Abstract
Parthenogenetically activated oocytes cannot develop to term in mammals owing to abnormal epigenetic modifications. Methylation of the N6 position of adenosine (m6A) is a post-transcriptional epigenetic modification of RNA. To investigate the role of m6A methylation in parthenogenetic (PA) embryonic development, we analyzed METTL3, METTL14, FTO, ALKBH5, YTHDF2, IGF2BP1, and IGF2BP2 expression by quantitative real-time PCR. These genes were found dynamically expressed during the 2-cell, 4-cell, 8-cell, and blastocyst stages of the embryo. Compared to normally fertilized embryos, the expression of these genes was perturbed in PA embryos, especially at the 8-cell stage. Furthermore, immunofluorescence was used to detect m6A expression. The results demonstrated that m6A expression decreased in the 2-cell stage, whereas it increased in the 8-cell stage of PA embryos. Taken together, these results suggest that the expression of RNA methylation-related genes was perturbed, leading to abnormal m6A modification during early development in PA embryos.
Keywords: m6A, gene expression, embryo development, parthenogenetic
Parthenogenetic (PA) embryos contain exclusively maternal genomes and have been used extensively to study epigenetic profiles and determine the expression patterns of imprinted genes in human diseases (Park et al., 2009; Zhang et al., 2015). However, due to abnormal epigenetic modification and the absence of paternal gene expression, mammalian PA embryos cannot develop to term (Zhu et al., 2003). Although previous studies have demonstrated perturbations in DNA methylation in PA embryos, studies of RNA methylation in these embryos are limited (Horii et al., 2008; Park et al., 2011).
A growing body of evidence indicates that global mRNA m6A levels are associated with embryonic development (Zhao et al., 2017; Bertero et al., 2018). Recent studies have indicated that m6A methylation is a post-transcriptional modification of RNA, which is regulated by adenosine methyltransferases and demethylases (Dominissini et al., 2012; Geula et al., 2015). The m6A methyltransferases include METTL3 (methyltransferase like 3) and METTL14 (Ping et al., 2014; Li et al., 2017). The m6A demethylases include FTO (fat mass and obesity-associated protein) and ALKBH5 (AlkB homolog 5) (Feng et al., 2014; Zheng et al., 2017). In addition, YTHDF2 (YTH N6-methyladenosine RNA binding protein 2) and IGF2BPs (insulin-like growth factor 2 mRNA-binding proteins) regulate mRNA fate by recognizing the consensus GG (m6A) C sequence (Nguyen et al., 2014; Wang and He, 2014; Huang et al., 2018).
In the present study, the expression patterns of METTL3, METTL14, FTO, ALKBH5, YTHDF2, IGF2BP1, and IGF2BP2 were determined by qRT-PCR during early embryonic development. Furthermore, immunofluorescence (IF) staining was used to compare m6A expression between normally fertilized and PA embryos. The experiments involving mice were carried out in accordance with the guidelines on animal care and use of animals in research and were approved by the Animal Care and Use Committee of Jilin University, Changchun, China (No. 201706005).
To analyze embryonic development, female ICR mice (6–8 weeks old) were obtained from the School of Medical Science, Jilin University. For the superovulation test, female mice were superovulated by an intraperitoneal injection of 10IU pregnant mare’s serum gonadotropin (PMSG; Merck Millipore) followed by an intraperitoneal injection of 10IU human chorionic gonadotropin (hCG; Sigma, USA) 48 h later. Subsequently, the female mice were individually mated with ICR males with proven fertility. The females were euthanized by cervical dislocation and oviducts were removed. Two-cell stage embryos were collected into droplets of pre-equilibrated M16 media (Sigma). The 2-cell stage embryos were then washed and cultured in M16 media overlaid with mineral oil and incubated at 37 °C in a humidified atmosphere of 5% CO2 in the air until they developed to the 4-cell, 8-cell, and blastula stages.
The production of PA embryos was previously described (Ma et al., 2005). Briefly, the oviducts of female mice were removed and cumulus-oocyte complexes (COCs) were collected, which were unfertilized oocytes after PMSG and hCG injection. The cumulus was removed and collected by briefly exposing MII oocytes to serum-free medium containing hyaluronidase (Sigma). The oocytes were treated with calcium ionophore (ionomycin Calcium salt; Sigma) for 5 min. Parthenogenesis was activated after incubation of the unfertilized oocytes in M16 medium containing 6-DMAP (2 mmol/L, Sigma) for 4h. Then, these unfertilized oocytes were transferred to fresh M16 medium. Parthenogenetic activation was confirmed by the presence of two pronuclei, which developed to the 2-cell stage. These parthenogenetic embryos (2-cell stage) were then transferred to fresh M16 medium and incubated until they reached the 4-cell, 8-cell, and blastula stages.
