Abstract
Atherosclerosis is the primary underlying cause of myocardial infarction, ischemic stroke, and peripheral artery disease. The disease preferentially occurs in arterial regions exposed to disturbed blood flow, in part, by altering expression of flow-sensitive coding- and non-coding genes. In this review, we summarize the role of noncoding RNAs, [microRNAs (miRNAs) and long noncoding RNAs(lncRNAs)], as regulators of gene expression and outline their relationship to the pathogenesis of atherosclerosis. While miRNAs are small noncoding genes that post-transcriptionally regulate gene expression by targeting mRNA transcripts, the lncRNAs regulate gene expression by diverse mechanisms, which are still emerging and incompletely understood. We focused on multiple flow-sensitive miRNAs such as, miR-10a, −19a, −23b, −17~92, −21, −663, −92a, −143/145, −101, −126, −712, −205, and −155 that play a critical role in endothelial function and atherosclerosis by targeting inflammation, cell cycle, proliferation, migration, apoptosis, and nitric oxide signaling. Flow-dependent regulation of lncRNAs is just emerging, and their role in vascular dysfunction and atherosclerosis is unknown. Here, we discuss the flow-sensitive lncRNA STEEL along with other lncRNAs studied in the context of vascular pathophysiology and atherosclerosis such as MALAT1, MIAT1, ANRIL, MYOSLID, MEG3, SENCR, SMILR, LISPR1, and H19. Also discussed is the use of these noncoding RNAs as potential biomarkers and therapeutics to reduce and regress atherosclerosis.
Keywords: microRNAs, Long noncoding RNAs, Atherosclerosis, Endothelial dysfunction, Disturbed blood flow, Gene expression, Vascular dysfunction
1. Introduction
Atherosclerosis is a chronic inflammatory disease of the arterial vessel which underlies the occurrence of myocardial infarction, ischemic stroke, and peripheral arterial disease [1]. The disease is characterized by the initial development of a lesion in the arterial wall called a “fatty streak,” which contains lipid-rich macrophages [2,3]. Sustained chronic exposure to pro-inflammatory molecules from leukocytes and lipids increases vascular dysfunction and the narrowing of the arterial lumen, leading to the development of an atheromatous plaque [4]. Interestingly, it has been observed that atherosclerotic plaques develop preferentially in regions of the vasculature exposed to disturbed blood flow (d-flow), typically at the branched and curved arterial regions. D-flow is characterized by flow patterns with low-magnitude and oscillatory shear stress (OS), whereas stable blood flow (s-flow) is characterized by high-magnitude, unidirectional laminar shear stress (LS) usually observed in the straight sections of the artery [5–8].
Endothelial cells (ECs) respond to shear stress primarily through numerous mechanosensors present on the cell surface, cell-cell junction, cell-matrix adhesion sites, and actin cytoskeletal structure, which transduce the mechanical cues into cell signaling events and ultimately changes in gene expression [5,9,10]. A large number of endothelial genes that change in response to flow are referred to as flow-sensitive genes, also known as mechanosensitive genes [11–18]. The majority of these flow-sensitive genes are protein-coding genes, such as the atheroprotective Klf2 [19], Klf4 [20], Timp3, and eNOS [18], which are upregulated by the stable flow. D-flow also upregulates a number of proatherogenic genes, including vascular cell adhesion molecule-1 (VCAM-1) [17,21], matrix metalloproteinases (MMPs) [22], and bone morphogenic protein-4 (BMP4), which mediate inflammatory, proliferative, and apoptotic responses in vascular endothelium [5,23,24].
While the role of flow-sensitive coding genes and proteins have been extensively studied for their role in atherosclerosis [5,11,12,15,25–32], the role of flow-sensitive non-coding genes is still emerging [33]. Noncoding genes are transcribed into functional RNAs; however, these RNAs do not code for proteins, i.e., non-coding RNAs (ncRNAs). Noncoding RNAs regulate a myriad of physiological process by acting as regulators of gene expression at the transcriptional, post-transcriptional, and epigenetic level [34–40]. In this review, we will discuss two main categories of ncRNAs; the short ncRNAs (< 30 nucleotides) and the long noncoding RNAs (> 200 nucleotides) or lncRNAs. Short ncRNAs consist of microRNAs (miRNAs), siRNAs, piRNAs, and other subgroups. Due to the limited information on other subgroups of the short ncRNAs, we will focus our discussion on the most well-studied group: miRNAs. Here, we review and update the current knowledge of flow-sensitive miRNAs and functionally essential lncRNAs for their role in vascular biology and atherosclerosis.
2. Biogenesis, transcription, and processing of miRNAs
miRNAs are short noncoding RNAs that regulate gene expression at the post-transcriptional level [41,42]. Typical miRNAs are transcribed by RNA polymerase II (RNA pol II) in the nucleus as pri-miRNAs, which are trimmed into 70–100 nucleotides (nt) hairpin-shaped precursor—pre-miRNA by the Drosha-DGCR8 complex [43–46]. Pre-miRNA is exported into the cytoplasm with the assistance of Ran-GTP and Exportin-5 complex [47,48]. After that, the pre-miRNA is cleaved by Dicer in association with its partners argonaute (AGO) and TransActivation Responsive RNA-Binding protein (TRBP) to produce a double-stranded 20–25 nt miRNA [49]. The fully processed miRNA duplex is then incorporated into a multicomponent protein complex known as an RNA-induced silencing complex (RISC). During this process, one strand of the miRNA duplex is selected as the mature miRNA (or miRNA-5p) while the other strand, known as miRNA* (passenger strand or miRNA-3p) which is typically rapidly degraded [50]. Mature miRNA-5p then further facilitates the cleavage of target mRNA and/or its translational repression via precise mechanisms [51–53]. The seed sequence (the nucleotides in position 2–8 of miRNA) binds to regions at the 3′UTR of its target mRNAs via complementary pairing [54]. In silico analyses have revealed that ~60% of protein-coding genes harbor miRNA target sites in their 3’UTR and that a single miRNA can typically modulate the expression of hundreds of genes. Not only are specific miRNA genes highly conserved in animals, but their target sites in the 3’UTR of genes are also under positive evolutionary selection. MiRNAs play an essential role in development and organogenesis and more importantly in vascular functions [55–59].
3. Role of miRNAs in vascular dysfunction and atherosclerosis
MiRNAs are frequently dysregulated in vascular pathologies such as atherosclerosis [60–64], and tremendous effort exists that aim to develop novel diagnostic markers and therapeutics for the treatment of atherosclerosis. Changes in miRNA expression levels due to blood flow have the potential to affect networks of genes regulating endothelial and vascular smooth muscle cell function, inflammation, and atherosclerosis.
The flow-sensitive microRNAs play essential roles in the regulation of vascular dysfunction and atherosclerosis. Here, we broadly categorize these miRNAs based on their response to flow: (1) miRNAs upregulated by s-flow or downregulated by d-flow and (2) miRNAs downregulated by s-flow or upregulated by d-flow (Table 1). MicroRNAs such as miR-10a, 23b, and 101 are either upregulated by s-flow/LS or downregulated by d-flow/OS, while microRNAs such as miR-17~92 cluster, 92a, 663, 712, and 205 are either upregulated by d-flow/OS or downregulated by s-flow/LS. Other microRNAs for which the information on flow-sensitivity is not explicit include miR-21, 155, 126, 143, and 145. Table 1 shows the flow sensitivity of the miRNA, its validated target genes, and its role in vascular dysfunction and atherosclerosis.
Table 1.
Role of flow-sensitive microRNAs in vascular dysfunction and atherosclerosis
| miRNAs | Response to flow | Direct gene targets | Pathway(s) regulated | Role in vascular dysfunction and atherosclerosis | Ref. |
|---|---|---|---|---|---|
| miR-10a | s-flow↑ | MAP3K7, β-TRC | VCAM-1, E-selectin and NFκB signaling↓ | Anti-inflammatory; Inhibit EC NFκB activation | [65] |
| miR-23b | LS ↑ | E2F1 | Cell cycle arrest (G1/S)/ Rb hypophosphorylation | Inhibits EC proliferation | [74] |
| FoxO4 | Increased regulation of smooth muscle actin (ACTA2) and MYH11 | Inhibits VSMC Switching and Proliferation | [70] | ||
| miR-101 | LS ↑ (* also upregulated by hypoxia) | mTOR | Cell cycle arrest (G1/S) | Inhibits EC proliferation, | [71] |
| Cul3 | Nrf2 translocation to nucleus | pro-angiogenic signaling | [72] | ||
| ABCA1 (in THP1 and hepatocytes) | Prevents Cholesterol Efflux | Cholesterol biosynthesis | [73 183] | ||
| miR-27b | LS ↑ | SEMA6A, SEMA6D | Endothelial-pericyte interaction | Vessel integrity | [74] |
| SPRY2, DLL4 | prevents vessel branching and regulate endothelial cell fate | Anti-angiogenic | [75] | ||
| Flt1 | prevents vascular sprouting | angiogenic function | [76] | ||
| TGFβ (* microvascular ECs and fibroblasts) | Endothelial-mesenchymal transition (EndMT) | cell differentiation | [77,78] | ||
| miR-126-5p | d-flow ↓ | Dlk1 | EC repair and proliferation (G2/M)↑ | endothelial miR-126-5p maintains a proliferative reserve and administration of miR-126-5p limits atherosclerosis | [82] |
| HMGB1 | Alleviates ox-LDL-induced HUVEC inflammation | dysregulated miR-126 expression regulates inflammation in diabetic vascular endothelium | [84] | ||
| FoxO3, BCL2, and IRS1 | Endothelial miR-126 increases SMC turnover | Systemic depletion of miR-126 in mice inhibited neointimal lesion formation of carotid arteries | [81] | ||
| PI3K/Akt/mTOR pathway | miR-126 mimics rescued ox-LDL-induced impaired autophagy flux through inhibiting PI3K/Akt/mTOR signaling | [83] | |||
| G protein signaling 16 | CXCL12/CXCR4 signaling axis | Administration of apoptotic bodies enriched in miR-126 or miR-126 mimic limited atherosclerosis | [193] | ||
| d-flow ↓ | increased VCAM1 and CCL2 | EC-leukocyte adhersion | Proinflammatory EC signaling | [85] | |
| LRP6 | VSMC proliferation | Neointimal formation post wire injury | [86] | ||
| s-flow ↓ | ABCA1 | increased hepatic and vascular ABCA1 expression | genetic knockdown of miR-143/145 in LDLR−/− mice reduced plaque size and macrophage infiltration | [90] | |
| s-flow ↓ | AMPKα2 and p53 | AMPK α2 suppresses endothelial ACE expression via the phosphorylation of p53 and upregulation of miR-143/145 | regulation of miR-143/145 may contribute to the vascular complications | [89] | |
| s-flow ↓ | KLF2 | ECs secrete microvesicles rich in miR-143/145 and regulate SMC activity | Extracellular vesicles derived from KLF2-expressing endothelial cells enriched in miR-143/145 reduced atherosclerotic lesion formation in the aorta of ApoE−/−mice | [87] | |
| miR-143/145 | KLF4, myocardin and Elk-1 | promote differentiation and repress proliferation of smooth muscle cells | miR-145 and miR-143 function to regulate the quiescent versus proliferative phenotype of smooth muscle cells | [88] | |
| Hexokinase II, Integrin β8 | angiogenic and vessel stabilization properties of Ecs | Regulate VSMC phenotypic switch and ABCA1 dependent cholesterol efflux in monocytes/macrophage | [91] | ||
| NT | KLF5 | Myocardin↑, VSMC contractile phenotype↑ | SMC-targeted miR-145 inhibits atherosclerosis | [226] | |
| miR-19a | s-flow ↓ | Cyclin D1 | Cell cycle arrest G1/S stage | Endothelial proliferation | [68] |
| OS ↑ | HMGB1 | CXCL1,MIF, TNFct, IL-6↑ | Promote immune response | [92,93] | |
| d-flow ↑ | HBP1 | Loss of HBP2 upregulates macrophage migration inhibiting factor (MIF) | In vivo delivery of anti-miR-19a decreases plaques in apoE-null mice | [92] | |
| miR-92a | (* also upregulated by hypoxia) | CXCL1 | increased proinflammatory cytokine release | Endothelial inflammation | [94] |
| LS ↓/ OS ↑ | KLF2 | eNOS expression, VCAM1 and MCP1 | thromboembolism | [97] | |
| LS ↓/ OS ↑ | KLF4 | eNOS expression, VCAM1 and MCP1 | endothelial inflammation (anti-miR-92a inhibits EC inflammation and atherosclerosis) | [97] | |
| d-flow ↑ | PPABP2B | myosin light-chain phosphorylation and intercellular gaps | anti-inflammatory phenotype and maintaining vascular integrity of endothelial monolayer under atheroprotective flow | [98] | |
| d-flow ↑ | ITGA5 | controls angiogenesis | overexpression of miR-92a in ECs blocks angiogenesis in vitro and in vivo | [96] | |
| miR-663 | d-flow ↑ | KLF2,KLF4 and SOCS5 | Suppressor of Cytokine Signaling 5 (SOCS5) | miR-92a blockage in Ldlr−/− mice reduced endothelial inflammation and atherosclerosis | [99] |
| KLF4, myocardin and Elk-1 | promote differentiation and repress proliferation of smooth muscle cells | miR-145 and miR-143 function to regulate the quiescent versus proliferative phenotype of smooth muscle cells | [88] | ||
| VEGF, ATF4 | unfolded protein response (UPR) in endothelial cells by oxidized phospholipids | knockdown of miR-663 reduced EC stress | [102] | ||
| KLF4 | expression of an inflammatory gene network in ECs | endothelial inflammation | [101] | ||
| miR-712 and miR-205 | d-flow ↑ and OS ↑ | TIMP3 | MMPs activity↑ | Anti-miR-712 prevents EC inflammation and atherosclerosis | [108] |
| Anti-miR-205 inhibits EC inflammation | [227] | ||||
| miR-21 | NT | PTEN, BCL2 | miR-21−/− inhibits neointimal formation in vein graft model | [228] | |
| OS ↑ | PPARα | Activates AP-1, monocyte adhesion | EC inflammation | [112] | |
| NT | Negative regulator of CD4 + CD25 + FOXP3 + regulatory T cells | [115] | |||
| LS ↑ | PTEN | eNOS phosphorylation and NO production↑ | Increases NO bioavailability and decreases EC apoptosis | [111] | |
| miR-155 | NT | Bcl6 | NFκB activation | Promotes atherosclerosis | [127] |
| NT | SOCS-1 | Cholesterol efflux↓, pro-inflammatory cytokine (TNFα, IL-6, IL-1β)↑ | miR-155−/− inhibits inflammation and atherosclerosis | [229,230] | |
| LS ↑ | eNOS, MYLK | RhoA↑ | Inhibits EC migration and proliferation | [123,129] | |
| NT | NT | Regulate monocyte subset | BMT with miR-155 knockout destabilizes plaque and stimulates atherosclerosis | [125,128] |
EC: endothelial cell; LS, laminar shear stress; OS, oscillatory shear stress; NT, not tested; d-flow, disturbed flow; KLF2 and 4, Krȕppel-like factor 2 and 4; ITGA5, integrin alpha 5; ACTA2, smooth muscle alpha (α)-2 actin; MYH11, myosin heavy chain 11; E2F1, E2F transcription factor 1; CDK, cyclin-dependent kinase; ABCA1, ATP-binding cassette transporter; AP-1, Activator protein 1; mTOR, mammalian target of rapamycin; ELK1, ETS transcription factor; MYOCD, Myocardin; PI3K, Phosphatidylinositol-4,5-bisphosphate 3-kinase; AKT/PKB, Protein kinase B; HMGB1, High mobility group box 1; TRAF7, TNF receptor associated factor 7; LRP6, Low-density lipoprotein receptor-related protein 6; HBP-1, HMG box-containing protein 1; IL-10, Interleukin 10; IL-6, Interleukin 6; MMP, Matrix metalloproteinase; CXCL1, chemokine (C-X-C motif) ligand 1; MIF, Macrophage migration inhibitory factor; TNFα, Tumor Necrosis Factor α; SIRT1, Sirtuin 1 (Silent Mating Type Information Regulation 2); FOSB, FBJ murine osteosarcoma viral oncogene homolog B; SLC7A5, Solute carrier family 7 member 5; NAV2, neuron navigator; TIMP3, tissue inhibitor of metalloproteinase 3; PPARα, Peroxisome proliferator-activated receptor alpha; PTEN, Phosphatase and tensin homolog; BCL6, B-cell lymphoma 6; SOCS-1, Suppressor of cytokine signaling 1; MYLK, Myosin light chain kinase; FOXO3, Forkhead box O3; FOXO4, Forkhead box O4 BCL2, B-cell lymphoma 2; IRS1, insulin receptor substrate 1; VCAM-1, Vascular cell adhesion protein 1; Dlk1, delta-like 1 homolog; MAP3K7, mitogen-activated protein kinase kinase kinase 7; β-TRC, β -transducin repeat-containing gene; AA : aortic arch; mTOR, Mammalian target of rapamycin; CROT, Carnitine O- Octanoyltransferase; ABCG1, ATP-binding cassette sub-family G member 1; CPT1A, Carnitine Palmitoyltransferase 1A; HADHB, Hydroxyacyl-CoA Dehydrogenase/3-Ketoacyl-CoA Thiolase/Enoyl-CoA Hydratase CAMK2d, Calcium/calmodulin-dependent protein kinase type II delta chain; SSH2, slingshot homolog 2; PHACTR4, phosphatase and actin regulator 4; FL1, Follicular lymphoma, susceptibility to 1.
