Abstract
Xenotransplantation of pig organs into people may help alleviate the critical shortage of donors which faces organ transplantation. Unfortunately, human antibodies vigorously attack pig tissues preventing the clinical application of xenotransplantation. The swine leukocyte antigens (SLA), homologs of human HLA molecules, can be xenoantigens. SLA molecules, encoded by genes in the pig major histocompatibility complex, contribute to protective immune responses in pig. Therefore, simply inactivating them through genome engineering, could reduce the ability of the human immune system to surveil transplanted pig organs for infectious disease or the development of neoplasms. A potential solution to this problem is to identify and modify epitopes in SLA proteins to eliminate their contribution to humoral xenoantigenicity while retaining their biosynthetic competence and ability to contribute to protective immunity. We previously showed that class II SLA proteins were recognized as xenoantigens and mutating arginine at position 55 to proline, in an SLA-DQ beta chain, could reduce human antibody binding. Here we extend these observations by creating several additional point mutants at position 55. Using a panel of monoclonal antibodies specific for class II SLA proteins, we show that these mutants remain biosynthetically competent. Examining antibody binding to these variants shows that point mutagenesis can reduce, eliminate, or increase antibody binding to class II SLA proteins. Individual mutations can have opposite effects on antibody binding when comparing samples from different people. We also performed a preliminary analysis of creating point mutants near to position 55 to demonstrate that manipulating additional residues also affect antibody reactivity.
Keywords: Xenotransplantation, Swine, Leukocyte Antigens, SLA-DQ, mutagenesis
Introduction
The presence of donor specific antibodies (DSA) has precluded the clinical application of xenotransplantation, but genome editing makes it possible to delete or alter xenoantigens so that the DSA no longer bind to the pig cells (Estrada 2015). The deletion of three glycan xenoantigens, α-gal, N-glycolylneuraminic acid, and the Sda has improved the crossmatch in all patients tested to the point where more than 30% of the patients tested have no detectable DSA (Martens 2017). While this is a very exciting development, there are still nearly 70% of patients with DSA present against this triple knockout (TKO) pig, making these patients likely to experience early antibody mediated rejection if they were to receive a TKO pig organ xenotransplant. Determining the identity of the remaining xenoantigens is critical to developing genome editing strategies that create new donor pigs for those patients who still have DSA to the TKO pig.
According to Organ Procurement and Transplantation Network, approximately 50% of the patients on the transplant waiting list are sensitized to HLA antigens, making it difficult to find acceptable human donor kidneys to which they have a negative crossmatch (https://optn.transplant.hrsa.gov/data/view-data-reports/national-data/). Some of these anti-HLA antibodies bind to SLA antigens present on the TKO pig (Mulder 2010; Varela 2003; Oostingh 2002; Naziruddin 1998; Taylor 1998; Ladowski 2018). The anti-HLA antibodies can bind to public SLA epitopes that are conserved across evolution so that they are present in all breeds of pig (Ladowski 2018). This means that it will not be feasible to simply choose a specific strain of pig that lacks these specific epitopes, which makes the SLA molecules potential targets for genome editing.
Anti-HLA class II antibodies play an important role in renal allograft failure, and are the most common cause of late graft loss (Devos 2012; Campos 2006; Langan 2007). Some anti-HLA class II antibodies bind to SLA class II molecules, since the class II HLA and SLA share common epitopes. Genome editing of SLA II could be used to eliminate binding of some anti-HLA class II cross-reactive antibodies. Our lab has developed tools to be able to examine the impact of genome editing particular epitopes on SLA class II.
Previously we showed that human IgG specific for HLA-DQ4, -DQ5, and -DQ6 molecules (HLA-DQ4,5,6) also cross-reacted with SLA-DQ proteins consisting of the SLA-DQα*0101 and SLA-DQβ1*0601 heterodimers (Ladowski 2018). As noted in the HLA epitope registry (http://www.epreqistrv.com.br/), this pattern of HLA reactivity suggests that antibodies recognize the 52PR epitope. This antigen contains a proline and arginine as amino acids 52 and 55 respectively in the resides in β chains of HLA-DQ4,5,6 and SLA-DQβ1*0601. We showed that human IgG binding could be affected by mutating amino acid 55 in SLA-DQβ1*0601 from an arginine to a proline (R55P) (Ladowski 2018). For 21 unique human sera exhibiting this pattern of reactivity, the R55P mutation eliminated IgG binding from three samples, increased binding of one sample, and partially reduced binding in the remaining 17 samples (Ladowski 2018). The origin of these antibodies, whether naturally occurring or the result of a sensitization event, were unknown. Here we extended these observations by examining antibody binding to SLA-DQ heterodimers having R55 replaced with amino acids other than proline. Eight other amino acids predicted to lie within 15Å radius of position 55 were also mutated in an effort to disrupt the entire antibody binding surface on SLA-DQ.
Materials and Methods
Research Oversight
All human sera were collected and used in accordance with IRB-approved protocols. Pig red blood cells were collected under IACUC approved protocols.
Culture of HEK 293T Cell Lines
HEK 293T cells (ATCC CRL-3216) were cultured in minimum essential media (MEM-α) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan UT) in collagen-I-coated plates (Becton Dickinson, Bedford, MA) at 37°C and 5% CO2. Cells were confirmed to be class II HLA negative by incubation with an anti-HLA-DR Ab (Clone L243) and anti-HLA-DQA1 (Clone DQA1) and analyzed using BD Accuri C6 Flow Cytometer (BD Biosciences, San Jose, CA). The cells were made class I HLA deficient as previously described by Ladowski et al ( Ladowski 2018).
Creation of β2m Deficient HEK 293T Cells Expressing Mutated SLA-DQ Molecules
The cells were generated as previously described with the SLA-DQα/β genes from the Hp-0.23 haplotye ( Ladowski 2018; Reyes 2014). Cells were made β2m-deficient in an effort to avoid “background” binding of antibodies that may be present in the samples and capable of interacting with class I HLA proteins. The bicistrionic expression vector, pBudCE4.1 (Thermo Fisher Scientific, Waltham, MA), was engineered to express cDNA encoding SLA-DQA1*0101 (NCBI accession KJ776451.1) behind the CMV promoter and SLA-DQB1*06:01 (NCBI accession KJ776454.1) behind the EF-1a promoter using restriction enzyme digestion and DNA ligation. A second plasmid, pCDNA3.1+/Hygro (Thermo Fisher Scientific, Waltham, MA) encoding the swine invariant chain (NCBI accession AB527073.1) was also created using restriction enzyme digestion and DNA ligation.
