Abstract
The endoplasmic reticulum (ER) is a continuous endomembrane system comprising the nuclear envelope, ribosome-studded sheets, dense peripheral matrices, and an extensive polygonal network of interconnected tubules. In addition to performing numerous critical cellular functions, the ER makes extensive contacts with other organelles, including endosomes and lysosomes. The molecular and functional characterization of these contacts has advanced significantly over the past several years. These contacts participate in key functions such as cholesterol transfer, endosome tubule fission, and Ca2+ exchange. Disruption of key proteins at the contact sites can result in often severe diseases, particularly those affecting the nervous system.
Keywords: lysosome, endosome, spasticity, paraplegia, genetics
1. Introduction
The endoplasmic reticulum (ER) is the largest continuous membrane organelle in the eukaryotic cell. It functions as a biosynthetic factory in cells [1, 2], responsible for lipid and protein synthesis and modification [3–5] as well as for protein quality control [6–8] and Ca2+ storage and signaling [9–11]. The ER is a highly-interconnected and dynamic organelle, comprising the double membrane of the nuclear envelope, stacked ribosome-studded sheets, and interconnected tubules extending to the cell periphery. The ER undergoes rapid structural changes, with constant growth, retraction, fusion, and rearrangement of its membrane network [12–16].
Structural and morphological changes of the ER have important functional implications, since the ER membrane dynamically contacts essentially every other cellular organelle – for instance endosomes, mitochondria, plasma membrane, and Golgi apparatus -- at specific membrane contact sites (MCSs) [17–20]. MCSs enable inter-organelle communication in a non-vesicular manner to facilitate the exchange of lipids and small molecules between organelles, which is crucial for maintaining cell homeostasis. MCSs are regions of close membrane apposition (10–30 nm) in which organelles are tethered by molecular bridges to one another, often reversibly, but not fused. These inter-organelle tethers are formed by a variety of molecular complexes composed of both integral membrane-associated and peripherally-associated proteins as well as specific lipids [21, 22]. MCSs between the ER and endocytic organelles are particularly abundant, and crosstalk between the ER and components of the endolysosomal pathway is important for coordinating the molecular activities among the different compartments -- including cholesterol transfer [23–28], endosome fission [29–31], endosome positioning [25, 32, 33], dephosphorylation of endosome membrane receptors [34–36], and Ca2+ exchange [37]. The coordinate regulation of lipid metabolism occurs between ER, which is the primary site for lipid synthesis; lipid droplets, an ER-derived organelle responsible for lipid storage and transport; endosomes, which are responsible for regulating cholesterol homeostasis; and lysosomes, which are responsible for lipid hydrolysis and recycling.
MCSs between the ER and endo-lysosomal organelles serve as functionally important microdomains that mediate distinct biological activities depending on the composition of tether/non-tether proteins and lipids present in the contacts. Defects in interactions among ER and endo-lysosomal compartments are associated with a variety of diseases including neurodegenerative Niemann-Pick diseases and hereditary spastic paraplegias (HSPs) [38, 39]. Here, we review the current understanding of how ER regulates endo-lysosomal pathways, the molecular composition and cellular function of ER-endo/lysosomal MCSs, and how changes in ER structure can compromise this interaction and contribute to disease states.
2. Structure and Functions of the Endoplasmic Reticulum
Each eukaryotic cell contains a continuous ER membrane network of interconnected sheets and tubules that extends throughout the cell and occupies a large fraction of the total cellular volume [14, 40, 41]. The continuity of the ER lumen has been experimentally demonstrated through fluorescence microscopy studies that injected lipophilic fluorescent dyes into live cells co-stained with ER markers [40, 42] and using GFP (green fluorescent protein) targeted to the ER and analyzed using FLIP (fluorescence loss in photobleaching) [43, 44]. Fragmentation of ER membranes, which has been reported during the cell cycle and fertilization, is rapidly counteracted by homotypic fusion [45, 46].
The structural complexity of the ER is only beginning to be full appreciated with the advent of super-resolution imaging techniques. These emerging approaches have revealed that the ER is comprised of heterogenous but interconnected membrane compartments, with highly complex architectures [16, 18, 47] that form distinct functional domains [1, 2]. These domains permit the ER to separate structurally and functionally its myriad biological activities, including protein and lipid synthesis, Ca2+ storage and signaling, vesicular transport, mitochondrial and endosomal fission, protein modification, protein quality control, lipid droplet/peroxisome/autophagosome biogenesis, and communication and regulation of other cellular organelles at MCSs – all while remaining continuous. The extensive ER membrane network comprises the nuclear envelope [48], helicoidal stacks of sheets in the perinuclear region [49], and a more peripheral network of numerous densely packed tubular structures (ER matrices), occasional cisternal structures, and an extensive polygonal network of interconnected tubules [16, 50–52] ranging from about 25–100 nm in diameter that extends to the cell boundary.
The ER in neurons is organized similarly to ER in non-neuronal cells and forms a continuous network of tubules and cisternae that extends promiscuously throughout all neuronal compartments, including into narrow regions such as the neck of dendritic spines and thin axonal terminals [51, 53–55]. The ER in dendrites is mostly tubular, with occasional small cisternae, while a single, continuous smooth ER tubule is observed in thin axonal segments [18, 51]. Functionally, stacked ER sheets as revealed by early electron micrographs were often studded with ribosomes and termed “rough ER,” while “smooth” tubules were shown to contain very few ribosomes [56, 57]. Later studies have largely confirmed that these juxtanuclear ER sheets are primary sites of protein translation, modification, and quality control, while ER tubules are principal sites for the synthesis and distribution of sterols and lipids, biogenesis of lipid droplets, and formation of MCSs with other organelles.
Shaping and maintaining these diverse ER morphologies requires numerous proteins and interactions with the cytoskeleton network. Wedge-shaped proteins containing intramembrane hydrophobic hairpin domains comprising members of the reticulon and REEP/DP1/Yop1p protein families are responsible for maintaining the high membrane curvatures found in ER tubules and at the edges of ER sheets and nuclear pores [58–60]. Atlastin/RHD3/Sey1p are membrane-bound GTPases that mediate membrane fusion to form three-way junctions between tubules, giving rise to the polygonal network so characteristic of the peripheral ER [61, 62]. Additionally, other tubular ER proteins containing hydrophobic transmembrane hairpin domains such the M1 isoform of the AAA ATPase spastin and REEPs interact with and remodel the microtubule cytoskeleton, which also helps to sculpt the tubular ER network [63]. A different set of integral membrane proteins including Climp63, p180, and kinectin regulate the shape of ER sheet-like structures through luminal spacing, cisternal stacking, and cytoskeletal interactions [14]. Overexpression or depletion of these proteins affects ER morphology by influencing the ratio of ER tubules to sheet-like structures [61] as well as their cellular distributions, and many of these proteins when mutated are associated with disease states, particularly affecting the nervous system [38].
In animal cells, the ER is intimately associated with the microtubule cytoskeleton [64–67], and this association is important for supporting the architecture of the ER network; as described above, several of the ER-shaping proteins harbor microtubule-binding domains. Although the microtubule cytoskeleton is not required for ER network formation in vitro, it is required to generate the characteristic shapes and dynamic movement seen in cells [58]. Depolymerization of microtubules using nocodazole results in retraction of the ER network and coalescing of ER membranes into sheet-like structures [67]. Nocodazole treatment also decreases the overall number of MCSs between ER and various other organelles, indicating that microtubules also play a vital role in establishing and maintaining inter-organelle contacts [20, 65, 68]. ER tubules can also be formed by sliding along microtubules via the action of motor proteins or else by attaching to polymerizing microtubules [64, 69]. Finally, ER tubules bound to other organelles such as mitochondria or endosomes can “hitch” a ride as these organelles are being transported along microtubules [65, 70].
