Ferrer-Picón et al. show that in active IBD patients, tumor necrosis factor alpha affects the response of the intestinal epithelium to bacteria-derived butyrate. This observation raises questions about the beneficial effects of butyrate supplementation during active inflammation.
Keywords: ulcerative colitis, Crohn’s disease, bacterial metabolites, TNFα, human organoids
Abstract
Background
Butyrate-producing gut bacteria are reduced in patients with active inflammatory bowel disease (IBD), supporting the hypothesis that butyrate supplementation may be beneficial in this setting. Nonetheless, earlier studies suggest that the oxidation of butyrate in IBD patients is altered. We propose that inflammation may decrease epithelial butyrate consumption.
Methods
Non-IBD controls and IBD patients were recruited for the study. Stool samples were used for short-chain fatty acid and bacterial butyryl CoA:acetate CoA-transferase quantification. Colonic biopsies and ex vivo differentiated epithelial organoids (d-EpOCs) treated with butyrate and/or tumor necrosis factor alpha (TNFα) were used for analyzing the expression of transporters MCT1 and ABCG2, metabolic enzyme ACADS, and butyrate receptor GPR43, and for butyrate metabolism and consumption assays.
Results
We observed that lower stool content of butyrate-producing bacteria in active IBD patients did not correlate with decreased butyrate concentrations. Indeed, the intestinal epithelial expression of MCT1, ABCG2, ACADS, and GPR43 was altered in active IBD patients. Nonetheless, d-EpOCs derived from IBD patients showed SLC16A1 (gene encoding for MCT1 protein), ABCG2, ACADS, and GPR43 expression levels comparable to controls. Moreover, IBD- and non-IBD-derived d-EpOCs responded similarly to butyrate, as assessed by transcriptional regulation. TNFα significantly altered SLC16A1, ABCG2, and GPR43 transcription in d-EpOCs, mimicking the expression profile observed in biopsies from active IBD patients and resulting in reduced butyrate consumption.
Conclusions
We provide evidence that the response to butyrate is not intrinsically altered in IBD patients. However, TNFα renders the epithelium less responsive to this metabolite, defeating the purpose of butyrate supplementation during active inflammation.
INTRODUCTION
Crohn’s disease (CD) and ulcerative colitis (UC) are considered the 2 major forms of chronic inflammatory bowel diseases (IBDs). Although their etiology is still poorly understood, growing evidence supports a role for defects within the epithelial cell compartment in driving disease.1–3 The intestinal epithelium is a single layer of specialized cells covering the mucosal surface that are organized into thousands of invaginations called crypts. At the base of each crypt, a small number of multipotent adult stem cells differentiate into the distinct epithelial lineages.4 The epithelial layer is crucial in maintaining intestinal homeostasis by interacting with both commensal and pathogenic bacteria, and with the underlying immune cells.
Gut microbial dysbiosis in IBD is a well-established observation.5 Indeed, several groups have reported important changes in the fecal and mucosa-associated microbial communities in different patient cohorts.6, 7 One commonly reported alteration associated with IBD is a decrease in short-chain fatty acid (SCFA)–producing bacteria, in particular butyrate-synthesizing species such as Roseburia and Faecalibacterium prausnitzii.8, 9 In agreement with this, other studies have noted a reduced capacity for butyrate synthesis based on stool samples taken from IBD patients. This was established by measuring the expression of Butyril-CoA:acetate CoA-transferase (BCoAT), 1 of the bacterial enzymes responsible for butyrate synthesis.8, 9 Alterations in stool SCFA concentrations have also been observed.10–12 In addition to changes in butyrate production, previous studies have also suggested potential alterations in butyrate oxidation in IBD patients,13 which could impair the ability of butyrate to exert its potentially beneficial effects14 beyond its contribution as an energy source for colonocytes.15 Thus, although butyrate supplementation has been proposed as a therapeutic approach in IBD,16–19 whether it could have any beneficial effects remains an open question.
Here, we take advantage of the access to human primary epithelial organoid cultures to test the effects of butyrate on healthy and IBD-affected intestinal epithelium. Using this ex vivo model, we also assess the impact of inflammation on the epithelial response to this bacterial metabolite.
METHODS
More detailed information is provided in Supplementary Methods, available online.
Patient Population
Patients with an established diagnosis of UC or CD of at least 3 months’ duration and non-IBD controls (Ctrl) were included in the study after obtaining written informed consent. Non-IBD controls were those subjects undergoing surgery for colorectal cancer (CRC), colonoscopy for mild gastrointestinal symptoms, or screening for CRC (none of these controls presented colonic lesions). Colonic biopsy samples were collected during routine colonoscopies from non-IBD controls and patients with IBD with no evidence of colitis-associated dysplasia or neoplasia, as assessed by chromoendoscopy. Biopsy samples from patients with IBD were collected from previously involved or mildly inflamed sigmoid mucosa. Surgical samples from patients with IBD were obtained after colectomies were performed for refractoriness to medical treatment. For samples from non-IBD controls who were undergoing surgery, a segment of healthy mucosa was collected at least 10 cm from the margin of the affected area. Supplementary Table 1 shows the clinical and demographic characteristics of the subjects included in the study. Samples were distributed among different subgroups on the basis of the experimental approaches used. Patients were recruited at the Department of Gastroenterology, Hospital Clinic Barcelona, and Hospital Mútua de Terrassa. The study protocol was approved by the ethics committees of both centers.
Assessment of Disease Activity
Endoscopic UC activity at the time of the colonoscopy was categorized according to the Mayo endoscopic subscore.20 Active UC was defined as a Mayo endoscopic subscore of ≥1, quiescent UC as a Mayo score of 0 in a previously involved segment. In CD patients, the Simple Endoscopic Score for Crohn’s Disease (SES-CD) was used21 for each segment. Active CD was defined as an SES-CD of ≥3, quiescent CD as an SES-CD score of ≤2 in a previously involved segment. For fecal samples (Group 5) (Supplementary Table 1), disease activity was defined using the Crohn’s Disease Activity Index (CDAI) as there was no endoscopic information available.
Fecal Butyryl CoA:Acetate CoA-Transferase Quantification
Feces from Ctrl and CD patients (Group 5) (Supplementary Table 1) were collected in Stool DNA Stabilizer (Stratec, Germany) and processed with the PSP Spin Stool DNA Kit (Stratec) following the manufacturer’s instructions. To amplify the bacterial butyryl CoA:acetate CoA-transferase (BCoAT) gene, previously designed primers were used (Sigma, Saint Louis, MO, USA).22 The ribosomal 16S gene was used as an internal control. Syber Green–based chemistry (Applied Biosystems) was used following the manufacturer’s instructions. Fluorescence was detected by the ABI PRISM 7500 Fast RT-PCR System (Applied Biosystems).