The 55Arg residue of the –DQB1*06:01 gene was previously converted to 55Pro as described by Ladowski et al (Ladowski 2018) using the QuickChange XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). The 55R residue was converted to 55E using the following primers: Forward: 5’-GTGAGTGACCCCGCTGGGGGAGCCGGACGCCGACTAC-3’ and Reverse: 5’- GTAGTCGGCGTCCGGCTCCCCCAGCGGGGTCAC-3’. Mutation of 55R to 55A used the following primers: Forward: 5’ -GTGAGTGACCCCGCTGGGGGCGCCGGACGCCGACTAC-3’ and Reverse: 5’- GTAGTCGGCGTCCGGCGCCCCCAGCGGGGTCAC-3’
The cells were transfected with the vector pCDNA3.1+/Hygro containing the swine invariant chain using a calcium phosphate protocol: 1 × 106 cells were plated into a 6-well dish and transfected by adding a cocktail of 214 uL water, 31 uL 2 M CaCI2, 2.5 ug DNA, and 250 uL of 2x HBS. SLA-DQ expression was maximized by co-expression of CD74 (Ladowski 2018). Negative control cells (−) were transfected with pig CD74 only. The cells were placed into selection with Hygromycin under normal culture conditions described above. Following stable selection, the cells were transfected with either the pBUDCE4.1-SLA-DQ R55E or R55A plasmids, grown in normal culture conditions and sorted at the UAB Comprehensive Flow Cytometry Core on a BD FACSAria II for functional class II SLA expression. The sorted cells were analyzed by a monoclonal anti-CD74 (Cerclip.1) antibody (Novus Biologicals, Littleton, Colorado, USA).
Monoclonal Antibody Binding to SLA-DQ Cells
A panel of monoclonal SLA-DQ antibodies were used to evaluate expression (Table 1: Clone PG-TH22A, PG-TH21A, P-TH81A5, and PG-H42A Washington State Monoclonal Antibody Center, Pullman, WA and Clone K274.3G8, Bio-Rad, Hercules, CA). The cells were stained in triplicate at an antibody dilution of 1:50 with traditional immunostaining technique: 200,000 cells in a 100 uL volume were incubated with 1% BSA in 1X PBS for five minutes to prevent non-specific binding. 10 uL of diluted monoclonal antibody stain was added to the cell volume and incubated for 30 min at 4°C. The cells were washed with three times with 1X PBS, and re-suspended in a 1:250 dilution of secondary antibody (Goat anti-Mouse IgG1 PE for mAb 4 or Goat anti-Mouse lgG2b Cross-adsorbed Secondary Antibody, Alexa Flour 647 for mAbs 1,2,3, and 5, Thermo Fisher Scientific, Waltham, MA), incubated at 4°C for 30 min, and washed three times with 1X PBS. Flow cytometry was performed as previously described by Ladowski et al at the UAB Comprehensive Flow Cytometry Core on a Thermo Fisher Scientific Attune Nxt ( Ladowski 2018).
Table 1:
Monoclonal Antibodies Specific for SLA-DQ
| # | Monoclonal Antibody |
|---|---|
| 1 | PG-TH22A |
| 2 | PG-TH21A |
| 3 | PG-TH81A5 |
| 4 | K274.3G8 |
| 5 | PG-H42A |
Class II Modelling and Selection of Residues for Mutation within a 15Å Distance of 55Arg
SLA class II molecule structure predictions were created in SWISS-MODEL (University of Basel, Basel, Switzerland) and visualized in UCSF Chimera (University of California, San Francisco, CA). The measure-by-distance function in UCSF Chimera was used to evaluate all amino acids within a 15Å radius of 55Arg (Pettersen 2004; Chen 2014). A BLAST analysis comparing SLA-DQB1*06:01, which exhibited the highest reactivity with human antibodies of the class II SLA that we previously studied (Ladowski 2018) and a second less reactive SLA-DQ molecule, SLA-DQB1*03:03, demonstrated 8 polymorphic residue differences within the 15Å radius between the two proteins. The amino acids were converted to more closely resemble the alternate -DQ molecule using the QuickChange XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). The cells were cultured and transfected as described above.
Human Antibody Binding to SLA-DQ Cells
22 human sera samples suspected to have anti-SLA-55Arg antibodies with sufficient volume for further analysis were tested for IgG and IgM antibody binding to the SLA-DQ 55R, 55P, 55E, and 55A cells. Sera treatment and methods of flow cytometric analyses were described previously (Ladowski 2018). Briefly sera were heat inactivated (57C for 30 minutes) and then absorbed on an equal volume of packed wild-type pig red blood cells to deplete non-SLA xenoreactive antibodies.
Statistical Analysis of Antibody Binding
Antibody binding results were analyzed by flow cytometry using FlowJoV10 software (FlowJo LLC, Ashland, GA) and reported as median fluorescence intensity (MFI) (Figs 1B and 3). Graph and data analyses were completed using Prism 7 for Macintosh (GraphPad Software Inc., La Jolla, CA, USA).
