Abstract
The oncogenic fusion protein RUNX1-ETO is a product of the t(8;21) translocation and consists of the hematopoietic transcriptional master regulator RUNX1 and the repressor ETO. RUNX1-ETO is found in 10–15% of acute myeloid leukemia and interferes with the expression of genes that are essential for myeloid differentiation. The neutrophil serine protease Cathepsin G is one of the genes suppressed by RUNX1-ETO, but little is known about its impact on the regulation of other lysosomal proteases. By lentiviral transduction of the t(8;21) positive cell line Kasumi-1 with an RUNX1-ETO specific shRNA, we analyzed long-term effects of stable RUNX1-ETO silencing on cellular phenotypes and target gene expression. Stable anti RUNX1-ETO RNAi reduces both proliferation and apoptosis in Kasumi-1 cells. In addition, long-term knockdown of RUNX1-ETO leads to an upregulation of proteolytic activity in Kasumi-1 cells, which may be released in vitro upon cell lysis leading to massive degradation of cellular proteins. We therefore propose that protein expression data of RUNX1-ETO-silenced Kasumi-1 cells must be analyzed with caution, as cell lysis conditions can heavily influence the results of studies on protein expression. Next, a mass spectrometry-based approach was used to identify protease cleavage patterns in RUNX1-ETO-depleted Kasumi-1 cells and Neutrophil Elastase has been identified as a RUNX1-ETO candidate target. Finally, proteolytic activity of Neutrophil Elastase and Cathepsin G was functionally confirmed by si/shRNA-mediated knockdown in Kasumi-1 cells.
Introduction
The translocation t(8;21) is found in 10–15% of acute myeloid leukemia (AML), representing one of the most prevalent chromosomal aberrations associated with AML. Clinically, AML with the translocation t(8;21) is associated with a relatively favorable prognosis at initial diagnosis but not at relapse [1,2]. The resulting oncogenic fusion protein RUNX1-ETO contains the N-terminal RUNT domain of RUNX1 (AML1) and the almost entire ETO (MTG8) protein [3–5]. The oncogenic potential of RUNX1-ETO is based on its ability to deregulate normal RUNX1-dependent gene expression, for which several mechanisms have been described. RUNX1-ETO acts as dominant-negative inhibitor of RUNX1-dependent gene expression by recruiting the corepressor proteins NCoR and SMRT bound to the ETO moiety of the fusion protein [6–8]. NCoR and SMRT can interact with mSin3a and histone deacetylases (HDAC) [9,10], assembling a repressor complex which leads to transcriptional silencing of RUNX1 target genes like MDR1 [7], FOS [11], CDKN2A [12] and BCL2 [13]. However, RUNX1-ETO can also activate gene expression. It recruits the histone acetyl transferase (HAT) p300/CBP complex, facilitating histone acetylation and, more importantly, the acetylation of RUNX1-ETO itself. This results in increased accessibility of regulatory elements and the recruitment of other activating transcription factors, and allows the transactivation of target genes, e.g. ID1, CDKN1A (p21) and EGR1 [14]. In addition, a mechanism by which RUNX1-ETO competes with RUNX1 for the binding to a negative regulatory element driving expression of the cell cycle regulator CCND2 has been recently proposed by Martinez-Soria et al. [15]. Furthermore, RUNX1-ETO can interact with hematopoietic transcription factors like PU.1, C/EBPα, GATA-1 and E2A thereby interfering with their regulatory functions [16–19]. Other binding partners of RUNX1-ETO include proteins of the HDAC, DNA methyltransferase (DNMT) and protein arginine methyltransferase (PRMT) families, which are involved in the modeling of chromatin structure [20–22], and genome-wide changes in transcription factor binding have been shown for the depletion of RUNX1-ETO in AML cells [23]. Despite its impact on transcriptional regulation, the expression of RUNX1-ETO is not sufficient for the induction of leukemia in transgenic mice, but additional mutations are indispensable for the onset of AML [24,25]. The relevance of RUNX1-ETO for cellular processes, including proliferation, differentiation and cell survival has been investigated in numerous studies. Loss-of-function studies have mostly been carried out by transient, RNAi-mediated silencing of RUNX1-ETO in t(8;21) positive cell lines. Transient depletion of RUNX1-ETO in Kasumi-1 and SKNO-1 has been shown to reduce the proliferation and colony formation, and to restore myeloid differentiation, as measured by expression of CD11B and C/EBPα [26]. Furthermore, Kasumi-1 cells display a block in transition from the G1- to S-phase of the cell cycle, accompanied by reduced apoptosis and the induction of cellular senescence, in response to RUNX1-ETO RNAi [27]. In line with these findings, Dunne et al. [28] have demonstrated the impact of RUNX1-ETO silencing on the expression of genes associated with proliferation and differentiation, such as Insulin-like growth factor-binding protein 7 (IGFBP7) and Cathepsin G (CTSG), in Kasumi-1 and patient derived AML blasts with t(8;21) by oligonucleotide array and qRT-PCR.
