Streptococcus pyogenes is a genetically diverse pathogen, with over 200 different genotypes defined by emm typing, but only a minority of these genotypes are responsible for the majority of human infection in high-income countries. Two prevalent genotypes associated with disease rose to international dominance following recombination of a toxin locus that conferred increased expression. Here, we found that recombination of this locus and promoter has occurred in other diverse genotypes, events that may allow these genotypes to expand in the population. We identified an association between the loss of hyaluronic acid capsule synthesis and high toxin expression, which we propose may be associated with an adaptive advantage. As S. pyogenes pathogenesis depends both on capsule and toxin production, new variants with altered expression may result in abrupt changes in the molecular epidemiology of this pathogen in the human population over time.
KEYWORDS: group A Streptococcus, NADase, Streptococcus pyogenes, streptolysin O, convergent evolution, homologous recombination, hyaluronic acid capsule, whole-genome sequencing
ABSTRACT
Gene transfer and homologous recombination in Streptococcus pyogenes has the potential to trigger the emergence of pandemic lineages, as exemplified by lineages of emm1 and emm89 that emerged in the 1980s and 2000s, respectively. Although near-identical replacement gene transfer events in the nga (NADase) and slo (streptolysin O) loci conferring high expression of these toxins underpinned the success of these lineages, extension to other emm genotype lineages is unreported. The emergent emm89 lineage was characterized by five regions of homologous recombination additional to nga-slo, including complete loss of the hyaluronic acid capsule synthesis locus hasABC, a genetic trait replicated in two other leading emm types and recapitulated by other emm types by inactivating mutations. We hypothesized that other leading genotypes may have undergone similar recombination events. We analyzed a longitudinal data set of genomes from 344 clinical invasive disease isolates representative of locations across England, dating from 2001 to 2011, and an international collection of S. pyogenes genomes representing 54 different genotypes and found frequent evidence of recombination events at the nga-slo locus predicted to confer higher toxin genotype. We identified multiple associations between recombination at this locus and inactivating mutations within hasAB, suggesting convergent evolutionary pathways in successful genotypes. This included common genotypes emm28 and emm87. The combination of no or low capsule and high expression of nga and slo may underpin the success of many emergent S. pyogenes lineages of different genotypes, triggering new pandemics, and could change the way S. pyogenes causes disease.
INTRODUCTION
The capacity for the bacterial human pathogen Streptococcus pyogenes to undergo genetic exchange, independent of known bacteriophages or mobile elements, is not well understood, yet recent evidence suggests it underpins the emergence of successful new variants that rapidly rise to international dominance. Homologous recombination of a chromosomal region encompassing the toxin genes nga (encoding NADase), ifs (encoding the inhibitor for NADase), and slo (encoding streptolysin O), which was dated to have occurred in the mid-1980s, is thought to have driven the rise of emm1 to almost global dominance (1). The homologous recombination event resulted in increased nga-slo expression compared to that of the previous variant, linked to the gain of a highly active nga-ifs-slo promoter in the new emm1 variant compared to that of the previous variant (2).
A very similar recombination event was recently identified in the genotype emm89. A new variant of emm89 sequence type (ST) 101 (also referred to as clade 3) emerged, having undergone six regions of predicted homologous recombination compared to its ST101 predecessor (also referred to as clade 2) (3, 4). One of the six regions encompassed the nga-ifs-slo locus, comprising a region almost identical to that of emm1, which conferred similarly high expression of nga and slo compared to that of the previous variant. Another recombination region within the emergent ST101 emm89 resulted in the loss of the hyaluronic acid capsule. We dated the emergence of this new acapsular high-toxin-expressing ST101 emm89 lineage to the mid-1990s, but there was a rapid increase and rise to dominance in the United Kingdom between 2005 and 2010 (3). The lineage is now the dominant form of emm89 in the United Kingdom as well as other parts of the world, including Europe, North America, and Japan (4–8).
Given that recombination associated with nga-ifs-slo can give rise to new successful S. pyogenes variants, we hypothesized that this may be a feature common to other successful emm types. To determine if this is the case, we sequenced the genomes of 344 S. pyogenes invasive disease isolates originating from hospitals across England between 2001 and 2011 and compared the data with other available historical and contemporary international S. pyogenes whole-genome sequence (WGS) data. We identified that recombination of the nga-ifs-slo locus has occurred in other leading emm types, supporting the hypothesis that it can underpin the emergence and success of new lineages. We also identified an association of nga-ifs-slo recombination toward a high-activity promoter variant with inactivating mutations within the capsule locus. This suggests that loss of capsule may also provide an advantage to certain genotypes, either through a direct effect on pathogenesis or an association with the process of recombination.
RESULTS
Genetic characterization of bacteremia isolates.
We performed whole-genome sequencing of 344 S. pyogenes invasive isolates collected from hospitals across England by the British Society for Antimicrobial Chemotherapy (BSAC) Bacteraemia Resistance Surveillance Program from 2001 to 2011. Forty-four different emm types were identified from de novo assembly, with the most common being emm1 (n = 64, 18.6%), emm12 (n = 34, 9.9%), emm89 (n = 32, 9.3%), emm3 (n = 28, 8.1%), emm87 (n = 22, 6.4%), and emm28 (n = 15, 4.4%) (see Fig. S1 in the supplemental material). Antimicrobial susceptibilities were typical for S. pyogenes, with 100% of isolates susceptible to penicillin and 22% resistant to clindamycin, erythromycin, and/or tetracycline; detailed susceptibilities and associated genotypes are reported in Data Set S1.
Number of isolates per emm type in the BSAC collection. Forty-four different genotypes were identified within the collection, but 16 were represented by single isolates (grey bars). Total number of isolates was 344. Download FIG S1, TIF file, 0.2 MB (172KB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Details of BSAC isolates and antimicrobial sensitivity testing. Download Data Set S1, XLSX file, 0.1 MB (46.1KB, xlsx) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
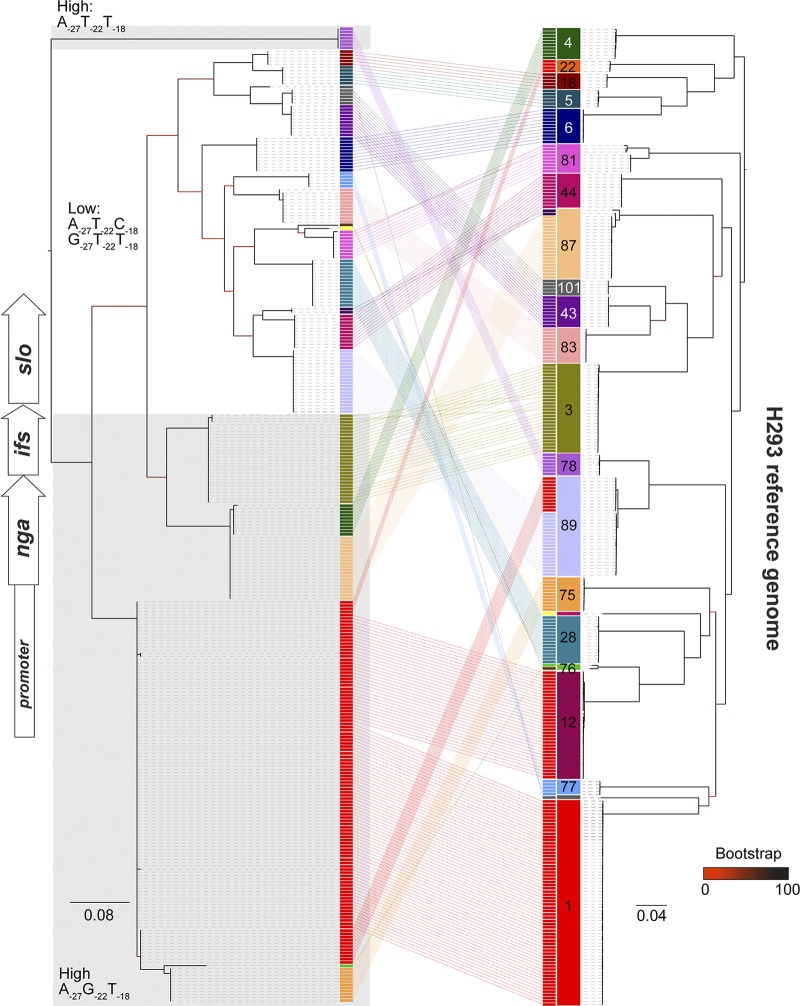
The phylogenetic distribution of the 344 isolates based on core genome variation revealed distinct clustering by emm type, each forming single lineages with the exceptions of emm44, emm90, and emm101, each of which formed two lineages (Fig. 1A). Pairwise distances between isolates gave a median of just 45 single nucleotide polymorphisms (SNPs) separating the genomes of isolates of the same emm genotype (range, 0 to 15,137 SNPs) compared to a median of 15,648 SNPs separating the genomes of isolates of different emm types (range, 5,312 to 18,317 SNPs) (Fig. 1B). The genotypes emm44, emm90, and emm101 gave the highest SNP distance for the intra-emm comparison (13,494 to 15,137 SNPs), which approaches the median level observed between emm types. This indicated that while other genotypes represent a relatively conserved chromosomal genetic background, the populations of emm44, emm90, and emm101 exhibit more diverse chromosomal backgrounds despite representing the same emm type, potentially due to emm gene switching.
FIG 1.
Low diversity within emm genotypes. (A) A maximum likelihood phylogenetic tree constructed from 113,805 core SNPs extracted after mapping all 344 BSAC isolates to the complete emm89 reference strain H293 (indicated by a star) identified that the majority of isolates cluster by emm genotype. Exceptions were emm44, emm90, and emm101 (highlighted with black dots), each of which were present as two separate lineages. Branches are colored based on bootstrap support (scale bar in figure). Boxes at branch tips are colored by emm type, and the emm-type numbers are provided outside the tree. (B) As reflected by the phylogenetic tree, the number of SNPs separating isolates was high (>5,000) when the genomes of isolates of different emm types were compared (black bars). This was much lower when comparisons were made between the genomes of isolates of the same emm type (red bars).
High level of variation within the nga-ifs-slo locus.
To identify the level of variation within the nga-ifs-slo locus, we extracted the sequence from the 3′ end of nusG (immediately upstream of nga) to the 3′ end of slo (P-nga-ifs-slo), comprising the entire locus and all of the upstream sequence, including the predicted ∼67-bp nga-ifs-slo promoter region (9). We constructed a phylogenetic tree from SNPs within the P-nga-ifs-slo region from the genomes of isolates belonging to the most common emm types and compared it to the phylogeny constructed with SNPs extracted from a whole-genome comparison to a reference emm89 genome, H293 (Fig. 2). Most emm genotypes were associated with a single P-nga-ifs-slo variant that was unique to that genotype. The main exception to this was the P-nga-ifs-slo variant found in modern (post-1980s M1T1) emm1, as this was also found in all emm12, all emm22 (a lineage known to be acapsular) isolates, and 11 of the 32 emm89 isolates. These 11 emm89 isolates represented the emergent acapsular ST101 variant, while the remaining 21 emm89 isolates represented the original encapsulated ST101 variant, with a different unique P-nga-ifs-slo as previously reported (3). The entire emm75 population and one of the two emm76 isolates were also associated with a P-nga-ifs-slo variant that was closely related to the emm1-like variant. All but two emm87 isolates had a P-nga-ifs-slo variant also found in the acapsular lineage emm4. The presence of multiple P-nga-ifs-slo variants within the emm76 and emm87 genotypes, where the core chromosome was otherwise relatively conserved, indicated that gene transfer and recombination are responsible for the P-nga-ifs-slo variation in these genotypes rather than extensive genome-wide divergence or emm “switching.”
FIG 2.
