Abstract
Purpose of review
Application of omics to study human health has created a new era of opportunities for epidemiology research. However, approaches to characterize exogenous health triggers have largely not leveraged advances in analytical platforms and big data. In this review, we highlight the exposome, which is defined as the cumulative measure of exposure and biological responses across a lifetime as a cornerstone for new epidemiology approaches to study complex and preventable human diseases.
Recent findings
While no universal approach exists to measure the entirety of the exposome, use of high-resolution mass spectrometry methods provide distinct advantages over traditional biomonitoring and have provided key advances necessary for exposome research. Application to different study designs and recommendations for combining exposome data with novel data analytic frameworks to study complex interactions of multiple stressors are also discussed.
Summary
Even though challenges still need to be addressed, advances in methods to characterize the exposome provide exciting new opportunities for epidemiology to support fundamental discoveries to improve public health.
Keywords: Exposome, Metabolome-wide association study, Precision medicine, High-resolution metabolomics, Environmental epidemiology, High-resolution exposomics
1. Introduction
Precision medicine provides a new paradigm for healthcare, where focus is shifted from treating a single disease phenotype to prevention and treatment strategies based on an individual’s unique characteristics [1, 2]. Currently, there are genetic tests for over 2,000 clinical conditions, and many more genetic markers are likely to be incorporated into emerging risk stratification models. However, disease etiology is multifactorial and driven by a combination of genetic, environment, nutritional and lifestyle factors, which represent key measures to evaluate disease risk [2, 3]. Thus, there is a clear need to improve assessment methods for exposures and to apply a broad approach in evaluating these exposures in order to fully incorporate the concept of the exposome into precision medicine.
2. The human exposome
To emphasize the importance of applying state-of-the-art and comprehensive approaches to evaluate the environment in studies of disease etiology, Christopher Wild introduced the concept of the exposome in 2005, which he defined as a framework for measuring environmental stressors that “encompasses life-course environmental exposures (including lifestyle factors), from the prenatal period onwards [4].” The exposome is envisioned as a complement to the genome, where an individual’s history of exposure and how these exposures interact with the genome defines risk for disease.
Unlike the genome, which remains stable over time, the exposome varies on timescales ranging from seconds to decades. As a result, exposures that occur episodically and/or that have relatively short biologic half-lives can be especially challenging to assess. A key approach to help address this is to conduct longitudinal studies that collect biological samples at multiple times in the life course. Further, in addition to directly measuring the actual exposure or its metabolites, metabolomics has the potential to measure patterns of exposure-specific biologic perturbations, including those that could persist even after the exposure ceases. For example, Miller and Jones defined the exposome as [5]: “The cumulative measure of environmental influences and associated biological responses throughout the lifespan, including exposures from environment, diet, behavior, and endogenous processes.” Within this framework, exposures include external stressors, processes internal to the body, socioeconomic influences and psychological factors [6]. By characterizing the exposome in terms of a cumulative measure of environment and biological response, this definition suggests that at least for some risk factors multiple measures of an individual’s exposure over the entire life course might not need to be measured, and that instead small molecules related to the exposure effect and maladaptation could be a surrogate for that exposure.
To date, no unified method exists to characterize the sum involvement of environment in disease etiology. Two main strategies are emerging from recent exposome research projects, one using personal sensors with geospatial monitoring and another using biological samples with broad measurement of biomarkers representing exposure, biological response and adverse effects [7, 8]. Both are enabled by computational power of big data and provide exciting new opportunities for implementing the exposome into environmental health and precision medicine research [9, 10].
3. Implementation of the exposome
One of the critical requirements for translating the exposome from concept to practice is the development of methods that allow measurement of exposures on the scale consistent with the chemical burden experienced by an individual over a lifetime. In the United States, close to 81,000 chemicals are registered with the Environmental Protection Agency (EPA) for manufacture, import, and use in commercial products, including 68,000 registered under the Toxic Substances Control Act and 13,000 chemicals declared exempt, while approximately 40,000 pesticide formulations, 100,000 phytochemicals, and 5,000 inert ingredients have been approved for use [11]. The majority of these are registered as the parent compound, and do not include abiotic and biotic transformation products that could occur during manufacture, commercial use, storage and environmental transport, or due to host biological processes. Current estimates suggest the potential for upwards of a million chemical exposures experienced over a lifetime [9, 12]. Common exposure assessment approaches are not capable of characterizing exposure on this scale.
A challenge for exposure science to address the exposome at this scale lies in the level of uncertainty which is acceptable for exposure assessment. Methods that provide exposure estimates for large populations, such as remote sensing or based upon geographical location, are limited by accuracy at the individual level. In contrast, targeted biomonitoring, which uses specific and sensitive methods to measure known biomarkers of exogenous chemicals, provides a direct estimate of internal dose [13]; however, the capability to expand biomonitoring beyond a few hundred chemicals to thousands or tens of thousands is cost- and resource-prohibitive, often resulting in underpowered studies that can only detect strong effects [14]. Recent advances in analytical chemistry approaches are beginning to provide the scale of biomonitoring needed to implement exposome research. The human metabolome, which contains all low-molecular weight (<2,000 dalton) chemicals present in a biological sample, has been identified as a key measure of the exposome. The metabolome includes all endogenous biological metabolites, the chemicals from human-environment interaction, and reactants arising from interaction of these compounds with enzymatic and bacterial processes [15]. Proteins, DNA, polymers and other large molecules are not considered components of the metabolome because they require different approaches for measurement; none-the-less, certain chemical modifications of these macromolecules can be detected as breakdown products [16, 17].
The metabolome, which includes chemicals from core nutrient metabolism, lipids, the microbiome, diet-derived chemicals, phytochemicals, pharmaceuticals, commercial products and environmental contaminants, can help integrate the environment and genetics, and can be directly studied for its impact on disease risk. Enzymes for the core pathways associated with basal metabolism are encoded within the genome and conserved across humans with metabolites present at tightly regulated physiological ranges. Characterizing the endogenous metabolites from these pathways is a functional measure of the genome, which is influenced in multiple ways by epigenetic and transcriptional mechanisms and also by distribution and post-translational modifications of proteins. Exogenous chemicals absorbed by a host, include compounds present in diet, drugs, microbiome, commercial products and environmental chemicals are detectable within the metabolome, either as the compound initially exposed to, or transformation products. These xenobiotics represent the exposome contribution to phenotype.
