Abstract
The genetic background of the Brazilian population is mainly characterized by three parental populations: European, African, and Native American. The aim of this study was to overview the genetic ancestry estimates for different Brazilian geographic regions and analyze factors involved in these estimates. In this systematic scoping review were included 51 studies, comprehending 81 populations of 19 states from five regions of Brazil. To reduce the potential of bias from studies with different sampling methods, we calculated the mean genetic ancestry weighted by the number of individuals. The weighted mean proportions of European, African, and Native American ancestries were 68.1%, 19.6%, and 11.6%, respectively. At the regional level, the highest European contribution occurred in the South, while the highest African and Native American contributions occurred in the Northeastern and Northern regions, respectively. Among states in the Northeast region, Bahia and Ceará showed significant differences, suggesting distinct demographic histories. This review contributes for a broader understanding of the Brazilian ancestry and indicates that the ancestry estimates are influenced by the type of molecular marker and the sampling method.
Keywords: Scoping review, genetic ancestry, Brazilian population, genetic admixture
Introduction
The Brazilian population has one of the most heterogeneous genetic constitutions in the world with a predominant tri-hybrid composition and an extensive admixture (Sans, 2000). This admixture is the result of more than 500 years of interethnic crosses between Native Americans, European colonizers, and African slaves. The study of the Native Americans genetic history is still quite controversial. Recent surveys of genetic diversity in Native Americans replaced the hypothesis of a single stream of migration to a complex scenario involving multiple sources (Reich et al., 2012). Most of Native American populations derived from the “First American” ancestral population, which crossed the Bering Strait more than 15,000 years ago (Tamm et al., 2007; Kitchen et al., 2008; Fagundes et al., 2008). However, other ancestral population streams were also detected with extensive gene flow among them (Lell et al., 2002; de Azevedo et al., 2011; Reich et al., 2012).
The estimated number of Native Americans living in Brazil was around 2.5 million when the first Portuguese colonizers arrived (Salzano and Freire-Maia, 1970; Bethell, 1999). The admixture, predominantly between European men and Native American women started immediately, however, conflicts and diseases contributed to a drastic reduction in the Native American population (Salzano and Freire-Maia 1967, 1970; Ribeiro, 1995). African people were introduced in Brazil as slaves for sugarcane farms and later for the gold and diamond mines and coffee plantations. During slave trade (1452 - 1870), it is estimated that around 4 million Africans arrived in Brazil, mainly from Guinea, Congo, Angola, Mozambique, and Nigeria (Salzano and Freire-Maia, 1967; Curtin, 1969; Ribeiro, 1995; Salzano and Bortolini, 2002). During colonization, more than 500,000 Portuguese people arrived in Brazil; when the ports were legally open, Italian, Spanish, and German also came to Brazil. Asian and Middle Eastern immigration started only in the 20th century, mainly from Japan, but also from Lebanon and Syria (Salzano and Freire-Maia, 1967). From 1500 to 1972, the immigrants to Brazil were 58% Europeans, 40% Africans, and only 2% Asian (Callegari-Jacques and Salzano, 1999).
Although the biological formation of the Brazilian people is due to the contribution of Native Americans, Europeans, and Africans, there may be a greater relative influence of one or another group depending on the geographical region. Variation in the process of colonization and occupation of the Brazilian regions created a diverse degree and extent of genetic admixture across the country. To understand the differences in the admixture among Brazilian individuals and estimate the contribution of these ancestral groups in populations from different geographical regions, several studies have been carried out. These studies have been performed using distinct panels of molecular markers, such as mitochondrial DNA (mtDNA) (Alves-Silva et al., 2000), Y-chromosome (Santos et al., 1996), short tandem repeats (STRs) (Callegari-Jacques et al., 2003), insertion/deletions (INDELs) (Santos et al., 2010), and single nucleotide polymorphisms (SNPs) (Lins et al., 2011a). Ancestry-informative markers (AIMs) are SNPs and INDELs selected to maximize the absolute difference in allele frequency between two ancestral populations, therefore they are powerful tools for inferring the genetic composition of admixed populations (Enoch et al., 2006; Kosoy et al., 2009). More recently, the development of high throughput genotyping technologies allowed genome-wide studies to infer the genomic ancestry using a large number of SNPs (Kehdy et al., 2015; Mychaleckyj et al., 2017). Studies using matrilineages (mitochondrial DNA) and patrilineages (Y-chromosome) reveal that in Brazil an asymmetric genetic sex-biased admixture has occurred, where males have, in general, higher European ancestry and females have higher African or Native American ancestry (Alves-Silva et al., 2000; Carvalho-Silva et al., 2001).
Using a set of 40 INDEL polymorphisms, Pena et al. (2011) demonstrated that genetic ancestry among Brazilian regions is more homogenous than previously expected, particularly concerning the high European ancestry observed in all regions. However, several studies have been highlighting the heterogeneous aspects of Brazilian regions. For instance, a large community-based study was carried out in three cities that represent different regions of Brazil and different scenarios of genetic structure (size, kinship, and inbreeding) (Kehdy et al., 2015). Another example is a study performed by Manta et al. (2013a) that included populations from all five Brazilian regions. In this scoping review, we conducted a robust review of studies estimating the contribution of the three parental populations (European, African, and Native American) in the genetic makeup of the Brazilian population. Here, we selected studies of 81 populations from different Brazilian localities for which genetic ancestry was estimated using nuclear autosomal makers. Considering the difficulty of real random sampling and the consequent potential bias introduced by studies with very different sample sizes, we estimated the genetic ancestry weighted by the number of individuals in each study. Moreover, we also analyzed other factors influencing the genetic ancestry estimates, such as type and number of molecular markers. Therefore, this review allows a more comprehensive understanding of the genetic ancestry estimation across the country and how the sampling and panel of markers have contributed to the current estimated pattern of admixture among Brazilian regions.
Materials and Methods
Study design
This study was based on Arksey and O’Malley’s scoping review framework (Arksey and O’Malley, 2005). The key steps followed were:
Identifying the research questions: What are the estimates of the genetic ancestry for Brazilian populations from different regions? How is the estimate of genetic ancestry affected by the panel of markers and sampling methods used for its identification?
Identifying relevant studies: The survey was performed based on articles published in two electronic databases/portals, PubMed and BVS (MEDLINE, LILACS, HISA, BBO, Coleciona SUS, Sec. Est. Saúde SP, IBECS, INDEX Psicologia) until November 1st 2017. The search terms used are shown in Table S1 (70KB, pdf) .
Study selection: Two members of our research group independently screened all retrieved titles, abstracts, or full-text articles for applicability to the review’s research questions. All citations were imported into an Excel worksheet and duplicate citations were manually removed. Articles were eligible if they met the inclusion criteria. Discrepancies resulting from the independent initial screening were discussed by the two research team members until consensus was reached. Articles that could not be obtained as full-text through online databases were also excluded from final analysis.
Charting the data: The data were entered onto a ‘data charting form’ using the Excel software to record key information from the included studies. The extracted data included the following sections: manuscript title, origin of human samples (region, state, and city, if available), number of individuals, number of markers, type of genetic markers, date of samples collection, and estimated percentage of European, African, and Native American ancestries.
Collecting, summarizing, and reporting the results - this process involved descriptive analyzes of all included studies.
Inclusion/exclusion criteria
Articles found in databases were examined for the following inclusion/exclusion criteria. Inclusion criteria: papers published until October 2017; studies using autosomal molecular markers to infer on genetic ancestry; studies that estimate European, African, and Native American genetic ancestries; studies that provided the number of individuals studied by each Brazilian State. Exclusion criteria: duplicated studies; full-text unavailable; studies based on individuals from other countries; studies using non-human samples; review article; other text formats different from research articles and theses; non-original articles describing the ancestry of the population estimated by others; without information about the state of origin of the sample individuals; lacking data on ancestry for the three ancestral populations (European, African, and Native American); case or family descriptions; studies based on related individuals; studies using blood group or uniparental markers; studies using only specific ethnic groups (for example, Afro-descendants or Caucasians). Studies with partially isolated populations, such as Native American tribes and Afro-descendants’ communities (Quilombos) were analyzed separately.
Statistical analysis
Data from the descriptive analyzes of the ancestry are presented as mean and standard deviation. The mean ancestry in cities, states, and regions and /or the molecular markers were weighted by the number of individuals included in each study by using Minitab 18.1 (State College, PA, USA).
Kruskal-Wallis test was performed and, if significance was found, Dunn’s multiple comparisons post-hoc test was used to define the differences among ancestry estimates based on type of molecular markers (INDEL, SNP, and STR) and to verify differences in ancestral estimates among regions and states. Comparisons among Brazilian regions and partially isolated afro-descendant communities were performed using Mann-Whitney U test. Correlation analysis (Spearman’s Rho test or Fischer’s test) was performed to verify associations between number of individuals or numbers of molecular markers and European, African, and Native American parental ancestries for each city. A p-value < 0.05 was considered significant. All statistical analyses were done using GraphPad Prism 5.0 (San Diego, California, USA) and Minitab 18.1. Triangular plots of the proportions of European, African, and Native American genetic ancestries were obtained using the Tri-Plot program (Graham and Midgley, 2000).