Figure 1:
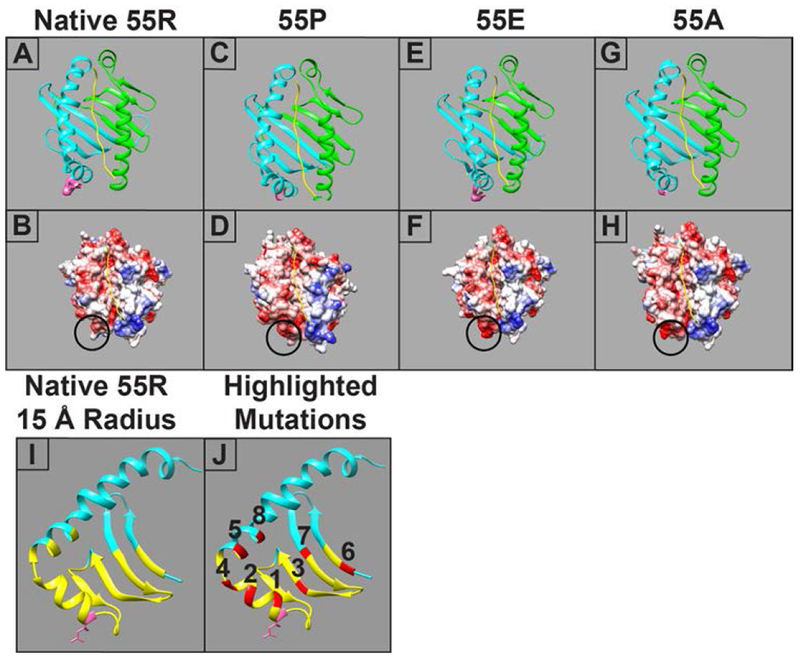
Molecular modeling of the SLA-DQα*0101 and SLA-DQβ1*0601 heterodimer. Four variants are shown, each having a unique amino acid at position 55 in the beta chain. Upper panels show ribbon models of SLA-DQ with the α chain in green, the DQ β chain in blue, and the peptide in yellow. The side chain of the amino acid 55 is shown in a space-filling representation and colored pink. The lower panels show models of surface electrostatic potential of the various SLA-DQ proteins ranging from −10 kcal/e.u (red) to 0 kcal/e.u (white) and +10 kcal/e.u (blue). The circle highlights the location of amino acid 55. Panels A,B: naturally occurring SLA-DQ molecule with arginine at position 55 in the SLA-DQβ1*0601 chain (55R). Panels C,D: SLA-DQβ1*0601 with position 55 mutated to proline (55P). Panels E,F: SLA-DQβ1*0601 with position 55 mutated to glutamate (55E). Panels G,H: SLA-DQβ1*0601 with position 55 mutated to alanine (55A). Panel I: A ribbon model showing the predicted structure of the natural variant of SLA-DQβ1*0601 chain having arginine at position 55 (pink amino acid with side chain). The yellow portions of the ribbon highlight a region of the protein within 15 Å of amino acid 55. Panel J: Eight amino acid positions, highlighted in red, lie within the 15 Å radius of amino acid 55 and were chosen for mutagenesis studies. The specific amino acid changes at each position are shown in table 2.
Figure 3:
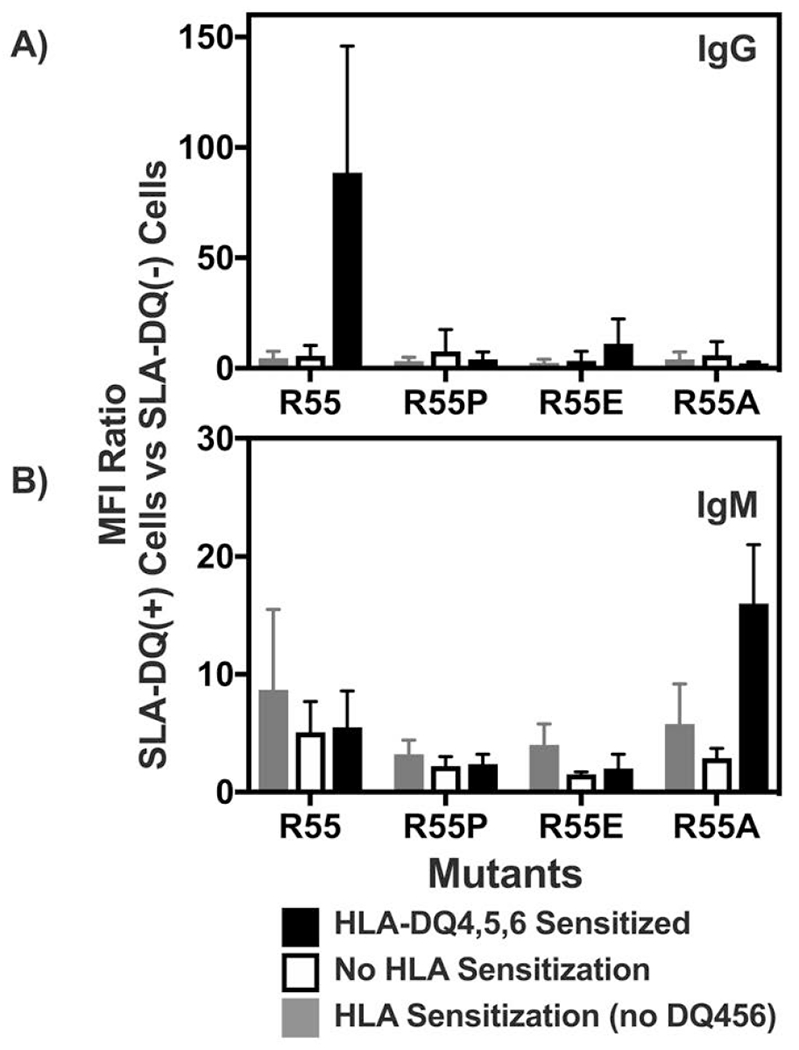
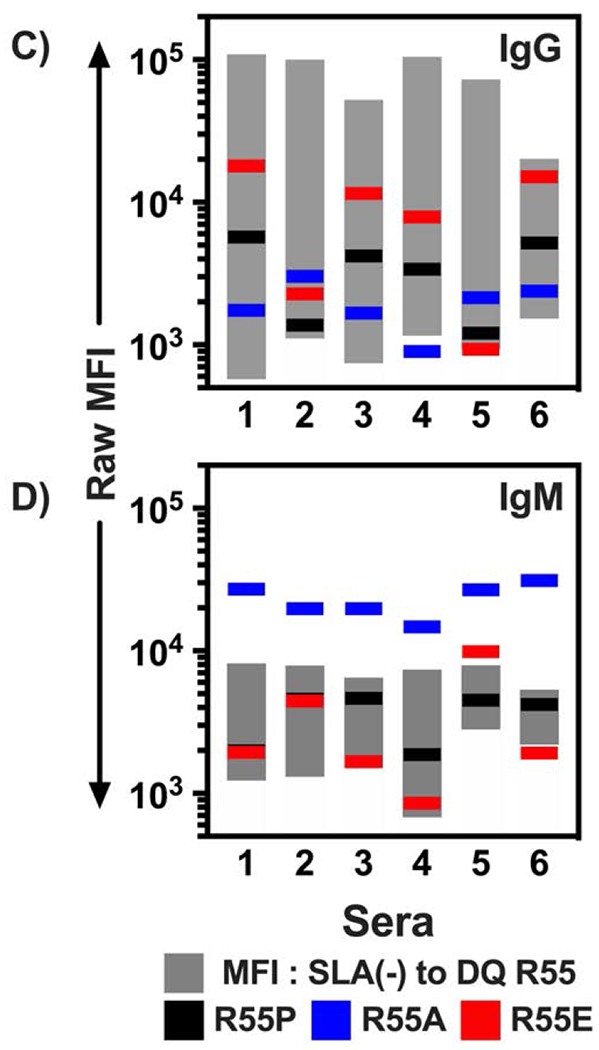
Flow cytometric analysis of human immunoglobulin binding to heterodimers of SLA-DQα*0101 and SLA-DQβ1*0601. The SLA-DQβ chain was mutated at position 55 to change arginine to either proline (R55P), alanine (R55A), or glutamate (R55E). Panels A and B: Human sera having no HLA sensitization (n=6, white bars), class II HLA sensitization, but not towards HLA-DQ4,5,6 (n=10; gray bars), or class II sensitization against HLA-DQ4,5,6 (n=6, black bars) were tested for reactivity with cells expressing individual SLA-DQ variants. Human IgG (panel A) and IgM (panel B) binding to each SLA-DQ variant was evaluated by flow cytometry. MFI of antibody binding to SLA-DQ expressing cells was divided by the MFI of the identical sample binding to cells lacking SLA-DQ expression (MFI Ratio). Means and standard deviations are shown. HLA-DQ456 sensitized