Abstract
Growing evidence points to an important role for commensal microbiota in susceptibility to food allergy. Epidemiologic studies demonstrate associations between exposures known to modify the microbiome and risk of food allergy. Direct profiling of the gut microbiome in human cohort studies has demonstrated that individuals with food allergy have distinct gut microbiomes compared to healthy controls, and dysbiosis precedes the development of food allergy. Mechanistic studies in mouse models of food allergy have confirmed that the composition of the intestinal microbiota can imprint susceptibility or resistance to food allergy on the host, and have identified a unique population of microbial-responsive RORγt+ Foxp3+ Tregs as critical for the maintenance of tolerance to foods. Armed with this new understanding of the role of the microbiota in food allergy and tolerance, therapeutics aimed at modifying the gastrointestinal microbiota are in development. In this manuscript, we review key milestones in the development of our current understanding of how the gastrointestinal microbiota contributes to food allergy and discuss our vision for the future of the field.
Keywords: food allergy, microbiome, microbiota, dysbiosis, prebiotic, probiotic, symbiotic, Tregs, short chain fatty acids
Introduction
The dynamics and functions of microbiota that inhabit our bodies contribute to both health and disease.1, 2 In recent years, there has been increasing interest in how dysregulation of resident microbial communities (i.e. dysbiosis) may be associated with allergy risk.3–7 The sum of microbes, their genomic elements, and their interactions in a given ecologic niche (i.e. microbiome) differs by body site.2 Accumulating evidence supports a potential role for the gut microbiome in the pathogenesis and course of food allergy.3–17 Next-generation sequencing, including 16S rRNA sequencing and shotgun metagenomic sequencing, has advanced microbiome research in recent years by enabling more comprehensive and culture-free profiling of taxa in a given sample.3, 4 Building on our previous addresses of this topic3–6, here we review the role of the gut microbiome in food allergy.
Observational human cohort studies support associations between microbial exposures and allergy risk
A potential role for microbiota in allergy risk was put forth as the “hygiene hypothesis” in 1989 with the epidemiologic observation that children from larger households had lower rates of allergic rhinitis and eczema.18 The hypothesis is that infection in early childhood as a result of contact with siblings or acquired prenatally by maternal contact with older siblings protects against allergy.18 Thinking on this has evolved over recent decades to suggest that the increased incidence of allergic disease may not be specifically due to reduced infections, but rather, due to the modern Western living environment, diet, and lifestyle where exposure to human microbiota are different from prior generations.19 Observational studies in support of this include those showing that maternal prenatal exposures to pets20, birth by vaginal vs. caesarian section delivery21, childhood in rural environments22, and childhood exposures to pets23 are associated with reduced allergy risk.3, 24 The “old friends” hypothesis and “biodiversity hypothesis” posit that the increase in allergies is from the loss of commensal relationships with parasites and bacteria that had previously exerted beneficial effects on human evolution.19, 25, 26
With regards to food allergy specifically, a few observational studies have provided concordant evidence that conditions leading to differential microbial exposures are associated with food allergy risk. An Australian birth cohort study of 5276 infants, of whom 453 were confirmed to be egg allergic by oral food challenge (OFC) or recent reaction plus evidence of IgE-mediated sensitization, found that having older siblings and dog ownership were each associated with decreased egg allergy risk (adjusted odds ratio (OR), 0.72; 95% confidence interval (95%CI), 0.62–0.83; and adjusted OR, 0.72; 95% CI, 0.52–0.99, respectively).27 Within a Norwegian birth cohort of 2803 infants, Cesarean delivery was not associated with confirmed reactions to egg (based on OFC or on history of reaction plus elevated egg specific IgE) by age 2.5 years, but stratified analyses demonstrated a four-fold increased risk (OR, 4.1; CI, 0.9–19; P =0.08) of confirmed egg allergy in children with atopic mothers.28 While conflicting results for delivery mode and food allergy risk do exist in the literature27–29, a meta-analysis of 6 studies on this topic showed an increased summary odds ratio of OFC-confirmed food allergy or food sensitizations (OR 1.32, 95% CI 1.12–1.55) in children who were born via Cesarean delivery.21 Collectively, these observational findings support the notion that differential microbial exposure and colonization in early childhood may modify risk for the development of food allergy.
Gut microbiota differ in human subjects with food allergy
While the hygiene, old friends, and biodiversity hypotheses and findings from epidemiologic studies support that circumstances linked to differential microbial exposures are associated with allergy and food allergy risk, studies that have directly measured microbial diversity and composition in populations with and without food allergy provide direct evidence that gut microbiota differ in subjects with food allergy (Figure 1). Culture-based studies of gut microbiota from subjects with and without food allergy provided early findings on this,13, 30 although because the vast majority of microbiota cannot be cultured, these cultured-based studies could provide only partial insight. More recent studies have employed next-generation sequencing, including 16S rRNA sequencing and shotgun metagenomic sequencing, which enable more comprehensive and culture-free profiling of taxa in a given sample.31 Sequencing of the 16S rRNA gene, which has highly conserved primer binding sites and contains hypervariable regions that can provide species-specific signature sequences for bacterial identification, allows for more comprehensive bacterial identification without the constraints of culture-based methods. Shotgun metagenomic sequencing, in which the total DNA of an ecosystem is sequenced, provides broad and deep characterization of all microbiota in addition to bacteria as well as better resolution at lower taxonomic levels and potential for functional annotation. However, shotgun metagenomic sequencing has not yet been widely implemented in studies of food allergy.3
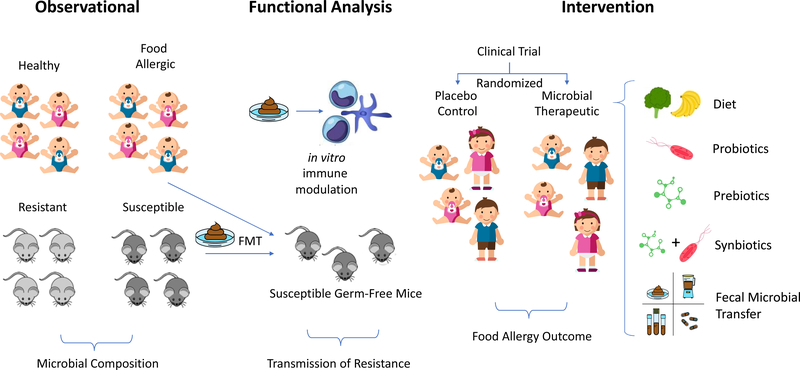
Figure 1: Pathways to development of microbial therapeutics for food allergy.