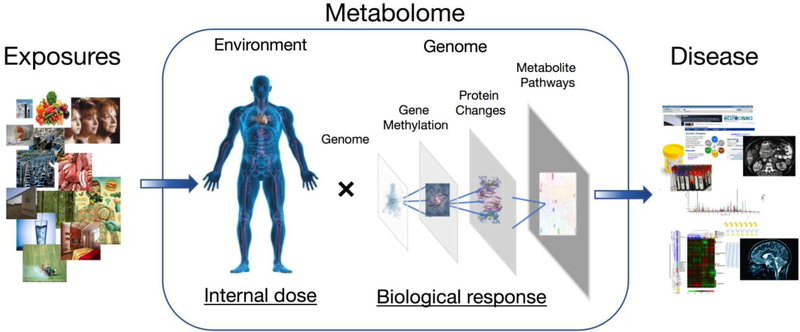
Xenobiotics influence biological processes through local and global changes within an organism, resulting in micro- and macroscale interactions between environmental chemicals and endogenous processes encoded by the genome. In some cases, toxicant-target interaction results in enhanced clearance of environmental chemicals and in other cases in the formation of reactive species that are more toxic than the original exposure. Genetic polymorphisms influence xenobiotic clearance and bioactivation, resulting in differences in metabolism and response from environmental exposures across a human population [18]. Metabolomics can be useful to characterize such interactions. Thus, the human metabolome can be used to assess the presence of an exposure and also to provide a framework for study of exposure-response relationships (Figure 1).
Figure 1:
Framework for the metabolome as a central measure for linking exposure to internal dose, biological response and disease. Environmental chemicals absorbed by a host, which are detected within the metabolome in either the parent form or as transformation products represent a measure of exposure internal dose and the exposome contribution to metabolic phenotype. These compounds can influence biological processes through local and global changes within an organism, resulting in micro- and macroscale interactions between environmental chemicals and endogenous processes encoded by the genome, the functional measures of these interactions can be detected as alterations to metabolic processes. By detecting metabolites from most metabolic pathways, metabolomic techniques allow evaluation of these biological changes, which represent markers of effective dose and response. Long-term shifts in metabolic processes accompany disease pathobiology and often represent distinct metabolic phenotypes from controls. Thus, the human metabolome can be used to assess the presence of an exposure and also to provide a framework for study of exposure-response and disease relationships. (Reprinted from Elsevier Books, Douglas I. Walker, Young-Mi Go, Ken Liu, Kurt D. Pennell, Dean P. Jones; Metabolic Phenotyping in Personalized and Public Healthcare, Pages 167–211; Jan 1, 2016; with permission from Elsevier) [111].
4. Recent advances in methods for measuring the human metabolome
Methods for characterizing the human metabolome were initially focused on development for precision medicine, disease biomarker discovery and nutrition [19, 20]. Application demonstrated these approaches not only provided measurement of endogenous metabolites, but were sensitive enough to detect exogenous chemicals and the metabolome was responsive to factors outside of the host, such as differences in diet or geographical location [21–23]. In these studies, untargeted approaches were found to be useful by not limiting measures to a priori selected analytical targets.
Targeted methods are developed to measure specific, known analytes in a population to test pre-defined hypotheses. As such, critical issues for sample collection and processing, chemical identification, quantification relative to authentic standards, and reproducibility, are addressed prior to analyses. Costs escalate in association with the number of targeted chemicals that are analyzed. Untargeted analysis uses methods that maximize the number of chemicals that can be measured in single sample and categorize their importance for identification using a metabolome-wide association study (MWAS) framework, which systematically evaluates association of each detected chemical with an outcome or disease. These analyses are analogous to GWAS, except the metabolic profile, rather than genetic variants, are tested as disease risk factors. As a result, untargeted metabolomics can be used to detect uncharacterized, unexpected and previously unknown exposures and metabolic products linked to disease. Because of the reliance on untargeted methods, analytical platforms that provide quantitative measures and the ability to identify molecules are required [24, 25].
While initial efforts in untargeted metabolomics were focused on using NMR, limited sensitivity to low-level compounds does not make these instruments useful for exposome applications [26]. In contrast, ultra-high-resolution mass spectrometers (UHRMS) and adaptive algorithms for processing complex mass spectral data now makes possible detection of over 100,000 chemical signals in blood, including environmental chemicals present at levels 100–1000 times lower than endogenous metabolites [27, 25, 9, 28]. The key advantage of available mass spectrometry methods is derived from the high mass resolution (>60,000) and mass accuracy, enabling separation of m/z differing by <2.5 parts-per-million. UHRMS profiling of blood plasma samples obtained from healthy individuals has indicated measure of metabolites from more than 80% of the pathways present in the KEGG database and detection of a broad spectrum of environmental chemicals [29]. Because of the high-mass accuracy and resolution, very low intensity peaks can be differentiated from background noise with high-sensitivity [27, 30–33]. For low abundance signals, analyses with multiple technical replicates improves confidence in detection as well as quantitative reliability [34].
An important limitation for current untargeted metabolomics methods is that analytical standards are not available for most detected chemicals and concentrations are reported as ion intensities. To make such data quantifiable in the future, an analytical strategy called reference standardization was developed using pooled samples analyzed within each batch [35, 27]. Known concentrations within the reference sample can then be used to determine a chemical response factor and calculate analytical sample concentrations based on single-point calibration. The benefits of this approach are that targeted quantification is only required in the reference sample, chemicals do not need to be selected a priori and population-wide estimates of chemical concentrations can be determined without having to re-analyze samples with chemical standards. By supporting quantification of large numbers of chemicals detected in human samples, reference standardization can provide the systematic biological and environmental chemical measurements required for risk assessment and harmonization across multiple laboratories.
The number and types of chemicals detected in biological samples is greatly expanded by combining complementary strategies for mass spectrometry. These include using alternate chromatography approaches, which are used to separate compounds before detection, and different types of mass spectrometers [23, 36–39]. Analysis by liquid chromatography (LC-UHRMS) and gas-chromatography (GC-UHRMS) provides the most comprehensive complementary platforms for metabolome- and exposome-wide association studies [40]. LC-UHRMS metabolomics platforms are best suited for measurement of polar molecules with specific functional groups, or large, non-polar molecules that contain these functional groups (such as lipids, fatty acids and sterols), making it useful for measures of endogenous metabolites, drug and environmental chemical metabolites. Many environmental chemicals, including volatile organic chemicals, brominated flame retardants, organohalogens, pesticides, hydrocarbons, perfumes and solvents are volatile enough to be introduced into the gas phase when heated and do not contain the functional groups that are required for detection on the LC-UHRMS. Thus, GC-UHRMS provides the best sensitivity and selectivity for these compounds. The combination of these platforms enables measure of both exposure and biological response, providing an integrated framework that can be used to link environmental exposures to internal dose, biological response and the metabolic changes of disease.