sera exhibited high levels of IgG that reacted with cells expressing the R55 variant of SLA-DQ. All three mutants of amino acid 55 significantly reduced IgG binding (R55 vs R55P, p = 0.047; R55 vs R55E, p = 0.047, and R55 vs R55A, p = 0.048). IgM binding results were similar across the different sera and mutants with one exception. In HLA-DQ456 sensitized sera, the R55A mutant increased IgM binding to SLA-DQ (R55 vs R55A, p = 0.001). Panels C and D show the raw MFI of IgG (panel C) and IgM (panel D) binding from each of the six HLA-DQ456 sensitized sera to all four variants of SLA-DQ and to negative control cells that are SLA deficient. The bottom of the gray bar represents the MFI of each sample binding to the class II SLA-deficient cells, and the top of the gray bar represents the binding to cells expression the R55 variant of SLA-DQ. The data in panels A and B were analyzed using one-way ANOVA and Dunnet’s correction for multiple comparisons.
Results
We created four variants of SLA-DQβ1*0601 to determine how different amino acids at residue 55 modify antigenicity of the SLA-DQ protein. Arginine is the 55th amino acid in the naturally occurring SLA-DQβ1*0601 (noted as R55). Three additional variants of this β chain included replacement of R55 with proline (R55P), glutamate (R55E) and alanine (R55A). Fig 1 shows a ribbon diagram and a surface charge model of the various SLA-DQ heterodimers. These highlight structural and electrostatic changes predicted to occur with each amino acid change.
HEK293 cells were used to express each of the four SLA-DQβ1*0601 variants in addition to SLA-DQα*0101. Cell surface expression of the different SLA-DQ proteins was examined using five different SLA-DQ specific monoclonal antibodies and flow cytometry (Table 1). Three of the monoclonal antibodies yielded uniform histogram distributions for the different variants allowing comparison of their expression intensity (Fig 2A, mAb1,2,4). Mab1 showed equivalent binding to R55, R55A, and R55E demonstrating identical expression of the variants by the HEK cells (Figs 2A and 2B). The R55E mutation reduced binding of mAb2 and mAb4 to SLA-DQ (Fig 2A, C, and D). R55P reduced binding of mAbs 1,2,3,5 and eliminated binding of mAb 4 (Fig 2A–D). Lastly, the alanine mutation reduced mAb4 binding, and the glutamate mutation eliminated recognition of SLA-DQ by mAb4 (Fig 2A–D).
