Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the etiologic agent of Kaposi’s sarcoma (KS), a vascular tumor frequently found in immunodeficient individuals. KSHV encodes 12 pre-microRNAs (pre-miRNAs), which are processed into 25 mature microRNAs (miRNAs). KSHV miRNAs maintain KSHV latency, enhance angiogenesis and dissemination of the infected cells, and interfere with the host immune system by regulating viral and cellular gene expression, ultimately contributing to KS development. In this review, we briefly introduce the biogenesis of miRNAs and then describe the recent advances in defining the roles and mechanisms of action of KSHV miRNAs in KS development.
KSHV and Its Related Malignancies
KSHV, also known as human herpesvirus 8 (HHV-8), is etiologically associated with the development of several malignancies, including KS, primary effusion lymphoma (PEL), and multicentric Castleman’s disease (MCD). Among them, KS is a tumor that frequently occurs in HIV-infected patients and immunocompromised individuals. KSHV-encoded genes, including KSHV-encoded miRNAs, contribute to the development of KS in multiple ways, including KSHV persistent infection in host cells, inhibition of the host immune system and direct manipulation of oncogenic and tumor suppressor pathways.
Similar to other herpesviruses, KSHV has two distinct phases of replication: latent and lytic replication. During the latent phase, a limited number of KSHV genes are expressed, including latency-associated nuclear antigen (LANA) encoded by ORF73, viral cyclin (vCyclin) encoded by ORF72, viral FLIP (vFLIP) encoded by ORF71, kaposin encoded by ORF-K12, and 25 mature viral miRNAs. These genes contribute to KSHV latent infection in host cells. Among them, miRNAs play major roles in KSHV latency and the development of KS. In this review, we summarize recent progress regarding the roles of KSHV miRNAs in KS development.
miRNAs and KSHV-Encoded miRNAs
The Biology of miRNAs
miRNAs are a group of noncoding small single-stranded RNAs of approximately 19 ~ 22 nucleotides (nt) in length. miRNAs usually inhibit target gene expression by binding to complementary regions, typically in the 3ʹ untranslated region (3ʹUTR) of the target gene. By binding to the 3ʹUTR of target genes, miRNAs post-transcriptionally compromise the expression of those target genes.
miRNAs are transcribed by RNA polymerase II (sometimes by Pol III) as pri-miRNAs, which are usually kilobases long and contain a stem–loop structure [1]. pri-miRNAs are then processed into ~65-nt-long hairpin pre-mRNA structures by a complex formed by the nuclear RNase III-type protein Drosha and the DiGeorge syndrome critical region gene 8 (DGCR8) proteins [1]. Pre-miRNAs are small hairpins with a ~2 nt 3′ overhang in structure which could be recognized by the nuclear export factor exportin 5 (EXP5), and transported into the cytoplasm [1].
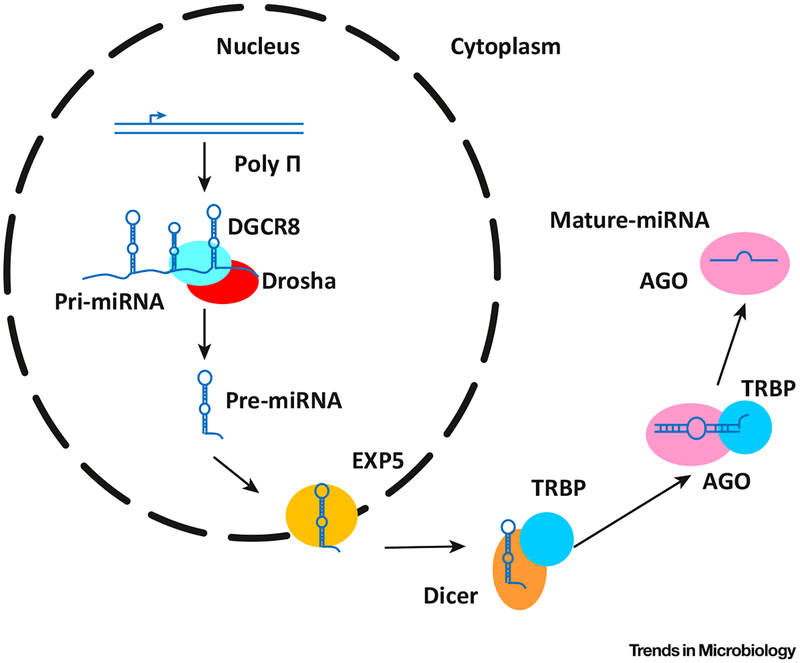
Once entering the cytoplasm, in cooperation with TRBP (TAR RNA-binding protein), another RNase III enzyme, Dicer, cleaves pre-miRNAs into ~22 nt miRNA duplexes [2,3]. The formation of Dicer and TRBP complex (RLC) triggers the binding of Argonaute 1–4 (AGO 1–4) to miRNAs, forming a RISC–miRNA complex [4]. Once released from Dicer, the stable end of the RNA duplexes is bound to TRBP in RLC while the other end interacts with the AGO protein, of which the RNA helicase activity removes the unselected strand, releasing the mature miRNA [1]. A diagram of miRNAs biogenesis is shown in Figure 1.
Figure 1. Biogenesis of miRNA and Mechanism of Function.
Pri-miRNAs are products of polymerase II (Poly II), and are usually kilobases long. Pri-miRNAs are further processed into much smaller RNAs by Drosha and transported into the cytoplasm by Exportin 5 (EXP5). In the cytoplasm, pre-miRNAs are selectively cleaved by Dicer and processed by RISC. The functions of mature miRNAs are executed by the RNA-induced silencing complex, or RISC.
The RISC–miRNA complex activates the formation of the GW body (a structure formed by multiple proteins, including GW182, which might function in miRNA binding), and binding to mRNA, initiated by RISC, triggers the enrichment of one or more heteromeric protein complexes, including GW182, on the mRNA [5]. The mRNA expression is inhibited, and at last degraded as a result of the binding of the heteromeric protein complexes [5].
During the biogenesis of miRNAs, the specificity of miRNAs is determined. The seed region (Box 1) of a mature miRNA is complementary to base pairs of target RNAs, including the coding and untranslated regions [1,6,7]. Inhibition of the target gene by an miRNA might be influenced by the translation process of the target gene.
Box 1. Seed Sequence of miRNAs.