4. miRNAs upregulated by s-flow or LS
These miRNAs are either increased by s-flow/LS (atheroprotective flow) or decreased by d-flow/OS (pro-atherogenic flow) in endothelial cells and are shown to reduce vascular inflammation and atherosclerosis (Table 1 and Figure 1).
Fig. 1.
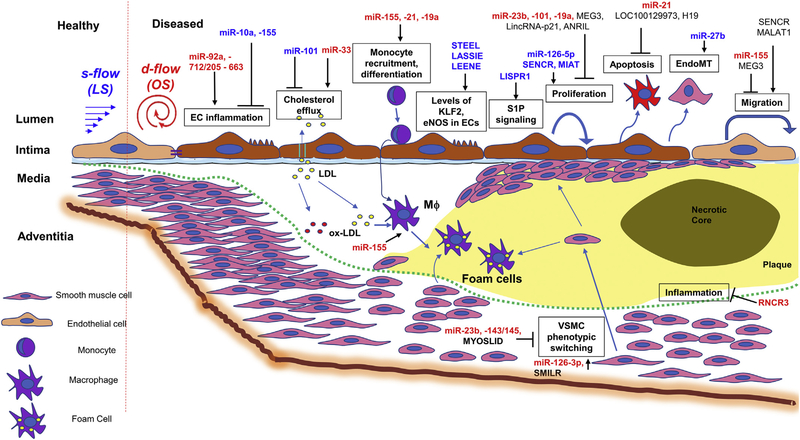
Flow-sensitive miRNAs and lncRNAs in vascular dysfunction and atherosclerosis: The microRNAs and long non-coding RNAs that are regulated by flow and are implicated in vascular dysfunction and atherosclerosis through regulating gene targets in endothelial cells, smooth muscle cells, and monocytes/macrophages are shown.
4.1. miR-10a
Stable blood flow upregulates the expression of miR-10a in the endothelium. Loss of miR-10a results in activation of NFκB via MAP3K7 and βTRC, both of which promote IκB degradation and p65 translocation, resulting in endothelial inflammation in a porcine model of atherosclerosis, suggesting that differential expression of miR-10a regulates a pro-inflammatory endothelial phenotype [65]. Expression of miR-10a in the athero-susceptible aorta has recently been shown to be rescued by retinoic acid receptor-α (RARα) and retinoid X receptor-α (RXRα) agonists, leading to inhibition of GATA6/VCAM-1 signaling and inflammatory cell infiltration [66]. Recently, it was shown that induction of miR-10a by administration of RARα/RXRα-specific agonists prevents inflammation and atherosclerosis in ApoE−/− mice, providing further support for the role of flow-sensitive miR-10a as an atheroprotective miRNA and potential therapeutic target [67].
4.2. miR-23b
Pulsatile LS or s-flow upregulates the expression of miR-23b in the endothelium. Increased levels of miR-23b suppressed endothelial proliferation by reducing E2F transcription factor 1 (E2F1) expression and Rb phosphorylation [68]. Also, miR-23b was shown to inhibit cyclin-dependent kinase-activating kinase (CAK) complex thereby suppressing cell cycle progression and reducing the basal transcription of RNA Pol II [69]. Furthermore, miR-23b was recently implicated as a novel regulator of vascular smooth muscle cell (VSMC) phenotype switching following vascular injury [70]. Here, miR-23b inhibited VSMC proliferation and migration while promoting expression of VSMC markers genes such as smooth muscle alpha (α)-2 actin (ACTA2) and smooth muscle myosin heavy chain 11 (MYH11). Transcription factor forkhead box O4 (FOXO4) was also identified as a direct target of miR-23b in VSMCs [70]. Together, these studies suggest miR-23b is a flow-sensitive miRNA involved in maintaining cellular quiescence, regulating cell identity and cell cycle in a flow-dependent context.
4.3. miR-101
LS increases the expression of miR-101, which then targets the mTOR gene, leading to cell cycle arrest in vascular endothelial cells [71]. Overexpression of miR-101 reduced the G1/S transition of the cell cycle in ECs subjected to LS [71]. However, expression of miR-101 is also regulated in other cell types and by various stress stimuli. For example, miR-101 is also upregulated in response to hypoxia, where it targets Cul3, a scaffold protein in the E3 ligase complex. Without Cul3, the transcription factor Nrf2 enters the nucleus and promote expression of angiogenesis-related genes [72]. In addition, in THP-1 monocytes and hepatocytes, miR-101 has also been reported to suppress ATP-binding cassette transporter A1 (ABCA1) expression under normal and inflammatory conditions [73]. Together, these findings suggest that miR-101 might regulate cellular functions in endothelium in a flow-dependent manner and may also play a context-specific role in other cell types.
4.4. miR-27b
miR-27b is upregulated by LS and plays a critical role in controlling angiogenesis by controlling the activators and suppressors of endothelial proliferation, migration, and differentiation [74]. Also, LS increased miR-27a and miR-27b expression in vitro and in ex vivo in mouse femoral artery explants and induced interaction of pericyte-endothelial cell interaction by repressing Semaphorin 6A (SEMA6A) and Semaphorin 6A (SEMA6D), resulting in vessel stability [75]. Under an antiangiogenic stimulus, down-regulation of miR-27b expression induces Spry2 and Dll4 expression, which prevents vessel branching and regulates endothelial cell fate, respectively [76]. In microvascular endothelial cells and fibroblasts, miR-27b regulates transforming growth factor-β (TGF-β) induced endothelial-mesenchymal transition (EndoMT) [77,78]. Taken together, miR-27b is a flow-sensitive miRNA that plays a critical role in the development and maintenance of blood vessels in healthy as well as pathophysiological conditions.
4.5. miR-126
The flow regulation and atherogenic effect of miR-126 are not entirely settled. However, the evidence favors that OS downregulates miR-126-5p expression and that it plays a role as an anti-atherogenic microRNA. MiR-126 (also referred to as miR-126-3p, miR-126*, and miR-126-5p) is highly abundant in ECs and regulates vascular integrity, angiogenesis [79], and inflammation [80]. The secretion of miR-126-3p into the conditioned media, but not its intracellular expression per se, was decreased by LS and increased by OS, respectively, in HUVECs [81]. Endothelial-derived miR-126-3p regulated SMC turnover in an EC-SMC co-culture system. Furthermore, genetic knockout of miR-126 inhibited neointimal formation in a complete carotid ligation model, while local reintroduction of miR-126 in the knockout mice enhanced neointimal formation [81], suggesting its role as a shear-sensitive proatherogenic miRNA. However, subsequent studies have since found miR-126 to have an anti-atherogenic effect mainly. First, Schober et al. demonstrated that d-flow decreased expression of both miR-126-5p and 126-3p. Further, they showed that treatment with miR-126-5p, but not miR-126-3p, reduced atherosclerotic lesion formation. The anti-atherogenic effect of miR-126-5p was mediated by targeting the Notch1 ligand, Delta-like 1 homolog (Dlk1), which promoted the proliferative potential of ECs [82]. Other studies showed that miR-126 protects ECs from inflammation and apoptosis. MiR-126 protected ECs from inflammation induced by oxidized LDL and hyperglycemic conditions [83,84]. These studies showed that miR-126 restored autophagy flux via repressing the PI3K/Akt/mTOR pathway [83] and targeted HMGB1 [84], respectively. Also, Cerutti et al. showed miR-126 targeted endothelial adhesion molecules, E-selectin and VCAM1 [85]. MiR-126 was also able to prevent palmitate-induced apoptosis by targeting TRAF7, a signal transducer for members of the tumor necrosis factor (TNF) receptor superfamily [86]. Interestingly, Jansen et al. showed that miR-126 transferred from ECs to VSMCs through microparticles reduces VSMC proliferation, migration, and subsequent neointima formation by inhibiting LDL Receptor-Related Protein 6 (LRP6) [86]. One significant potential source of conflicting results in many studies over the years on miR-126 appears to be the functional differences between miR-126, miR-126-3p, and miR-126-5p. Another possibility is due to differential expression of gene targets in the specific cells and tissues used. Taken together, mainly based on recent evidence, miR-126-5p appears to be an LS-inducible and OS-repressive anti-atherogenic miRNA.
4.6. miR-143/145 cluster
The miR-143/145 cluster expression is upregulated by stable flow and most, but not all, studies support their role as anti-atherogenic miRNAs. Cordes et al. first reported that expression of miR-143/145 is reduced in injured or atherosclerotic vessels [87,88]. The same study showed that these miRs promote SMC contractile phenotype while repressing proliferative response. In ECs, multiple studies showed that LS increases miR-143/145 expression by the mechanisms involving KLF2-dependent [87] and KLF2-independent mechanisms [89]. Hergenreider et al. demonstrated that LS increased expression of miR-143/145 by the KLF2-dependent manner. They further showed that these miRs were secreted as exosomes, which in turn could be transferred to VSMCs. Injection of these exosomes containing miR-143/145 inhibited atherogenesis by promoting VSMC differentiation and preventing proliferation in ApoE−/− mice [87,88]. Kohlstedt et al. also showed that LS increases miR-143/145 levels, but in a KLF2-independent mechanism by activating AMPKα2 [89]. Activated AMPKα2 then phosphorylated p53 tumor suppressor, which is known to regulate the miRNA processor drosha, thereby increasing mature miR-143/145 levels. They further demonstrated that LS inhibits expression of angiotensin-converting enzyme by the AMPKα2-dependent miR143/145 pathway [89]. In contrast to the above studies, Sala et al. found that miR-143/145 deficiency in LDLR−/− significantly reduced atherosclerosis in mice [90], suggesting their role as pro-atherogenic miRNAs. Interestingly, these miR-143/145 deficient LDLR−/− mice showed a reduction in plasma cholesterol levels. The authors attributed this result to the effect of miR-145 silencing the ABCA1, which plays an essential role in HDL biogenesis and cholesterol efflux in hepatocytes and macrophages, respectively. They also showed that miR-143/145 levels were increased in human carotid atherosclerotic plaques from symptomatic patients [90], suggesting the role of miR-143/145 as pro-atherogenic miRs. A more recent study showed that VSMCs could secrete miR-143/145, which were taken up by ECs and modulated angiogenic and vessel stability by regulating endothelial hexokinase II and integrin β8-2 [91]. Together, these results show that the role of miR-143/145 in atherosclerosis is not straightforward and may change depending on cell types, animal models, and pathobiological context.
5. miRNAs upregulated by d-flow or OS
These miRNAs are either increased by d-flow/OS or decreased by s-flow/LS in endothelial cells and associated with vascular dysfunction and pro-atherogenic responses.
5.1. miR-17~92
The miR-17~92a cluster comprises several miRs, including miR-17, 18a, 19a 19b, 20a, and 92a. These miRs have been studied extensively for flow-dependent regulation and their role in atherosclerosis. The miR-17~92 cluster is regulated by shear stress in that some members (miR-17, miR-19b, miR-20a, miR-92a) were downregulated by pulsatile LS [74]. Interestingly, s-flow increased the expression of miR-19a, and its transient overexpression leads to a significant decrease in the cyclinD1 mRNA and protein levels, leading to cell cycle arrest at the G1/S stage [68]. However, a recent study showed that miR-19a also has a pro-inflammatory phenotype by targeting the gene high mobility group box transcription protein 1 (HMGB1) in vitro and in vivo [92]. HMGB1 is a repressor of macrophage migration inhibiting factor (MIF), and increased expression of miR-19a under OS conditions. This leads to higher levels of MIF resulting in the release of pro-inflammatory cytokines TNFα and Interleukin-6 (IL-6) [92]. Furthermore, miR-19a is upregulated in CD19+ B-cells from patients with atherosclerosis, where it has been shown to suppress the anti-inflammatory cytokine interleukin-10 (IL-10) [93]. Interestingly, miR-19a was also shown to be regulated by hypoxic conditions. Hypoxia-inducible factor (HIF)-1α is increased in atherosclerotic lesions and is associated with atherosclerotic plaque inflammation. In vivo, endothelial HIF-1α promoted atherosclerosis by upregulating miR-19a, which indirectly caused an increase in expression of Chemokine Ligand 1 (CXCL1) [93,94]. Together, current evidence suggests that miR-19a regulates aspects of chemokine and cytokine expression under inflammatory and hypoxic conditions in both ECs and CD19+ B cells. Subsequent studies showed that miR-92a was downregulated by LS and was upregulated by OS [95]. These in vitro findings are consistent with in vivo studies showing that ECs in the athero-prone porcine aortic arch area have increased miR-92a levels as compared to those of the athero-resistant thoracic aorta [95]. Regarding its function, overexpression of miR-92a prevented angiogenesis in a mouse model of limb ischemia by targeting the integrin subunit alpha 5 [96]. Further studies demonstrate that miR-92a exerts its proatherogenic effect by inhibiting KLF2-mediated thromboembolic and eNOS expression, as well as KLF4-induced expression of E-selectin, eNOS, VCAM-1, and MCP1 [97]. Recently, miR-92a was shown to silence the integral membrane protein Phosphatidic Acid Phosphatase type 2B (PPAP2B) under OS conditions in vivo, resulting in an increased inflammatory response to circulating lipids [98]. Moreover, SOCS5 has been identified as a miR-92a target that is involved in the regulation of endothelial inflammation via activation of the JAK-STAT pathway [99]. Thus, miR-92a appears to target genes corresponding to several pro-angiogenic proteins [96] and inhibition of miR-92a in a preclinical study of porcine limb ischemia and myocardial infarction enhanced blood vessel growth and functional recovery of damaged tissue [100]. Taken together, inhibiting selected members of the miR-17~92 cluster can be an effective anti-atherogenic strategy.
5.2. miR-663
MiR-663 was identified as one of the most upregulated miRNAs from a microarray study using HUVECs exposed to OS conditions compared to the LS [101]. Overexpression of miR-663 induced EC inflammation, suggesting its potential pro-atherogenic role [101]. This study further identified several transcription factors including KLF4, CEBPB, and ATF3 as potential targets of miR-663 under the OS condition. These transcription factors regulate multiple genes involved in the inflammatory responses in endothelium. Furthermore, inhibiting miR-663 with miR-663-LNA restores KLF4 expression in ECs under OS condition, suggesting that d-flow induced miR-663 is critically involved in the tweaking of the KLF4 expression in endothelium [101]. Furthermore, miR-663 was upregulated in HUVECs exposed to proatherogenic oxidized phospholipids and was found to play a permissive role in the induction of VEGF and activation of ATF4 branch of unfolded protein response in ECs [102,103]. Interestingly, however, miR-663 regulates VSMC phenotypic switching by targeting the transcription factors JunB/Myosin Light chain 9 [104–106]. These results suggest that miR-663 has a context-dependent and cell type-specific role in the pathophysiological process, as overexpression of this human-specific microRNA using an adenoviral construct reduced neointimal formation in a mouse model of neointimal hyperplasia [107]. In order to be developed as a potential therapeutic candidate for atherosclerosis, future work is needed to delineate the cell-specific effects of miR-663.