Identification of mass spectral signals is one of the key challenges in untargeted chemical profiling. Many detected ions do not match metabolites listed in metabolomic or environmental chemical databases, and authentic standards are not available. Current efforts focused on leveraging computational approaches that assign annotation confidence have provided improved prediction of metabolites present in chemical databases [41–43]. For annotating unknown spectral peaks, numerous tools can be used for characterizing ion fragmentation patterns to predict possible identities and biotransformation products of parent metabolites [44–47]. Continued efforts focused on developing new chemical databases that house both environmental chemicals and endogenous metabolites as “MS-Ready” structures and development of new computational approaches is expected to rapidly improve annotation capabilities of untargeted mass spectrometry data in exposome research [48–50].
5. Exposure-response relationships in untargeted metabolomics analyses
Data structure and the relationships among features detected in untargeted metabolomic experiments allow a systems-biology approach to understanding molecular mechanisms [51, 52]. While single feature-outcome relationships can assist in identifying biomarkers of disease risk or exposures, they often do not adequately describe variability across a population spectrum and, as a result, suffer from reproducibility among populations and effectiveness as diagnostic or prognostic tools. Internal data structure, which can be represented using network-based topology approaches, has been used to characterize unknown features through relationship to known biological pathways, compare metabolism across species and assess systemic responses to environmental exposures and disease [35, 53–55, 36]. Combined with alternative omic data, such as methylation, gene expression, cell sub-populations or proteomics, a functional approach can be used to identify data-driven relationships across multiple phenotypic data sets, including exposure, and represents a key area of research for the exposome [51, 56, 10].
Conceptually, interconnecting pathways and networks are equivalent; the structure can be described by a set of edges connecting nodes that represent metabolites and enzymes. Thus, pathways can be mapped on top of correlation structure and metabolite subsets tested for pathway enrichment, which evaluates if more metabolites from a given pathway are present than would be expected by chance. An initial limitation of this approach is the need to assign metabolite identities prior to performing the enrichment tests, which is complicated by uncertainty in annotation of mass spectral data. This has been overcome by development of the pathway enrichment tool Mummichog, which was developed specifically for use with untargeted high-resolution mass spectrometry data and incorporates complexity of untargeted mass spectral data while isolating biological effects and reducing Type I error [57]. The algorithm has been applied to a range of studies examining metabolic effects of disease, drug and environmental exposures [36, 58, 59].
To date, this pathway enrichment approach has been used to identify alterations in endogenous metabolism. Studies of polycyclic aromatic hydrocarbon metabolites in US Armed Services Personnel showed that expected metabolites of environmental chemicals are detected and quantitatively related [60]. Thus, the feasibility is established to develop an identical approach for xenobiotic metabolite biomonitoring. With development of powerful new tools that predict in silico biotransformation and high-throughput multi-cellular exposome screening assays [61, 62], an exposure enrichment analysis can be used to mathematically evaluate greater exposures in one population relative to another. This has the potential to overcome many of the limitations of single biomarker approaches in epidemiology and improve reliability of environmental risks of disease identified in population studies.
Demonstration of how metabolomics can be applied using the framework described in Figure 1 to provide important insights into the underlying mechanism of exposure-disease relationships is demonstrated by a recent study of occupational exposure to the degreasing solvent, trichloroethylene (TCE) [63]. In this study, untargeted metabolomic profiling was used to evaluate early biological effect, of TCE exposure by comparing metabolic differences between 80 healthy workers using TCE and 95 unexposed, matched controls [63]. Full-shift TCE levels were evaluated for all workers, and blood samples were collected following completion of the work shift [64]. Using a MWAS, all detected mass spectral signals were tested for association with exposure. Metabolites associated with exposure included known TCE detoxification products, unidentifiable chlorinated compounds and endogenous metabolites. To elucidate biological response, pathway enrichment analysis was completed, and identified disruption to purine catabolism, decreases in sulfur amino acid and bile acid biosynthesis pathways; which are consistent with known toxic effects of TCE. Metabolites associated with exposure were also tested for their relationship with urinary TCE exposure biomarkers and physiologic endpoints, supporting known or suspected disease associations, including immune and renal effects. Thus, external exposure was linked to internal dose and biological response, providing insight into molecular mechanisms of exposure-related disease etiology. Most notably, TCE metabolites associated with physiologic endpoints included compounds that had not been previously described, identifying potential new mode-of-actions for TCE toxicity that would not have been detected if only targeted analyses were completed.
6. Metabolomic applications in epidemiology studies of disease
Due to the ability to characterize a diverse series of endogenous and exogenous metabolites in biological samples, metabolomic approaches have rapidly gained acceptance as an important tool in population health research. Results demonstrate application to measure the metabolome using small volumes of blood, urine, stool, saliva, exhaled breath condensate, cerebral spinal fluid, biopsies and other hard and soft tissues has the potential to inform on possible mechanisms underlying disease [65–70]. Furthermore, current analytical strategies are high-throughput and available at a relatively low cost, making possible analysis of large studies on the order of 1,000 to 10,000 samples [11, 60, 71, 72]. Thus, metabolomic methods are poised to provide a key analytical platform for exposome research in epidemiology.
A fundamental aspect of the exposome is to assess the occurrence and impacts of environmental exposures across the lifespan. The use of life course epidemiology approaches, which aim to elucidate biological and environmental processes that operate across an individual’s life course and how exposure at different periods influence disease risk [73], is of critical importance for exposome research. However, most existing evidence is currently from case-control or cross-sectional studies that do not allow establishing a clear temporal relationship between exposure, intermediate effect biomarkers and disease. Recently, metabolomics characterization of amniotic fluid, cord blood, and maternal/child urine or serum samples have been used to assess complex fetal-maternal exposures, and have potential to be linked to developmental problems [59, 74–77]. Newborn dried blood spots have also been proposed as a promising specimen for metabolomic profiling and have been used to identify metabolic biomarkers of future risk of cancer and other childhood diseases [78]. The integration of untargeted metabolomics in large-scale prospective pregnancy and childhood cohorts with continued follow-up of participants towards adulthood is a key requirement for the characterization of the exposome over the lifespan. As exogenous exposures and endogenous metabolites are time-varying, future study designs can be benefited by the integration of metabolomics at multiple time points; such approaches are currently lacking.
The use of occupational studies to understand the effect of workplace exposures on health can further provide key insight into disease risk factors and early biological effects of exposure. Metabolomic approaches have been applied to a range of occupational exposure studies [79, 80]. As indicated above, metabolomic associations with occupational TCE exposure demonstrates the power of using untargeted approaches to characterize the effects of chemical exposures [63]. Additional metabolomic studies of occupational exposures include welding fume exposures, metals, shiftwork, military deployments, farming, automotive exhaust and pesticide plant workers [81–86].