Comparison of the variation within the P-nga-ifs-slo region and core chromosome. A maximum likelihood phylogenetic tree was constructed from 205 SNPs extracted from an alignment of the nga-ifs-slo locus and associated upstream region to include the promoter (P-nga-ifs-slo) extracted from de novo assemblies of BSAC S. pyogenes collection (left tree). This was compared to the phylogenetic tree constructed using 75,851 SNPs across the entire core genome after mapping to the H293 reference genome (right tree). Only 20 of the most common emm genotypes were included: emm1, -3, -4, -5, -6, -12, -18, -22, -28, -43, -44, -75, -76, -77, -78, -81, -83, -87, -89, and -101 (n = 303 isolate genomes). Numbers and colored blocks on the right tree represent emm types. Variants of the P-nga-ifs-slo are of the same color as the emm type if unique to that emm type. The P-nga-ifs-slo variant found in emm1 (red) was common to other genotypes of emm12, emm22, and some emm89 isolates. The genotypes emm76, emm87, and emm89 were linked to more than one variant of P-nga-ifs-slo. Gray shading indicates high-expressing promoter variants: A−27T−22T−18 (top) or A−27G−22T−18 (bottom). Other nonshaded areas are low-expressing promoter variants A−27T−22C−18 or G−27T−22T−18. Scale bar represents substitutions per site. Bootstrap support values are provided on branches.
Variants of the nga-ifs-slo promoter associated with altered expression.
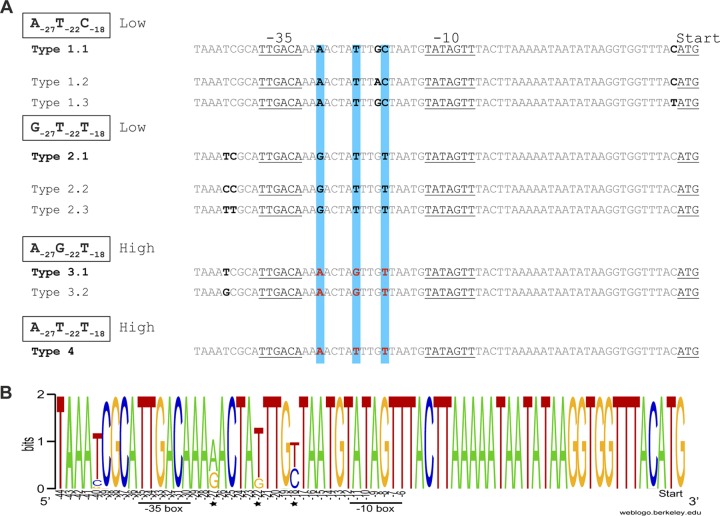
Recombination of P-nga-ifs-slo and surrounding regions in emm1 and emm89 conferred higher activity and expression of NGA (NADase) and SLO (1, 3, 10). This change in expression was linked to the combination of three key residues at −27, −22, and −18 within the nga-ifs-slo promoter. A−27G−22T−18 at these key sites was associated with high nga-ifs-slo promoter activity in emm1 and emergent emm89 following recombination (also referred to as Pnga3) compared to low promoter activity of historical emm1 and emm89, associated with the key site combinations A−27T−22C−18 and G−27T−22T−18, respectively (2) (Fig. 3A). We compared the ∼67-bp nga-ifs-slo promoter region of the 344 BSAC collection isolate genomes to identify different variants. We expanded the data analyzed by including assembled genome data from >5,000 isolates representing 54 different emm types: from Cambridge University Hospital (CUH) (11), from England and Wales collected by Public Health England (PHE) in 2014 and 2015 (PHE-2014/15) (12, 13), and from the United States collected by the Active Bacterial Core Surveillance System (ABCs) in 2015 (ABCs-2015) (14). We excluded 39 emm types represented by fewer than 3 isolates (Data Set S2).
FIG 3.
Variants of the nga-ifs-slo promoter. (A) The three key residues predicted to influence promoter activity are highlighted in blue, with those associated with high activity in red font. We identified four combinations of these residues (four promoter types) with subtype variants differing in residues other than −27, −22, and −18 (residue positions relative to the underlined −35 and −10 regions) in the predicted 67-bp promoter region (9). The combination of A−27T−22C−18 subtype 1.1 in historical emm1 and G−27T−22T−18 subtype 2.1 in older emm89 isolates has been shown to be associated with low-level promoter activity. A−27G−22T−18 subtype 3.1 promoter in modern emm1 and emergent variant emm89 has been shown to have high activity. A−27T−22T−18 subtype 4 promoter has also been shown to have high activity in emm28 (15). Subtypes 1.2, 1.3, and 2.3 were restricted to emm9, emm88, and emm32 strains, respectively. (B) WebLogo representation of the variability in the 67-bp promoter region of nga-ifs-slo within the 54 different emm types. Key residues −27, −22, and −18 are highlighted (star) and their positions are relative to the −35 and −10 boxes. Figure generated using WebLogo (http://weblogo.berkeley.edu).
Details of all isolates with assembly statistics, capsule gene mutations, and nga-ifs-slo promoter variants. Download Data Set S2, XLSX file, 0.6 MB (638.4KB, xlsx) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Four combinations of the −27, −22, and −18 residues were found across all 5,271 isolates (Table 1); variant 1 A−27T−22C−18 and variant 2 G−27T−22T−18 are associated with low promoter activity, while variant 3 A−27G−22T−18 and variant 4 A−27T−22T−18 are associated with high promoter activity. We also identified subtypes of the 67-bp promoter region which varied at bases other than −27, −22, and −18 (Fig. 3A and B; Table 1). A−27T−22C−18 variant subtype 1.1 and G−27T−22T−18 variant subtype 2.1 were both previously confirmed to have low promoter activity (2) and were the most common variants found across genotypes. Other subtypes of these variants were restricted to single genotypes except G−27T−22T−18 variant subtype 2.2, which differed by a single substitution of C for a T residue at −40 bp. Two subtypes of the high-activity variant A−27G−22T−18 were found, the most common being subtype 3.1, associated with emm1 and emergent emm89, and subtype 3.2, which was found predominantly in the genomes of emm4 and emm87 and differed from subtype 3.1 by a single substitution of G for T at −40 bp. We measured the activity of NADase in the culture supernatants of strains representing different promoter subtypes and found that the presence of T/G/C at −40 bp did not affect activity of the promoter (see Fig. S2). The fourth promoter variant, A−27T−22T−18, is also associated with high activity (15) and was identified in the genomes of emm28, emm75, and all emm78 isolates. Only three emm types were exclusively associated with the high-activity promoter variant A−27G−22T−18: emm1, emm3, and emm12. Other emm types with the high-activity promoter variant also had one or more of the other three promoter variants, suggesting a mixed population or, as in the case of emm89, an evolving population.
TABLE 1.
Three key residue variants within the nga-ifs-slo promoter
| Promoter variant | Type | Genotype (% of isolates)a |
|---|---|---|
| A−27T−22C−18 | 1.1 | 4 (1), 8 (100), 9 (92), 11 (100), 22 (3), 25 (33), 28 (87.7), 33 (100), 41 (100), 43 (100), 44 (9), 49 (100), 53 (100), 58 (15), 60 (100), 63 (100), 75 (9), 76 (41), 77 (29), 81 (23), 82 (1), 88 (33), 89 (1), 90 (4), 92 (100), 94 (6), 101 (100), 102 (50), 103 (17), 106 (100), 108 (89), 110 (100), 113 (100), 151 (100), 168 (100), 232 (100) |
| 1.2 | 9 (8) | |
| 1.3 | 88 (67) | |
| G−27T−22T−18 | 2.1 | 5 (100), 6 (100), 18 (100), 25 (67), 44 (28), 68 (100), 75 (1), 76 (5), 77 (1), 82 (1), 87 (2), 89 (6), 90 (96), 91 (100), 102 (50), 103 (83), 104 (100), 118 (100) |
| 2.2 | 2 (100), 27 (100), 44 (62), 58 (85), 59 (100), 73 (100), 76 (11), 77 (36), 82 (89), 83 (100) | |
| 2.3 | 32 (100) | |
| A−27G−22T−18 | 3.1 | 1 (100), 3 (100), 12 (100), 22 (97), 75 (90), 76 (43), 81 (77), 82 (9), 89 (93), 94 (94), 108 (11) |
| 3.2 | 4 (99), 28 (0.3), 77 (34), 87 (98) | |
| A−27T−22T−18 | 4 | 28 (12), 78 (100) |
Boldface font indicates genotypes with more than one variant within the population.
NADase activity of different promoter subtypes. The activity of NADase was measured in culture supernatants of BSAC isolates representing different promoter subtypes with predicted low (black) or high (red) activity. A−27T−22C−18 subtype 1.1 promoter had low activity in emm81 isolates, consistent with previous findings of this promoter in historical emm1. G−27T−22T−18 subtype 2.1 had low activity in older emm89 isolates, also consistent with previous findings, and subtype 2.2 in emm58 and emm77 also had low activity, as predicted despite the additional base change at −40 bp. Compared to G−27T−22T−18 subtype 2.1 (older emm89), significantly higher activity was detected in emm1, with A−27G−22T−18 subtype 3.1, and in emm4 and emm87, with subtype 3.2, also supporting a null effect of the base change at −40 bp. A−27T−22T−18 subtype 4 promoter in emm78 also had significantly higher activity. Isolates with mutations in regulators covRS or rocA were excluded as they influence the expression of nga. Data represent means + standard deviations (SDs) for emm1, n = 10; emm89, n = 11; emm58, n = 3; emm77, n = 3; emm4, n = 7; emm87, n = 17; emm78, n = 5; and emm81, n = 5. Statistical comparisons were made to emm89 subtype 2.1, for which we had the highest number of representative isolates and was previously confirmed to have low activity, using Kruskal-Wallis nonparametric multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant. Download FIG S2, TIF file, 0.1 MB (156.6KB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We sought evidence for acquisition of the high-activity-associated promoter A−27G−22T−18 variant by emm genotypes where the dominant or ancestral state was a low-activity-associated promoter; these included (in addition to the aforementioned emm89) emm75, emm76, emm77, emm81, emm82, emm87, emm94, and emm108, all of which are emm types frequently identified in the United Kingdom and the United States (12–14). Although one emm28 was found to carry the high-activity-associated A−27G−22T−18 promoter, the rest of the emm28 population was divided between either A−27T−22C−18 or A−27T−22T−18 variants. The data pointed to a switch in P-nga-ifs-slo in all cases rather than an emm switch, except for emm82, where the emm82 gene replaced the emm12 gene in an emm12 genetic background (14).
High level of mutations within the capsule locus leading to truncations of HasA or HasB.
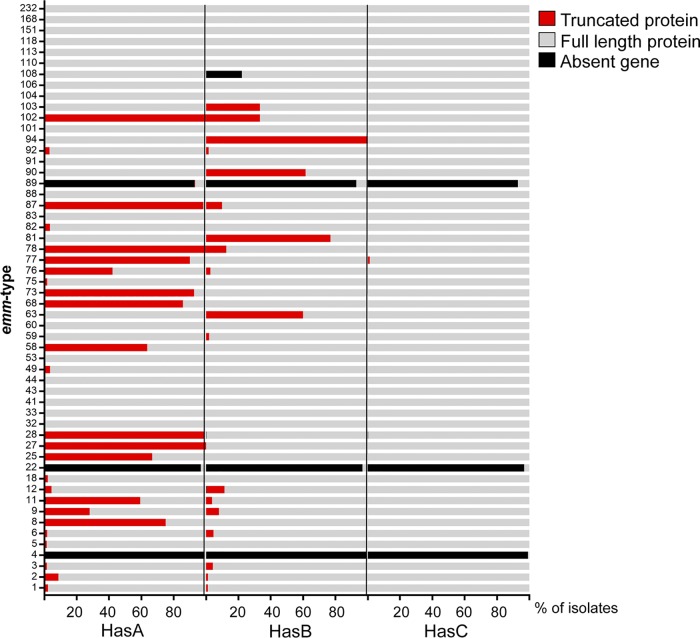
As well as recombination around the P-nga-ifs-slo region, the emergent ST101 variant of emm89 had also undergone recombination surrounding the hasABC locus, and, in place of the hasABC genes, there was a region of 156 bp that was not found in genotypes with the capsule locus but is found in the acapsular emm4 and emm22 isolates (3). To identify any similar events in other genotypes, we examined the sequences of hasA, hasB, and hasC in the assemblies of isolates from the BSAC collection as well as CUH (11), PHE-2014/15 (12, 13), and ABCs-2015 (14) collections for gene presence as well as premature stop codon mutations (Fig. 4). The hasABC locus was absent in the majority of emm89 isolates, consistent with the previous observations describing the recent emergence of the acapsular emm89 variant (3). Similarly, the hasABC genes were absent in all emm4 and emm22 isolates, as previously identified (16), except for two emm4 isolates and one emm22 isolate which had an intact hasABC locus predicted to encode full-length proteins. We confirmed the genotypes of these isolates by emm typing the assembled genomes. Multilocus sequence typing (MLST) and phylogenetic analysis indicated that they both had a very different genetic background to other emm4 or emm22 populations, suggesting that these were not typical of these emm types; therefore, they represent examples of emm switching. Interestingly, we also identified a similar replacement of hasABC for the 156-bp region in one emm28 isolate (PHE-2014/15, GASEMM1261 [13]), but phylogenetic analysis suggested this was highly divergent from the rest of the emm28 population, likely to represent another example of emm switching. Isolated examples of individual hasA or hasB gene loss were identified in the genomes of isolates belonging to emm1 (n = 1), emm3 (n = 1), emm11 (n = 1), emm12 (n = 4), and emm108 (n = 2).