Evidence for an important contribution of the gut microbiota to the pathogenesis of food allergy is derived from observational studies of distinct microbial composition in allergic and healthy human cohorts, and in mice with susceptibility versus resistance to food allergy. Evidence that changes in microbial composition lead to meaningful changes in host immunity come from studies utilizing fecal microbiota transplants (FMT) into germfree mice with susceptibility to food allergy. Additional functional evidence can be derived from in vitro culture systems examining the impact of stool extracts on immune phenotype. Based on the weight of this pre-clinical evidence, clinical trials utilizing defined microbial transfer, probiotics, prebiotics, synbiotics, and FMT are in process to determine efficacy of microbial therapeutics for the treatment of food allergy.
Findings from 16S rRNA sequencing-based studies have shown that children with food allergy have distinct gut microbiome compositions compared to those without food allergy (Figure 1). Further, results thus far suggest that gut microbiota may differ by individual food allergy. In a study of 141 children with egg allergy and non-food allergic controls from the multi-center, US-based Consortium for Food Allergy Research, where egg allergy was strictly defined based on OFC, convincing history and positive allergy testing, or flare of atopic dermatitis associated with egg ingestion and egg sIgE ≥ 2 kUA/L, genera from Lachnospiraceae, Streptococcaceae, and Leuconostocaceae were found to be differentially abundant in the gut microbiome of egg allergic vs. control children.9 Independent studies of gut microbiota in milk allergic individuals demonstrate overlapping as well as contrasting gut microbial compositions. A study of 39 Italian children showed that compared to healthy controls, the 19 milk allergic infants studied had a higher Shannon diversity index and gut microbial community structures dominated by Lachnospiraceae and Ruminococcaceae.14 Differences in the implicated taxa across these studies of egg and milk allergies may have been due to the individual food allergies targeted in each study, although additional explanations include differences in population characteristics, phenotyping approach, and/or analytic methodology. The characterization and comparison of microbial profiles of individual food allergies remains an area for continued exploration.
Gut microbiota are associated with the clinical trajectory of food allergy
Sequencing-based studies of gut microbiota in children with food allergy have shown that gut microbiome composition is associated with the clinical trajectory of food allergy, and the timing of host-microbe interactions in early life appears to be important. The gut microbiome changes during the course of life, with the most rapid changes occurring in early life.32 To examine the relationship between infant gut microbiome and the clinical course of food allergy, investigators studied 226 infants with milk allergy (strictly defined based on oral food challenge, convincing history and positive allergy testing, or flare of atopic dermatitis associated with milk ingestion and milk sIgE ≥ 5 kUA/L) from the multi-center Consortium for Food Allergy Research whose clinical courses were followed longitudinally up to age 8 years.8 The investigators found that taxa from the Firmicutes phylum including Clostridia were enriched in the gut microbiome of milk allergic infants age 3–6 months whose milk allergy later resolved by age 8 years, while this was not the case in same age milk allergic infants who continued to have persistent milk allergy.8 Among milk allergic infants >6 months age, there was no association between gut microbiome composition and later milk allergy resolution.8 Based on these results, it is possible that (1) early infancy (i.e. age 3–6 months) is a specific developmental window during which gut microbiota can shape the course of food allergy, and gut microbiota at later ages (e.g. > 6 months age) have little to no effect on food allergy course, and/or (2) gut microbiome composition may be associated with food allergy course later in life as well, but factors that appear during these older ages obscure this signal. Separately, a study of 56 Japanese infants found evidence that gut microbiota at age 2 months is associated with the development of self-reported food allergy by age 2 years. Specifically, among 14 infants who developed egg, cow’s milk, soy, and/or wheat allergy based on questionnaire by age 2 years, the investigators found that the genera Leuconostoc, Weissella, and Veillonella, were underrepresented in their fecal samples compared to 27 of their counterparts who did not develop any allergies by age 3 years.33 We note that while self-reported and/or questionnaire-based assessments of food allergy are often used in studies of food allergy, they are limited by the likelihood of reporting inaccuracy, and supervised oral challenge-based assessments of food allergy remain the gold standard for accurate diagnosis.34
Additional studies have similarly found associations between early-infancy gut microbiome composition and the development of food allergen sensitization, an intermediate phenotype to food allergy. Among children from the Canadian Healthy Infant Longitudinal Development cohort, each quartile increase in gut microbial richness at age 3 months was associated with 55% reduced risk for food sensitization (by skin prick test) by age 1 year adjusted OR, 0.45; 95% CI, 0.23, 0.87), while there was no association between gut microbial characteristics at age 12 months and food sensitization.35 In terms of gut microbiome composition, Enterobacteria were overrepresented and Bacteroidaceae were under-represented in food-sensitized infants at 3 months and 1 year.35 Separately, among 225 US children participating in the Vitamin D Antenatal Asthma Reduction Trial, lower relative abundances of Haemophilus, Dialister, Dorea, and Clostridium in stool samples collected at age 3–6 months was associated with sensitization (sIgE ≥ 0.10 kUA/L) to at least one food allergen among milk, egg, peanut, soy, and wheat.10
Findings from murine models also support age-sensitive contact with microbiota.36 Colonization of gnotobiotic mice with a diverse microbial population early, but not late, in life suppresses IgE and prevents mice from development of food allergy.37 These collective findings suggest that commensal microbial exposures during early immune system development could play a role in the natural history of food allergy.
Causation and mechanisms: lessons from mouse models of food allergy
Studies of human cohorts are observational, and even strong supporting evidence such as documented dysbiosis preceding development of food allergy does not demonstrate causation. Mechanistic studies in mice have been essential for supporting the link between the composition of intestinal microbiota and development of food allergy. The first approaches to test the role of the microbiota in food allergy used methods such as broad-spectrum antibiotics to reduce the bacterial load in the intestine or germ-free mice completely lacking a normal commensal microbiota. These tools showed that the microbiota provided a suppressive signal to prevent sensitization and development of peanut or milk allergy 38–41. In the absence of microbiota, there was a polyclonal hyper-IgE response that was CD4+ T cell dependent and was associated with elevated IL-4 production and IgE class switch in the Peyer’s patches of the small intestine 37 There was a need for diverse microbial colonization during a critical window in early life to suppress polyclonal IgE production and prevent susceptibility to allergic sensitization 37
Identification of protective organisms
Germ-free mice or antibiotic treatment demonstrated that the microbiota as a whole provided suppressive signals preventing food allergy, but these approaches do not model more subtle alterations in microbial composition that are observed in food allergy. Noval-Rivas et al used a mouse model susceptible to food allergy (the Il4raF709 mouse, which has a gain of function mutation in the IL-4 receptor) to study how the microbiota differs between mice with susceptibility or resistance to food allergy and anaphylaxis15. The composition of the fecal microbiota was significantly different between resistant (wild-type) and susceptible (Il4raF709) mice. Differences were observed between the strains at steady-state, and oral sensitization to the egg allergen OVA induced significant changes in the microbial composition of susceptible, but not resistant, mice. Bacterial families including Lachnospiraceae, Lactobacillaceae, Rikenellaceae, and Porphyromonadaceae were over-represented in the microbiota of food allergic mice. When fecal microbiota transplants (FMT) were performed from resistant or susceptible mice to germ-free mice, FMT from susceptible sensitized mice conferred greater susceptibility to food allergy than FMT from resistant sensitized mice onto the colonized mice. This was the first experimental model to show that food allergy susceptibility could be transmitted by the gastrointestinal microbiota15. It was not clear from these studies whether resistant mice harbored a protective microbe or susceptible mice harbored a microbe that could drive allergy susceptibility.
