Abstract
Brassica juncea is an important vegetable and condiment crop widely grown in Asia, and the yield and quality of its product organs are affected by flowering time. AGAMOUS-LIKE18-1 (AGL18-1) belongs to a member of MADS-domain transcription factors, which play vital roles in flowering time control, but the biological role of AGL18-1 in B. juncea (BjuAGL18-1) has not been thoroughly revealed in flowering regulatory network. In this study, BjuAGL18-1 expressed highly in inflorescence and flower, but slightly in root, stem and leaf. The sense and anti-sense transgenic lines of BjuAGL18-1 were generated and showed that BjuAGL18-1 functioned as a flowering inhibitor and depressed growth of lateral branching. During the vegetative phase, BjuAGL18-1 induced another flowering repressor AGAMOUS-LIKE15 (BjuAGL15) but inhibited the flowering signal integrator of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (BjuSOC1) in Brassica juncea. Whereas, during the flower developmental phase, both SOC1 and AGAMOUS-LIKE24 (AGL24) were down-regulated by BjuAGL18-1. By contrast, AGL15 was promoted by BjuAGL18-1, while SHORT VEGETATIVE PHASE (SVP) was independent of BjuAGL18-1. Additionally, HISTONE DEACETYLASE 9 (HDA9) was highly induced by BjuAGL18-1. These results will provide valuable information for clarifying the molecular mechanism of BjuAGL18-1 in mediating flowering time.
Keywords: AGL18-1, Brassica juncea, flowering related genes, flowering time
Introduction
MCM1-AGAMOUS-DEFICIENS-SRF (MADS) domain regulatory factors play essential roles in controlling the floral transition (Adamczyk et al. 2007; Borner et al. 2000; Hartmann et al. 2000; Lee et al. 2000; Yu et al. 2002). During the floral transition, MADS-domain proteins can act either as activators or repressors (Fernandez et al. 2014). In Arabidopsis, important floral activators include SOC1 and AGL24. Repressors of flowering include AGL15, AGL18 and SVP.
Transgenic plants of AGL15 overexpression produced a lot of phenotypic changes, including reduced fertility, delayed flowering time and inhibit senescence (Fernandez et al. 2000), as with the phenotypic of AGL18 overexpression lines (Adamczyk et al. 2007). AGL18 and AGL15 form a heterodimer to actively regulate the expression of miRNA156, which acted as a floral repressor (Serivichyaswat et al. 2015). In our previous studies, three members (AGL18-1, AGL18-2 and AGL18-3) of the AGL18 family, together with SOC1 and AGL24 were cloned from B. juncea (Li et al. 2018). However, only AGL18-1 protein interacted with promoters of SOC1 and AGL24 via yeast one-hybrid assays and Dual-Glo® luciferase assays (Li et al. 2018).
SOC1 was regarded as a pivotal flowering integrator in regulating flowering time (Immink et al. 2012; Melzer et al. 2008). Additionally, SOC1 involved in the regulation of leaf senescence (Chen et al. 2017), floral organ aging (Tan and Swain 2007) and fruit development (Papaefthimiou et al. 2012). SOC1 was regulated by the FT and CONSTANS (CO) genes (Helliwell et al. 2006), and was employed as downstream target of CO. CO regulated the expression of SOC1 gene by binding to the site of the SOC1 promoter region (Hepworth et al. 2002). Another crucial flowering integrator was AGL24 (Michaels et al. 2003), which highly expressed in shoot apices, leaves, stems, inflorescence and roots (Liu et al. 2008; Yu et al. 2002). AGL24 protein activated the expression of SOC1 in the flowering transition, and served as a positive regulator of SOC1 in Arabidopsis. Overexpression of AGL24 caused up-regulation of SOC1, and overexpression of SOC1 also led to up-regulation of AGL24. It identified that the SOC1 and AGL24 could be regulated with each other (Michaels et al. 2003).
