Abstract
Terbinafine N-dealkylation pathways result in formation of 6,6-dimethyl-2-hepten-4-ynal (TBF-A), a reactive allylic aldehyde, that may initiate idiosyncratic drug induced liver toxicity. Previously, we reported on the importance of CYP2C19 and 3A4 as major contributors to TBF-A formation. In this study, we expanded on those efforts to assess individual contributions of CYP1A2, 2B6, 2C8, 2C9, and 2D6 in terbinafine metabolism. The combined knowledge gained from these studies allowed us to scale the relative roles of the P450 isozymes in hepatic clearance of terbinafine including pathways leading to TBF-A, and hence, provide a foundation for assessing their significance in terbinafine-induced hepatotoxicity. We used in vitro terbinafine reactions with recombinant P450s to measure kinetics for multiple metabolic pathways and calculated contributions of all individual P450 isozymes to in vivo hepatic clearance for the average human adult. The findings confirmed that CYP3A4 was a major contributor (at least 30% total metabolism) to all three of the possible N-dealkylation pathways; however, CYP2C9, and not CYP2C19, played a critical role in terbinafine metabolism and even exceeded CYP3A4 contributions for terbinafine N-demethylation. A combination of their metabolic capacities accounted for at least 80% of the conversion of terbinafine to TBFA, while CYP1A2, 2B6, 2C8, and 2D6 made minor contributions. Computational approaches provide a more rapid, less resource-intensive strategy for assessing metabolism, and thus, we additionally predicted terbinafine metabolism using deep neural network models for individual P450 isozymes. Cytochrome P450 isozyme models accurately predicted the likelihood for terbinafine N-demethylation, but overestimated the likelihood for a minor N-denaphthylation pathway. Moreover, the models were not able to differentiate the varying roles of the individual P450 isozymes for specific reactions with this particular drug. Taken together, the significance of CYP2C9 and 3A4 and to a lesser extent, CYP2C19, in terbinafine metabolism is consistent with reported drug interactions. This finding suggests that variations in individual P450 contributions due to other factors like polymorphisms may similarly contribute terbinafine-related adverse health outcomes. Nevertheless, the impact of their metabolic capacities on formation of reactive TBF-A and consequent idiosyncratic hepatotoxicity will be mitigated by competing detoxification pathways, TBF-A decay, and TBF-A adduction to glutathione that remain understudied.
Keywords: terbinafine, reactivity, liver toxicity, bioactivation, TBF-A, model
1.). INTRODUCTION
Lamisil (terbinafine) is a fungicidal drug used for the treatment of infections in fingernails and toenails. The death of the fungus occurs due to inhibition of fungal squalene monooxygenase activity, resulting in an ergosterol deficiency and a toxic level of squalene that impairs cell membrane function and cell wall synthesis (Ryder, 1992). The effectiveness of this drug drives its extensive use. In the United States alone, 1.5 million prescriptions were filled in 2010 according to IMS Health data (Napodano, 2012); however, terbinafine use poses a rare risk of idiosyncratic liver toxicity. For one in 45,000–54,000 patients (“Terbianfine Product Monograph, Canada,” 1995), these adverse reactions can be severe and may lead to liver failure and transplantation (Perveze et al., 2007) or death (Song & Deresinski, 2016). Severe adverse drug events are reduced through precautionary liver function tests for patients after one month of treatment (Ajit, Zaeri, Munoz, & Suvannasankha, 2005). The detection of liver failure leads to discontinuance of the drug and typically, a return to normal liver function after 2–12 months (Choudhary, Kotecha, Saraf, Gautam, & Saigal, 2014). Currently, there are no predictive approaches for identifying patients at risk for idiosyncratic liver toxicity due to deficient knowledge of the underlying mechanism and factors impacting the onset and severity of terbinafine toxicity.
Initial studies on terbinafine metabolism revealed a complex metabolic profile (Battig, Nefzger, & Schulz, 1987) (Vickers, Sinclair, Zollinger, Heitz, Glänzel, et al., 1999). A secondary study by Iverson et al. lead to the detection of a metabolite with the potential to cause hepatocellular damage (Iverson & Uetrecht, 2001). Terbinafine N-dealkylation can yield a reactive aldehyde, 6,6-dimethyl-2-hepten-4-ynal (TBF-A) based on in vitro reactions in human liver microsomes. This reactive aldehyde can reversibly conjugate with glutathione through 1,6-Michael addition potentiating off-site toxicity. As reported for α-naphthyl isothiocyanate (Roth & Dahm, 1997), terbinafine induces hepatotoxicity likely through generation of a reactive metabolite (TBF-A) that binds glutathione to form a reversible adduct capable of transport into the bile duct. Once there, TBF-A adducts hepatobiliary proteins, such as transporters, to compromise bile acid transport resulting in cholestatic hepatitis (Iverson & Uetrecht, 2001). Knowledge of the pathways and enzymes responsible for generation of TBF-A and the subsequent capacity to drive this mechanism among patients remained unknown.
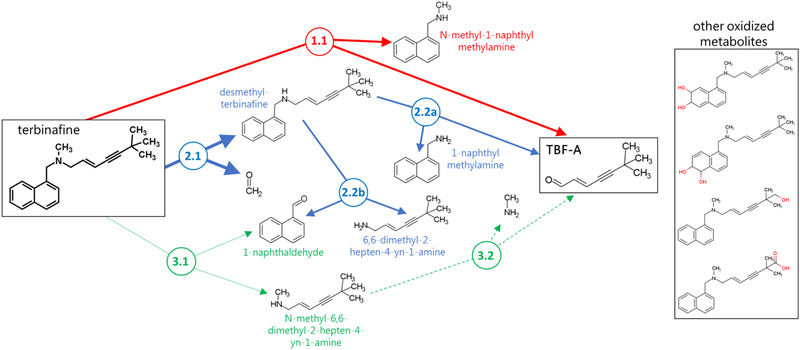
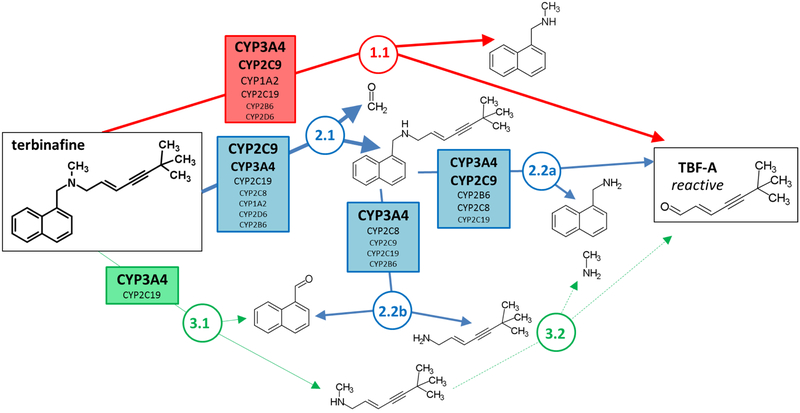
Recently, we identified two of three possible N-dealkylation pathways as significant contributors to TBF-A formation by reactions with human liver microsomes and through computational metabolic modeling (Pathways 1 and 2, Fig. 1) (Barnette et al., 2018). Pathway 1(red) led directly to TBF-A while Pathways 2 (blue) and 3 (green) required a two-step process for generation of TBF-A. A deep learning microsomal model predicted the preference for N-demethylation over N-denaphthylation but was not able to accurately predict the importance of direct TBF-A formation (Pathway 1). In a subsequent study (Davis et al., 2019), P450-specific chemical inhibitor phenotyping identified roles for eight P450 isozymes in one or more N-dealkylation pathways. CYP2C19 and 3A4 catalyzed the first step in all three pathways making them ideal for in depth steady-state analyses with recombinant isozymes. CYP2C19 and 3A4 similarly catalyzed N-dealkylation that directly yielded TBF-A (Pathway 1). Nevertheless, N-demethylation and other steps in Pathway 2 were all more efficiently catalyzed by CYP2C19 when compared to CYP3A4. Unlike microsomal studies, N-denaphthylation was surprisingly efficient for CYP2C19 and 3A4. Overall, CYP2C19 was the most efficient but CYP3A4 was more selective for steps leading to TBF-A. CYP3A4 was then more effective at terbinafine bioactivation based on analyses using metabolic split ratios for competing pathways. Computational model predictions do not extrapolate to quantitative kinetic constants, yet results for CYP3A4 agreed qualitatively with preferred reaction steps and pathways. The clinical relevance of CYP3A4 in terbinafine metabolism is bolstered with reports on drug interactions (“Lamisil,” 2004)(Rodrigues, 2008), while that for CYP2C19 remains understudied. CYP2C19 and 3A4 were chosen for in-depth analysis in the previous study because of their involvement in all three N-dealkylation pathways; however, the importance of the other six isozymes that only catalyze one or two of the pathways remains unclear.
Fig. 1. N-Dealkylation pathways of terbinafine leading to formation of reactive TBF-A.
Three N-dealkylation pathways for terbinafine yield TBF-A. Pathway 1 (red) is the dominant pathway and consists of a single reaction yielding TBF-A and N-methyl-1-naphthyl methylamine as a co-metabolite (Step 1.1). Pathway 2 (blue) is also a major pathway and is a two-step pathway yielding first desmethyl-terbinafine and formaldehyde via very efficient N-demethylation (Step 2.1), followed by less efficient generation of 1-naphthyl methylamine and TBF-A from desmethyl-terbinafine (Step 2.2a) and 1-naphthaldehyde through a competing reaction (Step 2.2b). Pathway 3 (green) has minor significance and is a two-step pathway first yielding naphthaldehyde and N-methyl-6,6-dimethyl-2-hepten-4-yn-1-amine (Step 3.1), which undergoes N-dealkylation to yield TBF-A (Step 3.2).
Herein, we combined experimental kinetics and computational modeling to assess the importance of CYP1A2, 2A6, 2B6, 2C8, 2C9, and 2D6 in N-dealkylation pathways for terbinafine leading to TBF-A. We determined steady-state kinetic mechanisms and constants for terbinafine metabolism to yield TBF-A by recombinant P450 Supersomes (Pathways 1, 2, and 3, Fig. 1). Experimental studies required significant time, effort and resources and thus, we explored more rapid, easily accessible model predictions for preferred TBF-A pathways among P450 isozymes (Zaretzki, Browning, Hughes, & Swamidass, 2015). Deep learning models predicted the likelihood for metabolic reactions to occur and when combined, provide insights into each isozyme preference for competing N-dealkylation pathways leading to TBF-A. We then compared model predictions to our experimental findings for assessing performance by individual P450 modeled reactions. Due to model scaling challenges, only in vitro kinetic data could be extrapolated to in vivo clearance and bioactivation of terbinafine. We leveraged kinetic information from this study and a previous one (Davis et al., 2019) to analyze the complete complement of hepatic P450 isozymes in terbinafine metabolism using isozyme abundance in human liver microsomes measured by others (Kawakami et al., 2011). Overall, this thorough assessment of the roles of P450 isozymes in TBF-A formation provides a basis for determining possible sources of variability in TBF-A bioaccumulation, which could account for the idiosyncrasy of terbinafine-induced hepatotoxicity events.