3. Endo-lysosomal Pathway
Endo-lysosomal organelles are components of the endocytic pathway, which is responsible for vesicular trafficking of cell surface cargoes (molecules, ligands, receptors) that are internalized from the plasma membrane [71–74]. Endocytic vesicles that form at the plasma membrane fuse with early endosomes and subsequently undergo a series of maturation steps as they traffic towards the cell center along microtubules, deciding the fate of the internalized cargoes. The early endosome maturation process involves acidification of the endosomal lumen, differential recruitment and activation of Rab family GTPases, and conversion of key phosphatidylinositol phospholipids via lipid kinases and phosphatases to generate sorting, recycling, and late endosomes as well as lysosomes [72, 75–78]. Cargo sorting initially occurs on Rab5-positive early endosomes; from there, cargoes are either sorted to the recycling pathways by trafficking back to the plasma membrane via Rab4-positive vesicles or Rab11-positive late recycling endosomes, sent to the trans-Golgi network (TGN) via the retrograde pathways, or else directed to lysosomes for degradation [79]. Cargoes destined for the lysosomal degradation pathway are retained in sorting endosomes that eventually mature into Rab7-positive late endosomes -- also known as multi-vesicular bodies (MVBs) -- that sequester the cargo into intraluminal vesicles (ILVs). Fusion of MVBs with lysosomes containing acidic hydrolases results in degradation of internalized cargoes.
Lysosomes are an important catabolic organelle of eukaryotic cells, containing a diverse repertoire of acidic hydrolases (luminal pH of 4.5–5.0) that can digest macromolecules such as lipids and proteins and even entire organelles. In mammals, there is a single lysosomal acid lipase (LAL) [80] that functions at acidic pH and mediates the breakdown of lipids and lipoproteins. This and other lysosomal enzymes are synthesized in the rough ER and imported from the Golgi apparatus via small vesicles that fuse with acidic (pH 5.5) late endosomes. Enzymes destined for lysosomes are specifically tagged with mannose 6-phosphate. Prior to the fusion of late endosomes with lysosomes, receptors for lysosomal hydrolases are sent back to the TGN via retrograde transport of tubular endosomal protrusions that bud off through endosomal fission. Tubular protrusions are generated on both early and late endosomes; certain cargoes destined for recycling, such as transferrin receptors (TfnRs) or mannose 6-phosphate receptors (M6PRs) for transport back to the TGN, accumulate in the narrow tubules, while those targeted for lysosomal degradation remain in the larger vacuolar portion [74, 81].
Cargo sorting to the recycling pathways and away from the lysosomal degradation pathway is primarily mediated by the evolutionarily-conserved retromer complex. Endosomal tubule formation is mediated by proteins that contain banana-shaped BAR (Bin-Amphiphysin-Rvs) domains -- such as sorting nexins (SNXs) -- that sense and induce membrane curvature [82, 83]. Cargo sorting into the endosomal protrusions involve the cargo-sorting retromer complex and the WASH (Wiskott-Aldrich syndrome protein and SCAR homologue) complex, which activates Arp2/3 to induces actin nucleation [84–86]. Recently, the retromer-independent cargo sorting machinery retriever has also been identified [87]. The pulling and pushing forces generated by the actin and microtubule cytoskeletons are also necessary to pull out the budding membranes [88]. Mutations in genes that disrupt lysosomal enzyme trafficking result in the accumulation of specific cargo substrates that cannot be degraded by lysosomes and give rise to several human genetic disorders, collectively termed lysosomal storage diseases (LDS) [89, 90]. For instance, LAL deficiency causes Wolman disease, a severe LSD characterized by the buildup of lipids in various organs, including the brain. Mutations in several genes of the endosomal sorting pathway are linked to inherited neurological disorders [38, 91]; VPS35, a subunit of the retromer complex causes autosomal dominant, late on-set Parkinson’s disease [92, 93], while mutations in the WASH complex component strumpellin/WASHC5 is associated with a type of HSP, SPG8 [94, 95].
Although these intracellular transport and sorting pathways are already complex, they are also highly regulated. Key components that mediate the sorting and maturation of endocytic organelles have been identified over the last several decades. These include endosomal sorting complexes required for transport (ESCRT) proteins (composed of four different protein complexes, ESCRTs −0, -I, -II and -III) that mediate cargo sorting into MVBs for lysosomal degradation [96], Rab GTPases involved in endosome fusion [76, 77, 97], motor proteins that regulate the transport of endosomes along microtubules [98–100], and specific MCSs between endosomes and the ER [101, 102].
4. Regulation of ER MCSs with Endo-lysosomal Organelles
The ER directly contacts endo-lysosomal organelles and regulates certain processes occurring on endo-lysosomal membranes [23, 101]. These highly-abundant MCSs generate microdomains between endosomes and the ER that exclude ribosomes and are typically associated with microtubules [23, 103]. Endosomal contacts with the ER also increase as the endosome matures; an estimated 50% of early endosomes, but >99% of late endosomes, are in contact with the ER [103]. Electron microscopy tomography studies reveal that the ER contacts ~5% of the endosome surface area through the formation of multiple contact sites on a single endosome. Furthermore, as endosomes are trafficked along microtubules via molecular motors, they pull ER tubules with them so that the two organelles remained attached, indicating that the organelles are tightly associated. MCSs are mediated by a variety of tethering complexes comprising protein-protein and/or protein-lipid interactions. Several interacting proteins have been identified that mediate the tethering between ER and endosomes, and many of these have a role in cholesterol exchange and endosome positioning (Table 1).
Table 1:
ER-endosome interacting proteins
| ER Protein | Endo-lysosomal proteins | Function | Reference |
|---|---|---|---|
| VAPA | ORP1L (late endosome) | Cholesterol sensing, endosome positioning | [25, 27] |
| VAPA/B | STARD3 / STARD3NL (late endosome) | Cholesterol sensing, endosome positioning | [23, 24] |
| VAPA/B | SNX2 (early and late endosome) | Budding dynamics and lipid modifications | [119] |
| Protrudin & VAPA | Rab7 & PtdIns(3)P (late endosome) | Endosome positioning | [33] |
| VPS13C | ? | ER-endosome tethering and lipid transport | [118] |
| S100A11 | Annexin A1 (early and late endosomes) | Receptor dephosphorylation and ILV formation | [26] |
| ORP5 | NPC1 (late endosome) | Cholesterol transfer | [28] |
| PTP1B | EGFR and other RTKs, G-CSFR and ESCRT-0 proteins HRS and STAM2 (early and late endosome/MVB) | Receptor dephosphorylation | [26, 34–36, 123] |
| M1 Spastin | IST1 (early and late endosome/MVB) | Endosome fission | [30, 39] |
| TMCC1 | Coronin 1C (early and late endosomes) | Endosome fission | [31] |
| M0SPD2 | STARD3 (late endosome) | ER-endosome tethering | [120] |
| M0SPD2 | STARD3NL (late endosome) | ER-endosome tethering | [120] |
| M0SPD2 | ORP1L (late endosome) | ER-endosome tethering | [120] |
By electron microscopy, electron-dense structures are often visible between the narrow gaps of opposing membranes of the ER and other organelles [104], and recently some of the responsible organelle-specific tethering proteins have been identified. Integral membrane ER proteins known as vesicle-associated membrane protein (VAMP)-Associated Proteins (VAPs; VAPA and VAPB in animals) are implicated in tethering ER with different types of organelles. ER-mitochondria MCSs are mediated by VAPB and PTP1P51, and this interaction has been demonstrated to regulate Ca2+ uptake by mitochondria from the ER [105, 106]. Importantly, missense mutation in VAPB (P56S) causes the fatal neurodegenerative disease amyotrophic lateral sclerosis type 8 (ALS8) [107]. Although transport of proteins between organelles requires vesicle trafficking, lipids can be transferred at MCSs [108, 109]. VAPs can also regulate lipid trafficking at several of these MCSs; the N-terminal major sperm protein (MSP) domain of VAPs interacts with the FFAT [two phenylalanines (FF) in an acidic tract] motif of several lipid-binding proteins found on the apposing membranes of other organelles, including those of the TGN: ceramide transferase 1 (CERT) [108], Nir2 [109], and oxysterol-binding protein (OSBP) [110]; and endosomes: ORP1L (OSBP-related protein 1L) [25, 111, 112], STARD3 (steroidogenic acute regulatory protein-related lipid transfer domain protein 3), and STARD3NL (STARD3 N-terminal like protein) [23, 24].