Interestingly, SVP had the highest homology with AGL24, but it showed the opposite biological role in the flowering process (Hartmann et al. 2000). SVP protein not only directly repressed the expression of flowering integrator SOC1 in shoot apices and leaves (Li et al. 2008), but also regulated the expression of flowering integrator FT to decay flowering (Lee et al. 2007). In addition, HDA9 was a member of histone deacetylase (HDAC) family in plants, which could delay the initiative of flowering by repressing the transcriptional factor AGL19 in Arabidopsis (Kang et al. 2015; Kim et al. 2013). The MADS transcription factor AGL18 directly interacted with the co-repressor TOPLESS (TPL) (Causier et al. 2012), which was regarded as a flowering repressor by recruiting histone deacetylase HDA19 (Krogan et al. 2012; Liu et al. 2009). Furthermore, the FUNCTION-RELATED 1(AFR1) protein and the FUNCTION-RELATED 2(AFR2) protein which periodic regulated HDAC, were ability to form complexes with AGL18 and HDAC in flowering transition (Gu et al. 2013).
In Arabidopsis, AGL18 acted as repressor of the floral transition, and redundancy with AGL15, Yeast two-hybrid assays reveal that AGL18 could interaction with AGL15 (Adamczyk et al. 2007). In yeast one-hybrid assays, AGL15 regulates SOC1 via binding to promoter of SOC1. Therefore, both AGL15 and AGL18 were acted as SOC1 targets (Immink et al. 2012). SOC1 directly interacts with AGL24 and forms a homodimer to integrate flowering signals. In addition, AGL24 and SOC1 affect expression of each other. The MADS-Domain factors AGL15 and AGL18 could along with SVP and AGL24 to inhibit expression of floral genes (Liu et al. 2008; Michaels et al. 2003; Yu et al. 2002).
However, it was still unknown how AGL18-1 functioned as a flowering regulation factor in B. juneca. Hence, the biological role of AGL18-1 in floral transition was identified in transgenic plants. Subsequently, the expression patterns of flowering regulatory genes, such as SOC1, AGL24, AGL15, SVP and HDA9, were investigated via qRT-PCR. This study will provide valuable information for elucidating regulatory mechanisms of AGL18-1 together with the above flowering-related genes in B. juncea.
Materials and methods
Transgenic plants and growth conditions
BjuAGL18-1 gene was cloned from B. juncea in our previous study (Li et al. 2018). Successively, the transgenic plants were generated in the background via agrobacterium-mediated method use recombinant plasmids pBI35S::sBjuAGL18-1 and pBI35S::aBjuAGL18-1, which constructed by inserted the full-length sequences of BjuAGL18-1 into the containing CaMV35S promoter plant binary vector pBI121 forwardly and reversely. Next, the above recombinants were transformed into B. juncea and Nicotiana tabacum to generate transgenic lines.
T1 transgenic plants were cultivated in the chamber under long-day conditions (16/8 h light/dark cycle at 25/20°C). The 35S::sBjuAGL18-1 transgenic lines were screened and identified via PCR with primer pairs of pBI121-F and AGL18-1-R (Supplementary Table S1), while the 35S::aBjuAGL18-1 lines were detected using primer pairs of pBI121-F and AGL18-1-F (Supplementary Table S1). Finally, 15 35S::sAGL18-1 plants and 12 35S::aAGL18-1 transgenic lines in B. juncea, 22 35S::sAGL18-1 plants and 19 35S::aAGL18-1 transgenic lines in Nicotiana tabacum were obtained.
Flowering time analysis
The number of leaves before bolting (LBB), the number of cauline leaves on lateral branches (CLLB) and the total leaf number (TL) of transgenic lines were regarded as a measure of flowering time during the bolting stage. The number of LBB was counted at the moment of the first plant bolting. The number of CLLB and TL were recorded at the tenth days after the first plant bolting. At least random 12 plants were chosen for each genotype and treatment.
gDNA and RNA extraction, cDNA Synthesis
Genomic DNA was extracted from the third and fourth attached rosette leaves of B. juncea using DNAsecure Plant Kit (TIANGEN, DP320-03). Total RNA was extracted from roots, stems, cauline leaves, flowering buds, sepals and petals of B. juncea by using of RNAprep Pure Plant Kit (TIANGEN, DP432). According to the manufacturer, s recommendations, cDNA was synthesized via PrimeScript™ RT reagent kit RR047A with gDNA Eraser (TaKaRa).