The aim of our study was to analyze phenotype, target gene expression and potential molecular mechanisms involved in Kasumi-1 cells upon stable long-term silencing of RUNX1-ETO. Unexpectedly, we observed RUNX1-ETO specific differential protein expression of target genes depending on cell lysis conditions. Focusing on this phenomenon, we demonstrate the induction of CTSG in response to RUNX1-ETO knockdown to be partially responsible for in vitro degradation of cellular proteins. We used this phenomenon to map the activation of proteases beside CTSG in RUNX1-ETO depleted Kasumi-1 cells, as CTSG is a well-known target of RUNX1-ETO but not much is known about the possible participation of other proteases. A liquid chromatography (LC)-mass spectrometry (MS)-based approach was applied to measure proteolytic cleavage in RUNX1-ETO silenced Kasumi-1 cells on a proteome-wide scale. The mapping of protease cleavage sites demonstrated a contribution of Neutrophil Elastase (ELANE) to the degradation of cellular proteins in RUNX1-ETO-depleted Kasumi-1 cells, which was functionally confirmed by ELANE-specific RNAi.
Materials and methods
Cell culture, lentiviral transduction and inhibitors
Kasumi-1 cells (Leibniz-Institute DSMZ, Braunschweig, Germany) are a myeloid cell line carrying the RUNX1-ETO t(8;21) translocation and were cultured in RPMI 1640 supplemented with 10% FCS (Sigma, St. Louis, MO, USA) and 1% Penicillin/Streptomycin (Gibco, Waltham, MA, USA). HEK293 cells (Leibniz-Institute DSMZ) for production of lentiviral particles were maintained in DMEM (Gibco), containing 10% FCS and 1% Penicillin/Streptomycin. Preparation of recombinant lentiviral supernatants and lentiviral transductions were performed as described earlier [29]. Transduction efficacy was determined by flow cytometry (Cytoflex, Beckman Coulter, Brea, CA, USA) based on expression of the lentivirally encoded EGFP or RFP reporters. FlowJo Single Cell Analysis Software v10 was used for data analysis. Cell lines were treated with different inhibitors as indicated. ABT-737 and Bortezomib were purchased from Selleckchem (Houston, TX, USA), Doxorubicin and Chloroquine from Sigma.
Lentiviral constructs and si/shRNA
The lentiviral transgene plasmids pdcH1-shRNA-SEW/SR and the control plasmids pdcH1-gl4-SEW/SR were cloned as described [29]. The sequence of RUNX1-ETO targeting siRNA was described by Heidenreich et al. in 2003 [26] and converted to an shRNA. ELANE-specific siRNA was obtained from Dharmacon (Lafayette, CO, USA). Cathepsin G targeting shRNA was cloned as described [29]. Oligonucleotides for Cathepsin G shRNA were purchased from BioSpring (Frankfurt am Main, Germany) and the sequences are listed in S1 Table.
