Abstract
MYB is a large family of plant transcription factors. Its function has been identified in several plants, while there are few reports in Medicago truncatula. In this study, we used RNA-seq data to analyze and identify R2R3-MYB genes in the genome of Medicago truncatula. Phylogenetic analysis classified 150 MtMYB genes into 21 subfamilies with homologs. Out of the 150 MtMYB genes, 139 were distributed among 8 chromosomes, with tandem duplications (TD) and segment duplications (SD). Microarray data were used for functional analysis of the MtMYB genes during growth and developmental processes providing evidence for a role in tissues differentiation, seed development processes, and especially the nodulation process. Furthermore, we investigated the expression of MtMYB genes in response to abiotic stresses using RNA-seq data, which confirmed the critical roles in signal transduction and regulation processes under abiotic stress. We used quantitative real-time PCR (qRT-PCR) to validate expression profiles. The expression pattern of M. truncatula MYB genes under different abiotic stress conditions suggest that some may play a major role in cross-talk among different signal transduction pathways in response to abiotic stresses. Our study will serve as a foundation for future research into the molecular function of M. truncatula R2R3-MYB genes.
Keywords: Medicago truncatula, MYB, abiotic stress, qRT-PCR
Introduction
Myeloblastosis (MYB) genes are one of the largest transcription factors families in the plant kingdom (Romero et al., 1998; Rosinski and Atchley, 1998). MYB typically contain an MYB-binding domain at the N-terminus, composed of 1-4 imperfect repeats, with approximately 51-52 amino acid residues of incomplete conserved peptides encoding three α-helices (Kanei-Ishii et al., 1990; Lipsick, 1996; Stracke et al., 2001). These three α-helices form a helix-turn-helix (HTH) structure and fold with three relatively conserved tryptophan residues, separated by 18-19 amino acid residues of regular arrangement, and further participate in the formation of hydrophobic interactions (Ogata et al., 1995). The structure of the MYB domain reveals that the HTH interacts with the major groove of DNA (Ogata et al., 1994). MYB-containing genes have a diverse number of MYB proteins containing incomplete repeats and are divided into four categories, 1R-MYB, R2R3-MYB, R1R2R3-MYB, and 4R-MYB (Jin and Martin, 1999).
In the MYB gene family, R2R3-MYB is the largest category in plants and yeast (Martin and Pazares, 1997; Jin and Martin, 1999). To date, 126 R2R3-MYB genes have been identified in Arabidopsis (Stracke et al., 2001), 157 in maize (Du et al., 2012a), 244 in soybean (Du et al., 2012b), 205 in Gossypium raimondii (He et al., 2016), and 166 in cassava (Liao et al., 2016). R2R3-MYB transcription factors are involved with abiotic stress response, reactive oxygen species signaling pathways, secondary metabolism, and hormone signaling pathways. AtMYB41 from Arabidopsis is induced in response to high salinity, drought, cold, and abscisic acid (Lippold et al., 2009). AtMYB102 are rapidly induced by osmotic stress and abscisic acid (ABA) treatment (Denekamp and Smeekens, 2003). OsMYB55 is induced in rice by high temperature and over-expression of OsMYB55 resulted in improved plant growth under high temperature (Elkereamy et al., 2012). Meanwhile, OsMYB91 plays a role in plant growth regulation and salt stress tolerance in rice (Zhu et al., 2015). TaMYB4 of wheat is induced by salicylic acid, ethylene, abscisic acid, and methyl jasmonate, demonstrating a role of TaMYB4 in response to biotic stress (Al-Attala et al., 2014). GbMYB5 from Gossypium barbadense is positively involved in response to drought stress during plant development (Chen et al., 2015). GbMYBFL, involving in flavonoid biosynthesis, is most closely related to R2R3-MYB and displays high similarity to MYB from other plants (Zhang et al., 2017).
Medicago truncatula is a model plant for genetic research of legume plants (Young and Udvardi, 2009), as it has a small genome, high genetic transformation efficiency, self-pollination, nitrogen fixing, along with high biological diversity. Crop improvement of legume breeding has become an important subject (Lee et al., 2017). MtERF and MtMAPKKK gene families of M. truncatula are valuable for characterizing molecular function to improve stress tolerance in plants (Li et al., 2016; Shu et al.. 2016). However, the R2R3-MYB family is poorly identified on a genomic level in M. truncatula.
In this study, we performed a genome-wide analysis of the R2R3-MYB gene family in M. truncatula, including multiple alignment analysis, phylogenetic analysis, chromosome localization, gene duplication analysis, and expression profiling. We used quantitative real-time reverse transcription (qRT-PCR) experiments to compare and analyze five stress treatments (ABA, cold, freezing, drought, and salt) with a control treatment. Our study will serve as a foundation for future research into the molecular function of M. truncatula R2R3-MYB genes.
Materials and Methods
Plant growth and treatment
M. truncatula (cv. JemalongA17) seeds were grown in a growth chamber (Conviron E15), between 18ºC (night) and 24ºC (day), with humidity ranging from 60% to 80%, and a 14/10 h light-dark period (light, 06:00–20:00). The seedlings were irrigated with half-strength Hoagland solution once every other day. At 8 weeks, M. truncatula were randomly divided into six groups for stress treatments. Plants treated with cold stress (B group) and freezing stress (C group) were transferred to chambers set at 4 ºC and -8 ºC, respectively. Plants treated with drought stress (D group) and salt stress (E group) were respectively treated with 300 mM mannitol and 200 mM NaCl solutions. Plants in the ABA (F group) treatment group were sprayed with 100 μM ABA solution. The control (untreated, A group) and treated (B-F groups) seedlings were harvested 3 h after treatment. For each group, five randomly chosen whole seedlings were pooled to form a biological replicate. All plant samples were frozen in liquid nitrogen and stored at -80 ºC until use.
Database search for MYB proteins in M. truncatula
M. truncatula genome and protein sequences were downloaded from JCVI (http://jcvi.org/medicago/, Mt4.0) (Young et al., 2011). R2R3-MYB protein sequences of Arabidopsis were obtained from TAIR (http://www.arabidopsis.org/) (Poole, 2007) and served as queries to search against the M. truncatula proteins using the BLASTP program with e-values of 1E-3. All BLAST hits were retrieved and searched using the Hidden Markov Model (HMM) profile of the R2R2-MYB DNA-binding domain (PF000249), which was downloaded from the Pfam website (http://pfam.xfam.org/) (Finn et al., 2014).
Phylogenetic analysis of the M. truncatula R2R3-MYB gene family
Phylogenetic analysis was performed using MEGA (Version 5.0), and constructed using neighbor-joining (NJ) methods. The NJ method used the following parameters: Poisson correction, pair-wise deletion, and 1000 bootstrap analysis for statistical reliability (Tamura et al., 2011).