FIG 4.
Nonfunctional mutations within the capsule locus genes. The hasABC genes were extracted from the assembled genomes of BSAC, CUH, PHE-2015/15, and ABCs-2015 isolate genome collections, and polymorphisms or indels leading to nonsense mutations and premature stop codons, as well as gene absence, were identified. The percentage of isolates with full-length (gray), truncated (red), or absent (black) HasA, HasB, or HasC is depicted for each of the 54 emm types. emm types with fewer than 3 isolates were excluded. N = 5,271 isolates genomes shown. Mutations in HasA were detected in more than 50% of isolates belonging to genotypes emm8 (n = 3/4), emm11 (n = 63/108), emm25 (n = 2/3), emm27 (n = 3/3), emm28 (n = 358/363), emm58 (n = 21/33), emm68 (n = 12/14), emm73 (n = 25/27), emm77 (n = 72/80), emm78 (n = 8/8), emm87 (n = 119/121), and emm102 (n = 6/6). Mutations in HasB were detected in 100% of emm94 isolates (n = 54/54) and 60% to 77% of emm63 (n = 3/5), emm81 (n = 50/65), and emm90 (n = 16/26) isolates.
The majority of genotypes (35/54 [65%]) had isolates without genes or truncation mutations in at least one of the hasABC genes (Fig. 4). Mutations in hasC were rare and only detected in one isolate, an emm77 isolate, which also had a mutation within hasA. Within seven of the eight emm types for which we identified potential P-nga-ifs-slo recombination, a high percentage of isolates had inactivating mutations in hasA and hasB, suggesting a possible association between an acapsular genotype/phenotype and recombination of P-nga-ifs-slo to gain a high-activity promoter. Including the previously identified emm1 and emm89 recombination events, P-nga-ifs-slo recombination to gain a high-activity promoter was detected in 10 genotypes, and in all 10 of these genotypes (100%) were isolates with hasAB gene mutations or gene absence. However, in the 44 genotypes that had not undergone P-nga-ifs-slo recombination to gain a high-activity promoter, significantly fewer (25/44 [57%]) had isolates with a hasAB gene mutation or gene absence (χ21df = 6.662, P = 0.0098).
Recombination of P-nga-ifs-slo and surrounding regions.
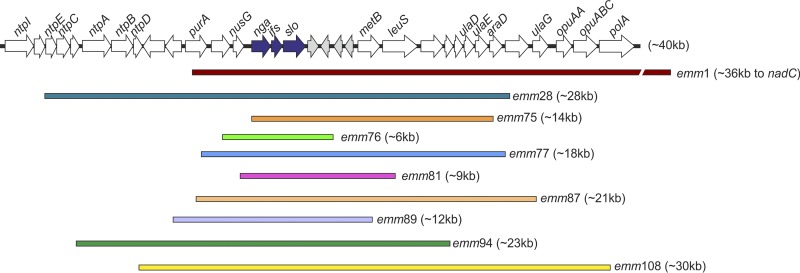
To confirm our prediction that genotypes emm28, emm75, emm76, emm77, emm81, emm87, emm94, and emm108 had undergone recombination around P-nga-ifs-slo, we mapped all the genome sequence data for each genotype to the emm89 reference genome H293. Gubbins analysis of SNP clustering predicted regions of recombination spanning the nga-ifs-slo region and varied in length in all eight genotypes (Fig. 5). To further analyze the recombination of these genotypes and potential capsule loss, we studied the population structure of each genotype individually.
FIG 5.
Regions of recombination spanning the P-nga-ifs-slo locus. Recombination across the nga, ifs, and slo genes (blue arrows) was identified in eight genotypes in addition to the previously described emm1 and emm89. Length of recombination, predicted by SNP cluster analysis, ranged from ∼6 kb to 36 kb. With the exception of emm75, all regions also encompassed the promoter of nga-ifs-slo. All regions are shown relative to an ∼40-kb region within the reference genome H293, and genes within this region are depicted as arrows. Recombination in emm1 extended beyond that depicted here and is shown as a broken line.
Recombination within emm28 and emm87 around P-nga-ifs-slo and the capsule locus.
The genotypes emm28 and emm87 were the sixth and fifth most common in the BSAC collection, respectively, and emm28 was previously noted to be a major cause of infection in high-income countries (17). We focused attention on emm28 and emm87, as there has been little genomic work on these genotypes so far.
All BSAC emm28 isolates carried the A−27T−22C−18 low-activity-associated promoter, but inclusion of international genomic data identified A−27T−22T−18 variant-carrying isolates. These two promoter variants were associated with different major lineages within the entire population of 379 international emm28 isolates, including one newly sequenced English isolate originally isolated in 1938. The majority of isolates (n = 373) clustered either with the reference MGAS6180 strain (United States) (18) or with the reference MEW123 strain (United States) (19) (Fig. 6A). Gubbins analysis for core SNP clustering predicted that the two lineages were distinguished by a single 28,200-bp region of recombination, between positions 142,426 bp (ntpE; M28_Spy0126) and 170,625 bp (M28_Spy0153) of the MGAS6180 chromosome. This suggests the emergence of one lineage from the other through a single recombination event followed by expansion of both lineages (Fig. 6B). Within the recombination region was the P-nga-ifs-slo locus, which differed between the two lineages; although unique in the MGAS6180-like lineage and with low-activity-associated promoter residues A−27T−22C−18, the MEW123-like lineage had a P-nga-ifs-slo identical to that found in emm78 isolates, with the three key residues of A−27T−22T−18. This is supported by recent findings identifying two main lineages within emm28 and that the A−27T−22T−18 promoter variant conferred greater toxin expression than A−27T−22C−18 (15).
FIG 6.
Recombination within the emm28 and emm87 populations. (A) Maximum likelihood phylogeny constructed with 33,537 core SNPs following mapping of all available emm28 genome data to the emm28 MGAS6180 reference genome (white square) (18). Modern UK isolates (red circles): BSAC (n = 15), CUH (n = 13 [11]), and PHE-2014/15 (n = 240 [12, 13]); one historical English isolate from 1938 (brown square). North American isolates (blue circles): ABCs-2015 (n = 95 [14]), Canada (2011 to 2013, n = 4 [47]), and completed genome strain HarveyGAS (United States, 2017 [48]). Other isolates: Lebanon (n = 1, orange circle [49]), Australia (n = 5, green circles [50]), and France (STAB10015 [51], M28PF1 [52], turquoise circles). Total number of isolate genomes was 379. Two lineages of emm28 were identified, one clustering with MGAS6180 (white square) and the other (shaded gray) clustering with MEW123 (2012 USA [19], white circle). (B) Regions of recombination were then identified within the emm28 genome alignment and removed before reconstructing a phylogenetic tree using 17,885 variable sites. (C) Maximum likelihood phylogeny constructed with 6,292 core SNPs following mapping of all available emm87 genome sequence data to the reference emm87 strain NGAS743 (Canada, white circle [53]). UK isolates (red circles): BSAC (2001 to 2011, n = 22), CUH (2008, n = 1 [11]), and PHE-2014/15 (n = 72, [12, 13]). North American isolates (blue circles): ABCs-2015 (n = 26 [14]), Canada (n = 23 [47, 53]), and Texas Children’s Hospital (2012 to 2016, n = 27 [54]). NCTC12065 (GenBank accession number GCA_900460075.1) isolate from ∼1970 to 1980 was also included (brown square). Total number of isolates was 173. Three isolates (shaded gray) were distinct from the main population. The branch was shortened for one isolate for presentation purposes. (D) Regions of recombination were identified within the emm87 genome alignment and removed before reconstructing a phylogenetic tree using 1,531 variable sites. Isolates indicated by an asterisk (*) in both emm28 and emm87 populations were predicted to have undergone recombination in regions surrounding the hasABC locus. Scale bars represent single nucleotide polymorphisms. PHE-2014/15 emm28 isolates GASEMM1261, GASEMM2648, GASEMM1396, and GASEMM1353 were removed for presentation purposes, as they represented highly divergent lineages.
Although we identified an A−27G−22T−18 high-activity variant of P-nga-ifs-slo within emm28, this was only associated with the highly divergent GASEMM1261 isolate that may represent an emm switching event. This isolate, along with three other PHE-2014/15 isolates (GASEMM2648, GASEMM1396, and GASEMM1353), also representing highly divergent lineages, were excluded from the phylogenetic analysis.
All emm28 isolates, regardless of lineage and including MGAS6180 (originally isolated in the 1990s), had the same insertion mutation within hasA of an A residue after 219 bp. This insertion was predicted to lead to a frameshift and a premature stop codon after 72 amino acids (aa) instead of full-length 420 aa, rendering hasA a pseudogene. Some isolates also had additional mutations in hasA: a deletion of an A residue in a septa-A tract leading to a frameshift and a stop codon after 7 aa (n = 1); a deletion of a T residue in a septa-T tract leading to a frameshift and a stop codon after 15 aa (n = 2); an insertion of an A residue after 57 bp leading to a frameshift and a stop codon after 46 aa (n = 3). The loss of full-length HasA would render the isolates acapsular.
In emm28, there were just two exceptions where hasA was found to be intact: the historical emm28 isolate from 1938 had an intact hasABC capsule operon, and BSAC_bs2099, which appeared to have undergone recombination to acquire a 22,316-bp region surrounding the hasABC genes that was 99% identical to the same region in emm2 isolate MGAS10270, suggesting emm2 might be the donor for this recombination. Both isolates were predicted to express full-length HasA and synthesize capsule. Taken together, in comparison with the oldest emm28 isolate, the data showed that post-1930s emm28 isolates became acapsular through mutation, but the contemporary population is divided into two major lineages, MEW123-like and MGAS6180-like lineages, that may differ in nga-ifs-slo expression. Additionally, there was evidence of geographical structure in the population: the MEW123-like lineage comprised mainly of North American isolates (39/44) and only five from England/Wales; isolates from Australia, France, and Lebanon were MGAS6180-like, along with the rest of the England/Wales isolates.
Phylogenetic analysis of the BSAC emm87 population was expanded and compared with publicly available emm87 genome sequence data, totaling 173 isolate genomes from the United Kingdom and North America, including one historical NCTC UK isolate from ∼1970 to 1980 (NCTC12065, GenBank accession number GCA_900460075.1). Gubbins analysis predicted a single 20,506-bp region of recombination surrounding the P-nga-ifs-slo region that distinguished the main population from the oldest BSAC isolates from 2001 and the historical 1970 to 1980 NCTC isolate (Fig. 6C). While the two 2001 BSAC isolates and the NCTC isolate had a P-nga-ifs-slo variant with low-activity-associated promoter residues, G27T−22T−18, all other emm87 isolates had a P-nga-ifs-slo region with high-activity-associated promoter residues, A−27G−22T−18, identical to that found in emm4 and some emm77 isolates. This suggested the emergence of a new lineage through a single recombination event followed by expansion within the population, redolent of that previously observed in emm89 (Fig. 6D).
Similar to emm28, all emm87 isolates, bar four, had an insertion of an A residue after 57 bp that resulted in a frameshift mutation in hasA and the introduction of a premature stop codon after 46 aa of HasA. This mutation was also identified within the historical NCTC isolate but was not found in the two 2001 BSAC isolates that had an intact hasABC locus. This mutation was also absent in two PHE-2014/15 isolates that had undergone an additional recombination event (32,243 bp) surrounding the hasABC locus; although, as this region shared 100% DNA identity to emm28 isolate MGAS6180, HasA is truncated. Overall the data showed that, like emm89 isolates, contemporary emm87 isolates are acapsular with a high-activity nga-ifs-slo promoter, suggesting that this emm lineage may have recently shifted toward this genotype/phenotype.
