Abstract
Grey mangrove (Avicennia marina) is a traditional medicine used for the treatment of various diseases, including rheumatism and ulcers; however, the compounds responsible for its curative effects remain largely unknown. Triterpenoids are a diverse group of plant-specialized metabolites derived from a common precursor, 2,3-oxidosqualene. Triterpenoids are potentially responsible for the beneficial effects of A. marina; however, the chemical profiles of triterpenoids in A. marina and their biosynthetic genes have not been identified. Cytochrome P450 monooxygenases (P450s) have key roles in the structural diversification of plant triterpenoids by catalyzing site-specific oxidation of triterpene scaffolds. Recent studies have revealed that the CYP716 family represents the most common clade of P450s involved in triterpenoid biosynthesis. In this study, we performed triterpenoid profiling and RNA sequencing of A. marina leaves. Mining of CYP716 family genes and enzymatic activity assays of encoded proteins revealed that CYP716A259 catalyzed oxidation at the C-28 position of the pentacyclic triterpene skeletons of β-amyrin, α-amyrin, and lupeol to produce oleanolic acid, ursolic acid, and betulinic acid, respectively. The other functionally defined P450, CYP716C53, catalyzed the C-2α hydroxylation of oleanolic acid and ursolic acid to produce maslinic acid and corosolic acid, respectively. The possible involvement of CYP716A259 and CYP716C53 in the biosynthesis of these health-benefiting compounds in A. marina leaves, and the possible contribution of the resulting compounds to the reported bioactivities of A. marina leaf extract, are discussed.
Keywords: Avicennia marina, corosolic acid, maslinic acid, P450, triterpenoid
Introduction
The gray mangrove, Avicennia marina (Acanthaceae) is widely distributed from coastal East Africa to the western Pacific and from northern New Zealand to Japan (Arnaud-Haond et al. 2006). Different parts of A. marina, including the leaf, bark, stem, and seeds, have traditional medicinal uses worldwide in the treatment of various diseases, such as rheumatism, small pox, and ulcers, by the local communities inhabiting the mangrove forests (Bandaranayake 1998, 2002; Thatoi et al. 2016).
A variety of natural products, including naphthalene derivatives, iridoid glucosides, phenylpropanoids, terpenoids, and steroids, have been detected in A. marina (Jain et al. 2014; Zhu et al. 2009). Recent pharmacological investigations of the extracts of this plant have revealed their medicinal or health-benefiting properties, including anti-inflammatory (Shafie et al. 2013), antiviral (Beula et al. 2012), anticancer (Sukhramani and Patel 2013), antidiabetic (Babuselvam et al. 2013), and anti-androgenic activities (Jain et al. 2014). Several metabolites that could be attributed to these desirable properties have been identified. For example, avicequinone C, a naphthoquinone derivative contained in extracts of A. marina heartwood and twigs, has been shown to have strong antiproliferative activities against L929 (mouse fibroblast) and K562 (human chronic myeloid leukemia) cell lines (Han et al. 2007), and anti-androgenic activity through inhibition of steroid type 1 5α-reductase, the enzyme responsible for the overproduction of 5α-dihydrotestosterone (5α-DHT), which causes androgenic alopecia (AGA) (Jain et al. 2014). A. marina leaf extracts have also shown antidiabetic activity in alloxan-induced diabetic rats (Babuselvam et al. 2013), cytotoxic activity against cancer cell lines (Sukhramani and Patel 2013), and antiviral activity against hepatitis B virus (Beula et al. 2012); however, it remains unknown which compounds are responsible for these activities.
Triterpenoids are a class of structurally diverse plant-specialized (secondary) metabolites commonly derived from 2,3-oxidosqualene (the precursor of sterols, steroids, and triterpenoids). Although the biological role of triterpenoids in plants remains largely unclear, these compounds have been shown to have various medicinal properties (see Furtado et al. 2017 and references therein). Triterpenoids are potentially responsible for the abovementioned bioactivities of A. marina; however, the chemical profiles of A. marina triterpenoids and their biosynthetic genes are largely unknown.
In this study, we performed triterpenoid profiling and RNA sequencing (RNA-Seq) analysis of A. marina leaves for transcriptome mining of genes encoding enzymes in triterpenoid biosynthesis. The biosynthetic pathway of triterpenoids is divided into two main parts, first involving 2,3-oxidosqualene, a linear compound of 30 carbon atoms produced via the mevalonate pathway, being subject to cyclization by oxidosqualene cyclase (OSC), thus generating various triterpene scaffolds; this is followed by site-specific oxidation, catalyzed by cytochrome P450 monooxygenases (P450s). Recent studies have revealed that P450s belonging to the CYP716 family represent the most common clade of P450s involved in triterpenoid biosynthesis in eudicots (Miettinen et al. 2017). Transcriptome mining for genes encoding P450 involved in the CYP716 family, and subsequent enzymatic activity assays with three triterpene scaffolds that have been identified in A. marina (Basyuni et al. 2007; Mahera et al. 2011, 2013), β-amyrin (oleanane-type), α-amyrin (ursane-type), and lupeol (lupane-type), as possible substrates revealed that CYP716A259 was able to catalyze a three-step oxidation reaction at C-28 on these substrates to produce oleanolic acid, ursolic acid, and betulinic acid, respectively. The other functionally defined P450, CYP716C53, was able to catalyze the C-2α hydroxylation of oleanolic acid and ursolic acid to produce maslinic acid and corosolic acid, respectively. The possible involvement of CYP716A259 and CYP716C53 in the biosynthesis of maslinic acid and corosolic acid in A. marina leaves, possible contribution of maslinic acid and corosolic acid to the reported bioactivities of A. marina leaf extract, and functional differentiation of CYP716 subfamilies in triterpenoid biosynthesis are discussed herein.