Proliferation and clonogenic assay
Transduced Kasumi-1 cells were cultured at 2x105 cells/ml in 24-well plates, and viable cells were counted by trypan blue exclusion to determine their proliferation rate. For colony assays, Kasumi-1 cells were seeded to 96-well plates in a limiting dilution of one cell per well, five days after lentiviral transduction. Colonies were counted on day 14 after transduction.
Flow cytometry for apoptosis and cell cycle analysis
Apoptotic cells were quantified by staining with 10 μg/ml Propidium iodide (PI, Serva, Heidelberg, Germany). PI positive cells were considered apoptotic and the percentage of PI positive cells was determined by flow cytometry. Cell cycle distribution of Kasumi-1 cells was analyzed by DNA content. Cells were fixed with 70% ethanol for 24 hours at -20°C and treated with 16.7 μg/ml PI and 20 μg/ml RNaseA in PBS for 30 minutes in the dark. Flow cytometry was performed on a FACSCalibur (Becton Dickinson, Heidelberg, Germany) and BD CellQuest™ Pro Software was used for data anlaysis.
MTS-Assay
The cytotoxicity of ABT-737 on transduced Kasumi-1 cells was determined by CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) according to the manufacturer´s instructions. Briefly, cells were seeded on 96-well plates to a concentration of 2x104/well in culture medium, with ABT-737 added to the indicated concentrations. After 48 hours, cells were incubated with MTS reagent for four hours, followed by measurement of the absorbance at 490 nm on a 96-well plate reader (Mithras LB940, Berthold Technologies, Bad Wildbad, Germany).
Electron microscopy
Preparation and imaging was done as described in Dawodu et al. [30].
Western Blot and Coomassie staining
For Western Blot analysis, whole cell lysates were prepared with high-salt lysis buffer (20 mM HEPES, pH 7.5, 0.4 M NaCl; 1 mM EDTA, 1 mM EGTA, 1 mM DTT), as recently described [31]. Alternatively, cells were pelleted and immediately boiled in SDS buffer (2x Laemmli: 62.5 mM Tris pH 6,8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.001% Bromophenol blue) for five minutes. Prior to boiling, cell lysates were kept on ice at all times. Proteins were separated by SDS-PAGE and transferred to Nitrocellulose (Thermo Fisher Scientific, Waltham, MA, USA) membrane at 800 mA for two hours. Membranes were blocked in 3% non-fat dry milk (Merck, Darmstadt, Germany) for one hour and incubated overnight at 4°C with primary antibodies, followed by one hour incubation with HRP-conjugated secondary antibodies at room temperature. Primary antibodies for RUNX1, GAPDH, LC3A/B and STAT3 were purchased from Cell Signaling Technology (Danvers, MA, USA), antibodies for PU.1 (T-21), STAT5A (L-20) and Ubiquitin from Santa Cruz Biotechnology (Dallas, TX, USA), Cathepsin G antibody from ProteinTech (Rosemont, IL, USA), anti-BCL2 from BD Pharmingen (Franklin Lakes, NJ, USA) and Human Neutrophil Elastase antibody (Clone #950317) from R&D Systems (Minneapolis, MN, USA). Secondary antibodies were purchased from Cell Signaling Technology (anti-rabbit), Santa Cruz (anti-mouse) and BD Pharmingen (anti-hamster). Proteins were visualized on Amersham Hyperfilm™ ECL (GE Healthcare) by chemiluminescence using Western Lightning Plus-ECL (Perkin Elmer, Waltham, MA, USA).
For Coomassie staining, proteins were separated by SDS-PAGE, incubated with Coomassie staining solution (0,25% Coomassie Brilliant Blue, 45,5% methanol, 9,2% acetic acid) for 15 minutes at room temperature and destained overnight in destaining solution (30% methanol, 10% acetic acid).