The seed region of an miRNA is a sequence of 7–8 nt at the 5′ end of the miRNA [81]. The seed region/sequence generally defines the specificity of an miRNA, and most of the time it is completely complementary to a specific region of a target gene transcript, including both coding region and untranslated regions [82]. Target sites in the ORF and 3′UTR of a single miRNA could have synergistic effects [34,83,84]. Multiple target sites of the same miRNA in the 3′UTR could significantly enhance the targeting efficiency of the miRNA [85]. Factors influencing miRNA targeting include the structure of target region and the distance between two seed sites, etc. [86,87]. While the seed region usually lies in the 5ʹ end of a miRNA, the 3′ end of an miRNA might also have a supplementary function [82]. miRNAs post-transcriptionally regulate the expression of genes, which include the levels of the transcripts and efficiencies of translation. Recent high-throughput approaches have shown that, in most cases, the levels of both protein and transcript of target genes are decreased by the miRNAs [88,89].
An mRNA could be targeted by multiple miRNAs. For instance, KSHV miR-K12–1, 3 and 4–3p, target the 3ʹUTR of caspase 3 (Casp3) [8], Epstein–Barr virus (EBV), and KSHV-encoded miRNAs share common targets, including BCL2L11 and BACH1 [9], and miR-155 or miR-K12–11 binds to the 3′UTR of BACH-1 [10]. Besides targeting mRNAs, miRNAs can also be transported by circulating lipid vesicles, which function in cell-to-cell communication and serve as cancer biomarkers [11]. The function and biogenesis of miRNAs can be influenced by naturally occurring single-nucleotide polymorphisms (SNPs), which can affect miRNA maturation and production [12].
Cellular miRNAs are of great significance in activities of cell, likewise, KSHV miRNAs also play significant roles in the infection of KSHV and the development of KSHV-related diseases.
KSHV-Encoded miRNAs
In 2005, three groups identified a total of 11 distinct pre-miRNAs encoded by KSHV, and the next year, another group found the 12th pre-miRNA [13–16]. These 12 pre-miRNAs can further evolve into 25 mature miRNAs. The 5′-proximal regions of most KSHV miRNAs are fixed, while the 3′ end usually varies, indicating that the 5′ end is significant for miRNA–mRNA recognition [17]. A list of 12 pre-miRNAs, 25 mature miRNAs and their corresponding sequences are shown in Table 1. However, analysis of miRNA sequences showed that miR-K10–3p has two distinct 5′ ends, which might affect its function [16,18].
Table 1.
KSHV-Encoded miRNAs.a
| pre-miRNAs | Pre-miRNA stem loop sequence | Mature miRNAs | miRNA sequences |
|---|---|---|---|
| miR-K12-1 | GGGUCUACUUCGCUAACUGUAGUCCGGGUCG- AUCUGAGCCAUUGAAGCAAGCUUCCAGAUCU- UCCAGGGCUAGAGCUGCCGCGGUGACACC |
miR-K1-5p | AUUACAGGAAACU- GGGUGUAAGC |
| miR-K1-3p | GCAGCACCUGUU- UCCUGCAACC |
||
| miR-K12-2 | GGGUCUACUUCGCUAACUGUAGUCCGGGUCGA- UCUGAGCCAUUGAAGCAAGCUUCCAGAUCUUC- CAGGGCUAGAGCUGCCGCGGUGACACC |
miR-K2-5p | AACUGUAGUCCG- GGUCGAUCUG |
| miR-K2-3p | GAUCUUCCAGG- GCUAGAGCUG |
||
| miR-K12-3 | GGCUAUCACAUUCUGAGGACGGCAGCGAC- GUGUGUCUAACGUCAACGUCGCGGUCA- CAGAAUGUGACACC |
miR-K3-5p | UCACAUUCUGAG- GACGGCAGCGA |
| miR-K3-3p | UCGCGGUCACAG- AAUGUGACA |
||
| miR-K12-4 | AUAACUAGCUAAACCGCAGUACUCUAGGG- CAUUCAUUUGUUACAUAGAAUACUGAG- GCCUAGCUGAUUAU |
miR-K4-5p | AGCUAAACCGCA- GUACUCUAGG |
| miR-K4-3p | UAGAAUACUGAGG- CCUAGCUGA |
||
| miR-K12-5 | UGACCUAGGUAGUCCCUGGUGCCCUAA- GGGUCUACAUCAAGCACUUAGGAUGCCU- GGAACUUGCCGGUCA |
miR-K5-5p | AGGUAGUCCCUG- GUGCCCUAAGG |
| miR-K5-3p | UAGGAUGCCUGG- AACUUGCCGGU |
||
| miR-K12-6 | CUUGUCCAGCAGCACCUAAUCCAUCGGCGG- UCGGGCUGAUGGUUUUCGGGCUGUUGAGCGAG |
miR-K6-5p | CCAGCAGCACCU- AAUCCAUCGG |
| miR-K6-3p | UGAUGGUUUUCG- GGCUGUUGAG |
||
| miR-K12-7 | GCGUUGAGCGCCACCGGACGGGGAUUUA- UGCUGUAUCUUACUACCAUGAUCCCAUG- UUGCUGGCGCUCACGG |
miR-K7-5p | AGCGCCACCGGA- CGGGGAUUUAUG |
| miR-K7-3p | UGAUCCCAUGUU- GCUGGCGC |
||
| miR-K12-8 | CGCGCACUCCCUCACUAACGCCCCGCUU- UUGUCUGUUGGAAGCAGCUAGGCGCG- ACUGAGAGAGCACGCG |
miR-K8-5p | ACUCCCUCACUA- ACGCCCCGCU |
| miR-K8-3p | CUAGGCGCGACU- GAGAGAGCA |
||
| miR-K12-9 | GGGUCUACCCAGCUGCGUAAACCCCG- CUGCGUAAACACAGCUGGGUAUACG- CAGCUGCGUAAACCC |
miR-K9-5p | ACCCAGCUGCGU- AAACCCCGCU |
| miR-K9-3p | CUGGGUAUACGC- AGCUGCGUAA |
||
| miR-K12-10a | CUGGAGGCUUGGGGCGAUACCACCACUC- GUUUGUCUGUUGGCGAUUAGUGUUGU- CCCCCCGAGUGGCCAG |
miR-K10a-5p | GGCUUGGGGCGA- UACCACCACU |
| miR-K10a-3p | UAGUGUUGUCCCC- CCGAGUGGC |
||
| miR-K12-10b | CUGGAGGCUUGGGGCGAUACCACCACU- CGUUUGUCUGUUGGCGAUUGGUGUU- GUCCCCCCGAGUGGCCAG |
miR-K10b-3p | UGGUGUUGUCC- CCCCGAGUGGC |
| miR-K12-11 | CGCUUUGGUCACAGCUUAAACAUUUC- UAGGGCGGUGUUAUGAUCCUUAA- UGCUUAGCCUGUGUCCGAUGCG |
miR-K11-5p | GGUCACAGCUUAA- ACAUUUCUAGG |
| miR-K11-3p | UUAAUGCUUAGCC- UGUGUCCGA |
||
| miR-K12-12 | GAUGGCCGGCACGCGGUGUCAACCAG- GCCACCAUUCCUCUCCGCAUUAAAGC- ACUCGGUGGGGGAGGGUGCCCUGGU- UGACACAAUGUGCCGCGCAUC |
miR-K12-5p | AACCAGGCCACCA- UUCCUCUCCG |
| miR-K12-3p | UGGGGGAGGGU- GCCCUGGUUGA |
Sequences were obtained from miRBase (http://www.mirbase.org/index.shtml).