Chromosomal location and gene duplication of R2R3-MYB gene family
The location of R2R3-MYB genes were mapped to different chromosomes using the Circos software (http://circos.ca/) (Krzywinski et al., 2009). If two genes with similarities of more than 85% were separated by four or fewer gene loci, they were identified as tandem duplications (TD). Otherwise, they were classified as segmental duplications (SD). The duplications with R2R3-MYB genes were identified by plant genome duplications (PGDD, http://chibba.agtec.uga.edu/duplication/) (Lee et al., 2012, Shu et al., 2016), and duplicated genes between different chromosomes or loci were linked in the diagrams.
Expression analysis of R2R3-MYB genes in plant growth and development using high throughput data
We studied the expression analysis of R2R3-MYB genes during growth and development and under different abiotic stress treatments. Gene expression data were downloaded from the Medicago truncatula Gene Expression Atlas (MtGEA) Project (MtGEA, http://mtgea.noble.org/v3/) (Benedito et al., 2008). Genome-wide transcriptome data from M. truncatula in different tissues during development were downloaded from the NCBI short read archive database (SRA database) (http://www.ncbi.nlm.nih.gov).
Expression analysis of R2R3-MYB genes under different abiotic stress treatment conditions
Under six different abiotic stress treatments, MtMYB gene expressional values were evaluated using the TopHat (Trapnell et al., 2009) and Cufflinks (Trapnell et al., 2012) software. The data were analyzed, clustered, and displayed using the ggplot2 of R software (Version 3.1.0), as our previous research described (Shu et al., 2016).
Quantitative reverse transcription PCR (qRT-PCR) analysis of R2R3-MYB genes expressed in M. truncatula
Ten R2R3-MYB genes were selected and quantitative primers were designed based on genes sequences, while the ACTIN and GAPDH genes served as reference genes (see Table S1 (98.3KB, pdf) ). Total RNA was extracted from M. truncatula grown under the six conditions (control, ABA, drought, salt, cold, and freezing) using a total RNA kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The RNA extracts were reverse transcribed to cDNA, using a PrimeScript RT reagent Kit (Toyobo, Shanghai, China). qRT-PCR was performed using the LightCycler®96 System (Roche, Rotkreuz, Switzerland) with SYBR Premix Ex TaqTM II (Toyobo, Shanghai, China). The experiments were repeated for three biological replicates and the PCR conditions were set as follows: 95 ºC for 2 min, 40 cycles of 95 ºC for 30 s, 55 ºC for 30 s, and 72 º for 1 min. The fold change value was calculated using the expression abundances, which based on the 2-ΔΔCT method.
Results
Identification and classification of R2R3-MYB genes in M. truncatula
To identify R2R3-MYB genes in M. truncatula, Arabidopsis R2R3-MYB gene sequences were used as queries with e-value set as 1E-3. The gene name, gene locus, chromosome location, amino acid sequence, introns, family groups, and isoelectric point (pI) are described in Table 1. Out of the 150 MtMYB genes, 139 genes were distributed across 8 chromosomes of M. truncatula. The remaining 11 MtMYB genes were MtMYB5, 6, 7, 13, 14, 16, 34, 35, 36, 37, and 52. The length of these 139 R2R3-MYB ranged from 194 to 1514 amino acids, with an intron distribution of 0-12, and the isoelectric point were distributed from 4.77 to 9.93.
Table 1. List of all MtMYB genes identified in the Medicago truncatula genome.
| Gene Name | Gene Locus | Chromosome Location | AA | Introns | Family Group | pI |
|---|---|---|---|---|---|---|
| MtMYB001 | Medtr3g039990 | chr3:13992780-13994617 | 312 | 2 | 1 | 5.44 |
| MtMYB002 | Medtr6g012180 | chr6:3615718-3617850 | 319 | 2 | 1 | 5.76 |
| MtMYB003 | Medtr7g010210 | chr7:2460324-2462646 | 272 | 3 | 1 | 8.94 |
| MtMYB004 | Medtr7g087130 | chr7:33927709-33929377 | 339 | 2 | 1 | 6.54 |
| MtMYB005 | Medtr0140s0030 | scaffold0140:9682-11671 | 335 | 2 | 1 | 6.6 |
| MtMYB006 | Medtr0489s0020 | scaffold0489:9594-10964 | 335 | 2 | 1 | 6.6 |
| MtMYB007 | Medtr0251s0050 | scaffold0251:9249-10577 | 273 | 1 | 2 | 5.09 |
| MtMYB008 | Medtr1g043050 | chr1:16126632-16128228 | 255 | 2 | 2 | 5.58 |
| MtMYB009 | Medtr1g043080 | chr1:16137369-16139502 | 256 | 2 | 2 | 5.6 |
| MtMYB010 | Medtr1g076150 | chr1:33730166-33733155 | 259 | 2 | 2 | 5.31 |
| MtMYB011 | Medtr7g096930 | chr7:38915313-38918018 | 279 | 2 | 2 | 5.35 |