Figure 2:
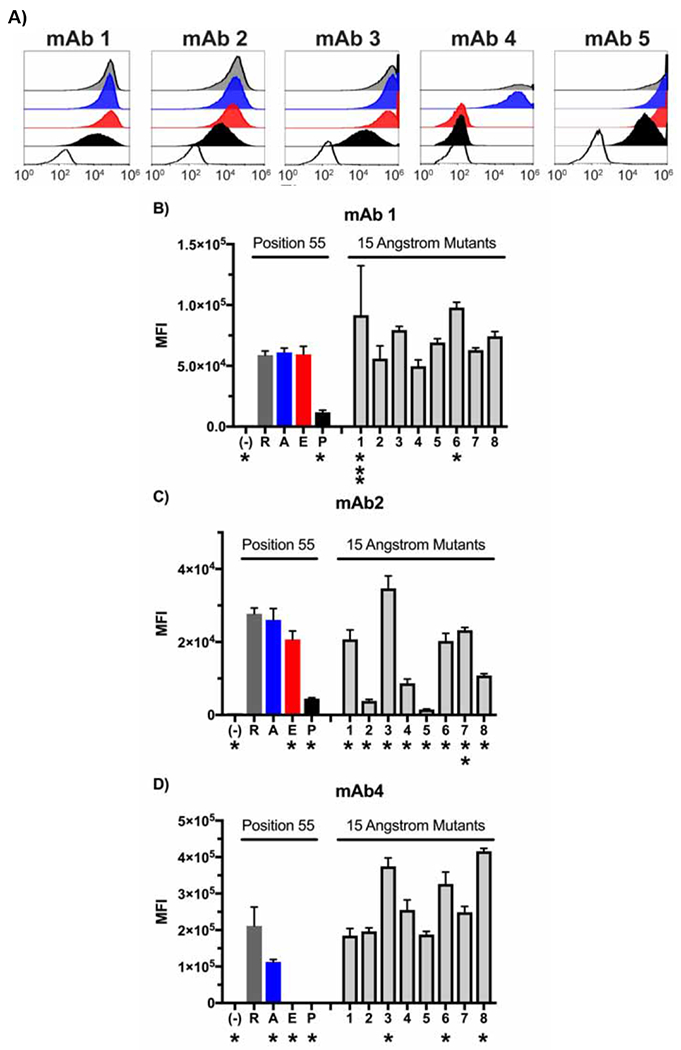
Expression analyses of point-mutant variants of the SLA-DQ protein. Panel A: HEK293 cells co-expressing SLA-DQα*0101 and SLA-DQβ1*0601 (R55, gray histograms) or position 55 mutants of SLA-DQβ1*0601. Cells were stained with five different monoclonal antibodies and analyzed by flow cytometry. Mutations analyzed included alanine (55A, blue histograms), glutamate (55E, red histograms), or proline (55P, black histograms). SLA-DQ deficient but pig CD74 positive cells were also stained with the monoclonal antibodies as a negative control (white histograms). Triplicate stainings of these cells were performed with panels B, C, and D showing the mean and standard deviation for the MFI of three monoclonal antibodies (mAb1,2, and 4). SLA-DQβ1*0601 mutants in eight amino acids within a 15 Angstrom radius of amino acid 55 were also tested. In panels B and C, asterisks note mutations that show significant alteration in SLA-DQ expression when compared to the naturally occurring SLA-DQ variant with arginine at position 55 of the beta chain (R). Binding comparisons were made using one-way ANOVA with Dunnet’s correction for multiple comparisons. One asterisk marks p-values less than 0.001. Two asterisks mark p value of 0.042, and three asterisk marks a p value of 0.023. No asterisk indicates p value of comparison greater than 0.05.
Fig 1I also shows a ribbon diagram of the SLA-DQβ1*0601 protein with yellow highlighting amino acids nearby the 55th amino acid (within 15Å). Eight residues in this radius, highlighted in red (Fig 1J), were mutated to determine how they affect antibody binding. Specific mutations were chosen based on two criteria: 1) proximity to amino acid 55, and 2) residues from SLA-DQB1*06:01 were replaced with the corresponding amino acids in SLA-DQB1*03:03 alleles in an effort to maintain the biosynthetic competence of the mutant SLA. Table 2 shows the exact amino acid changes made at each of these residues and their calculated distance from residue 55. The binding of mAb1,2, and 4 to these eight variants of SLA-DQ was evaluated (Fig 2B–D). 15 Å mutants having differential interaction with monoclonal antibodies relative to the 55R variant are marked by asterisks in Fig 2B–D. Some variants increased binding of monoclonal antibodies (Fig 2D, variant 8 as an example), while others reduced monoclonal antibody binding (Fig 2C, variant 5 as an example). Panel 2C shows that mAb2 appears most sensitive to 15Å variants 2 and 5. This reduced binding may reflect disruption of an epitope recognized by mAb2 because mAb1 indicates that the expression level of these mutants is similar to the naturally occurring SLA-DQ (Fig 2B position 55R vs 15Å mutants 2 and 5).
Table 2:
Epitope Mutants Near Amino Acid 55
| Variant | #Å | Amino Acid Change |
|---|---|---|
| 1 | 4Å | D57E |
| 2 | 5Å | Y60S |
| 3 | 7Å | V38L |
| 4 | 9Å | G63S |
| 5 | 11Å | F67V |
| 6 | 12Å | Y9V |
| 7 | 13Å | T28A |
| 8 | 15Å | T71K |
The cell lines expressing SLA-DQ with mutations at position 55 in the beta chain were then probed with various human sera. Three types of sera samples were tested in Fig 3: (i) Sera lacking HLA reactive IgG; (ii) sera having IgG that bound class II HLA molecules but not HLA-DQ4,5,6; or (iii) sera that contained HLA-DQ4,5,6 reactive IgG. As noted previously, a pattern of HLA-DQ4,5,6 binding strongly implicates arginine at position 55 in the beta chain as a key antigenic residue. IgG from the HLA-DQ456 sera demonstrated elevated binding to the R55 SLA-DQ variant. Mutating position 55 to alanine, glutamate, or proline significantly reduced the IgG reactivity of HLA-DQ456 sera towards SLA-DQ down to levels approximating the reactivity of IgG from the HLA unsensitized sera and the HLA-sensitized sera lacking antibodies towards HLA-DQ456 (Fig 3A). IgM binding across the different sera and SLA-DQ mutants exhibited limited variability with one notable exception. The R55A mutation caused an approximate 3-fold elevation of IgM binding to SLA-DQ, relative to R55, when analyzing sera having IgG specific for HLA-DQ456 (Fig 3B).
The raw data, from HLA-DQ456 sera used to create Figs 3A and 3B, are shown in Figs 3C and 3D to demonstrate how antibody binding to each SLA-DQ mutant varies from person to person. Typically, the trends are similar (i.e. if the mutation reduces IgG binding of one serum it reduces binding of all tested serum) though the magnitude of change can vary. Importantly, a single point mutation can eliminate nearly all serum IgG reactivity with SLA-DQ for humans. As noted in Fig 3B, the R55A mutation increased IgM binding for every serum sample tested despite markedly reducing IgG binding.
We next examined how the 15Å mutants (Fig 1 and Table 2) affected human IgG and IgM binding to SLA-DQ (Fig 4). Cells expressing the individual SLA-DQ molecules, one 15Å variant type per cell, were incubated with six human sera containing IgG specific for HLA-DQ456. These were the same serum samples tested in Fig 3. Relative to the R55 variant of SLA-DQ, the mutant T28A reduced IgG binding. Mutant Y9V slightly elevated human antibody binding and the remaining mutants exhibited statistically similar binding.