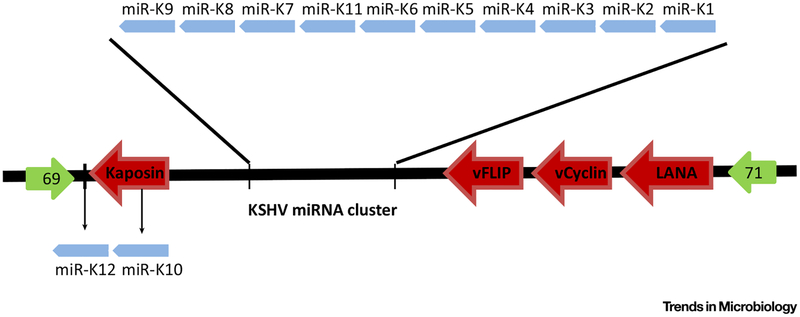
KSHV miRNAs are clustered together in the viral genome located in the latent locus. Of the 12 pre-miRNAs, 10 are located in the sequence between kaposin and ORF71, while miR-K10 is located within the ORF of kaposin and miR-K12 is mapped to the 3′UTR of kaposin [19]. A map of the relative position of KSHV miRNAs and latent genes is shown in Figure 2. Due to the compact structure of the viral genome, one ORF might localize in the 3′UTR of another gene. In the KSHV genome, ORF57/MTA is included in the 3′UTR of ORF56, and translation of ORF57/MTA might result in dispelling or compromising maximal miRNA recognition and regulation [7].
Figure 2. Map of the Relative Positions of Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) miRNAs and Viral Latent Genes in the KSHV Genome.
KSHV miRNAs are located in the latent cluster of the KSHV genome. Most of the KSHV miRNAs are clustered together while miR-K12 and −K10 lie in the 3ʹUTR and ORF of kaposin, respectively.
All known miRNAs are expressed during the viral latent phase. miRNAs deriving from pre-miR-K10 and pre-miR-K12 are further induced during the viral lytic phase [16,20]. In KSHV latently infected cells, as many as 2200 copies of an individual miRNA could be expressed in a single cell [15]. Expression of KSHV miRNAs is conservative among different cells examined. KSHV miRNAs are conservative in primary effusion lymphoma cell lines and in tissues from patients with KS or MCD [21–23]. Analysis of samples from KS patients revealed that some of KSHV miRNAs were significantly upregulated during lytic replication, and these results were consistent in studies done by two independent groups [24,25]. While all the KSHV miRNAs are expressed during viral latency, their activities and expression levels vary. For instance, in BC-1 and BCBL-1 cells, levels of miR-K5 are of great disparity, and levels of individual miRNAs differ from one another [26]. High-throughput sequencing indicates that miRNA-K9 is absent in BC-3 cells [17].
The expression of specific KSHV miRNAs is highly regulated and varies with different phases of the viral life cycle. A study by Happel et al. revealed a unique way of regulating the biogenesis by KSHV miRNAs [27]. During de novo infection of KSHV, expression of MCP-1-induced protein-1 (MCPIP1) is repressed, and knockdown of MCPIP1 in KSHV-infected cells induces the expression of IL-6 and KSHV miRNA [27]. Meanwhile, KSHV miRNA-K4–5p, 6–3p, and 10a inhibit the expression of MCPIP1 mRNA, while miRNA-K1, 6–3p, and 7 could bind to MCPIP1 3ʹUTR, and knockdown of these three miRNAs results in increased expression of MCPIP1 [27].
This study illustrates a novel strategy by which KSHV interacts with the host environment, which tightly regulates the expression of KSHV miRNAs.
KSHV miRNAs can be encapsidated into virions. In virions produced with Vero/Bac36, TRExBCBL-1-Rta, and BCBL-1 cells, at least 4 of the 25 miRNAs are detected [28]. These encapsidated miRNAs can be transported into host cells after KSHV reactivation, functioning in cell-to-cell communication [28]. Chugh et al. detected viral miRNAs in patient exosomes, and treatment of hTERT-(human umbilical vein endothelial cells(HUVECs) with patient-derived exosomes enhanced cell migration [29].
Similar to mammalian cells, KSHV miRNAs play important roles in the infection of KSHV and the development of KSHV-related diseases.
Methods for Identifying the Targets of miRNAs
To investigate the function of KSHV miRNAs in KSHV-induced malignancies, multiple methods were developed to predict and validate the targets of these miRNAs.
By bioinformatics analysis, mRNA sequences with complementary bases to the seed region of mature miRNAs have been identified [30]. Databases, including Targetscan, miRanda, DIANA Tool, and PicTar, can be used to predict target sites of miRNAs. However this approach may present with false-positive results, and further validation – such as by luciferase-report assays with the complementary sequences in the 3ʹUTR cloned in the reporters – is required to confirm the binding of miRNAs to their target regions.
High-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP or PAR-CLIP) is a method for delineating the genome-wide miRNA targets [18]. By using this method, Gottwein et al. revealed more than 2000 targets of KSHV miRNAs, and 58% of these mRNAs are also targeted by EBV miRNAs [18]. Gallaher et al. identified multiple human genes that are repressed at the protein level, but not at the mRNA level by using stable isotope labeling of amino acids in cell culture (SILAC) coupled with tandem mass spectrometry and microarray analysis [31]. Several other methods, such as cDNA microarray and protein profiling technologies, have been applied in miRNA research [32].
In studying the function of KSHV miRNAs, these methods are introduced to investigate the targets of KSHV miRNAs.
As miRNAs function through the RISC complex, an approach by combining RISC immunoprecipitation, microarray analysis, and dual luciferase-report assays identified NHP2L1, GEMIN8, LRRC8D, and CDK5RAP1 as targets of KSHV miRNAs [33]. The targeted regions of both LRRC8D and CDK5RAP1 are located in the 3ʹUTRs, while those of NHP2L1 and GEMIN8 are located in the coding region [24]. The biological consequences of these regulatory mechanisms remain to be explored.