| MtMYB012 | Medtr7g115650 | chr7:47812651-47813895 | 208 | 2 | 2 | 8.51 |
| MtMYB013 | Medtr0008s0390 | scaffold0008:236494-237588 | 273 | 1 | 2 | 5.55 |
| MtMYB014 | Medtr0008s0470 | scaffold0008:261364-262450 | 292 | 1 | 2 | 5.34 |
| MtMYB015 | Medtr2g067420 | chr2:28203900-28207822 | 330 | 2 | 3 | 4.74 |
| MtMYB016 | Medtr0063s0090 | scaffold0063:54059-56064 | 224 | 2 | 3 | 5.78 |
| MtMYB017 | Medtr4g073420 | chr4:27807701-27809440 | 286 | 1 | 4 | 8.65 |
| MtMYB018 | Medtr4g485530 | chr4:33337240-33340075 | 194 | 2 | 4 | 9.68 |
| MtMYB019 | Medtr5g079670 | chr5:34066783-34068495 | 214 | 2 | 4 | 9.14 |
| MtMYB020 | Medtr8g095390 | chr8:39916215-39917940 | 316 | 2 | 4 | 9.14 |
| MtMYB021 | Medtr4g100720 | chr4:41544004-41544940 | 176 | 3 | 5 | 8.25 |
| MtMYB022 | Medtr4g125520 | chr4:52070924-52072242 | 308 | 2 | 5 | 8.34 |
| MtMYB023 | Medtr8g020490 | chr8:7194946-7196540 | 255 | 2 | 5 | 8.55 |
| MtMYB024 | Medtr5g078800 | chr5:33679342-33680505 | 266 | 2 | 6 | 8.76 |
| MtMYB025 | Medtr5g078860 | chr5:33697936-33699039 | 307 | 2 | 6 | 8.76 |
| MtMYB026 | Medtr5g078910 | chr5:33733725-33736557 | 301 | 2 | 6 | 9.06 |
| MtMYB027 | Medtr5g078930 | chr5:33748446-33750303 | 220 | 2 | 6 | 9.74 |
| MtMYB028 | Medtr5g078950 | chr5:33762292-33763827 | 266 | 2 | 6 | 9.14 |
| MtMYB029 | Medtr5g079120 | chr5:33821199-33824299 | 307 | 3 | 6 | 9.12 |
| MtMYB030 | Medtr5g079220 | chr5:33865422-33867467 | 277 | 2 | 6 | 8.08 |
| MtMYB031 | Medtr5g079290 | chr5:33903955-33904999 | 265 | 2 | 6 | 9.11 |
| MtMYB032 | Medtr7g017260 | chr7:5468456-5471716 | 276 | 2 | 6 | 7.59 |
| MtMYB033 | Medtr8g060940 | chr8:21305461-21306435 | 245 | 2 | 6 | 5.83 |
| MtMYB034 | Medtr0001s0360 | scaffold0001:148343-149371 | 254 | 2 | 6 | 8.13 |
| MtMYB035 | Medtr0193s0090 | scaffold0193:23279-25596 | 208 | 2 | 6 | 9.27 |
| MtMYB036 | Medtr0197s0010 | scaffold0197:3158-6293 | 230 | 2 | 6 | 5.79 |
| MtMYB037 | Medtr0247s0040 | scaffold0247:19831-22448 | 180 | 2 | 6 | 9.56 |
| MtMYB038 | Medtr2g034790 | chr2:13342703-13345610 | 333 | 2 | 7 | 5.57 |
| MtMYB039 | Medtr4g121460 | chr4:50199327-50201231 | 340 | 2 | 7 | 5.21 |
| MtMYB040 | Medtr7g117730 | chr7:48891947-48893156 | 257 | 1 | 7 | 9.19 |
| MtMYB041 | Medtr8g027345 | chr8:9621235-9622899 | 301 | 1 | 7 | 7.03 |
| MtMYB042 | Medtr1g100653 | chr1:45593395-45595446 | 296 | 2 | 9 | 7.16 |
| MtMYB043 | Medtr4g082230 | chr4:31939369-31942857 | 408 | 2 | 9 | 6.02 |
| MtMYB044 | Medtr4g082290 | chr4:31999665-32001816 | 408 | 2 | 9 | 5.77 |
| MtMYB045 | Medtr6g012690 | chr6:3910120-3912657 | 358 | 2 | 9 | 5.54 |
| MtMYB046 | Medtr7g011170 | chr7:2948411-2950937 | 310 | 2 | 9 | 6.57 |
| MtMYB047 | Medtr7g076740 | chr7:28944308-28947368 | 305 | 2 | 9 | 8.15 |
| MtMYB048 | Medtr8g031360 | chr8:11745382-11748030 | 298 | 2 | 9 | 5.92 |
| MtMYB049 | Medtr4g478180 | chr4:30006885-30009067 | 354 | 3 | 10 | 6.36 |
| MtMYB050 | Medtr5g007370 | chr5:1317895-1319526 | 300 | 1 | 10 | 6.25 |
| MtMYB051 | Medtr5g029840 | chr5:12551795-12553088 | 335 | 1 | 10 | 6.26 |
| MtMYB052 | Medtr0008s0280 | scaffold0008:117749-119885 | 232 | 2 | 10 | 6.21 |
| MtMYB053 | Medtr2g011660 | chr2:2891380-2892991 | 346 | 2 | 11 | 5.85 |
| MtMYB054 | Medtr2g089450 | chr2:37807548-37809745 | 251 | 2 | 11 | 7.62 |
| MtMYB055 | Medtr2g089620 | chr2:37880385-37882262 | 327 | 2 | 11 | 6.45 |
| MtMYB056 | Medtr4g091490 | chr4:36245993-36247326 | 359 | 2 | 11 | 5.87 |
| MtMYB057 | Medtr1g085040 | chr1:37962367-37964302 | 370 | 2 | 13 | 7.1 |
| MtMYB058 | Medtr1g085640 | chr1:38268101-38269978 | 389 | 2 | 13 | 6.1 |
| MtMYB059 | Medtr1g112760 | chr1:51044088-51045093 | 231 | 2 | 13 | 8.61 |
| MtMYB060 | Medtr3g077110 | chr3:34625609-34627040 | 350 | 2 | 13 | 5.71 |
| MtMYB061 | Medtr4g105130 | chr4:43559175-43561153 | 362 | 2 | 13 | 5.51 |
| MtMYB062 | Medtr7g110830 | chr7:45437545-45439504 | 368 | 2 | 13 | 5.28 |
| MtMYB063 | Medtr7g111290 | chr7:45674622-45676610 | 382 | 2 | 13 | 6.26 |
| MtMYB064 | Medtr1g017000 | chr1:4609270-4612231 | 253 | 2 | 14 | 6.22 |
| MtMYB065 | Medtr1g017140 | chr1:4685126-4687287 | 342 | 2 | 14 | 7.21 |
| MtMYB066 | Medtr2g095520 | chr2:40818577-40820355 | 321 | 2 | 14 | 7.02 |
| MtMYB067 | Medtr3g103570 | chr3:47845667-47847265 | 348 | 2 | 14 | 6.26 |
| MtMYB068 | Medtr4g057635 | chr4:21173419-21175385 | 325 | 2 | 14 | 6.67 |
| MtMYB069 | Medtr4g097570 | chr4:40253351-40254860 | 294 | 2 | 14 | 8.04 |
| MtMYB070 | Medtr4g128670 | chr4:53556826-53558636 | 351 | 2 | 14 | 5.43 |