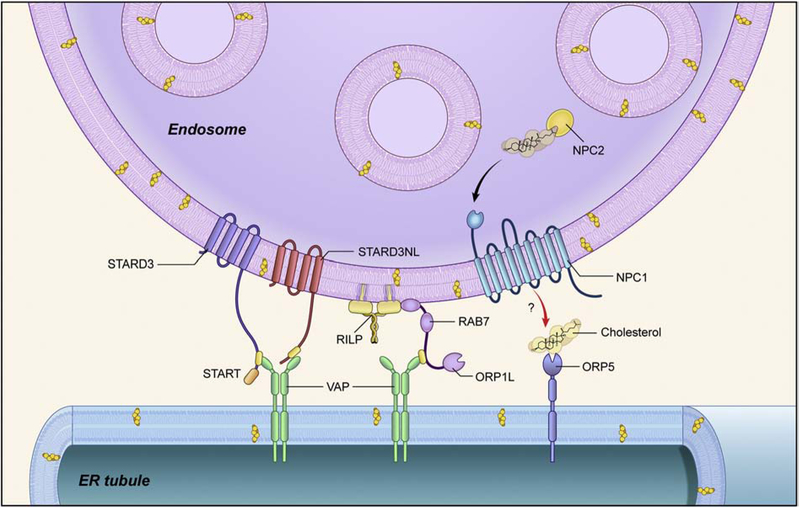
ORP1L is a cholesterol-binding protein that interacts with Rab7 and localizes to late endosomes containing the cholesterol transporter, Niemann-Pick Type C protein NPC1 (Figure 1). The ORP1L-VAPA interaction is dependent on low cholesterol conditions, resulting in ORP1L undergoing a conformational change that allows ORP1L’s FFAT domain to interact with VAPA [25]. Overexpression of an ORP1L mutant in which the cholesterol-binding domain is removed to mimic cholesterol-depleted conditions results in increased VAPA interactions and a consequent increase in both the number and size of ER-endosome MCSs. Two other endosomal proteins -- STARD3 and STARD3NL -- also contain FFAT domains that can interact with VAPA and similar results are seen upon overexpression of STARD3 or STARD3NL [23]. However, depletion of VAPA, ORP1L, or STARD3 does not significantly decrease the number of basal ER-endosome MCSs, suggesting that other protein-tethering complexes may be involved, or else that several tethering complexes work together to mediate these interactions.
Figure 1. ER tubule-endosome contact site.
Key proteins involved in functional ER-endosome contact sites are depicted schematically.
VAPA also interacts with the FFAT motif of protrudin, another integral membrane ER protein. Protrudin contains a FYVE domain and forms contact sites with late endosomes by interacting with Rab7 and phosphatidylinositol 3-phosphate (PI3P) phospholipids [113–116]. Expression of GFP-tagged protrudin showed extensive colocalization with LAMP1-positive late endosomes/lysosomes, while deletion of the FYVE domain prevented protrudin localization to late endosomes/lysosomes. By electron microscopy, extensive MCSs could be seen between the ER and late endosomes when protrudin and a GTPase-defective Rab7(Q67L) mutant were co-expressed [33]. VAPB interacts with VPS13C, a protein associated with early onset autosomal-recessive Parkinson’s disease [117], and localizes to contact sites between the ER and late endosomes/lysosomes and to lipid droplets [118]. VPS13C contains an N-terminal FFAT motif and a C-terminal domain that binds to late endosome/lysosomes, although the binding partner is not yet known. Based on structural studies, VSP13C contains a hydrophobic cavity to accommodate several lipid molecules, and in vitro assays have confirmed the ability of VSP13C to transfer fluorescently-labeled phospholipids between membranes [118], suggesting that VSP13C is a lipid transport protein between ER and endosomes.
VAPA and VAPB have also been shown to interact with the endosomal retromer protein SNX2 [119]. Immunofluorescence studies showed that SNX2 overexpression caused the normally diffusely-distributed VAPA and VAPB proteins to accumulate at ER focal points adjacent to SNX2 positive endosomes. Double-knockout of VAPA and VAPB had a major effect on phosphatidylinositol 4-phosphate (PI4P) accumulation on endosomes, suggesting that loss of VAP proteins disrupts the contact sites that mediate PI4P degradation via the ER-localized PI4P-phosphatase Sac1. Additionally, VAPA/B double-knockout cells showed accumulation of TGN proteins on endosomes, suggesting that sorting and fission mechanisms are also disrupted, possibly due to loss of ER-endosome MCSs that normally regulate these processes [119].
Recently, the VAP-related protein MOSPD2 (motile sperm domain-containing protein 2) was shown to function as an ER-tethering protein and to bind various proteins on other organelles containing a FFAT motif [120]. MOSPD2 is an ER-anchored-protein that interacts with endosomal proteins STARD3, STARD3NL, and ORP1L to form contact sites. Depletion of MOSPD2 resulted in significant loss of ER-endosome contacts, while depletion of VAPA and VAPB did not [120], suggesting that MOSPD2 may represent a more bona fide tethering protein for linking ER with endosomal compartments. However, MOSPD2 depletion also resulted in a significant increase in endosome-endosome contacts. This, it remains possible that changes to overall endosome organization contributed to the observed decrease in ER-endosome MCSs rather than just loss of tethering activity.
Other ER proteins have also been shown to interact with endosomal proteins, including ORP5 (oxysterol-binding protein-related protein 5), an ER transmembrane protein related to ORP1L; it binds the cholesterol transporter NPC1 on late endosomes [28]. Also, the ER protein PTP1B (protein tyrosine phosphatase 1B) interacts with endocytosed epidermal growth factor receptors (EGFRs) and other receptor tyrosine kinases [34, 36, 121, 122], granulocyte colony-stimulating factor receptors (G-CSFRs) [35], and the ESCRT-0 components HRS and STAM2 [34, 123]. Depletion of PTP1B or lack of stimulation by EGF did not decrease the number of basal contact sites formed, but expression of a substrate-trapping mutant of PTP1B (D181A) did cause formation of extended contacts between the ER and EGFR-containing endosomes [34, 36]. The interactions among PTP1B and various endosomal proteins are likely short-lived and transient and therefore are poorly suited as membrane tethers. However, it has been shown that PTP1B-EGFR interactions first requires the tethering activity of annexin A1, an endosomal protein that forms a Ca2+-dependent heterotetramer with ER-localized S100A11 [124]. Depletion of annexin A1 or S100A11 disrupts MCSs between ER and EGFR-containing endocytic compartments, but other endocytic compartments were not affected [26].
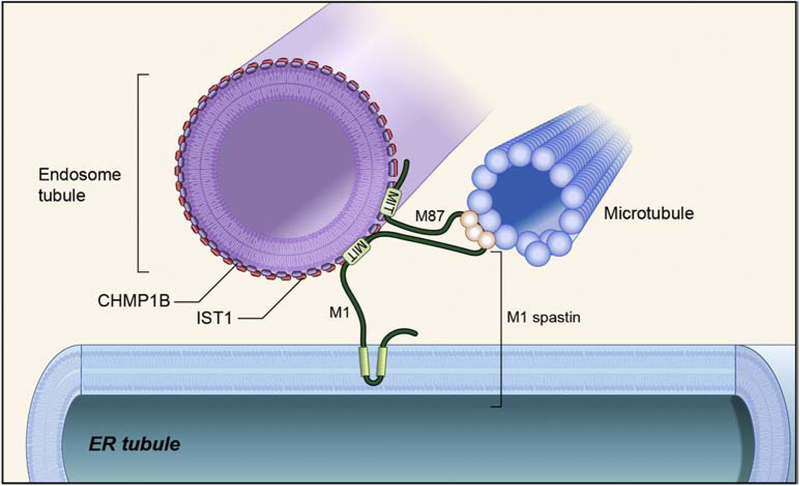
The membrane-bound ER-shaping protein M1 spastin (there is also a shorter, soluble form known as M87), a microtubule-severing AAA ATPase, interacts with the endosomal ESCRT-III component IST1 (increased sodium tolerance 1), and this interaction is required for efficient endosomal tubule fission [30, 39] (Figure 2). However, depleting spastin increases ER-endosomal membrane contacts. Although several interactions between endosome proteins and ER proteins have been identified and proposed to mediate ER-endosome membrane contacts, no single tethering complex has been identified that is necessary and sufficient for this process.
Figure 2. ER-endosome contacts mediated by M1 spastin.
The M1 spastin AAA ATPase localizes to ER and interacts with microtubules via its microtubule-binding domain and with endosome via interaction of its MIT domain with IST1 and CHMP1B on endosomes. See also Ref. 30.
