SUMMARY
The first immunization in a protein prime-boost vaccination is likely to be critical for how the immune response unfolds. Using fine needle aspirates (FNAs) of draining lymph nodes (LNs), we tracked the kinetics of the primary immune response in rhesus monkeys immunized intramuscularly (IM) or subcutaneously (s.c.) with an eOD-GT8 60-mer nanoparticle immunogen to facilitate clinical trial design. Significant numbers of germinal center B (BGC) cells and antigen-specific CD4 T cells were detectable in the draining LN as early as 7 days post-immunization and peaked near day 21. Strikingly, s.c. immunization results in 10-fold larger antigen-specific BGC cell responses compared to IM immunization. Lymphatic drainage studies revealed that s.c. immunization resulted in faster and more consistent axillary LN drainage than IM immunization. These data indicate robust antigen-specific germinal center responses can occur rapidly to a single immunization with a nanoparticle immunogen and vaccine drainage substantially impacts immune responses in local LNs.
Graphical Abstract
In Brief
The first immunization of protein prime-boost vaccination is likely critical but has been understudied in large animals and humans. Havenar-Daughton et al. use lymph node fine needle aspirates to determine primary germinal center response kinetics in rhesus monkeys immunized intramuscularly or subcutaneously with a clinical trial candidate nanoparticle immunogen.
INTRODUCTION
To induce immunity to difficult pathogens, vaccine technologies are becoming more sophisticated, including the development of structurally engineered immunogens (Correia et al., 2014; Sanders et al., 2013), germline-targeting concepts (Escolano et al., 2016; Jardine et al., 2016a; McGuire et al., 2014; Stamatatos et al., 2017; Steichen et al., 2016), replicating vectors (Barouch et al., 2018), and sophisticated vaccine delivery strategies (Moyer et al., 2016). Many of these approaches endeavor to generate protective antibody (Ab) responses by eliciting B cell responses that have particularly challenging characteristics, such as rare B cell precursors or high amounts of affinity maturation (Havenar-Daughton et al., 2017). Rational vaccine development depends on the ability to quantitatively and qualitatively measure multifaceted aspects of immune responses to candidate vaccines. This is essential to iterative design, which is a central tenet of successful engineering processes, instead of depending on “home run” outcomes (Burton, 2017; Kwong, 2017).
Engineered outer domain-germline targeting eight (eOD-GT8) 60-mer is a B cell receptor (BCR) germline-targeting immunogen specifically designed to activate human naive precursor B cells with epitope specificities similar to that of HIV VRC01-class broadly neutralizing antibodies (Jardine et al., 2016a, 2016b). eOD-GT8 60-mer immunization successfully primed inferred germline VRC01 BCR-transgenic B cells in mice (Abbott et al., 2018; Briney et al., 2016; Tian et al., 2016). A specific challenge for assessing the initial success of a germline-targeted vaccine candidate in humans is that the outcome is expansion of B cells with particular BCR sequence characteristics, rather than antigen (Ag)-specific serum Ab titers. BCR sequencing has not been previously used as a human vaccine clinical trial endpoint. In addition, key aspects of B cell responses are absent or poorly represented in blood. Most notably, germinal centers (GCs) are essential for almost all neutralizing Ab responses, but GCs, germinal center B (BGC) cells, and GC T follicular helper (GC-TFH) cells are present in LNs or spleen, not peripheral blood. Thus, human vaccine clinical trials to date have only been able to indirectly infer GC activity and BGC and GC-TFH specificities. This has been a critical knowledge gap. LN fine needle aspirates (LN FNAs) have a century-long history in the medical literature but have only been rarely used for research purposes (Xu et al., 2013). Recently, we used LN FNAs to serially monitor GC activity in the LNs of rhesus monkeys (RMs) after immunization with native-like HIV Env trimers (Cirelli et al., 2019; Havenar-Daughton et al., 2016a; Pauthner et al., 2017). By examining draining LNs by LN FNA after each immunization, we found that GC activity correlated with the generation of HIV-neutralizing Abs. The highest immunization-elicited neutralizing Ab responses were sufficient to protect RMs against repeated mid-dose rectal challenge with a Tier 2 simian/human immunodeficiency virus (SHIV) (Pauthner et al., 2019).
Here, we have tested whether LN FNAs can detect vaccine response outcomes after a single nanoparticle immunization in non-human primates (NHP) under conditions intended to model human immunization conditions to provide insights for clinical trial designs. The study included longitudinal assessment of GC activity in individual animals and quantitative assessment of Ag-specific BGC cell frequency and somatic hypermutation, providing high resolution of the B cell response to a candidate vaccine immunogen within a few weeks post-immunization.
RESULTS
Immunization Route and Adjuvant Impact Immunogen Drainage to Local LNs
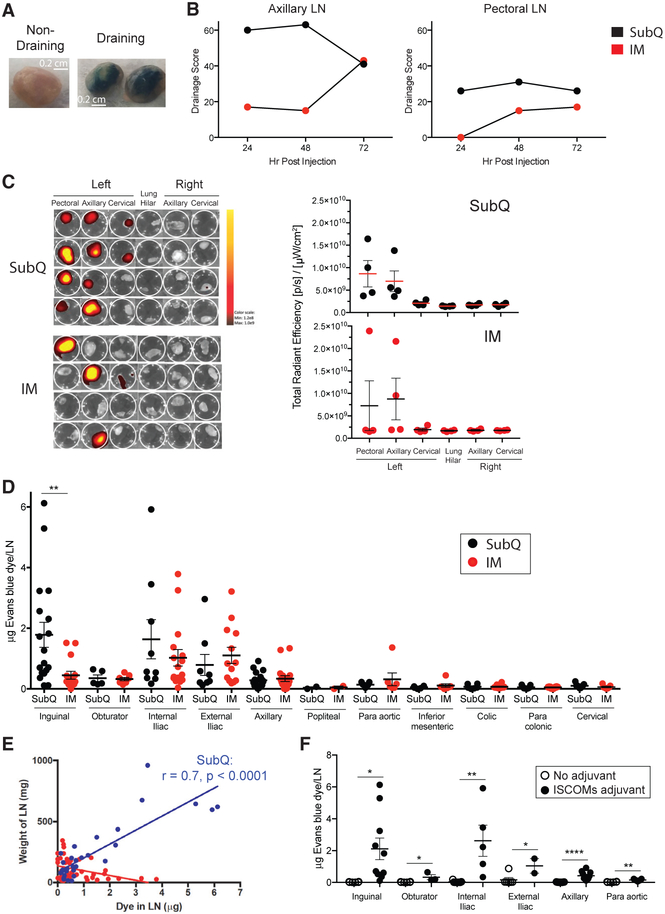
A primary goal of this project was to assess whether Ag-specific B cells could be identified in LNs after a single priming immunization with a protein nanoparticle in a strong adjuvant by using a Macaca mulatta (rhesus monkey, RM) NHP model as the closest available animal model to humans. A critical factor was the choice of LN(s) for FNA sampling. Protein vaccines in humans are conventionally administered by intramuscular (IM) injection in the upper deltoid. Subcutaneous (s.c. or SubQ) immunizations are administered in humans for several licensed vaccines (measles/mumps/rubella [MMR], yellow fever vaccine, and pneumococcal polysaccharide). In a direct comparison of s.c. and IM routes, s.c. immunization with SOSIP HIV Env trimers elicited higher autologous Tier 2 HIV neutralizing Ab responses than IM immunization (Pauthner et al., 2017). s.c. immunization appeared more immunogenic for GC responses than IM; however, it was unclear whether the GC differences observed were due to different LNs serving as the primary draining LNs after s.c. versus IM immunization (Pauthner et al., 2017). Thus, we performed lymphatic drainage experiments to assess 1° and 2° draining LNs upon IM or s.c. immunization in the upper deltoid region. A first study was done with Evans Blue (EB), which visibly stains draining LNs (Figure 1A). Examination of EB accumulation in LNs showed that dye rapidly accumulated in axillary and pectoral LNs following s.c. injection (Figure 1B). IM injection also led to dye uptake in these LNs but at less intensity and with slower kinetics (Figure 1B).
Figure 1. LN Drainage after Injection at the Deltoid or Quadricep.
(A) EB in draining and non-draining LNs.
(B) EB drainage score over time after bilateral s.c. or IM injection at the deltoid. Drainage to other LNs was not observed (listed in STAR Methods). n = 6, one per immunization condition per time point.
(C) IVIS fluorescent quantification of A647 labeled eOD-GT8 60-mer in LNs 48 h after injection at the left deltoid. Left, each row represents an individual animal. Right, each point represents an individual LN cluster from an individual animal shown at the left. n = 8, four per immunization condition. Mean and SEM shown.
(D) EB quantification of drainage after bilateral s.c. or IM injection at the quadricep in the presence of a soluble saponin-based ISCOMs-class adjuvant at 48 h. **p < 0.01 (unpaired t test, two tailed). n = 4 animals, two per immunization condition. Each point represents an individual LN. Mean and SEM shown.
(E) Correlation between the amount of EB dye (μg) and LN weight (mg). Subcutaneous injection (blue) and intramuscular infection (red) (unpaired t test, two tailed). Each point represents an individual LN.
(F) EB quantification of drainage after bilateral s.c. injection at the quadricep with or without a soluble saponin-based ISCOMs-class adjuvant at 48 h. *p < 0.05, **p < 0.01, ****p < 0.0001 (unpaired t test, one tailed). n = 4 animals, two per condition. Each point represents an individual LN. Mean and SEM shown
See also Figure S1.