Quantitative Real-Time PCR
The ACTIN2 gene was used as an internal reference and gene expression of BjuAGL18-1, BjuSOC1, BjuAGL24, BjuAGL15, BjuSVP and BjuHDA9 were detected in various tissues and developmental stages under inductive conditions. The qRT-PCR was performed in 96-well blocks using Bio-Rad CFX96 Real-Time PCR system with specific primers listed in Supplementary Table S1. The reaction system was as follows: 0.5 µl reverse primers (10 µmol/µl), 0.5 µl forward primers (10 µmol/µl), 2 µl cDNA template, 5 µl SsoFast™ EvaGreen® Supermix (Bio-Rad) and add ddH2O to 10 µl. The reactions procedure was carried out using conditions as follows: 95°C for 3 min followed by 40 cycles of 95°C for 10 s, 55–65°C for 30 s. All the qRT-PCR results were presented as means±SE of three biological replicates and each sample was quantified in triplicate. The relative expression level of genes were analyzed by the 2−ΔΔCt equation.
Results
Expression pattern of BjuAGL18-1 in B. juncea
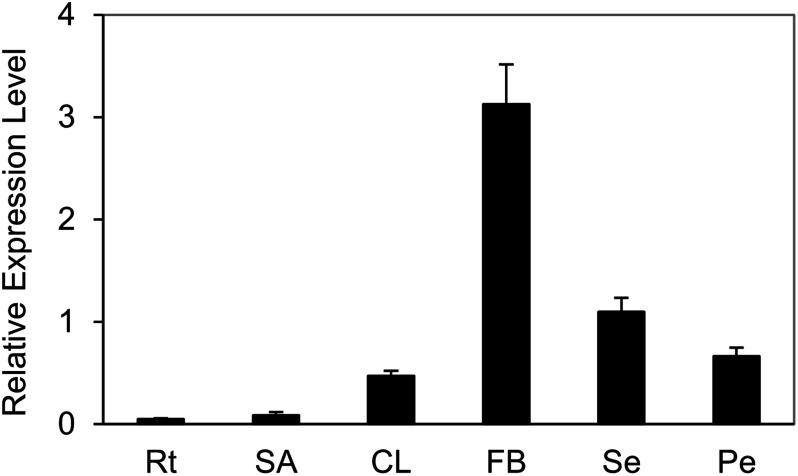
The qRT-PCR analysis indicated that transcript accumulation of BjuAGL18-1 were mainly found in flowering buds, sepals and petals, but barely in roots, shoot apex and cauline leaves of B. juncea. The highest expression of BjuAGL18-1 was detected in flowering buds (Figure 1). These results supported that a high transcript abundance of BjuAGL18-1 in flower organ, which might contribute to maintaining reproduction growth.
Figure 1. Expression of BjuAGL18-1 in main tissues under LD conditions in B. juncea. The expression patterns of BjuAGL18-1 in roots (Rt), shoot apex (SA), cauline leaves (CL), flowering buds (FB), sepals (Se) and petals (Pe). Total RNA was isolated from different tissues of two-month-old plants. Three technical and biological replicates were assessed. Bars represent means±SE.
Phenotype of 35S::sBjuAGL18-1 and 35S::aBjuAGL18-1 lines
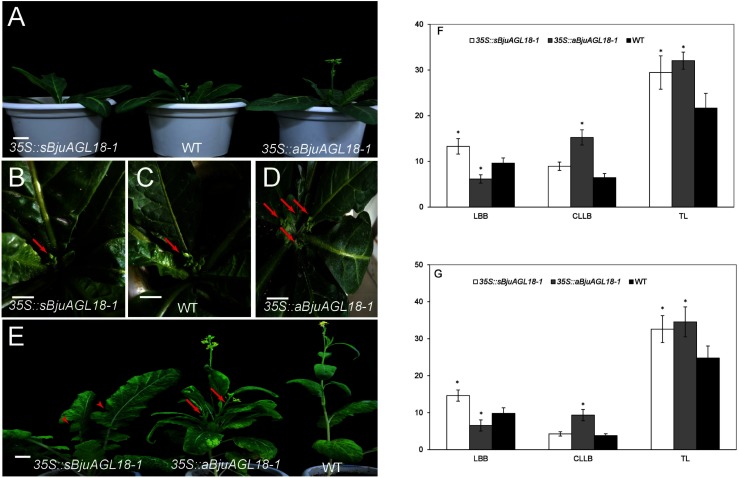
In this study, BjuAGL18-1 was transformed into Nicotiana tabacum by Agrobacterium-mediated method for identifying its biological role in the control flowering time. The results showed that two-month-old 35S::sBjuAGL18-1 lines exhibited a phenotype of delayed flowering time under inductive LD conditions. Conversely, 35S::aBjuAGL18-1 lines accelerated flowering compared with WT plants (Figure 2A). Except for flowering time, significant increase was found in the branch number between the 35S::aBjuAGL18-1 lines and the wild type or 35S::sBjuAGL18-1 lines (Figure 2B–D). Similar results also were observed in the transgenic B. juncea plants (Figure 2E). Furthermore, we investigated the number of leaves before bolting (LBB) which reflected the time of flowering, the number of cauline leaves on lateral branches (CLLB) which reflected the number of lateral branches and the total leaf number (TL) in transgenic B. juncea and Nicotiana tabacum. The LBB was the most in 35S::sBjuAGL18-1 lines and had significant difference with the wild type and 35S::aBjuAGL18-1. The 35S::aBjuAGL18-1 lines had significantly more CLLB than the wild-type plants and 35S::sBjuAGL18-1 lines. The TL in 35S::sBjuAGL18-1 and 35S::aBjuAGL18-1 lines was dramatically higher than of wild type (Figure 2F–G). It preliminarily suggested that BjuAGL18-1 probably served as a floral repressor in B. juncea and Nicotiana tabacum.