It remains unclear which of these tethering interactions are important in initiating MCS formation, and which are formed after the initial contacts are made to further stabilize tethering between ER and endosomal organelles. For example, depletion of spastin results in increased ER-endosome contacts, supporting the notion that M1 spastin-IST1 interactions do not create MCSs. It is also possible that the tethering complexes described above could mediate different types of ER-endosome MCSs for different functional exchanges. Alternatively, these complexes may form in order to mediate certain ER-endosome exchanges after tethering by yet to be identified proteins. For instance, despite the extensive role of VAPs in tethering ER to various organelles, cells depleted of VAPs remain viable, and MCSs between ER and endosomal compartments continue to persist [119]. One study also found that depletion of VAPA and VAPB using siRNAs resulted in no change to the number of ER contacts with MVBs containing EGFR, while MCSs with non-EGFR containing MVBs and lysosomes were reduced by 50% [26], supporting the notion that different tethering complexes are involved in different types of endosome-ER MCSs. Moreover, although VAP proteins have been identified as prominent tethers of the ER with various other organelles, they have many different binding partners and over a hundred proteins contain FFAT motifs [125]. Thus, it remains unclear what regulates the spatial and temporal formation of specific VAP contacts.
The recent finding that MOSPD2 is also a tethering protein of the ER, contains a FFAT motif-binding MSP domain, and interacts with many of the same binding partners as VAPs suggests that the formation of contact sites relies on redundant and conserved mechanisms. Furthermore, it is interesting to note that Rab7 is involved in both protrudin and ORP1L ER-endosome MCSs and that PTP1B-EGFR interactions occur on both early (Rab5-positive) and well as late (Rab7-positive) endosomes, suggesting that Rabs may be involved in the regulation of contact site formation. Although the molecular composition of ER contacts with the endocytic pathways remains poorly understood, it is likely that different tethering complexes are involved in different types of endosome-ER MCSs.
5. Cholesterol Transfer between Late Endosomes and ER
Cellular membranes in mammals are composed of phospholipids and sterols, and the ER is the main compartment for membrane lipid biogenesis. Further lipid modifications occur in the Golgi, mitochondria, and peroxisomes. Although the ER is the major site of lipid synthesis, the endocytic pathway plays a key role in cholesterol trafficking and many of the ER-endosome interacting proteins contain cholesterol-binding domains. In concert with de novo cholesterol synthesis, cells also acquire cholesterol though receptor mediated endocytosis of LDLs (low-density lipoproteins) synthesized by the liver. After internalization, LDLs dissociate from their receptors and are hydrolyzed by lipases in the lumen of late endosomes to release free cholesterol. Cholesterol is then transferred to the luminal endosomal protein NPC2 and incorporated into the membranes of late endosomes by the membrane-bound NPC1 protein (Figure 1).
It is estimated that ~30% of LDLs from endocytic compartments reach the ER via MCSs [126, 127]. ER-localized ORP5 contains a cytosolic cholesterol-binding domain, interacts with NPC1, and regulates cholesterol transfer from late endosome membranes to the ER [28, 128]. The depletion of ORP5 causes accumulation of LDLs on the limiting membranes of late endocytic compartments, while NPC1-depletion or depletion of both ORP5 and NPC1 results in luminal accumulation of cholesterol, indicating that NPC1 functions upstream of ORP5 to traffic cholesterol from late endosomes to the ER [28]. When cholesterol is low in endosomal membranes, ORP1L undergoes a conformational change to interact with VAPA and MOSPD2. Deletion of the late endosome cholesterol-binding protein ORP1L also results in accumulation of cholesterol in endo-lysosomal compartments, indicating a defect in endosomal cholesterol export, and disruption of cholesterol homeostasis in the ER [27, 129]. ORP1L has a N-terminal pleckstrin homology domain that binds phosphatidylinositol polyphosphates [130] and is both a cholesterol sensor and a cholesterol transporter; the sterol, PI4P, and the VAP-binding domains of ORP1L are required for cholesterol transfer activity from late endosome to ER to occur [27, 129]. ORP1L has also been shown to be a bidirectional transporter of cholesterol and can transport cholesterol from ER to endosomes. The ORP1L-VAPA dependent transport of ER-derived cholesterol to late endosomes/MVBs is necessary to support the EGF-stimulated formation of ILVs when endosomal cholesterol levels are low, in a population of ER-endosomes that are mediated by annexin A1 MCSs [26].
STARD3 is an endosome protein that contain a transmembrane MENTAL domain and a cholesterol-binding START (steroidogenic acute regulatory-related lipid transfer) domain. The START domain of STARD3 can bind cholesterol and transfer it between membranes. Mechanistically, the START domain contains a hydrophobic cavity that can harbor one lipid molecule and transport it across organelle membranes. The function for STARD3NL is not as well defined, since it contains a MENTAL domain but not a lipid-binding START domain. On the other hand, STARD3NL interacts with STARD3 and cholesterol and forms MCSs with the ER [23, 131]. In cells overexpressing STARD3, late endosomes accumulate a high level of fluorescently-labeled probes for cholesterol, and the ability to transfer cholesterol to late endosomes depends on both formation of ER-endosome contacts (via the FFAT motif) and cholesterol-transferring START domain [24]. When cells were grown in lipoprotein-deficient serum-containing media, the cholesterol accumulation phenotype persisted in cells overexpressing STARD3, suggesting that the source of cholesterol was from the ER. Therefore, STARD3 has the ability to transfer ER cholesterol towards endosomes. Cholesterol transfer by STARD3 to late endosomes also affects the formation of ILVs, with overexpression of STARD3 triggering more ILVs; depletion of STARD3 or VAPs results in fewer ILVs [24]. Many of these FFAT-containing proteins can bind VAPA and interact with membrane lipids in trans, thus mediating organelle tethering and lipid transfer between organelles (Figure 1). For instance, knockout of VAP proteins results in accumulation of PI4P phospholipids on endosomes due to impaired lipid transport between endosomes and the ER [119].
The ER converts excess cholesterol to sterol esters through conjugation with fatty acyl-CoA in a reaction catalyzed by acyl-CoA:cholesterol acyltransferase. Esterified cholesterol and other neutral lipids are stored in lipid droplets, which are lipid storage organelles that bud from the outer leaflet of the tubular ER bilayer [132]. Lipid droplets interact with other organelles including endosomes for lipid exchange and lysosomes via lipophagy for catabolic breakdown of cholesterol esters and other lipids to free fatty acids [133].
6. Other Types of Functional Crosstalk Among ER and Endo-Lysosomal Organelles
Additional functions of ER MCSs with endosomes include endosome fission, endosome positioning, dephosphorylation of endosome membrane receptors by a phosphatase residing on the ER membrane, and Ca2+ exchange. Formation of tubular protrusion on early or late endosomes partition certain cargo such as TfnRs or M6PRs away from the lysosomal degradation pathway. The positioning and timing of endosomal tubule fission is mediated by ER-endosome contacts. Although the exact molecular composition of these MCSs is not known, ER tubules have been well-documented to cross perpendicular to a site on tubular endosomes immediately prior to endosome fission [29, 39], analogous to the mitochondrial fission process [134]. Furthermore, the ER was shown to colocalize with FAM21/WASHC2, a component of the actin-regulatory WASH complex, after endosomal tubular protrusions have formed, indicating that the ER induces fusion after cargo sorting. Cells depleted of strumpellin, another WASH complex component, results in impaired endosomal tubule fission.
The ER-shaping protein M1 spastin has also been implicated in endosomal tubule fission [30, 39]. Loss of spastin results in impaired endosomal tubule fission, defects in M6PR sorting, and consequently defects in lysosomal enzyme trafficking and abnormal lysosomal morphology. Both M1 spastin and its smaller isoform M87 spastin interact with the endosomal ESCRT-III protein IST1; M1 spastin-IST1 interaction may function in mediating ER-endosome contacts important for ER-induced endosomal tubule fission. Cells lacking IST1 produced a similar effect in inhibiting endosome tubule fission. Spastin mutants that cannot bind IST1, cannot bind microtubules, or cannot hydrolyze ATP to sever microtubules were not able to function in endosome tubule fission, suggesting that microtubule severing mediated by spastin is important for this process.
