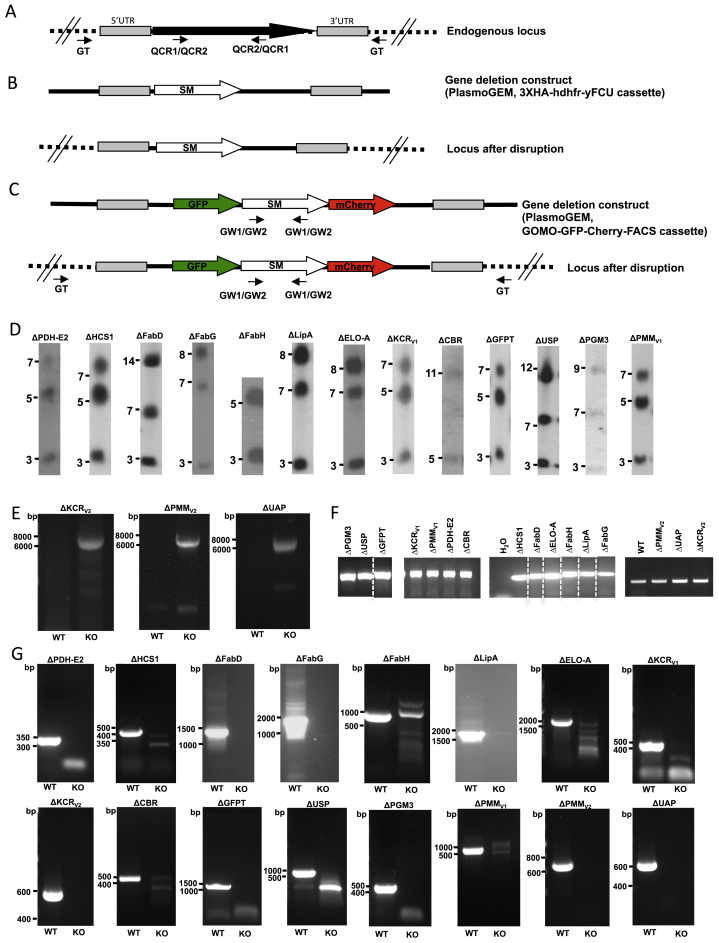
Figure S3.
Genotyping of Single Gene Knockout Parasite Lines, Related to Figures 5, 6, and 7
(A) Schematic representation of the endogenous gene of interest (GOI) locus, (B) the gene deletion targeting construct (PlasmoGEM) containing the 3xHA-hdhfr-yFCU cassette and the GOI locus after disruption following double homologous recombination. Such a replacement strategy was used in cases where no sexual stage crossing was performed to bypass lethal mosquito stage phenotypes. The positions of primers used in PCRs to confirm GOI deletion is indicated by arrows labeled QCR1 or QCR2 and QCR2 or QCR1. (C) Schematic representation of the endogenous gene of interest (GOI) locus, the gene deletion construct (PlasmoGEM) containing the GOMO-GFP-mCherry-FACS cassette and the GOI locus after disruption following double homologous recombination. PCR primers used to confirm successful integration of the construct are indicated by arrows GT and GW1 or GW2 and PCR primers used to confirm deletion of the GOI are indicated by arrows QCR1 or QCR2 and QCR2 or QCR1 respectively. All primer sequences are shown in Table S6. The gene replacement strategy with the GOMO-GFP-mCherry-FACS cassette was used to generate mutant parasites that were crossed with a WT parasite line for cases in which the sKO was blocked at the mosquito stage. (D) Pulse field gel electrophoresis (PFGE) for the following gene knockout parasite lines (chromosome location of replaced gene shown in brackets, knockouts generated via transfection with constructs containing the 3xHA-hdhfr-yFCU cassette): ΔPDH-E2 (5), ΔHCS1 (5), ΔFabD (14), ΔFabG (8), ΔFabH (3), ΔLipA (13), ΔELO-A (8), ΔKCRv1 (5), ΔCBR (11), ΔGFPT (5), ΔUSP (12), ΔPGM3 (9) and ΔPMMv1 (5). A probe was used that recognized the 3′UTR of the pbdhfr hybridized to the knockout cassette that replaced the above genes on the chromosomes listed above. A probe of ≈800 bp fragment of the 5′UTR of the PBANKA_0508000 gene located on the chromosome 5 was also used for the ΔFabH mutant parasite and a probe of ≈800 bp fragment of the 5′UTR of the PBANKA_0508000 gene located on the chromosome 5 was additionally used for the ΔCBR mutant parasite. All images have been cropped from PFGE images showing other parasite lines. (E) Diagnostic PCR of the single gene knockout parasite lines ΔKCRv2, ΔPMMv2 and ΔUAP using primers to test for the presence of the WT locus and successful integration at the kcr, pmm and uap loci, respectively (parasites generated by transfection with a construct containing the GOMO-GFP-mCherry-FACS cassette). GT and GW2 primers were used to show integration for ΔKCRv2 and ΔPMMv2 and GT and GW1 for ΔUAP. Parasites were generated with the aim of crossing with WT parasites to rescue the lethal mosquito stage phenotype. Lanes showing markers have been removed and also other PCR products from other clones. (F) Control PCR to show successful amplification from gDNA samples taken from all single gene knockout parasite lines generated in this study Dotted lines between lanes indicate the reordering of lane images from the same gel photo. A space between lanes indicates lane images taken from separate gels. (G) PCRs showing the presence of genes in WT parasites and absence in mutant parasites for all single gene knockout lines generated in this study. All primers used for genotyping PCRs are listed in Table S6. Lanes showing markers have been removed and also other PCR products from other clones.