2.). MATERIALS AND METHODS
2.1). Materials
All chemical solvents were purchased from Thermo Fisher Scientific (Waltham, MA). Substrate terbinafine hydrochloride and metabolites N-desmethyl-terbinafine hydrochloride, N-methyl-1-naphthyl methylamine hydrochloride, 1-naphthyl methylamine, naphthoic acid, and 1-napthaldehyde were purchased from Millipore-Sigma (Burlington, MA), while E-6,6-dimethylhept-2-ene-4-ynal (TBF-A) was obtained from Toronto Research Chemicals (Toronto, ON, Canada). Labeling agents dansyl chloride and dansyl hydrazine and internal standards fluoxetine hydrochloride and dimethyl benzaldehyde were purchased from Millipore-Sigma. Human liver microsomes pooled from 150 donors (HLM150) and 50 donors (HLM50) were purchased from Corning Gentest (Woburn, MA) and Sekisui XenoTech (Kansas City, KS), respectively. Recombinant Supersomes CYP1A2, 2A6, 2B6, 2C8, 2C9, and 2D6 were purchased from Corning Gentest. For NADPH regenerating system, glucose-6 phosphate dehydrogenase, glucose 6-phosphate, and NADP disodium salt were purchased from Sigma-Aldrich, and MgCl2 was purchased from Thermo Fisher Scientific. Isozyme inhibitors α-naphthoflavone, (+)-N-3-benzylnirvanol, and ketoconazole were also purchased from Millipore-Sigma.
2.2). Recombinant isozyme kinetic assays
In vitro reactions using recombinant CYP1A2, 2A6, 2B6, 2C8, 2C9, and 2D6 were carried out as described previously for CYP2C19 and 3A4 (Davis et al., 2019). Briefly, 100 nM recombinant Supersomes was pre-incubated with 3.125 to 500 μM terbinafine, desmethyl-terbinafine, or N-methyl-1-naphthyl methylamine in 50 mM potassium phosphate buffer pH 7.4 (2.5% volume methanol co-solvent) for 15 min at 37°C with 350 RPM shaking using a BMG Labtech THERMOstar incubator. Reactions were initiated by addition of an NADPH regenerating system (2 μU/μL 1-glucose-6 phosphate dehydrogenase, 10 mM glucose 6-phosphate, 2 mM MgCl2, 500 μM NADP+) and incubated for 30 min at 37°C with 350 rpm shaking. Negative control reactions included substrate/protein mixtures without the NADPH regenerating system. Reaction aliquots were quenched with an eight-fold volume of an ice-cold acetonitrile solution containing 200 μL fluoxetine and 0.5 μM dimethyl benzaldehyde (internal standards) and incubated on ice for five min to promote protein and phosphate buffer precipitation (Schellinger & Carr, 2004). Quenched reactions were centrifuged for 15 min at 4°C at 2500 RPM (2800×g) using a Beckman GPR Centrifuge. The supernatant was transferred to separate microplate wells. Each reaction aliquot was treated with dansyl hydrazine or dansyl chloride to label all aldehydes or amines, respectively, as previously described (Barnette et al., 2018).
Once complete, labeling reactions were dried down and then resuspended in mobile phase for UHPLC-MS analyses. For the dansyl hydrazine labeling reaction, dimethyl benzaldehyde present in the quench solution served as an internal standard and positive control (m/z 397). For the dansyl chloride labeling reaction, fluoxetine in the quench solution served as the internal standard (m/z 543). Signals for analytes were normalized to internal standards and corrected for background responses. The resultant values were then converted to analyte quantities based on serial dilution standard curves to calculate initial reaction rates. All steady-state reactions were performed in triplicate and each set was replicated two to three times. Initial rates were calculated and plotted against substrate concentration then fit to the Michaelis-Menten or allosteric sigmoidal model using GraphPad Prism 7.0 from GraphPad Software, Inc (San Diego, CA). The best-fit kinetic model and corresponding kinetic constants (Vmax, Km) were determined using the extra sum-of-squares F test. Overall efficiencies for each reaction were calculated as Vmax/Km. As we described previously (Barnette et al., 2018), Vmax/Km values were used to calculate a metabolic split ratio using the reaction efficiency at each nodal point of a metabolic pathway to describe the partitioning of metabolites. Through this approach, we were able to directly compare the relative significance of single and multistep pathways toward formation of TBF-A.
2.3). Recombinant isozyme reactions with inhibitors
In a set of control studies, we confirmed the capacity of recombinant CYP2C19 and 3A4 to generate significant levels of 1-naphthaldehyde during terbinafine metabolism. For those experiments, substrate reactions in recombinant CYP2C19 and 3A4 were carried out as described previously with the addition of selective inhibitors, 6 μM (+)-N-3-benzylnirvanol (BZV) and 1 μM ketoconazole (KCZ), respectively. Details of inhibitor stock preparation were described previously (Davis et al., 2019). Metabolite levels from inhibited reactions were compared to levels from control reactions without inhibitors (co-solvent only) to confirm the role of the P450s in the reaction as opposed to contaminating enzymes.
2.4). Human liver microsome reactions
Control studies using substrate reactions in human liver microsomes were conducted using our previously described methods (Davis et al., 2019). Briefly, Reactions contained 0.1 mg/mL protein (HLM150 and HLM50), 500 μM terbinafine or desmethyl-terbinafine, and 2.5% (final) methanol co-solvent due to solubility limits of substrate in 50 mM potassium phosphate buffer pH 7.4. After 15 min pre-incubation with shaking at 37 °C, reactions were initiated by addition of an NADPH regenerating system (2 μU/μL 1-glucose-6 phosphate dehydrogenase, 10 mM glucose 6-phosphate, 2 mM MgCl2, 500 μM NADP+) and incubated at 37°C with 350 RPM shaking. Reactions lacking the NADPH regenerating system served as negative controls for metabolite background signals. After 30 min, the reactions were quenched with an 8-fold volume of an ice-cold acetonitrile solution containing 0.5 μM dimethyl benzaldehyde (internal standard). Quenched reactions were centrifuged for 15 min at 4 °C and 2500 rpm (2800×g), and the supernatant was labeled with dansyl hydrazine, as described previously by our group (Barnette et al., 2018), for UHPLC-MS analysis. Each set of reactions was conducted in triplicate and replicated two to four times. Statistical difference was determined by the Mann-Whitney test (p-value = 0.05) using GraphPad Prism 7.0 from GraphPad Software, Inc. (San Diego, CA).
2.5). UHPLC-MS analysis of metabolic reactions
LC-MS analysis was used to resolve and quantitate analytes based on mass to charge ratio (m/z) and co-elution with authentic standards. Reaction metabolites were separated using a Cortecs C-18 2.7 μm column (4.6 × 50 mm) and a Waters Acquity Arc UHPLC system. Metabolites were detected by a Waters Acquity QDa single quadrupole MS system (Milford, MA). Total flow rate was 0.5 mL/min and total run time was 15 min. QDa cone voltage was 20V to detect m/z from 150 to 650 in positive ion mode (Vickers et al., 1999). The mobile phase comprised two solvents: ultrapure water with 0.01% formic acid (solvent A) and acetonitrile with 0.01% formic acid (solvent B). The gradient method began at 65% solvent A for 1 min, decreased to 20% over 5 min, and maintained at 20% for 2 min. Solvent A was then increased back to 65% over 1 min and maintained for remainder of run. Analyte peak areas were normalized to internal standards and when available, quantitated relative to authentic metabolite standards after correction with negative control reactions lacking NADPH. Targeted compounds were N-methyl-1-naphthyl methylamine (m/z 405, dansyl labeled), TBF-A (m/z 384, dansyl labeled), desmethyl-terbinafine formation (m/z 511, dansyl labeled), and 1-naphthaldehyde (m/z 404, dansyl labeled).
2.6). Deep learning model predictions of isozymes relevant to terbinafine metabolism
As an alternate, more rapid analysis of metabolism, we predicted the likelihood for reaction steps contributing to all three N-dealkylation pathways in Fig. 1 using deep learning metabolism models for individual P450 isozymes. As described previously, these models were trained using hundreds of molecules from literature-curated databases (Zaretzki et al., 2015). The models generate an output scaled from 0 to 1.0 such that the higher score corresponds to the likelihood for the reaction to occur at each atom of the molecule. How this modeling output describes and scales metabolism is different from the experimentally generated metabolic efficiencies, which are scaled based on kinetic constants and do not have an upper limit. As such, a direct comparison is not possible. As described previously (Barnette et al., 2018; Davis et al., 2019), we converted experimental metabolic efficiencies and computational predictions to split ratios for competing reactions as a strategy to allocate partitioning of substrate down pathways to a common endpoint, which was TBF-A in this study. In brief, the experimental efficiency of metabolism or model prediction for each reaction was divided by the total values for all possible reactions yielding its fractional contribution to metabolism, i.e. a metabolic or model split ratio. This approach made it possible to directly compare model predictions and experimental data at each nodal point of metabolic pathways in which more than one reaction is possible. Multiple nodal points occur along the terbinafine N-dealkylation pathways, yet for our comparison it was sufficient to focus only on the initial nodal point in which the three first-step reactions lead directly from terbinafine to the three N-dealkylation pathways (Pathway 1, Step 2.1, and Step 3.1).