Results
Summary of included studies
The scoping review search included papers published from 2002 to 2017. A total of 890 studies was identified in the electronic databases BVS (n = 804) and Pubmed (n= 86) with the selected search terms (Table S1 (70KB, pdf) ; Figure 1). After removing all duplicated documents, a total of 740 titles and abstracts were screened. From these, 70 studies were excluded (full-text not available; sample individuals were from countries other than Brazil; studies based on non-human samples; article’s format different than research papers). Finally, 670 full-text articles were assessed in detail for eligibility. Of these, 639 publications were excluded due to not completely fulfilling our inclusion criteria. We decided to analyze only studies of ancestry estimates based on autosomal markers and to exclude studies using classical markers or uniparental markers. As a result, 51 studies were selected, 31 identified by scoping review and 20 added from the references of the selected articles, comprehending a total of 81 populations (Table S2 (222.3KB, pdf) ). Moreover, the partially isolated populations, Native American tribes, and Afro-descendant communities were analyzed separately (Table S3 (122.4KB, pdf) ).
Figure 1. Flowchart of the literature search and screening process executed in this scoping review.
Ancestry by Brazilian geographic region
The genetic ancestry estimates of Brazilian populations identified in this review comprehended 19 states from the five political geographic regions (Table 1). All regions showed a higher prevalence of European ancestry (mean 62.4% for Brazil), which was significantly higher in the South and Southeast regions compared to the North and Northeast regions (Figure 2). African ancestry was the second most prevalent ancestral population in Brazil (22.6%) and was significantly lower in the South compared to Northeast and Southeast regions. Native American ancestry was the less prevalent in the Brazilian population (14.7%), and was significantly higher in the North region compared to the Northeast, South, and Southeast regions (Figure 2). The South region showed the most distinct profile of ancestry, with the highest number of significant differences compared to the other regions of the country.
Table 1. Characterization of the ancestry profile of Brazilian geographical regions based on all 51 manuscripts included in the scoping review.
| Geographical region | State | City | N | Mean± SD | Weighted Mean± weighted SD | Ref | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| EUR | AFR | AME | EUR | AFR | AME | |||||
| Center-West | Distrito Federal | Brasilia | 780 | 65.0± 10.58 | 22.0± 7.55 | 13.0± 3.61 | 63.0± 10.26 | 23.9± 7.65 | 13.1± 3.13 | 1 - 3 |
| Taguatinga# | 189 | 63.0 | 25.0 | 12.0 | 63.0 | 25.0 | 12.0 | 4 | ||
| Total/Mean of the state | 969 | 64.5± 8.70 | 22.8± 6.34 | 12.7± 2.98 | 63.0± 8.67 | 24.1± 6.49 | 12.9± 2.70 | |||
| Mato Grosso do sul | NI# | 84 | 59.0 | 26.0 | 15.0 | 59.0 | 26.0 | 15.0 | 5 | |
| Total/mean of the state | 84 | 59.0 | 26.0 | 15.0 | 59.0 | 26.0 | 15.0 | |||
| Total/mean of the region | 1053 | 63.4± 7.92 | 23.4± 5.68 | 13.2± 2.78 | 62.7± 8.15 | 24.2± 6.06 | 13.1± 2.59 | |||
| North | Pará | Belém | 1044 | 54.5± 10.19 | 18.33± 7.00 | 27.17± 4.17 | 55.5± 10.43 | 17.60± 7.06 | 26.9± 4.44 | 6 - 11 |
| Santarém# | 90 | 60.0 | 11.0 | 29.0 | 60.0 | 11.0 | 29.0 | 13 | ||
| Goianésia do Pará# | 273 | 44.0 | 32.0 | 25.0 | 44.0 | 32.0 | 25.0 | 14 | ||
| NI# | 210 | 45 | 23 | 32 | 45 | 23 | 32 | 12 | ||
| Total/mean of the state | 1617 | 52.9± 9.53 | 19.6± 7.86 | 27.56± 3.90 | 52.4± 9.88 | 20.4± 8.24 | 27.2± 4.18 | |||
| Amapá | Macapá | 437 | 48.0± 2.82 | 24.0± 7.07 | 28.0± 9.89 | 47.2± 2.59 | 22.0± 6.46 | 30.8± 9.05 | 15-16 | |
| Total/mean of the state | 437 | 48± 2.82 | 24± 7.07 | 28± 9.89 | 47.2± 2.59 | 22.0± 6.46 | 30.8± 9.05 | |||
| Amazonas | Manaus | 294 | 58.5± 17.68 | 14.0± 2.82 | 27.5± 14.85 | 67.4± 12.37 | 12.6± 1.98 | 20.0± 10.39 | 5,17 | |
| Rio Pardo# | 340 | 44.0 | 18.0 | 38.0 | 44.0 | 18.0 | 38.0 | 18 | ||
| Total/mean of the state | 634 | 53.7± 15.04 | 15.3± 3.05 | 31.0± 12.12 | 54.9± 16.06 | 15.5± 3.51 | 29.6± 12.6 | |||
| Rondônia | Porto Velho | 404 | 57.0± 4.24 | 24.0± 8.48 | 19.0± 12.02 | 55.8± 3.89 | 21.6± 7.79 | 22.9± 11.04 | 19-20 | |
| Total/mean of the state | 404 | 57.0± 4.25 | 24.0± 8.49 | 19.0± 12.03 | 55.8± 3.89 | 21.6± 7.79 | 22.9± 11.04 | |||
| Total/mean of the region | 3092 | 52.9± 9.27 | 19.9± 7.14 | 27.3± 7.41 | 52.6± 9.72 | 19.8± 6.95 | 27.7± 7.22 | |||
| Northeast | Ceará | Barbalha# | 60 | 68.0 | 19.0 | 13.0 | 68.0 | 19.0 | 13.0 | 13 |
| Fortaleza | 1712 | 55.4± 7.13 | 19.9± 3.8 | 24.7± 5.55 | 54.0± 7.33 | 18.7± 3.81 | 27.1± 7.00 | 21-22 | ||
| Total/mean of the state | 1772 | 57.0± 7.96 | 19.8± 3.53 | 23.2± 6.60 | 54.5± 7.63 | 18.7± 3.71 | 26.7± 7.34 | |||
| Bahia | Salvador | 4859 | 52.6± 14.15 | 39.8± 11.19 | 7.5± 4.11 | 47.9± 12.42 | 43.8± 9.21 | 8.3± 4.80 | 21,23-28 | |
| Ilheus | 256 | 61± 0.00 | 31.5± 2.12 | 7.5± 2.12 | 61± 0.00 | 31.3± 2.10 | 7.7± 2.10 | 7,29 | ||
| Jequie# | 20 | 44.0 | 42.0 | 11.0 | 44.0 | 42.0 | 11.0 | 30 | ||
| Total/mean of the state | 5135 | 53.4± 12.69 | 38.6± 10.03 | 7.8± 3.66 | 48.5± 12.23 | 43.18± 9.25 | 8.29± 4.60 | |||
| Alagoas | Maceió | 120 | 67 | 19 | 14 | 67 | 19 | 14 | 5,13 | |
| NI# | 104 | 55 | 27 | 19 | 55 | 27 | 19 | |||
| Total/mean of the state | 224 | 61.0± 8.49 | 23.0± 5.66 | 16.5± 3.55 | 61.4± 8.46 | 22.7± 5.64 | 16.3± 3.52 | |||
| Maranhão | São Luis# | 177 | 42.0 | 19.0 | 39.0 | 42.0 | 19.0 | 39.0 | 31 | |
| Total/mean of the state | 177 | 42.0 | 19.0 | 39.0 | 42.0 | 19.0 | 39.0 | |||
| Piauí | NI# | 204 | 60.0 | 22.0 | 18.0 | 60.0 | 22.0 | 18.0 | 32 | |
| Total/mean of the state | 204 | 60.0 | 22.0 | 18.0 | 60.0 | 22.0 | 18.0 | |||
| Pernambuco | Recife# | 192 | 60.0 | 23.0 | 17.0 | 60.0 | 23.0 | 17.0 | 33 | |
| NI# | 133 | 57.0 | 28.0 | 15.0 | 57.0 | 28.0 | 15.0 | 5 | ||
| Total/mean of the state | 325 | 58.5± 2.12 | 25.5± 3.54 | 16± 1.41 | 58.8± 2.08 | 25.0± 3.47 | 16.2± 1.39 | |||
| Total/mean of the region | 7837 | 55.3± 10.16 | 28.8± 11.31 | 15.8± 9.45 | 50.8± 11.09 | 35.2± 13.56 | 13.9± 9.99 | |||
| South | Rio Grande do Sul | Porto Alegre | 7197 | 81.7± 3.51 | 9.3± 4.72 | 8.3± 1.15 | 84.7± 1.49 | 4.5± 2.27 | 8.9± 0.43 | 7,17,34 |
| Pelotas# | 3736 | 76.0 | 16.0 | 8.0 | 76.0 | 16.0 | 8.0 | 25 | ||
| NI# | 81 | 95 | 1 | 4 | 95 | 1 | 4 | 9 | ||
| NI# | 23 | 73 | 14 | 13 | 73 | 14 | 13 | 5 | ||
| Total/mean of the state | 11037 | 81.5± 7.86 | 9.8± 5.98 | 8.3± 2.94 | 81.8± 4.82 | 8.4± 6.23 | 8.6± 0.75 | |||
| Paraná | NI# | 21 | 71.0 | 17.0 | 12.0 | 71.0 | 17.0 | 12.0 | 5 | |
| Total/mean of the state | 21 | 71.0 | 17.0 | 12.0 | 71.0 | 17.0 | 12.0 | |||
| Santa Catarina | NI# | 20 | 80.0 | 11.0 | 9.0 | 80.0 | 11.0 | 9.0 | 5 | |
| Total/mean of the state | 20 | 80.0 | 11.0 | 9.0 | 80.0 | 11.0 | 9.0 | |||
| Total/mean of the region | 11078 | 80.0± 7.60 | 10.9± 5.64 | 8.9± 2.80 | 81.8± 4.72 | 8.4± 6.08 | 8.6± 0.74 | |||
| Southeast | Espirito Santo | NI# | 92 | 74 | 13 | 13 | 74 | 13 | 13 | 5 |
| Total/mean of the state | 92 | 74 | 13 | 13 | 74 | 13 | 13 | |||
| Minas Gerais | Belo Horizonte | 324 | 70.5± 7.77 | 24.5± 14.14 | 5.5± 6.36 | 68.0± 6.97 | 28.7± 12.03 | 3.5± 5.70 | 35-36 | |