Materials and methods
Plant materials
The A. marina leaves used in this study were harvested at Xuan Thuy National Park (Nam Dinh Province, Vietnam) on June 27, 2015, and immediately frozen in liquid nitrogen.
Chemicals
β-Amyrin, erythrodiol, oleanolic acid, α-amyrin, uvaol, ursolic acid, and lupeol were purchased from Extrasynthese (Genay, France). Betulin, lanosterol, and β-sitosterol were purchased from Sigma-Aldrich (St. Louis, MO, USA). Betulinic acid was purchased from Tokyo Chemical Industry (Tokyo, Japan). Maslinic acid was purchased from the Cayman Chemical Company (Ann Arbor, MI, USA). Corosolic acid was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan).
Analysis of triterpenoids contained in A. marina leaves
Freeze-dried leaf tissues (40 mg) were extracted twice with methanol/chloroform (1 : 1, v/v). After the liquid had evaporated, 1 ml of methanol and 1 ml of 4 M hydrogen chloride were added to the residue and hydrolyzed at 80°C for 1 h. The hydrolyzed product was extracted twice with hexane/ethyl acetate (1 : 1, v/v), dried in vacuo, and resuspended in 400 µl of methanol/chloroform (1 : 1, v/v). Next, 100 µl of the solution was evaporated, resuspended in 50 µl of N,N-dimethylformamide, and trimethylsilylated with 50 µl of N,O-bis(trimethylsilyl)trifluoroacetamide+trimethylchlorosilane, 99 : 1 (Sigma-Aldrich), at 80°C for 30 min before gas chromatography-mass spectrometry (GC-MS) analysis.
GC-MS analysis
GC-MS analysis was performed using a 5977A mass-selective detector (MSD) (Agilent Technologies, Santa Clara, CA, USA) coupled with a 7890B GC system (Agilent Technologies) and a DB-1 ms capillary column (30 m×0.25 mm internal diameter, 0.25 µm film thickness; Agilent Technologies). The injection component and MSD transfer line were set at 250°C. To prepare leaf extracts, the oven temperature was programmed as follows: 80°C for 1 min, followed by an increase to 300°C at a rate of 20°C/min, followed by a constant 300°C for 28 min. The estimated content of each triterpenoid in leaves (Supplementary Table S1) was calculated based on peak areas from the mass chromatogram shown in Figure 1, by comparing the area of the authentic standard with known concentrations. For extracts from yeast (strains 1–18; Supplementary Table S2), the oven temperature was programmed in four steps. 1) The initial temperature of 80°C was held for 1 min. 2) The temperature was increased to 260°C at a rate of 20°C/min, and then held at 260°C for 10 min. 3) The temperature was increased to 280°C at a rate of 10°C/min, and held at 280°C for 20 min. 4) The temperature was increased to 320°C at a rate of 20°C/min, and held at 320°C for 16 min. For extracts from yeast (strains 19–27), the oven temperature was programmed as follows: 120°C for 1 min, followed by an increase to 300°C at a rate of 20°C/min, with temperature held at 300°C for 30 min.
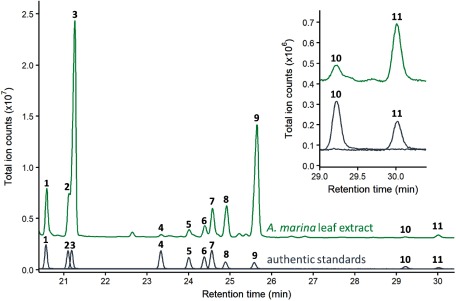
Figure 1. Gas chromatography-mass spectrometry (GC-MS) analysis of A. marina leaf extracts. GC-MS chromatogram of A. marina leaf extracts. Enlarged chromatograms are shown in the inset. Numbers in the chromatograms correspond to the compounds shown in Figure 3. The ion fragmentation pattern is shown in Supplementary Figure S1. Estimation of content of each triterpenoid was performed based on peak areas, by comparing the areas of corresponding authentic standards with known concentrations (Supplementary Table S1).
RNA extraction
Total RNA was extracted using PureLink Plant RNA Reagent (Thermo Fisher Scientific, Waltham, MA, USA) from frozen leaves and treated with recombinant DNaseI (RNase-free) (Takara Bio, Shiga, Japan), then purified using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the RNA clean-up protocol.