2.7). Scaling measured P450 kinetics to model liver clearance/bioactivation
We extrapolated in vitro kinetics from the P450 isozymes in this study and our previous study on CYP2C19 and 3A4 metabolism of terbinafine (Davis et al., 2019) to in vivo clearance and bioactivation of terbinafine using the average protein concentrations method described previously (Yu & Haining, 2001) (see Equation 1). In brief, we assumed low drug and metabolite concentration based on reported plasma terbinafine and metabolite levels not exceeding 3 μM (Vago et al., 1994)(Kovarik et al., 1995), which is lower than the Km for relevant N-dealkylation pathways by at least 10-fold. Under those conditions, the kinetics would be reflected by reaction metabolic efficiencies (Vmax/Km). The relative metabolic contributions for each P450 isozyme would then be dependent on the average hepatic levels of the enzymes. Consequently, we used values reported from quantitative mass spectral analysis of 11 P450 isozymes present in human liver microsomes pooled from 610 donors (Kawakami et al., 2011). Extrapolation calculations then involved multiplying reaction metabolic efficiencies for each P450 by their corresponding average protein concentration. The resulting scaled values were divided by the sum of scaled values for all contributing P450 isozymes to the reaction and multiplied by 100 to yield a value corresponding to the percent contribution of each isozyme to the reaction step.
| (Equation 1) |
3.). RESULTS
3.1). N-Dealkylation efficiencies among CYP1A2, 2B6, 2C9 and 2D6 were similar for Pathway 1.
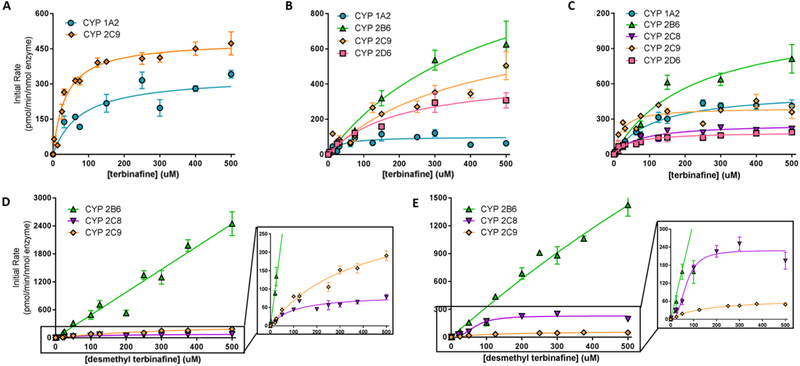
Kinetic parameters for terbinafine metabolism were measured for each N-dealkylation pathway and step using reactions by recombinant CYP1A2, 2B6, 2C8, 2C9, and 2D6 (Table 1). In this study, all P450 isozymes except CYP2A6 catalyzed direct formation of TBF-A from terbinafine but not breakdown of the co-metabolite N-methyl-1-naphthyl-methylamine. CYP2A6 was not able to generate metabolites for either reaction. For terbinafine N-dealkylation in Pathway 1, all kinetic data conformed to the Michaelis-Menten equation (Fig. 2, Table 1). The kinetic profiles for TBF-A yielded similar Km values and inconsistent Vmax values (Fig. 2A), which reflected a problem in baseline correction with Supersomes but not human liver microsomes as reported previously (Davis et al., 2019). This issue was not observed for the co-metabolite N-methyl-1-naphthyl-methylamine (Fig. 2B), making it a more reliable measure of TBF-A formation. CYP2B6 and 2C9 demonstrated the highest rates of turnover (Vmax), which were countered by poor affinities for the substrate (Km). By contrast, CYP1A2 bound substrate with the highest affinity while turning over substrate at the slowest rate. The kinetic constants for CYP2D6 were middling compared to values observed for the other isozymes. Despite differences, the resulting catalytic efficiencies for terbinafine N-dealkylation were comparable among CYP1A2, 2B6, 2C9, and 2D6. Their relevance in clearance and bioactivation would then depend on relative isozyme expression levels.
Table 1.
Michaelis-Menten kinetic constants for terbinafine N-dealkylation pathways.a
| P450 isozyme | Pathway | Step | Substrate | Steady-state kinetic constants for individual metabolites | |||||
|---|---|---|---|---|---|---|---|---|---|
| Vmaxb | Km (μM) | Vmax/Km | Vmaxb | Km (μM) | Vmax/Km | ||||
| CYP1A2d | Pathway 1 | 1 | terbinafine | TBF-Ac | N-methyl-1-naphthyl-methylamine | ||||
| - | 75 ± 42 | - | 100 ± 12 | 31 ± 17 | 3.2 | ||||
| Pathway 2 | 2.1 | terbinafine | formaldehyde | desmethyl-terbinafine | |||||
| high background masked rates | 530 ± 38 | 120 ± 23 | 4.4 | ||||||
| CYP2B6e | Pathway 1 | 1 | terbinafine | TBF-A | N-methyl-1-naphthyl-methylamine | ||||
| undetected | 1300 ± 280 | 470 ± 170 | 2.8 | ||||||
| Pathway 2 | 2.1 | terbinafine | formaldehyde | desmethyl-terbinafine | |||||
| high background masked rates | 1200± 150 | 220 ± 64 | 4.8 | ||||||
| 2.2a | desmethyl-terbinafine | TBF-Ac | 1-naphthyl-methylamine | ||||||
| - | 495 | - | linear | 4.8f | |||||
| 2.2b | desmethyl-terbinafine | 1-naphthaldehydeg | 6,6-dimethyl-2-hepten-4-yn-1-amine | ||||||
| 9100 ± 6200 | 2700 ± 2100 | 3.4 | undetected | ||||||
| CYP2C8e | Pathway 2 | 2.1 | terbinafine | formaldehyde | desmethyl-terbinafine | ||||
| high background masked rates | 260 ± 12 | 72 ± 12 | 3.6 | ||||||
| 2.2a | desmethyl-terbinafine | TBF-Ac | 1-naphthyl-methylamine | ||||||
| - | 133 | - | 85 ± 6.5 | 98 ± 23 | 0.87 | ||||
| 2.2b | desmethyl-terbinafine | 1-naphthaldehyde | 6,6-dimethyl-2-hepten-4-yn-1-amine | ||||||
| 300 ± 28 | 110 ± 28 | 2.9 | undetected | ||||||
| CYP2C9e | Pathway 1 | 1 | terbinafine | TBF-Ac | N-methyl-1-naphthyl-methylamine | ||||
| - | 35 ± 5.0 | - | 800 ± 170 | 380 ± 150 | 2.1 | ||||
| Pathway 2 | 2.1 | terbinafine | formaldehyde | desmethyl-terbinafine | |||||
| high background masked rates | 400 ± 19 | 20 ± 5.1 | 20 | ||||||
| 2.2a | desmethyl-terbinafine | TBF-Ac | 1-naphthyl-methylamine | ||||||
| - | 235 | - | 330 ± 41 | 380 ± 88 | 0.87 | ||||
| 2.2b | desmethyl-terbinafine | 1-naphthaldehyde | 6,6-dimethyl-2-hepten-4-yn-1-amine | ||||||
| 69 ± 4.4 | 140 ± 26 | 0.49 | undetected | ||||||
| CYP2D6d | Pathway 1 | 1 | terbinafine | TBF-A | N-methyl-1-naphthyl-methylamine | ||||
| undetected | 450 ± 74 | 200 ± 75 | 2.3 | ||||||
| Pathway 2 | 2.1 | terbinafine | formaldehyde | desmethyl-terbinafine | |||||
| high background masked rates | 190 ± 32 | 45 ± 19 | 4.2 | ||||||
Data fit best to the Michaelis-Menten equation over the Hill equation (P < 0.05) shown in Fig 2. Values shown with standard deviation from mean. Limit of quantitation calculated as standard deviation of response divided by slope of standard curve, whether metabolite was labeled or not. Pathway step 3.2 was not studied due to absence of authentic standards and low efficiency of previous step to obviate the significance of this pathway for TBF-A.
Units are pmol/min/nmol protein.
Vmax values highly variable so efficiency values could not be reliably calculated.
There were no detectable metabolites for Pathway Steps 1.2, 2.2a, 2.2b, or 3.
There were no detectable metabolites for Pathway Steps 1.2 or 3.
Efficiency expressed as slope of formation rate over substrate concentration due to lack of observed saturation.
Non-linear kinetic profile but poor fit to Michaelis-Menten equation due to inability to saturate enzyme, and thus reported kinetics are not highly reliable.
Fig. 2. Steady state kinetic profiles for N-dealkylation primary and secondary metabolites of terbinafine in recombinant P450s.
N-Dealkylation of terbinafine in CYP1A2 (blue circle), 2B6 (green triangle), 2C8 (purple inverted triangle), 2C9 (orange diamond), and 2D6 (pink square) yielded metabolic kinetics for one to three primary metabolites. The kinetic profiles include those for (A) TBF-A (dansyl hydrazine labeled) and (B) N-methyl-1-naphthyl methylamine (dansyl chloride labeled) from Pathway 1, and (C) desmethyl-terbinafine (dansyl chloride labeled) from Pathway 2. Secondary metabolites of terbinafine were formed from metabolism of desmethyl-terbinafine in Pathway 2, yielding (D) 1-naphthyl methylamine and (E) 1-naphthaldehyde. All sets of data were fit best to the Michaelis-Menten equation (p < 0.05), and the corresponding constants reported in Table 1. Six to nine experimental reactions were carried out and analyzed as described in Materials and Methods.
3.2). CYP2B6 was far more effective at catalyzing all reaction steps in Pathway 2.
The broad specificity of P450 isozymes led to most of them catalyzing the first step of Pathway 2 but subsequent steps were far more isozyme-dependent. With the exception of CYP2A6, all P450 isozymes in this study catalyzed N-demethylation of terbinafine to desmethyl-terbinafine (Step 2.1, Fig. 1). CYP2C9 was five-fold more efficient than the rest of the P450s (Fig. 2C, Table 1). CYP1A2, 2B6, 2C8, and 2D6 shared similar catalytic efficiencies yet differed significantly in the kinetic constants responsible for those reactions. CYP2D6 displayed the highest affinity for terbinafine based on the Km, but also had the lowest rate of turnover (Vmax). By contrast, CYP2B6 bound the substrate poorly, while catalyzing N-demethylation at the highest rate. The desmethyl-terbinafine metabolite from these reactions was subsequently metabolized only by CYP2B6, 2C8, and 2C9 down two competing pathways.
The Step 2.2a reaction yields TBF-A and 1-naphthyl-methylamine (Fig. 2D), but only kinetics for the amine co-metabolite were interpretable due to baseline correction issues with TBF-A as discussed previously. CYP2B6 kinetics did not saturate at the maximal concentration used in these studies based on solubility (500 μM), and so its efficiency value was estimated by the linear metabolite formation rate as a function of substrate concentration. This value was five-fold higher than the corresponding catalytic efficiencies for CYP2C8 and 2C9 generation of 1-naphthyl-methylamine from desmethyl-terbinafine. The kinetics for those P450 isozymes were fit best to the Michaelis-Menten equation yielding similar reaction efficiencies that were achieved through equally high or low sets of kinetic constants, respectively.