Quantitative real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen) and reverse transcription of 1 μg total RNA was performed with M-MLV reverse transcriptase (Invitrogen) and random hexamer primers (Thermo Scientific). SYBR green master mix or Taqman Universal master mix (Applied Biosystems, Foster City, CA, USA) was used for real-time PCR according to the manufacturer´s instructions and reactions were run on an ABI7500 cycler (Applied Biosystems). Expression of mRNA was normalized to 18S rRNA or β2M as housekeeping control and relative expression levels are shown as 2-ΔΔCTcompared to control cells. Primers and probes were synthetized by BioSpring, except for STAT5 primers and probe, which were purchased by MWG Biotech (Ebersberg, Germany). Taqman Assays were purchased from Applied Biosystems. Sequences of primers and probes are listed in S1 Table.
Sample preparation for MS analysis
Whole cell lysates from Kasumi-1/ctrl and Kasumi-1/shRE were prepared in RIPA buffer (50mM Tris HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS), supplemented with complete Protease Inhibitor Cocktail (Roche), on day twelve after lentiviral transduction. Protein concentration was determined by DC-Assay (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions and 50 μg protein per sample were boiled for five minutes in 2x NuPage LDS sample buffer (Invitrogen). Proteins were alkylated by acrylamide and further processed as described [32]. Peptide samples were analyzed with data dependent analysis in an LC-MS system (RSLC, LTQ Orbitrap Velos, both Thermo Fisher) as described recently [32]. Raw MS data were processed using Proteom discoverer 1.4 (Thermo Scientific) and Max Quant software (version 1.5, Cox and Mann 2008) and reviewed human and viral entries of the SwissProtUniprot database containing common contaminants. Proteins were stated identified by a false discovery rate of 0.01 on protein and peptide level.
Statistics
GraphPad Prism (version 7.04) has been used for statistical analyses by student´s t-test or 2-way ANOVA, as indicated. Statistical significance was considered for p<0.05.
Results and discussion
Stable long-term RUNX1-ETO silencing in Kasumi-1 cells
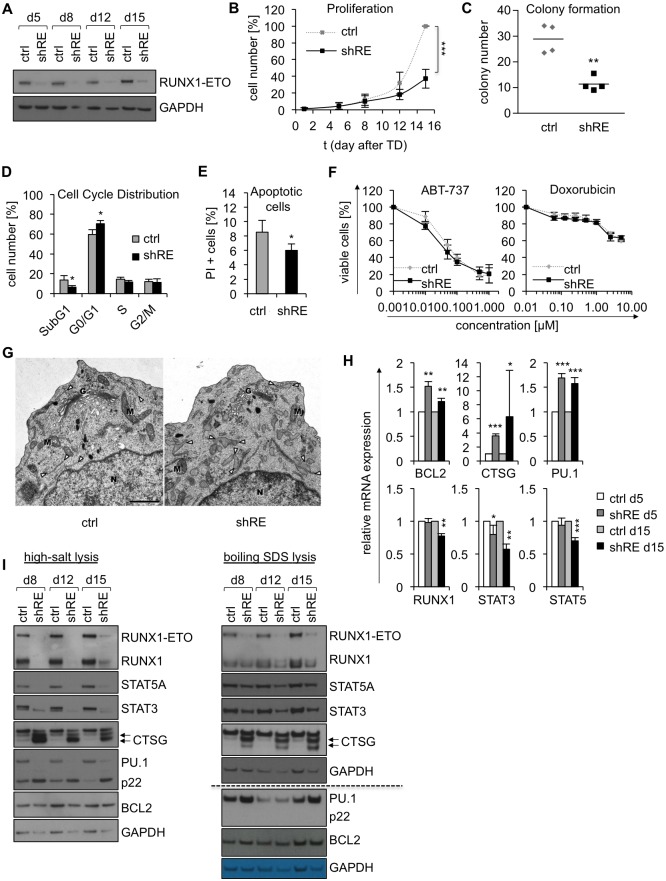
Stable knockdown of RUNX1-ETO was achieved by lentiviral transduction of Kasumi-1 cells with RUNX1-ETO specific shRNA. RUNX1-ETO-silenced (Kasumi-1/shRE) and control cells (Kasumi-1/ctrl) were monitored over 15 days for phenotypic changes and alterations in protein and mRNA expression. Transduction efficacy was determined based on eGFP reporter fluorescence and consistently reached >96% (Table A in S1 Fig). Stable depletion of RUNX1-ETO was verified by Western Blot (Fig 1A). Cell expansion of Kasumi-1/shRE cells was reduced as compared to Kasumi-1/ctrl cells (Fig 1B), and clonal expansion of Kasumi-1/shRE cells plated in limiting dilution was also decreased (Fig 1C), as described previously [26]. In line with the study of Martinez et al. 2004 [27], cell cycle analysis revealed a partial G0/G1 arrest and a smaller SubG1 fraction in Kasumi-1/shRE compared to Kasumi-1/ctrl cells (Fig 1D). These findings were supported by a reduced percentage of apoptotic Kasumi-1/shRE cells in comparison to control cells, as observed by PI staining (Fig 1E). However, RUNX1-ETO-depleted Kasumi-1 and control cells displayed equal sensitivity to the anthracycline doxorubicin and the BH3 mimetic ABT-737, which triggers mitochondrial apoptosis (Fig 1F).