Library construction
A 10-µg aliquot of total RNA was used to construct a cDNA library using an Illumina TruSeq Prep Kit v2 according to the manufacturer’s protocol (Illumina, San Diego, CA, USA). The resulting cDNA library was sequenced using HiSeq 2000 (Illumina) with 100-bp paired-end (PE) reads in high-output mode. The sequence reads were assembled using the CLC Genomics Workbench software (ver. 5.5.2; CLC Bio, Aarhus, Denmark) with a minimum contig length of 300 bp and the ‘perform scaffolding’ function. After the removal of adaptor sequences, low-quality reads, and redundant sequences using the TIGR Gene Indices clustering tools (Pertea et al. 2003), we assembled 46,833 contigs. We submitted the raw RNA-Seq reads obtained in this study to the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA) under the accession number DRA006410.
Functional annotation of assembled reads
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping was performed using the KEGG Automatic Annotation Server (http://www.genome.jp/kegg/kaas/) (Moriya et al. 2007) and the bi-directional best hit method against the ath dataset with the default threshold.
Cloning of candidate P450 genes
First-strand cDNA was synthesized from 1 µg total RNA prepared from the A. marina leaves using the SMARTer RACE cDNA Amplification Kit (Clontech/Takara Bio) according to the manufacturer’s instructions. Fragments containing the full-length coding sequence (CDS) of CYP716A259 or CYP716A260 were amplified by polymerase chain reaction (PCR) using primers 1 and 2 for CYP716A259, and primers 3 and 4 for CYP716A260. To obtain the missing 5′ sequences of GBIO01001098 and Contig00032757, 5′-rapid amplification of cDNA ends (RACE) PCR was performed using the SMARTer RACE cDNA Amplification Kit according to the manufacturer’s instructions, employing primers 5 (first) and 6 (nested) for GBIO01001098, and primers 7 (first) and 8 (nested) for Contig00032757. To obtain the missing 3′ sequences of Contig00031299, 3′-RACE PCR was performed using primers 9 (first) and 10 (nested) for Contig00031299. Using sequence information obtained by RACE-PCR, fragments containing the full-length CDS of CYP716D59, CYP716C53, or CYP716C54 were amplified by PCR using primers 11 and 12 for CYP716D59, primers 13 and 14 for CYP716C53, and primers 15 and 16 for CYP716C54. The sequences of the primers used in this study are shown in Supplementary Table S3. Amplified fragments were cloned into the pENTR-D-topo vector (Thermo Fisher Scientific) and sequenced. The nucleotide sequences isolated in this study have been submitted to DDBJ under the accession numbers LC325358 (CYP716C54), LC325359 (CYP716D59), LC325360 (CYP716A259), LC325361 (CYP716A260), and LC325362 (CYP716C53), respectively.
Generation of engineered yeast strains
Each P450 cDNA was transferred into a Gateway-compatible version of the pELC vector (Seki et al. 2011) using the Gateway LR Clonase II Enzyme mix (Thermo Fisher Scientific), to generate a construct for galactose-inducible dual expression of Lotus japonicas CPR1 and P450. Each P450 cDNA was also transferred into pYES-DEST52 (Thermo Fisher Scientific) using the Gateway LR Clonase II Enzyme mix to generate galactose-inducible expression of each P450. These constructs were introduced into Saccharomyces cerevisiae INVSc1 (MATa his3D1 leu2 trp1-289 ura3-52; Thermo Fisher Scientific) harboring pYES3-ADH-OSC1 (bAS), pYES-ADH-aAS, or pYES-ADH-LUS (Fukushima et al. 2011) using the Frozen-EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA, USA). The engineered yeast strains made for this study are listed in Supplementary Table S2.
In vivo P450 enzyme assays
Glycerol stock of each yeast strain was inoculated into 5 ml of appropriate synthetic defined (SD) medium containing 2% glucose and cultured overnight at 30°C with shaking at 200 rpm. The yeast cells were collected and resuspended in 10 ml of SD medium containing 2% galactose. These cells were then cultured at 30°C for a further 2 nights at 200 rpm. The yeast cultures were extracted three times with 6 ml of ethyl acetate. After the liquid had evaporated, the remaining residue was dissolved in 500 µl of methanol/chloroform (1 : 1, v/v). A total of 100 µl of this solution was allowed to evaporate and trimethylsilylated with 50 µl of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich) at 80°C for 30 min before analysis by GC-MS.
Phylogenetic analysis of P450
The full-length protein sequences of all CYP716 family enzymes functionally characterized as triterpenoid oxidases were collected from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (Supplementary Table S4) and aligned using MUSCLE (Edgar 2004). A rooted neighbor-joining tree was generated with 1,000 bootstrap replicates using MEGA 7 software (Kumar et al. 2016).
Results
Triterpenoid profile of A. marina leaves
We analyzed the triterpenoid profiles of A. marina leaves. Peaks 1–11 were detected in A. marina leaf extract (Figure 1). The retention times and mass spectra were in accordance with authentic standards of β-amyrin (1), α-amyrin (2), lupeol (3), erythrodiol (4), uvaol (5), betulin (6), oleanolic acid (7), betulinic acid (8), ursolic acid (9), maslinic acid (10), and corosolic acid (11) (Supplementary Figure S1). Among these compounds, β-amyrin (1) (Basyuni et al. 2007), α-amyrin (2), ursolic acid (9) (Mahera et al. 2013), lupeol (3), and betulinic acid (8) (Mahera et al. 2011) have been identified previously in A. marina. In contrast, this is the first report of detection of oleanolic acid (7), maslinic acid (10), and corosolic acid (11) in A. marina. The estimated content of each triterpenoid in leaves is shown in Supplementary Table S1.