For qPCR analysis, total RNA was extracted from each group of embryos (n = 10) using the AllPrep DNA/RNA Micro Kit (QIAGEN, Germany) following the manufacturer’s instructions. cDNA was synthesized using the First-Strand cDNA Synthesis kit (Promega, USA). Quantitative real-time PCR (qRT-PCR) was performed to determine METTL3, METTL14, FTO, ALKBH5, YTHDF2 IGF2BP1, and IGF2BP2 expression using the BioEasy SYBR Green I Real-Time PCR Kit on the BIO-RAD iQ5 Multicolor Real-Time PCR Detection System (Bioer Technology, China). The primer sequences used in this study are listed in Table S1 (162.6KB, pdf) . PCR was performed by initial denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s. The 2-DDCT method was used to determine relative gene expression, which was normalized to the transcript amount of the endogenous control gene, GAPDH. The experiments were performed at least in triplicate. The qRT-PCR data were analyzed by t-tests using SPSS 16.0 (SPSS Inc., Chicago, IL, USA). A p-value of <0.05 was considered statistically significant.
To detected m6A expression, IF was used. Briefly, the embryos were washed three times in PBS-PVA. Then, the thinning of the zona pellucida was performed using Tyrode’s Solution (Jisskang, China). The embryos werefixedwith 4% paraformaldehyde for 30 min at room temperature (RT). After fixation, the embryos were washed with PBS-PVA and permeabilized with PBS containing 0.2% Triton X-100 for 30 min. Embryos were then incubated in PBS containing 1% bovine serum albumin(BSA) for 1 h. Next, the embryos were probed with m6A (1:500, Abcam) antibodies and incubated at 4 °C overnight. The embryos were washed with PBS three times for 10 minutes each followed by incubation with Alexa Fluor 488-conjugated secondary (anti-rabbit) antibodies for 1 h at RT. DNA was stained with 10 ng/mL Hoechst 33342 (Thermo Scientific) for 15–20 min. The embryos were washed thrice with PBS-PVA for 10 min each, air dried, and mounted on a coverslip and a glass slide using an antifade mounting medium (BOSTER, China). Images of embryos were obtained using a confocal laser scanning microscope.
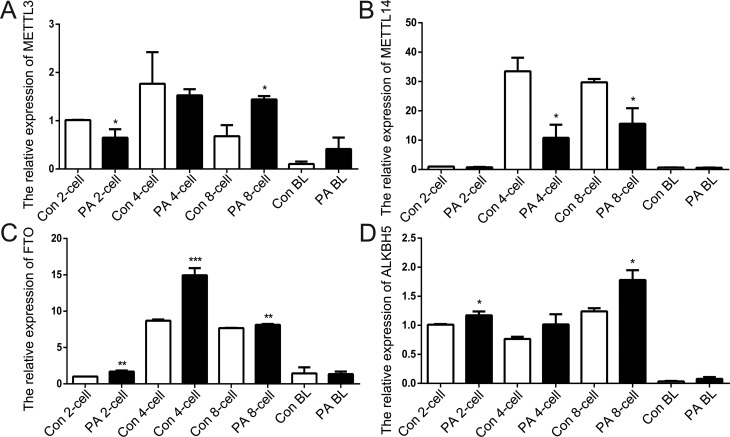
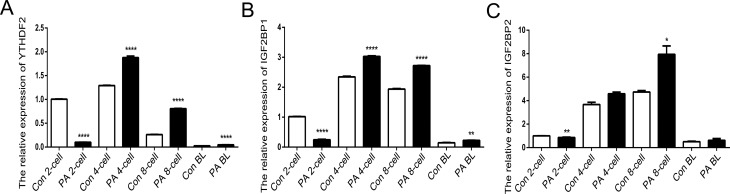
To better understand m6A modification in mice parthenogenesis, we analyzed the expression patterns of m6A-associated genes by qRT-PCR at the 2-cell, 4-cell, 8-cell, and blastocyst stages of the embryos. As m6A `writers’, METTL3 and METTL14 methylate the N6 position of adenosine on RNAs (Sledz and Jinek, 2016). METTL14 is believed to act synergistically with METTL3 (Wang et al., 2014). Moreover, the loss of METTL3 expression leads to embryonic lethality in mice (Geula et al., 2015). Our results showed that compared to the control (Con) embryos, METTL3 levels were lower in the 2-cell stage (p<0.05) and increased in the 8-cell stage (p<0.05) of PA embryos. However, METTL14 expression was low in the 4-cell (p<0.05) and 8-cell stage (p<0.05) of PA embryos (Figure 1A, B). FTO and ALKBH5 are demethylases that act as `erasers’ of the m6A modification (Huang et al., 2015). Previous studies of FTO and ALKBH5 have indicated the reversible nature of the RNA