A larger study was next done with eight animals to evaluate draining of Alexa647-labeled (A647) eOD-GT8 60-mer in the presence of adjuvant. Upon s.c. injection, eOD-GT8 60-mer-A647 rapidly drained to axillary LNs (4/4; Figure 1C). Consistent drainage to pectoral LNs was also observed (4/4; Figure 1C). eOD-GT8 60-mer-A647 was also observed to drain to cervical LNs in some animals (2/4; Figure 1C). In contrast, LN drainage was more sporadic after IM immunization; axillary LNs collected Ag in 2/4 animals, pectoral LNs collected Ag in 1/4 animals, and one animal had no positive LNs observed (Figure 1C). Thus, s.c. immunization consistently resulted in robust axillary and pectoral LN drainage, whereas IM immunization gave more sporadic results.
For comparison to injections at the deltoid, we reassessed s.c. and IM LN drainage upon injection in the anterior thigh (Pauthner et al., 2017). EB was injected together with a soluble saponin-based immune stimulating complex (ISCOM)-class adjuvant (Pauthner et al., 2019), and dye was extracted from excised LNs for quantification by fluorescence spectroscopy (Figures S1A and S1B) (Wang and Lai, 2014). At the 48-h time point, both s.c. and IM injections drained primarily to the inguinal and iliac LNs, with high levels of EB reliably detected following s.c. immunization in at least a portion of these LNs in all animals (Figure 1D). In contrast, IM immunization drained only sporadically to inguinal LNs but consistently to external and internal iliac LNs. Ag drainage positively correlated with adjuvant-induced swelling of LNs after s.c. immunization (s.c., p < 0.0001; Figure 1E). We then compared drainage in the presence or absence of adjuvant (Figure 1F). A striking enhancement of EB accumulation in the draining LNs occurred when administered in the presence of an ISCOM-class adjuvant (Figure 1F). We additionally noted an inverse correlation between animal weight and EB accumulation (Figures S1C). Drainage after injection in the exterior thigh region versus of the interior thigh yielded no clear difference in a small number of animals (Figures S1D). In summary, both the route of immunization and adjuvant have a dramatic impact on Ag trafficking to local LNs in NHPs.
Rapid and Robust B Cell Responses to a Single eOD-GT8 Nanoparticle Immunization
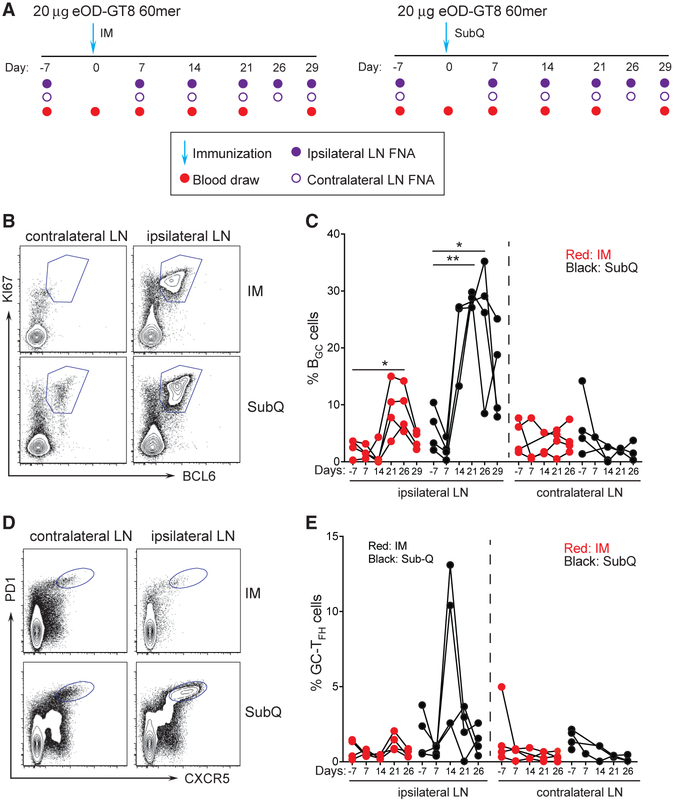
It was unclear whether measurable GC responses would occur after a single immunization of eOD-GT8 60-mer or what the kinetics of a GC response might be. This knowledge was important for understanding the likelihood of significant VRC01-class B cell affinity maturation in response to a priming immunization with eOD-GT8 60-mer (Abbott et al., 2018; Briney et al., 2016; Sok et al., 2016). It was also important for guidance of eOD-GT8 60-mer clinical trial design. Was it likely that eOD-GT8-specific BGC cells could be directly identified in a human clinical trial by axillary LN FNA? Therefore, we designed an NHP study to answer these questions, matching as closely as possible the likely immunization conditions for a human clinical trial: 20 μg of eOD-GT8 60-mer with a strong adjuvant administered IM in the left deltoid (Figure 2A). The adjuvant planned for the human clinical trial was not available for use in NHP studies, and therefore, we selected ISCOMATRIX as a biochemically related soluble adjuvant (Pauthner et al., 2017). Additionally, a second group of monkeys was immunized s.c. (Figure 2A). The primary samples were axillary LN FNAs from the same side as the immunization (ipsilateral) and the opposite side (contralateral). We posited that a dense (day −7, +7, +14, +21, +26, and +29) LN FNA sampling was needed to determine the kinetics of the primary GC response (Figure 2A). Given that ~3% of the LN is sampled per LN FNA (Havenar-Daughton et al., 2016a), BGC cells proliferate every 4–6 h (Tas et al., 2016), serial LN FNAs spaced weeks apart were shown to be well tolerated (Havenar-Daughton et al., 2016a), and weekly LN FNAs were unlikely to be disruptive to the overall scope of the immune response. LN FNA sampling was successful throughout this study. Average cell recovery was ~106 cells per LN FNA sample (Figure S2A); 95% of samples contained >1 × 105 cells and could be analyzed for BGC cells and GC-TFH cells by flow cytometry.
Figure 2. Longitudinal Sampling of GCs by LN FNA after eOD-GT8 60-mer Immunization.
(A) Schematic of LN FNA and blood sampling after IM (left) or s.c. (right) immunization.
(B) Flow cytometry identification of BGC cell frequencies in the ipsilateral (draining) and contralateral (non-draining) LNs after s.c. and IM immunization. BGC cells are KI67+BCL6+. Full gating is Figure S2.
(C) Weekly sampling of BGC cell frequency after one immunization. *p < 0.05, **p < 0.01 (paired t test, one tailed).
(D) Flow cytometry identification of GC-TFH cell frequencies in the ipsilateral (draining) and contralateral (non-draining) LNs after s.c. and IM immunization. GC-TFH cells are CXCR5+PD1+.
(E) Weekly sampling of GC-TFH cell frequency after one immunization. Gated on CD4+ T cells.
Each point represents an individual LN FNA sample. n = 8, four LN FNAs per immunization condition at each time point.
See also Figures S2 and S3.
BGC cell (CD20+BCL6+KI67+) and GC-TFH cell (CD4+ CXCR5+PD1high) frequencies were measured in ipsilateral and contralateral axillary LNs of IM immunized monkeys by flow cytometry at all time points (Figure S2B). No increases in contralateral axillary LN BGC cells was observed, as expected (Figure 2A and 2C). Ipsilateral axillary LN BGC cell frequencies were not increased at day 7 or day 14 after IM immunization but were increased at day 21 and significantly increased at day 26 post-immunization (p = 0.04; Figure 2C). GC-TFH cells were identified by flow cytometry (Figure 2D) but not statistically significantly increased (Figure 2E). While BGC cells depend on GC-TFH cells (Crotty, 2019), vaccine-specific GC-TFH cells are less abundant than BGC cells and, thus, vaccine-specific GC-TFH may not be easily distinguished from other GC-TFH cells without an Ag-specific assay. AIM assays measure Ag-specific GC-TFH cells (Dan et al., 2016; Havenar-Daughton et al., 2016b; Reiss et al., 2017), but insufficient cells were available from the LN FNAs for both Ag-specific GC-TFH and BGC cell assays; thus, Ag-specific BGC cell measurements took priority in this study due to their direct relevance as endpoints in the eOD-GT8 60-mer clinical trial.
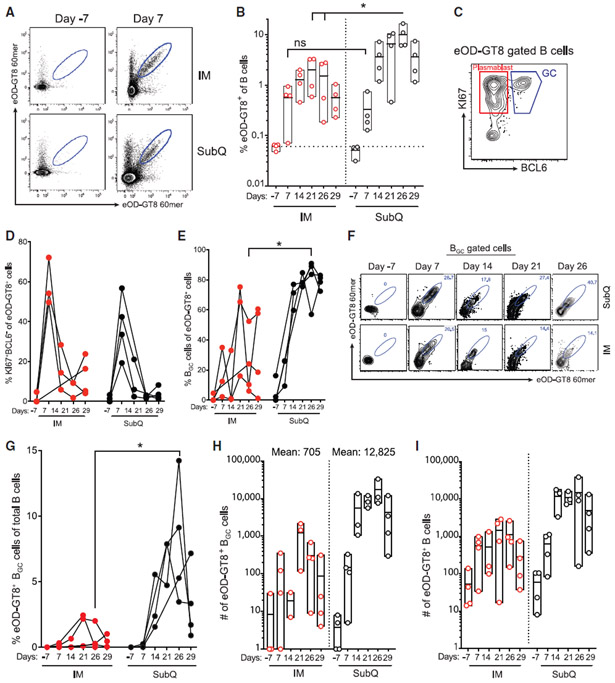
We established the use of eOD-GT8 60mers covalently labeled with small molecule fluorescent dyes as flow cytometry probes to identify eOD-GT8-specific B cells (Figure 3A; Figure S2C). The use of two separate probes with different fluorophores is preferable, as it substantially improves Ag-specific B cell signal-to-noise in flow cytometry (Havenar-Daughton et al., 2018). Ag-specific B cells (CD20+ eOD-GT8 60-mer-A488+ eOD-GT8 60-mer-A647+), Ag-specific plasmablast cells (CD20+BCL6−KI67+ eOD-GT8 60-mer-A488+ eOD-GT8 60-mer-A647+), and Ag-specific BGC cells (CD20+BCL6+KI67+ eOD-GT8 60-mer-A488+ eOD-GT8 60-mer-A647+) were measured in ipsilateral and contralateral axillary LNs after IM immunization. Few Ag-specific B cells were detected in LNs prior to immunization (≤0.1%), as expected. Strikingly, Ag-specific B cells were detected at a frequency of >0.5%, at day 7 in 3 of 4 animals (p = 0.014; Figure 3B). Ag-specific B cell frequency increased to 1% at day 14 (Figure 3B). Peak LN Ag-specific plasmablasts were detected at day 7 (Figure 3C and 3D). Ag-specific BGC cells were also detected at day 7 (Figure 3E, 3F, and S2D). Peak Ag-specific B cell responses (1%–2%) in LNs were detected at day 21, with similar frequencies at day 14 and day 26 (Figure 3B). Peak Ag-specific BGC cell responses in each individual animal constituted 20%–80% of the total Ag-specific B cells (Figure 3E). Thus, IM immunization with eOD-GT8 60mers resulted in rapid Ag-specific B cell responses in LNs, with detectable BGC cells in all immunized animals. Total Ag-specific B cell numbers recovered are also important from a clinical perspective, as a key endpoint in germline-targeting vaccine testing is BCR sequencing for B cells possessing desired VDJ gene combinations. PeakeOD-GT8-specific LN B cell numbers obtained by FNA at day 21 or day 26 from each animal were 714, 2,206, 2,611, and 2,849 cells. In conclusion, vaccine-specific B cell and BGC cell responses were consistently detected in draining LNs after a single IM immunization with eOD-GT8 60mers, with peak responses 21 days post-immunization.