For the competing N-denaphthylation reaction (Step 2.2b, Fig. 1), CYP2B6 kinetics showed a higher catalytic efficiency for the reaction when compared to the other P450 isozymes (Fig. 2E, Table 1). Substrate binding was poor for CYP2B6 so that the kinetic profile showed only the beginnings of curvature at higher desmethyl-terbinafine concentrations. For CYP2C8, the efficiency was initially low but increased as a function of substrate concentration so that the kinetic profile fit best to the Hill equation indicating significant positive cooperativity. CYP2C9 kinetics fit best to the Michaelis-Menten equation yielding a moderate Km and a very low Vmax. These findings contrasted with results on terbinafine metabolism (Step 3.1, Fig. 1) in which none of the P450 isozymes in this study catalyzed 1-naphthaldehyde (m/z 404, dansyl labeled) formation directly from terbinafine. The limit of detection for 1-naphthaldehyde was determined to be approximately 700 nM calculated by the slope divided by the standard deviation of an authentic standard curve. Overall, CYP2C9 favored shunting desmethyl-terbinafine toward TBF-A formation, while CYP2C8 preferred the competing detoxification pathway to 1-naphthaldehyde. CYP2B6 showed no clear preference for either pathway branch.
3.3). Multiple P450 isozymes catalyzed competing oxidative non-N-dealkylation pathways.
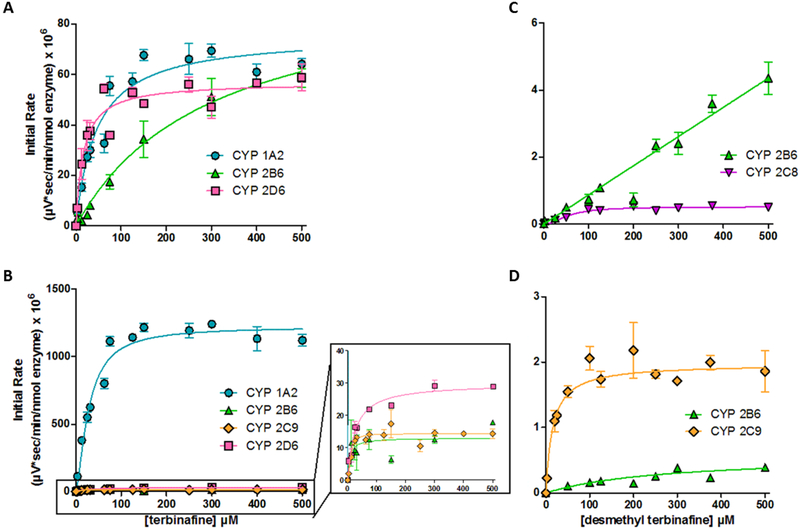
Two oxidation pathways compete with terbinafine N-dealkylation pathways:oxidation of (1) a terminal methyl group to form hydroxyterbinafine and (2) the naphthyl ring to form terbinafine dihydrodiol (Vickers, Sinclair, Zollinger, Heitz, Glänzel, et al., 1999). Due to the lack of authentic metabolite standards, we calculated initial rates based on normalized MS peak area as a function of time and then analyzed the resulting kinetic profiles to determine the metabolic mechanism and constants for the reaction (Table 2). The hydroxyterbinafine kinetic profiles for CYP1A2, 2B6, and 2D6 were best fit to the Michaelis-Menten model (Fig. 3A, Table 2). The Vmax values were comparable among all three isozymes. Nevertheless, CYP2B6 had the lowest affinity based on a Km six-fold higher than that observed for CYP1A2, while 2D6 had the highest affinity demonstrating a Km half that observed for CYP1A2. Consequently, substrate affinity was the biggest determining factor for differences in their relative efficiencies for hydroxyterbinafine formation.
Table 2.
Kinetic parameters for competing terbinafine oxidation pathways in recombinant P450s.a
| P450 isozyme | Substrate | Steady-state kinetic constants for individual metabolites | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vmaxb | Km or S50 (μM) | Hill slope (n)c | Vmax/Km | Vmaxb | Km or S50 (μM) | Hill slope (n)c | Vmax/Km | |||
| CYP1A2 | terbinafine | hydroxyterbinafine | terbinafine dihydrodiol | |||||||
| 76 ± 2.5 | 45 ± 5.7 | - | 1.7 | 1200 ± 31 | 130 ± 87 | 1.5 ± 0.20 | - | |||
| desmethyl-terbinafine | desmethyl hydroxyterbinafine | desmethyl-terbinafine dihydrodiol | ||||||||
| undetected | undetected | |||||||||
| CYP2B6 | terbinafine | hydroxyterbinafine | terbinafine dihydrodiol | |||||||
| 76 ± 2.598 ± 17 | 300 ± 100 | - | 0.32 | 14 ± 1.5 | 24 ± 10 | - | 1.7 | 0.5 | ||
| desmethyl-terbinafine | desmethyl hydroxyterbinafine | desmethyl-terbinafinedihydrodiol | ||||||||
| linear | - | - | 0.0081d | 0.59 ± 0.13 | 280 ± 130 | - | 0.0026 | |||
| CYP2C8 | terbinafine | hydroxyterbinafine | terbinafine dihydrodiol | |||||||
| undetected | undetected | |||||||||
| desmethyl-terbinafine | desmethyl hydroxyterbinafine | desmethyl-terbinafine dihydrodiol | ||||||||
| 0.61 ± 0.04 | 75 ± 18 | - | 0.0081 | undetected | ||||||
| CYP2C9 | terbinafine | hydroxyterbinafine | terbinafine dihydrodiol | |||||||
| undetected | 14 ± 0.6 | 72 ± 92 | 1.8 ± 0.49 | - | ||||||
| desmethyl-terbinafine | desmethyl hydroxyterbinafine | desmethyl-terbinafine dihydrodiol | ||||||||
| undetected | 2.0 ± 0.07 | 15 ± 3 | - | 0.13 | ||||||
| CYP2C19e | terbinafine | hydroxyterbinafine | terbinafine dihydrodiol | |||||||
| 210 ± 7 | 44 ± 2.9 | 1.40 ± 0.11 | 110 ± 17 | 110 ± 63 | 0.58 ± 0.08 | |||||
| desmethyl-terbinafine | desmethyl hydroxyterbinafine | desmethyl-terbinafine dihydrodiol | ||||||||
| 9.3 ± 0.33 | 52 ± 7.4 | - | 0.18 | 26 ± 1 | 68 ± 9.5 | - | ||||
| CYP2D6 | terbinafine | hydroxyterbinafine | terbinafine dihydrodiol | |||||||
| 57.0 ± 2.1 | 16 ± 3.2 | - | 3.6 | 30 ± 1.4 | 25 ± 4.1 | - | 3.8 | |||
| desmethyl-terbinafine | desmethyl hydroxyterbinafine | desmethyl-terbinafine dihydrodiol | ||||||||
| undetected | undetected | |||||||||
| CYP3A4e | terbinafine | hydroxyterbinafine | terbinafine dihydrodiol | |||||||
| 570 ± 20 | 25 ± 2.3 | 1.30 ± 0.16 | 150 ± 3.3 | 17.6 ± 1.3 | 2.0 ± 0.32 | |||||
| desmethyl-terbinafine | desmethyl hydroxyterbinafine | desmethyl-terbinafine dihydrodiol | ||||||||
| 0.53 ± 0.04 | 82 ± 20 | - | 0.0065 | 2.3 ± 0.17 | 98 ± 19 | - | 0.023 | |||
Data fit to the Michaelis-Menten equation or the Hill equation based on extra sum-of-squares F test (P < 0.05) using GraphPad Prism 7.0. Values shown with standard deviation from mean.
Due to absence of authentic standards, rates reflect normalized MS peak area for metabolite as a function of time, i.e. μV*sec/min/nmol. Final values were scaled x106.
Not shown in table: n=1 for Michaelis-Menten equation.
Efficiency expressed as slope of formation rate over substrate concentration due to lack of observed saturation.
Data taken from Davis et al. (2019). Normalized peak areas (μV*sec) reported from that study were converted to rates (μV*sec/min/nmol) by dividing by protein concentration (100 nM) and reaction time (30 min). This scaled conversion of the values does not change the shape of the plotted curve.
Fig. 3. Steady state kinetic profiles for metabolites of major oxidative non-N-dealkylation pathways of terbinafine in recombinant P450s.
Metabolites for major pathways (Vickers et al., 1999) (Kovarik et al., 1995) competing with N-dealkylation. (A) Hydroxyterbinafine and (B) terbinafine dihydrodiol from terbinafine, and (C) desmethyl hydroxyterbinafine and (D) desmethyl-terbinafine dihydrodiol from desmethyl-terbinafine were observed for CYP1A2 (blue circle), 2B6 (green triangle), 2C9 (orange diamond), and 2D6 (pink square). Compounds were not quantitated due to unavailability of standards, so initial rates are normalized to MS peak areas as a function of time. Data were fit best to the Michaelis-Menten equation or the Hill equation (p < 0.05), and the corresponding constants reported in Table 2. At least six experimental reactions per condition were carried out and analyzed as described in Materials and Methods.
CYP1A2, 2B6, 2C9, and 2D6 catalyzed dihydrodiol formation on the terbinafine naphthyl ring (Fig. 3B, Table 2). CYP2B6 and 2D6 followed the Michaelis-Menten model, demonstrating similar Km values, but the latter isozyme had a two-fold higher efficiency. The kinetic profiles for CYP1A2 and 2C9 were best described by the Hill equation indicating significant positive cooperativity, although the Vmax for the reaction by CYP2C9 was far lower than that observed for CYP1A2. Similarly, desmethylterbinafine undergoes these same competing oxidation pathways at the secondary step of Pathway 2. CYP2B6 and 2C8 metabolized the formation of desmethyl hydroxyterbinafine. The reaction by CYP2B6 did not saturate, while that for CYP2C8 followed Michaelis-Menten kinetics. Formation of the dihydrodiol was metabolized by CYP2B6 and 2C9 with both following Michaelis-Menten kinetics with 50-fold higher efficiency by CYP2C9 due to lower Km and higher Vmax.
3.4). Metabolic models predicted all steps in reaction pathways.