Recombination within different multilocus sequence types of emm75.
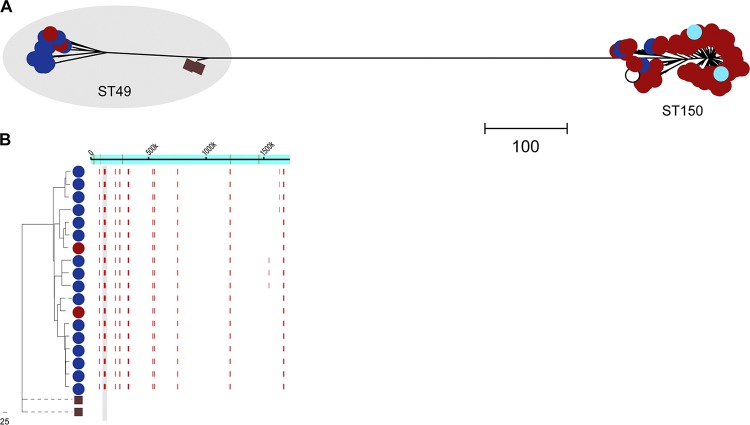
The emm75 genotype is of interest as a common cause of noninvasive infection in the United Kingdom; it is also used in models of nasopharyngeal infection (20, 21). Eleven emm75 isolates were present in the BSAC collection, all multilocus sequence type (ST) 150. When we incorporated other available genome sequence data for emm75 (n = 174), including two newly sequenced historical English emm75 isolates from 1937 and 1938, two major lineages were identified, characterized by two different MLSTs: ST49 or ST150 (Fig. 7A). Although the two historic English isolates were ST49, like the majority of modern North American isolates, the modern England/Wales isolates were predominantly ST150.
FIG 7.
Two lineages within emm75. (A) Maximum likelihood phylogeny constructed with 9,241 core SNPs following mapping of all available emm75 genome sequence data to the genome of French strain STAB090229 (white circle) (55). Modern UK collections (red circles): BSAC (n = 11), CUH (n = 6 [11]), and PHE-2014/15 (n = 141 [12, 13]; two English historical isolates (brown squares) from 1937 and 1938. North American isolates (blue circles): ABCs-2015 (n = 20 [14]), NGAS344, and NGAS604 from Canada 2011 to 2012 (53). French strains (turquoise circles): STAB120304 (2012) and STAB14018 (2014) (55). Total number of isolates was 185. Two lineages were identified, generally characterized by the MLSTs: ST49 (shaded gray) or ST150 (with minor MSLT variants ST788, ST851, and ST861 within these lineages). (B) Gubbins analysis identified ten regions of predicted recombination (red lines) in all modern ST49 compared to historical 1930s ST49 across the genome (indicated across the top). One region included P-nga-ifs-slo (shaded gray). The phylogenetic tree was constructed with 1,953 variable sites following removal of predicted regions of recombination. Scale bars represent single nucleotide polymorphisms. One PHE-2014/15 isolate (GASEMM1722) was excluded for presentation purposes, as it was highly divergent from the rest of the population.
Although these two ST lineages differed in the P-nga-ifs-slo region, there was a high level of predicted recombination across the genomes of both STs, perhaps indicative of historic emm switching or extensive genetic exchange. ST49 isolates had the subtype 1.1 low-activity A−27T−22C−18 promoter, whereas all ST150 isolates had the A−27G−22T−18 subtype 3.1 high-activity promoter variant, identical to that of emm1 and emm89. Modern ST49 isolates did, however, differ from historic 1930s isolates by ten distinct regions of predicted recombination (Fig. 7B), including a region spanning the nga-ifs-slo locus, although this did not include the promoter region. We did not detect any mutations affecting the capsule region in emm75. Taken together, emm75 was characterized by two major MLST lineages differing in P-nga-ifs-slo promoter activity genotypes but without evidence of recent recombination or loss of capsule.
Lineages associated with recombination in emm76, emm77, and emm81.
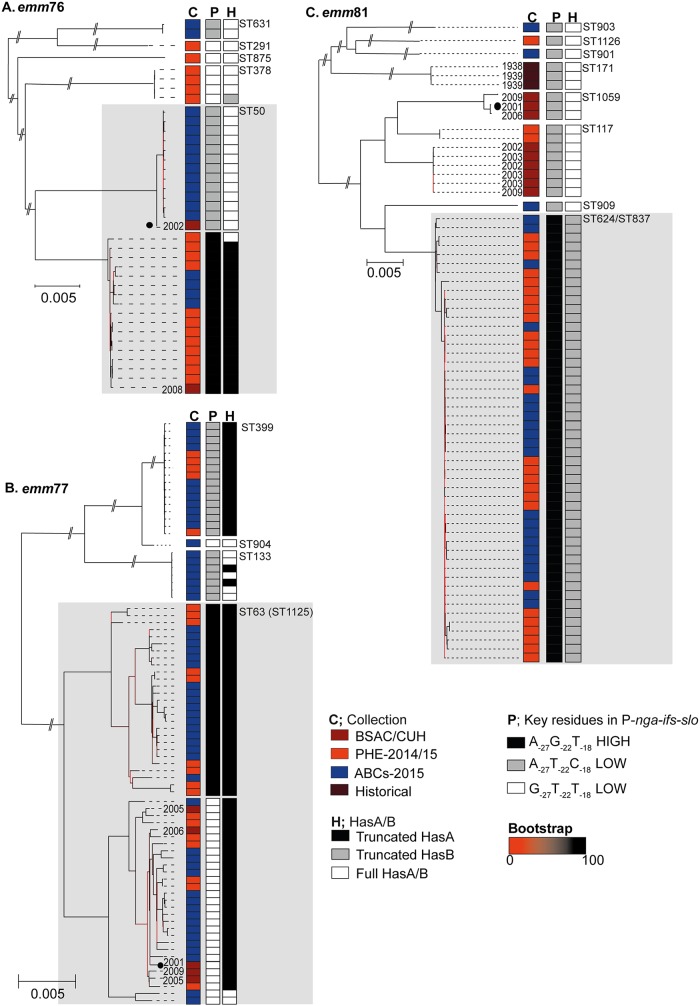
The phylogeny of all available genome data for emm76, emm77, and emm81 confirmed the presence of diverse lineages associated with different MLSTs (Fig. 8A to C). In all genotypes, however, there was a dominant MLST lineage representing the majority of isolates: ST50 emm76, ST63 emm77, and ST624 emm81. Within the dominant MLST lineages of emm76 and emm77, there were sublineages that were associated with different P-nga-ifs-slo variants as well as loss of functional HasA through mutation.
FIG 8.
Variants of P-nga-ifs-slo and capsule mutations associated with lineages of emm76, emm77, and emm81. Maximum likelihood phylogeny identified multiple sequence type (ST) lineages within the populations of emm76 (A), emm77 (B), and emm81 (C). Collection indicates either BSAC or CUH (dark red), PHE-2014/15 isolates (red), ABCs-2015 (blue), or English historical (brown). Dates for BSAC, CUH, or historical are shown; other isolates were from 2014/2015. STs are indicated on the right and major lineages are shaded gray. (A) Genome data for emm76 was mapped to the de novo assembled sequence of BSAC_bs448 from 2002, selected as the oldest isolate representing the genotype. Genome data from a total of 38 isolates were used: BSAC (n = 2), PHE-2014/15 (n = 18 [12, 13]), ABCs-2015 (n = 18 [14]). Predicted prophage regions were removed and a maximum likelihood phylogenetic tree constructed from 30,264 core SNPs. Five STs were identified (indicated on right of tree), but the main lineage was ST50. (B) All emm77 genome data were mapped to the de novo assembled sequence of BSAC_bs150 from 2001. Genome data from a total of 80 isolates were used: BSAC (n = 5), PHE-2014/15 (n = 21 [12, 13]), and ABCs-2015 (n = 54 [14]). Four STs were identified but the main lineage was ST63, with one isolate in this lineage being single locus variant ST1125. Predicted prophage regions were removed, and a maximum likelihood phylogenetic tree was constructed from 34,760 core SNPs. (C) All emm81 genome data were mapped to the de novo assembled seqeunce of BSAC_bs229 from 2001. Genome data from a total of 68 isolates were used: BSAC (n = 9), CUH (n = 1 [11]), PHE-2014/15 (n = 29 [12, 13]), ABCs-2015 (n = 26 [14]), and English historical 1930s (n = 3). Predicted prophage regions were removed, and a maximum likelihood phylogenetic tree was constructed from 42,258 core SNPs. Nine STs were identified but the main lineage was ST624 with and minor (single base change in recP) ST variant ST837. We identified variants of P-nga-ifs-slo (P) associated with one of three combinations of key promoter residues, including the high-activity-associated A−27G−22T−18 (P; black). For emm76 (A) and emm77 (B), mutations were detected in hasA predicted to truncate HasA (H; black). (C) All emm81 isolates were predicted to express full-length HasA, but the ST624/ST837 lineage carried a mutation within hasB leading to a truncated HasB (H; gray). Branches are colored based on bootstrap support (scale bar provided). Scale bars represent substitutions per site. Isolates used as references for mapping indicated with black circles. Branches for lineages outside main lineages were shortened for presentation purposes (indicated by line breaks). C; collection, P; promoter key residue combination, H; full-length or truncated HasA or HasB.
We identified five different MLSTs within emm76 (Fig. 8A), but the majority of isolates (30/38) belonged to ST50, including both BSAC isolates. Recombination analysis of the ST50 lineage identified a sublineage that differed from other ST50 isolates by 19 regions of recombination (see Fig. S3). One of these regions encompassed P-nga-ifs-slo, conferring a P-nga-ifs-slo variant closely related to that of modern emm1 and emm89 with an identical high-activity promoter (subtype 3.1). This sublineage was dominated by PHE-2014/15 isolates and also contained the more recent of the two BSAC isolates (2008). All isolates in this sublineage, except one, also had a nonsense mutation within hasA of a C-to-T change at 646 bp, resulting in a premature stop codon after 215 aa, likely to render the isolates acapsular. Only one ST50 isolate outside this sublineage had the same hasA C646T change. All other emm76 isolates would express full-length HasA.
Recombination within ST50 emm76. All sequence data for emm76 (n = 38) was mapped the de novo assembled sequence of BSAC_bs448 (bold). The majority of isolates were ST50 (ST; white) and within this ST were two sublineages; the lower sublineage was associated the high-activity promoter A−27G−22T−18 (P; black) and truncated HasA (HasA/B; black). Gubbins analysis (boxed region on right) of ST50 isolates identified 19 regions of recombination across the genome in all isolates (red vertical lines) belonging to the lower sublineage compared to the top sublineage. One of these regions (highlighted grey) surrounded the P-nga-ifs-slo locus conferring the high-activity-associated promoter (P; black) with residues A−27G−22T−18 to the lower sublineage compared to low-activity A−27T−22C−18 (P; grey) in the top sublineage. The presence (black) or absence (white) of mobile prophage-associated superantigens (speA, -C, -H, -I, -K, -L, -M, and ssa) and DNAses (sda, sdn, spd1, spd3, spd3v6, and spd4) as well as antimicrobial resistance genes and mutant variants of regulators CovR, CovS, and RocA was also determined for each isolate. All isolates within the lower ST50 sublineage carried a variant of the prophage-associated DNAse spd3 (spd3v6) (45) that is more divergent than other spd3 variants, including the spd3 variant carried by isolates belonging to the top ST50 sublineage. All lower sublineage isolates also carried the resistance gene ermB which was absent in other lineages, but they did not carry other antimicrobial elements found in the upper sublineage isolates. Sporadic truncated mutant variants of regulators CovR, CovS, and RocA (black) were also detected across the tree but were not associated with any specific lineages. Scale bar represents substitutions per site. Scale on boxed region represents position across the assembled BSAC_bs448 genome. Bootstrap values provided on major branches. Download FIG S3, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Two sublineages were also identified within the dominant emm77 lineage ST63 (Fig. 8B), and one was associated with the high-activity cluster P-nga-ifs-slo variant compared to predicted low-activity variants found in the other emm77 lineages. Recombination analysis predicted only two regions of recombination distinguishing the two sublineages: a region of 17,954 bp surrounding P-nga-ifs-slo, and a 173-bp region within a hypothetical gene (SPYH293_00394) (see Fig. S4). While all BSAC emm77 isolates (years 2001 to 2009) were ST63 with low-activity P-nga-ifs-slo, PHE isolates from 2014 to 2015 were almost evenly divided between the two sublineages, indicating a potential recent change in England/Wales. All ST63 isolates except two had a deletion of a T residue within a septa-poly(T) tract at 458 bp in hasA, predicted to truncate the HasA protein after 154 aa. The two exceptions were predicted to encode full-length HasA and were associated with low-activity P-nga-ifs-slo promoter variants. Although also not associated with high-activity P-nga-ifs-slo promoter variants, other lineages of emm77 also carried mutations within hasA that would truncate HasA; ST399 isolates carried an insertion of a T residue at 71 bp of the hasA gene resulting in a premature stop codon after 46 aa, and two ST133 isolates carried a G894A substitution resulting in a premature stop codon after amino acid residue 297.