Fig 1. Phenotype of RUNX1-ETO knockdown and kinetics of protein/mRNA expression in Kasumi-1.
A) Kasumi-1 cell lysates were prepared in SDS buffer at the indicated time points post transduction and RUNX1-ETO depletion was confirmed by Western Blot (n = 3). B) Cell expansion of Kasumi-1/ctrl and Kasumi-1/shRE was determined by counting and trypan blue exclusion, and growth curves depict cell numbers relative to control cells at day 15 after lentiviral transduction (n = 4). C) Number of Kasumi-1/ctrl and Kasumi-1/shRE colonies, as counted nine days after plating in limiting dilution (n = 4). Cell cycle distribution (D) of Kasumi-1/ctrl and Kasumi-1/shRE and the percentage of apoptotic cells (E) were determined by PI staining and subsequent flow cytometry at day twelve after transduction (n = 4). F) The response of Kasumi-1/ctrl and Kasumi-1/shRE to ABT-737 and Doxorubicin was measured by MTS Assay and PI staining, respectively, at day 14 post transduction (n = 3). The percentage of viable cells is shown relative to untreated cells after 48 hours of treatment with the indicated drug concentrations. G) Transmission electron microscopy of Kasumi-1/ctrl (left) and Kasumi-1/shRE (right) cells, representative for three independent experiments. N, nucleus; G, Golgi; M, mitochondria. Profiles of ER cisternae are highlighted by arrowheads. Note the flat and evenly formed ER cisternae in Kasumi-1/ctrl, compared to frequently dilated cisternae in Kasumi-1/shRE cells. Scale bar = 1 μm. Protein and mRNA expression of RUNX1-ETO target genes were analyzed at the indicated time points after lentiviral transfer of RUNX1-ETO-specific shRNA. H) mRNA expression was determined relative to control cells and normalized to housekeeping control. I) Whole-cell lysates of Kasumi-1/ctrl and Kasumi-1/shRE were prepared in high-salt lysis buffer (left, n = 4) or boiling SDS buffer (right, n = 3) and analyzed by Western Blot. Dashed line indicates separate membranes. Data are represented as mean +/- SD and p-values were calculated using 2-way ANOVA (B+F) or two-sided student´s t-test (C,D,E,H). *p<0.05, **p<0.01, ***p<0.001.
Electron microscopy revealed dilated cisternae in most of all cell profiles in Kasumi-1/shRE, in contrast to control cells (n = 3) (Fig 1G). Cisternae were identified as endoplasmatic reticulum (ER) by their arrangement, clearly discernable connections to not dilated parts of ER cisternae and by ribosomes on their surface. No other discernable alterations in cell morphology were detected, including Golgi apparatus, mitochondria, nuclei, cell size or other cellular components or ultrastructural parameters. ER dilation is recognized as a sign of ER stress [33–35] and has not yet been described in the context of RUNX1-ETO loss-of-function. However, the induction of granulocytic differentiation has been linked to ER stress, as a consequence of increased folding demands due to enhanced expression of secretory proteins, in acute promyelocytic leukemia (APL) cell lines and primary blasts [36]. The release of RUNX1-ETO suppression in Kasumi-1 has been shown to induce myeloid differentiation [26] and may present a conceivable explanation for the ER dilation, observed in our study.