Combining bioinformatics analysis, HITS-CLIP and PAR-CLIP, and luciferase-reporter assays, many targets of KSHV miRNAs have been identified and confirmed [34,35]. KSHV miRNAs can also target other regions in addition to the 3ʹUTR of an mRNA. This mechanism of targeting has also been identified for host miRNAs [36,37]. By mutagenesis analysis and luciferase-report assays, it was found that miR-K3 targets ORF31–33 transcripts via two binding sequences within the ORF33 coding region; miR-K10a-3p and miR-K10b-3p target the ORF71–72 transcripts by binding to the 5ʹ-distal ORFs and intergenic regions [38].
While different KSHV miRNAs might have the same targets or different targets in the same pathways [39], the gene regulatory networks of a specific miRNA in different cell lines usually have little overlap [40]. Ectopic expression of miR-K11 in BJAB (B-cell origin) and TIVE (vascular endothelial origin) resulted in differential regulation of gene expression networks [40]. Among the thousands of genes dysregulated by miR-K11 in BJAB and TIVE, only around 120 genes are common in these two cell lines [40]. Hence, KSHV miRNAs might function differently in different types of cell. A list of confirmed targets of KSHV miRNAs if given in Table 2.
Table 2.
KSHV miRNAs and Validated Targets.
| KSHV miRNAs | Targets of miRNAs | Functions | Refs |
|---|---|---|---|
| miR-K12-1 | Casp3 | Apoptosis | [8] |
| NF-κB signaling | KSHV latency | [41] | |
| THBS1 | Cell adhesion, migration, and angiogenesis | [32] | |
| p21 | Cell cycle arrest | [43] | |
| STAT3 | IκBα/NF-κB/IL-6 signaling pathway | [68] | |
| miR-K12-3 | Casp3 | Apoptosis | [8] |
| nuclear factor I/B | KSHV latency | [45,47,48] | |
| GRK2 | KSHV latency and angiogenesis, dissemination | [43,48] | |
| THBS1 | Cell adhesion, migration, and angiogenesis | [32] | |
| C/EBPβ p20 (LIP) | Influence the secretion of IL-8 and −10 and immune response | [60] | |
| miR-K12-4 | Casp3 | Apoptosis | [8] |
| Rbl2 | KSHV latency | [6] | |
| miR-K12-5 | BCLAF1 | KSHV latency | [49] |
| MYD88 | Regulating the TLR/IL-1R signaling cascade | [54] | |
| Tmskα1 | Apoptosis and angiogenesis | [57] | |
| miR-K12-6 | THBS1 | Cell adhesion, migration, and angiogenesis | [32] |
| Bcr | Angiogenesis | [58] | |
| SH3BGR | Angiogenesis and dissemination | [59] | |
| MAF | Influence the differentiation of infected cells | [69] | |
| miR-K12-7 | RTA | KSHV latency | [6,44,45] |
| MICB | Escape NK cell killing | [53] | |
| C/EBPβ p20 (LIP) | Influence the secretion of IL-8 & −10 and immune response | [60] | |
| miR-K12-9 | RTA | KSHV latency | [6,44,45] |
| BCLAF1 | KSHV latency | [49] | |
| IRAKI | Regulating the TLR/IL-1R signaling cascade | [54] | |
| miR-K12-10a | BCLAF1 | KSHV latency | [49] |
| TWEAK | Apoptosis | [56] | |
| TGF-β | KSHV infection and apoptosis | [71] | |
| miR-K12-10b | BCLAF1 | KSHV latency | [49] |
| TGF-β | KSHV infection and apoptosis | [71] | |
| miR-K12-11 | MYB | KSHV latency | [46] |
| C/EBPβ | Influences IL-6 mRNA expression | [51] | |
| Jarid2 | Specific function has not been studied. | [39] | |
| IKKε | Immune escape and KSHV latency | [51] | |
| THBS1 | Cell adhesion, migration, and angiogenesis | [32] | |
| BACH-1 | Cell sensitivity of KSHV infection | [63] | |
| MAF | Influence the differentiation of infected cells | [69] | |
| BMP | Facilitates viral infection and proliferation | [70] |
In order to uncover the functions of KSHV miRNAs in the context of viral infection, a reverse genetic system was used to generate mutants by knocking-out KSHV miRNAs in the viral genome initially with an entire cluster of 10 pre-miRNAs [41], then with individual miRNAs [42]. As of now, KSHV mutants with a knockout of individual or several miRNAs have been generated [42]. By using this method, the functions of KSHV miRNAs in cellular transformation and tumorigenesis have been defined [43]. Among the KSHV miRNAs, miR-K1 dysregulates the cell cycle and cell survival by inhibiting IκBα to activate the NF-κB pathway [43].
In summary, similar to cellular miRNAs, KSHV miRNAs function in a seed sequence recognition mode. Many methods and tools have been used to uncover the functions of KSHV miRNAs.
KSHV miRNAs Contribute to KS Development via Complex Mechanisms
KSHV miRNAs are of great importance in the tumorigenesis of KS. KSHV miRNAs regulate the expression of both viral and cellular genes. During the last decade, multiple targets of KSHV miRNAs have been found. By inhibiting the expression of target genes, KSHV miRNAs regulate the life cycle of KSHV, manipulate the host immune response, and contribute to the KSHV-induced angiogenesis and dissemination of KS.
KSHV miRNAs and the KSHV Life Cycle
KSHV miRNAs play significant roles in regulating the KSHV life cycle either by targeting key viral replication genes or cellular genes that regulate KSHV replication.
By interfering with the lytic switch protein, RTA, KSHV miRNAs help KSHV to maintain latent infection. Repression of RTA by miR-K9* (now known as miR-K9–5p) and miR-K7 has been reported in different cell systems [6,44,45]. Besides directly targeting viral genes, KSHV miRNAs also regulate the KSHV life cycle by targeting cellular genes, which, in turn, regulate viral genes. A luciferase-report assay revealed that, by targeting cellular transcription factor MYB, miR-K11 indirectly restrains the expression of RTA to maintain KSHV latency [46]. Through targeting nuclear factor I/B, an activator of RTA, miR-K3 inhibits RTA expression, contributing to KSHV latency [46,47]. MiR-K4–5p has a sequence targeting retinoblastoma (Rb)-like protein 2 (Rbl2), a suppressor of DNA methyl transferase 3a and 3b mRNA transcription [6]. By suppressing RbI2, the expression of DNMT1 was elevated, resulting in the methylation of the RTA promoter and maintenance of latent infection in host cells [6]. According to our recent study, overexpression of miR-K3 also inhibits lytic replication by targeting GRK2, which subsequently regulates the CXCR2/AKT pathway [48]. KSHV miR-K1 activates the NF-κB pathway by targeting IκBa, which results in the inhibition of lytic replication [41]. Tandem array-based expression identified Bcl-2-associated factor (BCLAF1) as the target of three KSHV miRNAs: miR-K5, −K9, and −K10a/b [49]. Knockdown of BCLAF1 resulted in increased viral lytic production [49].