5.3. miR-712 and miR-205 family
MiR-712 and miR-205 were identified as pro-atherogenic miRNAs induced by disturbed flow. MiR-712 was first reported from a miRNA array study using endothelial-enriched RNAs obtained from the mouse model of flow-induced atherosclerosis, known as the partial carotid ligation model [21]. Interestingly, miR-712 is murine-specific, and miR-205 was identified as its homolog in humans and other vertebrates from a seed-sequence matching study [108]. miR-205 shares the same “seed sequence” with miR-712 and both have increased expression under d-flow. The miR-712/205 family activates endothelial inflammation and permeability changes in a flow-dependent manner. The pro-inflammatory and pro-atherogenic effects of miR-712/miR-205 were mediated in large part by tissue inhibitor of metalloproteinase-3 (TIMP3) [108]. Loss of TIMP3 by miR-712 resulted in activation of matrix metalloproteinases (MMPs) and a disintegrin and metalloproteinases (ADAMs), ultimately leading to endothelial inflammation, hyperpermeability, and atherosclerosis [82] as well as in abdominal aortic aneurysm [109]. We also showed that targeting this miRNA using anti-miR-712 injected either systemically as the naked form was able to prevent atherosclerosis in a mouse model of atherosclerosis [108]. Furthermore, we demonstrated that anti-miR-712 could be delivered specifically to the inflamed artery endothelial cells in mice by encapsulating anti-miR-712 inside the cationic lipid nanoparticles decorated with the VCAMl-targeting peptide [110]. The VCAMl-targeting lipid nanoparticles containing anti-miR-712 was able to inhibit atherosclerosis in the partial ligation model of mouse atherosclerosis, compared to the control group demonstrating the specific effect of the nanoparticle approach as a targeted therapeutic method.
Interestingly, the mechanism of biogenesis for miR-712 (murine-specific miRNA) and miR-663 (primate-specific miRNAs) also share a familiar yet unexpected source, the internal transcribed spacer region of pre-ribosomal RNA (RN45s) gene [108], suggesting that mechanism of generation of flow-sensitive microRNAs is conserved in multiple species. It is interesting to note that two of the most disturbed flow/OS-induced miRNAs (miR-712 in mice and miR-663 in human ECs) are generated from this unusual, atypical source, especially what is considered to be a rubbish portion of RN45s region [108]. The pathobiological implication of this unusual biogenesis of this flow-sensitive miR-712 and miR-663 from the interspace translational regions of the ribosomal RNA gene is unknown at present. These miRNAs may be used as biomarkers and therapeutic candidates for atherosclerosis.
6. Other flow-sensitive microRNAs
Some flow-sensitive microRNAs, such as miR-21, and 155, have been implicated in both anti- and pro-atherogenic responses. This may reflect the fact that a single miRNA can target numerous target mRNAs, some of which mediate pro-atherogenic responses in one cell type or organ, while others act oppositely in different cell types, tissues or in various context-dependent manner. Therefore, the overall response of miRs in the whole body in vivo is likely to depend on cellular context, cell type, and environment [87].
6.1. miR-21
MiR-21, a flow-sensitive miRNA, has been extensively studied for its role in atherosclerosis. MiR-21 was initially shown to be upregulated by LS in HUVECs [111]. However, other study showed that the proatherogenic OS upregulated miR-21, which targeted peroxisome-proliferator-activated receptor α (PPARα) leading to the enhanced expression of the pro-inflammatory VCAM-1 [112]. Consistent with its role as a pro-atherogenic miRNA, miR-21 is up-regulated in human atherosclerotic plaques [113], arterial endothelium exposed to d-flow in the mouse PCL model [21,114], and in peripheral blood mononuclear cells from patients with coronary heart disease [115]. In coronary heart disease patients, miR-21 level negatively correlated with the number of circulating regulatory T (Treg) cells [115], which is known to play a protective role against atherosclerosis. Interestingly, miR-21 is induced by high glucose in macrophages and targets programmed cell death 4 (PDCD4), reducing macrophage apoptosis. Also, miR-21 is upregulated in tissues from patients with abdominal aortic aneurysm [116], serum from patients with cerebrovascular disease [117], and may also serve as a marker of plaque stability [63].
MiR-21 was the first miRNA shown to regulate VSMC growth and survival by silencing expression of phosphatase and tensin homolog (PTEN) and increased expression of B-cell leukemia/lymphoma 2 (BCL2), ultimately promoting cell survival and proliferation [118–120]. Further, it was shown that inhibition of PTEN by miR-21 upregulates AKT signaling and protects against ischemia-reperfusion and hypoxia-reperfusion-induced cardiomyocyte apoptosis, giving its clinical relevance [121]. Also, trimetazidine, an anti-ischemic and antioxidant agent, was shown to prevent the ischemia/reperfusion-induced cardiomyocyte apoptosis by up-regulating miR-21 [122]. Together, this evidence suggests that miR-21 seems to be upregulated by disturbed flow/OS and mostly plays a pro-atherogenic role in ECs. However, it also appears to serve a protective role for injured or diseased VSMCs and cardiomyocytes, highlighting the complex roles of miR-21 in cardiovascular pathobiology.
6.2. miR-155
MiR-155, a flow-sensitive miR, has been extensively studied in atherosclerosis and coronary artery disease, but its role has been reported as both pro- and anti-atherogenic. Initially, miR-155 expression was found to be abundantly expressed in the intima of the thoracic aorta, which is naturally exposed to s-flow in vivo [123]. It was also found to be increased by LS in HUVECs; suggesting it may be an anti-atherogenic flow-sensitive miRNA [123]. It was further shown to inhibit NF-kB signaling in HUVECs by targeting p65 under inflammatory conditions [124]. Consistent with this idea, hematopoietic deficiency of miR-155 induced atherosclerosis and decreased plaque stability by increasing myeloid inflammatory cell recruitment to the plaque regions [125]. Furthermore, miR-155 was shown to prevent pro-inflammatory signaling in macrophages, and dendritic cells via inhibition of MAPK310, a member of the pro-inflammatory Mitogen-Activated Protein-Kinase (MAPK) signaling pathway [126].
However, in contrast, other studies showed evidence suggesting that miR-155 mediates pro-atherogenic responses [127,128]. MiR-155 was shown to directly target eNOS mRNA in HUVECs and impair endothelium-dependent vascular relaxation in human arteries [129]. Leukocyte-specific miR-155 directly repressed expression of negative regulators of pro-inflammatory cytokine signaling such as B-cell Lymphoma 6 (BCL6), Suppressor of cytokine signaling (SOCS1) [86], and Src homology 2 domain-containing inositol-5-phosphatase (SHIP-1) [130,131]. Another study showed that genetic knockdown of miR-155 ameliorated atherogenesis in ApoE−/− mice by reducing inflammatory responses of macrophages and increasing macrophage cholesterol efflux [128]. Similar to miR-19a, miR-155 also promotes foam cell formation by targeting HMGB1 in macrophages [132]. Also, tissue-specific genetic knockdown of miR-155 in bone marrow-derived cells suppressed atherogenesis in ApoE−/− mice [128].
MiR-155 appears to play the pro-inflammatory role in macrophages but the anti-inflammatory role in ECs. It is interesting to note that miR-155 is induced by pro-inflammatory stimuli, such as TNFα [133] and oxidized LDL [132,134]. In these cases, it may function in a negative feedback loop to mitigate inflammation from pro-inflammatory stimuli. A recent study further showed that miR-155 targets calcium-regulated heat stable protein 1 (CARHSP1), which regulates the stability of TNFα [135], further reinforcing the role of miR-155 as a negative feedback regulator. Together, these conflicting results suggest that miR-155 has differential effects on atherosclerosis depending on the specific cell-types and pathobiological contexts. Cell-type and context-specific modulation of miR-155 by using appropriated targeted delivery methods [110,136] may be useful to overcome this conundrum.
Figure 1 shows a summary of the role of miRNAs in atherosclerosis
7. Other microRNA categories relevant to vascular dysfunction
There are other categories of miRNAs that are crucial for vascular dysfunction and atherosclerosis including but not limited to inflammation and NFκB regulators, lipid metabolism regulators, endothelial-to-mesenchymal regulators, TGFβ signaling, VEGF signaling and hypoxia- and ROS-related signaling. These miRNAs have been extensively reviewed elsewhere (for additional reviews on these and related subjects see references [124, 137–143].
8. Long non-coding RNAs (LncRNAs) in Atherosclerosis
The lncRNAs are a group of non-coding RNAs with a length of more than 200 nucleotides that play a role in the regulation of gene expression at the post-transcriptional, transcriptional, and chromatin levels [144,145]. Recently first flow-sensitive and endothelial-enriched lncRNA, STEEL, was reported [35], opening the possibility that there would be many other flow-sensitive lncRNAs that play essential roles in vascular pathobiology to be discovered. Given its relative paucity of data on flow-sensitive lncRNAs, here we review the emerging roles of other lncRNAs that have been studied in vascular dysfunction and atherosclerosis [146,147]. We also noted that Chen et al. performed a lncRNA and mRNA array study using HUVECs exposed to low shear stress (2 dyn/cm2) for a short duration (2 hours) [148]. Although the study reported 149 differentially expressed lncRNAs by the brief shear exposure, the findings from the study needs to be further validated.
8.1. STEEL
The spliced-transcript endothelial-enriched lncRNA (STEEL) is the first flow-sensitive lncRNA enriched in ECs. Recently, Man et al. showed that expression of STEEL is decreased by atheroprotective flow (steady LS) compared to the atheroprone flow (d-flow or OS) in a KLF2-dependent mechanism [35]. Functionally, they showed that endothelial expression of STEEL stimulated vessel network formation and maturation. Knockdown of STEEL decreased many shear sensitive genes including KLF2 and eNOS. Interestingly, however, overexpression of KLF2 downregulated STEEL expression, suggesting the feedback inhibition of STEEL by KLF2. Additional studies using chromatin immunoprecipation showed that STEEL is enriched in the nucleus where it binds to PARP1 at the KLF2 and eNOS genomic loci, suggesting its epigenetic regulatory mechanism [35]. Although the role of STEEL in atherosclerosis is yet to be studied, this finding opens the possibility that there may be many other flow-dependent lncRNAs waiting to be discovered for their roles in cardiovascular pathobiology.
8.2. LASSIE
A KLF2-dependent lncRNA Lassie (lncRNA activated by shear stress in the endothelium) was reported to be increased by shear stress in HUVECs [149]. This abstract reported that Lassie improved survival and angiogenic potential of endothelial cells; however, further studies are needed to validate the report and molecular mechanisms.
8.3. SENCR
SENCR was discovered as a new vascular cell-enriched lncRNA by Miano and colleagues by an RNA-Seq study [150,151]. It is abundantly expressed in both endothelial and smooth muscle cells, and no orthologues have been identified outside of human/chimp lineages. Studies using the SECNR knockdown approach in human coronary artery smooth muscle cells (HCASMCs) showed that its target genes include Myocardin, a critical contractile transcriptional factor, which in turn stabilizes contractile phenotype while inhibiting motile responses [150]. In embryonic stem cells, SENCR stimulates their commitment to ECs during development [151]. In HUVECs, SENCR increases angiogenic responses, which may be mediated by downregulating expression of migratory and angiogenic genes such as CCL5, CEACAM1, and CX3CL1, suggesting them as its target genes [151]. They further showed that SENCR expression was significantly reduced in EC isolated from patients with coronary artery disease compared to the healthy subjects [151], indicating its implication in atherosclerosis.
8.4. LEENE
An enhancer-associated lncRNA enhances eNOS expression (LEENE) has been shown to be co-regulated with eNOS. Mechanistically, LEENE facilitates the recruitment of RNA Pol II to the eNOS promoter to enhance eNOS transcription and regulates eNOS expression and EC function [152].
8.5. LOC100129973
This lncRNA has been shown to suppress apoptosis of vascular ECs. It was discovered as the most significantly upregulated lncRNA in a gene array study [153]. It is highly abundant in HUVECs, and its knockdown inhibits apoptosis [153]. LOC100129973 acted as a sponge for miRNA, thus suppressing the availability of target miRNAs. It has binding sites for miR-4707-5p and miR-4767, resulting in a decrease in these miR levels. MiR-4707-5p and miR-4767 promote apoptosis by downregulating two apoptosis inhibitors API5 and BCL2L12, which were inhibited by overexpression of LOC100129973 [153].
8.6. MEG3
EG3 was initially identified as a tumor suppressor gene in cancer [154], but its role in angiogenesis and atherosclerosis needs further clarification. Studies using human microvascular endothelial cells (HMEC-1) showed that Meg3 silencing induces angiogenic responses. Meg3 was shown to regulate vascular biology in part by targeting miR-21 and Notch signaling pathway [155]. Overexpression of Meg3 reduced miR-21 and its known target genes RhoB and PTEN, which led to the inhibition of endothelial cell migration and proliferation [155]. This study also showed that Meg3 negatively regulated the notch signaling pathway. Also, MEG3 was found to be downregulated in coronary artery disease tissues compared to healthy controls [155].
8.7. MALAT1
MALAT1 is a prominent lncRNA studied in the context of several pathologies including atherosclerosis, cancer, diabetes, and stroke. It is among the most abundant lncRNA transcripts in endothelial cells. Silencing of MALAT1 using GapmeRs induced pro-migratory phenotype in endothelium in vitro while decreasing endothelial cell proliferation [156]. Also, MALAT1 suppression led to a decrease in angiogenesis and capillary development (attributed to alteration of several cell cycle regulators in endothelial cells) in a hind limb ischemia mouse model. MALAT1 levels were reported to be lower in atherosclerotic plaques than in healthy vasculature [157]. A proposed mechanism for the protective mechanism of MALAT1 for the endothelium involves the elevated expression of MALAT1 in response to ox-LDL. MALAT1 then sponges miR-22-3p, which de-represses (upregulates) the expression of its target genes CXCR2 and AKT. This higher level of CXCR2 was shown to resist ox-LDL induced endothelial cell dysfunction via the AKT pathway [158]. In contrast, however, overexpression of MALAT1 was linked to increased inflammation and endothelial dysfunction in diabetic conditions [159], suggesting that inhibition of MALAT1 as a potential therapy. Therefore, the role of MALAT1 in atherosclerosis remains to be further determined.
8.8. LISPR1
A novel lncRNA LISPR1 has been recently reported to regulate S1P signaling pathway [160]. Interestingly, this lncRNA has been shown to be sensitive to shear stress in endothelium. The findings from the study reveal that laminar flow upregulates the lncRNA LISPR1 which enhances S1PR1 expression suggesting a vital role for the flow-sensitive lncRNA in endothelial biology and atherosclerosis [160].
8.9. H19
H19 is one of the first lncRNAs reported in smooth muscle cells and is implicated in vascular biology and atherosclerosis. H19 is expressed in embryonic vasculature stages and is subsequently suppressed in the adult vasculature [161]. Under stress conditions, such as atherosclerosis and vascular injury, expression of H19 is upregulated. A crucial development into understanding its function was the discovery of H19 modulation of the let-7 family of miRNA. H19 regulates let-7 miRNA expression by acting as a molecular sponge. Increased H19 decreases let-7g expression, which in turn led to attenuation of autophagy, apoptosis and reactive oxygen species [162]. Increased levels of H19 was associated with coronary artery disease in human population studies [163]. Moreover, H19 inhibition decreased endothelial growth by disrupting the cell cycle and also diminished angiogenic responses [34]. Similarly, ApoE−/− mice fed a Western diet express high levels of H19 [164]. Also, H19 was shown to promote atherosclerosis and inflammation via the MAPK and NF-kB signaling pathway [165].
8.10. LincRNA-p21
LincRNA-p21 is a VSMC-associated lncRNA that has also been implicated in atherosclerosis. LincRNA-p21 is significantly reduced in atherosclerotic plaques of ApoE−/− mice. Similarly, lincRNA-p21 levels are lower in coronary artery disease patients compared to controls [166,167]. In vivo lincRNA-p21 inhibition leads to neointimal hyperplasia in a carotid artery injury model. In VSMCs lincRNA-p21 was found to reduce cell proliferation and induce apoptosis. A genome-wide association study found this inhibition to correlate with the dysregulation of several p53 targets. LincRNA-p21 is a transcriptional target of p53, and also feeds forward and enhances p53 transcriptional activity via binding mouse double minute 2 (MDM2), an E3 ubiquitin-protein ligase. MDM2 reduces p53, so the ability of lincRNA-p21 to bind MDM2 allows for the release of additional p53 [166,167].