Untargeted metabolomic profiling of tissues, urine and blood in population studies can detect environmental chemical metabolites and previously uncharacterized biomarkers in human populations [87, 76, 31, 32, 27, 23]. Using MWAS, metabolic alterations can be associated with exposure levels and used to evaluate exposure-dependent relationships in biological pathways. To date, multiple exposures have been assessed, including air pollution, persistent organic pollutants, proximity to industrial operations, metals, perfluorinated substances and plasticizers [36, 88–95]. As with all observational studies it is important to control for confounding, which can be accomplished by study design ensuring comparability of “exposed” and “unexposed” subjects and by using questionnaire-based data and biological markers of known or suspected confounders. Replication of biologic response findings in other studies is critical to rule out false-positive associations. Further, where feasible, study designs that evaluate populations before and after an exposure takes place (e.g., before and after a large seasonal variation in an environmental exposure, or where appropriate, controlled low-level exposure studies) can be very useful to help established a causal relationship, as well as exposure intervention studies that evaluate subjects during the exposure and post-intervention when exposures are reduced. Finally, following up observed associations in experimental in vitro and animal studies can also help to support causal relationships in humans.
The application of metabolomics to the study of disease risk, screening, and treatment efficacy has generated some promising initial findings, although the field is still in its infancy. These include studies of neurodegenerative diseases [96], type II diabetes [97], cancer [98], human immunodeficiency virus (HIV) infection [53], tuberculosis [99], malaria [100] and cardiovascular disease [71, 72]. A critical next step in the application of metabolomics to the study of disease etiology and early disease detection will be the use of longitudinal studies, which have already shown their utility [101, 102, 72], and especially when repeat biological samples are collected and stored over many years. These studies will allow the direct measurement of exposure to exogenous and endogenous compounds at multiple points during the life course, which is especially important for environmental exposures that have relatively short half-lives and/or do not bioaccumulate, and the assessment of the trajectory of metabolic changes from those exposures leading to disease.
The use of untargeted metabolomic methods provides a systematic measure for conducting a metabolome- and exposome wide association study of disease. To understand the complexity of the human exposome, new data analytic strategies need to be adopted in epidemiology studies. Identifying relationships between environmental factors and disease, and establishing causality, will require strategies that incorporate multiple-levels of measurements that capture exposures, biological response and disease [103–105]. To avoid complication by factors related to reverse causality and identify exposures from the environment contributing to development of disease, so called “meet-in-the-middle” approaches hold promise for untargeted methods applied to human studies [8, 106]. When performing this type of data analysis, causal relationship between disease and environment is evaluated through a prospective search for intermediate biomarkers related to past exposure and associated with disease development. If overlapping associations are identified, it reinforces a potential causal interpretation of the exposure-disease association. As an additional step, the proportion of the association explained by intermediate biomarkers can be quantified using causal mediation analytical methods [107]. The associations identified using this framework can also be evaluated through animal exposure and disease models, which help establish biological plausibility. Target populations to support this approach are not limited to life-course studies, and existing studies of children or adults with previously collected biologic samples and questionnaire data can be used within this framework. For example, exposure can be tied to disease using other exposure assessment methods, such as geospatial data, exposure questionnaires, and remote sensing; exposure can be linked to intermediate biomarkers in existing occupational exposure studies that may reduce confounding due to additional exposures; and intermediate biomarkers can be linked to disease using already established or new prospective cohorts. Untargeted metabolomics can be integrated with each type of data to facilitate overall linkage of exposure to outcome.
7. Conclusions
Incorporation of the exposome into epidemiology research will improve the ability to understand the effect of environmental exposures on human health. In many cases, disease arises from a complex series of environmental, lifestyle and genetic factors that are not possible to elucidate using single biomarker approaches. While still rapidly developing, technology now exists to provide the functional measures of environment and biological response that potentially allows comprehension of the complex, human chemical experience, and efforts are underway to evaluate harmonization of these methods across laboratories [108–110]. Platforms based upon UHRMS now allow measurement of 10,000–100,000 chemical signals using minimal sample volumes and are cost-effective. While current technologies allow analysis of 40 samples per day (12,500 samplers per instrument-year) at a cost of approximately $100, with appropriate investments in development of automation, chromatography and bioinformatics, analysis of up to 500 samples-per-day (125,000 samples per instrument-year) and cost as low as $5 per sample may be possible. As a result, barriers to incorporate metabolomic approaches for measuring the exposome have been lowered and deserve consideration as a cornerstone in epidemiological biomarker studies. This technology, combined with complementary advancements in genetics, transcriptomics, epigenetics, proteomics, imaging and bioinformatic approaches for identifying patterns in this complex data provide exciting new opportunities for fundamental discoveries in human health.
Acknowledgements
This work was supported by funds received from the National Institute of Environmental Health Sciences (ES026561, ES023515, ES029328, ES019776, ES028903 and ES030163), the National Institute of Mental Health (MH107205), National Institutes of Health Office of the Director (OD018006) and intramural funds received from the National Cancer Institute. Funding sources did not direct the study.
Footnotes
Compliance with Ethical Standards Conflict of Interest
Douglas I. Walker, Damaskini Valvi, Nathaniel Rothman, Qing Lan and Dean P. Jones each declare no potential conflicts of interest. Gary W. Miller serves as Editor in Chief of the journal Toxicological Sciences and does receive renumeration for this, but it does not present a conflict.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
REFERENCES
- 1.Collins FS, Varmus H. A New Initiative on Precision Medicine. New England Journal of Medicine. 2015;372(9):793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. The New England journal of medicine. 2012;366(6):489–91. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 3.Rappaport SM. Genetic Factors Are Not the Major Causes of Chronic Diseases. PloS one. 2016;11(4):e0154387. doi: 10.1371/journal.pone.0154387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14(8):1847–50. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 5.Miller GW, Jones DP. The nature of nurture: refining the definition of the exposome. Toxicological sciences : an official journal of the Society of Toxicology. 2014;137(1):1–2. doi: 10.1093/toxsci/kft251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330(6003):460–1. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vrijheid M, Slama R, Robinson O, Chatzi L, Coen M, van den Hazel P et al. The human early-life exposome (HELIX): project rationale and design. Environmental health perspectives. 2014;122(6):535–44. doi: 10.1289/ehp.1307204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vineis P, Chadeau-Hyam M, Gmuender H, Gulliver J, Herceg Z, Kleinjans J et al. The exposome in practice: Design of the EXPOsOMICS project. International journal of hygiene and environmental health. 2017;220(2 Pt A):142–51. doi: 10.1016/j.ijheh.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uppal K, Walker DI, Liu K, Li S, Go YM, Jones DP. Computational Metabolomics: A Framework for the Million Metabolome. Chemical research in toxicology. 2016;29(12):1956–75. doi: 10.1021/acs.chemrestox.6b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang C, Wang X, Li X, Inlora J, Wang T, Liu Q et al. Dynamic Human Environmental Exposome Revealed by Longitudinal Personal Monitoring. Cell. 2018;175(1):277–91 e31. doi: 10.1016/j.cell.2018.08.060.•• This article provides one of the first demonstrations of comprehensive monitoring of the external exposome, including both chemical and biological exposures, and its inherent variability. The use of big-date and multi-omics are an important framework for future exposome studies.