Figure 3. Longitudinal eOD-GT8 60-mer-Specific BGC and Non-BGC Cell Responses in Draining LNs after a Single IM or s.c. Immunization.
(A) Flow cytometry identification of total eOD-GT8 60-mer-specific LN B cells before and after immunization (top and bottom samples pairs are from individual RMs). Gated on CD20+ B cells. Full gating in Figure S2.
(B) Frequency of total eOD-GT8 60-mer-specific B cells after IM or s.c. immunization (Mann-Whitney, one tailed).
(C) Flow cytometry identification of eOD-GT8 60-mer-specific plasmablast (KI67+BCL6−) or GC (KI67+BCL6+) B cells in the LN.
(D) Frequency of KI67+BCL6− plasmablasts among eOD-GT8 60-mer-specific B cells in the draining axillary LN over time.
(E) Frequency of BGC cells among total eOD-GT8 60-mer-specific B cells in the draining axillary LN over time (Mann-Whitney, two tailed).
(F) Representative flow cytometry gating of eOD-GT8 60-mer-specific B cells among BGC cells.
(G) Frequency of eOD-GT8 60-mer-specific BGC cells in the draining axillary LN over time (Mann-Whitney, two tailed).
(H) Total number of eOD-GT8 60-mer-specific BGC cells in the LN FNA sample of the draining axillary LN.
(I) Total number of eOD-GT8 60-mer-specific B cells in the LN FNA sample of the draining axillary LN.
*p < 0.05. Each point represents an individual LN FNA sample. n = 8, four LN FNAs per immunization condition at each time point.
See also Figures S2 and S3.
Vigorous GC Responses after s.c. Immunization with eOD-GT8 Nanoparticles
Longitudinal Ag-specific B cell responses were measured in LNs of s.c. immunized animals in parallel. One week post-immunization, Ag-specific B cell responses were detected in ipsolateral LNs of all s.c. immunized animals (p = 0.014), in a similar range to that observed after IM immunization (Figure 3B). Vaccine-specific BGC responses dramatically increased in s.c. immunized animals after day 7, with a mean Ag-specific B cell frequency of 9.4% of total B cells, and a mean Ag-specific BGC cell frequency of 7.5% of total B cells, between day 21 and day 26 (Figure 3B, 3F, and 3G). Vaccine-specific BGC cell frequencies were significantly greater in s.c. immunized animals (p = 0.03; Figure 3G), as were peak vaccine-specific total B cell frequencies (IM day 21 versus s.c. day 26, p = 0.03; IM day 26 versus s.c. day 26, p = 0.03; Figure 3B). Total GC responses were also significantly larger in s.c. immunized animals compared to IM (s.c.: day 7 versus day 21, p = 0.01; day 7 versus day 26, p = 0.009; Figure 2C). The B cell response was significantly more biased in favor of BGC cells after s.c. versus IM immunization (IM day 26 versus s.c. day 26, p = 0.03; Figure 3E). The magnitude of the eOD-GT8 response after s.c. immunization was also much larger by absolute cell numbers (Figure 3H). The mean number of vaccine-specific BGC cells recovered from s.c. immunized animals at day 21–26 was 12,825 cells versus 705 cells from IM immunized animals (p = 0.001). Thus, s.c. immunization elicited much more vigorous GC responses in draining axillary LNs than IM immunization.
Somatic Hypermutation of LN B Cell Responses to eOD-GT8 Nanoparticles
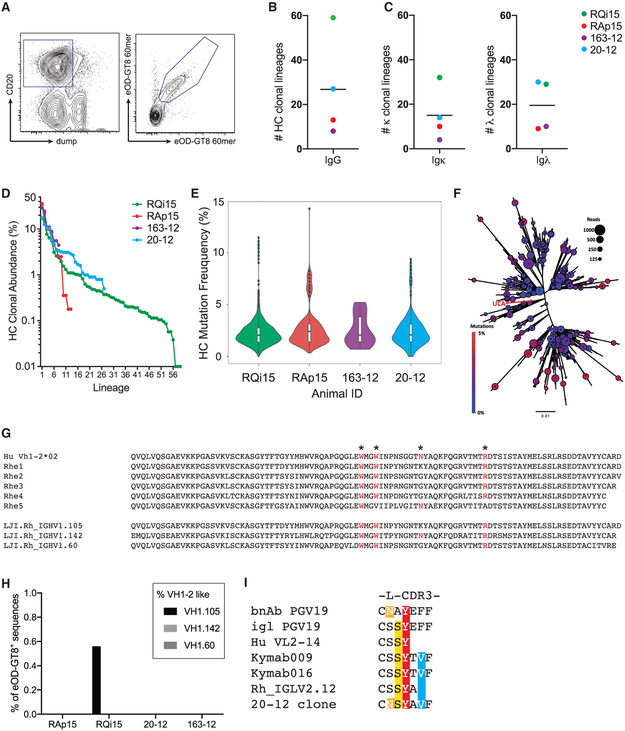
The robust GC responses led us to examine the quality of the eOD-GT8 60-mer specific B cells by BCR sequencing of Ag-specific B cells directly isolated from LN GCs by LN FNAs at 29 days post-immunization (Figure 4A). The BGC cell heavy chain (HC) sequences were frequently highly related, often separated by only a single nucleotide mutation, as expected when sequencing BGC cells (Abbott et al., 2018; Havenar-Daughton et al., 2016a; Kuraoka et al., 2016). Identical sequences were collapsed, and clonal lineages were defined based on (1) same V gene, (2) same J gene, (3) same CDR3 length, and (4) percentage identity of CDR3 nucleotide sequence of >85%. Eight to 59 HC eOD-GT8-specific BGC cell clonal lineages were isolated per LN FNA (Figure 4B). Clonal lineages were also determined for light chains (Figure 4C). Notably, in each LN sampled, a dominant B cell clonal lineage was identified comprising >10% of the cells (Figure 4D).
Figure 4. eOD-GT8 60-mer-Specific BGC Cell Lineages after s.c. Immunization.
(A) Representative sorting gate for eOD-GT8 60-mer-specific B cells at day 26 post-immunization.
(B) Number of heavy chain (HC) lineages present among eOD-GT8 60-mer specific B cells. Each point represents an individual RM. n = 4.
(C) Number of light chain kappa (κ) or lambda (λ) lineages present among eOD-GT8 60-mer specific B cells. Each point represents an individual RM. n = 4.
(D) Clonal abundance of individual lineages by HC from s.c. immunized RMs. Each point represents an individual B cell lineage.
(E) Nucleotide HC SHM frequency of eOD-GT8 specific B cells after one immunization for each immunized animals. Violin plot, individual points represent B cell lineages, white bars represent quartiles. n = 4.
(F) Example of the diversity found within a single eOD-GT8 60-mer-specific B cell lineage. The unmutated common ancestor (UCA) of the lineage is at the center of the plot indicated by the red arrow. Circle size indicates the number of reads for each variant, and circle color indicates the mutation count. Additional example lineages are shown in Figure S3.
(G) RM HC V gene with similarity to human VH1-2 identified in the LJI RM Ig annotation. Critical amino acids for eOD-GT8 binding are noted (*).
(H) Frequency of eOD-GT8 60-mer-specific B cell using huVH1-2 like V genes.
(I) A λ RM light chain sequence with a 5aa L-CDR3 similar to the VRC01-class bnAb PGV19 and Abs induced in human Ig loci transgenic mice immunized with eOD-GT8.
Each point represents an individual animal. n = 8, four LN FNAs per immunization condition at each time point.
See also Figures S4 and S5.
Substantial somatic mutation was present in the eOD-GT8 60-mer-specific BGC cells (Figure 4E). One clonal lineage from animal RQi contained 3,665 unique sequences, representing immense BCR diversification in a single lineage after one immunization (Figure 4F). Additional lineages are shown in Figure S3. The least-mutated common ancestor sequence (Figure 4F; UCA indicated by red arrow) identified in the dominant lineage from animal RQi15 was only 1 nucleotide different from the germline HC V gene sequence, indicative of robust BGC sequence sampling by LN FNA.