methylation process (Jia et al., 2011; Zheng et al., 2013). Unlike METTL3, the expression of both FTO and ALKBH5 increased in the 2-cell (p<0.05) and 8-cell stages (p<0.05) in the PA embryo (Figure 1C, D). YTHDF2, IGF2BP1, and IGF2BP2 are m6A `readers’ that recognize and bind to the m6A sites on target mRNAs (Ivanova et al., 2017; Huang et al., 2018). It has been demonstrated that m6A selectively binds to YTHDF2 and thus might contribute to the regulation of mRNA degradation (Chandola et al., 2015). IGF2BP1 and IGF2BP2 are critical for mRNA stability and translation through m6A modification (Huang et al., 2018). In this study, the expression of YTHDF2, IGF2BP1, and IGF2BP2 decreased in the 2-cell stage (p<0.01) and increased in the 8-cell stage (p<0.05) of PA embryos (Figure 2A–C). Thus, we demonstrate that METTL3, METTL14, FTO, ALKBH5, YTHDF2, IGF2BP1, and IGF2BP2, which are m6A methylation-related genes, are dynamically expressed during early embryonic development in mice (Kang et al., 2003). Furthermore, the abnormal expression of these genes might result in inappropriate m6A methylation in PA embryos.
Figure 1. Relative expression levels of RNA methylation-related genes in both normal and PA embryos. The expression levels of METTL3 (A), METTL14 (B), FTO (C), and ALKBH5 (D) were analyzed by qRT-PCR. The data are represented as means ± S.E.M. (n = 3). * p<0.05, ** p<0.01, *** p<0.005 indicate the respective statistically significant differences.
Figure 2. Relative expression levels of RNA methylation-related genes in normal and PA embryos. The expression levels of YTHDF2 (A), IGF2BP1 (B), and IGF2BP1 (C) were analyzed by qRT-PCR. The data are represented as the mean ± S.E.M. (n = 3). * p<0.05), ** p<0.01, **** p<0.001 indicate the respective statistically significant differences.
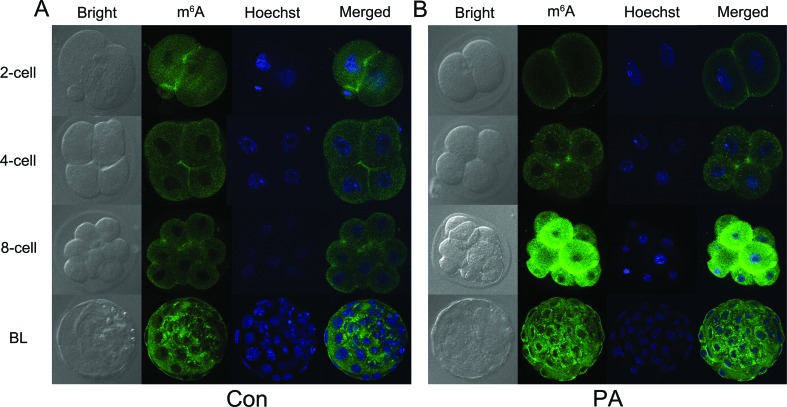
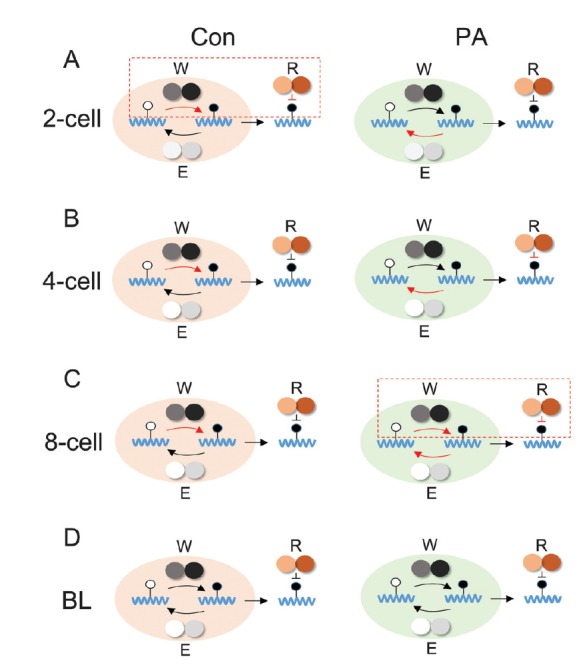
RNA m6A methylation affects RNA stability, which is crucial for embryonic development (Zhao et al., 2017). To further assess the effect of altered m6A methylation on PA embryonic development, we assessed m6A expression using IF staining. The results showed that compared to the Con group, m6A expression decreased in the 2-cell stage, whereas it increased in the 8-cell stage of the PA embryos (Figure 3), which is indicative of the abnormal expression of m6A in these embryos. Based on our results, we speculated that the increase in m6A expression in the 2-cell stage of control embryos was due to enhanced expression of the writer (METTL3) and reader (YTHDF2, IGF2BP1, and IGF2BP2), whereas m6A expression increased only in the 8-cell stage of the PA embryos (Figure 4). Indeed, the expression of both METTL3 and YTHDF2 were important for PA embryonic development. A previous study has suggested that YTHDF2 facilitates maternal mRNA clearance in the maternal-to-zygotic transition (MZT) (Zhao et al., 2017). Due to the lack of paternal genomes in PA embryos, MZT might be delayed compare to that in control embryos, which occurs at the 2-cell stage (Bouniol et al., 1995). Thus, the expression of METTL3 and YTHDF2 might be regulated by the m6A expression pattern in the 