Recombination within ST63 emm77. All sequence data for emm77 (n = 82) were mapped to the de novo assembled sequence of BSAC_bs150 (bold). The majority of isolates were ST63 (ST; white), or one single locus variant ST1125, and within this ST were two sublineages: the upper lineage associated with the high-activity promoter A−27G−22T−18 (P; black) and truncated HasA (H; black). Gubbins analysis (boxed region) of ST63 isolates identified two regions of recombination across the genome of all isolates (red vertical lines) belonging to the upper sublineage compared to the lower sublineage. One of these regions (highlighted grey) surrounded the P-nga-ifs-slo locus conferring the high-activity-associated promoter with residues A−27G−22T−18 (P; black) to the upper sublineage compared to low-activity G−27T−22T−18 (P; white) in the lower sublineage. The presence (black) or absence (white) of mobile prophage-associated superantigens (speA, -C, -H, -I, -K, -L, -M, and ssa) and DNAses (sda, sdn, spd1, spd3, spd3v6, and spd4) as well as antimicrobial resistance genes and truncated mutant variants of regulators CovR, CovS, and RocA was also determined for each isolate. The prophage-associated DNase spd3 was common to all upper sublineage ST63 isolates, and all except one of this sublineage carried the antimicrobial resistance gene ermTR. Sporadic truncated mutant variants of CovR, CovS, and RocA (black) were detected across the tree but were not associated with any specific lineages. Scale bar represents substitutions per site. Scale on boxed region represents position in the BSAC_bs150 assembly. Bootstrap values provided on main branches. Download FIG S4, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The emm81 population (n = 68) was more diverse with nine different sequence types (Fig. 8C), but the majority of isolates (41/68) were ST624 or the single locus variant ST837 (9/68; one SNP in recP allele) within the same lineage. ST171 was restricted to three historical isolates originally collected in 1938 and 1939. We did not detect any hasABC variations that would disrupt translation in emm81 lineages except for the dominant group of ST624/ST837, where we identified an A residue insertion at 128 bp in hasB resulting in a frameshift and premature stop codon after 50 aa. All ST624/ST837 isolates carried the high-activity cluster P-nga-ifs-slo variant identical to that seen in emm3 compared to all other lineages associated with other low-activity P-nga-ifs-slo variants. Recombination analysis identified extensive recombination had occurred within emm81 leading to the different levels of diversity, but we identified one region of recombination that distinguished the ST624/ST837 lineage from the closely related ST909 and ST117 populations (see Fig. S5). This region surrounded the P-nga-ifs-slo locus, suggesting ST624/ST837 gained the high-activity cluster P-nga-ifs-slo variant through recombination, like other emm types, potentially from emm3. The emergence of the high-activity P-nga-ifs-slo variant and truncated HasB ST624/ST837 lineage may be recent in England/Wales, as all BSAC isolates obtained prior to 2009 were outside this lineage.
Recombination within emm81. All sequence data for emm81 (n = 68) were mapped to the de novo assembled sequence of BSAC_bs229 (bold). The majority of isolates were ST624, with high-activity promoter A−27G−22T−18 (P; black) and truncated HasB (HasB; black). Gubbins analysis (boxed region) of ST624 isolates and closely related ST1059, ST117, ST909, and ST837, compared to BSAC_bs229, identified patterns of recombination across the genome in all isolates (red vertical lines, or blue vertical lines if unique to a single isolate). One of these regions (highlighted grey) surrounded the P-nga-ifs-slo locus conferring the high-activity-associated promoter with residues A−27G−22T−18 to the ST624/ST837 population compared to low-activity G−27T−22T−18 in all other isolates. The presence (black) or absence (white) of mobile prophage-associated superantigens (speA, -C, -H, -I, -K, -L, -M, and ssa) and DNAses (sda, sdn, spd1, spd3, spd3v6, and spd4) as well as antimicrobial resistance genes and truncated mutant variants of regulators CovR, CovS, and RocA was also determined for each isolate. The majority of all isolates carried the prophage-associated speH. Antimicrobial resistance genes were rarely detected in any ST. Scale bar represents substitutions per site. Scale on boxed region represents position in the BSAC_bs229 assembly. Bootstrap values provided on main branches. Download FIG S5, TIF file, 2.4 MB (3.3MB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
High-activity cluster P-nga-ifs-slo variants gained by recombination in emm94 and emm108.
Within emm94, we identified a P-nga-ifs-slo identical to that found in emm1 with high-activity promoter variant subtype 3.1. Phylogenetic analysis of 51 emm94 isolates identified a dominant lineage among England/Wales isolates separate to the single US isolate and two England/Wales isolates (see Fig. S6A) that belonged to ST89. Gubbins analysis predicted 11 regions of recombination in all lineage-associated isolates compared to the three outlying isolates, including one (22,648 bp) that encompassed P-nga-ifs-slo, transferring a high-activity A−27G−22T−18 P-nga-ifs-slo variant. All emm94 isolates contained an indel within hasB compared to the reference (H293), losing 6 bp and gaining 13 bp between 127 and 133 bp. This variation causes a frameshift and would truncate the HasB protein after 45 aa.
Recombination in emm94 and emm108. (A) In the PHE-2014/2015 (red) emm94 population, the majority (n = 51) form a lineage separate from two PHE-2014/2015 isolates and the single ABCs-2015 (blue) isolate. Gubbins analysis predicted 11 regions of recombination (red lines) in all the lineage associated isolates compared to the three other isolates. One of these regions (highlighted in grey) encompassed the P-nga-ifs-slo region. (B) Isolates of emm108 from the ABCs-2015 (blue) collection were of a different MLST (ST14) than PHE-2014/15 (red) (ST1088). The hasB gene was absent in the genomes of both ABCs-2015 isolates, and one had undergone recombination surrounding the P-nga-ifs-slo locus (shaded grey), as predicted by Gubbins analysis (shown on the right). Blue lines, predicted recombination unique to a single genome. Sequence data were mapped to the reference strain H293, also used as an outgroup for SNP cluster analysis. Scale bar represents SNPs. Download FIG S6, TIF file, 0.9 MB (961.2KB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We identified a similar high-activity cluster P-nga-ifs-slo variant within a single emm108 genome originating from the United States. Within the 9 isolates from PHE-2014/15 (n = 7) and ABCs-2015 (n = 2), there were two sequence types, ST1088 and ST14. ST14 was represented by the only two ABCs-2015 isolates, and we identified that both had lost the entire hasB gene, although hasA and hasC were still present (Fig. S6B). Additionally, one of the ABCs-2015 isolates had undergone recombination of a single ∼29,683-bp region surrounding the P-nga-ifs-slo, replacing P-nga-ifs-slo for one identical to that found in emm3 with high-activity promoter variant A−27G−22T−18 subtype 3.1.
Mobile genetic elements and antimicrobial resistance.
The acquisition of mobile genetic elements such as prophages and transposons may also be influenced by capsule expression and can also influence the expansion and success of new lineages. We therefore determined the presence of prophage-associated superantigen and DNase genes as well as antimicrobial resistance genes to estimate the number of mobile genetic elements present within each isolate of the genotypes emm28, emm75, -76, -77, -81, -87, -94, and -108 (Fig. S3 to S5; Data Set S3). On average, there were 4.4 elements present in isolates predicted to express full-length HasABC compared to 2.5 elements present in isolates with hasABC gene mutations or gene absence, suggesting that the presence of capsule does not hinder mobile genetic elements. We also detected no link between lineages within these genotypes that had undergone P-nga-ifs-slo recombination and mobile factors, except within emm76 and emm77. Isolates belonging to the emm76 ST50 sublineage associated with HasA mutation and P-nga-ifs-slo recombination all carried the prophage-associate superantigen genes speH and speI as well as a diverse variant of the DNase spd3 and the erythromycin resistance gene ermB (Fig. S3). This differed from the other ST50 isolates that carried another variant of spd3 and multiple different resistance genes. The sublineage of ST63 emm77 associated with P-nga-ifs-slo recombination also carried spd3, and all, except one isolate, carried the erythromycin resistance gene ermTR; both genes were not common in other ST63 emm77 isolates (Fig. S4).
Details of emm28, emm75, emm76, emm77, emm81, emm87, emm94, and emm108 isolates used in this study. Download Data Set S3, XLSX file, 0.2 MB (178.3KB, xlsx) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The emergence of new internationally successful lineages of S. pyogenes can be driven by recombination-related genome remodeling, as demonstrated by emm1 and emm89. The transfer of a P-nga-ifs-slo region conferring increased expression to the new variant was common to both genotypes. In the case of emm89, five other regions of recombination were identified in the emergent variant, one resulting in the loss of the hyaluronic acid capsule. Although, potentially, all six regions of recombination combined underpinned the success of the emergent emm89, we have shown here that recombination of P-nga-ifs-slo has occurred in other leading emm types as well as a high frequency of capsule loss through mutation. These data point to an association between genetic change affecting capsule and recombination affecting the P-nga-ifs-slo locus, conferring increased production of nga-ifs-slo; in some cases (notably emm87, emm89, and emm94), this has further been associated with an apparent fitness advantage and expansion within the population.
A number of genotypes were found to be associated with multiple variants of P-nga-ifs-slo. The majority of genotypes had P-nga-ifs-slo variants with the low-activity-promoter associated with three key residue variants: G−27T−22T−18 or A−27T−22C−18. Only emm1, emm3, and emm12 were exclusively associated with the high-activity A−27G−22T−18 variant. We have shown that the same high-activity promoter variant is present in isolates belonging to twelve other emm types, notably, emm76, emm77, emm81, emm87, and emm94, although this is not a consistent feature in these genotypes due to emm switching or recombination. We identified four combinations of the three key promoter residues and several subtypes of the 67-bp promoter that varied in bases other than those at the −27, −22, and −18 key positions. Although some subtypes were restricted to single genotypes, variation in the −40 base led to the subtype 2.2 of G−27T−22T−18 and subtype 3.2 of A−27G−22T−18. We measured the activity of NADase in representative strains and genotypes of these promoter variants and found that variation in the −40 base did not impact the activity conferred by the −27, −22, and −18 bases. Although we predicted the level of nga and slo expression based on the promoter variant, this may not relate to actual expression given the level of other genetic variation between genotypes. However, our consistent findings of lineages emerging following acquisition of the high-activity promoter variant supports the hypothesis that this confers some benefit that may relate to increased toxin expression.
Intriguingly, where we identified an acquisition of the high-activity promoter variant through recombination, these genotypes also had a genetic change in the capsule locus, likely rendering the organism unable to make capsule (hasA mutation) or only low levels of capsule (hasB mutation). To date, only emm4, emm22, and the emergent emm89 lineages are known to lack all three genes required to synthesize capsule. Here, we identified mutations that would truncate HasA and HasB in 35% of all isolates and 65% (35/54) of all genotypes. As the majority of isolates included in this study were invasive or sterile-site isolates, the findings further challenge the dogma that the hyaluronan capsule is required for full virulence of S. pyogenes and, in addition, lend credence to the possibility that the increased expression of NADase and SLO may in some way compensate for the lack of capsule (22). While capsule has been shown to underpin resistance to opsonophagocytic killing in the most constitutively hyperencapsulated genotypes such as emm18 (23, 24), there is less evidence that it contributes measurably to opsonophagocytosis killing resistance in other genotypes (3). Whether the loss of capsule synthesis is of benefit to S. pyogenes is uncertain; the capsule may shield several key adhesins used for interaction with host epithelium and fomites but may also act as a barrier to transformation with DNA. An accumulation of hasABC inactivating mutations has been identified during long-term carriage (25); although for some genotypes, capsule loss reduced survival in whole human blood, a high number of acapsular hasA mutants have also recently been found to be causing a high level of disease in children, including emm1, emm3, and emm12 (26).