Human VRC01-class B cells are minimally defined by the use of human VH1-2 and a light chain (LC) with a 5 amino acid (aa) CDR3 (Zhou et al., 2013). VRC01-class naive B cells were potentially extremely rare or non-existent in RMs. Human VH1-2 does not have an exact match in RMs; several cDNA sequences were identified as RM VH1-2 homologs, but they each lack aa residues known to be important for eOD-GT8 binding to human VH1-2+ B cells (Jardine et al., 2013) (Figure 4G). Additionally, RM B cells with 5aa L-CDR3s were found to be 10-fold less frequent than in humans (Figure S4A), which is in line with another report (Vigdorovich et al., 2016). Thus, it was unclear whether eOD-GT8 immunization would elicit RM VRC01-class B cells. A M. mulatta genome sequence (Cirelli et al., 2019) utilizing long read PacBio sequencing was used for a search for VH1-2 related M. mulatta V genes. The closest related V genes were VH1.105 (aka Rhe1), VH1.142, and VH1.60 (Figure 4G). Each lacked at least one key aa found in human VH1-2 (Figure 4G).
An analysis of the B cell HC sequences obtained from LN FNAs of eOD-GT8 60-mer immunized RMs showed that 0.5% of eOD-GT8 60-mer-specific B cells from one animal used VH1-2-like genes (Figure 4H). LCs were examined for clonal lineages with 5aa CDR3s. One 5aa L-CDR3 eOD-GT8 60-mer-specific B cell was identified. The L-CDR3 sequence was similar to that of VRC01-class bnAb PGV19 (Zhou et al., 2013) and sequences found in eOD-GT8 60-mer immunized Kymab mice (Sok et al., 2016) (Figure 4I). Synthetic immunoglobulin G (IgG) monoclonal Abs (mAbs) were made possessing this LC sequence with a 5aa CDR3 and candidate monkey VH1-2-like HC sequences, but the mAbs did not measurably bind eOD-GT8 (Figure S4B). Thus, VRC01-class B cells are rare or nonexistent in RMs. Overall, the experiments demonstrate that a single eOD-GT8 60-mer immunization prompted a multitude of B cell lineages to undergo somatic mutation in the draining LNs of NHPs.
Serum Antibody and Peripheral Blood CD4 T Cell Responses
Serum Ab titers were measured before and after immunization. Anti-eOD-GT8 60-mer IgG responses were present in all animals by day 7 post-immunization (Figure 5A). Anti-eOD-GT8-monomer IgG titers were also detectable after immunization (Figure 5A). The serum IgG titers were not significantly different between IM and s.c. immunized animals, but statistical power was limited by the group sizes (N = 4 each). Plasma eOD-GT8 avidity measurements were assessed by biolayer interferometry. A trend for lower avidity responses was detected in plasma at day 44 in IM immunized animals (Figure 5B). No neutralizing Ab responses were detected, as expected (Figure S4C).
Figure 5. Robust Systemic Antibody and CD4 T Cell Responses to OD-GT8 Nanoparticle Immunization.
(A) eOD-GT8 60-mer-specific (left) or eOD-GT8-monomer-specific (right) IgG response after one s.c. or IM immunization. Limit of detection (LOD) =1:100 plasma dilution. Geometric mean and geometric SD shown.
(B) Assessment of polyclonal avidity by biolayer interferometry. Faster dissociation rate (s−1) indicates lower avidity response. SD shown.
(C) eOD-GT8-specific CD4 T cell responses in PBMC (peripheral blood mononuclear cells) using the AIMOB assay before (day −7) and after (day 7, day 14) immunization. Gated on CD4 T cells. NS, not stimulated; eOD-GT8, eOD-GT8 peptide pool.
(D) Time course of the frequency (%) of eOD-GT8 AIMOB+ CD4 T cells after s.c. (black) or IM (red) immunization. Limit of detection (LOD) is the average frequency of AIMOB response in the unstimulated condition.
Each point represents an individual animal. n = 8, four per immunization condition at each time point.
See also Figures S5.
CD4 T cells are essential for GC responses and most neutralizing Ab responses to viruses (Crotty, 2019). We used AIM assays to quantify Ag-specific CD4 T cells in peripheral blood. Similar frequencies of eOD-GT8 60-mer-specific CD4 T cells were detected after both IM and s.c. immunization (day 7 and day 14; Figure 5C and 5D). eOD-GT8 60-mer-specific CXCR5+ CD4+ T cells were also present in peripheral blood (Figure S5). Thus, measurable Ab and CD4 T cell responses were elicited within 1 week of a primary immunization of NHPs with eOD-GT8 60-mer nanoparticles.
DISCUSSION
Rational vaccine development depends on the ability to measure multifaceted aspects of candidate vaccine immune responses that are likely to be involved in protective immunity and the ability to predict outcomes in human clinical trials. Here, we have tested whether LN FNAs can provide vaccine response outcomes and metrics in an NHP model at the level of longitudinal GC activity and quantitative assessment of Ag-specific BGC cells and somatic hypermutation (SHM) for a nanoparticle immunogen advancing to clinical trial. In this study, we show (1) the kinetics of a rapid GC response after a single nanoparticle immunization monitored by sequential LN FNAs; (2) LN drainage studies can provide assurance for accurately sampling LN immune response; (3) the induction of more vigorous BGC cell and GC-TFH cell responses after s.c. compared to IM immunization; and (4) induction of detectable systemic Ag-specific IgG and CD4 T cells responses after a single nanoparticle protein immunization. Overall, the robust CD4 T cell and BGC cell responses and SHM found here are encouraging for human eOD-GT8 60-mer clinical trial outcomes. Together with our recent report of HIV Env trimer immunized RMs (Cirelli et al., 2019), these studies show the analytical power of longitudinal LN FNAs for understanding and quantifying Ag-specific BGC cell responses.
Nanoparticle vaccines can enhance many aspects of immune responses (Kelly et al., 2019). Primary GC responses in the axillary LNs to the 60-mer nanoparticles after conventional IM immunization were substantial and lasted beyond day 21 in multiple animals. Primary GC responses after s.c. immunization were more impressive in magnitude. In the study reported here, the Ag-specific BGC cell response in axillary LNs was dramatically larger after s.c. immunization compared to IM immunization, with 18-fold more BGC cells (a 1,800% increase) generated in response to s.c. immunization.
In contrast, the peak circulating Ab response was similar between s.c. and IM immunized animals. This difference reveals an unexpected disconnect between LN GC activity and serum Ab concentrations. These results could suggest that important LN immune responses to IM deltoid immunization may occur outside of the axillary LN station. Alternatively, IM immunization may trigger a preferential induction of B cell plasma cell differentiation versus BGC cell differentiation. In addition, the differential B cell differentiation bias may be larger than measured in this study, due to the fact that as B cells transition to plasmablasts and then plasma cells, they downregulate surface BCRs; those Ag-specific cells may become undetected by flow cytometry. The data herein point to the importance of tracking the magnitude and kinetics of the plasma cell response in LNs in human clinical trials in addition to tracking GC responses.
GC-TFH cell responses peaked 14 days to 21 days after immunization, whereas Ag-specific CD4 T cells in the blood peaked 7 days after immunization. Immunization route did not seem to affect CD4 T cell responses after a single nanoparticle immunization. These results warrant further examination in future studies.
For the purposes of modeling clinical trial design and outcomes, the results from this study predict that a primary immunization of humans with eOD-GT8 60mers in a relatively strong adjuvant will likely elicit a significant serum IgG response and a measurable CD4 T cell response in peripheral blood. Moreover, these results also predict that Ag-specific GC responses will be detectable in axillary LNs for a period of multiple weeks. At face value, the BCR sequencing results indicate that, if a human VRC01-class B cell response occurs, it will be detectable in LNs if it represents more than 0.013% of the B cell response to eOD-GT8 60mers (see STAR Methods for calculation).
These predictions come with two caveats. First, a non-identical but saponin-containing adjuvant will be used in the human clinical trial. ISCOMATRIX induced strong GC and Ab responses in RMs (Havenar-Daughton et al., 2016a; Pauthner et al., 2017) and strong Ab responses in healthy human subjects (PMID: 19246990). AS01b induces potent Ab responses (Chlibek et al., 2013; Kester et al., 2009), but GC induction has not been characterized in NHPs or humans. Second, RM immunoglobulin gene loci are sufficiently divergent from humans that VRC01-class B cells were not observed. It is plausible that other aspects of the BCR repertoire are sufficiently different such that affinity or frequency difference in precursors of non-VRC01-class B cells would result in differences in immunodominant responses to eOD-GT8 60mers in humans compared to RMs (Abbott et al., 2018; Havenar-Daughton et al., 2017). Germline-targeting immunogens are only now being tested in humans. Therefore, there is currently no data for comparison between humans and RMs (Saunders et al., 2017), although immunodominance of Ab responses to influenza appear to be conserved between species (Angeletti et al., 2017).
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shane Crotty (shane@lji.org). All unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rhesus Macaques
Indian rhesus monkeys (RMs, Macaca mulatta) were used in this study. All animals were males between five and seven years of age, housed at the Yerkes National Primate Research Center, and maintained in accordance with NIH guidelines. These studies were approved by the Emory University Institutional Animal Care and Use Committee (IACUC). Animals were immunized with 100 μg of eOD-GT8 60-mer (Jardine et al., 2016a) and 75 U of ISCOMATRIX (CSL) in the left deltoid. Four animals received the immunization via intramuscular injection and the other four received a subcutaneous injection. The injections were in the same location in the deltoid are regardless of route of administration. At day 0 the immunization was unlabeled, and at week 6 the immunization the eOD-GT8 60-mer was labeled with alexa-647.
METHOD DETAILS
Fine needle aspirates of LNs and flow cytometry
Fine needle aspirates sampled both the right and left axillary LNs (LN) at weeks −2, 1, 2, 3, and 3.5, and 4. The skin over the LN region was clipped of hair and surgically scrubbed with 3 alternating applications of chlorhexidine or betadine scrub and alcohol. A palpable LN was stabilized and an FNA collected by passing a 22-gauge needle attached to a 3 mL syringe through the skin and into the LN 4 times. No suction was applied. The sample was expelled into media, and the needle was then flushed with media up to 3 times to collect all cells. Samples were kept on ice, centrifuged, and examined for blood contamination and if needed were treated with Ammonium-Chloride-Potassium (ACK) lysing buffer (Lonza). One day 21 ipsilateral axillary LN FNA failed in the SubQ animal group and therefore day 21 and day 26 B cell numbers were averaged for all animals in that group to obtain the mean calculations of Ag-specific BGC cell frequencies. LN FNA samples were stained phenotyping markers (Flow Cytometry Panel 1) for one hour before being washed, fixed with 1% formaldehyde, and acquired on a LSRII flow cytometer (BD Biosciences).