- 11.Niedzwiecki MM, Walker DI, Vermeulen R, Chadeau-Hyam M, Jones DP, Miller GW. The Exposome: Molecules to Populations. Annu Rev Pharmacol Toxicol. 2018. doi: 10.1146/annurev-pharmtox-010818-021315.•• This review summarizes the exposome as a key component of precision medicine. Approaches to characterize the human exposome are discussed in detail.
- 12.Athersuch T Metabolome analyses in exposome studies: Profiling methods for a vast chemical space. Archives of biochemistry and biophysics. 2016;589:177–86. doi: 10.1016/j.abb.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Dennis KK, Marder E, Balshaw DM, Cui Y, Lynes MA, Patti GJ et al. Biomonitoring in the Era of the Exposome. Environmental health perspectives. 2017;125(4):502–10. doi: 10.1289/EHP474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung MK, Buck Louis GM, Kannan K, Patel CJ. Exposome-wide association study of semen quality: Systematic discovery of endocrine disrupting chemical biomarkers in fertility require large sample sizes. Environment international. 2018. doi: 10.1016/j.envint.2018.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R et al. HMDB 4.0: the human metabolome database for 2018. Nucleic acids research. 2018;46(D1):D608–D17. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Adductomics: characterizing exposures to reactive electrophiles. Toxicology letters. 2012;213(1):83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Balbo S, Villalta PW, Hecht SS. Analysis of acrolein-derived 1, N(2)-propanodeoxyguanosine adducts in human lung DNA from smokers and non-smokers. Chemical research in toxicology. 2019. doi: 10.1021/acs.chemrestox.8b00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487–91. [DOI] [PubMed] [Google Scholar]
- 19.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453(7193):396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends in biotechnology. 2004;22(5):245–52. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. The Analyst. 2010;135(11):2864–70. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park YH, Lee K, Soltow QA, Strobel FH, Brigham KL, Parker RE et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012;295(1–3):47–55. doi: 10.1016/j.tox.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013;9(1 Suppl):S132–S43. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei Z, Huhman DV, Sumner LW. Mass spectrometry strategies in metabolomics. The Journal of biological chemistry. 2011;286(29):25435–42. doi: 10.1074/jbc.R111.238691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markley JL, Bruschweiler R, Edison AS, Eghbalnia HR, Powers R, Raftery D et al. The future of NMR-based metabolomics. Curr Opin Biotechnol. 2017;43:34–40. doi: 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappaport SM. Redefining environmental exposure for disease etiology. NPJ Syst Biol Appl. 2018;4:30. doi: 10.1038/s41540-018-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Go YM, Walker DI, Liang Y, Uppal K, Soltow QA, Tran V et al. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicological sciences : an official journal of the Society of Toxicology. 2015;148(2):531–43. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andra SS, Austin C, Patel D, Dolios G, Awawda M, Arora M. Trends in the application of high-resolution mass spectrometry for human biomonitoring: An analytical primer to studying the environmental chemical space of the human exposome. Environment international. 2017;100:32–61. doi: 10.1016/j.envint.2016.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Annual review of nutrition. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niedzwiecki MM, Samant P, Walker DI, Tran V, Jones DP, Prausnitz MR et al. Human Suction Blister Fluid Composition Determined Using High-Resolution Metabolomics. Analytical chemistry. 2018;90(6):3786–92. doi: 10.1021/acs.analchem.7b04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamin EL, Bonvallot N, Tremblay-Franco M, Cravedi JP, Chevrier C, Cordier S et al. Untargeted profiling of pesticide metabolites by LC-HRMS: an exposomics tool for human exposure evaluation. Analytical and bioanalytical chemistry. 2014;406(4):1149–61. doi: 10.1007/s00216-013-7136-2. [DOI] [PubMed] [Google Scholar]
- 32.Roca M, Leon N, Pastor A, Yusa V. Comprehensive analytical strategy for biomonitoring of pesticides in urine by liquid chromatography-orbitrap high resolution masss pectrometry. Journal of chromatography A. 2014;1374:66–76. doi: 10.1016/j.chroma.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Mol HG, Tienstra M, Zomer P. Evaluation of gas chromatography - electron ionization - full scan high resolution Orbitrap mass spectrometry for pesticide residue analysis. Analytica chimica acta. 2016;935:161–72. doi: 10.1016/j.aca.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 34.Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC bioinformatics. 2013; 14:15. doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Accardi CJ, Walker DI, Uppal K, Quyyumi AA, Rohrbeck P, Pennell KD et al. High Resolution Metabolomics for Nutrition and Health Assessment of Armed Forces Personnel. Journal of Occupational and Environmental Medicine. 2016;58:S80–S8. doi: 10.1097/jom.0000000000000770. [DOI] [PubMed] [Google Scholar]
- 36.Walker DI, Lane KJ, Liu K, Uppal K, Patton AP, Durant JL et al. Metabolomic assessment of exposure to near-highway ultrafine particles. Journal of exposure science & environmental epidemiology. 2018. doi: 10.1038/s41370-018-0102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Contrepois K, Jiang L, Snyder M. Optimized Analytical Procedures for the Untargeted Metabolomic Profiling of Human Urine and Plasma by Combining Hydrophilic Interaction (HILIC) and Reverse-Phase Liquid Chromatography (RPLC)-Mass Spectrometry. Molecular & cellular proteomics : MCP. 2015;14(6):1684–95. doi: 10.1074/mcp.M114.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu KH, Walker DI, Uppal K, Tran V, Rohrbeck P, Mallon TM et al. High-Resolution Metabolomics Assessment of Military Personnel: Evaluating Analytical Strategies for Chemical Detection. Journal of occupational and environmental medicine / American College of Occupational and Environmental Medicine. 2016;58(8 Suppl 1):S53–61. doi: 10.1097/JOM.0000000000000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiehn O Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr Protoc Mol Biol. 2016;114:30 4 1–4 2. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung A, Walker D, Juran B, McCauley B, Atkinson E, Schlicht E et al. High-Resolution Metabolomics and Exposomics in Primary Sclerosing Cholangitis and Primary Biliary Cholangitis Uncovers Novel, Disease-Specific Associations in Bile Acid and Amino Acid Metabolism and Environmental Toxicant Exposures Hepatology; November 9–13; San Francisco, CA: Wiley; 2018. p. 19A. [Google Scholar]