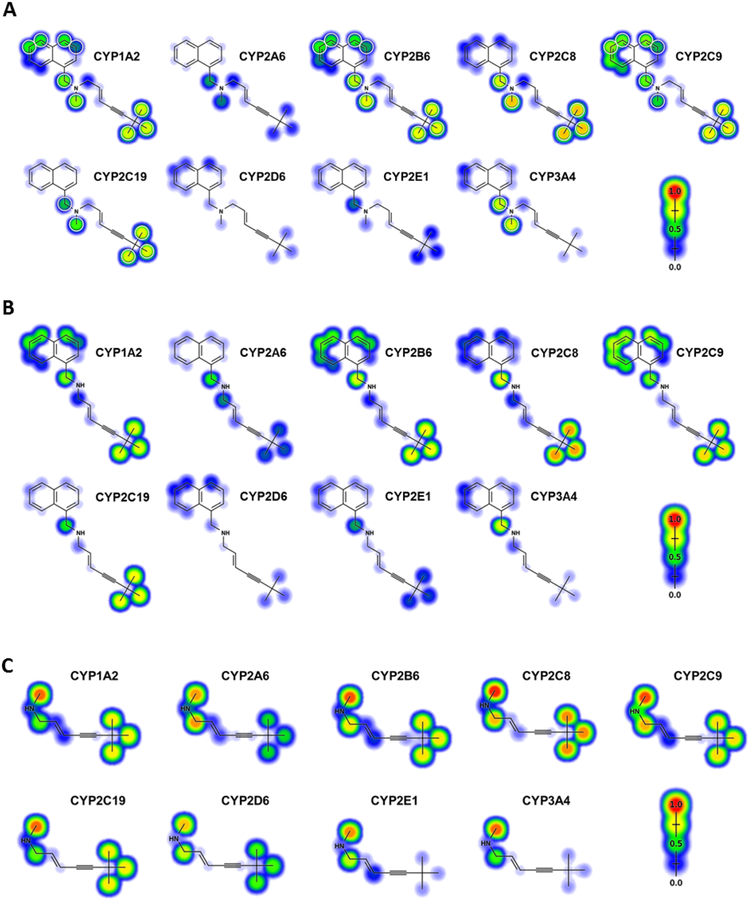
For a more rapid, less resource-intensive analysis of terbinafine metabolism, we predicted terbinafine oxidation using computational P450 isozyme models (Zaretzki et al., 2015). These efforts yielded predictions for all three N-dealkylation pathways along with those for the oxidation of terminal methyl and naphthyl groups for comparison to our experimental kinetic data (Fig. 4, Table 3). For each P450 isozyme, specific atoms were scored from 0 to 1.0 (low: 0–0.5, moderate: 0.5–0.75, high: 0.75–1.0) on the likelihood for oxidation to occur. The predictions for formation of TBF-A from the parent drug (Pathway 1) were consistently low for all P450s, whereby the highest value was approximately 0.25 for CYP2A6, 1A2, and 2C8. By contrast, the prediction for CYP2C8 N-demethylation of terbinafine (Step 2.1) yielded a high likelihood of occurrence while those for CYP2B6, 1A2, 3A4, and 2C19 were moderate. Model outcomes for CYP2C9 and 2A6 were low and prediction of metabolism by CYP2D6 and 2E1 was negligible for this reaction. Loss of the methyl group did not significantly alter predictions for secondary reactions with desmethyl-terbinafine (Steps 2.1a and 2.2b) with the exception of CYP2E1, which had a three-fold higher prediction for N-denaphthylation. For Pathway 3, terbinafine N-denaphthylation to 1-naphthaldehyde and N-methyl-6,6-dimethyl-2-hepten-4-yn-1-amine (Step 3.1) yielded essentially identical scores to the corresponding reaction for desmethyl-terbinafine (Step 2.2b) indicating very little effect of the methyl group on reaction likelihood. In contrast, loss of the naphthyl group greatly impacted predictions for secondary reactions. TBF-A formation from N-methyl-6,6-dimethyl-2-hepten-4-yn-1-amine (Step 3.2a) was highly predicted for CYP2A6, 2C8, 2C9, and 2B6, while others received moderate scores. Demethylation of N-methyl-6,6-dimethyl-2-hepten-4-yn-1-amine (Step 3.2b) yielded high outcomes for every P450. For other competing oxidative reactions, terminal hydroxylation of methyl groups was scored highly for CYP2C9 and 2C8, moderately for CYP2B6, 2C19, and 1A2 and poorly for CYP2E1. Terbinafine oxidation to the dihydrodiol was predicted moderately for CYP2C9, poorly for CYP1A2 and 2B6, and almost negligibly for the other studied isozymes. Overall, the denaphthylated metabolite had much higher scores for all subsequent reactions by the studied P450s, while loss of the methyl group had minimal effect on model predictions. CYP2C8 had the highest scores for most N-dealkylation reactions while CYP2D6 was generally the lowest and the other isozymes were in between.
Fig. 4. Metabolic model predictions for P450 catalyzed N-dealkylations.
Metabolic modeling was used to predict P450 catalyzed N-dealkylation of (A) terbinafine for all pathways (B) N-desmethyl-terbinafine for Pathway 2 secondary reactions, and (3) N-methyl-6,6-dimethyl-2-hepten-4-yn-1-amine for Pathway 3 secondary reactions. Predictions for metabolism at specific atomic locations were scored for increasing oxidation likelihood ranging from 0 – 1.0 (blue to red).
Table 3.
P450 model predictions for terbinafine N-dealkylation and competing oxidation pathways.
| Pathway | Step | Metabolites | Cytochrome P450 isozymes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A2 | 2A6 | 2B6 | 2C8 | 2C9 | 2C19 | 2D6 | 2E1 | 3A4 | |||
| 1 | 1.1 | TBF-A, N-methyl-1-naphthyl methylamine | 0.25 | 0.27 | 0.19 | 0.25 | 0.12 | 0.13 | 0.056 | 0.069 | 0.16 |
| 2 | 2.1 | desmethyl-terbinafine, formaldehyde | 0.66 | 0.38 | 0.74 | 0.80 | 0.46 | 0.56 | 0.086 | 0.15 | 0.73 |
| 2.2a | TBF-A, 1-naphthyl methylamine | 0.29 | 0.39 | 0.25 | 0.31 | 0.14 | 0.16 | 0.057 | 0.081 | 0.18 | |
| 2.2b | 1-napthaldehyde, 6,6-dimethyl-2-hepten-4-yn-1-amine | 0.63 | 0.51 | 0.72 | 0.75 | 0.61 | 0.49 | 0.14 | 0.42 | 0.72 | |
| 3 | 3.1 | 1-napthaldehyde, N-methyl-6,6-dimethyl-2-hepten-4-yn-1-amine | 0.61 | 0.40 | 0.68 | 0.71 | 0.60 | 0.56 | 0.10 | 0.36 | 0.70 |
| 3.2a | TBF-A, methyl amine | 0.59 | 0.85 | 0.77 | 0.82 | 0.82 | 0.60 | 0.70 | 0.69 | 0.56 | |
| 3.2b | 6,6-dimethyl-2-hepten-4-yn-1-amine, formaldehyde | 0.90 | 0.85 | 0.92 | 0.96 | 0.93 | 0.90 | 0.81 | 0.86 | 0.90 | |
| terminal hydroxylation | hydroxyterbinafine | 0.71 | 0.22 | 0.73 | 0.81 | 0.76 | 0.72 | 0.16 | 0.27 | 0.073 | |
| desmethyl hydroxyterbinafine | 0.73 | 0.33 | 0.75 | 0.84 | 0.79 | 0.75 | 0.17 | 0.32 | 0.082 | ||
| naphthyl oxidationb | terbinafine dihydrodiol | 0.48 | 0.062 | 0.49 | 0.21 | 0.57 | 0.070 | 0.18 | 0.11 | 0.14 | |
| desmethyl-terbinafine dihydrodiol | 0.51 | 0.098 | 0.53 | 0.28 | 0.63 | 0.097 | 0.23 | 0.15 | 0.17 | ||
Model output range and corresponding color: 0 – 0.24, light gray; 0.25 – 0.49, dark gray; 0.50 – 0.74, black; 0.75 – 1.0, bold black
Average prediction score for four carbon atoms on naphthyl group with highest likelihood for dihydrodiol formation.
3.5). Model predictions were more consistent with N-demethylations than other reactions in terbinafine metabolism.
Model outputs do not extrapolate to experimental kinetic information, so that a direct comparison of the results to assess model performance was not possible. Among the P450 isozymes, there was qualitative agreement between experimental findings and model predictions for some reactions but not others. For example, contrary to experimental data, none of the P450 isozyme models predicted the likelihood for direct formation of TBF-A in Pathway 1 but generally agreed with the ease of N-demethylation as the first step in Pathway 2. The model predictions for subsequent steps in Pathway 2 did not change significantly for desmethyl-terbinafine, while the kinetic data showed that demethylation of the parent drug greatly increased subsequent metabolic processes. Lastly, the moderate to high likelihoods for Pathway 3 reactions predicted by the models were only observed for CYP2C19 and 3A4 (Davis et al., 2019).
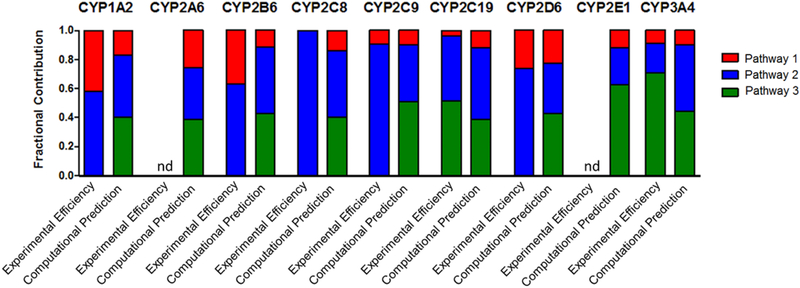
As an alternative, more direct comparison, we calculated partitioning of terbinafine (split ratios) down the initial steps of the three competing N-dealkylation pathways using either model outputs or kinetic efficiencies as a common metabolic readout for comparative purposes (Table 4, Fig 5). This analysis yielded four key features between model predictions and experimental data. First, CYP2A6 and 2E1 models predicted their metabolic capacity to catalyze all three initial N-dealkylation reactions for terbinafine, yet those enzymes demonstrated no measurable activity for those reactions. Second, all nine P450 isozyme models indicated a preference for N-demethylation in Pathway 2 over N-dealkylation in Pathway 1. Third, all nine models predicted a high likelihood for N-denaphthylation in Pathway 3 leading to almost equivalent partitioning of terbinafine down that pathway versus N-demethylation in Pathway 2; however, the experimental data only supported this level of competitiveness for CYP2C19 and 3A4. Fourth, the partitioning ratios revealed a consistent pattern among all nine P450 isozyme models in terms of the relative contributions of all three N-dealkylation pathways to one another. Pathways 2 and 3 were similarly more favored over Pathway 1. This observation contrasted with the high variability in specificity and efficiency of P450 isozymes catalyzing those reactions experimentally. The kinetics revealed opposite trends in partitioning and absence of some reaction pathways occurring altogether. In total, the models overpredicted the possibility of Pathway 3 and contributions of some P450s compared to experimental evidence. Thus we did not carry out metabolic flux analyses using the modeling predictions as shown for experimental data.