8.11. MIAT
MIAT is a conserved lncRNA implicated in several diseases including diabetic retinopathy, paranoid schizophrenia, myocardial infarction, and atherosclerosis. MIAT is highly expressed in endothelial and smooth muscle cells. MIAT regulates endothelial cell proliferation, migration and tube formation in vitro in rat retinal endothelial cells (RF/6A cells) [168,169]. In vivo MIAT knockdown improved diabetes-induced microvascular pathology. Also MIAT has been found to be highly upregulated in carotid atherosclerotic plaques compared to iliac artery controls [168,169]. Knockdown of MIAT via GapmeRs found decreased proliferation and migration of HCASMCs as well as increased apoptosis. MIAT inhibition reduced oxLDL uptake of murine peritoneal macrophages as well as human monocyte-differentiated macrophages in vitro. In clinical samples, a negative correlation between MIAT and miR-181b was found, MIAT expression was increased while miR-181b was decreased in atherosclerosis patients’ serum as well as in human coronary artery smooth muscle cells exposed to oxidized-LDL [168,169]. MIAT increased cell proliferation and cell cycle progression while inhibiting apoptosis in vitro. This effect was reduced by treatment with miR-181b. MIAT increased STAT3 expression by acting as a sponge for miR-181b. One mechanism of miR-181b’s reduction of cell growth via cell cycle arrest and increased apoptosis is via targeting STAT3 [169].
8.12. ANRIL
ANRIL is a lncRNA that has been previously associated with acute myocardial infarction and is an established vascular smooth muscle lncRNA (also referred to as CDKN2B-AS1) [170–172]. ANRIL has been linked to several cancers as well as coronary artery disease. This lncRNA slows cell cycle gene expression via the polycomb repressor complex. HCASMCs from patients with SNPs in the ANRIL locus have increased cell proliferation [173]. Another study found when comparing diabetic patients with, and without CAD, patients with CAD had significantly higher ANRIL expression in peripheral blood mononuclear cell samples [174].
8.13. MYOSLID
MYOSLID is a novel lncRNA discovered in VSMCs, but its role in ECs and atherosclerosis is unknown. The lncRNA was identified by using RNAseq in MYOCD overexpressing HCASMCs to find lncRNAs regulated by this group of proliferation genes [175]. MYOSLID was shown to be VSMC selective and a direct transcriptional product of the TGFB/SMAD and MYOCD/SRF pathways. In addition, MYOSLID activates the VSMC contractile phenotype. MYOSLID was lower in failed human arteriovenous fistula samples from patients with the end-stage renal disease compared to healthy control replacement veins, potentially promoting VSMC de-differentiation due to lack of MYOSLID. Knockdown of MYOSLID in HCASMCs resulted in significant decrease of VSMC contractile gene expression even under TGFB-1 induced conditions. Reduction of MYOSLID also promoted migration of HCASMCs.
8.14. SMILR
SMILR is another novel lncRNA discovered in VSMCs [176]. SMILR was discovered by RNAseq on human saphenous vein VSMCs post stimulation by interleukin-1a and platelet-derived growth factor. SMILR was increased by the stimulation in both the nucleus and cytoplasm and was detected in the media. SMILR reduced HAS2 expression and SMILR knockdown resulted in significantly reduced cell proliferation. In human samples of atherosclerotic plaques and plasma, SMILR was found to be increased in unstable atherosclerotic plaques and in the plasma of patients with high plasma C-reactive protein [176,177].
8.15. RNCR3
The lncRNAs RNCR3 (retinal non-coding RNA 3) is differentially expressed during atherosclerosis. It is upregulated in atherosclerotic VSMCs and ECs compared with non-atherosclerotic tissue in mice as well as humans [178]. Compared with control mice, downregulation of RNCR3 with shRNA aggravated atherosclerosis in thoracic aorta tissue and increased inflammatory factors in plasma, suggesting it serves an atheroprotective role. These findings suggest that targeting RNCR3 might be an attractive therapeutic intervention for atherosclerosis.
Table 2 and Figure 1 show a summary of lncRNAs and its physiological effects on the vessel wall.
Table 2.
Role of lncRNAs in vascular dysfunction and atherosclerosis
| LncRNA | Major Targets | Indirect targets & Signaling pathway | Pathophysiological Effects | Ref. |
|---|---|---|---|---|
| STEEL | eNOS, KLF2 | Regulates EC cell response to shear stress | [35] | |
| LASSIE | unknown | Angiogenesis, Cell survival | [149] | |
| SENCR | MYOCD? | CCL5, CEACAM1, and CX3CL1 (migratory and angiogenic genes) | Promotes migration, proliferation and tube formation of endothelial cells | [150,151] |
| LEENE | eNOS | Regulates eNOS expression and EC function | [152] | |
| LOC100129973 |
miR-4707-5p mir-4767 |
API5 and BCL2L12 | Suppress apoptosis of endothelial cells | [153] |
| MEG3 | miR-21 | RhoB and PTEN | Suppresses migration, proliferation and tube formation of endothelial cells | [155,231,232] |
| MALAT1 | miR-22-3p | CXCR2 and AKT, AKT pathway | Migratory behavior, increases AKT pathway behavior in endothelial cells | [155,156,158,233–238] |
| LISPR1 | S1PR1 | S1P signaling pathway | Angiogenesis, vascular stability and permeability | [160] |
| H19 | Let-7 miRNA family | MAPK and NF-kβ pathways | Reduces autophagy, apoptosis and reactive oxygen species in endothelial cells | [162,163,165,239–241] |
| LincRNA-p21 | MDM2 | p53 feed forward loop | Reduces cell proliferation and increases apoptosis | [167] |
| MIAT | miR-181b | STAT3 | Increases cell proliferation and cell cycle progression, inhibits apoptosis | [168,169,242] |
| ANRIL | Polycomb repressor complex | Slows cell cycle gene expression | [170,174,243–245] | |
| MYOSLID | unknown | TGFβ/SMAD, MYOCD/SRF pathways | Activates VSMC contractile phenotype | [175] |
| SMILR | unknown | HAS2 | VSMC proliferation | [177] |
| RNCR3 | unknown | Reduces inflammation in plasma, atheroprotective | [178] |
lncRNAs, long non-coding RNAs; VSMC, vascular smooth muscle cell; API5, Apoptosis inhibitor 5; BCL2L12, Bcl-2-like protein 12; CCL5, C-C motif chemokine 5 ligand 5; CEACAM1, Carcinoembryonic antigen-related cell adhesion molecule 1; CX3CL1, chemokine (C-X3-C motif); PTEN, Phosphatase And Tensin Homolog; MAPK, Mitogen-activated protein kinase; NF-kβ, Nuclear factor-κB; CXCR2, C-X-C chemokine receptor type 2; STAT3, Signal transducer and activator of transcription 3; TGFβ, Transforming growth factor β; MYOCD, Myocardin; SRF, Serum response factor; HAS2, Hyaluronan Synthase 2; MDM2, Mouse double minute 2 homolog; AKT/PKB, Protein kinase B.
9. Circulating miRNAs as Biomarkers for Atherosclerosis
A biomarker, or biological marker, typically refers to a quantifiable indicator of a biological state or pathological condition [179]. Circulating miRNAs have tremendous potential as a disease biomarkers as differential plasma miRNA levels have been described for many pathophysiological conditions, including atherosclerosis [180]. Here, we will review those studies that have shown evidence suggesting diagnostic and prognostic potentials for one or a few circulating miRNAs as specific biomarkers of atherosclerosis. Since multiple miRNAs are also secreted into the circulation, these can be used as potential circulating biomarkers. For example, miR-146a levels are increased in the serum of patients with acute coronary syndrome [181]. The peripheral blood levels of miR-92a, miR-126, and miR-222 in this study were markedly decreased in both atherosclerotic and pre-atherosclerotic patients compared to healthy controls, although the decease of miR-92a and miR-222 in pre-atherosclerotic patients was not as significant as that in atherosclerotic patients [181]. In patients with acute myocardial infarction (MI), circulating levels of muscle-enriched miRNAs (miR-1, −133a, and 499) and the cardiac-specific miR-208a were significantly increased in human patients [182]. This study especially suggested miR-208a as an early detection marker of acute MI. MiR-221/22 levels decrease while miR-21 and miR-130a increase in the plasma patients with peripheral arterial disease [183]. The opposite effects of miR-221/222 on the proliferation, migration, and apoptosis of endothelial cells and vascular smooth muscle cells may have significant therapeutic implications in many vascular diseases such as atherosclerosis and restenosis following angioplasty [184]. Interestingly, the plasma level of liver-specific miR-122 was increased significantly after acute MI or cardiogenic shock [185,186], suggesting their potential as a biomarker.
Circulating extracellular miRNAs are carried in various ways including exosomes, microvesicles, and complexes bound to low and high density lipoprotein particles as well as protein complexes such as AGO2-miRNAs [178,187–189]. These circulating miRNAs are biologically active, altering gene expression in recipient cells [190]. Therefore, miRNAs are considered as newly described forms of intercellular communication. For example, as discussed above, extracellular vesicles or exosomes enriched with miR-143/145, which are secreted from HUVECs in response to shear stress, can be taken up by VSMCs where the miRNAs regulate their target genes [191]. Conversely, ECs can be targeted by exogenous miRNAs secreted by other cells such as blood monocytes. For example, miR-150 containing microvesicles released from monocytes from atherosclerotic patients was shown to increase the proliferation of endothelial cells [192]. Similarly, apoptotic bodies enriched in miR-126 can be taken by HUVECs and promote atherosclerosis regression by inducing CXCL12 expression through CXCR4 [193]. There is a tremendous interest to understand the mechanisms underlying miRNA incorporation into exosomes, delivery, and targeting and recognition machinery.
It is important to note that circulating miRNAs will typically have systemic effects regulating many cells and tissues and are not likely to be specific for atherosclerosis. For example, miR-92a, miR-126, and miR-222 are not specific only for cardiovascular diseases. The levels of miR-92 also increase in the context of other diseases such as hypertension [194], colorectal cancer [195], and have been shown to play an essential role in diagnosis and prognosis of other diseases [196]. Similarly, upregulated levels of miR-126 are also associated with an immune imbalance in children with acute asthma [197], and exosomal miR-126 can be used as a circulating biomarker in non-small-cell lung cancer [198] and hepatocellular carcinoma [199]. Likewise, the potential of miR-222 is currently being explored as a biomarker in various diseases including cancer and inflammatory diseases [200–204]. Although many studies have shown one or a few circulating miRNAs as specific biomarkers of atherosclerosis, it is likely that a panel of circulating miRNAs instead of one or a few miRNAs, combined with other better-established biomarkers may be of better diagnostic and prognostic value. Also, specific and systematic large-scale clinical studies will be required to identify the real potential of these circulating miRNAs as diagnostic and prognostic biomarkers
The role of lncRNAs as potential biomarkers in cardiovascular disease especially in atherosclerosis is still in its infancy, and much work remains to be done. In diabetic patients, circulating levels of lncRNA predicting cardiac remodeling (LIPCAR) were associated with diastolic function [163]. Similarly, circulating levels of MIAT and SENCR were directly associated with left ventricular mass to the left ventricular end-diastolic volume ratio, a well-known cardiac remodeling marker suggesting that lncRNAs are independent predictors of diastolic function and cardiac remodeling [205]. Although a large number of studies suggest that circulating ncRNAs promise to serve as a minimally invasive diagnostic and prognostic biomarker for various types of cardiovascular disease, the lack of consistency in these studies has been a significant concern in this field [206–215]. Therefore, reliable, accurate, and sensitive detection of circulating ncRNAs and validation is a prerequisite for their use as a biomarker in clinical applications. Several methodological factors including sample collection and processing, as well as assay performance and ncRNA quantification, can influence the quality of the resulting data and need critical considerations. It is important to note that platelets and platelet microvesicles contain significant amounts of microRNAs and efficiently contribute to the pool of circulating miRNAs. These platelet-related miRNAs such as miR-223, miR-126, miR-197, miR-191, miR-21, miR-150, miR-155, miR-140, miR-96, miR-98 are a potential source of confusion in biomarker assays and need to be carefully excluded and analyzed [216,217]. Despite these potential technical limitations, It is an exciting future direction to determine the correlation between circulating levels of lncRNAs and atherosclerosis as well as the underlying mechanisms by which they are carried in circulation and delivered to target cells. These studies would reveal the potential of the circulating lncRNAs as diagnostic biomarkers and therapeutic targets.
10. MiRNAs and lncRNAs as therapeutic targets in atherosclerosis
Since multiple miRNAs and lncRNAs are involved in the regulation of several vital processes in every stage of atherosclerosis initiation and progression, and perhaps regression, modulation of miRNA expression could have beneficial effects in prevention, reduction, and regression of atherosclerosis. Both the gain-of-function and loss-of-function approaches are used to examine the role of miRNAs as anti-atherogenic therapeutics. MicroRNA overexpression (gain-of-function) has been used as an anti-atherogenic therapeutic strategy. MiR-145 overexpression in VSMCs promotes a reduction in atherosclerotic plaque size in the most common sites of plaque formation such as aortic sinuses, ascending aortas, and brachiocephalic arteries [218]. Furthermore, miR-145 enhances plaque stability through an increase in the number of VSMCs, collagen content and fibrous cap area associated with a decrease in the number of macrophages and necrotic area. In contrast, suppression of pro-atherogenic miRNAs is also of potential therapeutic strategy for atherosclerosis. Several studies used antagomiRs or anti-miRs (synthetic antisense oligonucleotides) to lower the expression of proatherogenic miRNAs, such as miR-712, miR-205, miR-92a, and miR-33 have demonstrated efficacy as anti-atherogenic therapies [97,99,108,219,220]. With the advancements in the synthesis of modified oligonucleotide technology, these miRNAs can be targeted with either miRNA mimics to increase repression of target genes or miRNA inhibitors (anti-miRs or antagomiRs) to prevent miRNA repressing its target gene(s). These approaches have shown great promise as potential therapies, and some of them are in clinical phase trial stages of drug development [221–223].
Although a growing number of lncRNAs are implicated in vascular function [33,140,176], it is unclear how they participate in the pathophysiological processes. Their potential as therapeutic targets has often been raised, and there are a few compelling examples of in vivo modulation of lncRNAs. However, modulating lncRNAs has been a challenging task to date. This is further complicated by the fact that many of the lncRNAs are not conserved between mice and humans. The widespread use of RNA-Sequencing methods reveals a rapidly expanding a number of potential new candidates for therapy. LncRNAs potentially represent a powerful tool for personalized medicine due to their specific expression patterns associated with distinct pathologies. There remain several limitations and challenges that need to be addressed before lncRNAs can genuinely reach the clinical application. Foremost is target specificity, given the pleiotropic implications of a single lncRNA in pathophysiological processes. Although lncRNAs may show dysregulation specific to certain diseases, they exhibit various functions in the organism and some lncRNAs may act through more than one mechanism [224]. Second, the low conservation of lncRNAs across evolution makes both the identification of human lncRNAs and their clinical testing challenges, because rodents may not be an adequate model for these studies.
A list of important datamining tools for miRNA and lncRNA research has been provided in Table 3
Table 3.
Important Data mining tools for miRNA/lncRNA research
| miRNA datamining tools/web servers | ||
|---|---|---|
| Name | Description | References |
| miRBase | A searchable database of published miRNA sequences and annotation. | [246–249] |
| miRTarBase | A searchable database of experimentally validated microRNA-target interactions | [250,251] |
| StarScan | StarScan is developed for scanning small RNA (miRNA, piRNA, siRNA) mediated RNA cleavage events in lncRNA, circRNA, mRNA and pseudo genes from degradome sequencing data. | [252] |
| TargetScan | Prediction program webserver that enlists biological targets of miRNAs by searching for the presence of binding sites that match the seed region of each miRNA. | [253] |
| TarBase | A comprehensive database of experimentally supported animal microRNA targets | [254] |
| Diana-microT | An algorithm based on several parameters provides conserved and non-conserved microRNA targets | [255–257] |
| miRwalk | Aggregates and compare results from other miRNA-to-mRNA databases | [258,259] |
| LncRNA data mining tools/web servers | ||
| deepBase | Identification, expression, evolution and function of lncRNAs from deep RNA sequencing data | [260–262] |
| LNCipedia | A compendium of human long non-coding RNAs | [263,264] |
| lncRNAdb | The reference database for functional lncRNAs | [265,266] |
11. Summary and perspectives
Here, we summarized the current knowledge of flow-sensitive miRNAs and lncRNAs, as well as their essential roles in the regulation of endothelial function and atherosclerosis. These studies demonstrate that lncRNAs and flow-sensitive microRNAs are crucial mediators of endothelial function and atherosclerosis. However, our knowledge of these noncoding RNAs, especially lncRNAs, their regulation, and their functions is still in its infancy and further investigation is warranted to identify other RNAs and explore their biological roles, mechanisms, and potential therapeutic applications. There have been some promising results from recent Phase II clinical trials indicating the safety, feasibility, and therapeutic potential of anti-miRs for the treatment of diseases.