More recently, a screen using BioID proximity-based labeling system found that the ER membrane protein TMCC1 was enriched at ER-endosome MCSs, and that depletion of TMCC1 impaired both endosomal tubule fission and cargo sorting to the TGN [31]. Moreover, the recruitment of TMCC1-labeled ER tubules was dependent on the endosome-associated actin regulator coronin 1C, further supporting a model positing that endosomal cargo sorting components could recruit the ER to endosomes thus temporally regulating the timing of endosomal tubule fission after sorting domains have been properly established. Disruption of ER morphology by overexpression of the tubular ER-shaping protein reticulon 4a (Rtn4a) [29] inhibited endosome tubule fission, and knockout of the ER-shaping protein REEP1 produced lysosomal abnormalities similar to those seen in spastin-depleted cells, suggesting that ER dynamics and shape affect the formation of MCSs and have functional consequences for endolysosomal organelles.
MCSs also facilitate the trafficking and maturation of endosomes; MCSs between endosomes and the ER increases as endosomes mature. ORP1L, in response to cholesterol levels, can affect endosome positioning by coordinating the association of late endosomes with motor proteins [25]. ORP1L contains an N-terminal ankyrin repeat region that targets it to late endosomes through binding with Rab7 [135]. When cholesterol levels are high, ORP1L interacts with a complex containing Rab7, RILP (Rab7-interacting lysosomal protein), the HOPS (homotypic fusion and vacuole protein sorting) complex, and the p150GLUED subunit of the dynactin complex, resulting in late endosome retrograde trafficking to the cell center where they can fuse with lysosomes. However, when cholesterol levels are low, ORP1L undergoes a conformational change resulting in interaction with VAPA, and disassociation from the Rab7-RILP-p150GLUED dynein motor interacting complex, disrupting retrograde trafficking and resulting in accumulation of late endosomes at the cell periphery.
STARD3 and STARD3NL may have a similar function in cholesterol sensing and late endosome trafficking. STARD3/STARD3NL can also bind cholesterol and interact with VAPA and varying their expression levels have been shown to affect endosome positioning [23]. Overexpression of STARD3 results in accumulation of late endosomes at the perinuclear region, while depletion results in accumulation at the cell periphery. The ER-endosome MCS protein protrudin also influences endosome positioning. Protrudin binds the kinesin-1 motor protein KIF5 and passes KIF5 to the late endosome protein FYOC1 (FYVE and coiled-coil domain-containing protein) for transport of late endosomes to the cell periphery, where they can undergo synaptotagmin VII-mediated fusion with the plasma membrane to form neurites [33, 136]. FYCO1, like protrudin, also binds to Rab7 and PI3P [137], possibly so that FYOC1 and protrudin localize to the same endosomal membranes for KIF5 exchange. Protrudin depletion results in both perinuclear accumulation of late endosomes and aberrant ER morphology of expanded sheets [113].
The endocytic pathway is important for the degradation of cell surface receptors. Lysosome-bound receptors, such as EGFR, can continue signaling from the limiting membrane of endosomes, even after the receptors have been endocytosed [138]. ER-endosome MCSs are implicated in the downregulation and sorting of cell surface receptors, including EGFRs [34] and G-CSFRs [35]. The peripherally-associated ER tyrosine phosphatase PTP1B dephosphorylates several receptors on ER membranes, preventing their aberrant signaling and facilitating their incorporation into ILVs of MVBs for lysosomal degradation. PTP1B-mediated dephosphorylation of EGFR is required for EGFR internalization into MVBs for degradation by lysosomes [34].
The ER is the main storehouse of Ca2+ within the cell, and Ca2+ signaling plays a key role in a wide-range of physiological processes [139]. Endocytic vesicles forming from the plasma membrane take up Ca2+ from the extracellular space but then it is quickly released, as Ca2+ levels have been reported to be very low (~0.5 μM in early endosomes) [140]. However, Rab7-positive late endosomes have a luminal Ca2+ concentration of about 2.5 μM [141, 142], similar to the luminal Ca2+ concentration of the ER, suggesting that MCSs between the ER and endolysosomal organelles results in Ca2+ uptake from the ER.
Ca2+ uptake and storage via endocytic compartments may be required for endosomal fusion and maturation [143]. Endocytic organelles can release Ca2+ stores through two-pore channels (TPCs), transient receptor channels, and mucolipin transient receptor potential (TRPML) channels [144–146]. Additionally, bidirectional communication between the ER and late endosomes/lysosomes has been reported; Ca2+ released by IP3Rs (inositol 1,4,5-trisphosphate receptors) and/or RyRs (ryanodine receptors) on the ER can trigger TRPML/TPCs on lysosomes. Signaling by NAADP (nicotinic acid adenine dinucleotide phosphate) Ca2+ messengers to TPCs can trigger IP3Rs/RyRs mediated Ca2+ release [147, 148]. Additionally, IP3Rs are found clustered at ER-lysosome contact sites and have been shown to deliver Ca2+ to lysosomes [37, 148]; however, IP3Rs are not required for MCS formation as the contact sites persist in cells without IP3Rs [37]. Furthermore, TPC1 was found localized to ER-endosome MCSs and depletion of TPC1 or inhibiting NAADP significantly decreased ER contact site formation with late endosomes and resulted in prolonged EGFR signaling, suggesting that local Ca2+ signals from endosomes may play a role in the formation of some MCSs with ER, possibly those regulated by the Ca2+ dependent annexin A1-S100A11 tethering activity [149]. Interestingly, depletion of TPC2 specifically reduces the number of MCSs between the ER and lysosomes [149], although the molecular composition of those contact sites are unknown.
7. Defects in ER-Endosome Contacts are Implicated in Disease States
ER-endosome contact sites are involved in many essential physiological functions. Abnormalities in ER membrane contacts are associated with various neurological disorders, and mutations in ER-shaping proteins themselves have been linked to human diseases. Mutations in the spastin (SPG4), REEP1 (SPG31), or strumpellin (SPG8) proteins implicated in ER-mediated endosomal tubule fission cause HSP, a large group of inherited diseases (SPG1–80, plus others) characterized by degeneration of the longest axons in the corticospinal tracts. Protrudin (SPG33) and KIF5A (SPG10) implicated in ER-endosome MCSs and endosomal positioning are also associated with HSPs, though the validity of the SPG33 assignment is in question. At least three other endo-lysosomal proteins, spatacsin (SPG11), spastizin (SPG15), and an AP5 complex subunit (SPG48) are implicated in HSP pathogenesis [150]. It is striking that many of the HSP mutations result in altered ER morphology, suggesting that changes to ER shape can disrupt MCS formation and function with endo-lysosomal and other organelles, given that the ER makes extensive contacts with virtually all cellular organelles. Disruption of M1 spastin-IST1 interactions that mediate endosome tubule fission impair sorting of mannose 6-phosphate receptors, leading to disrupted lysosomal enzyme trafficking and abnormal lysosomal morphology in neurons [39]. Furthermore, altering ER structure in it of itself can reduce endosome fission [29]. The importance of ER-mediated endosomal fission in lysosomal function is further supported by the findings that strumpellin SPG8 missense mutants show similar lysosomal abnormalities. The highly polarized organization of neurons requires the particularly delicate thin tubular ER of axons, which may be particularly vulnerable to ER-shaping defects and lead specifically to neurodegenerative disease.
The neurodegenerative Niemann-Pick type C disease is due to mutations in NPC1 and NPC2, resulting in a failure of cholesterol traffic to the ER and retention in late endosomes. NPC1 interacts with the ER protein ORP5 [28], and although NPC1 mutations have not been shown to alter ER-endosome contact site formation, NPC1 plays a key role in cholesterol transport between the two organelles. Furthermore, dysregulation of cholesterol homeostasis is associated with neurodegenerative disorders [151–153] and the importance of NPC1 in transfer of cholesterol to the ER via ORP5 is emphasized by the fatal nature of the neurodegenerative disease Neimann-Pick type C1 [154, 155].
Several other proteins highlighted in this article localized to ER-endo/lysosomal membrane contacts are linked to human diseases: VAPB in ALS [107]; VPS35 and VPS13C in Parkinson disease [92, 93, 117, 118]; and annexin A1 in cancer [156] (Table 2). Although it is known that MCSs are critical to normal cell physiology, whether defects in MCS formation and function directly cause these diseases remains to be investigated.