MiR-K11 shares 100% seed sequence homology with hsa-miR-155, a human oncomiR [50]. By mimicking the function of host miRNAs, miR-K11 enhances the proliferation of CD19+ B-cells in the spleen in a mouse model [51]. In this model, miR-K11 downregulates the expression of the basic region/leucine zipper motif transcription factor C/EBPβ, a regulator of interleukin-6, at the transcription level. Another group has also confirmed B-cell expansion in a transgenic mouse expressing miR-K11 [39]. In this study, besides C/EBPβ, Jarid2 is also used as a common target of KSHV miR-K11 and hsa-miR-155 [39]. Thus, miR-K11 modulates B-cells to facilitate KSHV latent infection, contributing to KSHV-induced malignancies.
By interfering with KSHV life cycle, KSHV miRNAs contribute to the development of KSHV-induced malignancies.
KSHV miRNAs and Host Immunity
KSHV miRNAs contribute to KHSV latent infection in host cells by interfering with immune surveillance as well. KSHV miR-K11 attenuates type I Interferon (IFN) signaling, the primary response in antiviral immunity, by targeting I-kappa-B kinase epsilon (IKKε), an activator of KSHV lytic production [52]. MiR-K7 directly binds to the 3ʹUTR of MICB, the stress-induced natural killer (NK) cell ligand; inhibition of MICB leads to reduced attack by NK cells [53]. By adopting these strategies, KSHV escapes from host immune responses and maintains viral latency, contributing to the development of KS.
KS lesions, including all subtypes of KS, present with excessive inflammatory cytokines. KSHV miRNAs also affect the secretion of inflammatory cytokines to facilitate KS development. Interleukin-1 receptor (IL-1R)-associated kinase (IRAK1) and myeloid differentiation primary response protein 88 (MYD88), components of the Toll-like receptor (TLR)/IL-1R signaling cascade, are the target of miR-K9 and K5, respectively [54]. In miR-K5 or K9 transfected and IL-1α stimulated HUVECs, expression of IL-6 and IL-8 mRNAs, and secretion of IL-6 and IL-8 in cell culture, are significantly reduced [54,55]. The function of proinflammation is not always clear. What is peculiar is that inhibition of the tumor necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) receptor by miR-K10a suppresses an immune response, leading to a reduction in the production of IL-8 and MCP-1 in primary HUVECs [56]. In conclusion, KSHV miRNAs target key genes and their signaling pathways, and manipulate host immune systems, hence contributing to the development of KS.
KSHV miRNAs and KS Dissemination and Angiogenesis
KS tumors often manifest abnormal angiogenesis and dissemination. KSHV miRNAs play important roles in KS angiogenesis and dissemination. In 293 and BJAB cells stably expressing KSHV miRNAs, thrombospondin 1 (THBS1), an antiangiogenic and antiproliferative protein, was significantly decreased [32]. Multiple miRNAs are involved in regulating THBS1, including miR-K1, miR-K3–3p, miR-K6–3p, and miR-K11 [32]. By inhibiting THBS1 expression, these miRNAs contribute to angiogenesis and proliferation of KSHV-infected cells.
Tumor suppressor protein tropomyosin 1 (TPM1) has multiple isoforms, including Tmskα1, TM3, and TM5. MiR-K5 represses Tmskα1 expression by targeting its 3ʹUTR; whereas miR-K2 may target TM3 outside the 3ʹUTR, both of which elevated the angiogenic activities of HUVECs [57]. By repressing the expression of the breakpoint cluster region (Bcr) protein, miR-K6–5p increased the tubulogenesis of HUVECs in vitro [58]. MiR-K6–3p inhibits SH3 domain-binding glutamate-rich protein (SH3BGR) by binding to the 3ʹUTR, which further activates the STAT3 pathway [59]. As a result, miR-K6–3p promotes cell migration and invasion, contributing to KS dissemination and angiogenesis [59].
MiR-K3 and −K7 both regulate C/EBPβ expression. By binding to the 3ʹUTR of C/EBPβ, these two miRNAs preferentially suppress the expression of C/EBPβ p20 (LIP), an isoform of C/EBPβ, which leads to the inhibition of secretion of IL-8 and IL-10 by macrophages, and immune response against infection [60].
Besides C/EBPβ, G-protein-coupled receptor kinase 2 (GRK2) is also a direct target of miR-K3. Inhibition of GRK2 expression resulted in increased expression of the chemokine receptor CXCR2, which induces angiogenesis, and promotes cell migration and invasion as well [48,61]. Meanwhile, inhibition of GRK2 expression by miR-K3 activates AKT signaling, which promotes cell migration and invasion [48,61].
Overall, by dysregulating key pathways and mRNAs, KSHV miRNAs contribute to KS development by inducing angiogenesis and dissemination.
Other Functions of KSHV miRNAs in KS Tumorigenesis
Despite their contribution in KS dissemination and angiogenesis, KSHV miRNAs also promote KS development by manipulating cell cycle arrest, cell survival, and cellular transformation. In a KSHV-induced cellular transformation model of primary cells, a KSHV mutant with a deletion of a cluster of 10 pre-miRNAs failed to transform primary cells. The miRNAs are essential for maintaining the homeostasis of the KSHV-infected cells by preventing cell cycle arrest and apoptosis by redundantly targeting multiple cancer-related pathways [43]. In this system, compared to the uninfected primary cells, the KSHV-transformed cells are hyperproliferative without depending on glucose [62]. Together with vFLIP, the cluster of KSHV miRNAs reprograms the cellular metabolic pathways to support cellular proliferation and transformation [62].
KSHV uses xCT as a coreceptor for entry into the cells [63]. By suppressing BACH-1, miR-K11 induces the expression of xCT, which renders macrophage and endothelial cells permissive to KSHV infection [63]. miR-K11, miR-K1 and −K9 also have a similar effect on macrophage and endothelial cells; however, the mechanisms of miR-K1 and −K9 regulating xCT remains unclear [64]. Overexpression of KSHV miRNAs promotes the secretion of reactive nitrogen species (RNS) by macrophages, which then conversely facilitates KSHV infection of macrophages [63].