In animal studies, some anti-miRs targeting flow-sensitive microRNAs such as miR-712, miR-205, or miR-155, have demonstrated their potential as anti-atherogenic therapies. This encourages the development of miRNA therapeutics for the treatment of atherosclerosis in humans. Despite these encouraging results, the development of miRNA therapeutics must overcome several significant challenges. First, there is a significant need to identify more athero-miRs (miRNAs with consistent pro- or anti-atherogenic effects) that could be used as reliable and safe anti-atherogenic therapeutic targets. Second, there is an urgent need to develop better and more specific miRNA modulators. The current methods to inhibit miRNAs include anti-miR/antagomiR and miR-sponge, both of which directly bind to miRNAs, thereby affecting all of their target genes (~ hundreds of genes) indiscriminately, potentially causing undesirable effects. Therefore, to minimize the potentially undesirable effects of anti-miR, antago-miR, or miR-sponge treatment, better strategies should be developed to deliver these inhibitors. These include targeted delivery of miRNA therapeutics specifically to cells and tissues of interest with minimal delivery to non-target cells to lower potential side effects. For example, we showed that anti-miR-712 could be delivered to inflamed endothelial cells by targeting VCAM1 on the endothelial cells by using nanoparticles coated with the VCAM1-targeting peptide [110]. Also, designing more specific inhibitors such as target site blockers that can specifically block a unique and desired mRNA-miRNA interaction without affecting the expression of off-target genes [225].
In summary, ncRNAs robustly regulate vascular physiology and pathophysiology. These non-coding RNAs also regulate several aspects of atherosclerosis including vascular dysfunction and inflammation and lipid metabolism. Although many lncRNAs have been identified til date, only a few of them have been studied for their association with atherosclerosis. Further advancements in the field of lncRNAs research will not only lead to identification of newer molecular pathways leading to disease process but also provide improved therapeutic candidates for targeting atherosclerosis.
Acknowledgments
This work was supported by funding from the National Institutes of Health grants HL119798 and HL095070 to HJ. HJ is John and Jan Portman Professor and would like to dedicate this paper in memory of late Mr. Portman for his generous support for the endowed Professorship.
Sources of funding
NIH grants HL095070, HL114772, and HL113451 to HJ.
Footnotes
Disclosures
None.
References
- [1].Geovanini GR, Libby P, Atherosclerosis and inflammation: overview and updates, Clin Sci (Lond) 132 (12) (2018) 1243–1252. [DOI] [PubMed] [Google Scholar]
- [2].Lusis AJ, Atherosclerosis 407 (6801) (2000) 233–241 Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Libby P, Coronary artery injury and the biology of atherosclerosis: inflammation, thrombosis, and stabilization, Am J Cardiol 86 (8B) (2000) 3J–8J discussion 8J-9J. [DOI] [PubMed] [Google Scholar]
- [4].Gimbrone MA Jr., Garcia-Cardena G, Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis, Circ Res 118 (4) (2016) 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chiu JJ, Chien S, Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives, Physiological reviews 91 (1) (2011) 327–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Heo K-S, Fujiwara K, Abe J, Disturbed-Flow-Mediated Vascular Reactive Oxygen Species Induce Endothelial Dysfunction, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Steinman DA, Taylor CA, Flow imaging and computing: large artery hemodynamics, Ann Biomed Eng 33 (12) (2005) 1704–1709. [DOI] [PubMed] [Google Scholar]
- [8].Davies PF, Polacek DC, Handen JS, Helmke BP, DePaola N, A spatial approach to transcriptional profiling: mechanotransduction and the focal origin of atherosclerosis, Trends Biotechnol 17 (9) (1999) 347–351. [DOI] [PubMed] [Google Scholar]
- [9].Davies PF, Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology, Nature clinical practice. Cardiovascular medicine 6 (1) (2009) 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao G, DeLisser H, Schwartz MA, A mechanosensory complex that mediates the endothelial cell response to fluid shear stress, Nature 437 (7057) (2005) 426–431. [DOI] [PubMed] [Google Scholar]
- [11].Simmons RD, Kumar S, Jo H, The role of endothelial mechanosensitive genes in atherosclerosis and omics approaches, Arch Biochem Biophys 591 (2016) 111–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kumar S, Kim CW, Son DJ, Ni CW, Jo H, Flow-dependent regulation of genome-wide mRNA and microRNA expression in endothelial cells in vivo, Scientific data 1 (2014) 140039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kumar S, Kim CW, Simmons RD, Jo H, Role of flow-sensitive microRNAs in endothelial dysfunction and atherosclerosis: mechanosensitive athero-miRs, Arterioscler Thromb Vasc Biol 34 (10) (2014) 2206–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Simmons RD, Kumar S, Thabet SR, Sur S, Jo H, Omics-based approaches to understand mechanosensitive endothelial biology and atherosclerosis, Wiley Interdiscip Rev Syst Biol Med 8 (5) (2016) 378–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dunn J, Thabet S, Jo H, Flow-Dependent Epigenetic DNA Methylation in Endothelial Gene Expression and Atherosclerosis, Arterioscler Thromb Vasc Biol 35 (7) (2015) 1562–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, Jang I, Son DJ, Kim D, Pan C, Fan Y, Jordan IK, Jo H, Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis, J Clin Invest 124 (7) (2014) 3187–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ni CW, Qiu H, Rezvan A, Kwon K, Nam D, Son DJ, Visvader JE, Jo H, Discovery of novel mechanosensitive genes in vivo using mouse carotid artery endothelium exposed to disturbed flow, Blood 116 (15) (2010) e66–e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Boo YC, Jo H, Flow-dependent regulation of endothelial nitric oxide synthase: role of protein kinases, American journal of physiology. Cell physiology 285 (3) (2003) C499–C508. [DOI] [PubMed] [Google Scholar]
- [19].van Thienen JV, Fledderus JO, Dekker RJ, Rohlena J, van Ijzendoorn GA, Kootstra NA, Pannekoek H, Horrevoets AJ, Shear stress sustains atheroprotective endothelial KLF2 expression more potently than statins through mRNA stabilization, Cardiovasc Res 72 (2) (2006) 231–240. [DOI] [PubMed] [Google Scholar]
- [20].Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK, Kruppel-like factor 4 regulates endothelial inflammation, J Biol Chem 282 (18) (2007) 13769–13779. [DOI] [PubMed] [Google Scholar]
- [21].Nam D, Ni CW, Rezvan A, Suo J, Budzyn K, Llanos A, Harrison D, Giddens D, Jo H, Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis, Am J Physiol Heart Circ Physiol 297 (4) (2009) H1535–H1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Magid R, Murphy TJ, Galis ZS, Expression of matrix metalloproteinase-9 in endothelial cells is differentially regulated by shear stress, Role of c-Myc, J Biol Chem 278 (35) (2003) 32994–32999. [DOI] [PubMed] [Google Scholar]
- [23].Chatzizisis YS, Coskun AU, Jonas M, Edelman ER, Feldman CL, Stone PH, Role of endothelial shear stress in the natural history of coronary atherosclerosis and vascular remodeling: molecular, cellular, and vascular behavior, Journal of the American College of Cardiology 49 (25) (2007) 2379–2393. [DOI] [PubMed] [Google Scholar]
- [24].Arroyo AG, Iruela-Arispe ML, Extracellular matrix, inflammation, and the angiogenic response, Cardiovasc Res 86 (2) (2010) 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kwak BR, Back M, Bochaton-Piallat ML, Caligiuri G, Daemen MJ, Davies PF, Hoefer IE, Holvoet P, Jo H, Krams R, Lehoux S, Monaco C, Steffens S, Virmani R, Weber C, Wentzel JJ, Evans PC, Biomechanical factors in atherosclerosis: mechanisms and clinical implications, Eur Heart J 35 (43) (2014) 3013–3020 (3020a-3020d). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tarbell JM, Shi ZD, Dunn J, Jo H, Fluid Mechanics, Arterial Disease, and Gene Expression, Annual review of fluid mechanics 46 (2014) 591–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Heo KS, Berk BC, Abe J, Disturbed Flow-Induced Endothelial Proatherogenic Signaling Via Regulating Post-Translational Modifications and Epigenetic Events, Antioxid Redox Signal 25 (7) (2016) 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Le NT, Sandhu UG, Quintana-Quezada RA, Hoang NM, Fujiwara K, Abe JI, Flow signaling and atherosclerosis, Cellular and molecular life sciences, CMLS 74 (10) (2017) 1835–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Davies PF, Endothelial transcriptome profiles in vivo in complex arterial flow fields, Ann Biomed Eng 36 (4) (2008) 563–570. [DOI] [PubMed] [Google Scholar]
- [30].Chiu JJ, Usami S, Chien S, Vascular endothelial responses to altered shear stress: pathologic implications for atherosclerosis, Annals of medicine 41 (1) (2009) 19–28. [DOI] [PubMed] [Google Scholar]
- [31].Bryan MT, Duckles H, Feng S, Hsiao ST, Kim HR, Serbanovic-Canic J, Evans PC, Mechanoresponsive networks controlling vascular inflammation, Arterioscler Thromb Vasc Biol 34 (10) (2014) 2199–2205. [DOI] [PubMed] [Google Scholar]
- [32].Abe J, Berk BC, Novel mechanisms of endothelial mechanotransduction, Arterioscler Thromb Vasc Biol 34 (11) (2014) 2378–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Simion V, Haemmig S, Feinberg MW, LncRNAs in vascular biology and disease, Vascul Pharmacol (2018), 10.1016/j.vph.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Voellenkle C, Garcia-Manteiga JM, Pedrotti S, Perfetti A, De Toma I, Da Silva D, Maimone B, Greco S, Fasanaro P, Creo P, Zaccagnini G, Gaetano C, Martelli F, Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing, Sci Rep 6 (2016) 24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Man HSJ, Sukumar AN, Lam GC, Turgeon PJ, Yan MS, Ku KH, Dubinsky MK, Ho JJD, Wang JJ, Das S, Mitchell N, Oettgen P, Sefton MV, Marsden PA, Angiogenic patterning by STEEL, an endothelial-enriched long noncoding RNA, Proceedings of the National Academy of Sciences of the United States of America 115 (10) (2018) 2401–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, Chun CZ, Samant GV, Zhao B, Garnaas MK, Horswill MA, Stanhope SA, North PE, Miao RQ, Wilkinson GA, Affolter M, Ramchandran R, A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo, Blood 115 (1) (2010) 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chowdhury TA, Koceja C, Eisa-Beygi S, Kleinstiver BP, Kumar SN, Lin CW, Li K, Prabhudesai S, Joung JK, Ramchandran R, Temporal and Spatial Post-Transcriptional Regulation of Zebrafish tie1 mRNA by Long Noncoding RNA During Brain Vascular Assembly, Arterioscler Thromb Vasc Biol 38 (7) (2018) 1562–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cheng HS, Njock MS, Khyzha N, Dang LT, Fish JE, Noncoding RNAs regulate NF-kappaB signaling to modulate blood vessel inflammation, Front Genet 5 (2014) 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bischoff FC, Werner A, John D, Boeckel JN, Melissari MT, Grote P, Glaser SF, Demolli S, Uchida S, Michalik KM, Meder B, Katus HA, Haas J, Chen W, Pullamsetti SS, Seeger W, Zeiher AM, Dimmeler S, Zehendner CM, Identification and Functional Characterization of Hypoxia-Induced Endoplasmic Reticulum Stress Regulating lncRNA (HypERlnc) in Pericytes, Circ Res 121 (4) (2017) 368–375. [DOI] [PubMed] [Google Scholar]
- [40].Neumann P, Jae N, Knau A, Glaser SF, Fouani Y, Rossbach O, Kruger M, John D, Bindereif A, Grote P, Boon RA, Dimmeler S, The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2, Nat Commun 9 (1) (2018) 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bartel DP, MicroRNAs: target recognition and regulatory functions, Cell 136 (2) (2009) 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Siomi H, Siomi MC, Posttranscriptional regulation of microRNA biogenesis in animals, Mol Cell 38 (3) (2010) 323–332. [DOI] [PubMed] [Google Scholar]
- [43].Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN, Posttranscriptional crossregulation between Drosha and DGCR8, Cell 136 (1) (2009) 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yeom KH, Lee Y, Han J, Suh MR, Kim VN, Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing, Nucleic Acids Res 34 (16) (2006) 4622–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN, Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex, Cell 125 (5) (2006) 887–901. [DOI] [PubMed] [Google Scholar]
- [46].Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN, The Drosha-DGCR8 complex in primary microRNA processing, Genes Dev 18 (24) (2004) 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim VN, MicroRNA precursors in motion: exportin-5 mediates their nuclear export, Trends Cell Biol 14 (4) (2004) 156–159. [DOI] [PubMed] [Google Scholar]
- [48].Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U, Nuclear export of microRNA precursors, Science 303 (5654) (2004) 95–98. [DOI] [PubMed] [Google Scholar]
- [49].Leuschner PJ, Martinez J, In vitro analysis of microRNA processing using recombinant Dicer and cytoplasmic extracts of HeLa cells, Methods 43 (2) (2007) 105–109. [DOI] [PubMed] [Google Scholar]
- [50].Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T, A uniform system for microRNA annotation, RNA 9 (3) (2003) 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R, Control of translation and mRNA degradation by miRNAs and siRNAs, Genes Dev 20 (5) (2006) 515–524. [DOI] [PubMed] [Google Scholar]
- [52].Saxena S, Jonsson ZO, Dutta A, Small RNAs with imperfect match to endogenous mRNA repress translation, Implications for off-target activity of small inhibitory RNA in mammalian cells, J Biol Chem 278 (45) (2003) 44312–44319. [DOI] [PubMed] [Google Scholar]
- [53].Hutvagner G, Zamore PD, A microRNA in a multiple-turnover RNAi enzyme complex, Science 297 (5589) (2002) 2056–2060. [DOI] [PubMed] [Google Scholar]
- [54].Lai EC, Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation, Nat Genet 30 (4) (2002) 363–364. [DOI] [PubMed] [Google Scholar]
- [55].Zhang C, MicroRNAs: role in cardiovascular biology and disease, Clin Sci (Lond) 114 (12) (2008) 699–706. [DOI] [PubMed] [Google Scholar]
- [56].Wu F, Yang Z, Li G, Role of specific microRNAs for endothelial function and angiogenesis, Biochem Biophys Res Commun 386 (4) (2009) 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Davis-Dusenbery BN, Wu C, Hata A, Micromanaging vascular smooth muscle cell differentiation and phenotypic modulation, Arterioscler Thromb Vasc Biol 31 (11) (2011) 2370–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Santoro MM, Fishing for endothelial microRNA functions and dysfunction, Vascul Pharmacol 55 (4) (2011) 60–68. [DOI] [PubMed] [Google Scholar]
- [59].Menghini R, Stohr R, Federici M, MicroRNAs in vascular aging and atherosclerosis, Ageing Res Rev 17 (2014) 68–78. [DOI] [PubMed] [Google Scholar]
- [60].Wojciechowska A, Braniewska A, Kozar-Kaminska K, MicroRNA in cardiovascular biology and disease, Advances in clinical and experimental medicine : official organ, Wroclaw Medical University 26 (5) (2017) 865–874. [DOI] [PubMed] [Google Scholar]
- [61].Mozos I, Malainer C, Horbanczuk J, Gug C, Stoian D, Luca CT, Atanasov AG, Inflammatory Markers for Arterial Stiffness in Cardiovascular Diseases, Frontiers in immunology 8 (2017) 1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kowara M, Cudnoch-Jedrzejewska A, Opolski G, Wlodarski P, MicroRNA regulation of extracellular matrix components in the process of atherosclerotic plaque destabilization, Clinical and experimental pharmacology & physiology 44 (7) (2017) 711–718. [DOI] [PubMed] [Google Scholar]