Whole LN biopsy tissue for eOD immunization study
At 48 hours post labeled immunization animals were sacrificed and lung hilar, pectoral, supratrochlear, cervical, and axillary LNs were collected. LN fat and connective tissue were removed and whole LN were shipped overnight at 4°C in R10 media RPMI+ 10% FBS + penicillin+ streptomycin+L-glutamine or PLP fixative. PLP fixative includes the following Soren’s phosphate buffer, 4% PFA, (Electron Microscopy Services) Lysine, and sodium periodate (Sigma).
Blood collection and processing
NaCitrate CPT tubes (BD Biosciences) were used for blood collection for PBMCs. Vacutainer serum separation tubes (SST; BD Biosciences) were used for serum collection.
LN drainage
LN drainage studies were conducted using two methods: EB and fluorescently labeled Ag. EB studies used 1 mL or 0.5 mL of 0.2% EB dye with or without 187.5 μg of an ISCOMs-class adjuvant was injected s.c. or i.m. in the deltoid or quadriceps regions. 24, 48 or 72 hours post injection draining and non-draining LN were removed and either photographed for analysis or prepared for dye extraction. For deltoid injections lung hilar, tracheobronchial supraclavicular, pectoral, supratrochlear, cervical, and inguinal axillary LNs were collected. For thigh injections inguinal, obturator iliac, internal iliac, external iliac, axillary, popliteal, para-aortic, inferior mesenteric, colic, para-colonic, cervical LNs were collected. Images of RM LNs were analyzed using computer program ImageJ. The total area of each LN was calculated by tracing along the perimeter of the lymphoid tissue with the Polygonal Lasso tool and using the Measure option to calculate the number of pixels in the enclosed area. The outline of the total drained area (stained dark blue) was similarly measured. The Drainage Score was calculated by dividing the sum of the measured drained areas by the sum of the total area of the corresponding LNs for each time point. LNs described above were harvested and fixed in freshly made 4% PFA for 24 h. Each LN was cut into small pieces and incubated in 0.5 mL of formamide for 24 h at 60 C to extract the dye. Fluorescence of the extracted dye in supernatant was measured (excitation at 620 nm, emission at 680 nm) using a Tecan Infinite M200 Pro plate reader and quantified using a standard curve. The extraction process recovered 77 ± 5% of LN-associated dye and the extraction efficiency is independent of amount of dye in LN.
Fluorescent Ag studies used 100 μg eOD-GT8 60-mer labeled with Alexa Fluor 647 and mixed with 187.5 μg of an ISCOMs-class adjuvant injected either intramuscularly or subcutanteously at the left deltoid in animals previous immunized with eOD-GT8 60-mer. LNs were removed 48h later and fluorescent quantification was determined by IVIS (Perkin Elmer). Left and right LNs were separately analyzed from the pectoral, axillary, cervical, and lung hilar sites.
Immunogen fluorescent labeling
eOD-GT8 60-mer nanoparticles were separately labeled with Amine Reactive Alexa Fluor 488 or Alexa Fluor 647 kits (Thermo Fisher Scientific).
Cell sorting of eOD-GT8 specific B cells
Previously frozen LN FNAs were thawed and washed in R10, then stained with fluorescently labeled eOD-GT8 in FACS buffer for 30 minutes at 4C (Flow Cytometry Panel 2). The samples were then stained with the remaining surface markers for an additional 30 minutes at 4C and washed with FACS buffer. 1,000 live B cells per sample that were positive for the eOD-GT8 probe in both colors were bulk sorted (Fusion, BD Biosciences) into 1ml of RPMI and spun at 600 × g for 10 minutes. The supernatant was removed and the cells vortexed with 350ul of 1% BME (Sigma-Aldrich, Cat #M3701) in RLT lysis buffer (QIAGEN, Cat #79216) before freezing at −80C.
AIM assay on PBMC and LN FNAs
Previously frozen LN FNAs or PBMCs were processed in the AIM assay as in Reiss et al. (2017). In brief, samples were thawed, LN FNAs were incubated with DNase for 15 minutes at 37°C, and washed with complete R10 media. All samples were divided into the following stimulation conditions: unstimulated (UN), stimulated with 5 μg/ml (per peptide) of 15-mer peptides spanning the eOD-GT8/lumazine construct, or stimulated with 100 pg/ml SEB for 18 hours at 37°C. The samples were then washed and stained with activation and phenotyping markers (Flow Cytometry Panel 3) for one hour before being washed twice, fixed with 1% formaldehyde, and acquired on a Fortessa flow cytometer (BD Biosciences).
Rhesus Immunoglobulin Repertoire Library Preparation and Sequencing
The protocol for RM repertoire sequencing was obtained by courtesy of Dr. Daniel Douek, NIAID/VRC. RNA was isolated using QIAGEN RNeasy kits (Valencia, CA). The input of cell for each animal was: RAp15: 500 cells, RQi15: 6400 cells, 20_12: 1700 cells, 163-12: 164 cells. Technical replicates were carried out for RQi15 and 20-12. Reverse transcription (RT) was performed using Clontech SMARTer cDNA template switching: 5′ CDS oligo(dT) (12 μM) was added to RNA and incubated at 72°C for 3 minutes and 4°C for at least 1 minute. The RT mastermix (5x RT Buffer (250 mM Tris-HCl (pH 8.3), 375 mM KCl, 30 mM MgCl2), Dithiothreitol, DTT (20 mM), dNTP Mix (10 mM), RNase Out (40U/μL), SMARTer II A Oligo (12 μM), Superscript II RT (200U/μL)) was added to the reaction and incubated at 42°C for 90 minutes and 70°C for 10 minutes. First-strand cDNA was purified using AMPure XP beads (catalog# A63882). Following RT, two PCR rounds were carried out to generate immunoglobulin amplicon libraries compatible with Illumina sequencing. All oligos were ordered from Integrated DNA Technologies. The first PCR amplification was carried out using KAPA Real-Time Library Amplification Kit (catalog# KK2702). cDNA was combined with master mix (2X KAPA PCR Master Mix, 12 μM μL 5PIIA and 5 μL IgG/IgK/IgL Constant Primer (2 μM)). The amplification was monitored using real-time PCR and was stopped during the exponential phase. The amplified products were again purified using AMPure XP beads. A second round of PCR amplification was carried out for addition of barcodes and Illumina adaptor sequences: master mix (2X KAPA PCR Master Mix 2x, SYBR Green 1:10K, Nuclease-free water), 10 μM of P5_Seq BC_XX 5PIIA, 10 μM of P7_ i7_XX IgG/IgK/IgL and were combined with amplified Immunoglobulin from the first round PCR and amplified using real-time PCR monitoring. The P5_Seq BC_XX 5PIIA primers contain a randomized stretch of four to eight random nucleotides followed by a barcode sequence. This was followed by purification with AMPure XP beads. A final PCR step was performed for addition of remaining Illumina adaptors by mixing master mix (2X KAPA PCR Master Mix, 10 μM P5_Graft P5_seq, Nuclease-free water), 10 μM of P7_ i7_XX IgG/IgK/IgL oligo and amplified products from the previous PCR step followed by purification with AMPure XP beads. The quality of library was assessed using Agilent Bio-analyzer. The amplicon libraries were pooled and sequenced on an Illumina MiSeq as a 309 paired-end run. Primers are listed in the Key Resources Table
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Fixable Viability Dye eFluor780 | Thermo Fisher Scientific | Cat #65-0865-18 |
| Viability Dye Aqua Blue | Invitrogen | Cat # L34965 |
| Mouse anti-human CD4 BV650 (clone: OKT-4) | BioLegend | Cat # 317436 |
| Mouse anti-human CD20 ECD (clone: B9E9) | Beckman Coulter | Cat # IM3607U |
| Mouse anti-human PD1 BV605 (clone: EH12.2H7) | BioLegend | Cat # 329924 |
| Mouse anti-human CD8a Qdot705 (clone: 3B5) | Thermo Fisher Scientific | Cat # Q10059 |
| Mouse anti-human Ki67 Alexa Fluor 700 (clone: B56) | BD Biosciences | Cat # 561277 |
| Mouse anti-human CXCR5 PE (clone: MU5UBEE) | Thermo Fisher Scientific | Cat # 12-9185-42 |