Atarashi et al had identified a group of Clostridia species, harvested as chloroform-resistant spores from the fecal contents of healthy mice, as being protective against colitis and IgE production when administered to mice with a normal microbiota 42 Stefka et al used this approach to demonstrate that Clostridia species are protective against experimental peanut allergy when transferred into germ-free mice 41. Monocolonization with a single Bacteroides species (B. uniformis) was not as effective as colonization with the Clostridia consortium in suppressing peanut-specific IgE and anaphylaxis, suggesting unique regulatory properties of the Clostridia consortium.
In order to translate findings to human disease, it was important to identify similarly protective organisms derived from human subjects, with a capacity to colonize the human gastrointestinal tract. Atarashi et al selected 17 strains of healthy human fecal-derived Clostridia based on their ability to colonize mice, induce Tregs, enhance IL-10 production, and increase ICOS expression on Tregs 43 The strains were derived from Clostridia clusters IV, XIVa, and XVIII, and lacked toxins or other virulence factors. This cocktail of Clostridia strains was able to suppress gastrointestinal manifestations of food allergy in mice 43
More recent approaches have focused on functional analyses of fecal organisms by performing FMT transferring samples derived from food allergic and healthy individuals to mice. Rodriguez was the first to report that FMT from a healthy infant could protect susceptible germ-free mice from milk allergy 16 Abdel-Gadir et al studied the stool microbial composition of 154 infants from 1–15 months, 56 with food allergy and 98 healthy controls 12 Significant differences were identified in 77 operational taxonomic units (OTU), including several species of Clostridiales. Functional implications of these differences were tested by transferring the microbiota into germfree susceptible (Il4raF709) mice. 7 allergic and 7 healthy control specimens were tested, and fecal microbiota from the healthy control, but not food allergic subjects protected these susceptible recipient mice from food allergy12.
A series of transfers with defined microorganisms was then performed to identify protective organisms. Germ-free mice colonized with a cocktail of 6 Clostridiales strains or 5 Bacteroidales strains including B. fragilis were protected from food allergy, while colonization with a cocktail of 4 Proteobacteria showed no protection. A single Clostridiales strain, S. variabile, was sufficient to suppress food allergy although less effectively than the 6-strain consortium. Importantly as a proof of concept for therapeutic use, these microbes were also protective in conventional mice having a normal microbiota that had been treated with antibiotics to facilitate microbial colonization 12 Feehley et al used a similar approach to test the functional consequences of compositional differences in the fecal microbiota of infants with or without milk allergy 44 They added a factor of dietary modification, which was necessary for maintaining the original microbial composition in the recipient mice. Mice receiving FMT from milk allergic donors were given hydrolyzed casein formula, while mice receiving FMT from healthy controls were given milk formula prior to sensitization to the milk allergen β-lactoglobulin. Mice receiving FMT from healthy controls plus milk formula were protected from milk allergy, while mice receiving FMT from milk allergic infants plus hydrolyzed casein formula developed symptoms to β-lactoglobulin after sensitization 44 The abundance of a Clostridiales species, A. caccae, was significantly associated with regulatory gene expression in the intestinal epithelium, and when this organism was administered to germ-free mice together with milk formula, was able to protect germ-free mice from milk allergy. With this design it is challenging to parse out the effects of the microbiota from the effects of dietary exposure. Germ-free mice receiving milk formula alone were not protected from food allergy, however oral tolerance to dietary antigens is known to be impaired in germ free mice 45–47.
Mechanisms of Protection Against Food Allergy by the Intestinal Microbiota
The weight of evidence indicates that the microbiota, and Clostridia strains in particular, protect against food allergy through the generation of peripheral antigen-experienced regulatory T cells. Monocolonization studies have shown that a diverse set of microbes can support the expansion of Foxp3+ Tregs in the mouse intestine 48 However, it is becoming clear that Tregs are a highly heterogenous population comprised of subsets that differ in regulatory activity. For example, a subset of Foxp3+ Tregs that express GATA-3 and IL-4 and have been referred to as “Th2 reprogrammed Tregs” are elevated in food allergy and have reduced suppressive activity 49 Ohnmacht et al identified a subset of antigen-experienced Foxp3+ Tregs that express the transcription factor RORγt+, are dependent on the presence of the intestinal microbiota and Vitamin A metabolism, and effectively suppress Th2 skewed immune responses 50 As shown by Abdel-Gadir et al, protection of susceptible germ-free mice from food allergy by defined microbial transfer was associated with expansion of RORγt+ Tregs 12 The effect of microbiota on Tregs was likely direct on the Tregs, as deletion of the bacterial sensing molecule MyD88 under control of the Foxp3 promoter (and therefore deleting MyD88 only in Tregs) abolished the protective effect of the microbiota in food allergy 12 RORγt+ Tregs express high levels of CTLA-4 and suppress Th2 immunity by modifying the phenotype of intestinal dendritic cells 50 Downstream of the induction of RORγt+ Tregs is suppression of IgE and enhancement of IgA responses. Intriguingly, an IgE response was detected against the gut microbiota in allergic humans and mice12, but the function of this antimicrobial IgE in food allergy pathogenesis or IgE persistence has not yet been explored. In addition to direct effects on Tregs, the microbiota can alter epithelial gene expression directly and via activation of type 3 innate lymphoid cells, leading to enhanced barrier function 41.
The link between the microbiota and protection from food allergy is not well understood. Short chain fatty acids (SCFA) including butyrate, propionate, and acetate are generated by the microbiota through the metabolism of dietary fiber. High fiber diet has been shown to protect mice from food allergy via SCFA acting on the receptors GPR43 and GPR109a 51. Feeding of exogenous SCFA to mice can expand RORγt+ Tregs 50, and can protect against food allergy 51 and atopic sensitization 52 However, not all studies have found that administration of exogenous SCFA is protective in the context of food allergy 12 In addition to SCFA, microbiota produce a wealth of products that are active on the human immune system. The monohydroxy fatty acid 12,13-diHOME is produced by intestinal bacteria, is elevated in neonates at risk of development of asthma, and drives Th2 skewing by acting on dendritic cells and T cells53, 54 The microbiota can metabolize tryptophan into ligands active on the aryl hydrocarbon receptor present on epithelial cells, intraepithelial lymphocytes, innate lymphoid cells, and dendritic cells 55 AhR ligands promote development of Tregs56, and suppress experimental peanut allergy57. The intestinal microbiota can also produce histamine that can act on histamine receptors on T cells, dendritic cells, and epithelial cells, and histamine-producing bacteria have been shown to be elevated in individuals with asthma 58, 59 Functional analysis of bacterial products identified via metabolomics will be an important parallel strategy alongside functional studies of candidate organisms to advance our understanding of the mechanisms of microbial regulation of food allergy risk. Figure 2 reviews the main immune mechanisms by which the healthy intestinal microbiota generates immune tolerance rather than allergic sensitization.