8-cell stage of PA embryos. In addition, m6A expression was higher in the blastocyst stage than in the 2-cell, 4-cell, and 8-cell stages, which might be due to the translation of accumulated mRNAs of METTL3 (Li et al., 2017). Indeed, numerous imprinting genes are expressed and genomic imprinting is established at the mouse 2-cell stage (Kamimura et al., 2014). Our results indicated that RNA methylation occurs in this stage. However, imprinted genes were abnormally expressed, and were delayed in PA embryos. This might affect RNA methylation established during PA embryonic development.
Figure 3. Immunofluorescence localization of m6A. The expression patterns of m6A in 2-cell, 4-cell, 8-cell, and blastocyst stages of normal (A) and PA (B) embryos. BL indicates the blastocyst stage of the embryo. Green indicates m6A. Blue indicates DNA stain.
Figure 4. Schematic representations of m6A expression patterns in 2-cell (A), 4-cell (B), 8-cell (C), and blastocyst stage (D) of normal and PA embryos. BL indicates the blastocyst stage of the embryo. W indicates writer (METTL3 and METTL14). E indicates eraser (FTO and ALKBH5). R indicates reader (YTHDF2, IGF2BP1, and IGF2BP2). Red arrow and inverted T indicate the over expression of genes. The red dotted line indicates enhance m6A expression.
The expression of imprinted genes and establishment of DNA methylation are dynamically regulated during early embryonic development (SanMiguel and Bartolomei, 2018). Our results confirmed that m6A methylation is perturbed in the PA genome (Sepulveda-Rincon et al., 2016). As the m6A methylation level is related to gene expression, it may be responsible for developmental failure during parthenogenesis.
In conclusion, our results suggest that m6A methylation-related gene expression changes dynamically and is perturbed during the development of PA embryos. Furthermore, m6A expression was lower in the 2-cell stage, whereas it increased in the 8-cell stage of PA embryos. Therefore, the findings of the present study suggested that the development of PA embryos is characterized by abnormal m6A expression, which is associated with altered levels of m6A methylation-related genes.
Acknowledgments
This work was supported by the National Natural Science Foundation of China under Grant 31601003; the Fundamental Research Funds for the Central Universities under Grant 2019JCKT-70, China Postdoctoral Science Foundation under Grant 2018T110250 and 2016M601384; and Jilin Scientific and Technological Development Program under Grant 20190103071JH, 20180101254JC, and 0170623093-TC.
Supplementary Material
The following online material is available for this article:
Footnotes
Associate Editor: Igor Schneider
Conflict of Interest
The authors declare that they have no competing interests.
Author contributions
Dongxu Wang designed the experiments and wrote the manuscript. Jindong Hao, Wei Gao and Jiaqi Wei performed cell experiment and gene expression analysis. Xianfeng Yu contributed reagents and materials. Minghui Qi, Liang Han and Shuming Shi carried out animal experiment. Chao Lin analyzed the data and prepared figures. All authors reviewed the manuscript.