- [63].Koroleva IA, Nazarenko MS, Kucher AN, Role of microRNA in Development of Instability of Atherosclerotic Plaques, Biochemistry. Biokhimiia 82 (11) (2017) 1380–1390. [DOI] [PubMed] [Google Scholar]
- [64].Dlouha D, Hubacek JA, Regulatory RNAs and cardiovascular disease - with a special focus on circulating microRNAs, Physiological research 66 (tableementum 1) (2017) S21–S38. [DOI] [PubMed] [Google Scholar]
- [65].Fang Y, Shi C, Manduchi E, Civelek M, Davies PF, MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro, Proceedings of the National Academy of Sciences of the United States of America 107 (30) (2010) 13450–13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lee DY, Lin TE, Lee CI, Zhou J, Huang YH, Lee PL, Shih YT, Chien S, Chiu JJ, MicroRNA-10a is crucial for endothelial response to different flow patterns via interaction of retinoid acid receptors and histone deacetylases, Proceedings of the National Academy of Sciences of the United States of America 114 (8) (2017) 2072–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lee DY, Yang TL, Huang YH, Lee CI, Chen LJ, Shih YT, Wei SY, Wang WL, Wu CC, Chiu JJ, Induction of microRNA-10a using retinoic acid receptor-alpha and retinoid x receptor-alpha agonists inhibits atherosclerotic lesion formation, Atherosclerosis 271 (2018) 36–44. [DOI] [PubMed] [Google Scholar]
- [68].Qin X, Wang X, Wang Y, Tang Z, Cui Q, Xi J, Li YS, Chien S, Wang N, MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells, Proceedings of the National Academy of Sciences of the United States of America 107 (7) (2010) 3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang KC, Nguyen P, Weiss A, Yeh YT, Chien HS, Lee A, Teng D, Subramaniam S, Li YS, Chien S, MicroRNA-23b regulates cyclin-dependent kinase-activating kinase complex through cyclin H repression to modulate endothelial transcription and growth under flow, Arterioscler Thromb Vasc Biol 34 (7) (2014) 1437–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Iaconetti C, De Rosa S, Polimeni A, Sorrentino S, Gareri C, Carino A, Sabatino J, Colangelo M, Curcio A, Indolfi C, Down-regulation of miR-23b induces phenotypic switching of vascular smooth muscle cells in vitro and in vivo, Cardiovasc Res 107 (4) (2015) 522–533. [DOI] [PubMed] [Google Scholar]
- [71].Chen K, Fan W, Wang X, Ke X, Wu G, Hu C, MicroRNA-101 mediates the suppressive effect of laminar shear stress on mTOR expression in vascular endothelial cells, Biochem Biophys Res Commun 427 (1) (2012) 138–142. [DOI] [PubMed] [Google Scholar]
- [72].Kim JH, Lee KS, Lee DK, Kim J, Kwak SN, Ha KS, Choe J, Won MH, Cho BR, Jeoung D, Lee H, Kwon YG, Kim YM, Hypoxia-responsive microRNA-101 promotes angiogenesis via heme oxygenase-1/vascular endothelial growth factor axis by targeting cullin 3, Antioxid Redox Signal 21 (18) (2014) 2469–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhang N, Lei J, Lei H, Ruan X, Liu Q, Chen Y, Huang W, MicroRNA-101 overexpression by IL-6 and TNF-alpha inhibits cholesterol efflux by suppressing ATP-binding cassette transporter A1 expression, Experimental cell research 336 (1) (2015) 33–42. [DOI] [PubMed] [Google Scholar]
- [74].Wang KC, Garmire LX, Young A, Nguyen P, Trinh A, Subramaniam S, Wang N, Shyy JY, Li YS, Chien S, Role of microRNA-23b in flow-regulation of Rb phosphorylation and endothelial cell growth, Proceedings of the National Academy of Sciences of the United States of America 107 (7) (2010) 3234–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Demolli S, Doddaballapur A, Devraj K, Stark K, Manavski Y, Eckart A, Zehendner CM, Lucas T, Korff T, Hecker M, Massberg S, Liebner S, Kaluza D, Boon RA, Dimmeler S, Shear stress-regulated miR-27b controls pericyte recruitment by repressing SEMA6A and SEMA6D, Cardiovasc Res 113 (6) (2017) 681–691. [DOI] [PubMed] [Google Scholar]
- [76].Melo SA, Kalluri R, Angiogenesis is controlled by miR-27b associated with endothelial tip cells, Blood 119 (11) (2012) 2439–2440. [DOI] [PubMed] [Google Scholar]
- [77].Suzuki HI, Katsura A, Mihira H, Horie M, Saito A, Miyazono K, Regulation of TGF-beta-mediated endothelial-mesenchymal transition by microRNA-27, Journal of biochemistry 161 (5) (2017) 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zeng X, Huang C, Senavirathna L, Wang P, Liu L, miR-27b inhibits fibroblast activation via targeting TGFbeta signaling pathway, BMC cell biology 18 (1) (2017) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN, The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis, Dev Cell 15 (2) (2008) 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ, MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1, Proceedings of the National Academy of Sciences of the United States of America 105 (5) (2008) 1516–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhou J, Li YS, Nguyen P, Wang KC, Weiss A, Kuo YC, Chiu JJ, Shyy JY, Chien S, Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microRNA-126: role of shear stress, Circ Res 113 (1) (2013) 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C, MicroRNA-126–5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1, Nature medicine 20 (4) (2014) 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Tang F, Yang TL, MicroRNA-126 alleviates endothelial cells injury in atherosclerosis by restoring autophagic flux via inhibiting of PI3K/Akt/mTOR pathway, Biochem Biophys Res Commun 495 (1) (2018) 1482–1489. [DOI] [PubMed] [Google Scholar]
- [84].Tang ST, Wang F, Shao M, Wang Y, Zhu HQ, MicroRNA-126 suppresses inflammation in endothelial cells under hyperglycemic condition by targeting HMGB1, Vascul Pharmacol 88 (2017) 48–55. [DOI] [PubMed] [Google Scholar]
- [85].Cerutti C, Edwards LJ, de Vries HE, Sharrack B, Male DK, Romero IA, MiR-126 and miR-126* regulate shear-resistant firm leukocyte adhesion to human brain endothelium, Sci Rep 7 (2017) 45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Wang Y, Wang F, Wu Y, Zuo L, Zhang S, Zhou Q, Wei W, Wang Y, Zhu H, MicroRNA-126 attenuates palmitate-induced apoptosis by targeting TRAF7 in HUVECs, Mol Cell Biochem 399 (1–2) (2015) 123–130. [DOI] [PubMed] [Google Scholar]
- [87].Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S, Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs, Nat Cell Biol 14 (3) (2012) 249–256. [DOI] [PubMed] [Google Scholar]
- [88].Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D, miR-145 and miR-143 regulate smooth muscle cell fate and plasticity, Nature 460 (7256) (2009) 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kohlstedt K, Trouvain C, Boettger T, Shi L, Fisslthaler B, Fleming I, AMP-activated protein kinase regulates endothelial cell angiotensin-converting enzyme expression via p53 and the post-transcriptional regulation of microRNA-143/145, Circ Res 112 (8) (2013) 1150–1158. [DOI] [PubMed] [Google Scholar]
- [90].Sala F, Aranda JF, Rotllan N, Ramirez CM, Aryal B, Elia L, Condorelli G, Catapano AL, Fernandez-Hernando C, Norata GD, MiR-143/145 deficiency attenuates the progression of atherosclerosis in Ldlr−/−mice, Thromb Haemost 112 (4) (2014) 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L, TGFbeta Triggers miR-143/145 Transfer From Smooth Muscle Cells to Endothelial Cells, Thereby Modulating Vessel Stabilization, Circ Res 116 (11) (2015) 1753–1764. [DOI] [PubMed] [Google Scholar]
- [92].Chen H, Li X, Liu S, Gu L, Zhou X, MircroRNA-19a promotes vascular inflammation and foam cell formation by targeting HBP-1 in atherogenesis, Sci Rep 7 (1) (2017) 12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ren ZQ, Liu N, Zhao K, Micro RNA-19a suppresses IL-10 in peripheral B cells from patients with atherosclerosis, Cytokine 86 (2016) 86–91. [DOI] [PubMed] [Google Scholar]
- [94].Akhtar S, Hartmann P, Karshovska E, Rinderknecht FA, Subramanian P, Gremse F, Grommes J, Jacobs M, Kiessling F, Weber C, Steffens S, Schober A, Endothelial Hypoxia-Inducible Factor-1alpha Promotes Atherosclerosis and Monocyte Recruitment by Upregulating MicroRNA-19a, Hypertension 66 (6) (2015) 1220–1226. [DOI] [PubMed] [Google Scholar]
- [95].Wu W, Xiao H, Laguna-Fernandez A, Villarreal G Jr., Wang KC, Geary GG, Zhang Y, Wang WC, Huang HD, Zhou J, Li YS, Chien S, Garcia-Cardena G, Shyy JY, Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a, Circulation 124 (5) (2011) 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S, MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice, Science 324 (5935) (2009) 1710–1713. [DOI] [PubMed] [Google Scholar]
- [97].Fang Y, Davies PF, Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium, Arterioscler Thromb Vasc Biol 32 (4) (2012) 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wu C, Huang RT, Kuo CH, Kumar S, Kim CW, Lin YC, Chen YJ, Birukova A, Birukov KG, Dulin NO, Civelek M, Lusis AJ, Loyer X, Tedgui A, Dai G, Jo H, Fang Y, Mechanosensitive PPAP2B Regulates Endothelial Responses to Atherorelevant Hemodynamic Forces, Circ Res 117 (4) (2015) e41–e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Loyer X, Potteaux S, Vion AC, Guerin CL, Boulkroun S, Rautou PE, Ramkhelawon B, Esposito B, Dalloz M, Paul JL, Julia P, Maccario J, Boulanger CM, Mallat Z, Tedgui A, Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice, Circ Res 114 (3) (2014) 434–443. [DOI] [PubMed] [Google Scholar]
- [100].Hinkel R, Penzkofer D, Zuhlke S, Fischer A, Husada W, Xu QF, Baloch E, van Rooij E, Zeiher AM, Kupatt C, Dimmeler S, Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model, Circulation 128 (10) (2013) 1066–1075. [DOI] [PubMed] [Google Scholar]
- [101].Ni CW, Qiu H, Jo H, MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells, Am J Physiol Heart Circ Physiol 300 (5) (2011) H1762–H1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Afonyushkin T, Oskolkova OV, Bochkov VN, Permissive role of miR-663 in induction of VEGF and activation of the ATF4 branch of unfolded protein response in endothelial cells by oxidized phospholipids, Atherosclerosis 225 (1) (2012) 50–55. [DOI] [PubMed] [Google Scholar]
- [103].Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ, Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides, Nature 478 (7369) (2011) 404–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Tili E, Michaille JJ, Adair B, Alder H, Limagne E, Taccioli C, Ferracin M, Delmas D, Latruffe N, Croce CM, Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD, Carcinogenesis 31 (9) (2010) 1561–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Sadeghi M, Ranjbar B, Ganjalikhany MR, F MK, Schmitz U, Wolkenhauer O, Gupta SK, MicroRNA and Transcription Factor Gene Regulatory Network Analysis Reveals Key Regulatory Elements Associated with Prostate Cancer Progression, PLoS One 11 (12) (2016) e0168760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Korff T, Pfisterer L, Schorpp-Kistner M, miR-663 and the miRaculous vascular smooth muscle phenotypic switch, Circ Res 113 (10) (2013) 1102–1105. [DOI] [PubMed] [Google Scholar]
- [107].Li P, Zhu N, Yi B, Wang N, Chen M, You X, Zhao X, Solomides CC, Qin Y, Sun J, MicroRNA-663 regulates human vascular smooth muscle cell phenotypic switch and vascular neointimal formation, Circ Res 113 (10) (2013) 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Son DJ, Kumar S, Takabe W, Kim CW, Ni CW, Alberts-Grill N, Jang IH, Kim S, Kim W, Won Kang S, Baker AH, Woong Seo J, Ferrara KW, Jo H, The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis, Nat Commun 4 (2013) 3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Kim CW, Kumar S, Son DJ, Jang IH, Griendling KK, Jo H, Prevention of abdominal aortic aneurysm by anti-microRNA-712 or anti-microRNA-205 in angiotensin II-infused mice, Arterioscler Thromb Vasc Biol 34 (7) (2014) 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kheirolomoom A, Kim CW, Seo JW, Kumar S, Son DJ, Gagnon MK, Ingham ES, Ferrara KW, Jo H, Multifunctional Nanoparticles Facilitate Molecular Targeting and miRNA Delivery to Inhibit Atherosclerosis in ApoE(−/−) Mice, ACS Nano 9 (9) (2015) 8885–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Weber M, Baker MB, Moore JP, Searles CD, MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity, Biochem Biophys Res Commun 393 (4) (2010) 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, Li JY, Chien S, MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation, Proceedings of the National Academy of Sciences of the United States of America 108 (25) (2011) 10355–10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kahonen M, Karhunen PJ, Laaksonen R, Lehtimaki T, miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study, Atherosclerosis 219 (1) (2011) 211–217. [DOI] [PubMed] [Google Scholar]
- [114].Nam D, Ni CW, Rezvan A, Suo J, Budzyn K, Llanos A, Harrison DG, Giddens DP, Jo H, A model of disturbed flow-induced atherosclerosis in mouse carotid artery by partial ligation and a simple method of RNA isolation from carotid endothelium, J Vis Exp (40) (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Li S, Fan Q, He S, Tang T, Liao Y, Xie J, MicroRNA-21 negatively regulates Treg cells through a TGF-beta1/Smad-independent pathway in patients with coronary heart disease, Cell Physiol Biochem 37 (3) (2015) 866–878. [DOI] [PubMed] [Google Scholar]
- [116].Kin K, Miyagawa S, Fukushima S, Shirakawa Y, Torikai K, Shimamura K, Daimon T, Kawahara Y, Kuratani T, Sawa Y, Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm, J Am Heart Assoc 1 (5) (2012) e000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Tsai PC, Liao YC, Wang YS, Lin HF, Lin RT, Juo SH, Serum microRNA-21 and microRNA-221 as potential biomarkers for cerebrovascular disease, Journal of vascular research 50 (4) (2013) 346–354. [DOI] [PubMed] [Google Scholar]
- [118].Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C, MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation, Circ Res 100 (11) (2007) 1579–1588. [DOI] [PubMed] [Google Scholar]
- [119].Harmalkar M, Upraity S, Kazi S, Shirsat NV, Tamoxifen-Induced Cell Death of Malignant Glioma Cells Is Brought About by Oxidative-Stress-Mediated Alterations in the Expression of BCL2 Family Members and Is Enhanced on miR-21 Inhibition, J Mol Neurosci 57 (2) (2015) 197–202. [DOI] [PubMed] [Google Scholar]
- [120].Li Y, Yan L, Zhang W, Wang H, Chen W, Hu N, Ou H, miR-21 inhibitor suppresses proliferation and migration of nasopharyngeal carcinoma cells through down-regulation of BCL2 expression, Int J Clin Exp Pathol 7 (6) (2014) 3478–3487. [PMC free article] [PubMed] [Google Scholar]
- [121].Yang Q, Yang K, Li A, microRNA-21 protects against ischemia-reperfusion and hypoxia-reperfusion-induced cardiocyte apoptosis via the phosphatase and tensin homolog/Akt-dependent mechanism, Mol Med Rep 9 (6) (2014) 2213–2220. [DOI] [PubMed] [Google Scholar]
- [122].Yang Q, Yang K, Li AY, Trimetazidine protects against hypoxia-reperfusion-induced cardiomyocyte apoptosis by increasing microRNA-21 expression, Int J Clin Exp Pathol 8 (4) (2015) 3735–3741. [PMC free article] [PubMed] [Google Scholar]
- [123].Weber M, Kim S, Patterson N, Rooney K, Searles CD, MiRNA-155 targets myosin light chain kinase and modulates actin cytoskeleton organization in endothelial cells, Am J Physiol Heart Circ Physiol 306 (8) (2014) H1192–H1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Wu XY, Fan WD, Fang R, Wu GF, Regulation of microRNA-155 in endothelial inflammation by targeting nuclear factor (NF)-kappaB P65, J Cell Biochem 115 (11) (2014) 1928–1936. [DOI] [PubMed] [Google Scholar]