Table 4.
Split ratiosa for terbinafine initial N-dealkylation reactions by P450s based on experimental kinetic efficiencies and computational predictions.
| Model | Reaction | Cytochrome P450 isozymes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1A2 | 2A6 | 2B6 | 2C8 | 2C9 | 2C19 | 2D6 | 2E1 | 3A4 | ||
| Experimental Kinetic Efficiency | 1.1 | 0.42 | NDb | 0.37 | ND | 0.10 | 0.04 | 0.26 | ND | 0.09 |
| 2.1 | 0.58 | ND | 0.63 | 1.00 | 0.90 | 0.44 | 0.74 | ND | 0.20 | |
| 3.1 | ND | ND | ND | ND | ND | 0.52 | ND | ND | 0.71 | |
| Computational Prediction | 1.1 | 0.17 | 0.26 | 0.12 | 0.14 | 0.10 | 0.12 | 0.23 | 0.12 | 0.10 |
| 2.1 | 0.43 | 0.36 | 0.46 | 0.46 | 0.39 | 0.50 | 0.35 | 0.26 | 0.46 | |
| 3.1 | 0.40 | 0.38 | 0.42 | 0.40 | 0.51 | 0.39 | 0.43 | 0.62 | 0.44 | |
Split ratio values calculated as described previously (Barnette et al., 2018) (Davis et al., 2019).
ND = metabolite not detected
Fig. 5. Comparison of experimental kinetics and modeling predictions for P450 catalyzed terbinafine N-dealkylations.
Modeling predictions compared to experimental kinetic parameters based on prediction scores (0–1.0) and kinetic efficiencies (Vmax/Km) scaled to fractional values. Fractional values were calculated based on predictions and efficiencies for metabolic split ratios of pathways within each individual P450. N-methyl-1-naphthyl methylamine, N-desmethyl-terbinafine, and 1-napthaldehyde were markers for the initial reactions of Pathways 1, 2, and 3, respectively. The graphs illustrate the calculated fractional values listed in Table 4. nd = no metabolite detected
3.6). Significant N-denaphthylation of terbinafine was limited to recombinant and not microsomal systems.
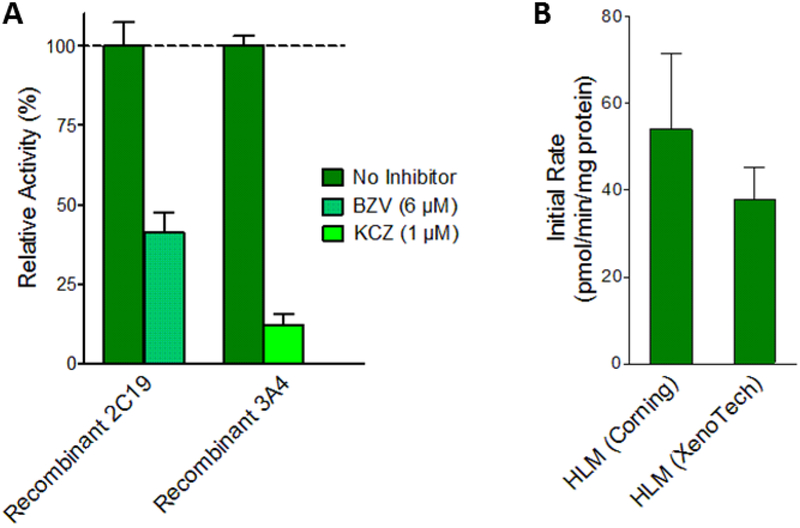
Coupled with previous kinetic experiments (Davis et al., 2019), completion of the current studies made possible the extrapolation of in vitro kinetics to in vivo hepatic clearance and bioactivation of terbinafine; however, there is a discrepancy between the kinetics for N-denaphthylation by recombinant P450 isozymes and those observed by human liver microsomes (Barnette et al., 2018). Kinetics for 1-naphthaldehyde formation by CYP2C19 and 3A4 Supersomes yielded relatively high rates for Step 3.1, which contrasted with the relatively low rates observed in the microsomal model. We previously showed that depletion of 1-naphthaldehyde by human liver microsomes was not sufficient to account for the difference in apparent rates of formation (Davis et al., 2019). In this study, we conducted more control experiments to resolve this inconsistency. First, we confirmed recombinant P450 isozymes contributed to N-denaphthylation by using selective inhibitors for the individual Supersomes. These reactions showed 60% inhibition of 1-naphthaldehyde formation for CYP2C19 and 90% inhibition for CYP3A4 (Fig. 6.A). Second, we validated that the lack of terbinafine N-denaphthylation by human liver microsomes was not due to the vendor. Triplicate reactions using pooled microsomes from Corning and XenoTech yielded 1-naphthaldehyde formation rates that were not statistically different based on the Student’s T test (p=0.214) (Fig. 6.B). Lastly, the lack of terbinafine N-denaphthylation in microsomal reactions cannot be explained by variations in P450 isozyme protein levels; CYP3A4 is the most abundant hepatic P450 isozyme as reflected in the reported levels by both Corning and XenoTech vendors for their pooled human liver microsomes. If microsomal CYP3A4 had the capacity to generate 1-naphthylaldehyde, it would have been easily measurable.
Fig. 6. Comparison of terbinafine metabolism in HLM and recombinant P450s.
Control experiments were carried out to confirm source and reliability of 1-naphthaldehyde formation rates measured in previous experiments. (A) 1-Naphthyaldehdye yields from recombinant CYP 2C19 and 3A4 measured in the presence of selective inhibitors. CYP2C19 was incubated with 6 μM (+)-N-3-benzylnirvanol (BZV) (yellow-green) and 3A4 was incubated with 1 μM ketoconazole (KCZ) (blue-green). Metabolite levels are reported as percentages of those from reactions without inhibitors (green). (B) Total 1-naphthaldehyde (pathway 3.1) formation rates were measured from Corning HLM150 and Xenotech HLM50 pooled microsomal systems. Statistically significant differences were determined by Student’s T-test. (C) Recombinant P450 reaction efficiencies (Vmax/Km) for each pathway were scaled to microsomal efficiencies for comparison. Scaling was achieved using the average percentage concentration scaling method (Yu & Haining, 2001) based on average percentages of specific P450s reported by others (Kawakami et al., 2011). Units, Vmax: pmol/min/mg protein, Km: μM
3.7). Based on in vitro to in vivo extrapolations, CYP2C9 and 3A4 were major determinants of terbinafine N-dealkylation.
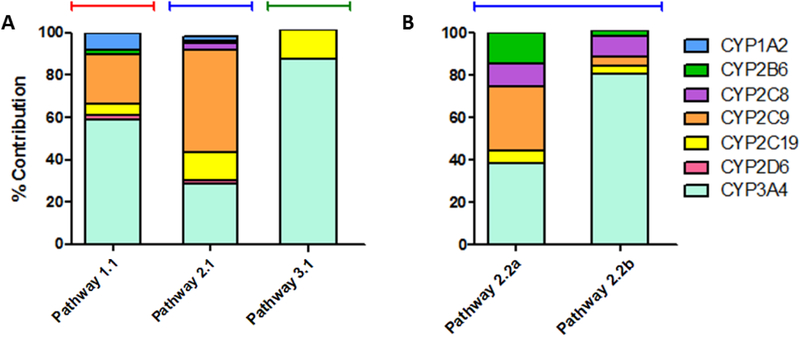
Contributions of individual P450 isozymes to each metabolic pathway under in vivo conditions depend on P450 isozyme kinetics and abundance in the liver. We experimentally measured kinetic efficiencies (Vmax/Km) for all tested P450s in this study (CYP1A2, 2B6, 2C8, 2C9, and 2D6) and our previous study (CYP2C19 and 3A4) (Davis et al., 2019) to model rates at clinically relevant low substrate concentrations (Kovarik et al., 1995). These values were scaled to relative abundances in human liver microsomes based on average isozyme concentrations measured by others using mass spectrometry in human liver microsomes pooled from 610 donors (Kawakami et al., 2011). All results were normalized to 100% for assessing the relevance of their roles in metabolism (Fig. 7, Table 5). For Pathway 1 (Fig. 8, Step 1.1), CYP3A4 was the dominant isozyme contributing 60% to the reaction generating N-methyl-1-naphthyl methylamine while CYP2C9 demonstrated moderate activity (23%) while CYP1A2, 2B6, 2D6, and 2C19 had minor activity (1–10%). For Pathway 2 (Fig. 8, Step 2.1), CYP2C9 was the major catalyst (50%) for the initial reaction with moderate contributions from CYP3A4 (30%) and 2C19 (13%). Activities by CYP1A2, 2B6, 2C8, and 2D6 were minor (<5%). The secondary reaction for Pathway 2 leading to TBF-A (Fig. 8, Step 2.2a) was primarily metabolized by CYP3A4 (40%) and 2C9 (30%) with minor contributions from CYP2B6, 2C8, and 2C19 (5–15%). The competing N-denaphthylation pathway for desmethyl-terbinafine was catalyzed primarily by CYP3A4 (80%) with CYP2B6, 2C8, 2C9, and 2C19 comprising the remaining 20% (Fig 8, Step 2.2b). For Pathway 3 (Fig. 8, Step 3.1), CYP3A4 was the major participant (86%), with CYP2C19 being the only other detectable contributor. Overall, the results demonstrated that CYP2C9 and 3A4 were the most important enzymes for terbinafine bioactivation due to their dominance in Pathways 1 and 2, the only relevant pathways for TBF-A (Barnette et al., 2018).
Fig. 7. In vitro to in vivo scaling of recombinant P450 kinetics to model hepatic clearance using average microsomal protein concentrations.
Efficiencies (Vmax/Km) for metabolite formation from substrate by recombinant P450s measured in this study (CYP1A2, 2B6, 2C8, 2C9, 2D6) and our previous study (CYP2C19, 3A4) (Davis et al., 2019) were scaled to model microsomal relative abundances using measured percentages of individual P450 isozymes reported by others (Kawakami et al., 2011). Calculated values for percent contributions for each P450 are listed in Table 5. (A) Percent contributions to primary reactions from terbinafine were calculated based on efficiency of N-methyl-1-naphthyl methylamine formation for Pathway 1, desmethylterbinafine formation for Pathway 2, and 1-naphthaldehyde formation for Pathway 3. (B) Percent contributions to secondary reactions from terbinafine were calculated based on efficiency of 1-naphthyl methylamine formation from desmethyl-terbinafine from Step 2.2a and 1-napthaldehyde formation from desmethyl-terbinafine for Step 2.2b.