References
- Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, Kadiwala J, Hubner NC, de los Mozos IR, Sadee C, et al. The SMAD2/3 interactome reveals that TGF beta controls m(6)A mRNA methylation in pluripotency. Nature. 2018;555:256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res. 1995;218:57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- Chandola U, Das R, Panda B. Role of the N6-methyladenosine RNA mark in gene regulation and its implications on development and disease. Brief Funct Genomics. 2015;14:169–179. doi: 10.1093/bfgp/elu039. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu W, Tong Y, Cheng C, Chen Z. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014;289:11571–11583. doi: 10.1074/jbc.M113.546168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Horii T, Kimura M, Morita S, Nagao Y, Hatada I. Loss of genomic imprinting in mouse parthenogenetic embryonic stem cells. Stem Cells. 2008;26:79–88. doi: 10.1634/stemcells.2006-0635. [DOI] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, Gan J, Jiang H, Jia GF, Luo C, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ, O’Carroll D. The RNA m(6)A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67:1059–1067. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia GF, Fu Y, Zhao X, Dai Q, Zheng GQ, Yang Y, Yi CQ, Lindahl T, Pan T, Yang YG, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura S, Hatanaka Y, Hirasawa R, Matsumoto K, Oikawa M, Lee J, Matoba S, Mizutani E, Ogonuki N, Inoue K, et al. Establishment of paternal genomic imprinting in mouse prospermatogonia analyzed by nuclear transfer. Biol Reprod. 2014;91:120. doi: 10.1095/biolreprod.114.120451. [DOI] [PubMed] [Google Scholar]
- Kang YK, Yeo S, Kim SH, Koo DB, Park JS, Wee G, Han JS, Oh KB, Lee KK, Han YM. Precise recapitulation of methylation change in early cloned embryos. Mol Reprod Dev. 2003;66:32–37. doi: 10.1002/mrd.10330. [DOI] [PubMed] [Google Scholar]
- Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SF, Liu XY, Miao DQ, Han ZB, Zhang X, Miao YL, Yanagimachi R, Tan JH. Parthenogenetic activation of mouse oocytes by strontium chloride: a search for the best conditions. Theriogenology. 2005;64:1142–1157. doi: 10.1016/j.theriogenology.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Nguyen LH, Robinton DA, Seligson MT, Wu LW, Li L, Rakheja D, Comerford SA, Ramezani S, Sun XK, Parikh MS, et al. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell. 2014;26:248–261. doi: 10.1016/j.ccr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Kim HS, Lee SG, Lee CK. Methylation status of differentially methylated regions at Igf2/H19 locus in porcine gametes and preimplantation embryos. Genomics. 2009;93:179–186. doi: 10.1016/j.ygeno.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Park CH, Uh KJ, Mulligan BP, Jeung EB, Hyun SH, Shin T, Ka H, Lee CK. Analysis of imprinted gene expression in normal fertilized and uniparental preimplantation porcine embryos. PLoS One. 2011;6:e22216. doi: 10.1371/journal.pone.0022216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel JM, Bartolomei MS. DNA methylation dynamics of genomic imprinting in mouse development. Biol Reprod. 2018;199:252–262. doi: 10.1093/biolre/ioy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda-Rincon LP, Solanas ED, Serrano-Revuelta E, Ruddick L, Maalouf WE, Beaujean N. Early epigenetic reprogramming in fertilized, cloned, and parthenogenetic embryos. Theriogenology. 2016;86:91–98. doi: 10.1016/j.theriogenology.2016.04.022. [DOI] [PubMed] [Google Scholar]
- Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. ELife. 2016;5:e18434. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. Rna Biol. 2014;11:669–672. doi: 10.4161/rna.28829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Tang B, Fan CL, Shi LX, Zhang XM, Sun LG, Li ZY. Effect of DNMT inhibitor on bovine parthenogenetic embryo development. Biochem Biophys Res Commun. 2015;466:505–511. doi: 10.1016/j.bbrc.2015.09.060. [DOI] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GQ, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat Immunol. 2017;18:1094–1103. doi: 10.1038/ni.3830. [DOI] [PubMed] [Google Scholar]
- Zhu J, King T, Dobrinsky J, Harkness L, Ferrier T, Bosma W, Schreier LL, Guthrie HD, DeSousa P, Wilmut I. In vitro and in vivo developmental competence of ovulated and in vitro matured porcine oocytes activated by electrical activation. Cloning Stem Cells. 2003;5:355–365. doi: 10.1089/153623003772032853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.