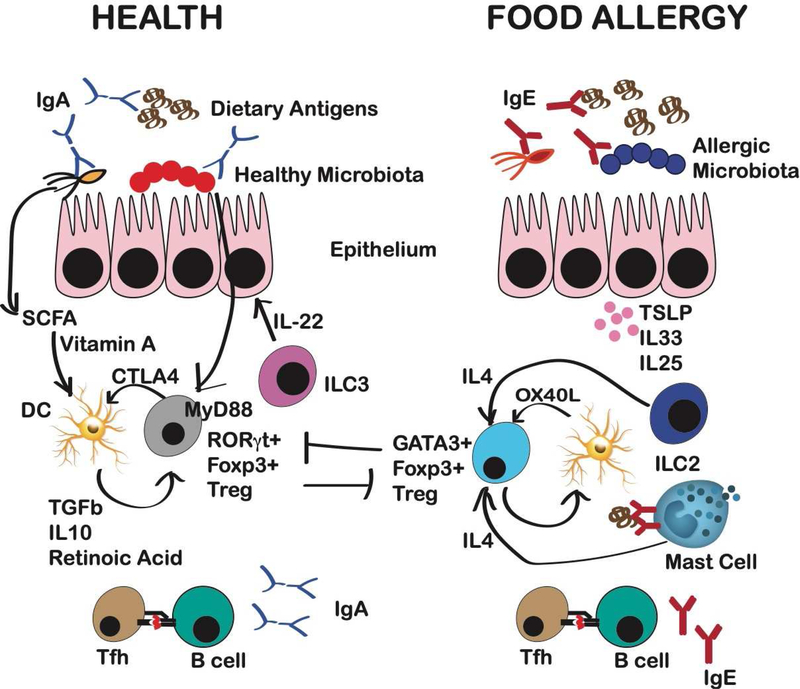
Figure 2: Contribution of the microbiota to immune mechanisms of tolerance and allergy to foods.
Healthy microbiota, or consortia composed of Clostridiales strains, Bacteroidales strains, or individual microorganisms including S. variabile or A. caccae, protect susceptible germ-free mice from food allergy. Mechanisms of protection include the generation of RORγt+ Foxp3+ Tregs by MyD88 signaling in Tregs, which enhances anti-microbial and anti-food allergen specific IgA production while suppressing IgE production, and suppresses generation of GATA3+ “Th2 reprogrammed” Tregs. Short chain fatty acids (SCFA) derived from the healthy microbiota drive a change in the mucosal DC phenotype to a more tolerogenic phenotype. RORgt+ Tregs suppress DCs via elevated CTLA4 expression to suppress costimulatory-driven priming. In food allergy, production of IL-4 from innate lymphoid cells type 2 (ILC2) and IgE-activated mast cells cooperates with DCs to promote the generation of Th2 reprogrammed Tregs that express GATA-3, produce IL-4, and contribute to rather than protect from food allergy. Epithelial-derived cytokines contribute to the immune milieu that drives this deviation of the normal regulatory response.
Microbial therapeutics for food allergy
The accumulating evidence from clinical and murine studies that gut microbiota may affect the pathogenesis and course of food allergy inspires the search for microbial therapeutics that might prevent and/or treat food allergy. Diet, probiotics, prebiotics, synbiotics, and fecal microbiota transplantation represent candidate interventions to shape the gut microbiome toward beneficial outcomes (Figure 1).4–6
Diet
The immune-modulating and allergy protective effects of many gut microbiota are thought to be mediated by metabolites produced from their fermentation of dietary fiber.51, 60, 61 These metabolites, such SCFAs, regulate the size and function of the colonic Treg pool.51, 60, 61 In human cohorts, significant associations have been reported between the composition of dietary intake and SCFA levels as measured in stool, suggesting that diet can be used to modulate microbial production of SCFA.52 For example, in a subset study of 301 children from the Protection against Allergy Study in Rural Environments birth cohort, consumption of yogurt, fish, vegetables, and fruits within the first year of life were each found to be associated with increased butyrate in stool samples at age 1 year.52 Further, children with stool butyrate and propionate levels ≥ 95th percentile at age 1 year had reduced sensitization to any environmental or food allergen between ages 3–6 years compared to those with levels <95th percentile (butyrate aOR 0.25, 95%CI 0.08–0.82, propionate aOR 0.20, 95%CI 0.05–0.74).52 Female BALB/c mice receiving oral administration of the SCFAs examined (acetate, propionate, or butyrate) during sensitization and challenge demonstrated reduced airway hyper-responsiveness following methacholine challenge compared to mice receiving vehicle.52 Administration of dietary fiber or exogenous SCFAs has been shown to suppress food allergy in mice in some studies51, while not in others.12 Clinical trials of fiber and/or SCFA supplementation for food allergy prevention and/or treatment have yet to be undertaken.
Conversely, other murine models have shown that a high-fat diet induces changes to gut microbiota that increase food allergy risk.62 Following a high fat diet, mice developed obesity, decreased gut bacterial diversity, and dysregulated intestinal effector mast cell responses.62 Transfer of the high fat diet-associated microbiome from the obese mice to germ-free mice conferred enhanced food allergy susceptibility to the recipient mice, suggesting a pathogenic role for the high fat-diet associated microbiota independent of obesity.62 If such results can be extrapolated to human biology, avoidance of high-fat diets common to Western lifestyles may have benefits to food allergy risk in addition to those for overall health.