Table 2:
ER-Endo/Lysosomal MCS Localized Proteins Linked to Human Diseases
| MCS Protein | Localization | Associated Disease | Reference |
|---|---|---|---|
| M1 Spastin | ER | HSP | [38, 150] |
| Protrudin | ER | HSP | [38, 150] |
| Strumpellin | ER | HSP | [38, 94, 95, 150] |
| VAPB | ER | ALS | [107] |
| NPC1 | Late endosomes | Niemann-Pick disease, type C1 (lysosome storage disease) | [28, 155] |
| Annexin A1 | Early and late endosomes | Cancer | [156] |
| VPS35 | Early and late endosomes | Parkinson’s Disease | [92][93] |
| VPS13C | MCSs of ER-late endosomes/lysosomes and lipid droplets | Parkinson’s Disease | [117, 118] |
8. Perspectives and Conclusions
The ER is the lipid and protein factory of the cell as well as the main conduit for organelle communication. Interactions between ER and endosomal compartments are functionally important to cellular homeostasis, and defects in proteins that mediate this interaction as associated with numerous, and often severe, disease states. As the character and composition of these contacts become more clear, new therapeutic approaches to these disorders will likely be identified.
Supplementary Material
Acknowledgments
The authors were supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This article is part of a Special Issue entitled: Endoplasmic Reticulum Platforms for Membrane Lipid Dynamics edited by Shamshad Cockcroft and Christopher Stefan.
References
- 1.Baumann O and Walz B, Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol, 2001. 205: p. 149–214. [DOI] [PubMed] [Google Scholar]
- 2.Lynes EM and Simmen T, Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER. Biochim Biophys Acta, 2011. 1813(10): p. 1893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helenius A, Marquardt T, and Braakman I, The endoplasmic reticulum as a protein-folding compartment. Trends Cell Biol, 1992. 2(8): p. 227–31. [DOI] [PubMed] [Google Scholar]
- 4.Reid DW and Nicchitta CV, Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol, 2015. 16(4): p. 221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fagone P and Jackowski S, Membrane phospholipid synthesis and endoplasmic reticulum function. J Lipid Res, 2009. 50 Suppl: p. S311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebert DN, Garman SC, and Molinari M, The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol, 2005. 15(7): p. 364–70. [DOI] [PubMed] [Google Scholar]
- 7.Braakman I and Hebert DN, Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol, 2013. 5(5): a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vincenz-Donnelly L and Hipp MS, The endoplasmic reticulum: A hub of protein quality control in health and disease. Free Radic Biol Med, 2017. 108: p. 383–93. [DOI] [PubMed] [Google Scholar]
- 9.Takei K, et al. , Ca2+ stores in Purkinje neurons: endoplasmic reticulum subcompartments demonstrated by the heterogeneous distribution of the InsP3 receptor, Ca(2+)-ATPase, and calsequestrin. J Neurosci, 1992. 12(2): p. 489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Put FH and Elliott AC, The endoplasmic reticulum can act as a functional Ca2+ store in all subcellular regions of the pancreatic acinar cell. J Biol Chem, 1997. 272(44): p. 27764–70. [DOI] [PubMed] [Google Scholar]
- 11.Carreras-Sureda A, Pihan P, and Hetz C, Calcium signaling at the endoplasmic reticulum: fine-tuning stress responses. Cell Calcium, 2018. 70: p. 24–31. [DOI] [PubMed] [Google Scholar]
- 12.Shibata Y, et al. , Mechanisms determining the morphology of the peripheral ER. Cell, 2010. 143(5): p. 774–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C and Chen LB, Dynamic behavior of endoplasmic reticulum in living cells. Cell, 1988. 54(1): p. 37–46. [DOI] [PubMed] [Google Scholar]
- 14.Goyal U and Blackstone C, Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta, 2013. 1833(11): p. 2492–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SH and Blackstone C, Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep, 2010. 11(7): p. 515–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nixon-Abell J, et al. , Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science, 2016. 354(6311): aaf3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips MJ and Voeltz GK, Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol, 2016. 17(2): p. 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, et al. , Contacts between the endoplasmic reticulum and other membranes in neurons. Proc Natl Acad Sci U S A, 2017. 114(24): p. E4859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefan CJ, Manford AG, and Emr SD, ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol, 2013. 25(4): p. 434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valm AM, et al. , Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature, 2017. 546(7656): p. 162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenberg-Bord M, et al. , A tether Is a tether Is a tether: tethering at membrane contact sites. Dev Cell, 2016. 39(4): p. 395–409. [DOI] [PubMed] [Google Scholar]
- 22.Ping HA, et al. , Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J Cell Biol, 2016. 213(5): p. 513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alpy F, et al. , STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J Cell Sci, 2013. 126(Pt 23): p. 5500–12. [DOI] [PubMed] [Google Scholar]
- 24.Wilhelm LP, et al. , STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J, 2017. 36(10): p. 1412–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocha N, et al. , Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol, 2009. 185(7): p. 1209–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eden ER, et al. , Annexin A1 Tethers Membrane Contact Sites that Mediate ER to Endosome Cholesterol Transport. Dev Cell, 2016. 37(5): p. 473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao K and Ridgway ND, Oxysterol-binding protein-related protein 1L regulates cholesterol egress from the endo-lysosomal system. Cell Rep, 2017. 19(9): p. 1807–18. [DOI] [PubMed] [Google Scholar]
- 28.Du X, et al. , A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol, 2011. 192(1): p. 121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowland AA, et al. , ER contact sites define the position and timing of endosome fission. Cell, 2014. 159(5): p. 1027–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allison R, et al. , An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J Cell Biol, 2013. 202(3): p. 527–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoyer MJ, et al. , A novel class of ER membrane proteins regulates ER-associated endosome fission. Cell, 2018. 175(1): p. 254–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jongsma ML, et al. , An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell, 2016. 166(1): p. 152–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raiborg C, et al. , Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature, 2015. 520(7546): p. 234–8. [DOI] [PubMed] [Google Scholar]
- 34.Eden ER, et al. , Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol, 2010. 12(3): p. 267–72. [DOI] [PubMed] [Google Scholar]
- 35.Palande K, et al. , Peroxiredoxin-controlled G-CSF signalling at the endoplasmic reticulum-early endosome interface. J Cell Sci, 2011. 124(Pt 21): p. 3695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haj FG, et al. , Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science, 2002. 295(5560): p. 1708–11. [DOI] [PubMed] [Google Scholar]
- 37.Atakpa P, et al. , IP3 receptors preferentially associate with ER-lysosome contact sites and selectively deliver Ca2+ to lysosomes. Cell Rep, 2018. 25(11): p. 3180–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackstone C, Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci, 2012. 35: p. 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allison R, et al. , Defects in ER-endosome contacts impact lysosome function in hereditary spastic paraplegia. J Cell Biol, 2017. 216(5): p. 1337–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terasaki M and Jaffe LA, Organization of the sea urchin egg endoplasmic reticulum and its reorganization at fertilization. J Cell Biol, 1991. 114(5): p. 929–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westrate LM, et al. , Form follows function: the importance of endoplasmic reticulum shape. Annu Rev Biochem, 2015. 84: p. 791–811. [DOI] [PubMed] [Google Scholar]
- 42.Terasaki M, et al. , Continuous network of endoplasmic reticulum in cerebellar Purkinje neurons. Proc Natl Acad Sci U S A, 1994. 91(16): p. 7510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dayel MJ, Hom EF, and Verkman AS, Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys J, 1999. 76(5): p. 2843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terasaki M, Dynamics of the endoplasmic reticulum and golgi apparatus during early sea urchin development. Mol Biol Cell, 2000. 11(3): p. 897–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S, et al. , Multiple mechanisms determine ER network morphology during the cell cycle in Xenopus egg extracts. J Cell Biol, 2013. 203(5): p. 801–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terasaki M, et al. , Structural change of the endoplasmic reticulum during fertilization: evidence for loss of membrane continuity using the green fluorescent protein. Dev Biol, 1996. 179(2): p. 320–8. [DOI] [PubMed] [Google Scholar]