Table 5.
Relative contributions of individual P450 isozymes to in vivo hepatic terbinafine clearance and/or bioactivation
| Pathway | Substrate | Metabolite | Percent contributions of individual isozymes for each reactiona | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CYP1A2 | CYP2B6 | CYP2C8 | CYP2C9 | CYP2C19b | CYP2D6 | CYP3A4b | |||
| 1.1 | terbinafine | N-methyl-1-naphthyl methylamine | 7.75 | 2.63 | - | 23.0 | 5.48 | 2.36 | 58.8 |
| 2.1 | terbinafine | desmethyl-terbinafine | 2.39 | 1.01 | 3.22 | 49.1 | 13.3 | 1.48 | 29.4 |
| 2.2a | desmethyl-terbinafine | 1-naphthyl-methylamine | - | 14.2 | 11.0 | 30.1 | 5.82 | - | 38.8 |
| 2.2b | desmethyl-terbinafine | 1-naphthaldehyde | - | 2.64 | 9.61 | 4.44 | 3.58 | - | 79.7 |
| 3.1 | terbinafine | 1-naphthaldehyde | - | - | - | - | 13.3 | - | 86.7 |
Calculations based on extrapolations of kinetic efficiencies and reported levels of P405 isozymes as described under Materials and Methods.
Contributions determined using catalytic efficiency values published previously (Davis et al., 2019).
Fig. 8. Relative P450 involvement in N-dealkylation pathways of terbinafine.
P450 isozymes contributing to reaction steps of each terbinafine N-dealkylation pathway have been identified and scaled based on reaction efficiency and relative abundance in human liver microsomes to model contributions in adult human liver conditions. Higher contributions for individual isozymes relative to others are indicated by larger font size and boldness for reactions in Pathway 1 (red), Pathway 2 (blue), and Pathway 3 (green).
4.). DISCUSSION
4.1). Seven P450s catalyzed two major pathways in TBF-A formation.
Early studies lacked an analysis of specific pathways determining terbinafine N-dealkylation, especially of those leading to generation of the reactive metabolite TBF-A (Vickers et al., 1999) (Fig. 1). Herein, we expanded on previous kinetic studies with CYP2C19 and 3A4 by our group (Davis et al., 2019) to include CYP1A2, 2B6, 2C8, 2C9, and 2D6 for a more complete analysis of possible N-dealkylation pathways and to serve as a foundation for in vitro to in vivo extrapolations to discern their relevance. Despite positive inhibitor phenotyping results for CYP2A6 and consistent with negative results for 2E1 (Davis et al., 2019), kinetic experiments for CYP2A6 and 2E1 in this study revealed no role for these isozymes in terbinafine metabolism. Thus, a total of seven P450 isozymes catalyze terbinafine N-dealkylation reactions. These findings are consistent with initial reports on terbinafine metabolism with the exception of CYP2D6 (Vickers et al., 1999). Previously, CYP2D6 was considered only to be a target of inhibition by terbinafine, which could lead to drug-drug interactions. This mechanism is likely still true given the low catalytic capacity of CYP2D6 in this study.
4.2). CYP2C19 and 3A4 remained dominant contributors to direct TBF-A formation in Pathway 1.
Our previous studies with human liver microsomes showed Pathways 1 and 2 are most important for TBF-A formation with only minor contributions through Pathway 3 (Fig. 1) (Barnette et al., 2018). The current kinetic studies with CYP1A2, 2B6, 2C8, 2C9, and 2D6 recapitulated this observation. For Pathway 1, CYP2C9 and 2D6 catalyzed direct formation of TBF-A with low efficiency, while CYP1A2 and 2B6 demonstrated higher efficiencies (~ 50% higher Vmax/Km). Nevertheless, those values were more than two-fold less than the values for the reaction catalyzed by CYP2C19 and 3A4 (Davis et al., 2019). The main driver in metabolism by most isozymes was the turnover number especially for CYP2B6; CYP1A2 was the exception given its low Km for the reaction.
4.3). For Pathway 2, terbinafine N-demethylation significantly impacted subsequent reactions by multiple P450 isozymes.
Surprisingly, loss of the methyl group in Pathway 2 significantly altered the capacity of P450 isozymes to catalyze metabolic reactions. The initial N-demethylation was measurable but relatively negligible by CYP1A2, 2B6, 2C8, and 2D6, while CYP2C9 was relatively efficient. In fact, that isozyme was more effective at initial N-demethylation than CYP3A4 (Davis et al., 2019). This broad specificity for the reaction likely reflected ease of access of the P450 isozymes to the methyl group to facilitate oxidation. Ranking of catalytic efficiencies yielded comparable findings to that reported by Vickers et al. (Vickers et al., 1999) with CYP1A2 and 2C8 being notable exceptions. Their importance in metabolism was anticipated to be higher in the previous report than in this study, which likely reflects differences in experimental design. The early studies considered only total N-demethylation without distinguishing individual pathways contributing. This approach would confound the interpretation of the reported kinetics. We avoided this confounding interpretation by measuring individual reactions to construct discreet pathway kinetics.
Desmethyl-terbinafine is a major metabolite of terbinafine (Kovarik et al., 1995), yet these studies and our previous work (Davis et al., 2019) are the only investigations of its metabolism by P450 isozymes. While a minor metabolizer of terbinafine, CYP2B6 was one of the most effective catalysts for either branch of the N-demethylation pathway (Steps 2.2a and 2.2b). Why the N-methyl group has such an impact on CYP2B6 metabolic efficiency is unclear. Only CYP2C8 and 2C9 were also capable of catalyzing those reactions, but CYP2C8 was the only relevant contributor given its efficiency for TBF-A formation, which was comparable to that of CYP2B6. These efficiencies remain two- to four-fold less than those observed for CYP2C19 and 3A4 (Davis et al., 2019). Consequently, the limited sequence differences among CYP2C family members are sufficient to differentiate their ability to catalyze terbinafine N-dealkylations.
4.4). Only CYP2C19 and 3A4 catalyzed terbinafine N-denaphthylation in Pathway 3.
In this study, the inability of P450 isozymes to N-denaphthylate terbinafine in Pathway 3 was consistent with previously reported inhibitor phenotyping studies (Davis et al., 2019). These findings contrasted with the N-denaphthylation of desmethylterbinafine by CYP2B6, 2C8 and 2C9 indicating the ability of all three isozymes to differentiate substrate specificity based on the presence of the methyl group. Their selectivity may arise from the slight difference in steric bulk and/or the change in the pKa of the nitrogen of substrate. These results demonstrated that only CYP2C19 and 3A4 were capable of this minor reaction in human liver microsomes (Davis et al., 2019)(Barnette et al., 2018).
4.5). Seven P450 isozymes catalyzed competing oxidative non-N-dealkylation pathways.
The same seven P450 isozymes (CYP1A2, 2B6, 2C8, 2C9, 2C19, 2D6, and 3A4) contributing to N-dealkylation were capable of catalyzing formation of a dihydrodiol and/or hydroxylation of the alkyl group terminus of terbinafine in both this study and the previous study (Davis et al., 2019). Unlike previous results, the loss of the N-methyl group led to an almost uniform, significant drop in activity for these reactions. Alternate binding interactions due to the change in structure is a possibility given that some N-dealkylation reactions were more efficient for desmethyl-terbinafine versus the parent drug. Similarly, substrate recognition was a determinant in whether Michaelis-Menten or cooperative mechanisms best described reaction kinetics. CYP1A2, 2C9, 2C19 and 3A4 displayed a preference for positive cooperative interactions with substrates, which is substantiated by our current knowledge of their penchant for non-Michaelis-Menten kinetics (Isin & Guengerich, 2016) (Niwa, Murayama, & Tamazaki, 2008). Importantly, positive cooperativity in metabolism likely diminishes their relative significance in terbinafine and desmethyl-terbinafine clearance. Despite multi-dosing, the parent drug and major metabolite achieve only low micromolar levels in plasma (Kovarik et al., 1995), such that observed reaction rates would derive from the less efficient form of the P450 isozymes at low substrate levels. By contrast, CYP2D6 metabolism involves Michaelis-Menten kinetics and a high affinity for substrate, and hence, its contributions would become more important despite a relatively low turnover number to other isozymes for these reactions. Taken together, CYP3A4 remains the dominant catalyzer for both pathways with CYP2C19 and 2D6 being secondary contributors.
4.6). Model predictions did not capture relative significance of pathways reflected by kinetic studies.
The detailed kinetic studies herein required significant time, effort and resources and thus, we explored computational models as an attractive alternative to predict terbinafine metabolism by P450 isozymes (Zaretzki et al., 2015). Model outputs are a single value scaled from 0 to 1.0 unlike the mechanism and corresponding constants generated from kinetic studies. Consequently, a direct comparison is not possible; however, we assessed the general patterns of metabolic predictions among reactions and respective P450 isozyme models as well as calculated partitioning of substrate down each metabolic pathway using computational and experimental data. A qualitative comparison of model predictions and reaction efficiencies revealed that models performed best for predicting loss of an N-methyl group and poorly for N-dealkylation and N-denaphthylation reactions. Additionally, the model did not predict the observed differences for non-N-dealkylation between the parent and demethylated metabolites. The partitioning results provided further insights on the computational model shortcomings. These outcomes may reflect higher representation of reactions with N-demethylations in the model training sets given their prevalence in drug metabolism. Since the models were developed in 2015 much more data is readily available for retraining models and may address the potential underrepresentation of more numerous and diverse N-dealkylation reactions.
The comparative analyses demonstrated another possible issue with the modeling. Despite variations in reaction outputs, the partitioning ratios for preferences for Pathways 1, 2, and 3 were nearly identical among all P450 isozyme models. Such a pattern and the inaccuracies were not apparent in our previously reported analysis of terbinafine metabolism by just two P450 models (Davis et al., 2019), yet the larger scope of the current study revealed this trend. This finding suggests a systematic problem with the model architecture in which it is not able to adequately distinguish among differences in substrate specificities among P450s isozymes observed experimentally – at least for terbinafine. Addressing the modeling concerns of scaling and predictability will allow for better leveraging of the advantages of the computational approaches. Models provide the capacity for high throughput and low cost predictions compound metabolism without the limits of experimental observability such as reactions for N-methyl-6,6-dimethyl-2-hepten-4-yn-1-amine in Pathway 3.