Probiotics
In theory, intake of beneficial microbiota would be a direct way to shape gut microbiota. Probiotics are live microbes whose intake is thought to be beneficial for health. A few clinical trials have examined probiotics for treatment of challenge-proven food allergy. Among 119 infants with cow’s milk allergy, supplementation with Lactobacillus casei and Bifidobacterium lactis for 12 months did not accelerate milk allergy resolution.63 However, a subsequent trial of 55 milk allergic children providing supplementation of extensively hydrolyzed casein formula with a different probiotic, Lactobacillus rhamnosus GG, demonstrated increased rates of milk allergy resolution after 6 and 12 months compared to a control group receiving the formula alone.64 Comparison of stool from 20 healthy infants and from 19 cow’s milk allergic infants before and after treatment with extensively hydrolyzed formula with (n=12) or without (n=7) Lactobacillus rhamnosus GG revealed that only gut microbe that differed in gut abundance between tolerant and allergic infants was Oscillospira, although Blautia and Roseburia were enriched in samples from tolerant infants with higher fecal butyrate.14 The same investigators followed 220 milk allergic infants randomized to extensively hydrolyzed formula with or without Lactobacillus rhamnosus GG for 3 years, finding that those receiving the formula plus probiotic combination had higher incidences of cow’s milk tolerance at ages 12, 24, and 36 months.65
Lactobacillus rhamnosus GG has also been studied as a microbial adjunct to peanut oral immunotherapy for peanut allergy. In a clinical trial where 62 children were randomized to receive Lactobacillus rhamnosus GG with peanut oral immunotherapy vs. placebo for 18 months, 89.7% of subjects who received the combination treatment achieved desensitization compared to 7.1% of those receiving placebo (P<0.001).66 However, effect due to the probiotic itself was not clear from this study given the absence of probiotic-only or oral immunotherapy-only control groups. Four years after treatment cessation, 67% of subjects who had received the combination treatment during the 18 months were eating peanut compared to 4% of those who had taken placebo (P = 0.001), and more participants from the treated group had smaller peanut skin prick test size (mean 8.1 mm vs. 13.3 mm, P = 0.035), and higher peanut sIgG4:sIgE ratios compared to placebo-treated controls (geometric mean 67.3 vs. 5.2, P = 0.031).67
While results from some of the above trials suggest that probiotics may be useful for food allergy, the most recent Cochrane review on this topic68 concluded that the data are insufficient thus far to recommend probiotic supplementation for food allergy. More recent meta-analyses on the subject69 as well as statements from the World Allergy Organization70,71 comment on the limited and “very low quality evidence” on this topic thus far and concur with the Cochrane conclusion.
Prebiotics
Another approach to shape gut microbiota is the provision of components that foster the growth of desired microbial community members. This is the idea behind prebiotics, nondigestible food components that specific commensal microbes can use to selectively foster their growth and activity. Frequently added to infant formulas, prebiotics are typically oligosaccharides that are not digested nor absorbed in the gastrointestinal tract before they reach the large intestine, where specific microbiota can use them for improved growth and activity.72 Although prebiotic supplementation in infants was found to be associated with decreased risk of asthma and eczema in some investigations,72–74 there have been no significant effects on the development of food allergy found thus far.72–74
Synbiotics
Synbiotics are combinations of probiotics and prebiotics. It is thought that they may be more effective than probiotics or prebiotics alone because the provision of both together puts forth a synergistic combination of desired microbes and the components they need to thrive and drive the host community. A prospective, randomized, doubleblind controlled study of 110 full term infants with cow’s milk allergy receiving either amino acid-based formula (AAF) or AAF with synbiotics showed that subjects in both arms have normal and similar growth.75 The synbiotics used in this trial included the probiotic Bifidobacterium breve M-16V in combination with the prebiotics oligofructose, long-chain inulin, and acidic oligosaccharides. The same company is now performing a clinical trial where 851 young infants at high risk for allergy have been randomized to take partially hydrolyzed infant formula with synbiotics vs. standard infant formula (NCT03067714). The children will be assessed for the primary outcome of fecal levels of Bifidobacteria at age 17 weeks and secondary outcomes of Bifidobacteria levels at age 1 year and IgE-mediated allergic manifestations, including food allergy, up to age 1 year (NCT03067714).
Fecal microbiota transplantation
Because microbes function not singly but as interacting members of communities, the provision of one or a few probiotic bacterial strains may be insufficient to meaningfully alter the human gut microbiome toward an observable phenotype (such as food allergy prevention). Transferring a microbial community associated with health may be more impactful. Fecal microbiota transplantation (FMT) refers to the transfer of communities of microbes from a donor to a recipient. FMT has been successfully used to treat patients with Clostridium difficile colitis, where communities from healthy donors are transferred to those suffering from the condition.76, 77 FMT is an intriguing gut microbiome manipulation strategy to consider, given findings from murine models supporting that fecal transfer is effective mode to alter allergy outcomes. A small Phase I open label trial to evaluate the safety and tolerability of oral encapsulated FMT administered open-label over 2 days for the treatment of peanut allergy in 10 adult subjects is underway (NCT02960074). The agent being used is a screened-donor inoculum of frozen fecal material.
A study of next-generation microbial transfer of rationally selected strains, based on preclinical findings by Atarashi et al,43 is in a Phase I clinical trial for treatment of peanut allergy (NCT03936998). VE416 is a combination of dormant bacteria given as a capsule to individuals pre-treated with vancomycin to facilitate colonization of the transferred bacteria. In this ongoing study, VE416 or placebo will be given with peanut OIT (NCT03936998).
The pending results from these interventional clinical studies will lend insight into FMT for food allergy treatment, although additional questions will remain to be answered about defining the optimal donor material or strains, methods to ensure establishment of the transferred community, and safety and acceptability of the procedure for food allergy treatment.
Future directions
Our understanding of the relationship between the gut microbiome and food allergy has grown in recent years with knowledge gained from observational human cohort studies, murine model experiments, and clinical trials. The aggregate evidence thus far supports that gut dysbiosis likely precedes the development of food allergy, and the timing of such dysbiosis is critical. Murine models have enabled valuable mechanistic insight into how microbiota modulate type 2 immunity.3, 4 With multiple clinical trials of microbial therapeutics underway, we are also likely to gain key information from these interventional studies that will inform next-level trials to design and execute. The challenges ahead are to build upon this knowledge base to delve deeper into current research foci and to pursue novel directions that will further our understanding, management, and treatment of food allergy.
While findings from human cohort studies support that food allergic individuals have distinct gut microbiota (that may further differentiate by individual food allergy) and the gut microbiome of early life has greatest effects on food allergy outcomes, additional cohort studies are needed to deepen our understanding of the temporality of such effects and to dissect the web of causal relationships between microbiota, host, environment, and food allergy outcomes. Importantly in this regard, human cohort studies with rigorous, longitudinal phenotyping, robust criteria for food allergy characterization, parallel immune profiling, and employment of best-use practices for microbiome profiling will allow for the most reliable results and optimal cross-study learning. Our current understanding of the gut microbiome in food allergy has been bacteria-centric, likely driven by the relative ease of profiling bacteria via culture-based methods in early days and by PCR subsequently and 16S rRNA sequencing more recently. It is possible that the virome and mycobiome have effects on food allergy course that we have not yet uncovered. Shotgun metagenomic sequencing, which is being increasingly used in prominent studies of the microbiome,78–80 can enable microbiome research beyond the bacterial microbiome while also offering better resolution at the species and strain levels and more accurate community diversity estimates than 16S rRNA sequencing, albeit at higher cost.31, 81 Additionally, studies to date have focused on the gut microbiome, but the role of microbiota at other sites such as skin and airway may be of importance due to the role of extra-intestinal sites in allergic sensitization 82.