Following KSHV infection, the cell cycle of the host cells is deregulated by KSHV-encoded genes [65]. KSHV miR-K1 participates in regulating the cell cycle by directly targeting the 3ʹUTR of p21, a key inducer of cell cycle arrest [66]. Inhibition of p21, as well as IκBα, by miR-K1 contributes to the homeostasis and survival of KSHV-transformed cells [43]. KSHV miR-K1, 3 and 4–3p target the 3ʹUTR of caspase 3 (Casp3), and inhibit Casp3-induced cell apoptosis [8]. Besides interfering with the the immune response, miR-K10a regulated TWEAK receptor downregulation also inhibits apoptosis [56]. In a most recent study, Liu et al. found that, by suppressing the expression of growth arrest DNA damage-inducible gene 45 beta (GADD45B), miRNA-K9 protects KSHV-infected cells from cell cycle arrest and apoptosis [67].
In addition to p21, miR-K1 also regulates the NF-κB and STAT3 pathways by targeting the IκBα/NF-κB/IL-6 signaling pathway, contributing to KSHV-related tumorigenesis [68]. KSHV miRNAs, especially miR-K11 and −K6, suppress the expression of the lymphatic endothelial cell (LEC)-specific transcription factor MAF, inhibiting LECs from converting to blood vessel endothelial cells and contributing to KSHV-induced oncogenesis [69]. The infamous miR-K11 targets the bone morphogenetic protein (BMP)-responsive transcriptional factor SMAD5, which then abates transforming growth factor β (TGF-β) signaling, and promotes KSHV viral infection and tumorigenesis [70]. KSHV miR-K10a and −10b promote cell survival and malignant cell transformation by targeting transforming growth factor β (TGF-β), a regulator for cell apoptosis [71]. miR-K10 also contributes to KSHV-induced cellular transformation. Ectopic expression of miR-K10 results in enhanced activity of cellular transformation by depressing several inhibitors of oncogenic transformation. These activities mimic those of its cellular ortholog miR-142–3p [72]. Numerous other KSHV miRNAs also mimic the functions of cellular miRNAs: 5′ nts 3 to 10 of miR-K6–5p are homologous to hsa-miR-15a/16 [73], miR-K3 and its variant miR-K3 + 1 (a variant with an additional adenosine (A) at the 5′ end) imitates miR-23 and targets miR-23’s targets, which favors KSHV infection [74].
KSHV miRNAs regulate cellular metabolism of infected cells, not individually, but as a whole cluster [75]. The KSHV miRNA cluster induces the Warburg effect by inhibiting EGLN2 and HSPA9, which are important for cell growth in a low-oxygen environment and latency maintenance [75]. Regulation of the metabolism of KSHV-infected cells is achieved through two independent pathways, activation of HIF1α and reduction of mitochondrial biogenesis [75]. However, in the model of KSHV-induced cellular transformation, KSHV suppresses aerobic glycolysis, which is essential for cell survival under nutrient deprivation conditions, which is commonly seen in the tumor microenvironment [62]. KSHV miRNAs contribute to the suppression of the Warburg effect by activation of the NF-κB pathway to suppress the glucose transporters 1 and 3 (GLUT1 and 3). As a result, the expression levels of GLUT1 and 3 are significantly lowered in KSHV-infected cells compared to the adjacent uninfected cells in KS tumors. Thus, manipulation and fine-tuning of metabolic pathways is essential for the proliferation and survival of KS cells [62].
The regulation network of KSHV miRNAs is complex. By regulating the activity of KSHV, or KSHV-infected cells, these unique miRNAs play significant roles in KSHV infection and the development of KSHV-induced malignancies. By studying the mechanism of KSHV miRNAs, more effective methods of treating KSHV-induced malignancies might be developed.
Concluding Remarks and Future Perspectives
Many functions of KSHV miRNAs have been uncovered in the past 10 years; however, more efforts are needed to fully de ne their functions in KSHV infection and KSHV-induced malignancies (see Outstanding Questions). Figure 3 (Key Figure) illustrates our current understanding of the functions of KSHV miRNAs. However, most studies have so far used artificial overexpression of individual KSHV miRNAs and the luciferase-report assay to identify miRNA binding sites, which do not reflect the physiological condition of cells and tissues in which KSHV miRNAs function. In investigating the mechanisms of KSHV miRNAs, studies so far show distinct regulation networks in different cell types and tissues. The lack of animal models is another problem encountered in studying KSHV miRNAs, which limits our efforts in exploring the functions of these miRNAs in vivo. Thus, targets and mechanisms identified in these studies may not reflect the situation in vivo. Future studies should address the issues of expression of individual miRNAs at a single cell level (copy number), cell type specificity of the targets, and the dynamic changes of expression levels of miRNAs in different phases of the KSHV life cycle.
Outstanding Questions.
Multiple KSHV microRNAs manipulate the KSHV life cycle, studies so far focus on the function of a single KSHV microRNA. How do these microRNAs work together as a whole, and does any one of these microRNAs play a leading role?
Do KSHV microRNAs regulate other non-coding RNA and contribute to the development of KS?
Could targets of KSHV miRNAs be used for developing vaccines or drugs?
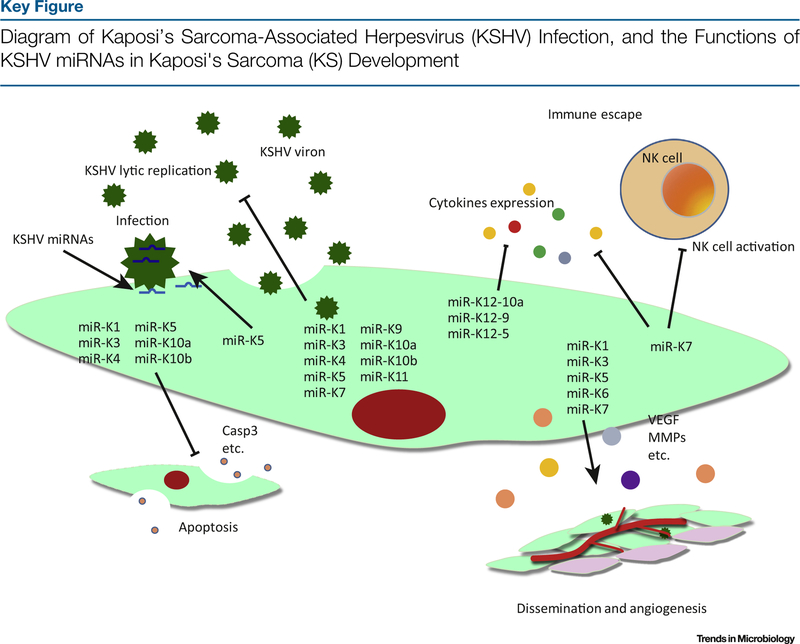
Figure 3.