Epithelial Organoid Culture
Biopsy or surgical specimens from non-IBD controls and patients with IBD were used for generating epithelial organoid cultures (EpOCs). Crypts embedded in Matrigel (BD Biosciences, San Jose, CA, USA) were cultured and passaged as previously described.23 Medium used for EpOC expansion contained Wnt3a-conditioned medium (produced using an L-Wnt3a cell line, ATCC CRL-2647) and Advanced DMEM/F12 medium (Invitrogen, Carlsbad, CA, USA) 50:50, 2 mM of Glutamax, 10 mM of HEPES, N-2 (1×; Gibco, Grand Island, NY, USA), B-27 without retinoic acid (1×; Gibco), 10 mM of nicotinamide (Sigma), 1 mM of N-Acetyl-L cysteine (Sigma), 500 ng/mL of R-spondin-1 (RSPO1, Sino Biologicals, Beijing, China), 50 ng/mL of human epidermal growth factor (EGF; Invitrogen), 100 ng/mL of human Noggin (Peprotech, Rocky Hill, NJ, USA), 1 μg/mL of Gastrin (Tocris Bioscience, Bristol, UK), 500 nM of LY2157299 (Axon MedChem, Groningen, the Netherlands), 10 μM of SB202190 (Sigma), and 0.01 μM of prostaglandin E2 (PGE2; Sigma). To induce EpOC differentiation into differentiated EpOCs (d-EpOCs), nicotinamide, SB202190, and Wnt3a-conditioned medium were removed from the culture medium. RSPO1 was reduced to 250 ng/mL during the first 4 days and then completely removed. To test the effect of butyrate on organoids, EpOCs and d-EpOCs were stimulated with 0.5 or 5 mM of sodium butyrate (Sigma). Incubation of d-EpOCs with 0–50 ng/mL of tumor necrosis factor alpha (TNFα; Gibco) was performed for 20 hours when indicated.
RNA Isolation
Total RNA from first-passage EpOCs and from biopsy samples (Group 1–3) (Supplementary Table 1) previously placed in RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany) was isolated using the RNeasy Kit (Qiagen).
Microarray Analysis
For microarray analysis, paired first-passage EpOCs and d-EpOCs generated from colonic samples of non-IBD controls, incubated with butyrate as indicated, were used. The derived cRNA was hybridized to high-density oligonucleotide Affymetrix GeneChip Human Genome U219 Array (Affymetrix, Santa Clara, CA, USA). Raw data were analyzed using Bioconductor tools in R (V.3.1.0). Microarray raw data (CEL files) and processed data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession No. GSE123553 ((https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123553; last accessed 12 June 2019).
Quantitative Real-time Polymerase Chain Reaction
Total RNA isolated from samples (Groups 1–3) (Supplementary Table 1) was transcribed to cDNA using the reverse transcriptase High Capacity cDNA Archive RT kit (Applied Biosystems, Carlsbad, CA, USA). Quantitative polymerase chain reaction (qPCR) was performed in the ABI PRISM 7500 Fast RT-PCR System (Applied Biosystems) using predesigned TaqMan Assays (Applied Biosystems).
Immunostaining
Immunohistochemical and immunofluorescent staining of tissue sections (Group 4) (Supplementary Table 1) was performed using commercially available antibodies: mouse anti-EpCAM (1:100; Dako), rabbit anti-MCT1 (1:200, NovusBio), mouse anti-ABCG2 (1:50, Abcam), rabbit anti-ACADS (1:50, Abcam), and rabbit anti-GPR43 (1:50, LSBio). Image acquisition and overlay were performed on an Olympus BX51 microscope (Tokyo, Japan) using CellF Software.
3H-Butyrate Beta-Oxidation Assay
To study the capacity of d-EpOCs to beta-oxidate butyrate, d-EpOCs were stimulated with 50 ng/mL of TNFα for 20 hours, and then with 5 mM of 3H-Butyrate (American Radiolabeled Chemicals, St. Louis, MO, USA) for an additional 24 hours. After incubation, 100 μL of supernatant was collected, and 900 μL of 0.6% phosphoric acid (Sigma) was added. The mixture was passed through a Bond Elut C18 column (Agilent), previously conditioned with 1 mL of 100% methanol (VWR Chemical) and 1 mL of 0.6% phosphoric acid. The resultant elute with 3H-H2O from 3H-Butyrate oxidation was measured with HIDEX 300SL (Scintillation counter).
Statistical Analysis
Quantitative data are expressed as the mean and range, and categorical variables as absolute frequencies. A Wilcoxon test was performed to examine statistically different expression patterns between the 2 groups, and a Kruskal-Wallis test was performed to examine statistical significance in multiple group data sets, followed by a pairwise Wilcoxon test with Bonferroni-Holm correction for multiple testing. A P value of <0.05 was considered statistically significant. Data were analyzed using R (version 3.1.0).
RESULTS
Low Content of Bacterial Butyryl CoA:Acetate CoA-Transferase in Crohn’s Disease Fecal Samples Does Not Result in Changes in Butyrate Concentration
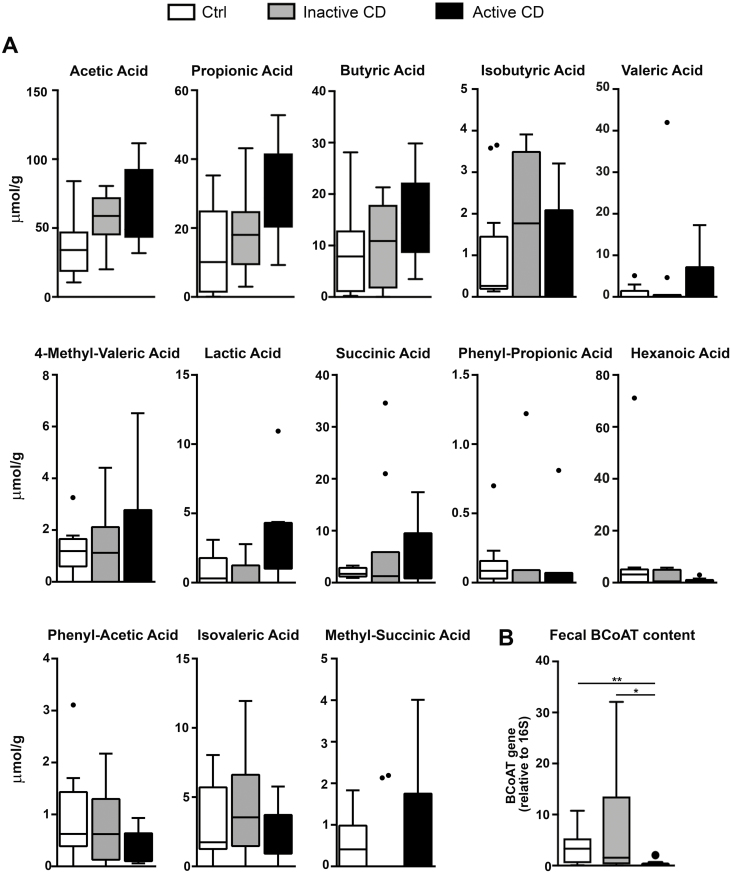
To study the possible differences in butyrate production by luminal bacteria in patients with CD, total SCFA concentrations were measured by HPLC in feces of active and inactive CD patients, as well as in non-IBD controls (Ctrl). In contrast to earlier studies,10–12 we observed no significant differences in fecal concentrations of SCFAs in CD patients, regardless of disease activity, compared with Ctrl (Fig. 1A).