Figure 2. Phenotype of 35S::sBjuAGL18-1 and 35S::aBjuAGL18-1 in transgenic Nicotiana tabacum and B. juncea. The flowering of 35S::sBjuAGL18-1, WT and 35S::aBjuAGL18-1 lines under LD conditions in Nicotiana tabacum (A). The phenotype of bolting appearance in Nicotiana tabacum (B–D), the arrows referred to the number of lateral branches produced when appearance of bolting. The flowering of 35S::sBjuAGL18-1, 35S::aBjuAGL18-1 and WT lines under LD conditions in B. juncea (E). The arrowheads in 35S::sBjuAGL18-1 lines indicated the serrated leaves, while the arrows in 35S::aBjuAGL18-1 lines referred the lateral branches. Bars=1 cm. Effect of transgenic lines on flowering time in B. juncea (F) and Nicotiana tabacum (G). The number of leaves before bolting (LBB), the number of cauline leaves on lateral branches (CLLB) and the total leaf number (TL) were used to investigate flowering time. Data were indicated with the means±SD (n≥12 plants). Asterisks indicated statistically significant differences in means between transgenic plants and wild-type. Three technical and biological replicates were assessed. Paired t-tests, * p<0.05, ** p<0.01.
Expression of BjuAGL18-1 and flowering-related genes in transgenic seedlings
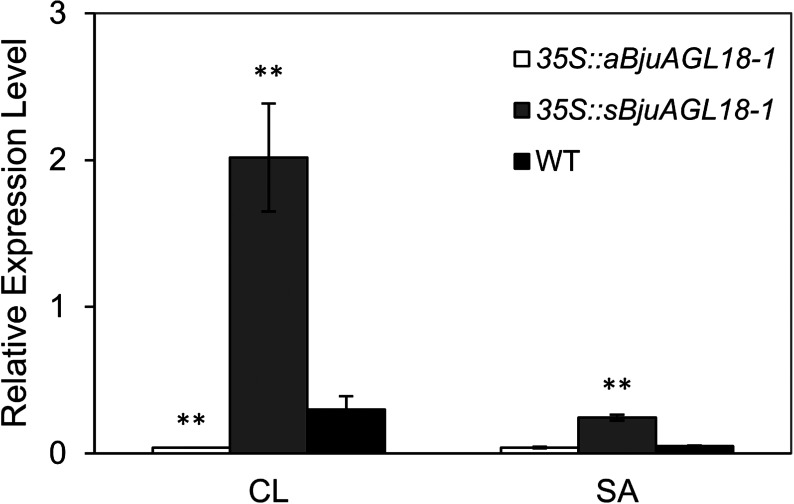
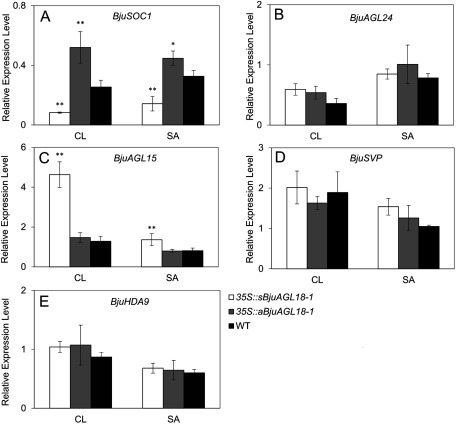
To further elucidate BjuAGL18-1 function of the regulation of flowering time, flowering signal perceptive tissues of cauline leaves and shoot apex were taken to measure transcript accumulation by qRT-PCR during the vegetative stage. The relative BjuAGL18-1 expression level in 35S::sBjuAGL18-1 lines was much higher than WT in cauline leaves and shoot apex of B. juncea seedlings. Conversely, we observed that 35S::aBjuAGL18-1 lines accumulated low levels of BjuAGL18-1 transcripts (Figure 3). Compared with WT, the 35S::sBjuAGL18-1 lines had a lower abundance of BjuSOC1 transcript in cauline leaves and shoot apex. Nevertheless, the transcript levels of BjuSOC1 was remarkable higher in 35S::aBjuAGL18-1 lines relative to WT (Figure 4A). The transcript accumulation of BjuAGL15 was substantially higher in 35S::sBjuAGL18-1 lines than in the WT. There was no major difference in BjuAGL15 expression levels detected between 35S::aBjuAGL18-1 lines and WT(Figure 4C). However, no significant expression changes were detected in either 35S::sBjuAGL18-1 lines or 35S::aBjuAGL18-1 lines compared to the WT plants in BjuAGL24 (Figure 4B), BjuSVP (Figure 4D), BjuHDA9 (Figure 4E).