KSHV contributes to KS development in multiple ways. In this diagram, we show the functions of KSHV miRNAs in KSHV infectivity, immune response, and tumor angiogenesis and dissemination. KSHV lytic replication is regulated by multiple KSHV miRNAs. miRNAs are encapsidated in the virions, which can be released into newly infected cells. miRNAs also regulate the expression of multiple cytokines, contributing to immune escape, cell proliferation, and tumor angiogenesis and dissemination.
With the recent development of models of KSHV-induced cellular transformation and tumorigenesis [76,77] combining the use of reverse genetics [78,79], defining the roles and identifying the mechanisms of action of KSHV miRNAs in KSHV infection and KSHV-associated malignancies in the context of KSHV infection has become possible. By using this approach, the functions of individual miRNAs can be studied in the context of KSHV infection. The recent development of the humanized mouse model also provides a new tool for defining the roles of KSHV miRNAs in KSHV persistent infection [80]. It is expected that novel therapeutic targets and prognostic markers would be identified through these efforts.
Trends.
miRNAs play significant roles in different diseases. By binding to target genes, miRNAs post-transcriptionally regulate gene expression. During viral infections, miRNAs manipulate the activities of viruses and host cells. Some viral miRNAs mimic cellular miRNAs, and interfere with cellular activities.
KSHV encodes 25 mature miRNAs and all play essential roles in the viral life cycle and cellular activities. Recent studies have focused on the roles of KSHV miRNAs in KSHV induced-tumorigenesis and KS development. Identification of the targets, and delineation of the functions of KSHV miRNAs, could provide novel strategies for treatment and prevention of KSHV-associated malignancies.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81361120387, 81371824, 81171552, 31270199, 81401662, and 81571984), grants from NIH (R01CA213275, R01CA177377 and R01CA132637), the PhD Programs Foundation of the Ministry of Education of China (20123234110006), and the Natural Science Youth Foundation of Jiangsu Province (BK20140908).
Footnotes
Supplemental Information
Supplemental information associated with this article can be found online at http://dx.doi.org/10.1016/j.tim.2017.02.002.
References
- 1.Kim VN (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 2.Ketting RF (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C elegans. Genes Dev. 15, 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutvagner G (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 [DOI] [PubMed] [Google Scholar]
- 4.Haase AD (2005) TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6, 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakymiw A (2007) The role of GW/P-bodies in RNA processing and silencing. J. Cell. Sci 120, 1317–1323 [DOI] [PubMed] [Google Scholar]
- 6.Lu F (2010) Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol 84, 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin HR and Ganem D (2011) Viral microRNA target allows insight into the role of translation in governing microRNA target accessibility. Proc. Natl. Acad. Sci. U. S. A 108, 5148–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suffert G (2011) Kaposi’s sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog. 7, e1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramalingam D (2012) Emerging themes from EBV and KSHV microRNA targets. Viruses 4, 1687–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skalsky RL (2007) Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol 81, 12836–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y (2015) Current state of circulating microRNAs as cancer biomarkers. Clin. Chem 61, 1138–1155 [DOI] [PubMed] [Google Scholar]
- 12.Sun G (2009) SNPs in human miRNA genes affect biogenesis and function. RNA 15, 1640–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundhoff A (2006) A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12, 733–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeffer S (2005) Identification of microRNAs of the herpesvirus family. Nat. Methods 2, 269–276 [DOI] [PubMed] [Google Scholar]
- 15.Cai X (2005) Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A 102, 5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samols MA (2005) Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J. Virol 79, 9301–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umbach JL and Cullen BR (2010) In-depth analysis of Kaposi’s sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol 84, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottwein E (2011) Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10, 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottwein E (2006) Expression and function of microRNAs encoded by Kaposi’s sarcoma-associated herpesvirus. Cold Spring Harbor Symp. Quant. Biol 71, 357–364 [DOI] [PubMed] [Google Scholar]
- 20.Lin YT (2010) Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA 16, 1540–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall V et al. (2007) Conservation of virally encoded micro-RNAs in Kaposi sarcoma–associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J. Infect. Dis 195, 645–659 [DOI] [PubMed] [Google Scholar]
- 22.Sullivan CS (2007) High conservation of Kaposi sarcoma-associated herpesvirus microRNAs implies important function. J. Infect. Dis 195, 618–620 [DOI] [PubMed] [Google Scholar]
- 23.Marshall V et al. (2010) Kaposi sarcoma (KS)-associated herpesvirus microRNA sequence analysis and KS risk in a European AIDS–KS case control study. J. Infect. Dis 202, 1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catrina AM (2014) MicroRNA expression profiles in Kaposi’s sarcoma. Pathol. Oncol. Res 20, 153–159 [DOI] [PubMed] [Google Scholar]
- 25.Wu X (2015) The expression profiles of microRNAs in Kaposi’s sarcoma. Tumor Biol. 36, 437–446 [DOI] [PubMed] [Google Scholar]
- 26.Gottwein E (2006) A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol 80, 5321–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Happel C (2016) Virus-mediated alterations in miRNA factors and degradation of viral miRNAs by MCPIP1. PLoS Biol. 14, e2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X (2012) MicroRNAs and unusual small RNAs discovered in Kaposi’s sarcoma-associated herpesvirus virions. J. Virol 86, 12717–12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chugh PE (2013) Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog. 9, e1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimson A (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallaher AM (2013) Proteomic screening of human targets of viral microRNAs reveals functions associated with immune evasion and angiogenesis. PLoS Pathog. 9, e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samols MA (2007) Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3, e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolken L (2010) Systematic analysis of viral and cellular micro-RNA targets in cells latently infected with human gamma-herpes-viruses by RISC immunoprecipitation assay. Cell Host Microbe 7, 324–334 [DOI] [PubMed] [Google Scholar]
- 34.Haecker I and Renne R (2014) HITS-CLIP and PAR-CLIP advance viral miRNA targetome analysis. Crit. Rev. Eukar. Gene 24, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quan L (2015) Identification of target genes regulated by KSHV miRNAs in KSHV-infected lymphoma cells. Pathol. Oncol. Res 21, 875–880 [DOI] [PubMed] [Google Scholar]