Figure 4:
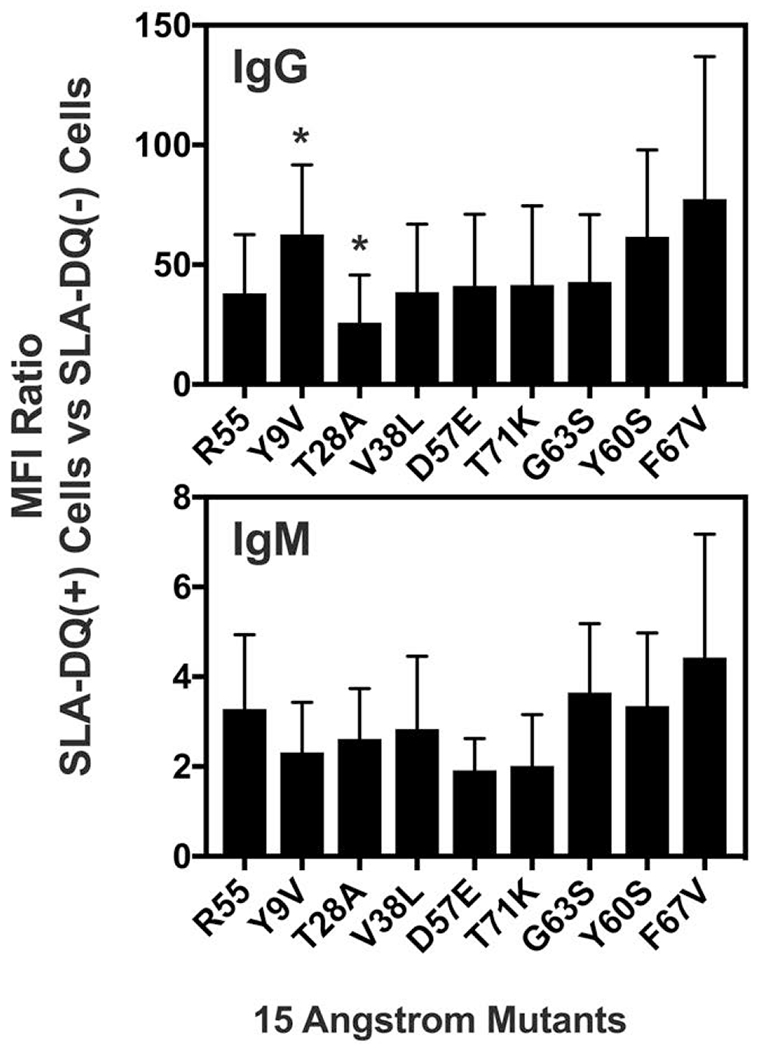
Binding of human antibodies to SLA-DQα*0101 and SLA-DQβ1*0601 heterodimers having beta chain point mutations within 15 angstroms of residue 55. Specific amino acid changes are noted as x-axis labels with as the single letter abbreviation of the original amino acid-the residue number-the mutant amino acid. Distances of each residue from position 55 are listed in table 2. Human IgM and IgG binding to cells expressing each variant of SLA-DQ were tested by flow cytometry with raw MFI values shown. Sera samples were collected from people having been sensitized to HLA-DQ456. These are the same samples tested in Fig 3. Cells devoid of SLA were used as negative controls. MFI ratios of IgG or IgM binding were SLA-DQ expressing cells vs SLA deficient cells were calculated. Means and standard deviations of these ratios for the six sera are shown. One-way Anova with Dunnet’s correction for multiple comparisons showed that the Y9V caused slight but significant elevations in IgG binding relative to the R55 SLA-DQ variant (p = 0.002). The T28A mutant slightly reduced IgG binding relative to the R55 SLA-DQ molecule (p = 0.020). All other IgG and IgM binding alterations did not reach statistical significance (p>0.05).
Discussion
Genetic engineering to eliminate xenoantigen expression is key to enabling the use of pig organs in human beings. To date, carbohydrate-based antigens can be eliminated simply by disrupting genes which guide their synthesis. Elimination of the carbohydratemodifying genes GGTA1, CMAH, and B4GalNT2 have reduced, and in some cases eliminated, human antibody binding to pig cells (Martens 2017). It has been possible to generate healthy pigs having these gene disruptions (Estrada 2015; Zhang 2018). The SLA proteins also contribute to xenoantigenicity in some people (Martens 2017; Mulder 2010; Varela 2003; Oostingh 2002; Naziruddin 1998; Taylor 1998; Ladowski 2018; Ladowski 2018). It is possible to create knockout animals lacking SLA gene expression and reduce human antibody binding to their cells (Martens 2017, Reyes 2014). However, given the central role of SLA in protective immunity, simply inactivating these genes may prove detrimental to animal health and possibly to immune surveillance of the transplanted organ. Therefore, we have been evaluating whether it is possible to eliminate SLA antigenicity while retaining SLA function. This report studies the effects of point mutations on class II SLA expression and interaction with human antibodies.
Our experimental system utilized human sera containing IgG specific for HLA-DQ456 molecules. This pattern of reactivity likely arises from antibodies recognizing a common epitope in the HLA-DQβ1 proteins of these molecules which consists of proline at amino acid 52 and arginine at position 55 (http://www.epregistry.com.br/). This same epitope, present in nearly every SLA-DQβ1 protein (https://www.ebi.ac.uk/ipd/mhc/group/SLA), may cause cross-reactivity of HLA-DQ-specific antibodies and SLA-DQ, with R55 being a key epitope residue (Fig 3; Ladowski 2018). The naturally occurring arginine at position 55 is likely to be positively charged at the typical extracellular pH of ~7.2. Therefore, we replaced it with the acidic amino acid glutamate (R55E) which is typically negatively charged at neutral pH. Next, we created a variant of SLA-DQβ1*0601 having an alanine at position 55 (R55A). The side chain of alanine is a methyl group which does not carry a charge. Lastly, we also made cells expressing the R55P variant of SLA-DQβ1*0601. With these variants, we hoped to examine the effects of structural disruption (R55P) and charge variability (R55, R55E, R55A) on antibody reactivity with SLA-DQ.