RNA-Seq analysis of A. marina leaves and functional annotation of contigs
To identify genes involved in the biosynthesis of triterpenoids in A. marina, total RNA extracted from A. marina leaves was sequenced using the Illumina HiSeq 2000 platform. We obtained 29,135,466 reads in total and assembled these into 46,833 contigs (Table 1).
Table 1. Summary of the RNA-Seq analysis of A. marina.
| Number of total reads used in the assembly | 29,135,466 |
| Number of contigs | 46,833 |
| N50 of contigs (bp) | 1,413 |
| Average length of contigs (bp) | 1,011 |
| Minimum length of contigs (bp) | 244 |
| Maximum length of contigs (bp) | 9,624 |
We assigned KEGG pathway annotations to the contigs. Transcripts were associated with metabolic pathways, such as flavonoid biosynthesis (16 contigs), flavone and flavonol biosynthesis (3 contigs), anthocyanin biosynthesis (1 contig), terpenoid backbone biosynthesis (40 contigs), monoterpenoid biosynthesis (1 contig), sesquiterpenoid and triterpenoid biosynthesis (6 contigs), and diterpenoid biosynthesis (8 contigs) (Supplementary Table S5) and mevalonate pathway and triterpenoid biosynthesis (19 contigs) (Supplementary Table S6). These results suggest that the transcriptome data may allow for identification of novel genes involved in specialized metabolic pathways in A. marina.
Selection of candidate P450s
The importance of P450s in the structural diversification of triterpenoids in plants has recently been demonstrated. Several different P450 (sub)families, including CYP88D, CYP72A, CYP93E, CYP51H, and CYP716, have been shown to be involved in triterpenoid biosynthesis in various plant species (reviewed in Ghosh 2017; Seki et al. 2015). Unlike CYP88Ds and CYP93Es, which are restricted to Fabaceae, or CYP51Hs, which are restricted to monocots, CYP716s have been identified in various plant species. The CYP716 family represents the most common clade of P450s shown to be involved in triterpenoid biosynthesis (Miettinen et al. 2017). Among these, CYP716A12, pentacyclic triterpenoid C28-oxidase, identified from the model legume Medicago truncatula, was the first CYP716 family enzyme shown to be involved in triterpenoid biosynthesis (Carelli et al. 2011; Fukushima et al. 2011).
To identify candidate P450s involved in triterpenoid biosynthesis in A. marina, a tblastn search was performed using the CYP716A12 protein sequence as a query in the in-house A. marina transcriptome database, established based on the RNA-seq data obtained in this study. We found that four contigs, Contig00009575, Contig00013756, Contig00031299, and Contig00032757, encoded proteins with more than 45% identity to CYP716A12 (Supplementary Table S7). In addition, we identified another partial CYP716 gene sequence (GBIO01001098) from the transcriptome shotgun assembly of A. marina deposited at DDBJ/EMBL/GenBank under the accession GBIO00000000 (Huang et al. 2014). Contig00013756 and Contig0009575 included the full-length CDS. We also obtained the full-length CDS of other candidates, Contig00031299, Contig00032757, and GBIO01001098 by RACE-PCR. The full-length cDNA sequences of these contigs were determined and the corresponding proteins were named by the P450 nomenclature committee: CYP716A259 (Contig00013756), CYP716A260 (Contig00009575), CYP716C53 (Contig00031299), CYP716C54 (GBIO01001098), and CYP716D59 (Contig00032757).
Enzymatic activities of CYP716A259 and CYP716C53
We expressed each of five candidate P450s, together with a cytochrome P450 reductase (CPR) as a redox partner, in a strain of engineered yeast that had been pre-transformed with β-amyrin synthase (bAS) and produced β-amyrin endogenously (strains 1–6; Supplementary Table S2). After culturing each yeast strain, the in vivo metabolites were extracted and analyzed by GC-MS.
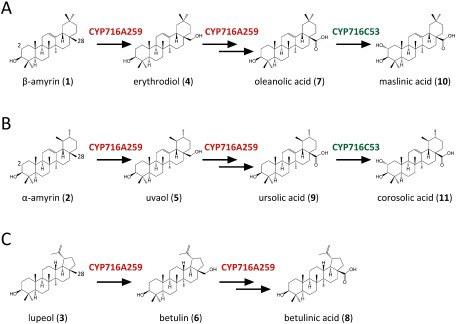
Among five candidate P450s, only CYP716A259 (76% amino acid identity with CYP716A12) was found to oxidize β-amyrin (1) (Figure 2A). In the bAS/CPR/CYP716A259-expressing yeast (strain 2), erythrodiol (4) and oleanolic acid (7) were detected by comparison with authentic standards (Supplementary Figure S2A). These compounds are oxidation products of β-amyrin (1) modified at C-28. Erythrodiol (4) is a reaction intermediate occurring between β-amyrin (1) and oleanolic acid (7) (Figure 3A). Therefore, CYP716A259 was identified as a β-amyrin 28-oxidase in A. marina.