- [125].Donners MM, Wolfs IM, Stoger LJ, van der Vorst EP, Pottgens CC, Heymans S, Schroen B, Gijbels MJ, de Winther MP, Hematopoietic miR155 deficiency enhances atherosclerosis and decreases plaque stability in hyperlipidemic mice, PLoS One 7 (4) (2012) e35877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Zhu J, Chen T, Yang L, Li Z, Wong MM, Zheng X, Pan X, Zhang L, Yan H, Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10, PLoS One 7 (11) (2012) e46551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, Weber C, Schober A, MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages, J Clin Invest 122 (11) (2012) 4190–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Du F, Yu F, Wang Y, Hui Y, Carnevale K, Fu M, Lu H, Fan D, MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice, Arterioscler Thromb Vasc Biol 34 (4) (2014) 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Sun HX, Zeng DY, Li RT, Pang RP, Yang H, Hu YL, Zhang Q, Jiang Y, Huang LY, Tang YB, Yan GJ, Zhou JG, Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase, Hypertension 60 (6) (2012) 1407–1414. [DOI] [PubMed] [Google Scholar]
- [130].O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D, Inositol phosphatase SHIP1 is a primary target of miR-155, Proceedings of the National Academy of Sciences of the United States of America 106 (17) (2009) 7113–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR, Shidham A, Croce CM, Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancerbinding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice, Blood 114 (7) (2009) 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Tian FJ, An LN, Wang GK, Zhu JQ, Li Q, Zhang YY, Zeng A, Zou J, Zhu RF, Han XS, Shen N, Yang HT, Zhao XX, Huang S, Qin YW, Jing Q, Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis, Cardiovasc Res 103 (1) (2014) 100–110. [DOI] [PubMed] [Google Scholar]
- [133].Wei Y, Schober A, Weber C, Pathogenic arterial remodeling: the good and bad of microRNAs, Am J Physiol Heart Circ Physiol 304 (8) (2013) H1050–H1059. [DOI] [PubMed] [Google Scholar]
- [134].Yan H, Wang S, Li Z, Zhao W, Wang Z, Sun Z, Pan Y, Zhu J, Upregulation of miRNA-155 expression by OxLDL in dendritic cells involves JAK1/2 kinase and transcription factors YY1 and MYB, International journal of molecular medicine 37 (5) (2016) 1371–1378. [DOI] [PubMed] [Google Scholar]
- [135].Li X, Kong D, Chen H, Liu S, Hu H, Wu T, Wang J, Chen W, Ning Y, Li Y, Lu Z, miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1, Sci Rep 6 (2016) 21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, Xing YP, Sager HB, Sahay G, Speciner L, Bader A, Bogorad RL, Yin H, Racie T, Dong YZ, Jiang S, Seedorf D, Dave A, Sandhu KS, Webber MJ, Novobrantseva T, Ruda VM, Lytton-Jean AKR, Levins CG, Kalish B, Mudge DK, Perez M, Abezgauz L, Dutta P, Smith L, Charisse K, Kieran MW, Fitzgerald K, Nahrendorf M, Danino D, Tuder RM, von Andrian UH, Akinc A, Panigrahy D, Schroeder A, Koteliansky V, Langer R, Anderson DG, In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight, Nature Nanotechnology 9 (8) (2014) 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Feinberg MW, Moore KJ, MicroRNA Regulation of Atherosclerosis, Circ Res 118 (4) (2016) 703–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Aryal B, Suarez Y, Non-coding RNA regulation of endothelial and macrophage functions during atherosclerosis, Vascul Pharmacol (2018), 10.1016/j.vph.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Nallamshetty S, Chan SY, Loscalzo J, Hypoxia: a master regulator of microRNA biogenesis and activity, Free Radic Biol Med 64 (2013) 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Nepal C, Coolen M, Hadzhiev Y, Cussigh D, Mydel P, Steen VM, Carninci P, Andersen JB, Bally-Cuif L, Muller F, Lenhard B, Transcriptional, post-transcriptional and chromatin-associated regulation of pri-miRNAs, pre-miRNAs and moRNAs, Nucleic Acids Res 44 (7) (2016) 3070–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Dykes IM, Emanueli C, Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA, Genomics Proteomics Bioinformatics 15 (3) (2017) 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q, LncRNADisease: a database for long-non-coding RNA-associated diseases, Nucleic Acids Res 41 (2013) D983–D986 Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Maass PG, Luft FC, Bahring S, Long non-coding RNA in health and disease, J Mol Med (Berl) 92 (4) (2014) 337–346. [DOI] [PubMed] [Google Scholar]
- [148].Z.M. S.L. Chen W, Hu ZY, Li B, Genome-wide analysis of differentially expressed long noncoding RNAs induced by low shear stress in human umbilical vein endothelial cells, Integr. Mol. Med (2015) 2. [Google Scholar]
- [149].Stanicek L, Lozano-Vidal N, Kwon H-B, Stainier D, Wittig I, Dimmeler S, Boon RA, Abstract 19863: The Shear Stress-induced Human Long Non-coding RNA Lassie Regulates Endothelial Cell-cell Junctions, Circulation 136 (Suppl. 1) (2017) A19863. [Google Scholar]
- [150].Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM, Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA, Arterioscler Thromb Vasc Biol 34 (6) (2014) 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Boulberdaa M, Scott E, Ballantyne M, Garcia R, Descamps B, Angelini GD, Brittan M, Hunter A, McBride M, McClure J, Miano JM, Emanueli C, Mills NL, Mountford JC, Baker AH, A Role for the Long Noncoding RNA SENCR in Commitment and Function of Endothelial Cells, Mol Ther 24 (5) (2016) 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Miao Y, Ajami NE, Huang TS, Lin FM, Lou CH, Wang YT, Li S, Kang J, Munkacsi H, Maurya MR, Gupta S, Chien S, Subramaniam S, Chen Z, Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function, Nat Commun 9 (1) (2018) 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lu W, Huang SY, Su L, Zhao BX, Miao JY, Long Noncoding RNA LOC100129973 Suppresses Apoptosis by Targeting miR-4707–5p and miR-4767 in Vascular Endothelial Cells, Sci Rep 6 (2016) 21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Zhou Y, Zhang X, Klibanski A, MEG3 noncoding RNA: a tumor suppressor, Journal of molecular endocrinology 48 (3) (2012) R45–R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Wu Z, He Y, Li D, Fang X, Shang T, Zhang H, Zheng X, Long noncoding RNA MEG3 suppressed endothelial cell proliferation and migration through regulating miR-21, Am J Transl Res 9 (7) (2017) 3326–3335. [PMC free article] [PubMed] [Google Scholar]
- [156].Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S, Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth, Circ Res 114 (9) (2014) 1389–1397. [DOI] [PubMed] [Google Scholar]
- [157].Arslan S, Berkan O, Lalem T, Ozbilum N, Goksel S, Korkmaz O, Cetin N, Devaux Y, n. Cardiolinc, Long non-coding RNAs in the atherosclerotic plaque, Atherosclerosis 266 (2017) 176–181. [DOI] [PubMed] [Google Scholar]
- [158].Tang Y, Jin X, Xiang Y, Chen Y, Shen CX, Zhang YC, Li YG, The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT, FEBS Lett. 589 (20 Pt B) (2015) 3189–3196. [DOI] [PubMed] [Google Scholar]
- [159].Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B, Jiang Q, Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus, Cell Death Dis 5 (2014) e1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Josipovic I, Pfluger B, Fork C, Vasconez AE, Oo JA, Hitzel J, Seredinski S, Gamen E, Heringdorf DMZ, Chen W, Looso M, Pullamsetti SS, Brandes RP, Leisegang MS, Long noncoding RNA LISPR1 is required for S1P signaling and endothelial cell function, J Mol Cell Cardiol 116 (2018) 57–68. [DOI] [PubMed] [Google Scholar]
- [161].Ballantyne MD, McDonald RA, Baker AH, lncRNA/MicroRNA interactions in the vasculature, Clin Pharmacol Ther 99 (5) (2016) 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y, The imprinted H19 lncRNA antagonizes let-7 microRNAs, Mol Cell 52 (1) (2013) 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Zhang Z, Gao W, Long QQ, Zhang J, Li YF, Liu DC, Yan JJ, Yang ZJ, Wang LS, Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population, Sci Rep 7 (1) (2017) 7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Bao MH, Luo HQ, Chen LH, Tang L, Ma KF, Xiang J, Dong LP, Zeng J, Li GY, Li JM, Impact of high fat diet on long non-coding RNAs and messenger RNAs expression in the aortas of ApoE(−/−) mice, Sci Rep 6 (2016) 34161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Pan JX, LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway, Eur Rev Med Pharmacol Sci 21 (2) (2017) 322–328. [PubMed] [Google Scholar]
- [166].Castellano JJ, Navarro A, Vinolas N, Marrades RM, Moises J, Cordeiro A, Saco A, Munoz C, Fuster D, Molins L, Ramirez J, Monzo M, LincRNA-p21 Impacts Prognosis in Resected Non-Small Cell Lung Cancer Patients through Angiogenesis Regulation, J Thorac Oncol 11 (12) (2016) 2173–2182. [DOI] [PubMed] [Google Scholar]
- [167].Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C, LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity, Circulation 130 (17) (2014) 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q, lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA, Circ Res 116 (7) (2015) 1143–1156. [DOI] [PubMed] [Google Scholar]
- [169].Zhong X, Ma X, Zhang L, Li Y, Li Y, He R, MIAT promotes proliferation and hinders apoptosis by modulating miR-181b/STAT3 axis in ox-LDL-induced atherosclerosis cell models, Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 97 (2018) 1078–1085. [DOI] [PubMed] [Google Scholar]
- [170].Thomas AA, Feng B, Chakrabarti S, ANRIL regulates production of extracellular matrix proteins and vasoactive factors in diabetic complications, American journal of physiology. Endocrinology and metabolism 314 (3) (2018) E191–E200. [DOI] [PubMed] [Google Scholar]
- [171].Holdt LM, Teupser D, From genotype to phenotype in human atherosclerosis-recent findings, Curr Opin Lipidol 24 (5) (2013) 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Aarabi G, Zeller T, Seedorf H, Reissmann DR, Heydecke G, Schaefer AS, Seedorf U, Genetic Susceptibility Contributing to Periodontal and Cardiovascular Disease, J Dent Res 96 (6) (2017) 610–617. [DOI] [PubMed] [Google Scholar]
- [173].Motterle A, Pu X, Wood H, Xiao Q, Gor S, Ng FL, Chan K, Cross F, Shohreh B, Poston RN, Tucker AT, Caulfield MJ, Ye S, Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells, Hum Mol Genet 21 (18) (2012) 4021–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Rahimi E, Ahmadi A, Boroumand MA, Mohammad Soltani B, Behmanesh M, Association of ANRIL Expression with Coronary Artery Disease in Type 2 Diabetic Patients, Cell J 20 (1) (2018) 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Zhao J, Zhang W, Lin M, Wu W, Jiang P, Tou E, Xue M, Richards A, Jourd’heuil D, Asif A, Zheng D, Singer HA, Miano JM, Long X, MYOSLID Is a Novel Serum Response Factor-Dependent Long Noncoding RNA That Amplifies the Vascular Smooth Muscle Differentiation Program, Arterioscler Thromb Vasc Biol 36 (10) (2016) 2088–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Deng L, Bradshaw AC, Baker AH, Role of noncoding RNA in vascular remodelling, Curr Opin Lipidol 27 (5) (2016) 439–448. [DOI] [PubMed] [Google Scholar]
- [177].Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, Joshi N, Dweck MR, Miano JM, McBride MW, Newby DE, McDonald RA, Baker AH, Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation, Circulation 133 (21) (2016) 2050–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Shan K, Jiang Q, Wang XQ, Wang YN, Yang H, Yao MD, Liu C, Li XM, Yao J, Liu B, Zhang YY, J Y, Yan B, Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction, Cell Death Dis 7 (6) (2016) e2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Madrigal-Matute J, Martin-Ventura JL, Blanco-Colio LM, Egido J, Michel JB, Meilhac O, Heat-shock proteins in cardiovascular disease, Adv Clin Chem 54 (2011) 1–43. [DOI] [PubMed] [Google Scholar]
- [180].Fichtlscherer S, Zeiher AM, Dimmeler S, Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol 31 (11) (2011) 2383–2390. [DOI] [PubMed] [Google Scholar]
- [181].Simionescu N, Niculescu LS, Carnuta MG, Sanda GM, Stancu CS, Popescu AC, Popescu MR, Vlad A, Dimulescu DR, Simionescu M, Sima AV, Hyperglycemia Determines Increased Specific MicroRNAs Levels in Sera and HDL of Acute Coronary Syndrome Patients and Stimulates MicroRNAs Production in Human Macrophages, PLoS One 11 (8) (2016) e0161201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q, Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans, Eur Heart J 31 (6) (2010) 659–666. [DOI] [PubMed] [Google Scholar]
- [183].Li T, Cao H, Zhuang J, Wan J, Guan M, Yu B, Li X, Zhang W, Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans, Clinica chimica acta; international journal of clinical chemistry 412 (1–2) (2011) 66–70. [DOI] [PubMed] [Google Scholar]
- [184].Liu X, Cheng Y, Yang J, Xu L, Zhang C, Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application, J Mol Cell Cardiol 52 (1) (2012) 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Cortez-Dias N, Costa MC, Carrilho-Ferreira P, Silva D, Jorge C, Calisto C, Pessoa T, Robalo Martins S, de Sousa JC, da Silva PC, Fiuza M, Diogo AN, Pinto FJ, Enguita FJ, Circulating miR-122–5p/miR-133b Ratio Is a Specific Early Prognostic Biomarker in Acute Myocardial Infarction, Circulation journal: official journal of the Japanese Circulation Society 80 (10) (2016) 2183–2191. [DOI] [PubMed] [Google Scholar]
- [186].Andersson P, Gidlof O, Braun OO, Gotberg M, van der Pals J, Olde B, Erlinge D, Plasma levels of liver-specific miR-122 is massively increased in a porcine cardiogenic shock model and attenuated by hypothermia, Shock (Augusta, Ga.) 37 (2) (2012) 234–238. [DOI] [PubMed] [Google Scholar]
- [187].Reiss AB, Vernice NA, Siegart NM, De Leon J, Kasselman LJ, Exosomes in Cholesterol Metabolism and Atherosclerosis, Cardiovascular & hematological disorders drug targets 17 (3) (2017) 185–194. [DOI] [PubMed] [Google Scholar]
- [188].Lu X, The Role of Exosomes and Exosome-derived microRNAs in Atherosclerosis, Current pharmaceutical design 23 (40) (2017) 6182–6193. [DOI] [PubMed] [Google Scholar]
- [189].Loyer X, Vion AC, Tedgui A, Boulanger CM, Microvesicles as cell-cell messengers in cardiovascular diseases, Circ Res 114 (2) (2014) 345–353. [DOI] [PubMed] [Google Scholar]
- [190].Sohel MH, Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges, Achievements in the Life Sciences 10 (2) (2016) 175–186. [Google Scholar]
- [191].Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT, MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins, Nat Cell Biol 13 (4) (2011) 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Boon RA, Vickers KC, Intercellular transport of microRNAs, Arterioscler Thromb Vasc Biol 33 (2) (2013) 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C, Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection, Science signaling 2 (100) (2009) ra81. [DOI] [PubMed] [Google Scholar]
- [194].Huang Y, Tang S, Ji-Yan C, Huang C, Li J, Cai AP, Feng YQ, Circulating miR-92a expression level in patients with essential hypertension: a potential marker of atherosclerosis, J Hum Hypertens 31 (3) (2017) 200–205. [DOI] [PubMed] [Google Scholar]
- [195].Faltejskova P, Bocanek O, Sachlova M, Svoboda M, Kiss I, Vyzula R, Slaby O, Circulating miR-17–3p, miR-29a, miR-92a and miR-135b in serum: Evidence against their usage as biomarkers in colorectal cancer, Cancer biomarkers : section A of Disease markers 12 (4) (2012) 199–204. [DOI] [PubMed] [Google Scholar]
- [196].Li M, Guan X, Sun Y, Mi J, Shu X, Liu F, Li C, miR-92a family and their target genes in tumorigenesis and metastasis, Experimental cell research 323 (1) (2014) 1–6. [DOI] [PubMed] [Google Scholar]