| Alfenas | 758 | 88.0± 1.41 | 10.0± 0.70 | 2.0± 0.70 | 87.8± 1.38 | 10.2± 1.38 | 2.0± 0 | 23,37 | ||
| Manhuaçu# | 30 | 73 | 19 | 8 | 73 | 19 | 8 | 39 | ||
| Montes Claros# | 24 | 73 | 19 | 8 | 73 | 19 | 8 | 39 | ||
| Ouro Preto# | 189 | 52 | 38 | 10 | 52 | 38 | 10 | 40 | ||
| Bambui# | 1442 | 79 | 14 | 7 | 79 | 14 | 7 | 26 | ||
| NI# | 88 | 59 | 29 | 12 | 59 | 29 | 12 | 5 | ||
| NI# | 291 | 58 | 34 | 4 | 58 | 34 | 4 | 38 | ||
| Total/mean of the state | 3146 | 68.0± 13.83 | 24.7± 11.42 | 6.8± 4.92 | 75.4± 12.04 | 18.3± 9.82 | 5.8± 3.95 | |||
| São Paulo | Araraquara# | 403 | 76 | 18 | 12 | 76 | 18 | 12 | 41 | |
| São Paulo | 2346 | 67.0± 8.4 | 21.8± 8.9 | 9.2± 3.11 | 68.0± 8.71 | 22.5± 7.08 | 8.8± 3.46 | 37,42-45 | ||
| Botucatu | 983 | 73.5± 9.19 | 12.0± 5.65 | 9.0± 3.53 | 74.8± 8.99 | 11.2± 5.53 | 8.9± 3.46 | 46-47 | ||
| Campinas# | 109 | 73 | 20 | 7 | 73 | 20 | 7 | 17 | ||
| Ribeirão Preto# | 448 | 86 | 9 | 4 | 86 | 9 | 4 | 48 | ||
| NI# | 49 | 63 | 25 | 12 | 63 | 25 | 12 | 5 | ||
| Total/mean of the state | 4338 | 70.9± 8.79 | 18.6± 7.90 | 8.54± 3.11 | 72.5± 9.08 | 18.1± 7.84 | 8.1± 3.20 | |||
| Rio de Janeiro | Rio de Janeiro | 880 | 67.0± 9.05 | 23.3± 5.44 | 10± 3.56 | 63.9± 10.20 | 25.3± 6.39 | 10.9± 3.78 | 7,13,49,50 | |
| NI# | 335 | 65 | 28 | 7 | 65 | 28 | 7 | 51 | ||
| Total/mean of the state | 1215 | 66.6± 7.89 | 24.2± 5.17 | 9.4± 3.36 | 64.2± 8.42 | 26.0± 5.44 | 9.9± 3.69 | |||
| Total/mean of the region | 8791 | 69.1± 10.46 | 21.7± 9.21 | 8.2± 3.97 | 72.3± 10.39 | 19.2± 8.49 | 7.6± 3.73 | |||
| Total/mean of the country | 31851 | 62.4± 13.07 | 22.6± 10.37 | 14.7± 9.63 | 68.1± 15.68 | 19.6± 13.64 | 11.6± 8.20 | |||
NI- city not identified; #Included in only one study. References: 1. Lins et al. 2011a; 2. Gontijo et al. 2014, 3. Benedet et al. 2012; 4. Lins et al, 2011b; 5. Manta et al. 2013b; 6. Cardoso et al. 2014, 7. Pena et al. 2011; 8. Pereira et al. 2012; 9.Santos et al. 2010; 10.Carvalho et al. 2015; 11.Vieira et al. 2015; 12.Vieira-Machado et al. 2016; 13.Brito et al. 2011; 14.Cassiano et al. 2015; 15.Francez et al. 2011; 16.Francez et al. 2012; 17.Callegari-Jacques et al. 2003; 18. Kano et al. 2016; 19. Krauze de França, 2005; 20.Tarazona-Santos et al. 2011; 21.Silva et al. 2015; 22.Mychaleckyj et al. 2017; 23.Aquino et al. 2014; 24.do Rego Borges et al. 2015; 25.Kehdy et al. 2015; 26.Teló 2010; 27.Felix et al. 2010; 28.Machado 2008; 29.Oliveira et al. 2016; 30.Nascimento et al. 2016; 31.Ferreira et al. 2002; 32. Lopes et al. 2014; 33.Coelho et al. 2015; 34.Wagner et al. 2016; 35.Scliar et al. 2009; 36.Rolim et al. 2016; 37.Brito et al. 2012; 38.Silva et al. 2011; 39.Silva et al. 2010; 40.Queiroz et al. 2013; 41.Martins et al. 2011; 42.Nastri et al. 2016; 43.Bermardez-Pereira et al. 2016; 44.Cardena et al. 2013; 45.Guindalini 2010; 46.D'Éllia et al. 2017; 47.Ramos et al. 2016; 48.Souza et al. 2015; 49. Manta et al.2013a; 50. Santos et al. 2009; 51.Suarez-Kurts et al. 2007.
Figure 2. Genetic ancestral estimates of studies from different geographic regions. Asterisks indicate the statistical differences (Kruskal-Wallis test, * p < 0.01; ** p < 0.001, *** p < 0.0001).
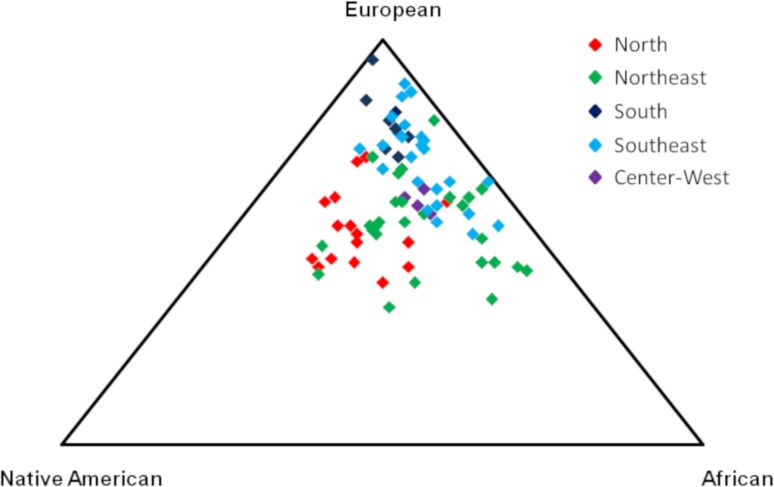
Triangular plot analysis was performed to visualize the spatial distribution of the populations from different Brazilian regions according to their ancestries. The triangular plot revealed that all populations are close to the European vertex (Figure 3). The populations of the South and Southeast regions are the closest to the European vertex. The Northeast populations showed a shift to the African and Native American ancestry vertices. The North populations showed more proximity to Native American ancestry vertex. The center-west populations showed an intermediate position overlapping with Southeast and Northeast population.
Figure 3. Tri-plot showing the clustering of the genetic ancestry inferred by studies from different regions of Brazil. Populations of the Center-West are represented in purple, North in red, Northeast in green, South in dark blue, and Southeast in light blue.
Local differences in ancestry estimates for Brazilian populations
Next, we evaluated the differences in ancestry estimates among populations of the different Brazilian states (Figure 4). Higher prevalence of European ancestry was observed for populations from all Brazilian states, and the highest was identified in Rio Grande do Sul (RS) (81.5%). On the contrary, the Maranhão state (MA) showed the lowest percentage of European ancestry (42.0%). Bahia (BA) showed the highest percentage of African ancestry (38.6%), while the lowest percentage was observed in Rio Grande do Sul (9.8%). Native American ancestry was highest in Maranhão (AM) (39%) and lowest in Minas Gerais state (MG) (6.8%).
Figure 4. Ancestral profiles of Brazilian states according to the scoping review. The graphs on the map represent the median of ancestry in Brazilian regions. The different sizes of the graphs reflect the sample sizes (scheme on the left).
Comparing the states from each region, significant differences were observed only in the Northeast region (Figure S1 (290.3KB, pdf) ). The African ancestry was significantly higher in Bahia (BA) than in Ceará (CE) (Kruskal-Wallis test, p=0.004) and the Native American ancestry was significantly lower in Bahia than in Ceará (Kruskal-Wallis test, p=0.001).
Influence of molecular markers in the genetic ancestry estimates
Studies were subdivided into three main categories according to the type of markers used in the analysis: those using INDELs, SNPs, or STR/VNTRs (Table 2). Most studies used INDELs (23 articles), followed by SNPs (15 articles), and STR (7 articles) to estimate genetic ancestry of Brazilian populations. The number of markers used in each study was highly variable, particularly for SNPs (ranging from 8 to 331,790). Comparisons of ancestry estimates for the same geographic region, but accessing different samples of individuals and using different molecular markers showed that only for Northeast region the European ancestry estimate was different using INDELs and SNPs (Table 2). The ancestry estimates using different molecular markers were also compared in studies sampling different individuals in the same city (Belém/PA, Macapá/AP, Manaus/AM, Porto Velho/RO, Fortaleza/CE, Salvador/BA, Porto Alegre/RS, São Paulo/SP and Belo Horizonte/MG) (Table S2 (222.3KB, pdf) ). For cities with more than one study for each type of marker, the comparison was performed using the mean. Significant differences (Chi-square test or Fisher’s test, p < 0.05) in ancestry estimates concerning molecular markers of distinct types were observed for Belem, Manaus, Porto Velho, Fortaleza, and Belo Horizonte (Table S2 (222.3KB, pdf) ). Comparison of ancestry estimates of the same sample of individuals using different markers was possible for two studies, Teló (2010) and Pedrosa (2006). Statistically significant differences were observed for a study in Salvador comparing STR and INDEL (Table S2 (222.3KB, pdf) , ref 26), and for Afro-descendant communities comparing STR, SNP/INDEL, and a combination of both (Table S3 (122.4KB, pdf) , ref 52).