| Mouse anti-human Bcl6 BV421 (Clone: K112-91) | BD Biosciences | Cat # 563363 |
| Mouse anti-human CD95 PE-Cy5 (clone: DX2) | BD Biosciences | Cat # 559773 |
| Mouse anti-human IgG PE-Cy7 (clone: G18-145) | BD Biosciences | Cat # 561298 |
| Mouse anti-human CD4 BV421 (clone: OKT-4) | BioLegend | Cat # 317434 |
| Mouse anti-human CD20 PE-Cy7 (clone: 2H7) | Thermo Fisher Scientific | Cat # 25-0209-42 |
| Mouse anti-human CD38 PE (Clone: OKT10) | NHP Reagents | Cat # PR-3802 |
| Mouse anti-human CD71 PE-CF594 (Clone: L01.1) | BD Biosciences | Custom conjugate |
| Mouse anti-human CD8 APC eFluor 780 (clone: RPA-T8) | Thermo Fisher Scientific | Cat # 47-0088-42 |
| Mouse anti-human CD20 BV650 (Clone: 2H7) | BioLegend | Cat # 302336 |
| Mouse anti-human PD1 BV785 (Clone: EH12.2H7) | BioLegend | Cat # 329930 |
| Mouse anti-human CXCR5 PE-Cy7 (clone: MU5UBEE) | Thermo Fisher Scientific | Cat # 25-9185-42 |
| Mouse anti-human CD25 FITC (Clone: BC96) | BioLegend | Cat # 302604 |
| Mouse anti-human OX40 PE (Clone: L106) | BD Biosciences | Cat # 340420 |
| Mouse anti-human 4-1BB APC (Clone:4B4-1) | BioLegend | Cat # 309810 |
| Goat anti-rhesus IgG (H+L) - HRP | Southern Biotech | Cat # 6200-05 |
| Recombinant Ab Rh60mer1 HC | Genscript | Custom |
| QVQLVQSGAEVKKPGSSVKVSCTASGYTFIDYYIHWLRQAPRQGLEWMGWINPYNGNTKYGQKFQDRVTMTRDTSTNTAYMELSSLRSEDTAVYYCARETWIQMQLRGFEYWGQGVLVTVSS | ||
| Recombinant Ab Rh60mer1 LC | Genscript | Custom |
| QAALTQSPSVSGSPGQSVTISCTGHGSNIGGYNRVSWYQQHPGKAPKLMIYEVSNRPSGVSDRFSGSTSGNTASLIISGLQAEDEADYYCNSYAVFGGGTRLTVL | ||
| Recombinant Ab Rh60mer2 HC | Genscript | Custom |
| QVQLVQSGAEVKKPGSSVKVSCTASGYTFIDYYIHWLRQAPRQGLEWMGWINPYNGNTKYGQKFQDRVTMTRDTSTNTAYMELSSLRSEDTAVYYCARETWIQMQLRGFEYWGQGVLVTVSS | ||
| Recombinant Ab Rh60mer2 LC | Genscript | Custom |
| QAALTQSPSVSGSPGQSVTISCTGTSSDIGGYNRVSWYQQHPGKAPKLMIYEVSKRPSGVSDRFSGSKSGNTASLTISGLQAEDEADYYCNSYAVFGGGTRLTVL | ||
| Recombinant Ab Rh60mer3 HC | Genscript | Custom |
| QVQLVQSGAEVKKPGASVKVSCKASGFTFGSYGINWVRQAPGQGLEWMGGIVPLVGVTNYAQKFQGRVTITADTSTRTAYMELSSLRSEDMAVYYCVRCGQYSSWSIWYFDLWGPGTPITISS | ||
| Recombinant Ab Rh60mer3 LC | Genscript | Custom |
| AALTQSPSVSGSPGQSVTISCTGHGSNIGGYNRVSWYQQHPGKAPKLMIYEVSNRPSGVSDRFSGSTSGNTASLIISGLQAEDEADYYCNSYAVFGGGTRLTVL | ||
| Recombinant Ab Rh60mer4 HC | Genscript | Custom |
| QVQLVQSGAEVKKPGASVKVSCKASGFTFGSYGINWVRQAPGQGLEWMGGIVPLVGVTNYAQKFQGRVTITADTSTRTAYMELSSLRSEDMAVYYCVRCGQYSSWSIWYFDLWGPGTPITISS | ||
| Recombinant Ab Rh60mer4 LC | Genscript | Custom |
| QAALTQSPSVSGSPGQSVTISCTGTSSDIGGYNRVSWYQQHPGKAPKLMIYEVSKRPSGVSDRFSGSKSGNTASLTISGLQAEDEADYYCNSYAVFGGGTRLTVL | ||
| Recombinant Ab Rh60mer5 HC | Genscript | Custom |
| QVQLVQSGAEITQPGASVKLSCKASGYTFTGYYMHWVRQAPGQGLEWIGLIYPYKGDKGYAQNFQGRVTITTDTSTSTGYMELSSLRSEDTAVYYCTRDRGLDGFDYWGQGVLVTVSS | ||
| Recombinant Ab Rh60mer5 LC | Genscript | Custom |
| AALTQSPSVSGSPGQSVTISCTGHGSNIGGYNRVSWYQQHPGKAPKLMIYEVSNRPSGVSDRFSGSTSGNTASLIISGLQAEDEADYYCNSYAVFGGGTRLTVL | ||
| Recombinant Ab Rh60mer6 HC | Genscript | Custom |
| QVQLVQSGAEITQPGASVKLSCKASGYTFTGYYMHWVRQAPGQGLEWIGLIYPYKGDKGYAQNFQGRVTITTDTSTSTGYMELSSLRSEDTAVYYCTRDRGLDGFDYWGQGVLVTVSS | ||
| Recombinant Ab Rh60mer6 LC | Genscript | Custom |
| QAALTQSPSVSGSPGQSVTISCTGTSSDIGGYNRVSWYQQHPGKAPKLMIYEVSKRPSGVSDRFSGSKSGNTASLTISGLQAEDEADYYCNSYAVFGGGTRLTVL | ||
| Bacterial and Virus Strains | ||
| HXB2 ENV HIV pseudovirus | NIH AIDS Reagent Program | Cat # 1069 |
| HXB2 N276A ENV HIV pseudovirus | Mutant made by Burton Lab; Sok et al., 2016 | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| eOD-GT8 60-mer | Produced inhouse; Jardine et al., 2016a | NA |
| ISCOMATRIX | CSL | N/A |
| Soluble saponin-based ISCOMs-class adjuvant | Adjuvant made by Irvine Lab; Pauthner et al., 2019 | NA |
| Alexa Fluor 488 Protein Labeling Kit | Thermo Fisher Scientific | Cat # A10235 |
| Alexa Fluor 647 Protein Labeling Kit | Thermo Fisher Scientific | Cat # A20173 |
| eOD-GT8 lumazine overlapping peptides | A&A Labs LLC | Cat # 17127902 |
| Staphylococcal enterotoxin B (SEB) | Toxin Technology Inc. | Cat # BT202 |
| TMB Substrate | Thermo Fisher Scientific | Cat # 34021 |
| Evan’s Blue Dye | Millipore Sigma | Cat # E2129 |
| RLT lysis buffer | QIAGEN | Cat #79216 |
| Critical Commercial Assays | ||
| eBioscience FoxP3/ Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat # 00-5523-00 |
| RNeasy kits | QIAGEN | Cat #74104 |
| KAPA Real-Time Library Amplification Kit | Roche | Cat # KK2702 |
| Deposited Data | ||
| eOD-GT8 60-mer specific Bulk BCR sequencing reads | NCBI SRA | SRA: SRP199928 |
| Experimental Models: Cell Lines | ||
| TZM-bl cells | NIH AIDS Reagent Program | Cat #8129 |
| Experimental Models: Organisms/Strains | ||
| Indian-origin rhesus macaques (outbred) | Yerkes National Primate Research Center | N/A |
| Olionucleotides | ||
| CDS Oligo (dT): TTTTTTTTTTTTTTTTTTTTTTTTVN | Integrated DNA Technologies, Inc. | N/A |
| SMARTer II A Oligo: AAGCAGTGGTATCAACGCAGAGTACATrGrGrG | Integrated DNA Technologies, Inc. | N/A |
| IgG Constant Primer: GCCAGGGGGAAGACCGATGGGCCCTTGGTGGA | Integrated DNA Technologies, Inc. | N/A |
| IgK Constant Primer: GCGGGAAGATGAAGACAGATGGTGCAGCCACAG | Integrated DNA Technologies, Inc. | N/A |
| IgL Constant Primer: GGCCTTGTTGGCTTGAAGCTCCTCAGAGGAGGG | Integrated DNA Technologies, Inc. | N/A |
| P5_Seq BC_XX 5PIIA: CACGACGCTCTTCCGATCT4-8xNAACCACTA AAGCAGTGGTATCAACGCAGAGT | Integrated DNA Technologies, Inc. | N/A |
| P7_ i7_XX IgG: CAAGCAGAAGACGGCATACGAGATTAGTGGTT GCCAGGGGGAAGACCGATGGGCCCTTGGTGGA | Integrated DNA Technologies, Inc. | N/A |
| P7_ i7_XX IgK: CAAGCAGAAGACGGCATACGAGATTAGTGGTT GCGGGAAGATGAAGACAGATGGTGCAGCCACAG | Integrated DNA Technologies, Inc. | N/A |
| P7_ i7_XX IgL: CAAGCAGAAGACGGCATACGAGATTAGTGGTT GGCCTTGTTGGCTTGAAGCTCCTCAGAGGAGGG | Integrated DNA Technologies, Inc. | N/A |
| P5_Graft P5_seq: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT | Integrated DNA Technologies, Inc. | N/A |
| Software and Algorithms | ||
| Prism v7.0/v8.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo v10.4 | FlowJo LLC | https://www.flowjo.com |
| ImageJ | National Institute of Health | https://imagej.nih.gov/ij/ |
| pRESTO | https://presto.readthedocs.io/en/latest/ | |
| ChangeO | https://changeo.readthedocs.io/en/stable/ | |
| Alakazam | https://alakazam.readthedocs.io/en/stable/ | |
| SHazaM | https://shazam.readthedocs.io/en/stable/ | |
| seqtk | https://github.com/lh3/seqtk | |
| MUSCLE | https://www.drive5.com/muscle/ | |
| EMBOSS 6.6.0.0 | ftp://emboss.open-bio.org/pub/EMBOSS/ | |
| CD-HIT | http://weizhongli-lab.org/cd-hit/ | |
| IgBLAST | https://ncbi.github.io/igblast/ | |
| blastn | ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ | |
| DomainGapAlign | IMGT® | http://www.imgt.org/IMGTindex/IMGTDomainGapAlign.php |
| parallel | https://www.gnu.org/software/parallel/ | |
| Other | ||
| Repertoire analysis and resources | This paper 10.5281/zenodo.3239472 |
https://github.com/BosingerLab/NHP_eOD-GT8_manuscript |
| Corning® 96 Well Half-Area Microplate | Sigma Millipore | Cat # CLS3690 |
Sequence analysis
The repertoire sequence analysis was carried out using the pRESTO 0.5.8, Change-O 0.4.1, Alakazam 0.2.11 and SHazaM 0.1.11 packages from the Immcantation pipeline. The pre-processing was performed using tools in the pRESTO package. The paired-end reads were first assembled with AssemblePairs tool. Reads with a mean quality score of less than 20 were filtered out using Filter-Seq. The MaskPrimers tool was used to remove the forward primers and the random nucleotides from the assembled sequences. The technical replicates were then combined. Duplicates were removed and the duplicate counts obtained for each sequence using CollapseSeq. SplitSeq was used to select sequences that had duplicate counts of at least two. Seqtk v1.2 was used to obtain fasta sequences from fastq files.