We evaluated the interaction of five SLA-DQ reactive monoclonal antibodies with the aforementioned mutants. This provides the simplest experimental system in which a single antibody interaction is evaluated against mutated SLA molecules. The binding of mAb1 indicated that the R55A and R55E cell surface expression levels closely matched the levels seen with the naturally occurring R55 molecule. This suggests that mAb1 may recognize an epitope that does not include amino acid 55. MAb4 recognized an epitope containing R55 because all three SLA-DQ mutants either decreased (R55E and R55P) or eliminated (R55A) its binding. MAb2 binding was slightly reduced by the R55E and R55P mutations suggesting this antibody may also recognize an epitope containing position 55 (Fig 2C). These results demonstrate that point mutagenesis of SLA-DQ can reduce or eliminate antibody binding while retaining normal levels of cell surface expression of the protein.
We also created mutants where residues within 15Å of amino acid 55 were altered. This was performed to determine whether manipulating other portions of the putative epitope surface could also modulate antibody binding. The 15Å mutants provide a second example that SLA-DQ humoral immunogenicity can be reduced by altering the amino acid sequence of SLA-DQ. Mutants 2 (Y60S) and 5 (F67V) markedly diminished mAb2 binding (Fig 2C). Binding of mAb1 and mAb4 was not altered to a level of statistical significance by these changes indicating that the expression of these molecules reasonably matched the naturally occurring R55 variant. In contrast, statistically significant loss of mAb2 binding arose from lack of an epitope rather than diminished cell surface expression of both mutants.
Once the monoclonal antibodies demonstrated that mutant SLA-DQ exhibit reduced xenoantigenicity while retaining the biosynthetic competence, we next tested if these variants affected binding of IgG and IgM from human serum (Fig 3). Sera may contain polyclonal immunoglobulin against an individual epitope or antibodies against multiple SLA-DQ epitopes. Therefore, it is possible that mutagenesis of SLA will have less effect on serologic antibody reactivity than on the monoclonal antibodies. Six sera were collected on the basis of their having IgG specific for HLA-DQ456 which suggests they may react with SLA-DQ containing 55R in the β chain. R55P and R55E mutations reduced human IgM binding. IgG reactivity against the R55E, R55P, and R55A variants was also diminished. These data suggest that amino acid residue 55 comprises an epitope in SLA-DQ that is targeted by antibodies generated in people who have been sensitized to HLA-DQ456. The fact that the R55A mutant increased IgM binding to SLA-DQ also supports this conclusion (Fig 3B and 3D). Why R55A increases IgM reactivity is unknown. This phenomenon does not occur when testing allosensitized sera containing IgG specific for class II HLA other than HLA-DQ456 (Fig 3B), though IgM HLA reactivity was unknown in these sera. IgG interaction with SLA-DQ was diminished by each mutation at position 55 in the sera containing HLA-DQ456 reactive antibodies (Fig 3A and 3C).
The six human serum samples containing HLA-DQ456 reactive IgG were also tested against the 15Å mutants. Relative to the R55 variant, the T28A (mutant 7) was the only molecule which exhibited reduced binding to IgG from human serum (Fig 4). This likely reflects an alteration in the epitope as mAb1 and mab4 staining indicated that T28A and R55 SLA-DQ proteins exhibited similar levels of cell surface expression (Fig 2B and 2D). As residue 28 lies in the peptide antigen groove, it probably does not directly interact with bound antibody. Therefore, its effects on humoral antigenicity potentially are exerted by altering peptides in the SLA-DQ molecule which in turn alters binding of some antibodies. The 15Å mutants were also tested for their pattern of IgM reactivity with these sera. No statistically meaningful changes in IgM binding were observed. It will be necessary to test additional amino acids at each position in the 15Å mutants to determine if there are specific amino acid changes which would more effectively reduce antibody binding.
This study demonstrates that mutagenesis reduces the xenoantigenicity of SLA-DQ. Developing pig tissues that are physiologically and immunologically compatible with humans is key to making xenotransplantation feasible. Humoral and complement-mediated destruction of pig tissues has been a major barrier to the success of xenotransplantation. The xenoantigenicity of SLA molecules offers several options to deal with the humoral challenge. One solution to this challenge is express transgenes encoding complement regulators to over-ride complement activation (Adams 2001; Chen 1999; Niemann 2001; Schurmann 2002; Cowan 2000) which may occur following antibody binding to SLA proteins (Ladowski 2018). Another solution could be to inactivate SLA genes to eliminate antibody binding. These approaches may be suboptimal as they could rob pig cells and tissues of protective immunity that would follow excessive complement inactivation or the deletion of SLA which are central to cell-mediated immunity. A third option which this study begins to address, is to mutagenize epitopes responsible for xenoantigenicity. If successful, a mutagenic approach could eliminate antibody binding and avoid humoral rejection of pig tissues, while leaving cell-mediated immune responses intact. Of the mutants tested so far, the R55E appears to yield the best results in that it does not diminish SLA-DQ expression, yet it reduces IgM and IgG binding for the majority of sera tested. R55P was suboptimal as it may impair the biosynthetic competence of SLA-DQ proteins. Though R55A maintained expression levels and markedly reduced IgG binding, it increased IgM binding. Clearly more work is needed to define the optimal mutagenesis strategy to achieve this goal.