Table 2. Description of molecular marker panels used to estimate the genetic ancestry in the selected studies from different Brazilian regions.
| Geographic region | Number of samples | Type of marker | Number of markers | Mean of Ancestry | REFERENCES | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| EUR | EUR (weighted) | AFR | AFR (weighted) | AME | AME (weighted) | |||||
| CW | 84 | INDEL | 46 | 59 | NA | 26 | NA | 15 | NA | 5 |
| 801 | SNP | 12 - 28 | 66 | 63.4 | 23 | 24.3 | 12 | 12.3 | 1,3,4 | |
| N | 1622 | INDEL | 40 - 48 | 53 | 53.3 | 19 | 20.0 | 28 | 26.7 | 5, 7,8,9,10,11,12,13 |
| 507 | SNP | 8 - 48 | 42 | 42.7 | 24 | 22.0 | 30 | 35.4 | 6, 18 | |
| 681 | STR | 4 - 12 | 59 | 57.8 | 20 | 18.4 | 21 | 24.0 | 16,17,19 | |
| NE | 2245 | INDEL | 9 - 46 | 63* | 62.9 | 25 | 28.3 | 11 | 8.9 | 5, 7, 13, 23, 24, 26 ,32 |
| 3528 | SNP | 8 - 331,790 | 49* | 48.7 | 29 | 34.5 | 21 | 16.7 | 21, 22, 25, 30, 33 | |
| 380 | STR | 4 - 8 | 51 | 51.1 | 26 | 26.5 | 24 | 22.4 | 26, 31 | |
| S | 7116 | INDEL | 40 - 48 | 80 | 84.8 | 10 | 4.3 | 9 | 8.9 | 5, 7, 9, 34 |
| 3736 | SNP | 331,790 | 76 | NA | 16 | NA | 8 | NA | 25 | |
| 226 | STR | 12 | 82 | NA | 11 | NA | 7 | NA | 17 | |
| SE | 4552 | INDEL | 40 - 61 | 71 | 72.7 | 19 | 17.8 | 9 | 8.4 | 5, 7, 13, 23, 36, 37, 44, 47, 48, 49, 50, 51 |
| 3202 | SNP | 14 - 331,790 | 65 | 73.4 | 25 | 18.5 | 8 | 7.1 | 25, 39, 40, 42, 43,45 | |
| 746 | STR | 18 - 34 | 71 | 72.1 | 24 | 23.3 | 5 | 4.6 | 17, 35, 41 | |
NA - not applicable. * Statistically significant differences in the ancestry estimates related to type of marker (Spearman’s correlation test, P < 0.05). References: 1. Lins et al., 2011a; 3. Benedet et al., 2012; 4. Lins et al., 2011b; 5. Manta et al., 2013b; 6. Cardoso et al., 2014; 7. Pena et al., 2011; 8. Pereira et al., 2012; 9. Santos et al., 2010; 10. Carvalho et al., 2015; 11. Vieira et al., 2015; 12. Vieira-Machado et al., 2016; 13. Brito et al., 2011; 14. Cassiano et al., 2015; 15. Francez et al., 2011; 16. Francez et al., 2012; 17. Callegari-Jacques et al., 2003; 18. Kano et al., 2016; 19. França, 2005; 21. Silva et al., 2015; 22. Mychaleckyj et al., 2017; 23. Aquino et al., 2014; 24. do Rego Borges et al., 2015; 25. Kehdy et al., 2015; 26. Teló, 2010; 30. Nascimento et al., 2016; 31. Ferreira et al., 2002; 32. Lopes et al., 2014; 33. Coelho et al., 2015; 34. Wagner et al., 2016; 35. Scliar et al., 2009; 36. Rolim et al., 2016; 37. Brito et al., 2012; 39. Silva et al., 2010; 40. Queiroz et al., 2013; 41. Martins et al., 2011; 42. Nastri et al., 2016; 43. Bermardez-Pereira et al., 2016; 44. Cardena et al., 2013; 45. Guindalini et al., 2010; 47. Ramos et al., 2016; 48. Souza et al., 2015; 49. Manta et al., 2013a; 50. Santos et al., 2009; 51. Suarez-Kurtz et al., 2007.
The association between the number of markers and ancestry estimates was not statistically significant for studies from the same city (data not shown). In this analysis, studies using genome-wide SNPs were excluded, thus analysis was performed for studies using samples from Belém, Salvador, Porto Alegre, São Paulo, and Rio de Janeiro.
Influence of sampling in the genetic ancestry estimates
To balance the ancestry estimates based on studies with different number of samples, we calculated a weighted mean. A slightly higher proportion of European ancestry (weighted mean 68.1%, mean 62.4%) and lower African (weighted mean 19.6%, mean 22.6%) and Native American ancestries (weighted mean 11.6%, mean 14.7%) were observed (Table 1). However, these differences between mean and weighted mean were not statistically significant (Mann Whitney, p=0.965 for European ancestry, p=0.974 for African ancestry, and p=0.990 for Native American ancestry). Concerning regions, the weighted mean showed also slightly different results, particularly for European and African ancestries for the Northeast region (Table 1).
Correlation analysis between number of individuals and ancestry estimated for each geographic region using the same type of marker did not show any significant result (Spearman’s correlation test, p > 0.05). Moreover, the analysis comparing different number of individuals from the same city was performed for Belem/PA, Salvador/BA, and Rio de Janeiro/RJ by INDEL; and Fortaleza/CE and São Paulo/SP by SNP ancestry estimates. No significant result was observed (data not shown).
Ancestry estimates among Afro-descendant communities and Native American tribes
Concerning studies of partially isolated ethnic populations included in this review, two focused in Native American tribes (Terena/MS and Santa Isabel/AM) and 15 in Afro-descendant communities from different geographical Brazilian regions (Table S3 (122.4KB, pdf) ). The two Native American tribes were studied by the same group, using the same panel of molecular markers and showed similar results of estimated ancestry (mean of 15% European, 8% African, and 77% Native American), despite the tribes being from different geographic regions. Among the studies that analyzed the Afro-descendant populations, the ancestry estimates were quite variable with the means of 32.7% (ranging from 4 to 74%) for European ancestry, 52.4% (26 to 89%) for African, and 14.8% (0 to 38%) for Native American. The ancestry estimates for both Afro-descendant communities and Native American tribes showed different means compared to other populations from the same region in Brazil (Table S3 (122.4KB, pdf) and Table 1).
Discussion
Several studies have pointed out the differences of genetic ancestry among the regions of Brazil (Callegari-Jacques et al., 2003; Godinho et al., 2008; Lins et al., 2010; Manta et al., 2013a). We performed an extensive review of the literature for a better understanding of the heterogeneity of the Brazilian population from different geographic regions and analyzed the influence of some factors, such as the molecular marker type and number of samples, in ancestry estimates. Nineteen states of Brazil were represented in this study and only seven were not included because studies about these states did not match our inclusion criteria or they did not have published information. We found that the mean proportions of European, African, and Native American ancestries in Brazil were 68.1%, 19.6%, and 11.6%, respectively. Prior to ours, Moura et al. (2015) performed a systematic review with meta-analysis of the genetic admixture studies of the Brazilian population compared to other countries in Latin America. Although that study included a lower number of populations from different Brazilian states (n=38, this study n=81), the authors found similar means of genetic ancestry (European – 62%, African – 21% and Native American – 17%).
A global overview of all studies showed a major European contribution across regions of the country. However, the distribution of the ancestral groups along the Brazilian territory did not occur homogeneously, differing significantly depending on the geographic region. This heterogeneity can be explained by the occupation history of each region. The Northeast was the cradle of Portuguese colonization, and it was there where they landed and established the great majority of African slaves (Holanda, 1995; Ribeiro, 1995; Schaan et al., 2017), explaining the great African ancestry observed in this region. We identified that Bahia and Ceará had distinct profiles of genetic ancestries within the Northeast region. This result has suggested that Salvador and Jequié, both cities in Bahia state, might have preserved the African ancestry from the high number of slaves that were brought there. Salvador is considered the most African city in Brazil, with around 80% of Afro-descendants (Azevedo et al., 1981; IBGE at https://ww2.ibge.gov.br/home/default.php). Moreover, the slaves arriving in Bahia were mainly originated from Nigeria, Ghana, and Benin, different from the slaves sent to other Brazilian States, who came mainly from Angola, Congo, and Mozambique (Verger, 1968; Kehdy et al., 2015). However, it is not clear whether the actual genetic profile of the populations from Bahia and Ceará were influenced by this differential slave trade. In the mid-seventeenth century, the decline of the sugar economy in the Northeast Region and the discovery of gold and precious stones in areas of the actual state of Minas Gerais caused a great migration of Africans to the Southeast (Krieger et al., 1965; Schwartz, 1988; Ribeiro, 1995). This migration caused a great genetic admixture in the southeastern populations, overlapping in the tri-plot with populations from the Northeast and South. In the North, due to the isolated geographic location and the Amazon forest, the occupation process occurred later and more effectively because of governmental development projects (Martine, 1990; de Castro et al., 2007). This region has a high number of Native Americans, and due to social policies that encouraged marriage between white men and Native American women, this region has a great influence of the Native American genetic ancestry (Benchimol, 1999; Carneiro-da-Cunha, 1992; Batista dos Santos et al., 1999). One of the highest proportions of Native Americans (38%) was observed in studies conducted in the Amazonas state (Manta et al., 2013a; Kano et al., 2016; 2018).