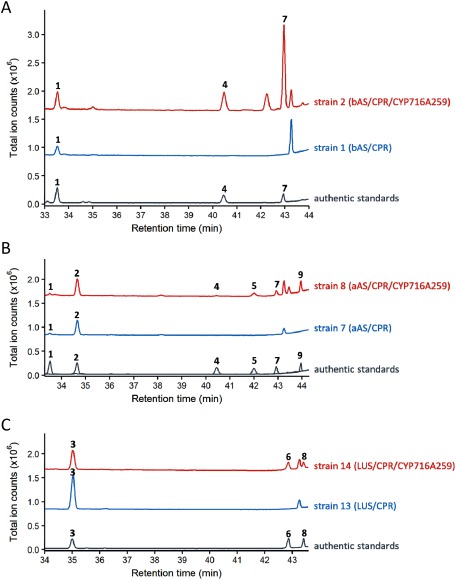
Figure 2. Enzyme assays of CYP716A259 in engineered yeast strains endogenously producing triterpene scaffold. The GC-MS total ion chromatograms of yeast extracts are shown. CYP716A259 was expressed in (A) β-amyrin-, (B) α-amyrin-, and (C) lupeol-producing yeast. The numbers in the chromatograms correspond to the compounds shown in Figure 3. The ion fragmentation patterns, of each product and the authentic standards, are shown in Supplementary Figure S2.
Figure 3. Reactions catalyzed by CYP716A259 and CYP716C53 in triterpene-producing engineered yeast. The arrows represent single oxidation reactions catalyzed by CYP716A259 and CYP716C53.
We also expressed each candidate P450 together with CPR in two strains of engineered yeast that had been pre-transformed with α-amyrin synthase (aAS) or lupeol synthase (LUS), which produced α-amyrin (strains 7–12) or lupeol (strains 13–18) endogenously. Again, only CYP716A259 was found to oxidize α-amyrin (2) and lupeol (3). In the aAS/CPR/CYP716A259-expressing yeast (strain 8), uvaol (5) and ursolic acid (9) were detected by comparison with authentic standards (Figure 2B; Supplementary Figure S2B). These compounds are oxidation products of α-amyrin (2) modified at C-28. Uvaol (5) is a reaction intermediate occurring between α-amyrin (2) and ursolic acid (9) (Figure 3B). It should be noted that the aAS used in this study was a multiproduct enzyme that produced not only α-amyrin, but also β-amyrin, and β-amyrin and its oxidized products were also detected in the aAS/CPR/CYP716A259-expressing yeast as minor peaks (1, 4, and 7) (Figure 2B).
In the LUS/CPR/CYP716A259-expressing yeast (strain 14), betulin (6) and betulinic acid (8) were detected by comparison with authentic standards (Figure 2C; Supplementary Figure S2C). These compounds are oxidation products of lupeol (3) modified at C-28. Betulin (6) is a reaction intermediate occurring between lupeol (3) and betulinic acid (8) (Figure 3C). These data indicate that, similar to most previously characterized CYP716A subfamily enzymes, CYP716A259 can catalyze a three-step oxidation reaction at C-28 of β-amyrin, α-amyrin, and lupeol to produce oleanolic acid, ursolic acid, and betulinic acid, respectively.
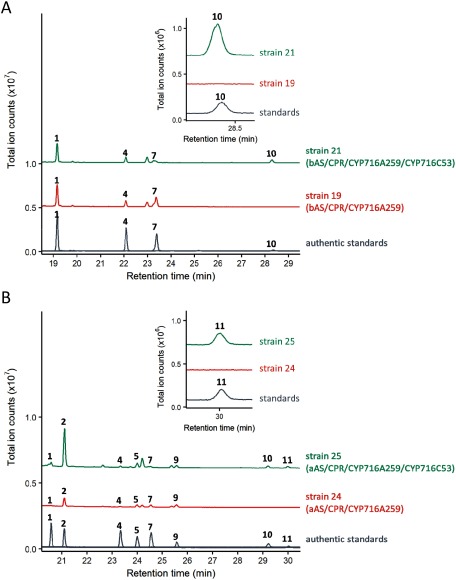
The remaining four P450s (CYP716A260, CYP716C53, CYP716C54, and CYP716D59) showed no catalytic activity on β-amyrin, α-amyrin, or lupeol. Nevertheless, it has been reported that although β-amyrin is not a substrate for CYP72A67 from M. truncatula or CYP716C11 from Centella asiatica, these enzymes showed C-2β and C-2α hydroxylase activity, respectively, for oleanolic acid, suggesting that the presence of a carboxylic group at C-28 is required in substrate recognition by these P450s (Fukushima et al. 2013; Miettinen et al. 2017). Therefore, to assess the potential activity of the four candidate P450s on oleanolic acid, we generated yeast strains expressing bAS, CPR, CYP716A259, and each of the four P450s together (strains 19–23). Only CYP716C53 (65% amino acid identity with CYP716C11) was found to oxidize oleanolic acid. As shown in Figure 4A, bAS/CPR/CYP716A259/CYP716C53-expressing yeast (strain 21) formed an additional peak (peak 10). The mass spectrum of peak 10 was an exact match with that of the authentic maslinic acid standard (10), corresponding to 2α-hydroxyoleanolic acid (Supplementary Figure S3A).