The process of recombination in S. pyogenes is not well understood, and natural competence has only been demonstrated once and under conditions of biofilm or nasopharyngeal infection (27). We do not know if the six regions of recombination that led to the emergence of the new ST101 emm89 variant occurred simultaneously, although no intermediate isolates have been identified. The loss of the hyaluronic acid capsule in the new emergent emm89, along with our consistent findings of inactivating mutations associated with P-nga-ifs-slo transfer, indicates either (i) the process of recombination requires the inactivation of capsule, (ii) capsule-negative S. pyogenes requires high expression of nga-ifs-slo for survival, or (iii) that a capsule-negative phenotype combined with high expression of nga-ifs-slo provides a greater selective advantage to S. pyogenes.
The phylogeny of emm28, emm87, emm77, emm94, and emm108 indicated that mutations in hasA or hasB occurred prior to recombination of P-nga-ifs-slo, supporting the first hypothesis that prior capsule inactivation is required for recombination. There is no evidence, however, to suggest this was required for recombination in the emm1 population. It could be hypothesized that capsule acts as a barrier to genetic exchange, but there has also been a positive genetic association of capsule to recombination rates (28). A positive association may, however, be related only to species expressing antigenic capsule, whereby recombination is required to introduce variation for immune escape.
The hasC gene is not essential for capsule synthesis (29), because a paralog of hasC exists within the S. pyogenes genome. A paralog for hasB (hasB.2) also exists elsewhere in the S. pyogenes chromosome and can act in the absence of hasB to produce low levels of capsule (30), but hasA is absolutely essential for capsule synthesis (29). The mutations in hasA in emm28 and emm87 have been previously noted and confirmed to render the isolates acapsular (26, 31). Not all acapsular isolates were found to carry the high-activity promoter of nga-ifs-slo, despite being invasive, perhaps refuting the hypothesis that the high-activity nga-ifs-slo promoter is essential for the survival of acapsular S. pyogenes. High expression of nga-ifs-slo may also occur through other mechanisms, for example, through mutation in regulatory systems. We looked at the sequences of covRS and rocA, known to negatively regulate nga-ifs-slo, in all isolates (see Data Set S2 in the supplemental material) and identified some emm-type specific variants, consistent with our previous findings (11). We did not identify any other genotypes where all isolates carried truncation mutations in rocA, such as emm3 and emm18 that were previously confirmed to affect function and increase expression of rocA-covR-regulated virulence factors (23, 32), consistent with other findings (14). It is unclear as to whether the amino acid changes in found in other genotypes would affect function of rocA as well as covR and covS, and this requires further work.
Interestingly, we identified that the capsule locus is also a target for recombination as, similarly to emm89, isolates within emm28 and emm87 had undergone recombination of this locus and surrounding regions, varying in length and restoring capsule synthesis in emm28. Isolated examples of hasA or hasB gene loss were identified in some genotypes, such as emm108, possibly due to internal recombination and deletion.
Only two emm4 isolates and one emm22 isolate were found to have P-nga-ifs-slo variants that were not A−27T−22G−18 high-activity promoter variants, and interestingly, these isolates carried the hasABC genes, typically absent in emm4 and emm22. The high genetic distance of these isolates to other emm4 and emm22 genomes indicated potential emm switching of the emm4 or emm22 genes into different genetic backgrounds. The single emm28 with a high-activity P-nga-ifs-slo variant may also be an example of this, and was one of four emm28 isolates that did not cluster with the two main emm28 lineages. Although we excluded them from our analysis, as we focused on recombination within the two main lineages, the presence of highly diverse variants within genotypes and the potential for emm-switching warrants further investigation, particularly as the most promising current vaccine is multivalent toward common M types (33).
All other genotypes carrying the high-activity P-nga-ifs-slo variant were found to have undergone recombination of this region: emm28, emm75, emm76, emm77, emm81, emm87, emm94, and emm108, as well as the previously described emm1 and emm89.
Within emm87, we identified three isolates outside the main population lineage that represented the oldest isolates in the collection: two from 2001 (different geographical locations within England) and one NCTC strain from ∼1970 to 1980 (NCTC12065). A single region of recombination surrounding the P-nga-ifs-slo locus distinguished the main population lineage from the three older isolates, consistent with a recombination event, but due to a lack of earlier isolates of emm87, we could not confirm a recombination-related shift in the population, as reported previously for emm89 and emm1.
The existence of two lineages within the contemporary emm28 suggests that one has not yet displaced the other, although the MEW123-like lineage was predominantly US isolates, consistent with recent findings (15). The P-nga-ifs-slo region with the high-activity-associated A−27T−22T−18 and acquired through recombination by the MEW123-like lineage was identical to that found in emm78, indicating emm78 as the potential genetic donor. We found emm78 to have high levels of NADase activity, as predicted, and interestingly, similarly to emm28, all eight emm78 isolates were acapsular due to a deletion within the hasABC promoter region extending into hasA. This again may support the hypothesis that capsule-negative S. pyogenes requires high expression of nga-ifs-slo for survival.
A strength of this study was the systematic longitudinal sampling over a 10-year period; as expected, this again identified the shift in the emm89 population. Other emm types exhibited lineages with different P-nga-ifs-slo variants, and those with the more active promoter variant did appear to become dominant over time, similarly to emm1 and the emergent emm89 lineages. For example, the high-activity P-nga-ifs-slo ST63 lineage of emm77 was not detected in England/Wales isolates prior to 2014 and 2015. Similarly, the high-activity P-nga-ifs-slo variant emm81 ST646/ST837 lineage was represented by only a single isolate (of six) collected between 2001 and 2009 but became dominant by 2014 to 2015 in England/Wales and the United States. emm75 was the 6th most common genotype in England/Wales in 2014 to 2015 and dominated by high-activity P-nga-ifs-slo variant ST150 lineage yet was less common in the United States, where ST49 with low-activity P-nga-ifs-slo is dominant. A high prevalence of emm94 was also found in England/Wales between 2014 and 2015 but was rare in the United States (only 1 isolate). Our analysis of this genotype indicated there has been a recombination-related change in the population, as we detected 11 regions of predicted recombination, including P-nga-ifs-slo, potentially conferring high toxin expression. The other ten regions of recombination may also provide advantages to this lineage along with a potential low level of capsule through hasB mutation.
Other factors may also contribute to the success of emergent new lineages, including mobile prophage-associated virulence factors and antimicrobial resistance genes. Acquisition of mobile genetic elements did not appear to be affected by capsule loss; indeed, fewer mobile genetic element-associated factors were detected in isolates with capsule gene mutations than in isolates with functional capsule genes. A number of bacteriophages that target S. pyogenes encode a hyaluronidase thought to allow the bacteriophage to access the bacterial surface by degrading the outer capsule layer (34); therefore, recombination of these elements is likely to be different from gene transfer of core genetic regions, such as P-nga-ifs-slo.
We did, however, identify an association in the lineages of emm76 and emm77 with prophage-associated virulence factors and antimicrobial resistance genes. It is possible that the superantigens speH, speI, and DNase spd3 may also have contributed to the success of the lineages that had undergone P-nga-ifs-slo recombination. Of concern is that both emm76 and emm77 carried genes for resistance to tetracycline and erythromycin, which were rarer in other genotypes. If the acapsular/high-toxin-expressing lineages do expand in the population, it will be important to monitor the levels of antimicrobial resistance in these lineages. This is also true for emm108, as tetM was detected in all isolates, but the presence of antimicrobial resistance genes was rare in emm28, emm75, emm81, emm87, and emm94, regardless of lineage.
The development and boosting of circulating antibodies to SLO is often used as a diagnostic biomarker of recent S. pyogenes infection and is known to be more specific to throat rather than skin infections. The genomic analysis provides explanation for this historic and well-recognized association between anti-streptolysin O (ASO) titers and disease patterns, due to known tissue tropism of S. pyogenes emm types. Whether the alteration of SLO activity in different S. pyogenes strains might render such a test more or less specific will be of interest, although it may explain observed differences in ASO titers between genotypes (35). There is also the possibility that other beta hemolytic streptococci might acquire similarly active SLO production, reducing the specificity of ASO titer to S. pyogenes.
Our genomic analysis has uncovered convergent evolutionary pathways toward capsule loss and recombination-related remodeling of the P-nga-ifs-slo locus in leading contemporary genotypes. This suggests that a combination of capsule loss and gain of high nga-ifs-slo expression provide a greater selective advantage than either of these phenotypes alone. Acquisition of the high-activity promoter led to pandemic emm1 and emm89 clones that are dominant and highly successful. Active surveillance of the lineages comprising emm76, emm77, emm81, emm87, emm94, and emm108 is required to determine if capsule loss/reduction and recombination of P-nga-ifs-slo toward high expression will trigger expansion toward additional pandemic clones in the next few years.
MATERIALS AND METHODS
Isolates.
Three hundred forty-four isolates of S. pyogenes associated with bloodstream infections and submitted to the British Society for Antimicrobial Chemotherapy (BSAC; http://www.bsacsurv.org) from 11 different sites across England between 2001 and 2011 were subjected to whole-genome sequencing (see Data Set S1 in the supplemental material). All BSAC isolates were tested for antibiotic susceptibility using the BSAC agar dilution method to determine MICs (36).
A further six isolates were sequenced from a historical collection of S. pyogenes originally collected in the 1930s from puerperal sepsis patients at Queen Charlottes Hospital, London, UK; one emm28 isolate from 1938 (ERR485803), two emm75 isolates from 1937 (ERR485807) and 1939 (ERR485820), and three emm81 isolates from 1938 (ERR485805) and 1939 (ERR485801, ERR485802).
Genome sequencing.
Streptococcal DNA was extracted using the QIAxtractor instrument according to the manufacturer’s instructions (Qiagen, Hilden, Germany) or manually using a phenol-chloroform method (37). DNA library preparation was conducted according to the Illumina protocol, and sequencing was performed on an Illumina HiSeq 2000 with 100-cycle paired-end runs.
Genomes were de novo assembled using Velvet with the pipeline and improvements found at https://github.com/sanger-pathogens/vr-codebase and https://github.com/sanger-pathogens/assembly_improvement (38). Annotation was performed using Prokka. emm genotypes were determined from the assemblies, and multilocus sequence types (MLSTs) were identified using the MLST database (https://pubmlst.org/spyogenes/) and an in-house script (https://github.com/sanger-pathogens/mlst_check). New MLSTs were submitted to the database (https://pubmlst.org/).
Genome sequence analysis.
Sequence reads were mapped using SMALT (https://www.sanger.ac.uk/science/tools/smalt-0) to the completed emm89 reference genome H293 (HG316453.2) (3), as this genome contains no known prophage regions. Other reference genomes were also used where indicated with predicted prophage regions (Table S1) excluded to obtain “core” SNPs. Maximum likelihood phylogenetic trees were generated from aligned core SNPs using RAxML (39) with the GTR substitution model and 100 bootstraps. Regions of recombination were predicted using Gubbins analysis with the default parameters (40). Branches of phylogenetic trees were colored according to bootstrap support using iTOL (41).
Reference genomes used for mapping in this study and excluded prophage regions. Download Table S1, DOCX file, 0.1 MB (12.9KB, docx) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Other genome sequence data were obtained from the short read archive. We combined data collected across England and Wales through Public Health England during 2014 and 2015 (PHE-2014/15) supplied by Kapatai et al. (13) and Chalker et al. (12) from invasive and noninvasive S. pyogenes isolates. We also used data supplied by Chochua et al. (14) collected by Active Bacterial Core Surveillance USA in 2015 (ABCs-2015) from invasive S. pyogenes isolates. ABCs-2015 sequence data were preprocessed by Trimmomatic (42) to remove adapters and low-quality sequences. PHE-2014/15 had already been preprocessed (12, 13). Genome data from these collections were assembled de novo using Velvet (assembly statistics provided in Data Set S2), and any isolates with greater than 2.2 Mbp total assembled length and/or more than 500 contig numbers were excluded. We also used data from Turner et al. (11) of invasive and noninvasive isolates from the Cambridgeshire region, UK, and collected through Cambridge University Hospital (CUH). We relied on the emm types determined during the original studies and excluded any data where the emm type was uncertain or negative. The genes hasA, hasB, hasC, covR, covS, and rocA, and the P-nga-ifs-slo were extracted from the assembled genome using in silico PCR (https://github.com/simonrharris/in_silico_pcr). Capsule locus and P-nga-ifs-slo variants were also confirmed through manual inspection of mapping data where genotype could not be accurately determined from the assembly.