FIGURE 1.
Feces from active CD patients present similar levels of fecal SCFAs and a decrease in fecal BCoAT content. A, Fecal SCFAs were measured by HPLC in Ctrl feces (n = 12) and feces from inactive (n = 11) and active (n = 10) CD patients. Values are expressed in µmol/g. B, The content of the fecal BCoAT gene was measured by qPCR in Ctrl feces (n = 12) and feces from inactive (n = 11) and active (n = 10) CD patients. BCoAT quantification was relative to the bacterial 16S ribosomal gene. Mean±SEMs. *P < 0.05; **P < 0.01 by 1-tailed Wilcoxon test.
BCoAT DNA expression in the same fecal samples was then measured as an indicator of the microbiota’s capacity to produce butyrate.8, 9, 24 We observed a significantly lower content of the BCoAT gene in the feces from active CD patients compared with Ctrl and CD patients in remission (Fig. 1B). Thus, we show a potentially reduced bacterial capacity to synthesize butyrate in active CD patients that does not result in reduced fecal concentrations of this metabolite.
Butyrate Regulates the Transcriptional Profile of Healthy Human Intestinal Epithelial Organoids
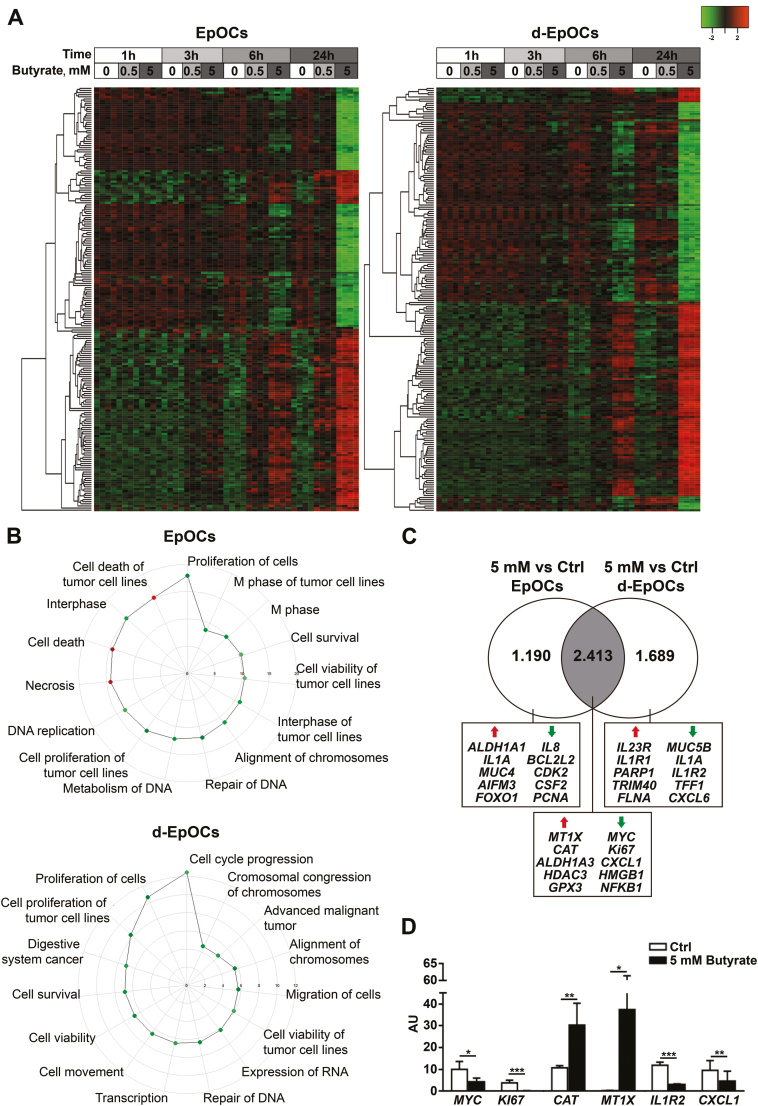
We hypothesize that the discrepancy between the enzymatic capacity to produce butyrate and its concentration in feces could be explained by defects in the consumption of this metabolite in the context of inflammation. To test this hypothesis, first we established the effects of butyrate in the healthy epithelium using ex vivo human primary organoid cultures derived from Ctrl individuals. EpOCs were generated from colonic crypts and differentiated in vitro into d-EpOCs. Analysis of microarray gene expression data identified 2014 genes differentially expressed (P ≤ 0.05; FDR ≤ 0.05) between EpOCs and d-EpOCs (Supplementary Fig. 1), which was in agreement with our previous results2 (GEO Series accession No. GSE75916; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse75916; last accessed 12 June 2019). The transcriptional signatures of EpOCs and d-EpOCs stimulated with 0.5 mM or 5 mM of butyrate for 1, 3, 6, and 24 hours were also analyzed. Principal component analysis (PCA) separated EpOC from d-EpOC populations (Supplementary Fig. 2). Butyrate induced a dose- and time-dependent transcriptional signature in both EpOCs and d-EpOCs, having a more profound effect in the latter (Supplementary Table 2 and Fig. 2A). Indeed, >4000 genes (~50% of them upregulated) were significantly modulated by 5 mM of butyrate at 24 hours on d-EpOCs (FDR ≤ 0.05; |FC| ≥ 1.5).
FIGURE 2.
Butyrate induces significant transcriptional changes in EpOCs and d-EpOCs. A, Heatmap representations of differentially expressed genes (FDR ≤ 0.05; |FC| ≥ 1.5) in EpOCs (n = 4 for each time and dose) and d-EpOCs (n = 4 for each time and dose) upon butyrate stimulation (0.5 mM and 5 mM). Differentially expressed genes in EpOCs and d-EpOCs are shown separately. Each row depicts 1 individual probe and each column an experimental sample. High expression levels are shown in red and low expression levels in green. An unsupervised hierarchical cluster method, using a Euclidean distance and average linkage method, was applied for both gene and sample classification. B, Polar graph showing the top 15 pathways altered in EpOCs and d-EpOCs stimulated with 5 mM of butyrate compared with the control group after 24 hours, using IPA. The downregulated pathways (z-score ≤ 2) are shown in green, the upregulated pathways (z-score ≥ 2) in red. C, Venn diagram showing the number of genes differentially expressed in EpOCs and d-EpOCs stimulated with 5mM of butyrate compared with the control group after 24 hours. The overlap, in gray, shows the differentially expressed genes shared between the 2 data sets. A selection of up- and downregulated genes is shown. D, Technical validation of selected butyrate target genes. Relative mRNA levels by qPCR of selected genes were found to be differentially expressed in the microarray analysis in d-EpOCs stimulated with 5 mM of butyrate after 24 hours (n = 6). Messenger RNA levels are relative to ACTB mRNA expression. Mean±SEMs. *P < 0.05; **P < 0.01; ***P < 0.001 by 1-tailed Wilcoxon test.