To parse the complex interactions between microbiota, our bodies’ molecular processes, and environmental exposures that influence food allergy risk, multidimensional profiling and systems biology approaches will be needed.31, 83, 84 Food allergy is a complex and heterogeneous disease, and it is unlikely that microbiota implicated in its pathobiology, or even the microbiome in its entirety, can capture the interdependent dynamics of the molecular networks involved in food allergy.31 Our understanding of food allergy has been advanced not only by studies of the microbiome, but also by findings from genome-wide association,85–88 transcriptome,89 epigenome,90, 91 and metabolomic 31, 92, 93 studies of food allergy. Integrating these types of system-wide data is critical if we are to construct models that are predictive of complex biological interactions and systems, a necessary step to developing a more complete understanding of food allergy.31, 83, 84 By moving forward research on the microbiome in food allergy and integrating our findings with our knowledge on food allergy, we can further our understanding of food allergy and derive new approaches for its prevention and therapy.
Key Concepts and Therapeutic Implications.
The gut microbiome likely plays a role in the pathogenesis and course of food allergy.
Individuals with food allergy have different gut microbiomes compared to healthy controls.
Imbalances in the gut microbial ecosystem precede the development of food allergy, and the timing of such dysbiosis is a key factor.
Microbial transfer studies in mice have demonstrated that food allergy-associated dysbiosis transmits susceptibility to food allergy.
Consortia composed of either Clostridiales or Bacteroidales strains inhibit food allergy in mice through induction of RORγt+ Tregs that suppress GATA3+ Tregs and IgE and enhance IgA
Diet, probiotics, prebiotics, synbiotics, and fecal microbiota transfer represent potential microbial therapeutics for food allergy prevention and treatment.
Future studies of the microbiome and food allergy are needed to deepen our understanding of the temporality the microbiome’s effects and to dissect the web of causal relationships between microbiota, host, environment, and food allergy outcomes.
Acknowledgments
Funding: SB is funded by the National Institutes of Health (NIH) R01AI147028, R01AI118833, and U19AI136053. CB is funded by the National Institutes of Health U24 AI118644 and U19AI136053.
Abbreviations
- CI
confidence interval
- DC
dendritic cell
- FMT
Fecal microbiota transplantation
- OFC
oral food challenge
- OR
odds ratio
- OTU
operational taxonomic unit
- rRNA
ribosomal RNA
- SCFA
short chain fatty acid
- sIgE
specific IgE
- SPT
skin prick test
- Treg
T regulatory cell
Glossary
- Microbiome
The sum of microbes and their genomic elements in a particular environment
- Microbiota
The community of microbes in a particular environment
- Dysbiosis
A state of imbalance in a microbial ecosystem
- Taxon or taxa
A group of organisms scientifically classified as a unit based on shared characteristics. The unit may be at any taxonomic level of the biological system of classification of organisms (e.g. phylum, order, family, genus, species)
- Operational taxonomic unit (OTU)
A group of organisms with similar sequence homology according to DNA sequence similarity of a specific taxonomic marker gene
- Richness
The number of taxa found in a given ecological niche
- Evenness
The relative distribution of taxa in a given ecological niche
- Diversity
A calculated index incorporating measures of richness and taxa distribution
- 16s rRNA sequencing
A method for culture-free classification and identification of bacteria present in a sample based on sequencing the hypervariable regions of the 16S ribosomal RNA gene that is present in all bacteria
- Metagenomic sequencing
Whole genome sequencing performed on genomic DNA from a mixed microbial community
- Probiotic
Live microorganisms with beneficial effects on the host
- Prebiotic
Nondigestible substrates that promote the growth and/or function of beneficial microorganisms
- Synbiotic
Products that contain both prebiotics and probiotics
- Fecal microbiota transplantation
The administration of fecal matter from a donor to a recipient
- Short chain fatty acid
Fatty acids with fewer than 6 carbon atoms
- Oral Tolerance
A state of local and systemic immune suppression to antigens introduced by the oral route, translating to a lack of clinical reactivity upon oral antigen introduction that is independent of both quantities of antigens ingested and routine antigen exposure
- RORγt+ Tregs
Also referred to as Type 3 Tregs, these Foxp3+ Tregs co-express the transcription factor RORγt. These cells are generated in response to the intestinal microbiota, and are essential for the suppression of type 2 immunity
- GATA3+ Tregs
Also referred to as Type 3 Tregs, these Foxp3+ Tregs co-express the transcription factor RORγt. These cells are generated in response to the intestinal microbiota, and are essential for the suppression of type 2 immunity
- Desensitization
A temporary increase in reactivity threshold to provide a measure of safety that is dependent on continued treatment exposure.
- Sustained Unresponsiveness
A state of clinical-unresponsiveness to food allergen that persists despite discontinuation of treatment exposure.
Footnotes
Conflicts of interest: SB: no conflicts of interest. MCB serves on the advisory board for ProtaTherapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ 2017; 356:j831. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017; 550:61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho HE, Bunyavanich S. Role of the Microbiome in Food Allergy. Curr Allergy Asthma Rep 2018; 18:27. [DOI] [PubMed] [Google Scholar]
- 4.Zhao W, Ho HE, Bunyavanich S. The gut microbiome in food allergy. Ann Allergy Asthma Immunol 2019; 122:276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunyavanich S. Food allergy: could the gut microbiota hold the key? Nat Rev Gastroenterol Hepatol 2019; 16:201–2. [DOI] [PubMed] [Google Scholar]
- 6.Ho HE, Bunyavanich S. Microbial Adjuncts for Food Allergen Immunotherapy. Curr Allergy Asthma Rep 2019; 19:25. [DOI] [PubMed] [Google Scholar]
- 7.Huang YJ, Marsland BJ, Bunyavanich S, O’Mahony L, Leung DY, Muraro A, et al. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol 2017; 139:1099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, Dawson P, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazlollahi M, Chun Y, Grishin A, Wood RA, Burks AW, Dawson P, et al. Early-life gut microbiome and egg allergy. Allergy 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savage JH, Lee-Sarwar KA, Sordillo J, Bunyavanich S, Zhou Y, O’Connor G, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee-Sarwar K, Kelly RS, Lasky-Su J, Moody DB, Mola AR, Cheng TY, et al. Intestinal microbial-derived sphingolipids are inversely associated with childhood food allergy. J Allergy Clin Immunol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, et al. Microbiota therapy acts via a regulatory T cell MyD88/RORgammat pathway to suppress food allergy. Nat Med 2019; 25:1164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A. Changes in faecal microbiota of infants with cow’s milk protein allergy--a Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol 2010; 21 :e394–400. [DOI] [PubMed] [Google Scholar]
