/j.resuscitation.2016.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin C., Gu J., Liu R. Release of mitochondrial DNA correlates with peak inflammatory cytokines in patients with acute myocardial infarction. Anatol J Cardiol. 2017;17(3):224–228. doi: 10.14744/AnatolJCardiol.2016.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marenzi G., Cosentino N., Boeddinghaus J. Diagnostic and prognostic utility of circulating cytochrome c in acute myocardial infarction. Circ Res. 2016;119(12):1339–1346. doi: 10.1161/CIRCRESAHA.116.309792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohamed A.A., Ragab A.S., Rashed R.A. Plasma mitochondrial DNA at admission can predict the outcome of acute trauma patients admitted to ICU. Egypt J Anaesth. 2016;32(4):565–571. [Google Scholar]
- 32.Timmermans K., Kox M., Scheffer G.J., Pickkers P. Plasma nuclear and mitochondrial DNA levels, and markers of inflammation, shock, and organ damage in patients with septic shock. Shock. 2016;45(6):607–612. doi: 10.1097/SHK.0000000000000549. [DOI] [PubMed] [Google Scholar]
- 33.Omura T., Kushimoto S., Yamanouchi S., Kudo D., Miyagawa N. High-mobility group box 1 is associated with neurological outcome in patients with post-cardiac arrest syndrome after out-of-hospital cardiac arrest. J Intensive Care. 2016;4:37. doi: 10.1186/s40560-016-0161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin C., Gu J., Qian H., Meng W. Analysis of circulatory mitochondrial DNA level after cardiac surgery with cardiopulmonary bypass and potential prognostic implications. Indian Heart J. 2016;68(3):389–390. doi: 10.1016/j.ihj.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Caro V., Walko T.D., III, Bola R.A. Plasma mitochondrial DNA—a novel DAMP in pediatric sepsis. Shock. 2016;45(5):506–511. doi: 10.1097/SHK.0000000000000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schäfer S.T., Franken L., Adamzik M. Mitochondrial DNA: an endogenous trigger for immune paralysis. Anesthesiology. 2016;124(4):923–933. doi: 10.1097/ALN.0000000000001008. [DOI] [PubMed] [Google Scholar]
- 37.Timmermans K., Kox M., Vaneker M. Plasma levels of danger-associated molecular patterns are associated with immune suppression in trauma patients. Intensive Care Med. 2016;42(4):551–561. doi: 10.1007/s00134-015-4205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krychtiuk K.A., Ruhittel S., Hohensinner P.J. Mitochondrial DNA and Toll-like receptor-9 are associated with mortality in critically ill patients. Crit Care Med. 2015;43(12):2633–2641. doi: 10.1097/CCM.0000000000001311. [DOI] [PubMed] [Google Scholar]
- 39.Bhagirath V.C., Dwivedi D.J., Liaw P.C. Comparison of the proinflammatory and procoagulant properties of nuclear, mitochondrial, and bacterial DNA. Shock. 2015;44(3):265–271. doi: 10.1097/SHK.0000000000000397. [DOI] [PubMed] [Google Scholar]
- 40.McIlroy D.J., Bigland M., White A.E. Cell necrosis-independent sustained mitochondrial and nuclear DNA release following trauma surgery. J Trauma Acute Care Surg. 2015;78(2):282–288. doi: 10.1097/TA.0000000000000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H.C., Lin Y.J., Tsai N.W. Serial plasma deoxyribonucleic acid levels as predictors of outcome in acute traumatic brain injury. J Neurotrauma. 2014;31(11):1039–1045. doi: 10.1089/neu.2013.3070. [DOI] [PubMed] [Google Scholar]
- 42.Yamanouchi S., Kudo D., Yamada M., Miyagawa N., Furukawa H., Kushimoto S. Plasma mitochondrial DNA levels in patients with trauma and severe sepsis: time course and the association with clinical status. J Crit Care. 2013;28(6):1027–1031. doi: 10.1016/j.jcrc.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Simmons J.D., Lee Y.L., Mulekar S. Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects. Ann Surg. 2013;258(4):591–596. doi: 10.1097/SLA.0b013e3182a4ea46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H.C., Yang T.M., Lin W.C. The value of serial plasma and cerebrospinal fluid nuclear and mitochondrial deoxyribonucleic acid levels in aneurysmal subarachnoid hemorrhage. J Neurosurg. 2013;118(1):13–19. doi: 10.3171/2012.8.JNS112093. [DOI] [PubMed] [Google Scholar]
- 45.Wang H.C., Lin Y.J., Lin W.C. The value of serial plasma nuclear and mitochondrial DNA levels in acute spontaneous intra-cerebral haemorrhage. Eur J Neurol. 2012;19(12):1532–1538. doi: 10.1111/j.1468-1331.2012.03761.x. [DOI] [PubMed] [Google Scholar]