- 47.Schroeder LK, et al. , Dynamic nanoscale morphology of the ER surveyed by STED microscopy. J Cell Biol, 2019. 218(1): p. 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hetzer MW, The nuclear envelope. Cold Spring Harb Perspect Biol, 2010. 2(3): p. a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terasaki M, et al. , Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell, 2013. 154(2): p. 285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Busnadiego R, Saheki Y, and De Camilli P, Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc Natl Acad Sci U S A, 2015. 112(16): p. E2004–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terasaki M, Axonal endoplasmic reticulum is very narrow. J Cell Sci, 2018. 131(4): jcs210450. [DOI] [PubMed] [Google Scholar]
- 52.Puhka M, et al. , Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol, 2007. 179(5): p. 895–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spacek J and Harris KM, Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J Neurosci, 1997. 17(1): p. 190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martone ME, et al. , Three-dimensional visualization of the smooth endoplasmic reticulum in Purkinje cell dendrites. J Neurosci, 1993. 13(11): p. 4636–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renvoisé B and Blackstone C, Emerging themes of ER organization in the development and maintenance of axons. Curr Opin Neurobiol, 2010. 20(5): p. 531–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palade GE and Porter KR, Studies on the endoplasmic reticulum. I. Its identification in cells in situ. J Exp Med, 1954. 100(6): p. 641–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palade GE, Studies on the endoplasmic reticulum. II. Simple dispositions in cells in situ. J Biophys Biochem Cytol, 1955. 1(6): p. 567–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voeltz GK, et al. , A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell, 2006. 124(3): p. 573–86. [DOI] [PubMed] [Google Scholar]
- 59.Shibata Y, et al. , The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem, 2008. 283(27): p. 18892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu J, et al. , Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science, 2008. 319(5867): p. 1247–50. [DOI] [PubMed] [Google Scholar]
- 61.Hu J, et al. , A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell, 2009. 138(3): p. 549–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orso G, et al. , Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature, 2009. 460(7258): p. 978–83. [DOI] [PubMed] [Google Scholar]
- 63.Park SH, et al. , Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest, 2010. 120(4): p. 1097–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterman-Storer CM and Salmon ED, Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol, 1998. 8(14): p. 798–806. [DOI] [PubMed] [Google Scholar]
- 65.Friedman JR, et al. , ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol, 2010. 190(3): p. 363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wozniak MJ, et al. , Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci, 2009. 122(Pt 12): p. 1979–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terasaki M, Chen LB, and Fujiwara K, Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol, 1986. 103(4): p. 1557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Forges H, Bouissou A, and Perez F, Interplay between microtubule dynamics and intracellular organization. Int J Biochem Cell Biol, 2012. 44(2): p. 266–74. [DOI] [PubMed] [Google Scholar]
- 69.Grigoriev I, et al. , STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr Biol, 2008. 18(3): p. 177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zajac AL, et al. , Local cytoskeletal and organelle interactions impact molecular-motor- driven early endosomal trafficking. Curr Biol, 2013. 23(13): p. 1173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katzmann DJ, Odorizzi G, and Emr SD, Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol, 2002. 3(12): p. 893–905. [DOI] [PubMed] [Google Scholar]
- 72.Huotari J and Helenius A, Endosome maturation. EMBO J, 2011. 30(17): p. 3481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waterman H and Yarden Y, Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett, 2001. 490(3): p. 142–52. [DOI] [PubMed] [Google Scholar]
- 74.Mellman I, Endocytosis and molecular sorting. Annu Rev Cell Dev Biol, 1996. 12: p. 575–625. [DOI] [PubMed] [Google Scholar]
- 75.Mellman I, Fuchs R, and Helenius A, Acidification of the endocytic and exocytic pathways. Annu Rev Biochem, 1986. 55: p. 663–700. [DOI] [PubMed] [Google Scholar]
- 76.Zerial M and McBride H, Rab proteins as membrane organizers. Nat Rev Mol Cell Biol, 2001. 2(2): p. 107–17. [DOI] [PubMed] [Google Scholar]
- 77.Stenmark H, Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol, 2009. 10(8): p. 513–25. [DOI] [PubMed] [Google Scholar]
- 78.Di Paolo G and De Camilli P, Phosphoinositides in cell regulation and membrane dynamics. Nature, 2006. 443(7112): p. 651–7. [DOI] [PubMed] [Google Scholar]
- 79.Sonnichsen B, et al. , Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol, 2000. 149(4): p. 901–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lubke T, Lobel P, and Sleat DE, Proteomics of the lysosome. Biochim Biophys Acta, 2009. 1793(4): p. 625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hsu VW, Bai M, and Li J, Getting active: protein sorting in endocytic recycling. Nat Rev Mol Cell Biol, 2012. 13(5): p. 323–8. [DOI] [PubMed] [Google Scholar]
- 82.van Weering JR, et al. , Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J, 2012. 31(23): p. 4466–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Weering JR, Verkade P, and Cullen PJ, SNX-BAR-mediated endosome tubulation is coordinated with endosome maturation. Traffic, 2012. 13(1): p. 94–107. [DOI] [PubMed] [Google Scholar]
- 84.Puthenveedu MA, et al. , Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell, 2010. 143(5): p. 761–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Derivery E, et al. , The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell, 2009. 17(5): p. 712–23. [DOI] [PubMed] [Google Scholar]
- 86.Gautreau A, Oguievetskaia K, and Ungermann C, Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb Perspect Biol, 2014. 6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McNally KE, et al. , Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat Cell Biol, 2017. 19(10): p. 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hunt SD, et al. , Microtubule motors mediate endosomal sorting by maintaining functional domain organization. J Cell Sci, 2013. 126(Pt 11): p. 2493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parkinson-Lawrence EJ, et al. , Lysosomal storage disease: revealing lysosomal function and physiology. Physiology (Bethesda), 2010. 25(2): p. 102–15. [DOI] [PubMed] [Google Scholar]
- 90.Platt FM, Boland B, and van der Spoel AC, The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J Cell Biol, 2012. 199(5): p. 723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neefjes J and van der Kant R, Stuck in traffic: an emerging theme in diseases of the nervous system. Trends Neurosci, 2014. 37(2): p. 66–76. [DOI] [PubMed] [Google Scholar]
- 92.Vilariño-Güell C, et al. , VPS35 mutations in Parkinson disease. Am J Hum Genet, 2011. 89(1): p. 162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zimprich A, et al. , A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet, 2011. 89(1): p. 168–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Valdmanis PN, et al. , Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet, 2007. 80(1): p. 152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Bot ST, et al. , Pure adult-onset spastic paraplegia caused by a novel mutation in the KIAA0196 (SPG8) gene. J Neurol, 2013. 260(7): p. 1765–9. [DOI] [PubMed] [Google Scholar]
- 96.Raiborg C and Stenmark H, The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature, 2009. 458(7237): p. 445–52. [DOI] [PubMed] [Google Scholar]
- 97.Jordens I, et al. , Rab proteins, connecting transport and vesicle fusion. Traffic, 2005. 6(12): p. 1070–7. [DOI] [PubMed] [Google Scholar]
- 98.Hollenbeck PJ and Swanson JA, Radial extension of macrophage tubular lysosomes supported by kinesin. Nature, 1990. 346(6287): p. 864–6. [DOI] [PubMed] [Google Scholar]
- 99.Brown CL, et al. , Kinesin-2 is a motor for late endosomes and lysosomes. Traffic, 2005. 6(12): p. 1114–24. [DOI] [PubMed] [Google Scholar]
- 100.Jordens I, et al. , The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol, 2001. 11(21): p. 1680–5. [DOI] [PubMed] [Google Scholar]
- 101.Honscher C and Ungermann C, A close-up view of membrane contact sites between the endoplasmic reticulum and the endolysosomal system: from yeast to man. Crit Rev Biochem Mol Biol, 2014. 49(3): p. 262–8. [DOI] [PubMed] [Google Scholar]
- 102.van der Kant R and Neefjes J, Small regulators, major consequences - Ca(2)(+) and cholesterol at the endosome-ER interface. J Cell Sci, 2014. 127(Pt 5): p. 929–38. [DOI] [PubMed] [Google Scholar]