Ingenuity pathway analysis (IPA) of genes significantly regulated by 5 mM of butyrate at 24 hours revealed modulation of mechanisms related to cell proliferation and survival, which were downregulated by butyrate in both organoid systems (Fig. 2B). In fact, butyrate significantly decreased the expression of proliferation markers (MYC and KI67) (Fig. 2C). In contrast, the regulation of pathways leading to cell death and necrosis was only confirmed in EpOCs (Fig. 2B), but not in d-EpOCs. More specifically, butyrate increased pro-apoptotic AIFM3 and FOXO1 transcription and decreased the mRNA levels of the anti-apoptotic gene BCL2L2 in EpOCs (Fig. 2C). At 24 hours of butyrate stimulation, both EpOCs and d-EpOCs downregulated the expression of inflammatory markers (CXCL1, HMGB1, and NFKB1), whereas they upregulated genes associated with the defense against oxidative stress (MT1X, CAT, and GPX3). HDAC3 mRNA levels also increased upon butyrate stimulation in both systems, in agreement with histone deacetylase inhibition.25 In addition, butyrate increased ALDH1A3 transcriptional levels, 1 of the enzymes that participates in acid retinoic formation, a factor involved in the differentiation of gut-homing Treg cells26 and IgA-secreting B cells.27 Six of the genes most significantly regulated by 5 mM of butyrate in d-EpOCs at 24 hours and involved in the proliferation and response to oxidative stress and immune functions were validated by real-time qPCR (Fig. 2D).
Given that d-EpOCs mimic the phenotype of differentiated epithelial cells residing on the luminal (upper) side of the crypt, which is directly exposed to bacteria and its metabolites, we decided to limit our study to the effects of butyrate on the d-EpOCs system.
Butyrate Response in IBD-Derived Differentiated Epithelial Organoids Is Comparable to That of Controls
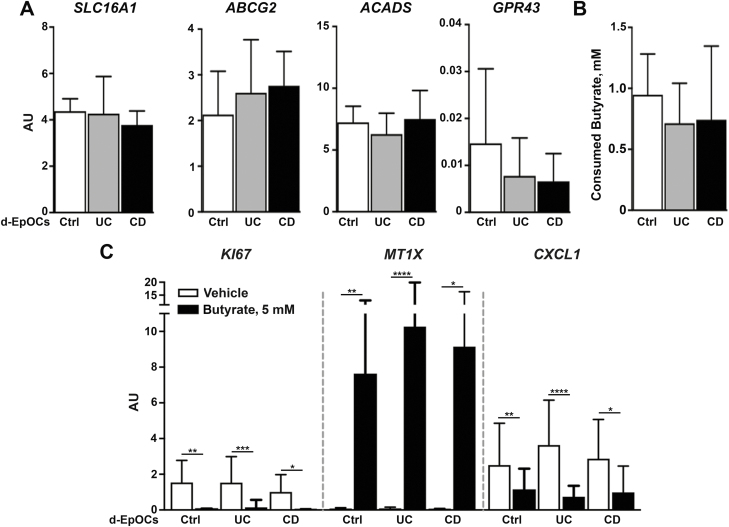
Next, we asked if epithelium derived from IBD patients responded differently to butyrate. We generated d-EpOCs from colonic biopsies of Ctrl and IBD patients with inactive disease. First, we analyzed a panel of genes associated with butyrate uptake and metabolism. We found no significant differences between Ctrl, UC, and CD organoids in the mRNA transcription of SLC16A1 and ABCG2, transporters, ACADS (an enzyme that participates in butyrate intracellular degradation), and the butyrate receptor GPR43 (Fig. 3A). Second, we examined the ability of d-EpOCs from Ctrl and IBD patients to consume butyrate by measuring butyrate concentrations in culture supernatants after 24 hours of 5-mM butyrate supplementation. No differences were found in butyrate consumption, regardless of the origin of the sample (Fig. 3B). In addition, the modulation of butyrate-dependent genes (KI67, MT1X, and CXCL1) proved similar in Ctrl and UC- and CD-derived d-EpOCs (Fig. 3C). Overall, these data suggest that under steady-state conditions and in the absence of a concomitant pro-inflammatory stimulus, the intestinal epithelium from IBD patients presents the same intrinsic capacity to consume and respond to butyrate as that of Ctrl individuals.
FIGURE 3.
Responses to butyrate of d-EpOCs derived from Ctrl and IBD patients are comparable. A, Relative mRNA expression by qPCR of SLC16A1, GPR43, ABCG2, and ACADS of Ctrl-derived (n = 10), UC-derived (n = 13), and CD-derived (n = 7) d-EpOCs. B, Butyrate consumption after 24 hours’ stimulation with 5 mM of butyrate in d-EpOCs derived from Ctrl and IBD patients. C, Relative mRNA expression by qPCR of previously described (Fig. 2) butyrate target genes in Ctrl-derived (n = 10), UC-derived (n = 13), and CD-derived (n = 7) d-EpOCs upon 5-mM butyrate stimulation during 24 hours. Gene expression is relative to ACTB mRNA. Mean±SEMs. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-tailed Wilcoxon test.
The Inflamed Mucosa of IBD Patients Presents Alterations in Epithelial Butyrate Transport and Metabolism
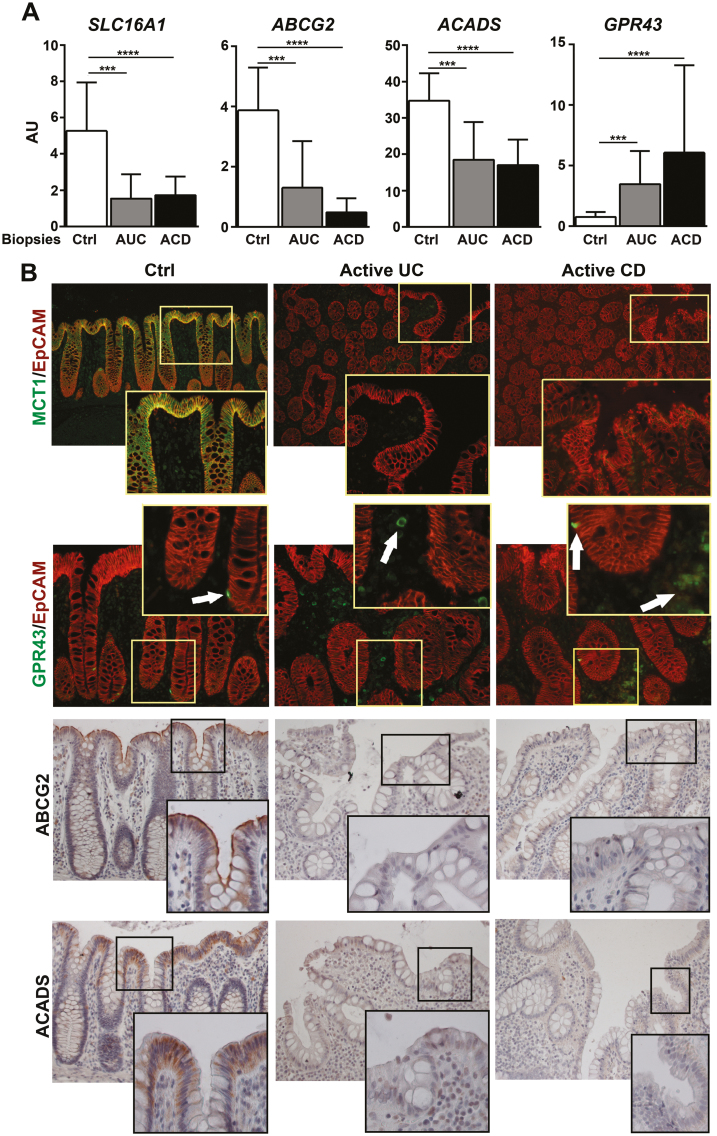
Next, we asked whether active inflammation could affect butyrate consumption. To answer this question, we first measured the expression of the butyrate transporters SLC16A1 and ABCG2, along with the enzyme ACADS. All 3 transcripts were significantly reduced in colonic biopsies from active UC (AUC) and active CD (ACD) patients (Fig. 4A), in agreement with previous reports.28–32 Notably, MCT1 (encoded by SLC16A1), ABCG2, and ACADS proteins primarily localized in the apical epithelium of Ctrl mucosa but remained undetectable in the inflamed mucosa of both UC and CD patients (Fig. 4B). In contrast, expression of GPR43 was limited to a small fraction of cells within the epithelial layer and the lamina propria in healthy mucosa (Fig. 4B). Remarkably, both GPR43 mRNA and protein expression (predominantly within the lamina propria compartment) were increased in the inflamed IBD mucosa. GPR43+ cells may be newly recruited monocytes and neutrophils, which are known to express this butyrate receptor.33 Thus, active inflammation in both UC and CD patients is associated with significant changes in the expression of those proteins involved in butyrate transport and metabolism.