In the South there was an intense migratory flow of Europeans, especially in the 18th and 19th centuries (Holanda, 1995; Carvalho-Silva et al., 2001; Marrero et al., 2005; Pena, 2005; Pena et al., 2011). In this region, the first group of colonizers consisting of Portuguese immigrants arrived in 1808, and later the arrival of Europeans for wage labor was encouraged by the government (Salzano and Freire-Maia, 1967; Ribeiro, 1995; Alves-Silva et al., 2000; Marrero et al., 2005; Pena et al., 2011). This region was sparsely inhabited by Native Americans due to the attacks of Portuguese colonizers (Leite, 1996; Ramos, 2006; Ribeiro, 1995). In addition, the area received small numbers of African slaves (Levy, 1974). This immigration process explains why this region showed the most different proportions of genetic ancestries, with the highest contribution of European ancestry (81.8%) in Brazil.
For the Central-West region only five studies were selected, and further studies are needed to better understand the genetic makeup of this population. The occupation of this region occurred due to the expansion of the agricultural frontier and the construction of the city of Brasília in the 1970’s (Cunha, 2002, 2006). This region received migrants from all parts of Brazil, showing a strong overlap in the tri-plot with Southeast and Northeast populations.
The estimates of genetic ancestry depend on the selected molecular marker panel, method used to infer population admixture, and the sampling method. For this study, we selected only studies that used nuclear autosomal markers. We excluded the studies that used classical markers, published between the years 1957-2000, which are based indirectly on variation in the products of genes. Despite the unquestionable value of classical markers for human ancestry understanding, as summarized by Cavalli-Sforza et al. (1994), we chose to include only nuclear markers. These markers allow to infer the individual ancestry and are widely used in genetic association studies to control for statistical biases related to population stratification between cases and controls or as a strategy to map susceptibility alleles associated with diseases that differ between recently admixed populations (Seldin, 2007; Tian et al., 2008). Uniparental genetic markers, despite being good lineage markers, the maternal lineage (mitochondrial inheritance), and the paternal lineage (Y chromosome) were also excluded because they represent only the contribution of one side of the ancestry history. Comparing the ancestry estimates for each geographic region, the only significant difference was in the average of European ancestry in the Northeast region estimated using INDELs compared to the estimate using SNPs.
Two studies carried out in the Amazonas state showed a highly consistent estimate of all three ancestries independent of the type of molecular markers (INDELs (Manta et al., 2013a) or SNPs (Kano et al., 2016). However, another study using STRs showed different estimates of ancestry for a population from the same state (Callegari-Jacques et al., 2003). Half of the studies based on samples from same cities showed significant differences in ancestry estimates using distinct types of molecular markers. The difference in ancestry estimates was found between INDEL and SNP (e.g., Belem and Fortaleza), and between INDEL and STR (e.g., Manaus and Porto Velho). However, these comparisons must be viewed with caution, since studies using different sample of individuals from different cities or even from the same city may differ in the level of admixture. The influence of molecular marker panels in the estimates of ancestry is clearer in studies based on the same sample using different maker panels. For instance, Pedrosa (2006) showed different ancestry estimates for the same sample of individuals analyzed using three different panels of markers (8 STRs, 9 SNP/INDELs, and a combination of both). STR panel showed the most distinct results compared to the other two panels (Table S3 (122.4KB, pdf) ). Teló (2010) also reported different ancestry estimates using INDELs and STRs for the same sample of individuals from Salvador/BA (Table 1). Microsatellite markers, frequently used in individual identification, as forensic markers used in all studies included in this review, were not able to detect differences among populations and, consequently, among ethnic groups (Barnholtz-Sloan et al., 2005; Pedrosa, 2006). For microsatellites, it is not possible to claim that similar allelic frequencies in specific loci are directly related to common ancestries or share the same evolutionary history (Santos and Tyler-Smith, 1996). They are markers of fast evolution with high rates of mutation (Ellegren, 2004) with different allelic frequencies in admixture and in parental populations, distorting the ancestry profile of the population. On the other hand, SNPs and INDELs are markers of slow evolution (Stoneking, 2001; Weber et al., 2002). Studies have highlighted that some microsatellites could be more informative than SNPs (Ruiz Linares, 1999; Pfaff et al., 2004; Tang et al., 2005). Therefore, more important than the type of marker is the selection of markers based on their selectivity for ancestral populations (Parra et al., 2001).
An important aspect to be considered for genetic ancestry estimates is the minimum number of markers needed to determine the proportions of parental ancestry within a population. This analysis was possible only for studies based on samples from the same city using different numbers of the same type of marker. Here, only studies using INDELs were compared, without any significant difference between ancestry estimates and number of markers. Ding et al. (2011) showed that using the top 20 ranked informative markers accurately classified all ancestral populations. Lao et al. (2006) suggested that as few as 10 highly informative SNP markers were able to differentiate individuals from Africa, Europe, Asia, and America. Kosoy et al. (2009) also have shown that a few dozen well-selected markers have a similar power of discrimination to sets of larger numbers of random markers. Therefore, the number of markers is highly dependent of the informativeness of the panel as discussed above. The number of markers for ancestry estimates will also depend on the level of genetic differentiation of the admixture populations and the desired stringency of assignment (Baye et al., 2011).
One of the factors of great importance for studies on population ancestry is the nature of the sample. Many studies performed the ancestry estimates using selected samples from hospitals, clinics, universities, diagnostic units, or specific areas of the city, which may not correctly represent the ancestral estimates of the entire population. Moreover, it is very difficult to obtain a truly random sample of the population to be studied. Here, we determined the mean of ancestry weighted by the number of samples to balance potential bias in the sampling. For instance, in the study of Kehdy et al. (2015) the sample from Salvador/BA was from poor areas of the city, with a strong bias for higher African ancestry than the city as a whole (Barreto et al., 2006). Moreover, we did not identify any statistically significant correlation among the number of individuals in the studies performed from the same region or the same cities using the same type of molecular marker and the ancestry estimates. Therefore, the number of individuals per se did not seem to influence the ancestry estimates.
Studies that selected only a few ethnic subgroups of the population were excluded since they might not represent the ancestry estimates of the whole population, such as skin color subgroups studied by Pena et al. (2011). Similarly, some studies were excluded as they included relatives who may cause bias in ancestry estimates (Leite et al., 2011; Giolo et al., 2012). Moreover, studies of specific ethnic communities that have little contact with the dominant civilization, such as Native American tribes and communities of Afro-descendants originally formed by fugitive slaves, the Quilombos, were analyzed separately. The Native American tribes showed differences in estimated average ancestries compared to the Brazilian population. The Afro-descendant communities showed a highly diverse estimate of ancestry with significant differences only for the mean of African and European ancestries compared to the average of other Brazilian populations. This variability reflects the history of each community with different levels of admixture, and the ability of molecular markers to identify genetic ancestries in admixture population, as discussed above for the data from Pedrosa (2006).
The history of the formation of quilombos was quite variable across the country, with distinct levels of interaction with Europeans and Native Americans. The distribution of Native American tribes and Afro-descendants’ communities are asymmetrical over the Brazilian territory. Consistent with the highest African ancestry, the Northeast region also has the highest number of Afro-descendant communities (n=1,724). Moreover, the North region has the highest Native American ancestry and also higher number of Native American individuals (305,873 individuals - 37.4% of the total indigenous population in the country) (www.funai.gov.br/index.php/indios-no-brasil/so-sao).
The correlation between genetic ancestry and ethno-racial classification in Brazilian individuals was not the aim of this review. However, it is important to mention the recent study by Ruiz-Linares et al. (2014) that analyzed more than 7,000 individuals from Latin America including 1,594 Brazilians and showed a significant correlation between genetic ancestry and the self-perception of ancestry. Despite the correlation found, the authors suggested that some physical appearance traits and other factors, possibly non-biological factors, biased the self-perception of ancestry. Lima-Costa et al. (2015) also showed a significant correlation that is most consistent with the extremes of African proportions.
Several studies have pointed out high gene flow and genetic heterogeneity among regions of Brazil. In this study, we scanned the literature to obtain a robust review of the Brazilian population’s ancestry. In 81 populations included in this review there was a predominance of European ancestry, which reflects the process of colonization and the relatively recent European mass immigration to Brazil. In the general population, we identified a low contribution of Native American ancestry that may be due in part to the mass extermination of this population during the colonial period. In areas where slaves were widely used as cheap labor, as in the Northeast and Southeast regions, we found a greater African ancestry. Of importance, this review contributes for a broader understanding of the genetic makeup of the Brazilian population and the roles of marker types and sampling method in the genetic ancestry estimates.
Acknowledgments
CFAB and TNS thank CNPq for researcher scholarships and AMS thanks Fapemig for the doctorate scholarship. The authors thank Daniela Rezende Garcia Junqueira, Fabian Rodrigues dos Santos (Faculdade de medicina da UFMG), and Glaucia Fernandes Cota for their assistance in the scoping review methodology.
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Francisco Mauro Salzano
Conflict of Interest
The authors declare no conflict of interests.