- 36.Kang JG (2011) Kaposi’s sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J. Virol 85, 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang JG (2011) Kaposi’s sarcoma-associated herpesviral IL-6 and human IL-6 open reading frames contain miRNA binding sites and are subject to cellular miRNA regulation. J. Pathol 225, 378–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai Z (2013) Genomewide mapping and screening of Kaposi’s sarcoma-associated herpesvirus (KSHV) 3’ untranslated regions identify bicistronic and polycistronic viral transcripts as frequent targets of KSHV microRNAs. J. Virol 88, 377–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahlke C (2012) A microRNA encoded by Kaposi sarcoma-associated herpesvirus promotes B-cell expansion in vivo. PLoS One 7, e49435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y (2014) A systems biology approach identified different regulatory networks targeted by KSHV miR-K12–11 in B cells and endothelial cells. BMC Genomics 15, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei X et al. (2010) Regulation of NF-κB inhibitor IκBα and viral replication by a KSHV microRNA. Nat. Cell Biol. 12, 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain V (2016) A toolbox for herpesvirus miRNA research: construction of a complete set of KSHV miRNa deletion mutants. Viruses 8, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moody R (2013) KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways. PLoS Pathog. 9, e1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellare P and Ganem D (2009) Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6, 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin X (2011) miR-K12-7-5p encoded by Kaposi’s sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS One 6, e16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plaisance-Bonstaff K (2014) KSHV miRNAs decrease expression of lytic genes in latently infected PEL and endothelial cells by targeting host transcription factors. Viruses 6, 4005–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu C (2010) MicroRNAs encoded by Kaposi’s sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. 11, 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W (2016) A KSHV microRNA enhances viral latency and induces angiogenesis by targeting GRK2 to activate the CXCR2/AKT pathway. Oncotarget 7, 32286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziegelbauer JM (2008) Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet 41, 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gottwein E (2007) A viral microRNA functions as an orthologue of cellular miR-155. Nature 450, 1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boss IW (2011) A Kaposi’s sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2R null mice. J. Virol 85, 9877–9886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lan DLYG (2011) A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKε. Cell Res. 21, 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nachmani D (2009) Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5, 376–385 [DOI] [PubMed] [Google Scholar]
- 54.Abend JR (2012) Kaposi’s sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the Toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J. Virol 86, 11663–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tudor S (2014) Cellular and Kaposi’s sarcoma-associated herpes virus microRNAs in sepsis and surgical trauma. Cell Death Dis. 5, e1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abend JR (2010) Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi’s sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J. Virol 84, 12139–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kieffer-Kwon P (2015) KSHV MicroRNAs repress tropomyosin 1 and increase anchorage-independent growth and endothelial tube formation. PLoS One 10, e135560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramalingam D (2015) Kaposi’s sarcoma-associated herpesvirus microRNAs repress breakpoint cluster region protein expression, enhance rac1 activity, and increase in vitro angiogenesis. J. Virol 89, 4249–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W (2016) The SH3BGR/STAT3 pathway regulates cell migration and angiogenesis induced by a Gammaherpesvirus microRNA. PLoS Pathog. 12, e1005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin Z (2010) Kaposi’s sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J. Leuk. Biol 87, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu M et al. (2015) A KSHV microRNA directly targets G protein-coupled receptor kinase 2 to promote the migration and invasion of endothelial cells by inducing CXCR2 and activating AKT signaling. PLoS Pathog. 11, e1005171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Y (2016) An oncogenic virus promotes cell survival and cellular transformation by suppressing glycolysis. PLoS Pathog. 12, e1005648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qin Z (2010) Upregulation of xCT by KSHV-encoded microRNAs facilitates KSHV dissemination and persistence in an environment of oxidative stress. PLoS Pathog. 6, e1000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaleeba JA and Berger EA (2006) Kaposi’s sarcoma-associated herpesvirus fusion-entry receptor: cystine transporter xCT. Science 311, 1921–1924 [DOI] [PubMed] [Google Scholar]
- 65.Wei F (2016) Cell cycle regulatory functions of the KSHV oncoprotein LANA. Front. Microbiol 7, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gottwein E and Cullen BR (2010) A human herpesvirus micro-RNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J. Virol 84, 5229–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu X (2016) KSHV microRNAs target GADD45B to protect infected cells from cell cycle arrest and apoptosis. J. Virol 91, e02045–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen M (2016) Kaposi’s sarcoma herpesvirus (KSHV) micro-RNA K12–1 functions as an oncogene by activating NF-κB/IL-6/STAT3 signaling. Oncotarget 7, 33363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen A (2010) KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 24, 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y (2012) Kaposi’s sarcoma-associated herpesvirus-encoded microRNA miR-K12–11 attenuates transforming growth factor beta signaling through suppression of SMAD5. J. Virol 86, 1372–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lei X et al. (2012) A Kaposi’s sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor β pathway to promote cell survival. J. Virol 86, 11698–11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forte E (2015) MicroRNA-mediated transformation by the Kaposi’s sarcoma-associated herpesvirus kaposin locus. J. Virol 89, 2333–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Skalsky RL (2007) Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol 81, 12836–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manzano M (2013) Kaposi’s sarcoma-associated herpesvirus encodes a mimic of cellular miR-23. J. Virol 87, 11821–11830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yogev O (2014) Kaposi’s sarcoma herpesvirus microRNAs induce metabolic transformation of infected cells. PLoS Pathog. 10, e1004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones T (2012) Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J. Clin. Invest 122, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee MS et al. (2016) Human mesenchymal stem cells of diverse origins support persistent infection with Kaposi’s sarcoma-associated herpesvirus and manifest distinct angiogenic, invasive, and transforming phenotypes. mBio 7, e2109–e2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou FC (2002) Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol 76, 6185–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brulois KF (2012) Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol 86, 9708–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang LX (2014) Humanized-BLT mouse model of Kaposi’s sarcoma-associated herpesvirus infection. Proc. Natl. Acad. Sci. U. S. A 111, 3146–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doench JG (2004) Specificity of microRNA target selection in translational repression. Genes Dev. 18, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stromberg AJ (2010) Individual microRNAs (miRNAs) display distinct mRNA targeting ‘rules’. RNA Biol. 7, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Selbach M (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 84.Baek D (2008) The impact of microRNAs on protein output. Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fang Z and Rajewsky N (2011) The impact of miRNA target sites in coding sequences and in 3’UTRs. PLoS One 6, e18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Long D (2007) Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol 14, 287–294 [DOI] [PubMed] [Google Scholar]
- 87.Saetrom P (2007) Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 35, 2333–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo H (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baek D (2008) The impact of microRNAs on protein output. Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]