| MtMYB071 | Medtr5g014990 | chr5:5113035-5114677 | 310 | 2 | 14 | 7.49 |
| MtMYB072 | Medtr6g090405 | chr6:34357171-34358762 | 256 | 2 | 14 | 5.53 |
| MtMYB073 | Medtr7g102110 | chr7:41207062-41209098 | 273 | 2 | 14 | 6.13 |
| MtMYB074 | Medtr1g100667 | chr1:45604161-45606367 | 273 | 2 | 16 | 9.4 |
| MtMYB075 | Medtr6g055910 | chr6:20067285-20071069 | 284 | 2 | 16 | 6.55 |
| MtMYB076 | Medtr1g083630 | chr1:37214098-37220601 | 631 | 10 | 18 | 8.87 |
| MtMYB077 | Medtr1g085770 | chr1:38342281-38444471 | 308 | 3 | 18 | 7.64 |
| MtMYB078 | Medtr1g085880 | chr1:38446474-38447221 | 194 | 1 | 18 | 9.25 |
| MtMYB079 | Medtr2g088170 | chr2:37135155-37138193 | 460 | 2 | 18 | 5.04 |
| MtMYB080 | Medtr5g062790 | chr5:26032596-26033729 | 305 | 2 | 18 | 4.67 |
| MtMYB081 | Medtr5g088080 | chr5:38180518-38181517 | 267 | 2 | 18 | 5.06 |
| MtMYB082 | Medtr5g088150 | chr5:38189614-38190613 | 267 | 2 | 18 | 5.06 |
| MtMYB083 | Medtr5g088610 | chr5:38446993-38448346 | 356 | 2 | 18 | 5.03 |
| MtMYB084 | Medtr5g088640 | chr5:38455878-38456905 | 276 | 2 | 18 | 5.06 |
| MtMYB085 | Medtr5g488170 | chr5:38207742-38208675 | 245 | 2 | 18 | 8.27 |
| MtMYB086 | Medtr5g488210 | chr5:38216839-38217838 | 267 | 2 | 18 | 5.06 |
| MtMYB087 | Medtr6g006030 | chr6:968935-973160 | 210 | 2 | 19 | 6.79 |
| MtMYB088 | Medtr7g035075 | chr7:13292852-13298285 | 209 | 2 | 19 | 6.07 |
| MtMYB089 | Medtr1g086510 | chr1:38706430-38707982 | 266 | 2 | 20 | 6.71 |
| MtMYB090 | Medtr1g086530 | chr1:38719605-38720569 | 279 | 1 | 20 | 5.57 |
| MtMYB091 | Medtr1g110460 | chr1:49847994-49849320 | 312 | 2 | 20 | 6.97 |
| MtMYB092 | Medtr2g033170 | chr2:12558787-12561118 | 330 | 2 | 20 | 5.54 |
| MtMYB093 | Medtr4g123040 | chr4:50776703-50778712 | 316 | 2 | 20 | 6.9 |
| MtMYB094 | Medtr4g082040 | chr4:31789875-31791702 | 428 | 2 | 21 | 5.48 |
| MtMYB095 | Medtr5g041570 | chr5:18244022-18246575 | 439 | 2 | 21 | 5.81 |
| MtMYB096 | Medtr6g027340 | chr6:9347619-9348708 | 206 | 2 | 21 | 9.43 |
| MtMYB097 | Medtr6g027370 | chr6:9359727-9361707 | 353 | 2 | 21 | 9.13 |
| MtMYB098 | Medtr6g074860 | chr6:27791872-27795423 | 242 | 2 | 21 | 8.94 |
| MtMYB099 | Medtr7g086960 | chr7:33824449-33826853 | 357 | 2 | 21 | 8.87 |
| MtMYB100 | Medtr3g101290 | chr3:46598679-46599616 | 227 | 0 | 22 | 6.39 |
| MtMYB101 | Medtr4g094982 | chr4:39426938-39428438 | 272 | 0 | 22 | 5.76 |
| MtMYB102 | Medtr5g016510 | chr5:5923364-5924314 | 317 | 0 | 22 | 8.35 |
| MtMYB103 | Medtr5g082910 | chr5:35747490-35748793 | 289 | 1 | 22 | 8.33 |
| MtMYB104 | Medtr6g015455 | chr6:5126272-5138191 | 1514 | 9 | 22 | 6.6 |
| MtMYB105 | Medtr4g019370 | chr4:6040082-6046125 | 351 | 1 | 23 | 6.1 |
| MtMYB106 | Medtr3g074520 | chr3:33695057-33696802 | 347 | 2 | 24 | 5.76 |
| MtMYB107 | Medtr1g026870 | chr1:8820780-8826887 | 984 | 10 | 25 | 5.45 |
| MtMYB108 | Medtr1g045610 | chr1:17117606-17119422 | 259 | 2 | 25 | 9.28 |
| MtMYB109 | Medtr1g057980 | chr1:25521596-25523733 | 298 | 2 | 25 | 6.68 |
| MtMYB110 | Medtr1g062940 | chr1:27632125-27633748 | 219 | 3 | 25 | 5.7 |
| MtMYB111 | Medtr1g073170 | chr1:32447073-32448262 | 258 | 2 | 25 | 7.64 |
| MtMYB112 | Medtr2g064160 | chr2:27171680-27173646 | 314 | 2 | 25 | 6.14 |
| MtMYB113 | Medtr2g096380 | chr2:41173925-41175393 | 237 | 2 | 25 | 6.2 |
| MtMYB114 | Medtr2g097910 | chr2:41830130-41832026 | 320 | 1 | 25 | 5.63 |
| MtMYB115 | Medtr2g099740 | chr2:42787607-42789410 | 402 | 1 | 25 | 6.27 |
| MtMYB116 | Medtr3g065440 | chr3:29570010-29573532 | 312 | 3 | 25 | 4.81 |
| MtMYB117 | Medtr3g077650 | chr3:34915543-34917386 | 448 | 2 | 25 | 6.65 |
| MtMYB118 | Medtr3g083540 | chr3:37705148-37706353 | 304 | 1 | 25 | 6.83 |
| MtMYB119 | Medtr3g097450 | chr3:44673726-44675570 | 298 | 2 | 25 | 8.71 |
| MtMYB120 | Medtr3g110028 | chr3:50889343-50895792 | 904 | 11 | 25 | 5.08 |
| MtMYB121 | Medtr3g461490 | chr3:24436965-24438138 | 326 | 2 | 25 | 6.31 |
| MtMYB122 | Medtr4g063100 | chr4:23343932-23345888 | 254 | 2 | 25 | 5.44 |
| MtMYB123 | Medtr4g065017 | chr4:24357511-24360058 | 333 | 1 | 25 | 5.31 |
| MtMYB124 | Medtr4g088015 | chr4:34653348-34657396 | 443 | 2 | 25 | 9.15 |
| MtMYB125 | Medtr4g102380 | chr4:42421826-42423447 | 273 | 1 | 25 | 5.26 |
| MtMYB126 | Medtr4g105660 | chr4:43865250-43867027 | 431 | 2 | 25 | 6.39 |
| MtMYB127 | Medtr5g010020 | chr5:2590454-2592700 | 340 | 2 | 25 | 5.8 |
| MtMYB128 | Medtr5g010650 | chr5:2899487-2902910 | 477 | 7 | 25 | 8.32 |
| MtMYB129 | Medtr5g038910 | chr5:17123381-17129369 | 391 | 10 | 25 | 6.19 |