Figure 3. BjuAGL18-1 relative expression in cauline leaves (CL) and shoot apex (SA) of transcript B. juncea lines and WT. Total RNA was isolated from different tissues of 40-day-old plants. Asterisks indicated statistically significant differences in means between transgenic plants and wild-type. Three technical and biological replicates were assessed. Paired t-tests, * p<0.05, ** p<0.01.
Figure 4. The transcript abundance of BjuSOC1 (A), BjuAGL24 (B), BjuAGL15 (C), BjuSVP (D) and BjuHDA9 (E) in different tissues of B. juncea seedlings grown under LD conditions. The expression was measured in cauline leaves (CL), shoot apex (SA) via qRT-PCR. The means±SE were shown. Total RNA was isolated from different tissues of 40-day-old plants. Asterisks indicated statistically significant differences in means between transgenic plants and wild-type. Three technical and biological replicates were assessed. Paired t-tests, * p<0.05, ** p<0.01.
BjuSOC1 and BjuAGL24 were repressed by AGL18-1 during flower developmental stage
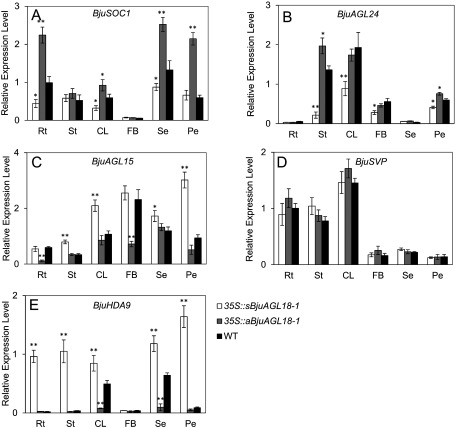
To elucidate the function of AGL18-1 of B. juncea during flower developmental phase, the transcript accumulation of BjuSOC1 and BjuAGL24 were assessed in different tissues of transgenic AGL18-1 lines via qRT-PCR. BjuSOC1 transcript abundance increased in roots, cauline leaves, sepals and petals of 35S::aBjuAGL18-1 lines. On the contrary, the expression of these tissues in 35S::sBjuAGL18-1 lines decreased except in the petals. Nevertheless, no significant changes were detected in either 35S::sBjuAGL18-1 lines or 35S::aBjuAGL18-1 lines compared to the stems and flowering buds of WT plants (Figure 5A).The abundance of the BjuAGL24 transcript were principally detected in stems, cauline leaves, flowering buds and petals, but rarely expressed in roots and sepals regardless of transgenic lines or WT plants. BjuAGL24 transcript accumulation at elevated levels relative to the WT in stems, cauline leaves, flowering buds and petals of 35S::sBjuAGL18-1 lines (Figure 4B). These results revealed that BjuAGL18-1 repressed the expression of flowering-activator BjuSOC1 and BjuAGL24 during flower developmental phase.