FIGURE 4.
Patients with active IBD present a deregulation in those genes associated with butyrate absorption, metabolism, and response. A, Relative mRNA expression of SLC16A1, GPR43, ABCG2, and ACADS by qPCR in colonic biopsies from non-IBD controls (n = 12), active UC (n = 10), and active CD (n = 10) patients. Messenger RNA levels are relative to ACTB expression. Mean±SEMs. *P < 0.05; **P < 0.01; ***P < 0.001 by pairwise Wilcoxon test with Bonferroni-Holm correction for multiple testing. B, Representative images and immunofluorescent immunohistochemical staining of tissue sections of colonic samples from Ctrl (n = 6) and active UC (n = 6) and CD (n = 7) patients. Protein expression was determined by immunofluorescent staining using a combination of MCT1 and GPR43 proteins (in green) with the epithelial marker EpCAM (in red). White arrows indicate GPR43-positive cells. Expression of ABCG2 and ACADS was detected by immunochemical staining. Images were taken with a ×20 objective lens and magnified with a ×40 objective lens, except images of for MCT1, which were taken with a ×10 objective lens and magnified with a ×20 objective lens.
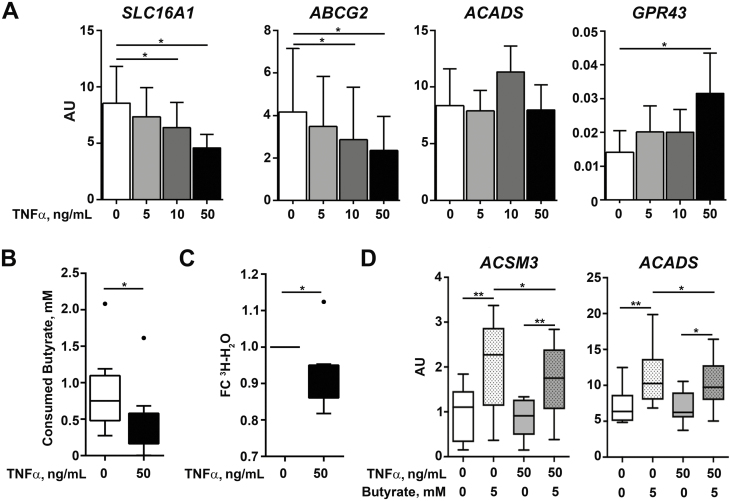
TNFα Affects Butyrate Uptake and Response by the Intestinal Organoids
To mimic the effects of inflammation in the epithelium, d-EpOCs from healthy donors were stimulated with increasing doses of TNFα, a cytokine involved in the physiopathology of IBD. TNFα stimulation induced a significant dose-dependent decrease of SLC16A1 and ABCG2, as well an increase of GPR43 mRNA expression. Remarkably, TNFα had no effect on the transcription of ACADS (Fig. 5A). Next, we asked whether anti-TNF treatment could restore the expression of these genes. To this end, we examined the published findings on IBD patients with whole-transcriptome data available from biopsies before and after 8 weeks of infliximab induction therapy (series accession number GSE16879; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse16879; last accessed 12 June 2019).34 Expression of SLC16A1 and ABCG2 was significantly increased, whereas GPR43 transcripts were significantly decreased after infliximab treatment (Supplementary Fig. 3), thus supporting a role for TNFα in the deregulation of these genes.
FIGURE 5.
TNFα reduces butyrate consumption in d-EpOCs. A, Relative mRNA expression by qPCR of SLC16A1, GPR43, ABCG2, and ACADS in d-EpOCs (n = 8) upon TNFα stimulation at increasing doses for 24 hours. Gene expression is relative to the ACTB gene. B, Butyrate consumption after 24 hours’ stimulation with 5 mM of butyrate in d-EpOCs stimulated with 50 ng/mL of TNFα. C, 3H-H2O production after 24 hours’ stimulation with 5 mM of 3H-Butyrate in the presence of TNFα. Values are represented as the fold-change (FC) with respect to the Ctrl. Mean±SEMs. *P < 0.05 by 1-tailed Wilcoxon test. D, Relative mRNA expression by qPCR of ACSM3 and ACADS in d-EpOCs upon TNFα and butyrate stimulation (n = 8). Gene expression is relative to the ACTB gene. Mean±SEMs. *P < 0.05; **P < 0.01 by 1-tailed Wilcoxon test.
These data suggest that the changes observed in the expression of genes related to butyrate metabolism in inflamed IBD mucosa could be driven by increased levels of TNFα.
We subsequently determined the effects of TNFα on butyrate metabolism. First, levels of beta-hidroxybutyrate and acetoacetate, 2 ketone bodies produced upon butyrate consumption, were measured after sequential TNFα and butyrate stimulation. No differences were found in either metabolic product after TNFα stimulation (Supplementary Fig. 4). However, ketone body production possibly resulted from butyrate-independent cellular metabolism and may be affected by TNFα itself. Therefore, we measured butyrate consumption in the culture supernatants of d-EpOCs and found a significant decrease in butyrate intake in cells previously exposed to TNFα (Fig. 5B). Moreover, direct beta-oxidation of butyrate was measured by the production of 3H-H2O following incubation with 3H-Butyrate. In agreement with the overall decrease in butyrate uptake, beta-oxidation of butyrate was significantly decreased after TNFα stimulation of d-EpOCs (Fig. 5C).