Mapping of emm76, emm77, and emm81 sequence data was performed using de novo assembled genome data from one BSAC collection isolate representing the equivalent genotype. Prophage regions were predicted using PHASTER (43) and removed before SNP extraction.
Antimicrobial resistance genes were identified by srst2 (44) using the ARG-ANNOT database (ARGannot_r2.fasta) (45). The presence of prophage-associated superantigen genes speA, speC, speH, speI, speL, speM, and ssa was determined using srst2 and the feature database previously used by Chochua et al. (14) available at https://github.com/BenJamesMetcalf. The presence of prophage-associated DNAses genes sda, sdn, spd1, spd3, spd3v6, and spd4 was also determined using srst2 by adding regions of these genes to the feature database. Representative alleles of these DNase genes were taken from previous analysis (46) to identify regions that would detect all variants of each DNase, except we included spd3v6 separate from spd3 as it represents a divergent allele to spd3. Sequences used are available at Mendeley (https://data.mendeley.com/datasets/hzwjkj2gtp/1).
NADase activity.
Activity of NADase was measured in culture supernatants as previously described (3). Activity was determined as the highest dilution capable of hydrolyzing NAD+. Isolates were selected from the BSAC collection to represent different promoter variants for which there were three or more isolates available that were lacking mutations in regulatory genes.
Data availability.
Sequence data have been submitted to the European Nucleotide Archive (ENA) (www.ebi.ac.uk/ena) as accession numbers ERS361826 to ERS379364, ERR1359331 to ERR485881, ERS361826 to ERS379364, and SRR5853328 to SRR5858742 (listed in Data Sets S1 and S2 in the supplemental material).
ACKNOWLEDGMENTS
This publication presents independent research supported by the Health Innovation Challenge Fund (WT098600, HICF-T5-342), a parallel funding partnership between the Department of Health and the Wellcome Trust. The work was also funded by the UK Clinical Research Collaboration (UKCRC, National Centre for Infection Prevention & Management) and the National Institute for Health Research Biomedical Research Centre awarded to Imperial College London.
The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health, NIHR, or Wellcome Trust. C.E.T. was an Imperial College Junior Research Fellow and is a Royal Society & Wellcome Trust Sir Henry Dale Fellow (208765/Z/17/Z). S.J.P. is a consultant to Specific and Next Gen Diagnostics.
Footnotes
Citation Turner CE, Holden MTG, Blane B, Horner C, Peacock SJ, Sriskandan S. 2019. The emergence of successful Streptococcus pyogenes lineages through convergent pathways of capsule loss and recombination directing high toxin expression. mBio 10:e02521-19. https://doi.org/10.1128/mBio.02521-19.
REFERENCES
- 1.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. 2014. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A 111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, Kristinsson KG, Gottfredsson M, Porter AR, DeLeo FR, Musser JM. 2015. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest 125:3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner CE, Abbott J, Lamagni T, Holden MT, David S, Jones MD, Game L, Efstratiou A, Sriskandan S. 2015. Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. mBio 6:e00622-15. doi: 10.1128/mBio.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu L, Olsen RJ, Nasser W, de la Riva Morales I, Musser JM. 2015. Trading capsule for increased cytotoxin production: contribution to virulence of a newly emerged clade of emm89 Streptococcus pyogenes. mBio 6:e01378-15. doi: 10.1128/mBio.01378-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beres SB, Olsen RJ, Ojeda Saavedra M, Ure R, Reynolds A, Lindsay DSJ, Smith AJ, Musser JM. 2017. Genome sequence analysis of emm89 Streptococcus pyogenes strains causing infections in Scotland, 2010-2016. J Med Microbiol 66:1765–1773. doi: 10.1099/jmm.0.000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friaes A, Machado MP, Pato C, Carrico J, Melo-Cristino J, Ramirez M. 2015. Emergence of the same successful clade among distinct populations of emm89 Streptococcus pyogenes in multiple geographic regions. mBio 6:e01780-15. doi: 10.1128/mBio.01780-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latronico F, Nasser W, Puhakainen K, Ollgren J, Hyyrylainen HL, Beres SB, Lyytikainen O, Jalava J, Musser JM, Vuopio J. 2016. Genomic characteristics behind the spread of bacteremic group A Streptococcus type emm89 in Finland, 2004–2014. J Infect Dis 214:1987–1995. doi: 10.1093/infdis/jiw468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasegawa T, Hata N, Matsui H, Isaka M, Tatsuno I. 2017. Characterisation of clinically isolated Streptococcus pyogenes from balanoposthitis patients, with special emphasis on emm89 isolates. J Med Microbiol 66:511–516. doi: 10.1099/jmm.0.000460. [DOI] [PubMed] [Google Scholar]
- 9.Kimoto H, Fujii Y, Yokota Y, Taketo A. 2005. Molecular characterization of NADase-streptolysin O operon of hemolytic streptococci. Biochim Biophys Acta 1681:134–149. doi: 10.1016/j.bbaexp.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis 192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 11.Turner CE, Bedford L, Brown NM, Judge K, Torok ME, Parkhill J, Peacock SJ. 2017. Community outbreaks of group A Streptococcus revealed by genome sequencing. Sci Rep 7:8554. doi: 10.1038/s41598-017-08914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalker V, Jironkin A, Coelho J, Al-Shahib A, Platt S, Kapatai G, Daniel R, Dhami C, Laranjeira M, Chambers T, Guy R, Lamagni T, Harrison T, Chand M, Johnson AP, Underwood A, Scarlet Fever Incident Management Team. 2017. Genome analysis following a national increase in scarlet fever in England 2014. BMC Genomics 18:224. doi: 10.1186/s12864-017-3603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapatai G, Coelho J, Platt S, Chalker VJ. 2017. Whole genome sequencing of group A Streptococcus: development and evaluation of an automated pipeline for emm gene typing. PeerJ 5:e3226. doi: 10.7717/peerj.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chochua S, Metcalf BJ, Li Z, Rivers J, Mathis S, Jackson D, Gertz RE Jr, Srinivasan V, Lynfield R, Van Beneden C, McGee L, Beall B. 2017. Population and whole-genome sequence based characterization of invasive group A streptococci recovered in the United States during 2015. mBio 8:e01422-17. doi: 10.1128/mBio.01422-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kachroo P, Eraso JM, Beres SB, Olsen RJ, Zhu L, Nasser W, Bernard PE, Cantu CC, Saavedra MO, Arredondo MJ, Strope B, Do H, Kumaraswami M, Vuopio J, Grondahl-Yli-Hannuksela K, Kristinsson KG, Gottfredsson M, Pesonen M, Pensar J, Davenport ER, Clark AG, Corander J, Caugant DA, Gaini S, Magnussen MD, Kubiak SL, Nguyen HAT, Long SW, Porter AR, DeLeo FR, Musser JM. 2019. Integrated analysis of population genomics, transcriptomics and virulence provides novel insights into Streptococcus pyogenes pathogenesis. Nat Genet 51:548–559. doi: 10.1038/s41588-018-0343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores AR, Jewell BE, Fittipaldi N, Beres SB, Musser JM. 2012. Human disease isolates of serotype m4 and m22 group A Streptococcus lack genes required for hyaluronic acid capsule biosynthesis. mBio 3:e00413-12. doi: 10.1128/mBio.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. 2009. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis 9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 18.Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, LeFebvre RB, Musser JM. 2005. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis 192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- 19.Jacob KM, Spilker T, LiPuma JJ, Dawid SR, Watson ME Jr.. 2016. Complete genome sequence of emm28 type Streptococcus pyogenes MEW123, a streptomycin-resistant derivative of a clinical throat isolate suitable for investigation of pathogenesis. Genome Announc 4:e00136-16. doi: 10.1128/genomeA.00136-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam FM, Turner CE, Smith K, Wiles S, Sriskandan S. 2013. Inactivation of the CovR/S virulence regulator impairs infection in an improved murine model of Streptococcus pyogenes naso-pharyngeal infection. PLoS One 8:e61655. doi: 10.1371/journal.pone.0061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osowicki J, Azzopardi KI, McIntyre L, Rivera-Hernandez T, Ong CY, Baker C, Gillen CM, Walker MJ, Smeesters PR, Davies MR, Steer AC. 2019. A controlled human infection model of group A Streptococcus pharyngitis: which strain and why? mSphere 4:e00647-18. doi: 10.1128/mSphere.00647-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sierig G, Cywes C, Wessels MR, Ashbaugh CD. 2003. Cytotoxic effects of streptolysin O and streptolysin S enhance the virulence of poorly encapsulated group A streptococci. Infect Immun 71:446–455. doi: 10.1128/iai.71.1.446-455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynskey NN, Goulding D, Gierula M, Turner CE, Dougan G, Edwards RJ, Sriskandan S. 2013. RocA truncation underpins hyper-encapsulation, carriage longevity and transmissibility of serotype M18 group A streptococci. PLoS Pathog 9:e1003842. doi: 10.1371/journal.ppat.1003842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moses AE, Wessels MR, Zalcman K, Alberti S, Natanson-Yaron S, Menes T, Hanski E. 1997. Relative contributions of hyaluronic acid capsule and M protein to virulence in a mucoid strain of the group A Streptococcus. Infect Immun 65:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores AR, Jewell BE, Olsen RJ, Shelburne SA III, Fittipaldi N, Beres SB, Musser JM. 2014. Asymptomatic carriage of group A Streptococcus is associated with elimination of capsule production. Infect Immun 82:3958–3967. doi: 10.1128/IAI.01788-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores AR, Chase McNeil J, Shah B, Van Beneden C, Shelburne SA III. 2018. Capsule-negative emm types are an increasing cause of pediatric group A streptococcal infections at a large pediatric hospital in Texas. J Pediatric Infect Dis Soc 8:244–250. doi: 10.1093/jpids/piy053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marks LR, Mashburn-Warren L, Federle MJ, Hakansson AP. 2014. Streptococcus pyogenes biofilm growth in vitro and in vivo and its role in colonization, virulence, and genetic exchange. J Infect Dis 210:25–34. doi: 10.1093/infdis/jiu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rendueles O, de Sousa JAM, Bernheim A, Touchon M, Rocha E. 2018. Genetic exchanges are more frequent in bacteria encoding capsules. PLoS Genet 14:e1007862. doi: 10.1371/journal.pgen.1007862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashbaugh CD, Alberti S, Wessels MR. 1998. Molecular analysis of the capsule gene region of group A Streptococcus: the hasAB genes are sufficient for capsule expression. J Bacteriol 180:4955–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole JN, Aziz RK, Kuipers K, Timmer AM, Nizet V, van Sorge NM. 2012. A conserved UDP-glucose dehydrogenase encoded outside the hasABC operon contributes to capsule biogenesis in group A Streptococcus. J Bacteriol 194:6154–6161. doi: 10.1128/JB.01317-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tagini F, Aubert B, Troillet N, Pillonel T, Praz G, Crisinel PA, Prod'hom G, Asner S, Greub G. 2017. Importance of whole genome sequencing for the assessment of outbreaks in diagnostic laboratories: analysis of a case series of invasive Streptococcus pyogenes infections. Eur J Clin Microbiol Infect Dis 36:1173–1180. doi: 10.1007/s10096-017-2905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynskey NN, Turner CE, Heng LS, Sriskandan S. 2015. A truncation in the regulator RocA underlies heightened capsule expression in serotype M3 group A streptococci. Infect Immun 83:1732–1733. doi: 10.1128/IAI.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steer AC, Carapetis JR, Dale JB, Fraser JD, Good MF, Guilherme L, Moreland NJ, Mulholland EK, Schodel F, Smeesters PR. 2016. Status of research and development of vaccines for Streptococcus pyogenes. Vaccine 34:2953–2958. doi: 10.1016/j.vaccine.2016.03.073. [DOI] [PubMed] [Google Scholar]
- 34.McShan WM, Nguyen SV. 2016. The bacteriophages of Streptococcus pyogenes In Ferretti JJ, Stevens DL, Fischetti VA (ed), Streptococcus pyogenes: basic biology to clinical manifestations. University of Oklahoma Health Sciences Center, Oklahoma City, OK: https://www.ncbi.nlm.nih.gov/books/NBK333409/ [PubMed] [Google Scholar]