4.7). In vivo extrapolation of P450 isozyme kinetics implicated significant roles for CYP2C9 and 3A4 in hepatic terbinafine metabolism.
The determination of metabolic kinetics by recombinant P450 isozymes provides an opportunity to extrapolate their relative roles in hepatic clearance. Prior studies involved scaling overall N-demethylation and deamination reactions and relied on isozyme relative activity factors (Vickers et al., 1999). The combination of reactions yielding those metabolites confounds the ability to ascribe each P450 isozyme to individual pathways or pathway steps, and hence, cannot predict factors impacting critical metabolic reactions like the formation of TBF-A. Moreover, the use of relative activity factors may compromise the value of extrapolations of metabolism given that activity scaling factors can be highly depend on probe selection and may result in incorrect predictions (Siu & Lai, 2017). Thus, we scaled our P450 isozyme efficiencies for terbinafine reactions based on average protein concentrations (Yu & Haining, 2001) using reported values in human liver microsomes pooled from 610 donors (Kawakami et al., 2011). From this analysis, the combination of CYP2C9 and 3A4 accounted for over 60% of every metabolic step. CYP3A4 contributed the most to direct TBF-A formation (Pathway 1.1), while CYP2C9 dominated the two-step Pathway 2 to the reactive metabolite. In the clinic, multi-dosing of terbinafine results in comparable levels of desmethyl-terbinafine to the parent drug (Kovarik et al., 1995), such that CYP2C9 contributions would become more significant in TBF-A formation.
4.8). Reported drug-drug interactions consistent with important roles for CYP2C9 and 3A4 in terbinafine metabolism.
The current studies highlight the potential clinical relevance of CYP2C9 and 3A4 in terbinafine clearance and metabolism into the reactive metabolite TBF-A. The critical role of CYP3A4 in those processes was implicated by our previous kinetic studies with the isozyme (Davis et al., 2019) as well as reported drug-drug interactions (“Lamisil,” 2004) (Rodrigues, 2008). Nevertheless, our previous work also implicated CYP2C19 based on its high efficiencies for reactions; however, that study only assessed the enzymatic efficiencies along the pathways. A comprehensive analysis of relative isozyme dominance in the N-dealkylation pathways must also consider the average concentration of each isozyme in the human liver. We accounted for these scaling factors in the current study using estimations based on average protein concentration in human liver microsomes. The participation of CYP2C19 in terbinafine bioactivation is dwarfed due to higher CYP2C9 protein levels and moderate efficiencies for these reactions as suggested by the extrapolation of the kinetic data to hepatic clearance. In fact, evidence for the clinical relevance of CYP2C9 includes reported drug-drug interactions such as an increased risk of bleeding following warfarin and terbinafine co-administration (Gantmacher, Mills-Bomford, & Williams, 1998). Polymorphisms may further alter CYP2C9-mediated pathways and can significantly modulate terbinafine bioactivation likelihoods or clearance rates (Lee, Goldstein, & Pieper, 2002). Collectively, these effects on CYP2C9 and 3A4 metabolism could alter their ability to generate TBF-A as a causative factor in terbinafine-associated idiosyncratic liver toxicity (Iverson & Uetrecht, 2001).
4.9). Concluding Remarks
Terbinafine undergoes metabolism by seven P450 isozymes, with CYP2C9 and 3A4 playing a dominant role. These isozymes are responsible for 80% of all TBF-A formation from the parent drug. This knowledge facilitates further investigation into what alters their roles in metabolism and how these alterations may impact toxicity, whether they be through isozyme genetic alleles, drug-drug interactions, or other environmental factors. Nevertheless, these mechanisms of potential patient sensitization still depend on other factors such as TBF-A decay (Barnette et al., 2018) and adduction of TBF-A with glutathione (Iverson & Uetrecht, 2001) that remain understudied. The collective information gained from addressing those gaps in the field would facilitate development of a more predictive approach to reveal and stratify patient vulnerability to terbinafine-induced toxicity.
Acknowledgments:
Research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under award numbers R01LM012222 and R01LM012482. Davis is partially supported by the National Institute of General Medical Sciences under grant number GM106999. Computations were performed using the facilities of the Washington University Center for High Performance Computing that was partially funded by National Institutes of Health (NIH) grant numbers 1S10RR022984–01A1 and 1S10OD018091–01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- Ajit C, Zaeri N, Munoz SJ, & Suvannasankha A (2005). Terbinafine-associated hepatotoxicity. Curr Opin Investig Drugs, 6(2), 107–177. Retrieved from https://www.amjmedsci.org/article/S0002-9629(15)34282-8/fulltext [Google Scholar]
- Barnette DA, Davis MA, Dang NL, Pidugu AS, Hughes T, Swamidass SJ, … Miller GP (2018). Lamisil (terbinafine) toxicity : Determining pathways to bioactivation through computational and experimental approaches. Biochemical Pharmacology, 156(August), 10–21. 10.1016/j.bcp.2018.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battig FA, Nefzger M, & Schulz G (1987). Major biotransformation routes of some Allylamine antimycotics. Recent Trends in the Discovery, Development, and Evaluation of Antifungal Agents, 479–495. [Google Scholar]
- Choudhary NS, Kotecha H, Saraf N, Gautam D, & Saigal S (2014). Terbinafine Induced Liver Injury : Journal of Clinical and Experimental Hepatology, 4(3), 264–265. 10.1016/j.jceh.2014.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Barnette DA, Flynn NR, Pidugu AS, Swamidass SJ, Boysen G, & Miller GP (2019). CYP2C19 and 3A4 Dominate Metabolic Clearance and Bioactivation 2 of Terbina fi ne Based on Computational and Experimental 3 Approaches 1. Chemical Research in Toxicology. 10.1021/acs.chemrestox.9b00006 [DOI] [PMC free article] [PubMed]
- Gantmacher J, Mills-Bomford J, & Williams T (1998). Interaction between warfarin and oral terbinafine. BMJ : British Medical Journal, 317(7152), 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isin EM, & Guengerich FP (2016). Substrate Binding to Cytochromes P450. Anal Bioanal Chem, 392(6), 1019–1030. 10.1007/s00216-008-2244-0.Substrate [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson SL, & Uetrecht JP (2001). Identification of a reactive metabolite of terbinafine: Insights into terbinafine-induced hepatotoxicity. Chemical Research in Toxicology. 10.1021/tx0002029 [DOI] [PubMed]
- Kawakami H, Ohtsuki S, Kamiie J, Suzuki T, Abe T, & Terasaki T (2011). Simultaneous Absolute Quantification of 11 Cytochrome P450 Isoforms in Human Liver Microsomes by Liquid Chromatography Tandem Mass Spectrometry with In Silico Target Peptide Selection. Journal of Pharmaceutical Sciences, 100(1), 341–352. 10.1002/jps.22255 [DOI] [PubMed] [Google Scholar]
- Kovarik JM, Mueller EA, Zehender H, Denoue J, Caplain H, & Millerioux L (1995). Multiple-Dose Pharmacokinetics and Distribution in Tissue of Terbinafine and Metabolites, 39(12), 2738–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamisil. (2004). Novartis Pharmaceuticals Corporation: East Hanover, NJ. [Google Scholar]
- Lee CR, Goldstein JA, & Pieper JA (2002). Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics, 12(3), 251–263. [DOI] [PubMed] [Google Scholar]
- Napodano J (2012). Anacor looks right for a long-term play. Seeking Alpha, 1–9. [Google Scholar]
- Niwa T, Murayama N, & Tamazaki H (2008). Heterotropic Cooperativity in Oxidation Mediated by Cytochrome P450. Current Drug Metabolism, 9(5), 453–462. [DOI] [PubMed] [Google Scholar]
- Perveze Z, Johnson MW, Rubin RA, Sellers M, Zayas C, Jones JL, … Shrestha R (2007). Terbinafine-Induced Hepatic Failure Requiring Liver Transplantation, 162–164. 10.1002/lt. [DOI] [PubMed] [Google Scholar]
- Rodrigues AD (2008). Drug-Drug Interactions (2nd ed.). CRC Press: Boca Raton, FL, 2008. [Google Scholar]
- Roth RA, & Dahm LJ (1997). Neutrophil- and glutathione-mediated hepatotoxicity of alpha-naphthylisothiocyanate. Drug Metabolism Reviews, 29(1–2), 153–165. [DOI] [PubMed] [Google Scholar]
- Ryder N (1992). Terbinafine: mode of action and properties of the squalene epoxidase inhibition. Br. J. Dermatol, 126(Suppl 39), 2–7. [DOI] [PubMed] [Google Scholar]
- Schellinger AP, & Carr PW (n.d.). Solubility of Buffers in Aqueous–Organic Eluents for Reversed-Phase Liquid Chromatography, 3. [Google Scholar]
- Siu YA, & Lai WG (2017). Impact of Probe Substrate Selection on Cytochrome P450 Reaction Phenotyping Using the Relative Activity Factor s, 183–189. [DOI] [PubMed] [Google Scholar]
- Song J, & Deresinski S (2016). Hepatotoxicity of antifungal agents, (March 2005). [PubMed] [Google Scholar]
- Terbianfine Product Monograph, Canada. (1995). Dorval, QC: Sandoz, Canada. [Google Scholar]
- Vago T, Baldi G, Colombo D, Barbareschi M, Norbiato G, Dallegri F, & Bevilacqua M (1994). Effects of Naftifine and Terbinafine, Two Allylamine Antifungal Drugs, on Selected Functions of Human Polymorphonuclear Leukocytes, 38(11), 2605–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers AEM, Sinclair JR, Zollinger M, Heitz F, Glänzel U, Johanson L, & Fischer V (1999). Multiple cytochrome P-450s involved in the metabolism of terbinafine suggest a limited potential for drug-drug interactions. Drug Metabolism and Disposition. [PubMed] [Google Scholar]
- Yu A, & Haining RL (2001). Comparative contribution to dextromethorphan metabolism by cytochrome P450 isoforms in vitro: Can dextromethorphan be used as a dual probe for both CYP2D6 and CYP3A activities? Drug Metabolism and Disposition, 29(11), 1514–1520. [PubMed] [Google Scholar]
- Zaretzki JM, Browning MR, Hughes TB, & Swamidass SJ (2015). Structural bioinformatics Extending P450 site-of-metabolism models with region-resolution data, 31(February), 1966–1973. 10.1093/bioinformatics/btv100 [DOI] [PubMed] [Google Scholar]