Disclosures:
G.R.M. is supported by a research scholarship from the American College of Surgeons. A.J.T. is the founder of Xenobridge LLC, and holds and applied for patents related to xenotransplantation. Portions of this work have been funded by University of Alabama at Birmingham, United Therapeutics, and the American College of Surgeons.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References:
- Adams DH, Kadner A, Chen RH, Farivar RS, Logan JS, Diamond LE. Human membrane cofactor protein (MCP, CD 46) protects transgenic pig hearts from hyperacute rejection in primates. Xenotransplantation. 2001. February;8(1):36–40. [DOI] [PubMed] [Google Scholar]
- Campos EF, Tedesco-Silva H, Machado PG, Franco M, Medina-Pestana JO, Gerbase-DeLima M. Post-transplant anti-HLA class II antibodies as risk factor for late kidney allograft failure. American Journal of Transplantation. 2006. October;6(10):2316–20. [DOI] [PubMed] [Google Scholar]
- Chen JE, Huang CC, Ferrin TE. RRDistMaps: a UCSF Chimera tool for viewing and comparing protein distance maps. Bioinformatics. 2014. December 24;31(9):1484–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Naficy S, Logan JS, Diamond LE, Adams DH. Hearts from transgenic pigs constructed with CD59/DAF genomic clones demonstrate improved survival in primates. Xenotransplantation. 1999. August;6(3):194–200. [DOI] [PubMed] [Google Scholar]
- Cowan PJ, Aminian A, Barlow H, Brown AA, Chen CG, Fisicaro N, Francis DM, Goodman DJ, Han W, Kurek M, Nottle MB. Renal xenografts from triple-transgenic pigs are not hyperacutely rejected but cause coagulopathy in non-immunosuppressed baboons. Transplantation. 2000. June 27;69(12):2504–15. [DOI] [PubMed] [Google Scholar]
- DeVos JM, Gaber AO, Knight RJ, Land GA, Suki WN, Gaber LW, Patel SJ. Donor-specific HLA-DQ antibodies may contribute to poor graft outcome after renal transplantation. Kidney international. 2012. September 1;82(5):598–604. [DOI] [PubMed] [Google Scholar]
- Estrada JL, Martens G, Li P, Adams A, Newell KA, Ford ML, Butler JR, Sidner R, Tector M, Tector J. Evaluation of human and non-human primate antibody binding to pig cells lacking GGTA 1/CMAH/β4Gal NT 2 genes. Xenotransplantation. 2015. May;22(3):194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladowski JM, Reyes LM, Martens GR, Butler JR, Wang ZY, Eckhoff DE, Tector M, Tector AJ. Swine Leukocyte Antigen Class II Is a Xenoantigen. Transplantation. 2018. February 1;102(2):249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladowski JM, Martens GR, Reyes LM, Wang ZY, Eckhoff DE, Hauptfeld-Dolejsek V, Tector M, Tector AJ. Examining the Biosynthesis and Xenoantigenicity of Class II Swine Leukocyte Antigen Proteins. The Journal of Immunology. 2018. March 14:ji1800022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan LL, Park LP, Hughes TL, Irish A, Luxton G, Witt CS, Christiansen FT. Post-transplant HLA class II antibodies and high soluble CD30 levels are independently associated with poor kidney graft survival. American journal of transplantation. 2007. April;7(4):847–56. [DOI] [PubMed] [Google Scholar]
- Martens GR, Reyes LM, Li P, Butler JR , Ladowski JM, Estrada JL, Sidner RA, Eckhoff DE, Tector M, and Tector AJ, Humoral reactivity of renal transplant-waitlisted patients to cells from GGTA1/CMAH/B4GalNT2, and SLA class I knockout pigs. Transplantation. 2017. April 1;101(4):e86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder A, Kardol MJ, Arn JS, Eijsink C, Franke ME, Schreuder GM, Haasnoot GW, Doxiadis II, Sachs DH, Smith DM, Claas FH. Human monoclonal HLA antibodies reveal interspecies crossreactive swine MHC class I epitopes relevant for xenotransplantation. Molecular immunology. 2010. January 1;47(4):809–15. [DOI] [PubMed] [Google Scholar]
- Naziruddin B, Durriya S, Phelan D, Duffy BF, Olack B, Smith D, Howard T, Mohanakumar T. HLA Antibodies Present In The Sera Of Sensitized Patients Awaiting Renal Transplant Are Also Reactive To Swine Leukocyte Antigens1, 2. Transplantation. 1998. October 27;66(8):1074–80. [DOI] [PubMed] [Google Scholar]
- Niemann H, Verhoeyen E, Wonigeit K, Lorenz R, Hecker J, Schwinzer R, Hauser H, Kues WA, Halter R, Lemme E, Herrmann D. Cytomegalovirus Early Promoter Induced Expression Of hcd59 In Porcine Organs Provides Protection Against Hyperacute Rejection1. Transplantation. 2001. December 27;72(12):1898–906. [DOI] [PubMed] [Google Scholar]
- Oostingh GJ, Davies HF, Tang KC, Bradley JA, Taylor CJ. Sensitisation to swine leukocyte antigens in patients with broadly reactive HLA specific antibodies. American Journal of Transplantation. 2002. March;2(3):267–73. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004. October;25(13):1605–12. [DOI] [PubMed] [Google Scholar]
- Reyes LM, Blosser RJ, Smith RF, Miner AC, Paris LL, Blankenship RL, Tector MF, Tector AJ. Characterization of swine leucocyte antigen alleles in a crossbred pig to be used in xenotransplant studies. Tissue Antigens. 2014. November;84(5):484–8. [DOI] [PubMed] [Google Scholar]
- Reyes LM, Estrada JL, Wang ZY, Blosser RJ, Smith RF, Sidner RA, Paris LL, Blankenship RL, Ray CN, Miner AC, Tector M. Creating class I MHC-null pigs using guide RNA and the Cas9 endonuclease. The Journal of Immunology. 2014. October 22:1402059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurman HJ, Pino-Chavez G, Phillips MJ, Thomas L, White DJ, Cozzi E. Incidence of hyperacute rejection in pig-to-primate transplantation using organs from hDAF-transgenic donors. Transplantation. 2002. April 15;73(7):1146–51. [DOI] [PubMed] [Google Scholar]
- Taylor CJ, Tang KG, Smith SI, White DJ, Davies HF. HLA-specific antibodies in highly sensitized patients can cause a positive crossmatch against pig lymphocytes. Transplantation. 1998. June 27;65(12):1634–41. [DOI] [PubMed] [Google Scholar]
- Varela ID, Mozo PS, Cortés AC, Blanco CA, Cañedo FV. Cross-Reactivity between Swine Leukocyte Antigen and Human Anti-HLA-Specific Antibodies in Sensitized Patients Awaiting Renal Transplantation. Journal of the American Society of Nephrology. 2003. October 1;14(10):2677–83. [DOI] [PubMed] [Google Scholar]
- Zhang R, Wang Y, Chen L, Wang R, Li C, Li X, Fang B, Ren X, Ruan M, Liu J, Xiong Q. Reducing immunoreactivity of porcine bioprosthetic heart valves by genetically-deleting three major glycan antigens, GGTA1/β4GalNT2/CMAH. Acta biomaterialia. 2018. May 1;72:196–205. [DOI] [PubMed] [Google Scholar]