Author contributions
CFAB and TNS conceived and designed the study, AMS conducted the experiments, AMS and SSR analyzed the data, AMS and CFADB wrote the manuscript, all authors read and approved the final version.
References
- Alves-Silva J, da Silva Santos M and Guimaraes PE, Ferreira ACS, Bandelt HJ, Pena SDJ, Prado VF. The ancestry of Brazilian mtDNA lineages. Am J Hum Genet. 2000;67:444–461. doi: 10.1086/303004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino SN, Messetti AC, Hoshi RAL. Analysis of susceptibility polymorphisms for nonsyndromic cleft lip with or without cleft palate in the Brazilian population. Birth Defects Res A Clin Mol Teratol. 2014;100:36–42. doi: 10.1002/bdra.23204. [DOI] [PubMed] [Google Scholar]
- Arksey H, O’Malley L. Scoping studies: Towards a methodological framework. Int J Social Res Methodol. 2005;8:19–32. [Google Scholar]
- Azevedo ES, Silva KMC, Silva MCBO, Lima AMVMD, Fortuna CMM, Santos MG. Genetic anthropological studies in the island of Itaparica, Bahia, Brazil. Hum Hered. 1981;31:353–357. doi: 10.1159/000153237. [DOI] [PubMed] [Google Scholar]
- Barnholtz-Sloan JS, Pfaff CL, Chakraborty R, Long JC. Informativeness of the CODIS STR loci for admixture analysis. J Forensic Sci. 2005;50:1322–1326. [PubMed] [Google Scholar]
- Barreto ML, Cunha SS, Alcântara-Neves N, Carvalho LP, Cruz AA, Stein RT, Genser B, Cooper PJ, Rodrigues LC. Risk factors and immunological pathways for asthma and other allergic diseases in children: background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (Salvador-SCAALA study) BMC Pulm Med. 2006;6:15. doi: 10.1186/1471-2466-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista dos Santos SE, Rodrigues JD, Ribeiro-dos-Santos AK, Zago MA. Differential contribution of indigenous men and women to the formation of an urban population in the Amazon region as revealed by mtDNA and Y-DNA. Am J Phys Anthropol. 1999;109:175–180. doi: 10.1002/(SICI)1096-8644(199906)109:2<175::AID-AJPA3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Benchimol S. Amazônia: Formação social e cultural. Valer; Manaus: 1999. p. 480. [Google Scholar]
- Benedet AL, Moraes CF, Camargos EF, Oliveira LF, Souza VC, Lins TC, Henriques AD, Carmo DG, Machado-Silva W, Araújo CN, et al. Amerindian genetic ancestry protects against Alzheimer’s disease. Dement Geriatr Cogn Disord. 2012;33:311–317. doi: 10.1159/000339672. [DOI] [PubMed] [Google Scholar]
- Bernardez-Pereira S, Gioli-Pereira L, Marcondes-Braga FG, Santos PC, Spina JM, Horimoto AR, Santos HC, Bacal F, Fernandes F, Mansur AJ, et al. Genomic ancestry as a predictor of haemodynamic profile in heart failure. Open Heart. 2016;3:e000434. doi: 10.1136/openhrt-2016-000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethell L. Notas sobre as populações americanas ás vesperas das invasões européias. In: Bethell L, editor. História da America Latina: A América Colonial. Vol. 22. Edusp, São Paulo; Fundação Alexandre de Gusmão; Brasília: 1999. pp. 129–131. [Google Scholar]
- Baye TM, He H, Ding L, Kurowski BG, Zhang X, Martin LJ. Population structure analysis using rare and common functional variants. BMC Proc. 2011;5:S8. doi: 10.1186/1753-6561-5-S9-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito LA, Cruz LA, Rocha KM, Barbara LK, Silva CB, Bueno DF, Aguena M, Bertola DR, Franco D, Costa AM, et al. Genetic contribution for non-syndromic cleft lip with or without cleft palate (NS CL/P) in different regions of Brazil and implications for association studies. Am J Med Genet A. 2011;155A:1581–1587. doi: 10.1002/ajmg.a.34036. [DOI] [PubMed] [Google Scholar]
- Brito LA, Paranaiba LM, Bassi CF, Masotti C, Malcher C, Schlesinger D, Rocha KM, Cruz LA, Bárbara LK, Alonso N, et al. Region 8q24 is a susceptibility locus for nonsyndromic oral clefting in Brazil. Birth Defects Res A Clin Mol Teratol. 2012;94:464–468. doi: 10.1002/bdra.23011. [DOI] [PubMed] [Google Scholar]
- Callegari-Jacques SM, Salzano FM. Brazilian Indian/non indian interactions and their effects. Ciênc Cult. 1999;51:166–174. [Google Scholar]
- Callegari-Jacques SM, Grattapaglia D, Salzano FM, Salamoni SP, Crossetti SG, Ferreira ME, Hutz MH. Historical genetics: Spatiotemporal analysis of the formation of the Brazilian population. Am J Hum Biol. 2003;15:824–834. doi: 10.1002/ajhb.10217. [DOI] [PubMed] [Google Scholar]
- Cardena MM, Ribeiro-Dos-Santos A, Santos S, Mansur AJ, Pereira AC, Fridman C. Assessment of the relationship between self-declared ethnicity, mitochondrial haplogroups and genomic ancestry in Brazilian individuals. PLoS One. 2013;8:e62005. doi: 10.1371/journal.pone.0062005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso GL, Diniz IG, Silva NA, Cunha DA, Silva JS, Junior, Uchôa CT, Santos SE, Trindade SM, Cardoso MS, Guerreiro JF. DNA polymorphisms at BCL11A, HBS1L-MYB and Xmn1-HBG2 site loci associated with fetal hemoglobin levels in sickle cell anemia patients from Northern Brazil. Blood Cells Mol Dis. 2014;53:176–179. doi: 10.1016/j.bcmd.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Carneiro-da-Cunha M. História dos Índios no Brasil. 2nd edition. Companhia das Letras; São Paulo: 1992. p. 658. [Google Scholar]
- Carvalho DC, Wanderley AV, Amador MA, Fernandes MR, Cavalcante GC, Pantoja KB, Mello FA, de Assumpção PP, Khayat AS, Ribeiro-Dos-Santos Â, et al. Amerindian genetic ancestry and INDEL polymorphisms associated with susceptibility of childhood B-cell Leukemia in an admixed population from the Brazilian Amazon. Leuk Res. 2015;39:1239–1245. doi: 10.1016/j.leukres.2015.08.008. [DOI] [PubMed] [Google Scholar]
- Carvalho-Silva DR, Santos FR, Rocha J, Pena SD. The phylogeography of Brazilian Y-chromosome lineages. Am J Hum Genet. 2001;68:281–286. doi: 10.1086/316931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiano GC, Santos EJ, Maia MH, Furini Ada C, Storti-Melo LM, Tomaz FM, Trindade PC, Capobianco MP, Amador MA, Viana GM, et al. Impact of population admixture on the distribution of immune response co-stimulatory genes polymorphisms in a Brazilian population. Hum Immunol. 2015;76:836–842. doi: 10.1016/j.humimm.2015.09.045. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press; Princeton: 1994. p. 438. [Google Scholar]
- Coelho AV, Moura RR, Cavalcanti CA, Guimarães RL, Sandrin-Garcia P, Crovella S, Brandão LA. A rapid screening of ancestry for genetic association studies in an admixed population from Pernambuco, Brazil. Genet Mol Res. 2015;14:2876–2884. doi: 10.4238/2015.March.31.18. [DOI] [PubMed] [Google Scholar]
- Cunha JMP. A migração do Centro-Oeste brasileiro no período 1970-96: O esgotamento de um processo de ocupação. Unicamp; Campinas: 2002. p. 169. [Google Scholar]
- Cunha JMP. Dinamica migratória e o processo de ocupação do Centro-Oeste brasileiro: O caso de Mato Grosso. Rev Bras Estud Popul. 2006;23:87–107. [Google Scholar]
- Curtin PD. The Atlantic slave trade: A census. Am Hist Rev. 1969;75:2011. [Google Scholar]
- De Azevedo S, Nocera A, Paschetta C, Castillo L, González M, Gonzalez-José R. Evaluating microevolutionary models for the early settlement of the New World: the importance of recurrent gene flow with Asia. Am J Phys Anthropol. 2011;146:539–552. doi: 10.1002/ajpa.21564. [DOI] [PubMed] [Google Scholar]
- De Castro MC, Sawyer DO, Singer BH. Spatial patterns of malaria in the Amazon: Implications for surveillance and targeted interventions. Health Place. 2007;13:368–380. doi: 10.1016/j.healthplace.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Ding L, Wiener H, Abebe T, Altaye M, Go RC, Kercsmar C, Grabowski G, Martin LJ, Khurana Hershey GK, Chakorborty R, et al. Comparison of measures of marker informativeness for ancestry and admixture mapping. BMC Genomics. 2011;12:622. doi: 10.1186/1471-2164-12-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Rego Borges RA, Sá J, Hoshi R, Viena CS, Mariano LC, de Castro Veiga P, Medrado AP, Machado RA, de Aquino SN, Messetti AC, et al. Genetic risk factors for nonsyndromic cleft lip with or without cleft palate in a Brazilian population with high African ancestry. Am J Med Genet A. 2015;167A:2344–2349. doi: 10.1002/ajmg.a.37181. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nature Reviews Genetics. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20:19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- Fagundes NJ, Kanitz R, Eckert R, Valls AC, Bogo MR, Salzano FM, Smith DG, Silva WA, Jr, Zago MA, Ribeiro-dos-Santos AK, et al. Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. Am J Hum Genet. 2008;82:583–592. doi: 10.1016/j.ajhg.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LAS, Pimentel BJ, Almeida de Azevedo D. Allele frequencies of nine STR loci- D16S539, D7S820, D13S317, CSF1PO, TPOX, TH01, F13A01, FESFPS and vWA - in the population from Alagoas, northeastern Brazil. Forens Sci Int. 2002;130:187–188. doi: 10.1016/s0379-0738(02)00353-5. [DOI] [PubMed] [Google Scholar]
- França AK. Freqüências alélicas de quatro locos SRTs: HUMCSF1PO, HUMTH01, HUMTPOx e vWA — em duas populações do município de Porto Velho, RO. M. Sc. Thesis. Universidade Federal de Rondônia; Porto Velho: 2005. [Google Scholar]