| MtMYB130 | Medtr5g049190 | chr5:21549233-21551259 | 243 | 2 | 25 | 6.36 |
| MtMYB131 | Medtr5g070020 | chr5:29670807-29672960 | 356 | 2 | 25 | 8.58 |
| MtMYB132 | Medtr5g078140 | chr5:33378172-33380732 | 321 | 2 | 25 | 5.12 |
| MtMYB133 | Medtr6g009430 | chr6:2790201-2792486 | 337 | 2 | 25 | 5.59 |
| MtMYB134 | Medtr7g035350 | chr7:13467452-13470094 | 451 | 2 | 25 | 5.66 |
| MtMYB135 | Medtr7g037130 | chr7:13641861-13644153 | 359 | 2 | 25 | 8.62 |
| MtMYB136 | Medtr7g037260 | chr7:13704241-13706602 | 359 | 2 | 25 | 8.99 |
| MtMYB137 | Medtr7g061330 | chr7:22157000-22164437 | 531 | 7 | 25 | 9.15 |
| MtMYB138 | Medtr7g061550 | chr7:22184367-22187445 | 360 | 1 | 25 | 9.39 |
| MtMYB139 | Medtr7g450950 | chr7:17066918-17069310 | 339 | 2 | 25 | 5.53 |
| MtMYB140 | Medtr7g451170 | chr7:17173579-17176966 | 413 | 2 | 25 | 9.22 |
| MtMYB141 | Medtr7g461410 | chr7:22216202-22220641 | 567 | 6 | 25 | 8.75 |
| MtMYB142 | Medtr8g006470 | chr8:571945-573793 | 288 | 2 | 25 | 6.22 |
| MtMYB143 | Medtr8g017340 | chr8:5841430-5843448 | 493 | 2 | 25 | 4.92 |
| MtMYB144 | Medtr8g017350 | chr8:5847584-5850190 | 644 | 3 | 25 | 4.79 |
| MtMYB145 | Medtr8g017390 | chr8:5885779-5887719 | 476 | 2 | 25 | 5.34 |
| MtMYB146 | Medtr8g017440 | chr8:5919956-5921521 | 342 | 2 | 25 | 6.45 |
| MtMYB147 | Medtr8g017500 | chr8:5958601-5960679 | 349 | 2 | 25 | 5.96 |
| MtMYB148 | Medtr8g017540 | chr8:5983119-5985752 | 395 | 3 | 25 | 6.07 |
| MtMYB149 | Medtr8g098860 | chr8:41349431-41351751 | 321 | 2 | 25 | 5.85 |
| MtMYB150 | Medtr8g468380 | chr8:24758342-24766275 | 437 | 12 | 25 | 8.72 |
Phylogenetic analysis of the R2R3-MYB genes in M. truncatula
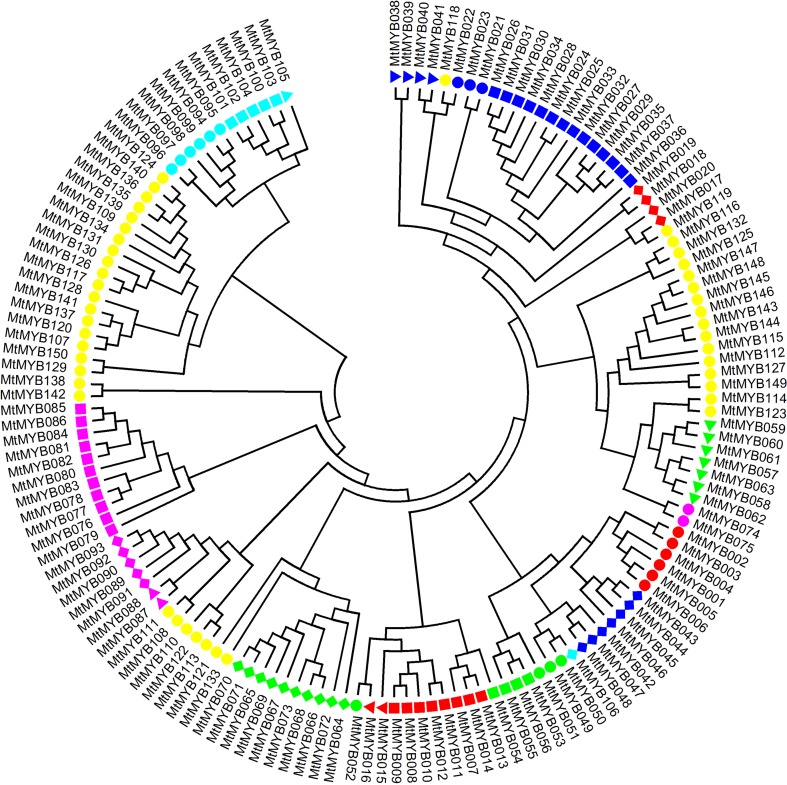
To investigate the evolutionary relationships of R2R3-MYB genes, we performed multiple sequence alignment and phylogenetic analysis. MtMYB transcription factors were queried against AtMYB transcription factors using BLASTP, and we constructed phylogenetic trees of the MtMYB transcription factor family using ClustalW2 and MEGA. MtMYB genes were divided into 21 subfamilies (Figure 1) consistent with the distribution of AtMYB gene family members. The subfamily 1 has six members, subfamily 6 has 14 members, subfamily 13 has seven members, subfamily 14 has ten members, and subfamily 18 has eleven members. This is similar to the distribution of the Arabidopsis subfamily members. R2R3-MYB transcription factors appeared to be conserved in plants as the evolution of M. truncatula is present in the evolution of Arabidopsis.
Figure 1. Phylogenetic tree analysis of the R2R3-MYB transcription factors in Medicago truncatula. Red circle: subgroup 1; red square: subgroup 2; red triangle: subgroup 3; red diamond: subgroup 4; blue circle: subgroup five; blue square subgroup six; blue indicates subgroup seven; blue diamond subgroup nine; green circular subgroup ten; green square subgroup 11; green triangle: subgroup 13; green diamond: subgroup 14; pink circle: subgroup 16; pink square: subgroup 18; pink triangle: subgroup 19; pink diamond: subgroup 20; cyan circle: subgroup 21; cyan square: subgroup 22; cyan triangle: subgroup 23; cyan diamond: subgroup 24; yellow circle: subgroup 25.
Chromosomal location and gene duplication of R2R3-MYB gene family
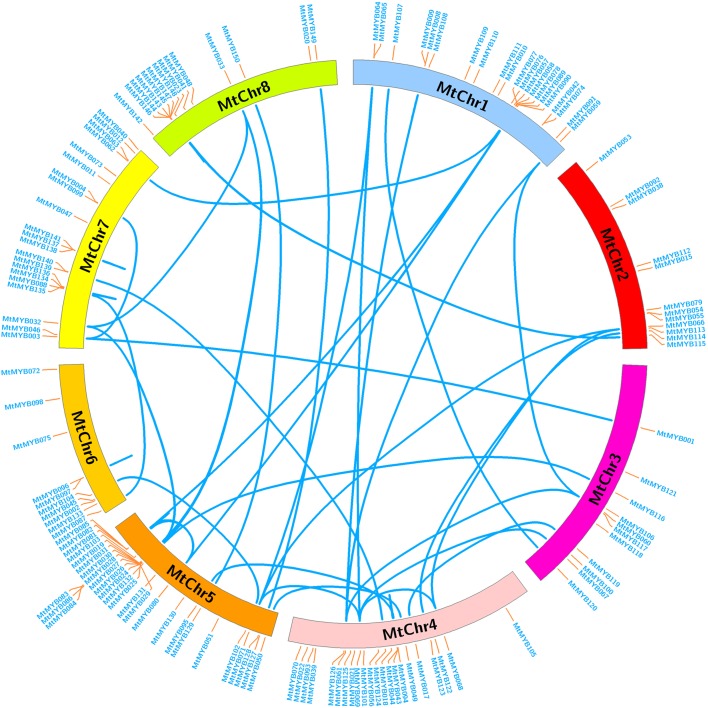
To determine the evolution and expansion of MYB genes, we used Circos software to construct the distribution of MYB genes across chromosomes (Figure 2). Out of the 150 R2R3-MYB genes, 139 were distributed across 8 chromosomes, mainly chromosome 1, 4, 5, and 7 (MtChr1, MtChr4, MtChr5 and MtChr7, respectively). There were 28 R2R3-MYB genes located on MtChr5 alone. The fewest number of genes were found on MtChr6 with ten R2R3-MYB genes. Using sequence alignment, mainly through gene duplication, 128 out of 139 genes were duplicated and divided into two categories. There were 68 segment duplications (SD) caused by the amplification of R2R3-MYB transcription factor members on different chromosomes and 60 tandem duplications (TD) resulting from the generation of R2R3-MYB transcription factor gene clusters. The SD and TD genes were mainly found on MtChr4, MtChr5, MtChr6, MtChr7, and MtChr8, while other chromosomes only contained SD genes. The regions containing TD genes were hot regions of gene distribution. These duplications may have led to the expansion of the MtMYB gene family in the M. truncatula genome.