- 103.Friedman JR, et al. , Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell, 2013. 24(7): p. 1030–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Csordas G, et al. , Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol, 2006. 174(7): p. 915–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Vos KJ, et al. , VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum Mol Genet, 2012. 21(6): p. 1299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stoica R, et al. , ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun, 2014. 5: p. 3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nishimura AL, et al. , A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet, 2004. 75(5): p. 822–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hanada K, et al. , Molecular machinery for non-vesicular trafficking of ceramide. Nature, 2003. 426(6968): p. 803–9. [DOI] [PubMed] [Google Scholar]
- 109.Peretti D, et al. , Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell, 2008. 19(9): p. 3871–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mesmin B, et al. , A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell, 2013. 155(4): p. 830–43. [DOI] [PubMed] [Google Scholar]
- 111.van der Kant R, et al. , Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci, 2013. 126(Pt 15): p. 3462–74. [DOI] [PubMed] [Google Scholar]
- 112.van der Kant R, et al. , Cholesterol-binding molecules MLN64 and ORP1L mark distinct late endosomes with transporters ABCA3 and NPC1. J Lipid Res, 2013. 54(8): p. 2153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chang J, Lee S, and Blackstone C, Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation. Proc Natl Acad Sci U S A, 2013. 110(37): p. 14954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shirane M and Nakayama KI, Protrudin induces neurite formation by directional membrane trafficking. Science, 2006. 314(5800): p. 818–21. [DOI] [PubMed] [Google Scholar]
- 115.Saita S, et al. , Promotion of neurite extension by protrudin requires its interaction with vesicle-associated membrane protein-associated protein. J Biol Chem, 2009. 284(20): p. 13766–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gil JE, et al. , Phosphoinositides differentially regulate protrudin localization through the FYVE domain. J Biol Chem, 2012. 287(49): p. 41268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lesage S, et al. , Loss of VPS13C Function in autosomal-recessive parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-dependent mitophagy. Am J Hum Genet, 2016. 98(3): p. 500–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kumar N, et al. , VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol, 2018. 217(10): p. 3625–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dong R, et al. , Endosome-ER contacts control actin nucleation and retromer function through VAP-Dependent Regulation of PI4P. Cell, 2016. 166(2): p. 408–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Di Mattia T, et al. , Identification of MOSPD2, a novel scaffold for endoplasmic reticulum membrane contact sites. EMBO Rep, 2018. 19(7): e45453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Romsicki Y, et al. , Protein tyrosine phosphatase-1B dephosphorylation of the insulin receptor occurs in a perinuclear endosome compartment in human embryonic kidney 293 cells. J Biol Chem, 2004. 279(13): p. 12868–75. [DOI] [PubMed] [Google Scholar]
- 122.Sangwan V, et al. , Regulation of the Met receptor-tyrosine kinase by the protein-tyrosine phosphatase 1B and T-cell phosphatase. J Biol Chem, 2008. 283(49): p. 34374–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stuible M, et al. , PTP1B targets the endosomal sorting machinery: dephosphorylation of regulatory sites on the endosomal sorting complex required for transport component STAM2. J Biol Chem, 2010. 285(31): p. 23899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rety S, et al. , Structural basis of the Ca2+-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure, 2000. 8(2): p. 175–84. [DOI] [PubMed] [Google Scholar]
- 125.Murphy SE and Levine TP, VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta, 2016. 1861(8 Pt B): p. 952–61. [DOI] [PubMed] [Google Scholar]
- 126.Mobius W, et al. , Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic, 2003. 4(4): p. 222–31. [DOI] [PubMed] [Google Scholar]
- 127.Neufeld EB, et al. , Intracellular trafficking of cholesterol monitored with a cyclodextrin. J Biol Chem, 1996. 271(35): p. 21604–13. [DOI] [PubMed] [Google Scholar]
- 128.Subramanian K and Balch WE, NPC1/NPC2 function as a tag team duo to mobilize cholesterol. Proc Natl Acad Sci U S A, 2008. 105(40): p. 15223–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dong J, et al. , Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2. Nat Commun, 2019. 10(1): 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Johansson M and Olkkonen VM, Assays for interaction between Rab7 and oxysterol binding protein related protein 1L (ORP1L). Methods Enzymol, 2005. 403: p. 743–58. [DOI] [PubMed] [Google Scholar]
- 131.Alpy F, et al. , MENTHO, a MLN64 homologue devoid of the START domain. J Biol Chem, 2002. 277(52): p. 50780–7. [DOI] [PubMed] [Google Scholar]
- 132.Walther TC, Chung J, and Farese RV Jr., Lipid droplet biogenesis. Annu Rev Cell Dev Biol, 2017. 33: p. 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gao Q and Goodman JM, The lipid droplet-a well-connected organelle. Front Cell Dev Biol, 2015. 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Friedman JR, et al. , ER tubules mark sites of mitochondrial division. Science, 2011. 334(6054): p. 358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Johansson M, et al. , The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell, 2005. 16(12): p. 5480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Matsuzaki F, et al. , Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Mol Biol Cell, 2011. 22(23): p. 4602–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pankiv S, et al. , FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol, 2010. 188(2): p. 253–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sorkin A and von Zastrow M, Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol, 2009. 10(9): p. 609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Berridge MJ, Bootman MD, and Roderick HL, Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol, 2003. 4(7): p. 517–29. [DOI] [PubMed] [Google Scholar]
- 140.Gerasimenko JV, et al. , Calcium uptake via endocytosis with rapid release from acidifying endosomes. Curr Biol, 1998. 8(24): p. 1335–8. [DOI] [PubMed] [Google Scholar]
- 141.Christensen KA, Myers JT, and Swanson JA, pH-dependent regulation of lysosomal calcium in macrophages. J Cell Sci, 2002. 115(Pt 3): p. 599–607. [DOI] [PubMed] [Google Scholar]
- 142.Lloyd-Evans E, et al. , Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med, 2008. 14(11): p. 1247–55. [DOI] [PubMed] [Google Scholar]
- 143.Pryor PR, et al. , The role of intraorganellar Ca2+ in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J Cell Biol, 2000. 149(5): p. 1053–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Abe K and Puertollano R, Role of TRP channels in the regulation of the endosomal pathway. Physiology (Bethesda), 2011. 26(1): p. 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lelouvier B and Puertollano R, Mucolipin-3 regulates luminal calcium, acidification, and membrane fusion in the endosomal pathway. J Biol Chem, 2011. 286(11): p. 9826–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ruas M, et al. , Purified TPC isoforms form NAADP receptors with distinct roles for Ca2+ signaling and endolysosomal trafficking. Curr Biol, 2010. 20(8): p. 703–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Morgan AJ, et al. , Bidirectional Ca2+ signaling occurs between the endoplasmic reticulum and acidic organelles. J Cell Biol, 2013. 200(6): p. 789–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lopez-Sanjurjo CI, et al. , Lysosomes shape Ins(1,4,5)P3-evoked Ca2+ signals by selectively sequestering Ca2+ released from the endoplasmic reticulum. J Cell Sci, 2013. 126(Pt 1): p. 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kilpatrick BS, et al. , An endosomal NAADP-sensitive two-pore Ca2+ channel regulates ER-endosome membrane contact sites to control growth factor signaling. Cell Rep, 2017. 18(7): p. 1636–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Blackstone C, Converging cellular themes for the hereditary spastic paraplegias. Curr Opin Neurobiol, 2018. 51: p. 139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vivas O, et al. , Niemann-Pick type C disease reveals a link between lysosomal cholesterol and PtdIns(4,5)P2 that regulates neuronal excitability. Cell Rep, 2019. 27(9): p. 2636–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Abdel-Khalik J, et al. , Defective cholesterol metabolism in amyotrophic lateral sclerosis. J Lipid Res, 2017. 58(1): p. 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Di Paolo G and Kim TW, Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci, 2011. 12(5): p. 284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yamamoto T, et al. , NPC1 gene mutations in Japanese patients with Niemann-Pick disease type C. Hum Genet, 1999. 105(1–2): p. 10–6. [DOI] [PubMed] [Google Scholar]
- 155.Millat G, et al. , Niemann-Pick C1 disease: the I1061T substitution is a frequent mutant allele in patients of Western European descent and correlates with a classic juvenile phenotype. Am J Hum Genet, 1999. 65(5): p. 1321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Foo SL, et al. , Annexin-A1 - a Blessing or a curse in cancer? Trends Mol Med, 2019. 25(4): p. 315–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.