Figure 4. Enzyme assays of CYP716C53 in engineered yeast strains endogenously producing oleanolic acid and ursolic acid. The GC-MS chromatograms of the extracts of (A) bAS/CPR/CYP716A259/CYP716C53-expressing yeast and (B) aAS/CPR/CYP716A259/CYP716C53-expressing yeast are shown. Enlarged chromatograms are shown in the inset. The numbers in the chromatograms correspond to the compounds shown in Figure 3. The ion fragmentation patterns of peak 10 and peak 11 are shown in Supplementary Figure S3.
To examine the activities of CYP716C53 against ursolic acid and betulinic acid, we generated yeast strains expressing aAS or LUS, CPR, CYP716A259, and CYP716C53. As shown in Figure 4B, aAS/CPR/CYP716A259/CYP716C53-expressing yeast (strain 25) formed an additional peak (peak 11). The mass spectrum of peak 11 was an exact match with that of the authentic corosolic acid standard (11), corresponding to 2α-hydroxyursolic acid (Supplementary Figure S3B). These results indicate that CYP716C53 can catalyze the C-2α hydroxylation of oleanolic acid and ursolic acid to produce maslinic acid and corosolic acid, respectively (Figure 3A, B). However, no oxidation activity of CYP716C53 on betulinic acid was detected.
Discussion
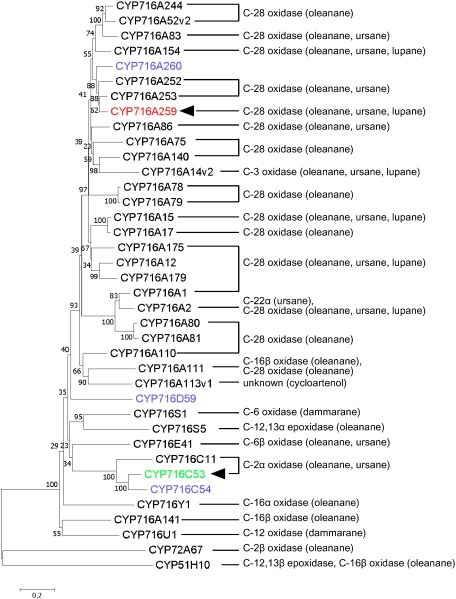
We identified two P450s, CYP716A259 and CYP716C53, which function as triterpene oxidases in A. marina. CYP716 family enzymes, divided into five subfamilies (A, C, E, S, and Y), are widely distributed among eudicots and considered to be a major contributor to the diversification of eudicot triterpenoid biosynthesis (Miettinen et al. 2017). Most CYP716A subfamily enzymes have been identified to catalyze oxidation at the C-28 position of pentacyclic triterpene skeletons (Figure 5), such as β-amyrin, α-amyrin, and lupeol, except for several CYP716As, including CYP716A14v2 from Artemisia annua catalyzing oxidation at the C-3 position (Moses et al. 2015) and CYP716A141 from Platycodon grandiflorus catalyzing oxidation at the C-16β position (Miettinen et al. 2017; Tamura et al. 2017). In accordance with these observations, CYP716A259 catalyzed oxidation at C-28 in β-amyrin, α-amyrin, and lupeol (Figure 3).
Figure 5. Phylogenetic tree of previously characterized CYP716 enzymes and the A. marina CYP716s identified in this study. The numbers above the branches indicate bootstrap values for 1,000 replicates. The scale bar shows the amino acid substitution ratio. The P450s isolated in this study are denoted by color text: CYP716A259 in red, CYP716C53 in green, and P450s without significant activity in purple. The filled arrowheads indicate the P450s defined functionally in this study. The determined biochemical activities of the P450s are indicated on the right, with their substrate triterpene skeletons or compounds shown in parentheses. CYP72A67 and CYP51H10 were used as outgroups. The Origin and GenBank accession numbers of P450 sequences used for the phylogenetic analysis are shown in Supplementary Table S4.
In contrast to well-characterized CYP716A subfamily enzymes, the enzymatic function of the CYP716C subfamily remains largely unknown. Only CYP716C11 from C. asiatica (Apiaceae) has been characterized and shown to catalyze C-2α hydroxylation of oleanolic acid (Figure 5) (Miettinen et al. 2017). In this study, we identified CYP716C53 as a second C-2α hydroxylase of oleanolic acid and ursolic acid (Figure 3), suggestive of the conserved function of the CYP716C subfamily P450s in C-2α oxidation of pentacyclic triterpenes. Interestingly, both CYP716C11 and CYP716C53 showed C-2α hydroxylase activity against oleanolic acid, but not β-amyrin, suggesting that a C-28 carboxy group is a prerequisite for recognition as a substrate by CYP716C subfamily proteins.