In addition, the expression of 2 enzymes involved in butyrate degradation (ACSM3 and ACADS) was measured after sequential TNFα and butyrate stimulation. TNFα alone did not regulate the transcription of either of the 2 genes (as seen for ACADS in Fig. 5A); however, both transcripts were significantly upregulated by butyrate (Fig. 5D). This upregulation was significantly decreased by the previous exposure of d-EpOCS to TNFα.
Taken together, these results suggest that TNFα decreases butyrate consumption by the epithelium, potentially by deregulating the mechanisms of uptake and degradation.
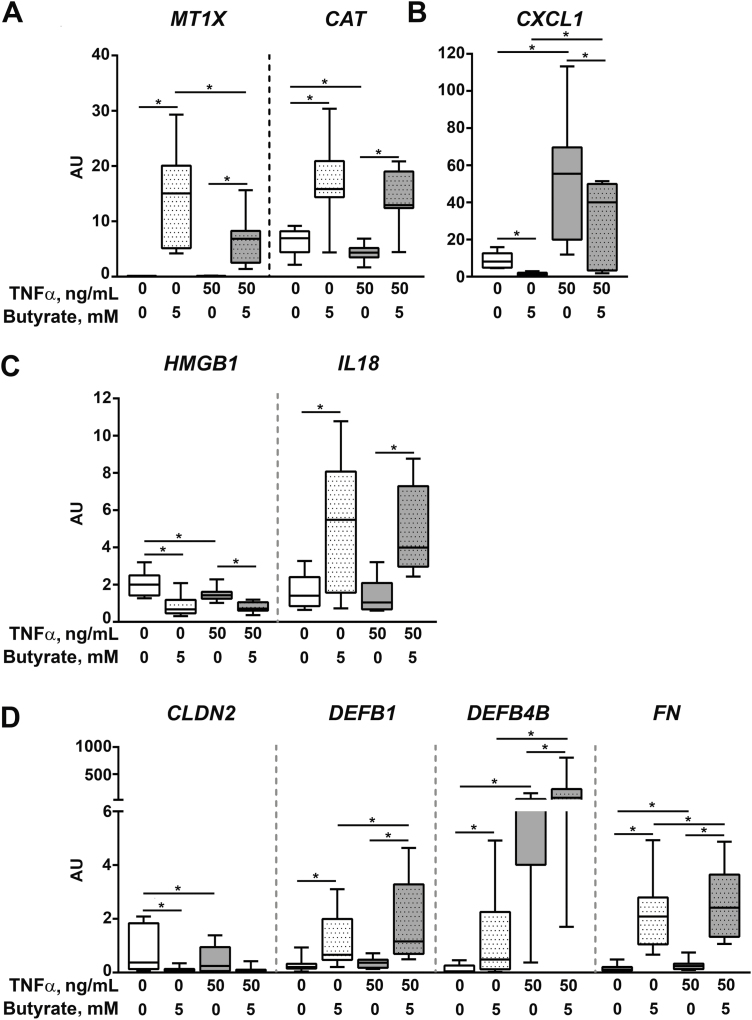
We therefore sought to determine whether this decreased uptake of butyrate in TNFα-stimulated d-EpOCs could affect the transcriptional regulation of some of the main butyrate-dependent genes identified above. In particular, we looked at genes involved in key epithelial pathways such as the response to oxidative stress (Fig. 6A), immune response and inflammasome (Fig. 6B and C, respectively), and structural and antimicrobial responses (Fig. 6D). Overall, TNFα stimulation did not markedly alter the quality of the response of d-EpOCs to butyrate, although for some genes it did modulate the magnitude of the change (ie, it attenuated the butyrate-dependent upregulation of MTX1 (Fig. 6A) while further increasing the transcription of DEFB1, DEFB4B, and FN (Fig. 6D)).
FIGURE 6.
mRNA expression levels of selected butyrate and TNFα response genes. Relative mRNA expression by qPCR of genes associated with oxidative stress (A), inflammation (B), inflammasome (C), and epithelial integrity and damage (D) in d-EpOCs stimulated with TNFα and butyrate (n = 8). Gene expression is relative to the ACTB gene. Mean±SEMs. *P < 0.05 by 1-tailed Wilcoxon test.
Overall, we show that although TNFα does reduce the ability of the intestinal epithelium to uptake and metabolize butyrate, d-EpOCs continue to respond to SCFAs, which promote an antioxidative, antimicrobial, and overall protective epithelial response.
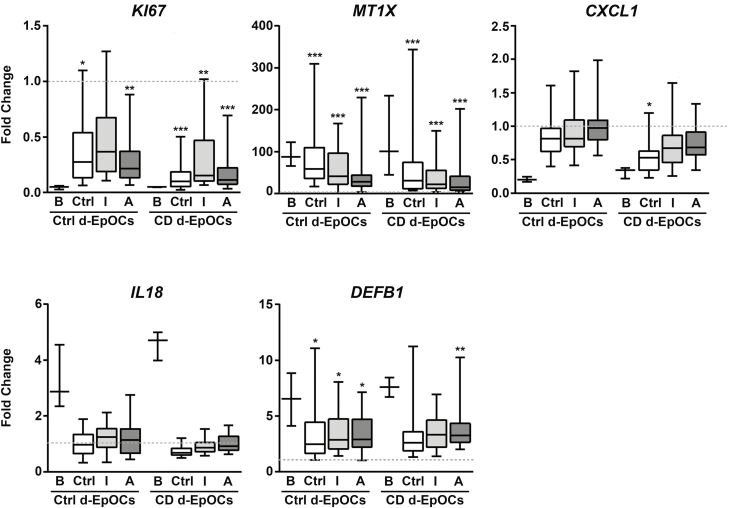
Ctrl- and CD-Derived Epithelium Respond Similarly to Fecal SCFAs Extracts
Although butyrate is 1 of the most abundant SCFAs in the gut, the intestinal lumen contains other abundant SCFAs that may exert some potentially biological impact on IBD. To reproduce the effects of this complex environment on the epithelium, we obtained extracts of SFCAs from the feces of Ctrl and CD patients. SCFA concentrations from fecal extracts are detailed in Supplementary Table 3. SCFA fecal extracts from Ctrl feces (n = 12) and feces from inactive (n = 11) and active (n = 11) CD or vehicle (organic solvent extract) patients were then used at a 1:50 dilution to stimulate Ctrl- (n = 3) and CD-derived (n = 3) d-EpOCs. d-EpOCs stimulated with 5 mM of butyrate were also included for comparison. Changes in expression were calculated for all stimulated conditions compared with vehicle-treated d-EpOCs (Fig. 7). The effects of fecal extracts containing a complex mixture of SCFAs were similar but overall less pronounced, compared with 5 mM of butyrate alone. Indeed, fecal extracts from Ctrl, inactive (I), or active (A) CD patients induced a significant decrease in the proliferation marker KI67 and an increase in MT1X and DEFB1 mRNA expression in both Ctrl and CD d-EpOCs (Fig. 7). Conversely, no or mild effects on IL18 and CXCL1 expression were observed by incubation of both Ctrl and CD d-EpOCs with SCFA extracts.