Figure 5. The transcript accumulation of BjuSOC1 (A), BjuAGL24 (B), BjuAGL15 (C), BjuSVP (D) and BjuHDA9 (E) in different tissues of B. juncea during flowering phase under LD conditions. The expression were measured in roots (Rt), stems (St), cauline leaves (CL), flowering buds (FB), sepals (Se) and petal (Pe) via qRT-PCR. The means±SE were shown. Total RNA was isolated from different tissues of two-month-old plants. Asterisks indicated statistically significant differences in means between transgenic plants and wild-type. Three technical and biological replicates were assessed. Paired t-tests, * p<0.05, ** p<0.01.
BjuAGL15 was regulated by BjuAGL18-1 during flowering phase
Expression of BjuAGL15 increased everywhere except in a portion of the roots and flowering buds in 35S::sBjuAGL18-1 lines, compared to the wild-type plants. In addition, BjuAGL15 transcript level considerably declined in roots, and flowering buds of 35S::aBjuAGL18-1 lines. No obvious expression differences of BjuAGL15 were detected in cauline leaves, sepals and petals relative to WT (Figure 5C). It suggested that BjuAGL18-1 regulated the expression of inhibitor BjuAGL15during flower developmental stage.
BjuSVP was independent of BjuAGL18-1 during flowering phase
BjuSVP ubiquitously expressed in all organs. Higher BjuSVP accumulation was observed in vegetative organs, compared with floral tissues. However, there was no significant expression difference of BjuSVP among 35S::sBjuAGL18-1 lines, 35S::sBjuAGL18-1 lines and wild-type (Figure 5D), implying that BjuAGL18-1 could not probably affect the expression of BjuSVP gene during flowering phase.
BjuHDA9 was highly induced by BjuAGL18-1 during flowering phase
The transcript level of BjuHDA9, a histone deacetylase, was evaluated via qRT-PCR in B. juncea transgenic lines. The results showed that the transcript of BjuHDA9 was highly induced in 35S::sBjuAGL18-1 lines except in a portion of the flowering buds, There was no detectable significant expression difference of BjuHDA9 in roots, stems, flowering buds and petal between 35S::aBjuAGL18-1 lines and wild type, except in cauline leaves and sepals (Figure 5E). It inferred that the high transcript abundance of BjuHDA9 was probably responsible for the flower development.
Discussion
Biology role of AGL18-1 in flowering time regulation
In this study, the phenotypes of sense and anti-sense BjuAGL18-1 transgenic lines were investigated and indicated that BjuAGL18-1 not only delayed flowering time but also inhibited growth of lateral branching in B. juncea and Nicotiana tabacum. It suggested that BjuAGL18-1 was a flowering time repressor in B. juncea, consistent with the previous reports in Arabidopsis (Adamczyk et al. 2007; Fernandez et al. 2014; Serivichyaswat et al. 2015). Whereas, BjuAGL18-1 also acted as a branching regulator in B. juncea, which was quite different from Arabidopsis. Furthermore, previous studies showed that AGL18 expressed highly in roots, inflorescences and mature flowers, but slightly in stems and leaves in Arabidopsis (Adamczyk et al. 2007). In contrast, the expression of BjuAGL18-1 was barely detected in roots of B. juncea in this study.
Flowering activators BjuSOC1 and BjuAGL24 were down-regulated by BjuAGL18-1
Indeed, one finding from our previous studies indicated that BjuAGL18-1 protein could bind to promoters of BjuSOC1 and BjuAGL24 (Li et al. 2018). Here, during the vegetative phase, BjuAGL18-1 significantly repressed the expression of BjuSOC1, but not BjuAGL24. In addition, during the flowering phase, BjuAGL18-1 could negatively regulate the expression of flowering signal integrators of BjuSOC1 and BjuAGL24 in some organs, while the mRNA levels and expression patterns were different between BjuSOC1 and BjuAGL24 in some tissues of transgenic lines. It inferred that there might be different regulation mechanisms or redundant functions between BjuSOC1 and BjuAGL24 in floral transition of B. juncea. In Arabidopsis, the AGL18 was related to flowering time via regulation of SOC1, and it could inhibit the expression of SOC1. In addition, the SOC1, AGL24, AGL15 and AGL18 acted partially redundantly (Fernandez et al. 2014).