In this study, we detected maslinic acid (2α-hydroxyoleanolic acid) and corosolic acid (2α-hydroxyursolic acid) in A. marina leaf extract (Figure 1). The biochemical functions of CYP716A259 and CYP716C53 (Figure 3), together with the fact that both maslinic acid and corosolic acid were detected in A. marina leaf extract, strongly suggest the involvement of CYP716A259 and CYP716C53 in maslinic acid and corosolic acid biosynthesis in planta. In contrast to CYP716A259 and CYP716C53, the remaining three P450s showed no catalytic activity on the potential substrates tested in this study (β-amyrin, α-amyrin, lupeol, and oleanolic acid). Among these P450s, CYP716A260 and CYP716D59 showed expression levels comparable to those of CYP716A259 and CYP716C53, respectively, in leaves as estimated by their RPKM values (Supplementary Table S7), and no contigs corresponding to CYP716C54 were detected among our RNA-seq data. It is possible that CYP716A260 and CYP716D59 use different substrates from CYP716A259 and CYP716C53, for example, different triterpene scaffolds and/or their derivatives in planta. Further enzyme assays using various triterpenoids substrates are required to clarify their enzyme activities.
Rheumatoid arthritis is a chronic disease that produces joint inflammation symptoms. Recently, it was demonstrated that oral administration of maslinic acid suppressed arthritis, accompanied by the repression of inflammatory mediator gene expression in mice (Fukumitsu et al. 2016). As mentioned earlier, A. marina has long been used in the treatment of rheumatism (Bandaranayake 1998, 2002). In addition, A. marina leaf extract treatment reduced rheumatoid arthritis symptoms in a rat model of rheumatoid arthritis (Shafie et al. 2013). These observations imply that maslinic acid may contribute to the anti-arthritic effects of A. marina leaf extract.
Corosolic acid was another triterpenoid detected for the first time in A. marina. The increasing incidence of diabetes has become a concern in developed countries. Frequent insulin treatment for diabetes is expensive and may increase anti-insulin antibody production, which can have undesirable side effects (Sivakumar et al. 2009). Corosolic acid, which is a “phyto-insulin”, has recently attracted attention for its insulin-like properties, but without induction of anti-insulin antibodies (Sivakumar et al. 2009). A. marina leaf extracts have also shown antidiabetic activity in alloxan-induced diabetic rats (Babuselvam et al. 2013), suggesting a contribution of corosolic acid to the antidiabetic activity of A. marina leaf extracts. Although many P450s involved in triterpenoid biosynthesis have been identified, the identification of CYP716A259 and CYP716C53 is important in terms of their possible use in synthetic biology to produce triterpenoids with beneficial bioactivities, such as maslinic acid, corosolic acid, and their derivatives.
Finally, we generated a transcriptome dataset of A. marina leaves, and validated the utility of the transcriptome data for gene discovery in metabolic pathways by identifying two novel P450s involved in triterpenoid biosynthesis. As mentioned earlier, a variety of specialized metabolites potentially associated with the beneficial properties of A. marina leaf extract have been identified. Our transcriptome data will provide valuable resource for further discovery of biosynthetic genes of valuable metabolites in A. marina.
Acknowledgments
We thank Dr. Yoshinori Sumimura, at the Global Collaboration Center, Osaka University, for comprehensive support of our activities. We also thank Dr. Ery Odette Fukushima (Osaka University) and Dr. Shuhei Yasumoto (Osaka University) for helpful discussions. We also thank Mr. Tsutomu Hosouchi and Ms. Sayaka Shinpo (Kazusa DNA Research Institute) for technical support with the Illumina sequencing. This work was supported by a bilateral joint research project between Japan and Vietnam from the Japan Society for the Promotion of Science (JSPS-collaboration project code: VAST.HTQT.Nhat Ban.02/15-17), and JSPS KAKENHI grant Nos. JP17K07754 and JP17H05442 to H. Seki.
Abbreviations
- aAS
α-amyrin synthase
- bAS
β-amyrin synthase
- CAS
cycloartenol synthase
- CDS
coding sequence
- CPR
cytochrome P450 reductase
- LUS
lupeol synthase
- OSC
oxidosqualene cyclase
Supplementary Data
References
- Arnaud-Haond S, Teixeira S, Massa SI, Billot C, Saenger P, Coupland G, Duarte CM, Serrao EA (2006) Genetic structure at range edge: Low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol Ecol 15: 3515–3525 [DOI] [PubMed] [Google Scholar]
- Babuselvam M, Abideen S, Gunasekaran T, Beula JM, Dhinakarraj M (2013) Bioactivity of Avicennia marina and Rhizophora mucronata for the management of diabetes mellitus. WJPR 3: 11–18 [Google Scholar]
- Bandaranayake WM (1998) Traditional and medicinal uses of mangroves. Mang Salt Marsh 2: 133–148 [Google Scholar]
- Bandaranayake WM (2002) Bioactivities, bioactive compounds and chemical constituents of mangrove plants. Wet Eco Man 10: 421–452 [Google Scholar]