FIGURE 7.
mRNA expression levels of selected butyrate response genes upon stimulation with SCFA fecal extracts. d-EpOCs (Ctrl, n = 3; CD, n = 3) were stimulated with SCFA fecal extracts from Ctrl feces (n = 12) and feces from inactive (n = 11) and active (n = 11) CD patients at a 1:50 dilution. Results are represented as relative mRNA expression fold-change between SCFA fecal extract and vehicle stimulation. The line intersecting the ratio of 1 refers to the basal mRNA expression in d-EpOCs stimulated with the vehicle. B refers to 5-mM butyrate stimulation. Statistical analysis was performed comparing the SCFA fecal extract with the vehicle stimulation. *P < 0.05; **P < 0.01; ***P < 0.001 by 1-tailed Wilcoxon test.
Overall, SCFAs containing fecal extracts from Ctrls and CD patients show qualitatively similar, though milder, effects when compared with 5 mM of butyrate on d-EpOCs. This could, at least in part, be due to the presence of lower SCFA concentrations in the extracts. Importantly, d-EpOCs from both Ctrl and CD patients show overall comparable behavior in response to SCFA fecal extracts. This further supports our initial observation that d-EpOCs from CD patients inherently respond to fecal metabolites.
DISCUSSION
In this study, we propose that the inflammation present in the intestinal mucosa of IBD patients can alter the capacity of the epithelium to metabolize and respond to butyrate. Indeed, based on BCoAT PCR analysis,22 our results show that although the abundance of butyrate-metabolizing bacteria was decreased in active CD stool samples, this did not correlate with a concomitant reduction of butyrate concentration or of any other analyzed SCFAs. Although earlier studies showed a significant decrease of fecal butyrate in IBD patients,10–12 most included small patient samples, did not properly stratify patients by disease activity, or used a variety of analytical techniques, thus making any comparisons of the results challenging. The SCFA extracts prepared from the samples of our study also had similar metabolite concentrations throughout groups and induced comparable responses in human epithelial organoid cultures accordingly. Nonetheless, measuring fecal SCFA content provides an incomplete measurement of the luminal metabolite content, as SCFAs are primarily consumed by the colonic epithelium and only around 5% of its production is estimated to be found in feces.35 Thus, large fluctuations in gut butyrate-producing bacteria may not necessarily result in dramatic changes in fecal butyrate concentrations. Regardless of the concentrations of luminal butyrate, we believe that the lack of any correlation between the abundance of butyrate-producing species and luminal butyrate concentrations in IBD patients may reflect its impaired uptake and oxidation by inflamed colonic cells. This is in line with previous reports that suggest altered butyrate oxidative metabolism in UC patients.13 Moreover, a decrease in MCT1, the protein that acts as a transporter for butyrate and that is encoded by SLC16A1,28 along with expression of the enzymes involved in butyrate oxidation,36 are downregulated in IBD active mucosa. Both TNFɑ and interferon (IFN) ɣ have been shown to downregulate SLC16A1 expression in the human epithelial cell line HT-29 and to result in a decrease in butyrate uptake and oxidation,28 suggesting a potential role of inflammation in decreasing butyrate uptake and metabolism. However, the effects of inflammation on the other proteins involved in butyrate consumption, specifically in the context of primary human cells, have not been previously reported.
Our study takes advantage of patient-derived organoid cultures to maintain human primary epithelium in culture, in lieu of immortalized cell lines. Human organoids have shown some similarities to cell lines in their response to butyrate (inhibition of proliferation, activation of reactive oxygen species detoxification pathways, enhanced epithelial barrier function, promotion of retinoic acid synthesis, and regulation of the inflammatory response),37–39 but also striking differences. For instance, HT-29 cells have been shown to produce TGFβ in response to butyrate,40 whereas our whole-genome analysis shows no induction of TGFβ or TGFβ target genes when using primary epithelial organoids (data not shown), which indicates important differences between primary and immortalized cell systems. We also provide evidence that the intestinal stem and differentiated epithelium are highly sensitive to the presence of this bacterial metabolite, with both systems containing a large number of commonly regulated pathways. Remarkably, butyrate regulated genes that promote cell apoptosis in stem-enriched, but not differentiated, epithelial organoids. These results are in line with a previous study in mice that suggested that the potential detrimental effects of high butyrate concentrations on the stem compartment are avoided by the crypt architecture.41
Ours is the first study to compare the intrinsic ability of human epithelial organoids from UC and CD patients to uptake, metabolize, and respond to butyrate, along with the complex SCFA mixtures extracted from fecal samples. We clearly show that organoids from noninflamed IBD mucosa respond to butyrate similarly to controls, which is consistent with recent observations by Vancamelbeke et al.,42 who found no differences in the response of control and UC cultures to butyrate, alone or in combination with TNFɑ/IFNγ. On the other hand, we confirmed in both ex vivo epithelial cultures and whole mucosal samples that the inflammatory milieu, in particular the presence of TNFɑ, does impact butyrate uptake and oxidation. Indeed, TNFɑ may be responsible for the downregulation of butyrate transporters in the human intestinal epithelium observed in patient biopsies, and that may result in decreased butyrate consumption. In d-EpOCs, TNFɑ had no effect on the expression of ACADS or ACSM3, 2 enzymes involved in butyrate metabolism that are instead upregulated by butyrate itself. Both transcripts, as shown here and elsewhere,13 are downregulated in active IBD biopsies, which could be explained by the decrease in butyrate uptake in the context of inflammation. In fact, using organoids, we also show that pretreatment with TNF decreases the butyrate-induced upregulation of both genes.
Overall our data confirm, using patient-derived organoids, the potent effects of butyrate on the intestinal epithelium and the impact that inflammation, at least in part mediated by the presence of TNFɑ, has on the ability of epithelial cells to uptake, metabolize, and respond to this bacterial metabolite.
Moreover, we provide evidence that response to butyrate is not intrinsically altered in IBD patients under quiescent conditions, which thus supports the beneficial effects of butyrate in the absence of inflammation. Nonetheless, as the downregulation of butyrate transporters and enzymes is associated with both active UC and CD patients, supplementation of active patients with butyrate or SCFA extracts may not provide any beneficial effects as long as inflammation persists.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Biobank Facility at the Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS) for their support, the Endoscopy Department at Hospital Clínic Barcelona and Hospital Universitari Mútua Terrassa for providing us with the samples required to conduct this study, and our patients for their selfless participation. We thank Carles Cristòfol (Universitat Autònoma de Barcelona) for technical support. We thank Montse Arroyes for administrative and technical support. We are indebted to Joe Moore for editorial assistance.
Supported by: This work was supported by grant RTI2018-096946-B-I00 from the Ministerio de Ciencia, Innovación y Universidades, Spain, and grant 2015PG-IBD005 from the Leona and Harry Helmsley Charitable Trust. E.F.P. was a recipient of an FI-DGR 2015 fellowship from AGAUR (Generalitat de Catalunya). A.M. has received funding from the Programa Estatal de Investigación, Desarrollo e Innovación del Ministerio de Economía, Industria y Competitividad, co-funded by the European Social Fund (ESF) under grant agreement number FPI BES-2016–076642. M.E. is supported by the Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd).