The pre-processed sequences were then annotated using IgBLAST v1.6.1. Since the IMGT database (Brochet et al., 2008) is lacking several V genes, a custom IgBLAST database was created for V genes by combining sequences from IMGT database and previously published studies (Cirelli et al., 2019; Corcoran et al., 2016; Sundling et al., 2012), IMGT database and sequences from the PacBio assembly. The protein sequences for V genes from all these datasets were using EMBOSS 6.6.0.0 and combined. The sequences were aligned using MUSCLE v3.8.1551 and only the V genes with complete sequence and no unknown amino acid (X) were selected. The corresponding nucleotide sequences of these V genes were clustered using CD-HIT v4.7 to remove 100% redundant sequences. The protein sequences for this non-redundant set were submitted to the IMGT DomainGapAlign tool to obtain gapped V sequences. Corresponding gaps were introduced in the nucleotide sequences and the positions for framework (FR) and complementarity-determining regions (CDR) regions determined using custom scripts. These sequences were used to create the IgBLAST database for V genes. The family for V genes was based on the previous annotation or the closes V gene family of the closest hit. The names were assigned arbitrarily based on clustering at 95% sequence identity. The databases for J and D genes was obtained from the IgBLAST ftp site (ftp://ftp.ncbi.nih.gov/blast/executables/igblast/release/internal_data/rhesus_monkey/). The annotations from IgBLAST were saved into a Change-O database and functional sequences were selected. The functional sequences were assigned to a clone based on the following criteria: (i) same V gene, (ii) same J gene, (iii) same CDR3 length and (iv) percentage identity of CDR3 nucleotide sequence > 85%. The gene usage and clonal frequencies were obtained from the Alakazam package and SHM was obtained from the SHazaM package. Blastn v2.6.0 was used to obtain sequences in the repertoire that were similar to the VH1 family genes. The gnu parallel tool was used for running parallel processes.
Lineage assignment was performed, as in Cirelli et al. (2019) by clustering on the similarity of the inferred germline sequence, as well as the mature sequence itself. Two sequences potentially share a lineage when two criteria are met: 1) When their inferred UCA sequences (masking away the junction and D region) are 99% identical (exploiting a kmer-based distance approximation for efficiency). This allows the clustering to tolerate mistaken V or J assignments, if the assigned V or J is close enough to the correct one. 2) When the length-normalized Levenshtein distance between their junction+D sequences is within 10%. The clustering algorithm maintains a set of candidate lineages, storing all sequences for each lineage, and each additional sequence is added to the lineage where the largest proportion of sequences match the above two criteria. If no existing candidate cluster matches a sequence with > 50% of its reads, then that sequence is used to seed a new candidate cluster. Where members of a lineage have different inferred UCA sequences, the most frequent among them was chosen as the UCA for the entire lineage. This lineage clustering algorithm was implemented in the Julia language for scientific computing (v0.6.2). Lineages were each aligned with MAFFT, and maximum likelihood phylogenetic trees were inferred using FastTree2. Phylogenies were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/), using automated annotation routines implemented in Julia.
eOD-GT8 ELISA
96 well half area plates (Corning) were coated overnight at 4C with 2.5 μg/ml eOD-GT8 60-mer in PBS. The following morning, plates were washed 5 times with wash buffer (0.1% Tween 20 in PBS), then filled with 100 μL per well of blocking buffer (3% BSA in PBS; Sigma-Aldrich) for one hour at room temperature. Three-fold serial dilutions of RM plasma from weeks −2, 1, 2, 3, 3.5, and 4 weeks post immunization and at necropsy were prepared, beginning at 1:100 in blocking buffer. Blocking buffer was removed from the plate and 50 μL of each serial dilution was added per well for 2 hours at room temperature. The plates were then washed 5 times with wash buffer. Anti-rhesus IgG (H+L)-HRP secondary antibody (Southern Biotech) was diluted 1:10,000 in blocking buffer and incubated in the plate for 1 hour at room temperature. The plate was then washed 5 times, then 50 μL of Pierce TMB substrate kit was added to each well. After 10 minutes the reaction was stopped with 50 μl/well of 2N H2SO4 and the plate was analyzed on a Spectramax plate reader at 450nm. Limit of detection was set at 1:100 the lowest plasma dilution tested.
Biolayer Interferometry
RM plasma was heat inactivated at 37°C for 30 minutes, then analyzed with a ForteBio Octet RED96 System. 10 μg/ml of biotinylated eOD-GT8 monomer was immobilized on streptavidin biosensors for 300 s. After immobilization, baseline interference was read for 300 s in kinetic buffer (0.01% BSA and 0.02% Tween-20 in PBS). The sensors were then exposed to RM plasma diluted 1:20 in kinetic buffer for 900 s, then transferred to kinetic buffer and all kinetic interactions were measured over the course of 1800 s. Dissociation curves were background subtracted with readings from a buffer-only reference well and aligned to the dissociation step, with interstep correction using Octet data analysis software (Klasse, 2014). Observed binding curves were fit globally to a 1:1 binding model and the equilibrium dissociation constant (KD) was calculated from the fitted curves.
Envelope pseudovirus production
Envelope pseudoviruses were generated through the cotransfection of the pSG3ΔEnv backbone plasmid (obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) and a plasmid encoding the full Env gp160 in a 3:1 ratio in HEK293T cells (ATCC) using the PEIMAX transfection reagent (Polysciences). Following 48 hours, the media was filtered through a 0.45 μm Steriflip unit (EMD Millipore), aliquoted, frozen and titrated.
Neutralization assay
The neutralization assay (Sarzotti-Kelsoe et al., 2014) was performed with DMEM (Corning) supplemented with 10% FBS (Corning), 1% L-glutamine (Corning), 0.5% Gentamicin (Sigma), 2.5% HEPES (GIBCO) and all incubations were performed at 37°C, 5% CO2. On day one, 25 μL diluted pseudovirus mixed with 25 μL of 1:3 serially-diluted serum or control antibody was incubated for 1 hour, followed by the addition of 20 μL of TZM-bl cells at a concentration of 500,000 cells/mL with DEAE-Dextran at a final concentration of 40 μg/mL. On day two, following incubation for 24 hours, 130 μL of supplemented DMEM was added and the samples which then incubated overnight. Finally, on day three after another 24 hours, the media was completely removed and 60 μL lysis buffer together with 60 μL luciferase substrate (Bright-Glo, Promega) was added per well, and luminescence was measured on the Synergy2 plate reader (BioTek). Neutralization data is reported as ID50 or IC50 (μg/mL) values which was calculated as the dilution or concentration at which a 50% reduction in infection was observed. Neutralization assays were performed in duplicate and SEM is reported. The TZM-bl cell line engineered from CXCR4-positive HeLa cells to express CD4, CCR5, and a firefly luciferase reporter gene (under control of the HIV-1 LTR) was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (developed by Dr. John C. Kappes, and Dr. Xiaoyun Wu.
QUANTIFICATION AND STATISTICAL ANALYSIS
Calculations
LN FNAs recover an average of 1 million cells (Figure S2A). 50% are B cells so 500,000 total B cells. 2% of B cells are eOD-GT8+ after one IM immunization, so 10,000 eOD-GT8 specific B cells. Bulk sequencing was 79% efficient for recover of HC sequences compared to B cell input numbers. Therefore, if VRC01-class B cells are present in the LN post immunization at greater than 0.013%, we would expect to find this sequence.
Statistical analyses
Prism 7 (GraphPad) was used for all statistical analyses. Two-tailed unpaired t tests were used to compare differences between LN draining data. Two-tailed paired t tests were used to compare differences in response at various time points before and after immunization within the IM or SubQ group. One-tailed paired t tests were used to compare differences in response at various time points between the no adjuvant and ISCOMATRIX adjuvant groups. Paired T one-tailed tests were used to test for increases in GC and CD4 T cell responses after immunization. Mann-Whitney two-tailed tests were used to compare differences in responses between IM and SubQ immunized groups.
DATA AND CODE AVAILABILITY
The accession number for the BCR sequencing datasets reported in this paper are available at NCBI Sequence Read Archive: SRP199928.