- 46.Puskarich M.A., Shapiro N.I., Trzeciak S., Kline J.A., Jones A.E. Plasma levels of mitochondrial DNA in patients presenting to the emergency department with sepsis. Shock. 2012;38(4):337–340. doi: 10.1097/SHK.0b013e318266a169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kung C.T., Hsiao S.Y., Tsai T.C. Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. J Transl Med. 2012;10:130. doi: 10.1186/1479-5876-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGill M.R., Sharpe M.R., Williams C.D., Taha M., Curry S.C., Jaeschke H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J Clin Invest. 2012;122(4):1574–1583. doi: 10.1172/JCI59755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrabou G., Morén C., López S. The effects of sepsis on mitochondria. J Infect Dis. 2012;205(3):392–400. doi: 10.1093/infdis/jir764. [DOI] [PubMed] [Google Scholar]
- 50.Lu C.H., Chang W.N., Tsai N.W., Chuang Y.C., Huang C.R., Wang H.C. The value of serial plasma nuclear and mitochondrial DNA levels in adult community-acquired bacterial meningitis. QJM. 2010;103(3):169–175. doi: 10.1093/qjmed/hcp201. [DOI] [PubMed] [Google Scholar]
- 51.Chou C.C., Fang H.Y., Chen Y.L., Wu C.Y., Siao F.Y., Chou M.J. Plasma nuclear DNA and mitochondrial DNA as prognostic markers in corrosive injury patients. Dig Surg. 2008;25(4):300–304. doi: 10.1159/000152846. [DOI] [PubMed] [Google Scholar]
- 52.Timmermans K., Kox M., Gerretsen J. The involvement of danger-associated molecular patterns in the development of immunoparalysis in cardiac arrest patients. Crit Care Med. 2015;43(11):2332–2338. doi: 10.1097/CCM.0000000000001204. [DOI] [PubMed] [Google Scholar]
- 53.Fernández-Ruiz I., Arnalich F., Cubillos-Zapata C. Mitochondrial DAMPs induce endotoxin tolerance in human monocytes: an observation in patients with myocardial infarction. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0095073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu X., Yao Y., Wu G., Lv T., Luo L., Song Y. The plasma mitochondrial DNA is an independent predictor for post-traumatic systemic inflammatory response syndrome. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0072834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnalich F., Codoceo R., López-Collazo E., Montiel C. Circulating cell-free mitochondrial DNA: a better early prognostic marker in patients with out-of-hospital cardiac arrest. Resuscitation. 2012;83(7):e162–e163. doi: 10.1016/j.resuscitation.2012.03.032. [DOI] [PubMed] [Google Scholar]
- 56.Tsai N.W., Lin T.K., Chen S.D. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin Chim Acta. 2011;412(5-6):476–479. doi: 10.1016/j.cca.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 57.Lam N.Y., Rainer T.H., Chiu R.W., Joynt G.M., Lo Y.M. Plasma mitochondrial DNA concentrations after trauma. Clin Chem. 2004;50(1):213–216. doi: 10.1373/clinchem.2003.025783. [DOI] [PubMed] [Google Scholar]
- 58.Hotz M.J., Qing D., Shashaty M.G.S. Red blood cells homeostatically bind mitochondrial DNA through TLR9 to maintain quiescence and to prevent lung injury. Am J Respir Crit Care Med. 2018;197(4):470–480. doi: 10.1164/rccm.201706-1161OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiu R.W., Chan L.Y., Lam N.Y. Quantitative analysis of circulating mitochondrial DNA in plasma [published correction appears in Clin Chem. 2004;50(2):461] Clin Chem. 2003;49(5):719–726. doi: 10.1373/49.5.719. [DOI] [PubMed] [Google Scholar]
- 60.Amsterdam E.A., Wenger N.K., Brindis R.G. 2014 AHA/ACC guideline for the management of patients with non-ST-elevation acute coronary syndromes: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines [published correction appears in J Am Coll Cardiol. 2014;64(24):2713-2714] J Am Coll Cardiol. 2014;64(24):e139–e228. doi: 10.1016/j.jacc.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 61.Siontis G.C., Tzoulaki I., Castaldi P.J., Ioannidis J.P. External validation of new risk prediction models is infrequent and reveals worse prognostic discrimination. J Clin Epidemiol. 2015;68(1):25–34. doi: 10.1016/j.jclinepi.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Gagnier J.J., Moher D., Boon H., Beyene J., Bombardier C. Investigating clinical heterogeneity in systematic reviews: a methodologic review of guidance in the literature. BMC Med Res Methodol. 2012;12:111. doi: 10.1186/1471-2288-12-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.