Conflicts of interest: None.
Author contributions: E.F.P. designed and conducted experiments, acquired and analyzed data, and wrote the manuscript. A.M.C. performed biostatistics analysis. A.M. designed and conducted experiments. M.E. collected samples and provided technical support. A.C., M.E., M.C.M., E.T., and E.R. recruited patients and/or collected samples. J.C.P. designed experiments. A.S. and I.D. designed the study, supervised experiments, analyzed data, and wrote the manuscript.
REFERENCES
- 1. Mora-Buch R, Dotti I, Planell N, et al. Epithelial IL-1R2 acts as a homeostatic regulator during remission of ulcerative colitis. Mucosal Immunol. 2016;9:950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dotti I, Mora-Buch R, Ferrer-Picón E, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Planell N, Lozano JJ, Mora-Buch R, et al. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62:967–976. [DOI] [PubMed] [Google Scholar]
- 4. Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639–3649. [DOI] [PubMed] [Google Scholar]
- 5. Ni J, Wu GD, Albenberg L, et al. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khan S, Imran A, Malik A, et al. Bacterial imbalance and gut pathologies: association and contribution of E. coli in inflammatory bowel disease. Crit Rev Clin Lab Sci. 2019;56:1–17. [DOI] [PubMed] [Google Scholar]
- 7. Kabeerdoss J, Jayakanthan P, Pugazhendhi S, et al. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res. 2015;142:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laserna-Mendieta EJ, Clooney AG, Carretero-Gomez JF, et al. Determinants of reduced genetic capacity for butyrate synthesis by the gut microbiome in Crohn’s disease and ulcerative colitis. J Crohns Colitis. 2018;12:204–216. [DOI] [PubMed] [Google Scholar]
- 9. Mottawea W, Chiang CK, Mühlbauer M, et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat Commun. 2016;7:13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huda-Faujan N, Abdulamir AS, Fatimah AB, et al. The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J. 2010;4:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marchesi JR, Holmes E, Khan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. [DOI] [PubMed] [Google Scholar]
- 12. Bjerrum JT, Wang Y, Hao F, et al. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics. 2015;11:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Preter V, Geboes KP, Bulteel V, et al. Kinetics of butyrate metabolism in the normal colon and in ulcerative colitis: the effects of substrate concentration and carnitine on the β-oxidation pathway. Aliment Pharmacol Ther. 2011;34:526–532. [DOI] [PubMed] [Google Scholar]
- 14. Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. [DOI] [PubMed] [Google Scholar]
- 15. Donohoe DR, Garge N, Zhang X, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Di Sabatino A, Morera R, Ciccocioppo R, et al. Oral butyrate for mildly to moderately active Crohn’s disease. Aliment Pharmacol Ther. 2005;22:789–794. [DOI] [PubMed] [Google Scholar]
- 17. Cummings JH. Short-chain fatty acid enemas in the treatment of distal ulcerative colitis. Eur J Gastroenterol Hepatol. 1997;9:149–153. [DOI] [PubMed] [Google Scholar]
- 18. Segain JP, Raingeard de la Blétière D, Bourreille A, et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut. 2000;47:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Canani RB, Costanzo MD, Leone L, et al. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol. 2011;17:1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625–1629. [DOI] [PubMed] [Google Scholar]
- 21. Daperno M, D’Haens G, Van Assche G, et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc. 2004;60:505–512. [DOI] [PubMed] [Google Scholar]
- 22. Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73:2009–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jung P, Sato T, Merlos-Suárez A, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. [DOI] [PubMed] [Google Scholar]
- 24. Louis P, Young P, Holtrop G, et al. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010;12:304–314. [DOI] [PubMed] [Google Scholar]
- 25. Lukovac S, Belzer C, Pellis L, et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio. 2014;5:e01438–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mora JR, Bono MR, Manjunath N, et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424:88–93. [DOI] [PubMed] [Google Scholar]
- 27. Mora JR, Iwata M, Eksteen B, et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314:1157–1160. [DOI] [PubMed] [Google Scholar]
- 28. Thibault R, De Coppet P, Daly K, et al. Down-regulation of the monocarboxylate transporter 1 is involved in butyrate deficiency during intestinal inflammation. Gastroenterology. 2007;133:1916–1927. [DOI] [PubMed] [Google Scholar]
- 29. Deuring JJ, de Haar C, Koelewijn CL, et al. Absence of ABCG2-mediated mucosal detoxification in patients with active inflammatory bowel disease is due to impeded protein folding. Biochem J. 2012;441:87–93. [DOI] [PubMed] [Google Scholar]
- 30. Thibault R, Blachier F, Darcy-Vrillon B, et al. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflamm Bowel Dis. 2010;16:684–695. [DOI] [PubMed] [Google Scholar]
- 31. Englund G, Jacobson A, Rorsman F, et al. Efflux transporters in ulcerative colitis: decreased expression of BCRP (ABCG2) and Pgp (ABCB1). Inflamm Bowel Dis. 2007;13:291–297. [DOI] [PubMed] [Google Scholar]
- 32. Gutmann H, Hruz P, Zimmermann C, et al. Breast cancer resistance protein and P-glycoprotein expression in patients with newly diagnosed and therapy-refractory ulcerative colitis compared with healthy controls. Digestion. 2008;78:154–162. [DOI] [PubMed] [Google Scholar]
- 33. Brown AJ, Goldsworthy SM, Barnes AA, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. [DOI] [PubMed] [Google Scholar]
- 34. Arijs I, De Hertogh G, Lemaire K, et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One. 2009;4:e7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064. [DOI] [PubMed] [Google Scholar]
- 36. De Preter V, Arijs I, Windey K, et al. Impaired butyrate oxidation in ulcerative colitis is due to decreased butyrate uptake and a defect in the oxidation pathway. Inflamm Bowel Dis. 2012;18:1127–1136. [DOI] [PubMed] [Google Scholar]
- 37. Rosignoli P, Fabiani R, De Bartolomeo A, et al. Protective activity of butyrate on hydrogen peroxide-induced DNA damage in isolated human colonocytes and HT29 tumour cells. Carcinogenesis. 2001;22:1675–1680. [DOI] [PubMed] [Google Scholar]
- 38. Schilderink R, Verseijden C, Seppen J, et al. The SCFA butyrate stimulates the epithelial production of retinoic acid via inhibition of epithelial HDAC. Am J Physiol Gastrointest Liver Physiol. 2016;310:G1138–G1146. [DOI] [PubMed] [Google Scholar]
- 39. Inan MS, Rasoulpour RJ, Yin L, et al. The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology. 2000;118:724–734. [DOI] [PubMed] [Google Scholar]
- 40. Martin-Gallausiaux C, Béguet-Crespel F, Marinelli L, et al. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep. 2018;8:9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaiko GE, Ryu SH, Koues OI, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vancamelbeke M, Laeremans T, Vanhove W, et al. Butyrate does not protect against inflammation-induced loss of epithelial barrier function and cytokine production in primary cell monolayers from patients with ulcerative colitis. J Crohns Colitis. 2019;jjz064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.