- 41.Uppal K, Walker DI, Jones DP. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Analytical chemistry. 2017;89(2):1063–7. doi: 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nash WJ, Dunn WB. From mass to metabolite in human untargeted metabolomics: Recent advances in annotation of metabolites applying liquid chromatography-mass spectrometry data. TrAC Trends in Analytical Chemistry. 2018. doi: 10.1016/j.trac.2018.11.022. [DOI] [Google Scholar]
- 43.Wang X, Jones DR, Shaw TI, Cho JH, Wang Y, Tan H et al. Target-Decoy-Based False Discovery Rate Estimation for Large-Scale Metabolite Identification. Journal of proteome research. 2018;17(7):2328–34. doi: 10.1021/acs.jproteome.8b00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen F, Pon A, Wilson M, Greiner R, Wishart D. CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic acids research. 2014;42(Web Server issue):W94–9. doi: 10.1093/nar/gku436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerlich M, Neumann S. MetFusion: integration of compound identification strategies. Journal of mass spectrometry : JMS. 2013;48(3):291–8. doi: 10.1002/jms.3123. [DOI] [PubMed] [Google Scholar]
- 46.Ma Y, Kind T, Yang D, Leon C, Fiehn O. MS2Analyzer: A software for small molecule substructure annotations from accurate tandem mass spectra. Analytical chemistry. 2014;86(21):10724–31. doi: 10.1021/ac502818e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N et al. Molecular Networking as a Dereplication Strategy. Journal of natural products. 2013;76(9):1686–99. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warth B, Spangler S, Fang M, Johnson CH, Forsberg EM, Granados A et al. Exposome-Scale Investigations Guided by Global Metabolomics, Pathway Analysis, and Cognitive Computing. Analytical chemistry. 2017;89(21):11505–13. doi: 10.1021/acs.analchem.7b02759. [DOI] [PubMed] [Google Scholar]
- 49.McEachran AD, Mansouri K, Grulke C, Schymanski EL, Ruttkies C, Williams AJ. “MS-Ready” structures for non-targeted high-resolution mass spectrometry screening studies. J Cheminform. 2018;10(1):45. doi: 10.1186/s13321-018-0299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform. 2017;9(1):61. doi: 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Sullivan NL, Rouphael N, Yu T, Banton S, Maddur MS et al. Metabolic Phenotypes of Response to Vaccination in Humans. Cell. 2017;169(5):862–77 e17. doi: 10.1016/j.cell.2017.04.026.• Big data and multi-omics will be critical for understanding how the exposome contributes to human health. In this article, Li et al. describe an approach to integrate mutiple omic datasets to understand biological response to vaccine.
- 52.Rosato A, Tenori L, Cascante M, De Atauri Carulla PR, Martins Dos Santos VAP, Saccenti E. From correlation to causation: analysis of metabolomics data using systems biology approaches. Metabolomics. 2018;14(4):37. doi: 10.1007/s11306-018-1335-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cribbs SK, Uppal K, Li S, Jones DP, Huang L, Tipton L et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome. 2016;4(1):3. doi: 10.1186/s40168-016-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uppal K, Ma C, Go YM, Jones DP, Wren J. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics. 2018;34(4):701–2. doi: 10.1093/bioinformatics/btx656.• New tools are needed to handle the complex data generated in exposome studies. Uppal et al. developed xMWAS, which provides a means of integrating up to four complex datasets using network analysis. Class comparisons and modularity analysis can be used to assess clusters of biological response to exposures.
- 55.Uppal K, Soltow QA, Promislow DE, Wachtman LM, Quyyumi AA, Jones DP. MetabNet: An R Package for Metabolic Association Analysis of High-Resolution Metabolomics Data. Frontiers in bioengineering and biotechnology. 2015;3:87. doi: 10.3389/fbioe.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yugi K, Kubota H, Hatano A, Kuroda S. Trans-Omics: How To Reconstruct Biochemical Networks Across Multiple ‘Omic’ Layers. Trends in biotechnology. 2016;34(4):276–90. doi: 10.1016/j.tibtech.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 57.Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA et al. Predicting network activity from high throughput metabolomics. PLoS computational biology. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker DI, Perry-Walker K, Finnell RH, Pennell KD, Tran V, May RC et al. Metabolome-wide association study of anti-epileptic drug treatment during pregnancy. Toxicology and applied pharmacology. 2018;363:122–30. doi: 10.1016/j.taap.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson O, Keski-Rahkonen P, Chatzi L, Kogevinas M, Nawrot T, Pizzi C et al. Cord Blood Metabolic Signatures of Birth Weight: A Population-Based Study. Journal of proteome research. 2018;17(3):1235–47. doi: 10.1021/acs.jproteome.7b00846. [DOI] [PubMed] [Google Scholar]
- 60.Walker DI, Mallon CT, Hopke PK, Uppal K, Go YM, Rohrbeck P et al. Deployment-Associated Exposure Surveillance With High-Resolution Metabolomics. Journal of occupational and environmental medicine / American College of Occupational and Environmental Medicine. 2016;58(8 Suppl 1):S12–21. doi: 10.1097/JOM.0000000000000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Djoumbou-Feunang Y, Fiamoncini J, Gil-de-la-Fuente A, Greiner R, Manach C, Wishart DS. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J Cheminform. 2019;11(1):2. doi: 10.1186/s13321-018-0324-5.• Many of the chemical signatures detected using untargeted metabolomic methods do not match compounds present in chemical databases. BioTransformer provides a software tool that can predict potential metabolic products using a multi-component biotransformation algorithm, improving annotation of uncharacterized environmental chemicals.
- 62.Walker D, Kalia V, Uppal K, Li S, Miller G, Jones D. Metabolome Wide Association Study and the Exposome: Linking Exposure to Internal Dose, Biological Response and Disease. Annual Meeting of the International Society for Environmental Epidemiology; August 26–30; Ottawa, CA2018. [Google Scholar]
- 63.Walker DI, Uppal K, Zhang L, Vermeulen R, Smith M, Hu W et al. High-resolution metabolomics of occupational exposure to trichloroethylene. International journal of epidemiology. 2016;45(5):1517–27. doi: 10.1093/ije/dyw218.• This study provides one of the first demonstrations showing the metabolome can be used as a single measure to link external exposures to internal dose, biological response and disease.