- 35.Johnson DR, Kurlan R, Leckman J, Kaplan EL. 2010. The human immune response to streptococcal extracellular antigens: clinical, diagnostic, and potential pathogenetic implications. Clin Infect Dis 50:481–490. doi: 10.1086/650167. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds R, Hope R, Williams L, BSAC Working Parties on Resistance Surveillance. 2008. Survey, laboratory and statistical methods for the BSAC resistance surveillance programmes. J Antimicrob Chemother 62(Suppl 2):ii15–ii28. doi: 10.1093/jac/dkn349. [DOI] [PubMed] [Google Scholar]
- 37.Pospiech A, Neumann B. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet 11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 38.Page AJ, De Silva N, Hunt M, Quail MA, Parkhill J, Harris SR, Otto TD, Keane JA. 2016. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom 2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remmington A, Turner CE. 2018. The DNases of pathogenic Lancefield streptococci. Microbiology 164:242–250. doi: 10.1099/mic.0.000612. [DOI] [PubMed] [Google Scholar]
- 47.Athey TB, Teatero S, Li A, Marchand-Austin A, Beall BW, Fittipaldi N. 2014. Deriving group A Streptococcus typing information from short-read whole-genome sequencing data. J Clin Microbiol 52:1871–1876. doi: 10.1128/JCM.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long SW, Kachroo P, Musser JM, Olsen RJ. 2017. Whole-Genome sequencing of a human clinical isolate of emm28 Streptococcus pyogenes causing necrotizing fasciitis acquired contemporaneously with Hurricane Harvey. Genome Announc 5:e01269-17. doi: 10.1128/genomeA.01269-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ibrahim J, Eisen JA, Jospin G, Coil DA, Khazen G, Tokajian S. 2016. Genome analysis of Streptococcus pyogenes associated with pharyngitis and skin infections. PLoS One 11:e0168177. doi: 10.1371/journal.pone.0168177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben Zakour NL, Venturini C, Beatson SA, Walker MJ. 2012. Analysis of a Streptococcus pyogenes puerperal sepsis cluster by use of whole-genome sequencing. J Clin Microbiol 50:2224–2228. doi: 10.1128/JCM.00675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Andrade Barboza S, Meygret A, Vincent P, Moullec S, Soriano N, Lagente V, Minet J, Kayal S, Faili A. 2015. Complete genome sequence of noninvasive Streptococcus pyogenes M/emm28 strain STAB10015, isolated from a child with perianal dermatitis in French Brittany. Genome Announc 3:e00806-15. doi: 10.1128/genomeA.00806-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Longo M, De Jode M, Plainvert C, Weckel A, Hua A, Chateau A, Glaser P, Poyart C, Fouet A. 2015. Complete Genome Sequence of Streptococcus pyogenes emm28 Clinical Isolate M28PF1, Responsible for a Puerperal Fever. Genome Announc 3. doi: 10.1128/genomeA.00750-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Athey TB, Teatero S, Sieswerda LE, Gubbay JB, Marchand-Austin A, Li A, Wasserscheid J, Dewar K, McGeer A, Williams D, Fittipaldi N. 2016. High incidence of invasive group A Streptococcus disease caused by strains of uncommon emm types in Thunder Bay, Ontario, Canada. J Clin Microbiol 54:83–92. doi: 10.1128/JCM.02201-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flores AR, Luna RA, Runge JK, Shelburne SA III, Baker CJ. 2017. Cluster of fatal group A streptococcal emm87 infections in a single family: molecular basis for invasion and transmission. J Infect Dis 215:1648–1652. doi: 10.1093/infdis/jix177. [DOI] [PubMed] [Google Scholar]
- 55.Rochefort A, Boukthir S, Moullec S, Meygret A, Adnani Y, Lavenier D, Faili A, Kayal S. 2017. Full sequencing and genomic analysis of three emm75 group A Streptococcus strains recovered in the course of an epidemiological shift in French Brittany. Genome Announc 5:e00957-17. doi: 10.1128/genomeA.00957-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of isolates per emm type in the BSAC collection. Forty-four different genotypes were identified within the collection, but 16 were represented by single isolates (grey bars). Total number of isolates was 344. Download FIG S1, TIF file, 0.2 MB (172KB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Details of BSAC isolates and antimicrobial sensitivity testing. Download Data Set S1, XLSX file, 0.1 MB (46.1KB, xlsx) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Details of all isolates with assembly statistics, capsule gene mutations, and nga-ifs-slo promoter variants. Download Data Set S2, XLSX file, 0.6 MB (638.4KB, xlsx) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NADase activity of different promoter subtypes. The activity of NADase was measured in culture supernatants of BSAC isolates representing different promoter subtypes with predicted low (black) or high (red) activity. A−27T−22C−18 subtype 1.1 promoter had low activity in emm81 isolates, consistent with previous findings of this promoter in historical emm1. G−27T−22T−18 subtype 2.1 had low activity in older emm89 isolates, also consistent with previous findings, and subtype 2.2 in emm58 and emm77 also had low activity, as predicted despite the additional base change at −40 bp. Compared to G−27T−22T−18 subtype 2.1 (older emm89), significantly higher activity was detected in emm1, with A−27G−22T−18 subtype 3.1, and in emm4 and emm87, with subtype 3.2, also supporting a null effect of the base change at −40 bp. A−27T−22T−18 subtype 4 promoter in emm78 also had significantly higher activity. Isolates with mutations in regulators covRS or rocA were excluded as they influence the expression of nga. Data represent means + standard deviations (SDs) for emm1, n = 10; emm89, n = 11; emm58, n = 3; emm77, n = 3; emm4, n = 7; emm87, n = 17; emm78, n = 5; and emm81, n = 5. Statistical comparisons were made to emm89 subtype 2.1, for which we had the highest number of representative isolates and was previously confirmed to have low activity, using Kruskal-Wallis nonparametric multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant. Download FIG S2, TIF file, 0.1 MB (156.6KB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombination within ST50 emm76. All sequence data for emm76 (n = 38) was mapped the de novo assembled sequence of BSAC_bs448 (bold). The majority of isolates were ST50 (ST; white) and within this ST were two sublineages; the lower sublineage was associated the high-activity promoter A−27G−22T−18 (P; black) and truncated HasA (HasA/B; black). Gubbins analysis (boxed region on right) of ST50 isolates identified 19 regions of recombination across the genome in all isolates (red vertical lines) belonging to the lower sublineage compared to the top sublineage. One of these regions (highlighted grey) surrounded the P-nga-ifs-slo locus conferring the high-activity-associated promoter (P; black) with residues A−27G−22T−18 to the lower sublineage compared to low-activity A−27T−22C−18 (P; grey) in the top sublineage. The presence (black) or absence (white) of mobile prophage-associated superantigens (speA, -C, -H, -I, -K, -L, -M, and ssa) and DNAses (sda, sdn, spd1, spd3, spd3v6, and spd4) as well as antimicrobial resistance genes and mutant variants of regulators CovR, CovS, and RocA was also determined for each isolate. All isolates within the lower ST50 sublineage carried a variant of the prophage-associated DNAse spd3 (spd3v6) (45) that is more divergent than other spd3 variants, including the spd3 variant carried by isolates belonging to the top ST50 sublineage. All lower sublineage isolates also carried the resistance gene ermB which was absent in other lineages, but they did not carry other antimicrobial elements found in the upper sublineage isolates. Sporadic truncated mutant variants of regulators CovR, CovS, and RocA (black) were also detected across the tree but were not associated with any specific lineages. Scale bar represents substitutions per site. Scale on boxed region represents position across the assembled BSAC_bs448 genome. Bootstrap values provided on major branches. Download FIG S3, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombination within ST63 emm77. All sequence data for emm77 (n = 82) were mapped to the de novo assembled sequence of BSAC_bs150 (bold). The majority of isolates were ST63 (ST; white), or one single locus variant ST1125, and within this ST were two sublineages: the upper lineage associated with the high-activity promoter A−27G−22T−18 (P; black) and truncated HasA (H; black). Gubbins analysis (boxed region) of ST63 isolates identified two regions of recombination across the genome of all isolates (red vertical lines) belonging to the upper sublineage compared to the lower sublineage. One of these regions (highlighted grey) surrounded the P-nga-ifs-slo locus conferring the high-activity-associated promoter with residues A−27G−22T−18 (P; black) to the upper sublineage compared to low-activity G−27T−22T−18 (P; white) in the lower sublineage. The presence (black) or absence (white) of mobile prophage-associated superantigens (speA, -C, -H, -I, -K, -L, -M, and ssa) and DNAses (sda, sdn, spd1, spd3, spd3v6, and spd4) as well as antimicrobial resistance genes and truncated mutant variants of regulators CovR, CovS, and RocA was also determined for each isolate. The prophage-associated DNase spd3 was common to all upper sublineage ST63 isolates, and all except one of this sublineage carried the antimicrobial resistance gene ermTR. Sporadic truncated mutant variants of CovR, CovS, and RocA (black) were detected across the tree but were not associated with any specific lineages. Scale bar represents substitutions per site. Scale on boxed region represents position in the BSAC_bs150 assembly. Bootstrap values provided on main branches. Download FIG S4, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombination within emm81. All sequence data for emm81 (n = 68) were mapped to the de novo assembled sequence of BSAC_bs229 (bold). The majority of isolates were ST624, with high-activity promoter A−27G−22T−18 (P; black) and truncated HasB (HasB; black). Gubbins analysis (boxed region) of ST624 isolates and closely related ST1059, ST117, ST909, and ST837, compared to BSAC_bs229, identified patterns of recombination across the genome in all isolates (red vertical lines, or blue vertical lines if unique to a single isolate). One of these regions (highlighted grey) surrounded the P-nga-ifs-slo locus conferring the high-activity-associated promoter with residues A−27G−22T−18 to the ST624/ST837 population compared to low-activity G−27T−22T−18 in all other isolates. The presence (black) or absence (white) of mobile prophage-associated superantigens (speA, -C, -H, -I, -K, -L, -M, and ssa) and DNAses (sda, sdn, spd1, spd3, spd3v6, and spd4) as well as antimicrobial resistance genes and truncated mutant variants of regulators CovR, CovS, and RocA was also determined for each isolate. The majority of all isolates carried the prophage-associated speH. Antimicrobial resistance genes were rarely detected in any ST. Scale bar represents substitutions per site. Scale on boxed region represents position in the BSAC_bs229 assembly. Bootstrap values provided on main branches. Download FIG S5, TIF file, 2.4 MB (3.3MB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombination in emm94 and emm108. (A) In the PHE-2014/2015 (red) emm94 population, the majority (n = 51) form a lineage separate from two PHE-2014/2015 isolates and the single ABCs-2015 (blue) isolate. Gubbins analysis predicted 11 regions of recombination (red lines) in all the lineage associated isolates compared to the three other isolates. One of these regions (highlighted in grey) encompassed the P-nga-ifs-slo region. (B) Isolates of emm108 from the ABCs-2015 (blue) collection were of a different MLST (ST14) than PHE-2014/15 (red) (ST1088). The hasB gene was absent in the genomes of both ABCs-2015 isolates, and one had undergone recombination surrounding the P-nga-ifs-slo locus (shaded grey), as predicted by Gubbins analysis (shown on the right). Blue lines, predicted recombination unique to a single genome. Sequence data were mapped to the reference strain H293, also used as an outgroup for SNP cluster analysis. Scale bar represents SNPs. Download FIG S6, TIF file, 0.9 MB (961.2KB, tif) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Details of emm28, emm75, emm76, emm77, emm81, emm87, emm94, and emm108 isolates used in this study. Download Data Set S3, XLSX file, 0.2 MB (178.3KB, xlsx) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reference genomes used for mapping in this study and excluded prophage regions. Download Table S1, DOCX file, 0.1 MB (12.9KB, docx) .
Copyright © 2019 Turner et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequence data have been submitted to the European Nucleotide Archive (ENA) (www.ebi.ac.uk/ena) as accession numbers ERS361826 to ERS379364, ERR1359331 to ERR485881, ERS361826 to ERS379364, and SRR5853328 to SRR5858742 (listed in Data Sets S1 and S2 in the supplemental material).