Figure 2. Chromosome distribution and expansion analysis of R2R3-MYB transcription factors in Medicago truncatula. The genome locations of R2R3-MYB transcription factors were retrieved from Medicago genome website, and the duplications between R2R3-MYB genes identified using software PGDD and BLAST analysis.
Expression analysis of R2R3-MYB genes in growth and development
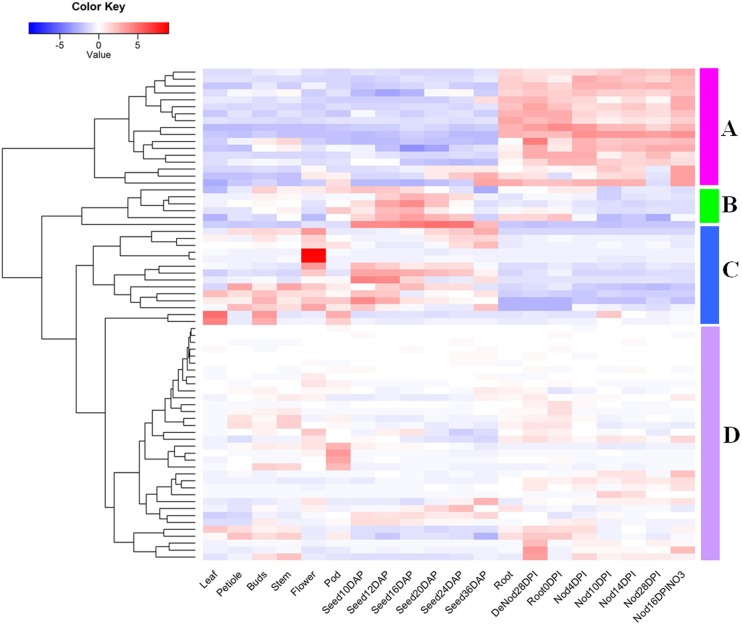
The expression information of 71 R2R3-MYB transcription factors were extracted and analyzed by cluster analysis (Figure 3 and Table S2 (270.9KB, pdf) ) using the annotation information of microarray data based on MtGEA. We clustered the 71 R2R3-MYB transcription factors into four groups (A-D). Group A was mainly concentrated in the roots and nodules of M. truncatula. There was only low expression in other tissues and during developmental processes. Group B was mainly expressed in the development of seed. Group C genes were expressed in various organ tissues. These results indicated that they were both involved in tissues construction process of M. truncatula. Finally, Group D genes were expressed at a low level in various tissues and during developmental processes.
Figure 3. Microarray expression data of R2R3-MYB transcription factors in Medicago truncatula. The heatmap was generated using R gplots package. The expressional values of 71 R2R3-MYB genes were retrieved from MtGEA, and they were normalized and used as input, red represents high expressional levels, while blue represents low expressional level.
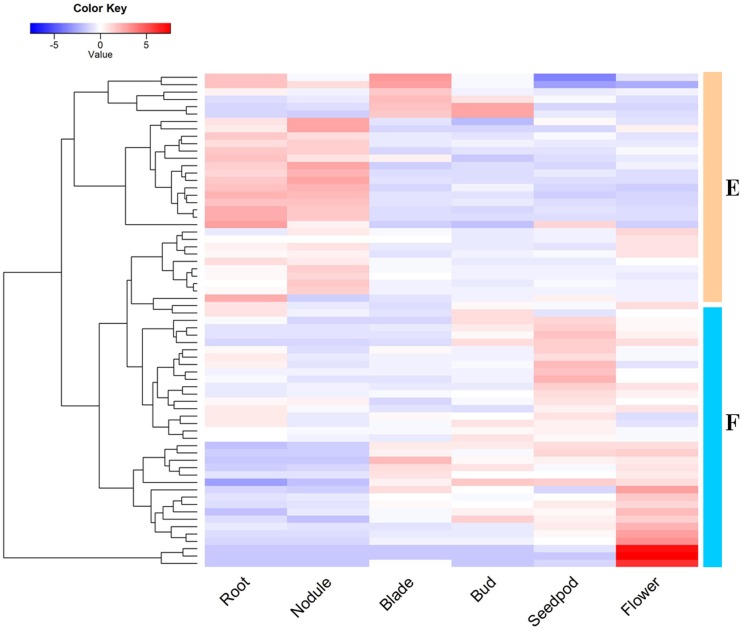
The transcriptome sequencing data of M. truncatula were downloaded from the NCBI SRA database, and the gene expression level was obtained using TopHat2 and Cufflinks analyses. We extracted 67 R2R3-MYB transcription factors (Figure 4 and Table S3 (144KB, pdf) ). Multiple genes with high expression in the roots and nodules were classified into Group E, while Group F contained numbers of paralog genes, which were mainly up-regulated in the flower, the carps, and inflorescence.
Figure 4. RNA-seq analysis of R2R3-MYB transcription factors in Medicago truncatula. The heatmap was generated using R gplots package, and the FPKM values of Medicago truncatula genes were evaluated and normalized based RNA-seq data from NCBI SRA database. The plot data included expressional profiles of 67 R2R3-MYB genes in six tissues, and red represents high expressional levels, while blue represents low expressional level.