- [197].Tian M, Ji Y, Wang T, Zhang W, Zhou Y, Cui Y, Changes in circulating microRNA-126 levels are associated with immune imbalance in children with acute asthma, Int. J. Immunopathol. Pharmacol 32 (2018) (2058738418779243). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Grimolizzi F, Monaco F, Leoni F, Bracci M, Staffolani S, Bersaglieri C, Gaetani S, Valentino M, Amati M, Rubini C, Saccucci F, Neuzil J, Tomasetti M, Santarelli L, Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression, Sci Rep 7 (1) (2017) 15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Qi J, Wang J, Katayama H, Sen S, Liu SM, Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma, Neoplasma 60 (2) (2013) 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Wei T, Ye P, Peng X, Wu LL, Yu GY, Prognostic Value of miR-222 in Various Cancers: a Systematic Review and Meta-Analysis, Clinical laboratory 62 (8) (2016) 1387–1395. [DOI] [PubMed] [Google Scholar]
- [201].Song J, Ouyang Y, Che J, Li X, Zhao Y, Yang K, Zhao X, Chen Y, Fan C, Yuan W, Potential Value of miR-221/222 as Diagnostic, Prognostic, and Therapeutic Biomarkers for Diseases, Frontiers in immunology 8 (2017) 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Mirzaei HR, Sahebkar A, Mohammadi M, Yari R, Salehi H, Jafari MH, Namdar A, Khabazian E, Jaafari MR, Mirzaei H, Circulating microRNAs in Hepatocellular Carcinoma: Potential Diagnostic and Prognostic Biomarkers, Current pharmaceutical design 22 (34) (2016) 5257–5269. [DOI] [PubMed] [Google Scholar]
- [203].Fiorino S, Bacchi-Reggiani ML, Visani M, Acquaviva G, Fornelli A, Masetti M, Tura A, Grizzi F, Zanello M, Mastrangelo L, Lombardi R, Di Tommaso L, Bondi A, Sabbatani S, Domanico A, Fabbri C, Leandri P, Pession A, Jovine E, de Biase D, MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis B- and C-related-hepatocellular-carcinoma, World journal of gastroenterology 22 (15) (2016) 3907–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Ding S, Huang H, Xu Y, Zhu H, Zhong C, MiR-222 in Cardiovascular Diseases: Physiology and Pathology, Biomed Res Int 2017 (2017) 4962426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, Smit JW, Lamb HJ, Llorente-Cortes V, Thum T, Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes, Sci Rep 6 (2016) 37354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Han D, Gao Q, Cao F, Long noncoding RNAs (LncRNAs) - The dawning of a new treatment for cardiac hypertrophy and heart failure, Biochim Biophys Acta 1863 (8) (2017) 2078–2084. [DOI] [PubMed] [Google Scholar]
- [207].Meng F, Yan J, Ma Q, Jiao Y, Han L, Xu J, Yang F, Liu J, Expression status and clinical significance of lncRNA APPAT in the progression of atherosclerosis, PeerJ 6 (2018) e4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Gangwar RS, Rajagopalan S, Natarajan R, Deiuliis JA, Noncoding RNAs in Cardiovascular Disease: Pathological Relevance and Emerging Role as Biomarkers and Therapeutics, Am J Hypertens 31 (2) (2018) 150–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Buhrke A, Bar C, Thum T, Non-coding RNA : Innovative regulators with therapeutic perspective, Herz 43 (2) (2018) 115–122. [DOI] [PubMed] [Google Scholar]
- [210].Bitarafan S, Yari M, Broumand MA, Ghaderian SMH, Rahimi M, Mirfakhraie R, Azizi F, Omrani MD, Association of Increased Levels of lncRNA H19 in PBMCs with Risk of Coronary Artery Disease, Cell J 20 (4) (2019) 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Yan Y, Zhang B, Liu N, Qi C, Xiao Y, Tian X, Li T, Liu B, Circulating Long Noncoding RNA UCA1 as a Novel Biomarker of Acute Myocardial Infarction, Biomed Res Int 2016 (2016) 8079372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Viereck J, Thum T, Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury, Circ Res 120 (2) (2017) 381–399. [DOI] [PubMed] [Google Scholar]
- [213].Gao L, Liu Y, Guo S, Yao R, Wu L, Xiao L, Wang Z, Liu Y, Zhang Y, Circulating Long Noncoding RNA HOTAIR is an Essential Mediator of Acute Myocardial Infarction, Cell Physiol Biochem 44 (4) (2017) 1497–1508. [DOI] [PubMed] [Google Scholar]
- [214].Cai Y, Yang Y, Chen X, Wu G, Zhang X, Liu Y, Yu J, Wang X, Fu J, Li C, Jose PA, Zeng C, Zhou L, Circulating ‘lncRNA OTTHUMT00000387022’ from monocytes as a novel biomarker for coronary artery disease, Cardiovasc Res 112 (3) (2016) 714–724. [DOI] [PubMed] [Google Scholar]
- [215].Bazan HA, Hatfield SA, Brug A, Brooks AJ, Lightell DJ Jr., Woods TC, Carotid Plaque Rupture Is Accompanied by an Increase in the Ratio of Serum circR-284 to miR-221 Levels, Circ Cardiovasc Genet 10 (4) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Mitchell AJ, Gray WD, Hayek SS, Ko YA, Thomas S, Rooney K, Awad M, Roback JD, Quyyumi A, Searles CD, Platelets confound the measurement of extracellular miRNA in archived plasma, Sci Rep 6 (2016) 32651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Buschmann D, Haberberger A, Kirchner B, Spornraft M, Riedmaier I, Schelling G, Pfaffl MW, Toward reliable biomarker signatures in the age of liquid biopsies - how to standardize the small RNA-Seq workflow, Nucleic Acids Res 44 (13) (2016) 5995–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Zhao W, Zhao SP, Zhao YH, MicroRNA-143/−145 in Cardiovascular Diseases, Biomed Res Int (2015) (2015) 531740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C, Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice-brief report, Arterioscler Thromb Vasc Biol 33 (8) (2013) 1973–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K, MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− mice, J Am Heart Assoc 1 (6) (2012) e003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Ling H, Non-coding RNAs: Therapeutic Strategies and Delivery Systems, Advances in experimental medicine and biology 937, 2016, pp. 229–237. [DOI] [PubMed] [Google Scholar]
- [222].Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS, Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine, Molecular therapy. Nucleic acids 8 (2017) 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Bernardo BC, Ooi JY, Lin RC, McMullen JR, miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart, Future medicinal chemistry 7 (13) (2015) 1771–1792. [DOI] [PubMed] [Google Scholar]
- [224].Gomes CPC, Spencer H, Ford KL, Michel LYM, Baker AH, Emanueli C, Balligand JL, Devaux Y, n. Cardiolinc, The Function and Therapeutic Potential of Long Non-coding RNAs in Cardiovascular Development and Disease, Molecular therapy. Nucleic acids 8 (2017) 494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Hartmann P, Zhou Z, Natarelli L, Wei Y, Nazari-Jahantigh M, Zhu M, Grommes J, Steffens S, Weber C, Schober A, Endothelial Dicer promotes atherosclerosis and vascular inflammation by miRNA-103-mediated suppression of KLF4, Nat Commun 7 (2016) 10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Zhang YN, Xie BD, Sun L, Chen W, Jiang SL, Liu W, Bian F, Tian H, Li RK, Phenotypic switching of vascular smooth muscle cells in the ‘normal region’ of aorta from atherosclerosis patients is regulated by miR-145, J Cell Mol Med 20 (6) (2016)1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Kim CW, Kumar S, Son DJ, Jang IH, Griendling KK, Jo H, Prevention of abdominal aortic aneurysm by anti-miRNA-712 or anti-miR-205 in Angiotensin II infused mice, Arterioscler Thromb Vasc Biol 34 (2014) 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].McDonald RA, White KM, Wu J, Cooley BC, Robertson KE, Halliday CA, McClure JD, Francis S, Lu R, Kennedy S, George SJ, Wan S, van Rooij E, Baker AH, miRNA-21 is dysregulated in response to vein grafting in multiple models and genetic ablation in mice attenuates neointima formation, Eur Heart J 34 (22) (2013) 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Zhang H, Zhao Z, Pang X, Yang J, Yu H, Zhang Y, Zhou H, Zhao J, Genistein Protects Against Ox-LDL-Induced Inflammation Through MicroRNA-155/SOCS1-Mediated Repression of NF-kB Signaling Pathway in HUVECs, Inflammation 40 (4) (2017) 1450–1459. [DOI] [PubMed] [Google Scholar]
- [230].He S, Yang L, Li D, Li M, Kruppel-Like Factor 2-Mediated Suppression of MicroRNA-155 Reduces the Proinflammatory Activation of Macrophages, PLoS One 10 (9) (2015) e0139060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Liu X, Wang TT, Li Y, Shi MM, Li HM, Yuan HX, Mo ZW, Chen J, Zhang B, Chen YX, Wang JF, Dai WP, Xu YQ, Wang ZP, Zhang X, Ou ZJ, Ou JS, High density lipoprotein from coronary artery disease patients caused abnormal expression of long non-coding RNAs in vascular endothelial cells, Biochem. Biophys. Res. Commun 487 (3) (2017) 552–559. [DOI] [PubMed] [Google Scholar]
- [232].Zhang CY, Yu MS,Li X, Zhang Z, Han CR, Yan B, Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway, Tumour Biol. 39 (6) (2017) (1010428317701311). [DOI] [PubMed] [Google Scholar]
- [233].Xin JW, Jiang YG, Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells, Exp Ther Med 13 (4) (2017) 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Tee AE, Liu B, Song R, Li J, Pasquier E, Cheung BB, Jiang C, Marshall GM, Haber M, Norris MD, Fletcher JI, Dinger ME, Liu T, The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression, Oncotarget 7 (8) (2016) 8663–8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S, Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells, J Cell Mol Med 19 (6) (2015) 1418–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Luo F, Sun B, Li H, Xu Y, Liu Y, Liu X, Lu L, Li J, Wang Q, Wei S, Shi L, Lu X, Liu Q, Zhang A, A MALAT1/HIF-2alpha feedback loop contributes to arsenite carcinogenesis, Oncotarget 7 (5) (2016) 5769–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Man HS, Yan MS, Lee JJ, Marsden PA, Epigenetic determinants of cardiovascular gene expression: vascular endothelium, Epigenomics 8 (7) (2016) 959–979. [DOI] [PubMed] [Google Scholar]
- [238].Deng QJ, Xie LQ, Li H, Overexpressed MALAT1 promotes invasion and metastasis of gastric cancer cells via increasing EGFL7 expression, Life Sci 157 (2016) 38–44. [DOI] [PubMed] [Google Scholar]
- [239].Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai Y, Wu D, Wang Y, Zhuang Z, Xia H, Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells, J Neurosurg 2016 (1) (2016) 129–136. [DOI] [PubMed] [Google Scholar]
- [240].Jia P, Cai H, Liu X, Chen J, Ma J, Wang P, Liu Y, Zheng J, Xue Y, Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a, Cancer Lett 381 (2) (2016) 359–369. [DOI] [PubMed] [Google Scholar]
- [241].Gao W, Zhu M, Wang H, Zhao S, Zhao D, Yang Y, Wang ZM, Wang F, Yang ZJ, Lu X, Wang LS, Association of polymorphisms in long non-coding RNA H19 with coronary artery disease risk in a Chinese population, Mutat Res 772 (2015) 15–22. [DOI] [PubMed] [Google Scholar]
- [242].Li Y, Jin H, Perisic L, Chernogubova E, Hansson GK, Hedin U, Bergmark C, Maegdefessel L, Abstract 15313: Long Non-Coding RNA MIAT Regulates Smooth Muscle Cell Plasticity and Macrophage Activity in Advanced Atherosclerosis Lesions, Circulation 136 (Suppl. 1) (2017) (A15313). [Google Scholar]
- [243].Bochenek G, Hasler R, El Mokhtari NE, Konig IR, Loos BG, Jepsen S, Rosenstiel P, Schreiber S, Schaefer AS, The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10, Hum Mol Genet 22 (22) (2013) 4516–4527. [DOI] [PubMed] [Google Scholar]
- [244].Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gabel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D, Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks, PLoS Genet 9 (7) (2013) e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, Gan LM, Cao H, Liang Z, Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-kappaB pathway, RNA Biol 13 (1) (2016) 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Griffiths-Jones S, miRBase: the microRNA sequence database, Methods Mol Biol 342 (2006) 129–138. [DOI] [PubMed] [Google Scholar]
- [247].Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ, miRBase: microRNA sequences, targets and gene nomenclature, Nucleic Acids Res 34 (2006) D140–D144 Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Griffiths-Jones S, miRBase: microRNA sequences and annotation, Curr Protoc Bioinformatics Chapter 12 (12) (2010) 9 1–10) Unit. [DOI] [PubMed] [Google Scholar]
- [249].Kozomara A, Griffiths-Jones S, miRBase: annotating high confidence microRNAs using deep sequencing data, Nucleic Acids Res 42 (Database issue) (2014) D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, Chiew MY, Tai CS, Wei TY, Tsai TR, Huang HT, Wang CY, Wu HY, Ho SY, Chen PR, Chuang CH, Hsieh PJ, Wu YS, Chen WL, Li MJ, Wu YC, Huang XY, Ng FL, Buddhakosai W, Huang PC, Lan KC, Huang CY, Weng SL, Cheng YN, Liang C, Hsu WL, Huang HD, miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions, Nucleic Acids Res 46 (D1) (2018) D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP, Huang HD, miRTarBase: a database curates experimentally validated microRNA-target interactions, Nucleic Acids Res 39 (2011) D163–D169 Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Liu S, Li JH, Wu J, Zhou KR, Zhou H, Yang JH, Qu LH, StarScan: a web server for scanning small RNA targets from degradome sequencing data, Nucleic Acids Res 43 (W1) (2015) W480–W486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Agarwal V, Bell GW, Nam JW, Bartel DP, Predicting effective microRNA target sites in mammalian mRNAs, Elife 4 (2015) e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Sethupathy P, Corda B, Hatzigeorgiou AG, TarBase: A comprehensive database of experimentally supported animal microRNA targets, RNA 12 (2) (2006) 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG, DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows, Nucleic Acids Res 41 (2013) W169–W173 Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T, Hatzigeorgiou AG, DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association, Nucleic Acids Res 39 (Web Server issue) (2011) W145–W148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG, DIANA-microT web server: elucidating microRNA functions through target prediction, Nucleic Acids Res 37 (Web Server) (2009) W273–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Dweep H, Gretz N, Sticht C, miRWalk database for miRNA-target interactions, Methods Mol Biol 1182 (2014) 289–305. [DOI] [PubMed] [Google Scholar]
- [259].Dweep H, Sticht C, Pandey P, Gretz N, miRWalk-database: prediction of possible miRNA binding sites by “walking” the genes of three genomes, J Biomed Inform 44 (5) (2011) 839–847. [DOI] [PubMed] [Google Scholar]
- [260].Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL, Zhou H, Yang JH, Qu LH, deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data, Nucleic Acids Res 44 (D1) (2016) D196–D202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Yang JH, Qu LH, DeepBase: annotation and discovery of microRNAs and other noncoding RNAs from deep-sequencing data, Methods Mol Biol 822 (2012) 233–248. [DOI] [PubMed] [Google Scholar]
- [262].Yang JH, Shao P, Zhou H, Chen YQ, Qu LH, deepBase: a database for deeply annotating and mining deep sequencing data, Nucleic Acids Res 38 (Database issue) (2010) D123–D130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P, An update on LNCipedia: a database for annotated human lncRNA sequences, Nucleic Acids Res 43 (8) (2015) 4363–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P, LNCipedia: a database for annotated human lncRNA transcript sequences and structures, Nucleic Acids Res 41 (2013) D246–D251 Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME, lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs, Nucleic Acids Res 43 (Database issue) (2015) D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS, lncRNAdb: a reference database for long noncoding RNAs, Nucleic Acids Res 39 (2011) D146–D151 Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]