- 14.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J 2016; 10:742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol 2013; 131:201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez B, Prioult G, Hacini-Rachinel F, Moine D, Bruttin A, Ngom-Bru C, et al. Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T-cell response. FEMS Microbiol Ecol 2012; 79:192–202. [DOI] [PubMed] [Google Scholar]
- 17.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strachan DP. Hay fever, hygiene, and household size. BMJ 1989; 299:1259–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambrecht BN, Hammad H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol 2017; 18:1076–83. [DOI] [PubMed] [Google Scholar]
- 20.Aichbhaumik N, Zoratti EM, Strickler R, Wegienka G, Ownby DR, Havstad S, et al. Prenatal exposure to household pets influences fetal immunoglobulin E production. Clin Exp Allergy 2008; 38:1787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy 2008; 38:634–42. [DOI] [PubMed] [Google Scholar]
- 22.von Mutius E, Radon K. Living on a farm: impact on asthma induction and clinical course. Immunol Allergy Clin North Am 2008; 28:631–47, ix-x. [DOI] [PubMed] [Google Scholar]
- 23.Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA 2002; 288:963–72. [DOI] [PubMed] [Google Scholar]
- 24.Kim H, Sitarik AR, Woodcroft K, Johnson CC, Zoratti E. Birth Mode, Breastfeeding, Pet Exposure, and Antibiotic Use: Associations With the Gut Microbiome and Sensitization in Children. Curr Allergy Asthma Rep 2019; 19:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rook GA. Hygiene hypothesis and autoimmune diseases. Clin Rev Allergy Immunol 2012; 42:5–15. [DOI] [PubMed] [Google Scholar]
- 26.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci U S A 2012; 109:8334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koplin JJ, Dharmage SC, Ponsonby AL, Tang ML, Lowe AJ, Gurrin LC, et al. Environmental and demographic risk factors for egg allergy in a population-based study of infants. Allergy 2012; 67:1415–22. [DOI] [PubMed] [Google Scholar]
- 28.Eggesbo M, Botten G, Stigum H, Nafstad P, Magnus P. Is delivery by cesarean section a risk factor for food allergy? J Allergy Clin Immunol 2003; 112:420–6. [DOI] [PubMed] [Google Scholar]
- 29.Metsala J, Lundqvist A, Kaila M, Gissler M, Klaukka T, Virtanen SM. Maternal and perinatal characteristics and the risk of cow’s milk allergy in infants up to 2 years of age: a case-control study nested in the Finnish population. Am J Epidemiol 2010; 171:1310–6. [DOI] [PubMed] [Google Scholar]
- 30.Bjorksten B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy 1999; 29:342–6. [DOI] [PubMed] [Google Scholar]
- 31.Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases: a multiscale approach. J Allergy Clin Immunol 2015; 135:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature 2012; 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka M, Korenori Y, Washio M, Kobayashi T, Momoda R, Kiyohara C, et al. Signatures in the gut microbiota of Japanese infants who developed food allergies in early childhood. FEMS Microbiol Ecol 2017; 93. [DOI] [PubMed] [Google Scholar]
- 34.Sampson HA, Gerth van Wijk R, Bindslev-Jensen C, Sicherer S, Teuber SS, Burks AW, et al. Standardizing double-blind, placebo-controlled oral food challenges: American Academy of Allergy, Asthma & Immunology-European Academy of Allergy and Clinical Immunology PRACTALL consensus report. J Allergy Clin Immunol 2012; 130:1260–74. [DOI] [PubMed] [Google Scholar]
- 35.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy 2015; 45:632–43. [DOI] [PubMed] [Google Scholar]
- 36.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 2013; 14:559–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez B, Prioult G, Bibiloni R, Nicolis I, Mercenier A, Butel MJ, et al. Germ-free status and altered caecal subdominant microbiota are associated with a high susceptibility to cow’s milk allergy in mice. FEMS Microbiol Ecol 2011; 76:133–44. [DOI] [PubMed] [Google Scholar]
- 39.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol 2004; 172:6978–87. [DOI] [PubMed] [Google Scholar]
- 40.Hazebrouck S, Przybylski-Nicaise L, Ah-Leung S, Adel-Patient K, Corthier G, Wal JM, et al. Allergic sensitization to bovine beta-lactoglobulin: comparison between germ-free and conventional BALB/c mice. Int Arch Allergy Immunol 2009; 148:65–72. [DOI] [PubMed] [Google Scholar]
- 41.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A 2014; 111:13145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331:337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232–6. [DOI] [PubMed] [Google Scholar]
- 44.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med 2019; 25:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wannemuehler MJ, Kiyono H, Babb JL, Michalek SM, McGhee JR. Lipopolysaccharide (LPS) regulation of the immune response: LPS converts germfree mice to sensitivity to oral tolerance induction. J Immunol 1982; 129:959–65. [PubMed] [Google Scholar]
- 46.Maeda Y, Noda S, Tanaka K, Sawamura S, Aiba Y, Ishikawa H, et al. The failure of oral tolerance induction is functionally coupled to the absence of T cells in Peyer’s patches under germfree conditions. Immunobiology 2001; 204:442–57. [DOI] [PubMed] [Google Scholar]
- 47.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol 1997; 159:1739–45. [PubMed] [Google Scholar]
- 48.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe- host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med 2014; 6:220ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noval Rivas M, Burton OT, Wise P, Charbonnier LM, Georgiev P, Oettgen HC, et al. Regulatory T Cell Reprogramming toward a Th2-Cell-like Lineage Impairs Oral Tolerance and Promotes Food Allergy. Immunity 2015; 42:512–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015; 349:989–93. [DOI] [PubMed] [Google Scholar]
- 51.Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE, et al. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep 2016; 15:2809–24. [DOI] [PubMed] [Google Scholar]
- 52.Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019; 74:799–809. [DOI] [PubMed] [Google Scholar]
- 53.Levan SR, Stamnes KA, Lin DL, Panzer AR, Fukui E, McCauley K, et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016; 22:1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018; 23:716–24. [DOI] [PubMed] [Google Scholar]
- 56.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 2008; 453:65–71. [DOI] [PubMed] [Google Scholar]
- 57.Schulz VJ, Smit JJ, Willemsen KJ, Fiechter D, Hassing I, Bleumink R, et al. Activation of the aryl hydrocarbon receptor suppresses sensitization in a mouse peanut allergy model. Toxicol Sci 2011; 123:491–500. [DOI] [PubMed] [Google Scholar]
- 58.Barcik W, Pugin B, Westermann P, Perez NR, Ferstl R, Wawrzyniak M, et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol 2016; 138:1491–4 e7. [DOI] [PubMed] [Google Scholar]