- 64.Lan Q, Zhang L, Tang X, Shen M, Smith MT, Qiu C et al. Occupational exposure to trichloroethylene is associated with a decline in lymphocyte subsets and soluble CD27 and CD30 markers. Carcinogenesis. 2010;31(9):1592–6. doi: 10.1093/carcin/bgq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C et al. The human urine metabolome. PloS one. 2013;8(9):e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S et al. The human serum metabolome. PloS one. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bessonneau V, Pawliszyn J, Rappaport SM. The Saliva Exposome for Monitoring of Individuals’ Health Trajectories. Environmental health perspectives. 2017;125(7):077014. doi: 10.1289/EHP1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karu N, Deng L, Slae M, Guo AC, Sajed T, Huynh H et al. A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Analytica chimica acta. 2018;1030:1–24. doi: 10.1016/j.aca.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 69.Wishart DS, Lewis MJ, Morrissey JA, Flegel MD, Jeroncic K, Xiong Y et al. The human cerebrospinal fluid metabolome. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2008;871(2):164–73. doi: 10.1016/j.jchromb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Brown MV, McDunn JE, Gunst PR, Smith EM, Milburn MV, Troyer DA et al. Cancer detection and biopsy classification using concurrent histopathological and metabolomic analysis of core biopsies. Genome Med. 2012;4(4):33. doi: 10.1186/gm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ganna A, Salihovic S, Sundstrom J, Broeckling CD, Hedman AK, Magnusson PK et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS genetics. 2014;10(12):e1004801. doi: 10.1371/journal.pgen.1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wurtz P, Havulinna AS, Soininen P, Tynkkynen T, Prieto-Merino D, Tillin T et al. Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation. 2015;131(9):774–85. doi: 10.1161/CIRCULATIONAHA.114.0m16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuh D, Ben-Shlomo Y, Lynch J, Hallqvist J, Power C. Life course epidemiology. J Epidemiol Community Health. 2003;57(10):778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perng W, Rifas-Shiman SL, McCulloch S, Chatzi L, Mantzoros C, Hivert MF et al. Associations of cord blood metabolites with perinatal characteristics, newborn anthropometry, and cord blood hormones in project viva. Metabolism. 2017;76:11–22. doi: 10.1016/j.metabol.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lau CE, Siskos AP, Maitre L, Robinson O, Athersuch TJ, Want EJ et al. Determinants of the urinary and serum metabolome in children from six European populations. BMC medicine. 2018;16(1):202. doi: 10.1186/s12916-018-1190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang A, Gerona RR, Schwartz JM, Lin T, Sirota M, Morello-Frosch R et al. A Suspect Screening Method for Characterizing Multiple Chemical Exposures among a Demographically Diverse Population of Pregnant Women in San Francisco. Environmental health perspectives. 2018;126(7):077009. doi: 10.1289/EHP2920.•• Untargeted methods have the potential to identify unknown and unexpected chemical exposures. In this article, the authors use an untargeted, high-resolution mass spectrometry method to detect exposure biomarkers, demonstrating these platforms provide sensitivity for human biomonitoring.
- 77.Eguchi A, Sakurai K, Watanabe M, Mori C. Exploration of potential biomarkers and related biological pathways for PCB exposure in maternal and cord serum: A pilot birth cohort study in Chiba, Japan. Environment international. 2017;102:157–64. doi: 10.1016/j.envint.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 78.Petrick L, Edmands W, Schiffman C, Grigoryan H, Perttula K, Yano Y et al. An untargeted metabolomics method for archived newborn dried blood spots in epidemiologic studies. Metabolomics. 2017;13(3). doi: 10.1007/s11306-016-1153-z.• Stored dried-blood spots are a key resource for linking metabolomic alterations to risk of childhood and adult. This article demonstrates usefulness of these samples for untargeted metabolomic profiling.
- 79.Vermeulen R The Use of High-Resolution Metabolomics in Occupational Exposure and Health Research. Ann Work Expo Health. 2017;61(4):395–7. doi: 10.1093/annweh/wxx016. [DOI] [PubMed] [Google Scholar]
- 80.Bonvallot N, David A, Chalmel F, Chevrier C, Cordier S, Cravedi J-P et al. Metabolomics as a powerful tool to decipher the biological effects of environmental contaminants in humans. Current Opinion in Toxicology. 2018;8:48–56. doi: 10.1016/j.cotox.2017.12.007. [DOI] [Google Scholar]
- 81.Pradhan SN, Das A, Meena R, Nanda RK, Rajamani P. Biofluid metabotyping of occupationally exposed subjects to air pollution demonstrates high oxidative stress and deregulated amino acid metabolism. Sci Rep. 2016;6:35972. doi: 10.1038/srep35972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dudka I, Kossowska B, Senhadri H, Latajka R, Hajek J, Andrzejak R et al. Metabonomic analysis of serum of workers occupationally exposed to arsenic, cadmium and lead for biomarker research: a preliminary study. Environment international. 2014;68:71–81. doi: 10.1016/j.envint.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 83.Saberi Hosnijeh F, Pechlivanis A, Keun HC, Portengen L, Bueno-de-Mesquita HB, Heederik D et al. Serum metabolomic pertubations among workers exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Environ Mol Mutagen. 2013;54(7):558–65. doi: 10.1002/em.21802. [DOI] [PubMed] [Google Scholar]
- 84.Guardiola JJ, Beier JI, Falkner KC, Wheeler B, McClain CJ, Cave M. Occupational exposures at a polyvinyl chloride production facility are associated with significant changes to the plasma metabolome. Toxicology and applied pharmacology. 2016;313:47–56. doi: 10.1016/j.taap.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Accardi CJ, Walker DI, Uppal K, Quyyumi AA, Rohrbeck P, Pennell KD et al. High-Resolution Metabolomics for Nutrition and Health Assessment of Armed Forces Personnel. Journal of occupational and environmental medicine / American College of Occupational and Environmental Medicine. 2016;58(8 Suppl 1):S80–8. doi: 10.1097/JOM.0000000000000770. [DOI] [PubMed] [Google Scholar]
- 86.Skene DJ, Skornyakov E, Chowdhury NR, Gajula RP, Middleton B, Satterfield BC et al. Separation of circadian- and behavior-driven metabolite rhythms in humans provides a window on peripheral oscillators and metabolism. Proc Natl Acad Sci U S A. 2018;115(30):7825–30. doi: 10.1073/pnas.1801183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rotander A, Karrman A, Toms LM, Kay M, Mueller JF, Gomez Ramos MJ. Novel fluorinated surfactants tentatively identified in firefighters using liquid chromatography quadrupole time-of-flight tandem mass spectrometry and a case-control approach. Environmental science & technology. 2015;49(4):2434–42. doi: 10.1021/es503653n. [DOI] [PubMed] [Google Scholar]