Supplementary Material
FLOW CYTOMETRY PANEL 1 FOR LN FNA STAINING
| Marker | Color | Clone | Company |
|---|---|---|---|
| eOD-GT8 60-mer | Ax647 | N/A | N/A |
| eOD-GT8 60-mer | Ax488 | N/A | N/A |
| CD4 | BV650 | OKT-4 | Biolegend |
| CD20 | ECD | B9E9 | Beckman Coulter |
| PD-1 | BV605 | EH12.2H7 | Biolegend |
| CD8 | Qdot 705 | 3B5 | Invitrogen |
| Viability Dye | Aqua Blue | N/A | Invitrogen |
| KI67 | Ax700 | B56 | BD Bioscience |
| BCL6 | BV 421 | K112-91 | BD Bioscience |
| CXCR5 | PE | Mu5UBEE | eBioscience |
| CD95 | PE-Cy5 | DX2 | BD Bioscience |
| IgG | PE-Cy7 | G18-145 | BD Bioscience |
FLOW CYTOMETRY PANEL 2 FOR B CELL SORTING
| Marker | Color | Clone | Company |
|---|---|---|---|
| eOD-GT8 60-mer | Ax647 | N/A | N/A |
| eOD-GT8 60-mer | Ax488 | N/A | N/A |
| CD4 | BV421 | OKT-4 | BioLegend |
| CD20 | PE-Cy7 | 2H7 | Thermo Fisher |
| CD38 | PE | OKT10 | NHP Resource |
| CD71 | PE-CF594 | L10.1 | BD Bioscience |
| Viability Dye | APCe780 | N/A | Thermo Fisher |
| CD8 | APCe780 | RPA-T8 | Thermo Fisher |
FLOW CYTOMETRY PANEL 3 FOR AIM ASSAY
| Marker | Color | Clone | Company |
|---|---|---|---|
| CD4 | BV421 | OKT-4 | BioLegend |
| CD20 | BV650 | 2H7 | BioLegend |
| PD-1 | BV785 | EH12.2H7 | BioLegend |
| CXCR5 | PE-Cy7 | MU5UBEE | Thermo Fisher |
| CD8 | APCe780 | RPA-T8 | Thermo Fisher |
| Viability Dye | APCe780 | N/A | Thermo Fisher |
| OX40 | PE | L106 | BD Bioscience |
| 4-1BB | APC | 4B4-1 | BioLegend |
| CD25 | FITC | BC96 | BioLegend |
Highlights.
Rapid immunogen-specific lymph node responses detected at day 7 post-immunization
Lymph node immune response kinetics can be longitudinally tracked
Greater germinal center responses after subcutaneous versus intramuscular immunization
Immunization injection method substantially impacts immune responses in local LNs
ACKNOWLEDGMENTS
We thank Jennifer Wood and Yerkes veterinary staff for their excellent technical assistance and LN fine needle aspirate procedures. We thank the Flow Cytometry Core at the La Jolla Institute for Immunology for expert cell sorting. This work was supported by the NIH NIAID 1UM1Al100663; Scripps CHAVI-ID and UM1AI44462; Scripps CHAVD (S.C., G.S., D.J.I., W.R.S., and D.R.B.); National Primate Research Center funding (P51 RR000165/OD011132 to the Yerkes National Primate Research Center); and NIH NIAID P01A048240 to D.J.I. The FACSAria II Cell Sorter was acquired through the NIH Shared Instrumentation Grant (SIG) Program (S10 RR027366). D.J.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.008.
DECLARATIONS OF INTEREST
IAVI and the Scripps Research Institute have filed a patent relating to the eOD-GT8 immunogens in this manuscript, which included inventor W.R.S. W.R.S. is a cofounder and stockholder in CompuVax, Inc., which has programs in non-HIV vaccine design that might benefit indirectly from this research.
REFERENCES
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, et al. (2017). Defining B cell immunodominance to viruses. Nat. Immunol 18, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Tomaka FL, Wegmann F, Stieh DJ, Alter G, Robb ML, Michael NL, Peter L, Nkolola JP, Borducchi EN, et al. (2018). Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19). Lancet 392, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, Jacak R, Kalyuzhniy O, de Val N, Sesterhenn F, et al. (2016). Tailored Immunogens Direct Affinity Maturation toward HIV Neutralizing Antibodies. Cell 166, 1459–1470.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc M-P, and Giudicelli V (2008). IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–W508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR (2017). What Are the Most Powerful Immunogen Design Vaccine Strategies? Reverse Vaccinology 2.0 Shows Great Promise. Cold Spring Harb. Perspect. Biol 9, a030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlibek R, Bayas JM, Collins H, de la Pinta MLR, Ledent E, Mols JF, and Heineman TC (2013). Safety and Immunogenicity of an AS01-adjuvanted Varicella-zoster Virus Subunit Candidate Vaccine Against Herpes Zoster in Adults ≥50 Years of Age. J. Infect 208, 1953–1961. [DOI] [PubMed] [Google Scholar]
- Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, Enemuo CA, Gebru EH, Choe Y, Viviano F, et al. (2019). Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell 177, 1153–1171.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran MM, Phad GE, Vázquez Bernat N, Stahl-Hennig C, Sumida N, Persson MAA, Martin M, and Karlsson Hedestam GB (2016). Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat. Commun 7, 13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, et al. (2014). Proof of principle for epitope-focused vaccine design. Nature 507, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2019). T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 50, 1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar-Daughton C, Reiss SM, Brigger M, Bothwell M, Sette A, and Crotty S (2016). A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. J. Immunol 197, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, et al. (2016). Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell 166, 1445–1458.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Carnathan DG, Torrents de la Peña A, Pauthner M, Briney B, Reiss SM, Wood JS, Kaushik K, van Gils MJ, Rosales SL, et al. (2016a). Direct Probing of Germinal Center Responses Reveals Immunological Features and Bottlenecks for Neutralizing Antibody Responses to HIV Env Trimer. Cell Rep. 17, 2195–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Reiss SM, Carnathan DG, Wu JE, Kendric K, Torrents de la Peña A, Kasturi SP, Dan JM, Bothwell M, Sanders RW, et al. (2016b). Cytokine-Independent Detection of Antigen-Specific Germinal Center T Follicular Helper Cells in Immunized Nonhuman Primates Using a Live Cell Activation-Induced Marker Technique. J. Immunol 197, 994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Lee JH, and Crotty S (2017). Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem. Immunol. Rev 275, 49–61. [DOI] [PubMed] [Google Scholar]
- Havenar-Doughton C, Abbott RK, Schief WR, and Crotty S (2018). When designing vaccines, consider the starting material: the human B cell repertoire. Curr. Opin. Immunol 53, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien J-P, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang P-S, MacPherson S, Jones M, et al. (2013). Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ererño-Orbea J, Kalyuzhniy O, Deresa I, et al. (2016a). HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Sok D, Julien J-P, Briney B, Sarkar A, Liang C-H, Scherer EA, Henry Dunand CJ, Adachi Y, Diwanji D, et al. (2016b). Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design. PLoS Pathog. 12, e1005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly HG, Kent SJ, and Wheatley AK (2019). Immunological basis for enhanced immunity of nanoparticle vaccines. Expert Rev. Vaccines 18, 269–280. [DOI] [PubMed] [Google Scholar]
- Kester KE, Cummings JF, Ofori-Anyinam O, Ockenhouse CF, Krzych U, Moris P, Schwenk R, Nielsen RA, Debebe Z, Pinelis E, et al. (2009). Randomized, Double-Blind, Phase 2a Trial of Falciparum Malaria Vaccines RTS,S/AS01B and RTS,S/AS02A in Malaria-Naive Adults: Safety, Efficacy, and Immunologic Associates of Protection. J., Infect 200, 337–346. [DOI] [PubMed] [Google Scholar]
- Klasse PJ (2014). Neutralization of Virus Infectivity by Antibodies: Old Problems in New Perspectives. Adv. Biol 2014, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, and Kelsoe G (2016). Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity 44, 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD (2017). What Are the Most Powerful Immunogen Design Vaccine Strategies? A Structural Biologist’s Perspective. Cold Spring Harb. Perspect. Biol 9, a029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, and Stamatatos L (2014). HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science 346, 1380–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TJ, Zmolek AC, and Irvine DJ (2016). Beyond antigens and adjuvants: formulating future vaccines. J. Clin. Invest 126, 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity 46, 1073–1088.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prévost J, Nedellec R, von Bredow B, Abbink P, et al. (2019). Vaccine-Induced Protection from Homologous Tier 2 SHIV Challenge in Nonhuman Primates Depends on Serum-Neutralizing Antibody Titers. Immunity 50, 241–252.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S, Baxter AE, Cirelli KM, Dan JM, Morou A, Daigneault A, Brassard N, Silvestri G, Routy J-P, Havenar-Daughton C, et al. (2017). Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS One 12, e0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al. (2013). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin C-L, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, et al. (2014). Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders KO, Verkoczy LK, Jiang C, Zhang J, Parks R, Chen H, Housman M, Bouton-Verville H, Shen X, Trama AM, et al. (2017). Vaccine Induction of Heterologous Tier 2 HIV-1 Neutralizing Antibodies in Animal Models. Cell Rep. 21, 3681–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Briney B, Jardine JG, Kulp DW, Menis S, Pauthner M, Wood A, Lee E-C, Le KM, Jones M, et al. (2016). Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Pancera M, and McGuire AT (2017). Germline-targeting immunogens. Immunol. Rev 275, 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, et al. (2016). HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity 45, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O’Dell S, Feng Y, Mascola JR, Wyatt RT, and Karlsson Hedestam GB (2012). High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Science Translational Medicine 4, 142ra96–142ra96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas JM, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill JC, Reynaud CA, Browne EP, et al. (2016). Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Cheng C, Chen X, Duan H, Cheng H-L, Dao M, Sheng Z, Kimble M, Wang L, Lin S, et al. (2016). Induction of HIV Neutralizing Antibody Lineages in Mice with Diverse Precursor Repertoires. Cell 166, 1471–1484.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdorovich V, Oliver BG, Carbonetti S, Dambrauskas N, Lange MD, Yacoob C, Leahy W, Callahan J, Stamatatos L, and Sather DN (2016). Repertoire comparison of the B-cell receptor-encoding loci in humans and rhesus macaques by next-generation sequencing. Clin. Transl. Immunology 5, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-L, and Lai TW (2014). Optimization of Evans blue quantitation in limited rat tissue samples. Sci. Rep 4, 6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Fernandez C, Alcantara S, Bailey M, De Rose R, Kelleher AD, Zaunders J, and Kent SJ (2013). Serial study of lymph node cell subsets using fine needle aspiration in pigtail macaques. J. Immunol. Methods 394, 73–83. [DOI] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang G-Y, Joyce MG, et al. ; NISC Comparative Sequencing Program (2013). Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the BCR sequencing datasets reported in this paper are available at NCBI Sequence Read Archive: SRP199928.