- 59.Barcik W, Wawrzyniak M, Akdis CA, O’Mahony L. Immune regulation by histamine and histamine-secreting bacteria. Curr Opin Immunol 2017; 48:108–13. [DOI] [PubMed] [Google Scholar]
- 60.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T- cell generation. Nature 2013; 504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hussain M, Bonilla-Rosso G, Kwong Chung CKC, Bariswyl L, Rodriguez MP, Kim BS, et al. High dietary fat intake induces a microbiota signature that promotes food allergy. J Allergy Clin Immunol 2019; 144:157–70 e8. [DOI] [PubMed] [Google Scholar]
- 63.Hol J, van Leer EH, Elink Schuurman BE, de Ruiter LF, Samsom JN, Hop W, et al. The acquisition of tolerance toward cow’s milk through probiotic supplementation: a randomized, controlled trial. J Allergy Clin Immunol 2008; 121:1448–54. [DOI] [PubMed] [Google Scholar]
- 64.Berni Canani R, Nocerino R, Terrin G, Coruzzo A, Cosenza L, Leone L, et al. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: a randomized trial. J Allergy Clin Immunol 2012; 129:580–2, 2 e1–5. [DOI] [PubMed] [Google Scholar]
- 65.Berni Canani R, Di Costanzo M, Bedogni G, Amoroso A, Cosenza L, Di Scala C, et al. Extensively hydrolyzed casein formula containing Lactobacillus rhamnosus GG reduces the occurrence of other allergic manifestations in children with cow’s milk allergy: 3-year randomized controlled trial. J Allergy Clin Immunol 2017; 139:1906–13 e4. [DOI] [PubMed] [Google Scholar]
- 66.Tang ML, Ponsonby AL, Orsini F, Tey D, Robinson M, Su EL, et al. Administration of a probiotic with peanut oral immunotherapy: A randomized trial. J Allergy Clin Immunol 2015; 135:737–44 e8. [DOI] [PubMed] [Google Scholar]
- 67.Hsiao KC, Ponsonby AL, Axelrad C, Pitkin S, Tang MLK, Team PS. Long-term clinical and immunological effects of probiotic and peanut oral immunotherapy after treatment cessation: 4-year follow-up of a randomised, double-blind, placebo-controlled trial. Lancet Child Adolesc Health 2017; 1:97–105. [DOI] [PubMed] [Google Scholar]
- 68.Osborn DA, Sinn JK. Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst Rev 2007:CD006475. [DOI] [PubMed] [Google Scholar]
- 69.Cuello-Garcia CA, Brozek JL, Fiocchi A, Pawankar R, Yepes-Nunez JJ, Terracciano L, et al. Probiotics for the prevention of allergy: A systematic review and meta-analysis of randomized controlled trials. J Allergy Clin Immunol 2015; 136:952–61. [DOI] [PubMed] [Google Scholar]
- 70.Fiocchi A, Pawankar R, Cuello-Garcia C, Ahn K, Al-Hammadi S, Agarwal A, et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ J 2015; 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fox A, Bird JA, Fiocchi A, Knol J, Meyer R, Salminen S, et al. The potential for pre-, pro- and synbiotics in the management of infants at risk of cow’s milk allergy or with cow’s milk allergy: An exploration of the rationale, available evidence and remaining questions. World Allergy Organ J 2019; 12:100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Osborn DA, Sinn JK. Prebiotics in infants for prevention of allergy. Cochrane Database Syst Rev 2013:CD006474. [DOI] [PubMed] [Google Scholar]
- 73.Grimshaw K, Logan K, O’Donovan S, Kiely M, Patient K, van Bilsen J, et al. Modifying the infant’s diet to prevent food allergy. Arch Dis Child 2017; 102:179–86. [DOI] [PubMed] [Google Scholar]
- 74.Wopereis H, Sim K, Shaw A, Warner JO, Knol J, Kroll JS. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J Allergy Clin Immunol 2018; 141:1334–42 e5. [DOI] [PubMed] [Google Scholar]
- 75.Burks AW, Harthoorn LF, Van Ampting MT, Oude Nijhuis MM, Langford JE, Wopereis H, et al. Synbiotics-supplemented amino acid-based formula supports adequate growth in cow’s milk allergic infants. Pediatr Allergy Immunol 2015; 26:316–22. [DOI] [PubMed] [Google Scholar]
- 76.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407–15. [DOI] [PubMed] [Google Scholar]
- 77.Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA 2014; 312:1772–8. [DOI] [PubMed] [Google Scholar]
- 78.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018; 359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, et al. Strain-Level Analysis of Mother-to-Child Bacterial Transmission during the First Few Months of Life. Cell Host Microbe 2018; 24:146–54 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018; 24:133–45 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tessler M, Neumann JS, Afshinnekoo E, Pineda M, Hersch R, Velho LFM, et al. Large-scale differences in microbial biodiversity discovery between 16S amplicon and shotgun sequencing. Sci Rep 2017; 7:6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tordesillas L, Berin MC, Sampson HA. Immunology of Food Allergy. Immunity 2017; 47:32–50. [DOI] [PubMed] [Google Scholar]
- 83.Knight R, Callewaert C, Marotz C, Hyde ER, Debelius JW, McDonald D, et al. The Microbiome and Human Biology. Annu Rev Genomics Hum Genet 2017; 18:65–86. [DOI] [PubMed] [Google Scholar]
- 84.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 2016; 535:94–103. [DOI] [PubMed] [Google Scholar]
- 85.Marenholz I, Grosche S, Kalb B, Ruschendorf F, Blumchen K, Schlags R, et al. Genome-wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy. Nat Commun 2017; 8:1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asai Y, Eslami A, van Ginkel CD, Akhabir L, Wan M, Ellis G, et al. Genome-wide association study and meta-analysis in multiple populations identifies new loci for peanut allergy and establishes C11orf30/EMSY as a genetic risk factor for food allergy. J Allergy Clin Immunol 2018; 141:991–1001. [DOI] [PubMed] [Google Scholar]
- 87.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. letters Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nature Genetics 2012; 44:1222–6. [DOI] [PubMed] [Google Scholar]
- 88.Hong X, Hao K, Ladd-Acosta C, Hansen KD, Tsai HJ, Liu X, et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat Commun 2015; 6:6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watson CT, Cohain AT, Griffin RS, Chun Y, Grishin A, Hacyznska H, et al. Integrative transcriptomic analysis reveals key drivers of acute peanut allergic reactions. Nat Commun 2017; 8:1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hong X, Ladd-Acosta C, Hao K, Sherwood B, Ji H, Keet CA, et al. Epigenome- wide association study links site-specific DNA methylation changes with cow’s milk allergy. J Allergy Clin Immunol 2016; 138:908–11 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martino D, Joo JE, Sexton-Oates A, Dang T, Allen K, Saffery R, et al. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics 2014; 9:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Steinke JW, Pochan SL, James HR, Platts-Mills TAE, Commins SP. Altered metabolic profile in patients with IgE to galactose-alpha-1,3-galactose following in vivo food challenge. J Allergy Clin Immunol 2016; 138:1465–7 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kong J, Chalcraft K, Mandur TS, Jimenez-Saiz R, Walker TD, Goncharova S, et al. Comprehensive metabolomics identifies the alarmin uric acid as a critical signal for the induction of peanut allergy. Allergy 2015; 70:495–505. [DOI] [PubMed] [Google Scholar]