Expression analysis of R2R3-MYB genes under different abiotic stress treatment conditions
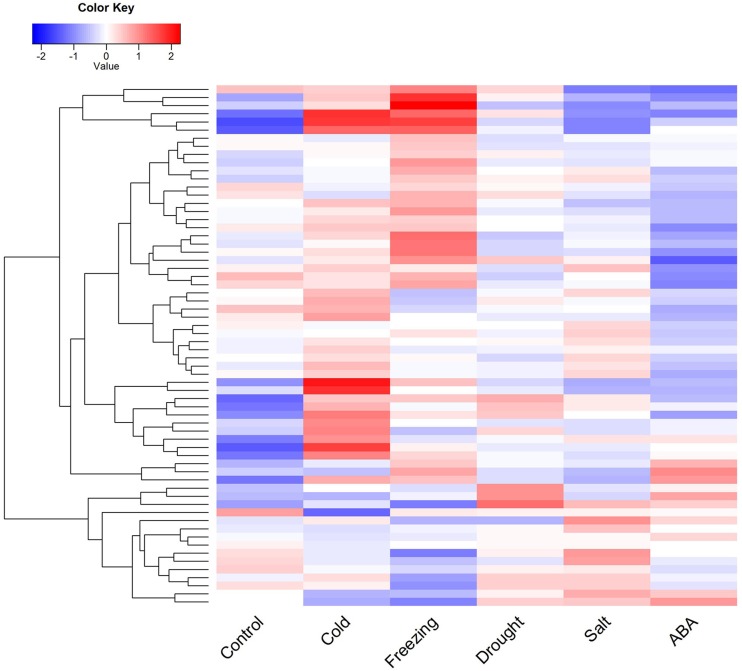
RNA-seq was used to investigate the expression of R2R3-MYB transcription factors in response to abiotic stress of M. truncatula. Out of the 150 MtMYB genes, 64 were expressed in response to abiotic stress (cold, freezing, drought, salt and ABA) (Figure 5 and Table S4 (143.3KB, pdf) ). In the control treatment, multiple genes were down-regulated or showed no change in expression, yet few genes were up-regulated. We compared genes regulated in response to abiotic stress to the control treatment and found genes highly up-regulated in response to cold and freezing stresses (50/64, 78.1%). These genes interacted with their paralog genes showing co-expression. In response to drought, high salt, and ABA stresses, multiple genes showed opposing expression and co-expression of the pairs of paralog genes. Significantly, most of these genes showed only low expression or no expression. In particular, some genes were down-regulated and co-expressed in response to ABA stress. It was suggested that the expression of these transcription factors are induced by ABA hormones, which regulate the downstream response genes and affect the response of plants to drought stress and salt stress.
Figure 5. Expression profile analysis of R2R3-MYB transcription factors in response to abiotic stresses in Medicago truncatula. The heatmap was generated using R gplots package, and the FPKM values of Medicago truncatula genes were evaluated and normalized based RNA-seq data from NCBI SRA database. The plot data included expressional profiles of 64 R2R3-MYB genes in response to abiotic stress, and red represents high expressional levels, while blue represents low expressional level.
qRT-PCR validation of R2R3-MYB genes expression
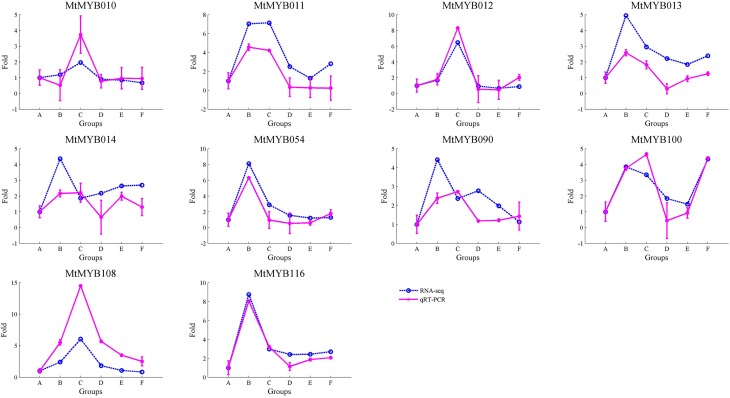
To validate the reliability of the RNA-seq data under abiotic stress in M. truncatula, we selected 10 R2R3-MYB transcription factors (MtMYB010, MtMYB011, MtMYB012, MtMYB013, MtMYB014, MtMYB054, MtMYB090, MtMYB100, MtMYB108, and MtMYB116) from M. truncatula for qRT-PCR validation. The expression levels of the RNA-seq data and qRT-PCR gene expression analysis resulted in a correlation coefficient of 0.85 (Figure 6). These results suggest the RNA-seq data are highly reliable.
Figure 6. qRT-PCR validation of R2R3-MYB transcription factors in Medicago truncatula. All expressional levels of each R2R3-MYB genes were normalized with control expression value (group A) set as one. The groups B (cold stress), C (freezing stress), D (drought stress), E (salt stress), and F (ABA treatment) were compared with control group, and fold-changes were calculated. The fold-change data was used to create the plot, and the blue indicates RNA-seq results, while pink indicates qRT-PCR analysis results.
Discussion
To date, R2R3-MYB transcription factors have been identified in plants, including Arabidopsis (126) (Yanhui et al., 2006), maize (157) (Du et al., 2012a), rice (102) (Yanhui et al., 2006), Populus (192) (Wilkins et al., 2009), and cassava (166) (Liao et al., 2016). The M. truncatula genome has been sequenced, yet the R2R3-MYB transcription factors have not been researched. This study performed genome-wide analysis of the R2R3-MYB transcription factors in M. truncatula. We identified 150 R2R3-MYB genes in M. truncatula and compared these with other plants. The R2R3-MYB genes of M. truncatula are similar to their Arabidopsis, maize, and cassava counterparts. The results indicated that R2R3-MYB transcription factors are highly conserved in plants.
Cannon et al. (2004) have shown gene duplication was closely related to plant evolution and played an important role in the gene amplification. Gene duplication analysis showed the same subfamily members were located on different chromosomes, such as the 21 subfamily gene members. These genes were distributed across all chromosomes and occurred as TD and SD. These results were consistent with the results of the phylogenetic tree. Furthermore, distribution of other subfamily members was confirmed based on clustering in the phylogenetic tree. These results suggest that the R2R3-MYB genes in M. truncatula are highly conserved as a result of gene duplication. The duplication pattern of R2R3-MYB genes, are consistent with the expression of R2R3-MYB genes in tomato (Zhao et al., 2014) and Populus (Wilkins et al., 2009). Tandem gene duplication was a major driver of gene expansion in M. truncatula.
MYB genes play a role in plant development and stress tolerance (Martin and Pazares, 1997). We have shown that R2R3-MYB genes were typically expressed in the roots, nodules, seedpods, and flowers. Compared with soybean and Arabidopsis, differential expression was observed during flower development, root formation, and seed development for R2R3-MYB genes (Du et al., 2012b). Nodulation is the result of a symbiosis between legumes and rhizobial bacteria in soil. Libault et al. (2009) have reported a gene named Control of Nodule Development (CND), encoding an MYB transcription factor gene. When the CND gene is silenced, nodulation is reduced (Libault et al., 2009). These results indicate that the MYB transcription factors may play a major role in regulation of legume-specific nodulation.