- 88.van Veldhoven K, Kiss A, Keski-Rahkonen P, Robinot N, Scalbert A, Cullinan P et al. Impact of short-term traffic-related air pollution on the metabolome - Results from two metabolome-wide experimental studies. Environment international. 2019;123:124–31. doi: 10.1016/j.envint.2018.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vlaanderen JJ, Janssen NA, Hoek G, Keski-Rahkonen P, Barupal DK, Cassee FR et al. The impact of ambient air pollution on the human blood metabolome. Environ Res. 2017;156:341–8. doi: 10.1016/j.envres.2017.03.042. [DOI] [PubMed] [Google Scholar]
- 90.Carrizo D, Chevallier OP, Woodside JV, Brennan SF, Cantwell MM, Cuskelly G et al. Untargeted metabolomic analysis of human serum samples associated with exposure levels of Persistent organic pollutants indicate important perturbations in Sphingolipids and Glycerophospholipids levels. Chemosphere. 2017;168:731–8. doi: 10.1016/j.chemosphere.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z, Zheng Y, Zhao B, Zhang Y, Liu Z, Xu J et al. Human metabolic responses to chronic environmental polycyclic aromatic hydrocarbon exposure by a metabolomic approach. Journal of proteome research. 2015;14(6):2583–93. doi: 10.1021/acs.jproteome.5b00134. [DOI] [PubMed] [Google Scholar]
- 92.Yuan TH, Chung MK, Lin CY, Chen ST, Wu KY, Chan CC. Metabolic profiling of residents in the vicinity of a petrochemical complex. Sci Total Environ. 2016;548–549:260–9. doi: 10.1016/j.scitotenv.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 93.Baker MG, Simpson CD, Lin YS, Shireman LM, Seixas N. The Use of Metabolomics to Identify Biological Signatures of Manganese Exposure. Ann Work Expo Health. 2017;61(4):406–15. doi: 10.1093/annweh/wxw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salihovic S, Fall T, Ganna A, Broeckling CD, Prenni JE, Hyotylainen T et al. Identification of metabolic profiles associated with human exposure to perfluoroalkyl substances. Journal of exposure science & environmental epidemiology. 2018. doi: 10.1038/s41370-018-0060-y. [DOI] [PubMed] [Google Scholar]
- 95.Maitre L, Robinson O, Martinez D, Toledano MB, Ibarluzea J, Marina LS et al. Urine Metabolic Signatures of Multiple Environmental Pollutants in Pregnant Women: An Exposome Approach. Environmental science & technology. 2018;52(22):13469–80. doi: 10.1021/acs.est.8b02215.•• The metabolome can provide an important measure of biological response to environmental exposures. By linking urinary endogenous metabolites to environmental exposures, this article shows how metabolomics can be incorporated into exposome studies and provides important insight into the role of exposures in pregnancy.
- 96.Roede JR, Uppal K, Park Y, Lee K, Tran V, Walker D et al. Serum metabolomics of slow vs. rapid motor progression Parkinson’s disease: a pilot study. PloS one. 2013;8(10):e77629. doi: 10.1371/journal.pone.0077629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guasch-Ferre M, Hruby A, Toledo E, Clish CB, Martinez-Gonzalez MA, Salas-Salvado J et al. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes care. 2016;39(5):833–46. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wishart DS, Mandal R, Stanislaus A, Ramirez-Gaona M. Cancer Metabolomics and the Human Metabolome Database. Metabolites. 2016;6(1). doi: 10.3390/metabo6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frediani JK, Jones DP, Tukvadze N, Uppal K, Sanikidze E, Kipiani M et al. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PloS one. 2014;9(10):e108854. doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Collins JM, Walker DI, Jones DP, Tukvadze N, Liu KH, Tran VT et al. High-resolution plasma metabolomics analysis to detect Mycobacterium tuberculosis-associated metabolites that distinguish active pulmonary tuberculosis in humans. PloS one. 2018;13(10):e0205398. doi: 10.1371/journal.pone.0205398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rogers AJ, McGeachie M, Baron RM, Gazourian L, Haspel JA, Nakahira K et al. Metabolomic derangements are associated with mortality in critically ill adult patients. PloS one. 2014;9(1):e87538. doi: 10.1371/journal.pone.0087538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Playdon MC, Ziegler RG, Sampson JN, Stolzenberg-Solomon R, Thompson HJ, Irwin ML et al. Nutritional metabolomics and breast cancer risk in a prospective study. Am J Clin Nutr. 2017;106(2):637–49. doi: 10.3945/ajcn.116.150912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vineis P Exposomics: mathematics meets biology. Mutagenesis. 2015. doi: 10.1093/mutage/gev068. [DOI] [PubMed] [Google Scholar]
- 104.Vineis P, Perera F. Molecular epidemiology and biomarkers in etiologic cancer research: the new in light of the old. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16(10):1954–65. doi: 10.1158/1055-9965.EPI-07-0457. [DOI] [PubMed] [Google Scholar]
- 105.Escher BI, Hackermuller J, Polte T, Scholz S, Aigner A, Altenburger R et al. From the exposome to mechanistic understanding of chemical-induced adverse effects. Environment international. 2017;99:97–106. doi: 10.1016/j.envint.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chadeau-Hyam M, Athersuch TJ, Keun HC, De Iorio M, Ebbels TM, Jenab M et al. Meeting-in-the-middle using metabolic profiling - a strategy for the identification of intermediate biomarkers in cohort studies. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals. 2011;16(1):83–8. doi: 10.3109/1354750X.2010.533285. [DOI] [PubMed] [Google Scholar]
- 107.Bind MA, Vanderweele TJ, Coull BA, Schwartz JD. Causal mediation analysis for longitudinal data with exogenous exposure. Biostatistics. 2016;17(1):122–34. doi: 10.1093/biostatistics/kxv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ulrich EM, Sobus JR, Grulke CM, Richard AM, Newton SR, Strynar MJ et al. EPA’s non-targeted analysis collaborative trial (ENTACT): genesis, design, and initial findings. Analytical and bioanalytical chemistry. 2019;411(4):853–66. doi: 10.1007/s00216-018-1435-6.• New initiatives are underway to evaluate reproduciability and harmonization of untargeted methods across laboratories. This article details the ENTACT study, which is a multi-laboratory round-robin trial to investigate the ability of untargeted methods to detect environmental chemicals in complext mixtures.
- 109.Sobus JR, Wambaugh JF, Isaacs KK, Williams AJ, McEachran AD, Richard AM et al. Integrating tools for non-targeted analysis research and chemical safety evaluations at the US EPA. Journal of exposure science & environmental epidemiology. 2018;28(5):411–26. doi: 10.1038/s41370-017-0012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schymanski EL, Singer HP, Slobodnik J, Ipolyi IM, Oswald P, Krauss M et al. Non-target screening with high-resolution mass spectrometry: critical review using a collaborative trial on water analysis. Analytical and bioanalytical chemistry. 2015;407(21):6237–55. doi: 10.1007/s00216-015-8681-7. [DOI] [PubMed] [Google Scholar]
- 111.Walker DI, Go Y, Liu K, Pennell K, Jones D, editors. Population Screening for Biological and Environmental Properties of the Human Metabolic Phenotype: Implications for Personalized Medicine Metabolic Phenotyping in Personalized and Public Healthcare: Elsevier; 2016. [Google Scholar]