- Francez CPA, Rodrigues EM, Frazao GF, Borges NDR, Batista dos Santos SE. Allelic frequencies and statistical data obtained from 12 codis STR loci in an admixed population of the Brazilian Amazon. Genet Mol Biol. 2011;34:35–39. doi: 10.1590/S1415-47572011000100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez PA, Ribeiro-Rodrigues EM, dos Santos SE. Allelic frequencies and statistical data obtained from 48 AIM INDEL loci in an admixed population from the Brazilian Amazon. Forensic Sci Int Genet. 2012;6:132–135. doi: 10.1016/j.fsigen.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Giolo SR, Soler JM, Greenway SC, Almeida MA, de Andrade M, Seidman JG, Seidman CE, Krieger JE, Pereira AC. Brazilian urban population genetic structure reveals a high degree of admixture. Eur J Hum Genet. 2012;20:111–116. doi: 10.1038/ejhg.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho NMO, Gontijo CC, Diniz MECG, Falcão-Alencar G, Dalton GC, Amorim CEG, Barcelos RSS, Klautau-Guimarães MN, Oliveira SF. Regional patterns of genetic admixture in South America. Forensic Sci Int. 2008;1:329–330. [Google Scholar]
- Guindalini C, Colugnati FA, Pellegrino R, Santos-Silva R, Bittencourt LR, Tufik S. Influence of genetic ancestry on the risk of obstructive sleep apnoea syndrome. Eur Respir J. 2010;36:834–841. doi: 10.1183/09031936.00146809. [DOI] [PubMed] [Google Scholar]
- Graham DJ, Midgley NG. Graphical representation of particle shape using triangular diagrams: an Excel spreadsheet method. Earth Surface Processes Landforms. 2000;25:1473–1477. [Google Scholar]
- Holanda SB. Raízes do Brasil. 26ª edição. Companhia das Letras; São Paulo: 1995. p. 224. [Google Scholar]
- Kano FS, de Souza AM, Torres LM, Costa MA, Souza-Silva FA, Sanchez BAM, Fontes CJF, Soares IS, de Brito CFA, Carvalho LH, et al. Susceptibility to Plasmodium vivax malaria associated with DARC (Duffy antigen) polymorphisms is influenced by the time of exposure to malaria. Sci Rep. 2018;8:13851. doi: 10.1038/s41598-018-32254-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano FS, Souza-Silva FA, Torres LM, Lima BA, Sousa TN, Alves JR, Rocha RS, Fontes CJ, Sanchez BA, Adams JH, et al. The presence, persistence and functional properties of Plasmodium vivax Duffy Binding Protein II antibodies are influenced by HLA Class II allelic variants. PLoS Negl Trop Dis. 2016;10:e0005177. doi: 10.1371/journal.pntd.0005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehdy FS, Gouveia MH, Machado M, Magalhães WC, Horimoto AR, Horta BL, Moreira RG, Leal TP, Scliar MO, Soares-Souza GB, et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci U S A. 2015;112:8696–8701. doi: 10.1073/pnas.1504447112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen A, Miyamoto MM, Mulligan CJ. A three-stage colonization model for the peopling of the Americas. PLoS One. 2008;3:e1596. doi: 10.1371/journal.pone.0001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger H, Morton NE, Mi MP, Azevêdo E, Freire-Maia A, Yasuda N. Racial admixture in north-eastern Brazil. Ann Hum Genet. 1965;29:113–125. doi: 10.1111/j.1469-1809.1965.tb00507.x. [DOI] [PubMed] [Google Scholar]
- Lao O, van Duijn K, Kersbergen P, de Knijff P, Kayser M. Proportioning whole-genome single-nucleotide-polymorphism diversity for the identification of geographic population structure and genetic ancestry. Am J Hum Genet. 2006;78:680–690. doi: 10.1086/501531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite IB. Negros no Sul do Brasil. Letras Contemporâneas; Florianópolis: 1996. p. 143. [Google Scholar]
- Leite TK, Fonseca RM, de Franca NM, Parra EJ, Pereira RW. Genomic ancestry, self-reported “color” and quantitative measures of skin pigmentation in Brazilian admixed siblings. PLoS One. 2011;6:e27162. doi: 10.1371/journal.pone.0027162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC. The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MSF. The role of international migration on the evolution of the Brazilian population (1872 to 1972) Rev Saude Publ. 1974;8:49–90. [PubMed] [Google Scholar]
- Lima-Costa MF, Rodrigues LC, Barreto ML, Gouveia M, Horta BL, Mambrini J, Kehdy FS, Pereira A, Rodrigues-Soares F, Victora CG, et al. Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative) Sci Rep. 2015;5:9812. doi: 10.1038/srep09812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins TC, Vieira RG, Abreu BS, Grattapaglia D, Pereira RW. Genetic composition of Brazilian population samples based on a set of twenty-eight ancestry informative SNPs. Am J Hum Biol. 2010;22:187–192. doi: 10.1002/ajhb.20976. [DOI] [PubMed] [Google Scholar]
- Lins TC, Vieira RG, Grattapaglia D, Pereira RW. Population analysis of vitamin D receptor polymorphisms and the role of genetic ancestry in an admixed population. Genet Mol Biol. 2011a;34:377–385. doi: 10.1590/S1415-47572011000300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins TC, Vieira RG, Abreu BS, Gentil P, Moreno-Lima R, Oliveira RJ, Pereira RW. Genetic heterogeneity of self-reported ancestry groups in an admixed Brazilian population. J Epidemiol. 2011b;21:240–245. doi: 10.2188/jea.JE20100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes TR, Santos S, Ribeiro-Dos-Santos A, Resque RL, Pinto GR, Yoshioka FK. Population data of the 46 insertion-deletion (INDEL) loci in population in Piaui State, Northeastern Brazil. Forensic Sci Int Genet. 2014;9:e13–15. doi: 10.1016/j.fsigen.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Manta FSN, Pereira R, Vianna R, Araújo ARB, Gitaí DLG, Silva DA, Wolfgramm EV, Pontes IM, Aguiar JI, Moraes MO, et al. Revisiting the genetic ancestry of Brazilians using autossomal AIM-INDELs. PloS One. 2013a;8:e75145. doi: 10.1371/journal.pone.0075145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manta FS, Pereira R, Caiafa A, Silva DA, Gusmão L, Carvalho EF. Analysis of genetic ancestry in the admixed Brazilian population from Rio de Janeiro using 46 autosomal ancestry- informative indel markers. Ann Hum Biol. 2013b;40:94–98. doi: 10.3109/03014460.2012.742138. [DOI] [PubMed] [Google Scholar]
- Marrero AR, Leite FPN, Carvalho BA, Peres LM, Kommers TC, Cruz IM, Salzano FM, Ruiz-Linares A, Silva WA, Júnior, Bortolini MC. Heterogeneity of the genome ancestry of individuals classified as White in the state of Rio Grande do Sul Brazil. Am J Hum Biol. 2005;17:496–506. doi: 10.1002/ajhb.20404. [DOI] [PubMed] [Google Scholar]
- Martine G. Rondônia and the fate of small producers. In: Goodman D, Hall AL, editors. The future of Amazonia: Destruction or Sustainable Development. Macmillan Publ; Basingstoke: 1990. pp. 23–48. [Google Scholar]
- Martins JA, Figueiredo RF, Yoshizaki CS, Paneto GG, Cicarelli RM. Genetic data of 15 autosomal STR loci: an analysis of the Araraquara population colonization (São Paulo, Brazil) Mol Biol Rep. 2011;38:5397–5403. doi: 10.1007/s11033-011-0693-6. [DOI] [PubMed] [Google Scholar]
- Moura RR, Coelho AV, Balbino VQ, Crovella S, Brandão LA. Meta-analysis of Brazilian genetic admixture and comparison with other Latin America countries. Am J Hum Biol. 2015;27:674–680. doi: 10.1002/ajhb.22714. [DOI] [PubMed] [Google Scholar]
- Mychaleckyj JC, Havt A, Nayak U, Pinkerton R, Farber E, Concannon P, Lima AA, Guerrant RL. Genome-wide analysis in Brazilians reveals highly differentiated Native American genome regions. Mol Biol Evol. 2017;34:559–574. doi: 10.1093/molbev/msw249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AF, Oliveira JS, Silva JC, Junior, Barbosa AA. Genomic ancestry evaluated by ancestry-informative markers in patients with sickle cell disease. Genet Mol Res. 2016;15:15017604. doi: 10.4238/gmr.15017604. [DOI] [PubMed] [Google Scholar]
- Nastri AC, Malta FM, Diniz MA, Yoshino A, Abe-Sandes K, Santos SE, Lyra AC, Carrilho FJ, Pinho JR. Association of IFNL3 and IFNL4 polymorphisms with hepatitis C virus infection in a population from southeastern. Brazil Arch Virol. 2016;161:1477–1484. doi: 10.1007/s00705-016-2809-8. [DOI] [PubMed] [Google Scholar]
- Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garvey WT, Jin L. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pedrosa MAF. Composição Genética de quatro populações remanescentes de quilombos do Brasil com base em microssatélitese marcadores de ancestralidade. M. Sc. Thesis. Universidade de Brasília; Brasília: 2006. p. 143. [Google Scholar]
- Pena SD. Reasons for banishing the concept of race from Brazilian medicine. Hist Cienc Saude Manguinhos. 2005;12:321–346. doi: 10.1590/s0104-59702005000200006. [DOI] [PubMed] [Google Scholar]
- Pena SD, Di Pietro G, Fuchshuber-Moraes M, Genro JP, Hutz MH, Kehdy FS, Kohlrausch F, Magno LA, Montenegro RC, Moraes MO, et al. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS One. 2011;6:e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R, Phillips C, Pinto N, Santos C, Santos SE, Amorim A, Carracedo Á, Gusmão L. Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS One. 2012;7:e29684. doi: 10.1371/journal.pone.0029684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff CL, Barnholtz-Sloan J, Wagner JK, Long JC. Information on ancestry from genetic markers. Genet Epidemiol. 2004;26:305–315. doi: 10.1002/gepi.10319. [DOI] [PubMed] [Google Scholar]
- Queiroz EM, Santos AM, Castro IM, Machado-Coelho GL, Cândido AP, Leite TM, Pereira RW, Freitas RN. Genetic composition of a Brazilian population: The footprint of the Gold Cycle. Genet Mol Res. 2013;12:5124–5133. doi: 10.4238/2013.October.29.6. [DOI] [PubMed] [Google Scholar]
- Ramos AD. A formação histórica dos municípios da região das Missões do Brasil. 2006. http://www.urisan.tche.br/~iphan/upload/downloads/file1.pdf.