Flowering repressor AGL15 was promoted by BjuAGL18-1, and another flowering repressor SVP was independent of BjuAGL18-1
In this study, the expression level of BjuAGL15 was dramatically higher in stems, leaves, petals, and sepals of the 35S::sBjuAGL18-1 lines than in the wild type, while there was no significant expression change in some organs of 35S::aBjuAGL18-1 lines. In previous studies, AGL15 and AGL18 were regarded as flowering inhibitors of the upstream of FLOWERING LOCUS T (FT) (Becker and Theissen 2003), and acted in a functional redundancy in regulating floral transition of Arabidopsis (Adamczyk et al. 2007). Therefore, we speculated that BjuAGL15 and BjuAGL18-1 probably acted redundantly as flowering inhibitors in B. juncea. However, as reported previously, AGL18-containing complexes could work independently with the SVP-containing complexes (Adamczyk et al. 2007). In this study, the expression pattern of BjuSVP in transgenic lines also demonstrated that BjuSVP was probably independent of BjuAGL18-1 to affect flowering in B. juncea.
HDA9 was highly induced by BjuAGL18-1
In Arabidopsis, the expressions of SOC1 and AGL24 were substantially enhanced in hda9 mutants relative to the wild-type (Kang et al. 2015). Our previous studies showed that BjuHDA9 protein interacted with promoters of BjuSOC1 and BjuAGL24 in B. juncea (Jiang et al. 2018). Here, the expression of BjuHDA9 was significantly affected by BjuAGL18-1 in transgenic lines during flowering phase, suggesting that histone deacetylase BjuHDA9 was probably involved in flowering control through interacting with BjuAGL18-1 and other flowering factors in B. juncea. Similarly, ternary protein complexes AGL18/AFR/HDAC were capable of inhibiting expression of FT and then regulated the flowering (Gu et al. 2013). Hence, protein complexes AGL18-1/AFR/HDA9 or more higher-order complexes were proposed to regulate SOC1 and AGL24 in the regulation of flowering of B. juncea. Based on the above results, we proposed a regulatory network of AGL18-1 involved in flowering control of B. juncea (Supplementary Figure S1).
Acknowledgments
This work was supported by grants from the Fundamental Research Funds for the Central Universities (Grant number XDJK2017B036; XDJK2017D089).
Supplementary Data
References
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B (2012) The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhu X, Ren J, Qiu K, Li Z, Xie Z, Gao J, Zhou X, Kuai B (2017) Suppressor of overexpression of CO 1 negatively regulates dark-induced leaf degreening and senescence by directly repressing pheophytinase and other senescence-associated genes in Arabidopsis. Plant Physiol 173: 1881–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC (2000) The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12: 183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Wang CT, Zheng YM, Adamczyk BJ, Singhal R, Hall PK, Perry SE (2014) The MADS-domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol 165: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Wang Y, He Y (2013) Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol 11: e1001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Pose D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk AD, Schmid M, et al. (2012) Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wei D, Zhou W, Wang Z, Tang Q (2018) HDA9 interacts with the promoters of SOC1 and AGL24 involved in flowering time control in Brassica juncea. Biochem Biophys Res Commun 499: 519–523 [DOI] [PubMed] [Google Scholar]
- Kang MJ, Jin HS, Noh YS, Noh B (2015) Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol 206: 281–294 [DOI] [PubMed] [Google Scholar]
- Kim W, Latrasse D, Servet C, Zhou DX (2013) Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem Biophys Res Commun 432: 394–398 [DOI] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA (2012) APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC, Ma GP, Xie T, Chen J, Wang ZM, Song M, Tang QL (2018) SOC1 and AGL24 interact with AGL18-1, not the other family members AGL18-2 and AGL18-3 in Brassica juncea. Acta Physiol Plant 40: 3 [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet 40: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33: 867–874 [DOI] [PubMed] [Google Scholar]
- Papaefthimiou D, Kapazoglou A, Tsaftaris AS (2012) Cloning and characterization of SOC1 homologs in barley (Hordeum vulgare) and their expression during seed development and in response to vernalization. Physiol Plant 146: 71–85 [DOI] [PubMed] [Google Scholar]
- Serivichyaswat P, Ryu HS, Kim W, Kim S, Chung KS, Kim JJ, Ahn JH (2015) Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol Cells 38: 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FC, Swain SM (2007) Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol Plant 131: 481–495 [DOI] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP (2002) AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci U S A 99: 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.