- Basyuni M, Oku H, Baba S, Takara K, Iwasaki H (2007) Isoprenoids of okinawan mangroves as lipid input into estuarine ecosystem. J Oceanogr 63: 601–608 [Google Scholar]
- Beula M, Gnanadesigan JM, Banerjee PR, Ravikumar S, Anand M (2012) Antiviral antioxidant and toxicological evaluation of mangrove plant from south east coast of India. Asian Pac J Trop Biomed 1: 570–573 [Google Scholar]
- Carelli M, Biazzi E, Panara F, Tava A, Scaramelli L, Porceddu A, Graham N, Odoardi M, Piano E, Arcioni S, et al. (2011) Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23: 3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumitsu S, Villareal MO, Fujitsuka T, Aida K, Isoda H (2016) Anti-inflammatory and anti-arthritic effects of pentacyclic triterpenoids maslinic acid through NF-κB inactivation. Mol Nutr Food Res 60: 399–409 [DOI] [PubMed] [Google Scholar]
- Fukushima EO, Seki H, Ohyama K, Ono E, Umemoto N, Mizutani M, Saito K, Muranaka T (2011) CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol 52: 2050–2061 [DOI] [PubMed] [Google Scholar]
- Fukushima EO, Seki H, Sawai S, Suzuki M, Ohyama K, Saito K, Muranaka T (2013) Combinatorial biosynthesis of legume natural and rare triterpenoids in engineered yeast. Plant Cell Physiol 54: 740–749 [DOI] [PubMed] [Google Scholar]
- Furtado NAJC, Pirson L, Edelberg H, Miranda LM, Loira-Pastoriza C, Preat V, Larondelle Y, Andre CM (2017) Pentacyclic triterpene bioavailability: An overview of in vitro and in vivo studies. Molecules 22: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S (2017) Triterpene structural diversification by plant cytochrome P450 enzymes. Front Plant Sci 8: 1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Huang X, Dahse HM, Moellmann U, Fu H, Grabley S, Sattler I, Lin W (2007) Unusual naphthoquinone derivatives from the twigs of Avicennia marina. J Nat Prod 70: 923–927 [DOI] [PubMed] [Google Scholar]
- Huang J, Lu X, Zhang W, Huang R, Chen S, Zheng Y (2014) Transcriptome sequencing and analysis of leaf tissue of Avicennia marina using the illumina platform. PLoS One 9: e108785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Monthakantirat O, Tengamnuay P, De-Eknamkul W (2014) Avicequinone C isolated from Avicennia marina exhibits 5α-Reductase-Type 1 inhibitory activity using an androgenic alopecia relevant cell-based assay system. Molecules 19: 6809–6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahera SA, Ahmad VU, Saifullah SM, Mohammad FV, Ambreen K (2011) Steroids and triterpenoids from grey mangrove Avicennia marina. Pak J Bot 43: 1417–1422 [Google Scholar]
- Mahera SA, Saifullah SM, Ahmad VU, Mohammad FV (2013) Phytochemical studies on mangrove Avicennia marina. Pak J Bot 45: 2093–2094 [Google Scholar]
- Miettinen K, Pollier J, Buyst D, Arendt P, Csuk R, Sommerwerk S, Moses T, Mertens J, Sonawane PD, Pauwels L, et al. (2017) The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis. Nat Commun 8: 14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35(Web Server): W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses T, Pollier J, Shen Q, Soetaert S, Reed J, Erffelinck ML, Van Nieuwerburgh FC, Vanden Bossche R, Osbourn A, Thevelein JM, et al. (2015) OSC2 and CYP716A14v2 catalyze the biosynthesis of triterpenoids for the cuticle of aerial organs of Artemisia annua. Plant Cell 27: 286–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, et al. (2003) TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652 [DOI] [PubMed] [Google Scholar]
- Seki H, Sawai S, Ohyama K, Mizutani M, Ohnishi T, Sudo H, Fukushima EO, Akashi T, Aoki T, Saito K, et al. (2011) Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 23: 4112–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Tamura K, Muranaka T (2015) P450s and UGTs: Key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol 56: 1463–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafie M, Forghani A, Moshtaghiyan J (2013) Anti-inflammatory effects of hydro-alcoholic extracts of mangrove (Avicennia marina) and vitamin C on arthritic rats. Bull Environ Pharmacol Life Sci 2: 32–37 [Google Scholar]
- Sivakumar G, Vail DR, Nair V, Fabricio Medina-Bolivar F, Lay JO (2009) Plant-based corosolic acid: Future anti-diabetic drug? Biotechnol J 4: 1704–1711 [DOI] [PubMed] [Google Scholar]
- Sukhramani PS, Patel PM (2013) Biological screening of Avicennia marina for anticancer activity. Pharm Sin 4: 125–130 [Google Scholar]
- Tamura K, Teranishi Y, Ueda S, Suzuki H, Kawano N, Yoshimatsu K, Saito K, Kawahara N, Muranaka T, Seki H (2017) Cytochrome P450 monooxygenase CYP716A141 is a unique β-amyrin C-16β oxidase involved in triterpenoid saponin biosynthesis in Platycodon grandiflorus. Plant Cell Physiol 58: 874–884 [DOI] [PubMed] [Google Scholar]
- Thatoi H, Samantaray D, Das SK (2016) The genus Avicennia, a pioneer group of dominant mangrove plant species with potential medicinal values: A review. Front Life Sci 9: 267–291 [Google Scholar]
- Zhu F, Xin C, Yihua Y, Meizhen H, Huili S, Wenzhou X (2009) The chemical investigations of the mangrove plant Avicennia marina and its endophytes. Open Nat Prod J 2: 24–32 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.