MYB transcription factor genes have been investigated as regulators for plant responses (Ambawat et al., 2013). Their results show that MYB genes are involved in response to various abiotic stresses in higher plants. Herein, we found that MYB genes were not only down-regulated, but in some cases up-regulated in response to drought and salt stress. A positive response of MYB genes following drought stress, salt stress, and ABA-induced stress has been observed in Arabidopsis (AtMYB002, AtMYB060/AtMYB094, AtMYB044, and AtMYB096) (Cominelli et al., 2005; Jung et al., 2008; Seo et al., 2009; Urao and Shinozaki, 1993), Boea crassifolia (BcMYB1) (Chen et al., 2005), and Saccharum officinarum (ScMYBAS1) (Prabu and Prasad, 2012). However, MtMYB genes were up-regulated in response to cold and freezing stresses, while the opposite is observed in Arabidopsis. Interestingly, our result agrees with the expression profile of cotton in response to drought and salt stress (He et al., 2016). Both cotton and M. truncatula are diploid and their duplication can be divided into TD and SD. The expression pattern of M. truncatula MYB genes under different abiotic stress conditions suggest that some may play a major role in cross-talk among different signal transduction pathways in response to abiotic stresses.
CONCLUSIONS
In summary, we have identified 150 MYB genes in M. truncatula, which were classified into 21 subfamilies based on phylogenetic analysis. Meanwhile, their expression profiles were investigated using microarray and RNA-seq. The results revealed regulatory roles in plant growth and tissue development, and especially nodule development. In addition, we explored the role of the MYB genes in response to abiotic stresses. Our results suggested MYB transcription factors broadly participate in abiotic stress response of M. truncatula, whose function can be carefully explored in future.
Acknowledgments
We thank Miss Jun Zhang and Dr. Lili Song (Harbin Normal University) for their helpful technical supports in plant growth and treatment. The data analysis work was supported by high performance computing center of Harbin Normal University. This work was supported by grants from the National Natural Science Foundation of China (31302019 and 31470571), the China Postdoctoral Science Foundation (2016T90307 and 2015M571430), the University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYST-2016071), the MOST 863 Project (2013AA102607-5), and the Heilongjiang Province Postdoctoral Science Foundation (LBH-Z14126).
Supplementary material
The following online material is available for this article
Associate Editor: Hong Luo
Author Contributions
YS and CG conceived and designed the experiments; WL, YL, JZ, and ZX performed the experiments; YS, WL, JZ, and ZX analyzed the data; YS, WL, and CG wrote the paper. All authors read and approved the final version.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Al-Attala MN, Wang X, Abou-Attia MA, Duan X, Kang Z. A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Mol Biol. 2014;84:589–603. doi: 10.1007/s11103-013-0156-7. [DOI] [PubMed] [Google Scholar]
- Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013;19:307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. A gene expression atlas of the model legume Medicago truncatula . Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana . BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Wang Y, Hu YL, Wu Q, Pin ZP. Cloning and characterization of a drought inducible MYB gene from Boea crassifolia . Plant Sci. 2005;168:493–500. [Google Scholar]
- Chen T, Li W, Hu X, Guo J, Liu A, Zhang B. A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 2015;56:917–929. doi: 10.1093/pcp/pcv019. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhartd N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Denekamp M, Smeekens SC. Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol. 2003;132:1415–1423. doi: 10.1104/pp.102.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Feng BR, Yang SS, Huang YB, Tang YX. The R2R3-MYB transcription factor gene family in maize. PLoS One. 2012a;7:e37463. doi: 10.1371/journal.pone.0037463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX. Genome-wide analysis of the MYB transcription factor superfamily in soybean . BMC Plant Biol. 2012b;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkereamy A, Bi YM, Ranathunge K, Beatty PH, Good AG, Rothstein SJ. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS One. 2012;7:e52030. doi: 10.1371/journal.pone.0052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Jones DC, Li W, Xie F, Ma J, Sun R, Wang Q, Zhu S, Zhang B. Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii . Sci Rep. 2016;6:22980. doi: 10.1038/srep22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis . Plant Physiol. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei-Ishii C, Sarai A, Sawazaki T, Nakagoshi H, He DN, Ogata K, Nishimura Y, Ishii S. The tryptophan cluster: A hypothetical structure of the DNA-binding domain of the myb protooncogene product. J Biol Chem. 1990;265:19990–19995. [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Yu D, Choi HK, Kim RW. Reconstruction of a composite comparative map composed of ten legume genomes. Genes Genom. 2017;39:111. doi: 10.1007/s13258-016-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2012;41:D1152–D1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Liu Y, Song L, Guo C, Shu Y. Bioinformatics analysis of MAPKKK family genes in Medicago truncatula . Genes. 2016;7:E13. doi: 10.3390/genes7040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Yang Y, Li Y, Wang Ga, Peng M. Genome-wide identification of cassava R2R3 MYB family genes related to abscission zone separation after environmental-stress-induced abscission. Sci Rep. 2016;6:32006. doi: 10.1038/srep32006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Joshi T, Takahashi K, Hurley-Sommer A, Puricelli K, Blake S, Finger RE, Taylor CG, Xu D, Nguyen HT, et al. Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development. Plant Physiol. 2009;151:1207–1220. doi: 10.1104/pp.109.144030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis . Plant Physiol. 2009;149:1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- Martin C, Pazares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ischii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Hojo H, Yoshimura S, Zhang R, Aimoto S, Ametani Y, Hirata Z, Sarai A, et al. Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nat Struct Biol. 1995;2:309–320. doi: 10.1038/nsb0495-309. [DOI] [PubMed] [Google Scholar]
- Poole RL. The TAIR database. Methods Mol Biol. 2007;406:179–212. doi: 10.1007/978-1-59745-535-0_8. [DOI] [PubMed] [Google Scholar]
- Prabu G, Prasad DT. Functional characterization of sugarcane MYB transcription factor gene promoter (PScMYBAS1) in response to abiotic stresses and hormones. Plant Cell Rep. 2012;31:661–669. doi: 10.1007/s00299-011-1183-y. [DOI] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. More than 80 regulatory genes in the genome of Arabidopsis thaliana . Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol. 1998;46:74–83. doi: 10.1007/pl00006285. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Parket JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis . Plant Physiol. 2009;151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Liu Y, Zhang J, Song L, Guo C. Genome-Wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula . Front Plant Sci. 2016;6:1247. doi: 10.3389/fpls.2015.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana . Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149:981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Udvardi M. Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol. 2009;12:193–201. doi: 10.1016/j.pbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Zhang W, Xu F, Cheng S. Characterization and functional analysis of a MYB gene (GbMYBFL) related to flavonoid accumulation in Ginkgo biloba . Genes Genomics. 2017;49:49–61. doi: 10.1007/s13258-017-0609-5. [DOI] [PubMed] [Google Scholar]
- Zhao P, Li Q, Li J, Wang L, Ren Z. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum . Mol Genet Genomics. 2014;289:1183–1207. doi: 10.1007/s00438-014-0879-4. [DOI] [PubMed] [Google Scholar]
- Zhu N, Cheng S, Liu X, Du H, Dai M, Zhou DX, Yang W, Zhao Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015;236:146–156. doi: 10.1016/j.plantsci.2015.03.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.