- Ramos BR, Mendes ND, Tanikawa AA, Amador MA, dos Santos NP, dos Santos SE, Castelli EC, Witkin SS, da Silva MG. Ancestry informative markers and selected single nucleotide polymorphisms in immunoregulatory genes on preterm labor and preterm premature rupture of membranes: a case control study. BMC Pregnancy Childbirth. 2016;16:30. doi: 10.1186/s12884-016-0823-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro D. O povo brasileiro: A formação e o sentido do Brasil. Companhia das Letras; São Paulo: 1995. p. 477. [Google Scholar]
- Roberts DF, Hiorns RW. The dynamics of racial admixture. Am J Hum Genet. 1962;14:261–277. [PMC free article] [PubMed] [Google Scholar]
- Rolim H, Cronemberger S, Rangel H, Batista WD, Bastos-Rodrigues L, De Marco L. The role of genetic ancestry in Brazilian patients with primary congenital glaucoma. J Glaucoma. 2016;25:e24–28. doi: 10.1097/IJG.0000000000000167. [DOI] [PubMed] [Google Scholar]
- Ruiz-Linares A. Microsatellites and the reconstruction of the history of human populations. In: Goldstein DB, Schlötterer C, editors. Microsatellites — Evolution and Applications. Oxford University Press; Oxford: 1999. pp. 183–197. [Google Scholar]
- Ruiz-Linares A, Adhikari K, Acuña-Alonzo V, Quinto-Sanchez M, Jaramillo C, Arias W, Fuentes M, Pizarro M, Everardo P, de Avila F, et al. Admixture in Latin America: Geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10:e1004572. doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzano FM, Bortolini MC. The evolution and genetics of Latin American population. Cambridge University Press; Cambridge: 2002. p. 512. [Google Scholar]
- Salzano FM, Freire-Maia N. Populações brasileira, aspectos demográficos, genéticos e antropológicos. Companhia Editora Nacional; São Paulo: 1967. p. 177. [Google Scholar]
- Salzano FM, Freire-Maia N. Problems in human biology: A study of Brazilian populations. Wayne State University Press; Detroit: 1970. p. 200. [Google Scholar]
- Sans M. Admixture studies in Latin America: From the 20th to the 21st century. Hum Biol. 2000;72:155–177. [PubMed] [Google Scholar]
- Santos FR, Tyler-Smith C. Reading the human Y chromosome: The emerging DNA markers and human genetic history. Braz J Genet. 1996;19:665–670. [Google Scholar]
- Santos FR, Bianchi NO, Pena SD. Worldwide distribution of human Y- hromosome haplotypes. Genome Res. 1996;6:601–611. doi: 10.1101/gr.6.7.601. [DOI] [PubMed] [Google Scholar]
- Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK, Pereira R, Gusmão L, Amorim A, Guerreiro JF, Zago MA, Matte C, Hutz MH, et al. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31:184–190. doi: 10.1002/humu.21159. [DOI] [PubMed] [Google Scholar]
- Santos RV, Fry PH, Monteiro S, Maio MC, Rodrigues JC, Bastos-Rodrigues L, Pena SD. Color, race, and genomic ancestry in Brazil: Dialogues between anthropology and genetics. Curr Anthropol. 2009;50:787–819. doi: 10.1086/644532. [DOI] [PubMed] [Google Scholar]
- Schaan AP, Costa L, Santos D, Modesto A, Amador M, Lopes C, Rabenhorst SH, Montenegro R, Souza BDA, Lopes T, et al. mtDNA structure: The women who formed the Brazilian Northeast. BMC Evol Biol. 2017;17:185. doi: 10.1186/s12862-017-1027-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schwartz SB. Recent trends in the study of slavery in Brazil. Luso-Braz Rev. 1988;25:1–25. [Google Scholar]
- Scliar MO, Vaintraub MT, Vaintraub PM, Fonseca CG. Admixture analysis with forensic microsatellites in Minas Gerais, Brazil: The ongoing evolution of the capital and of an African-derived community. Am J Phys Anthropol. 2009;139:591–595. doi: 10.1002/ajpa.21046. [DOI] [PubMed] [Google Scholar]
- Seldin MF. Admixture mapping as a tool in gene discovery. Curr Opin Genet Dev. 2007;17:177–181. doi: 10.1016/j.gde.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Zuccherato LW, Soares-Souza GB, Vieira ZM, Cabrera L, Herrera P, Balqui J, Romero C, Jahuira H, Gilman RH, et al. Development of two multiplex mini-sequencing panels of ancestry informative SNPs for studies in Latin Americans: An application to populations of the State of Minas Gerais (Brazil) Genet Mol Res. 2010;9:2069–2085. doi: 10.4238/vol9-4gmr911. [DOI] [PubMed] [Google Scholar]
- Silva MT, Sandhya Rani MR, Costa GNO, Figueiredo MA, Melo PS, Nascimento JF, Molyneaux ND, Barreto ML, Reis MG, Teixeira MG, et al. The correlation between ancestry and color in two cities of Northeast Brazil with contrasting ethnic compositions. Eur J Hum Genet. 2015;23:984–989. doi: 10.1038/ejhg.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza ES, Araújo LF, Alencar DO, Santos SEB, Silva WA, Jr., Ferreira CA, Baddini-Martinez J. Does ethnic ancestry play a role in smoking? An Acad Bras Cienc. 2015;87:447–453. doi: 10.1590/0001-3765201520140187. [DOI] [PubMed] [Google Scholar]
- Stoneking M. Single nucleotide polymorphisms. From the evolutionary past.. Nature. 2001;409:821–822. doi: 10.1038/35057279. [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G, Vargens DD, Struchiner CJ, Bastos-Rodrigues L, Pena SD. Self-reported skin color, genomic ancestry and the distribution of GST polymorphisms. Pharmacogenet Genomics. 2007;17:765–771. doi: 10.1097/FPC.0b013e3281c10e52. [DOI] [PubMed] [Google Scholar]
- Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martinez-Labarga C, Khusnutdinova EK, et al. Beringian standstill and spread of Native American founders. PLoS One. 2007;2:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- Teló EP. Estimativa de mistura étnica avaliada por Marcadores Informativos de Ancestralidade (AIMs) e Microssatélites. D. Sc. Thesis. Centro de Pesquisas Gonçalo Moniz; Salvador: 2010. p. 46. [Google Scholar]
- Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: Population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger P. Flux et Reflux de la Traite des Nègres Entre le Golfe de Benin et Bahia de Todos os Santos. Mouton Press; Paris: 1968. p. 720. [Google Scholar]
- Vieira PC, Burbano RM, Fernandes DC, Montenegro RC, Santos SE, Sortica VA, Assumpção PP, Ribeiro-Dos-Santos ÂK, Carvalho AA, Santos NP. Population stratification effect on cancer susceptibility in an admixed population from Brazilian Amazon. Anticancer Res. 2015;35:2009–2014. [PubMed] [Google Scholar]
- Vieira-Machado CD, Carvalho FM, Silva LCS, Santos SE, Martins C, Poletta FA, Mereb JC, Vieira AR, Castilla EE, Orioli IM. Analysis of the genetic ancestry of patients with oral clefts from South American admixed populations. Eur J Oral Sci. 2016;124:406–411. doi: 10.1111/eos.12275. [DOI] [PubMed] [Google Scholar]
- Wagner SC, Lindenau JD, Castro SM, Santin AP, Zaleski CF, Azevedo LA, Santos ÂKR, Santos SE, Hutz MH. High frequency of Hb E-Saskatoon (HBB: c.67G > A) in Brazilians: A new genetic origin? Hemoglobin. 2016;40:228–230. doi: 10.1080/03630269.2016.1189433. [DOI] [PubMed] [Google Scholar]
- Weber JL, David D, Heil J, Fan Y, Zhao C, Marth G. Human diallelic insertion/deletion polymorphisms. Am J Hum Genet. 2002;71:854–862. doi: 10.1086/342727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.