Next, we analyzed the kinetics of mRNA and protein expression of RUNX1-ETO target genes up to 15 days after lentiviral transduction of Kasumi-1 cells. As expected from the mode of RUNX1-ETO action, the well-known RUNX1-ETO targets PU.1, BCL2 and CTSG [16,13,28] were upregulated on both mRNA and protein level on day five after transduction (Fig 1H and S1 Fig). Surprisingly, Western Blot analysis of cellular extracts, prepared in high-salt lysis buffer, showed a strong reduction of RUNX1-ETO target protein expression at later time points post transduction (Fig 1I, left), while the mRNA levels remained stable (Fig 1H, upper panel). In contrast, non RUNX1-ETO target genes, such as RUNX1, STAT5A and STAT3 showed a consistent reduction of both mRNA (Fig 1H, lower panel) and protein levels (Fig 1I) throughout the time course. However, such a reduction in protein expression upon stable anti RUNX1-ETO RNAi has not yet been described. In addition to reduced protein expression, we observed a lower molecular weight isoform of PU.1 (referred to as PU.1 p22), immunoreactive with a C-terminal PU.1 antibody, as detected by Western Blot (Fig 1I, left). Expression of PU.1 p22 strongly increased after RUNX1-ETO knockdown in parallel to reduced expression of wildtype PU.1, suggesting proteolytic cleavage of PU.1. However, treatment of the cells with inhibitors of proteasomal (Bortezomib) and lysosomal (Chloroquine) degradation did not rescue protein expression of RUNX1-ETO target genes or reduce the occurrence of PU.1 p22 (S2 Fig). To evaluate the significance of these observations, we analyzed possible cell lysis-dependent effects, applying alternative protocols for the lysis of Kasumi-1/shRE and control cells (S2 Fig). Indeed, preparation of cellular lysates in boiling SDS sample buffer did not result in the observed loss of protein expression and PU.1 p22 was not detected. The resulting protein expression kinetics matched the kinetics of mRNA expression (Fig 1I, right), indicating protein degradation in Kasumi-1/shRE during the preparation of whole cell lysates under high-salt conditions.
The lysosomal protease CTSG is a well-known RUNX1-ETO target [28] and was strongly induced in our experiments (Fig 1H+1I), representing an obvious candidate for the degradation of proteins under cell lysis conditions in our study. CTSG has been shown to degrade RUNX1-ETO in vitro and in vivo [37]. This process has been discussed as a cellular mechanism to selectively remove aberrant proteins, and the suppression of CTSG by RUNX1-ETO could promote leukemogenesis by evading this intracellular surveillance system [37]. Furthermore, Schuster et al. identified CTSG as the STAT5 protease [38], which has been ascribed to the generation of C-terminally truncated isoforms of STAT5A and STAT5B, referred to as STAT5γ [39–41]. Truncated isoforms have also been observed for STAT3 [42–44] and STAT6 [45–47]. The relevance of STAT5γ has been discussed in the context of apoptosis, myeloid differentiation and AML [48–52]. However, Schuster et al. [38] clearly demonstrated that the generation of STAT5γ does not happen in vivo, but is an artifact of cell lysis. This finding might also question the significance of CTSG-mediated cleavage of RUNX1-ETO, which has been discussed by Jin et al. [37]. Based on the above-mentioned studies, the strong induction of CTSG and the loss of detectable STAT3 and STAT5 protein in Kasumi-1/shRE cells (Fig 1H+1I) suggest a contribution of CTSG to the decrease of protein expression in our study.
Mapping of protease cleavage patterns in RUNX1-ETO silenced Kasumi-1 cells by LC-MS-based peptide analysis
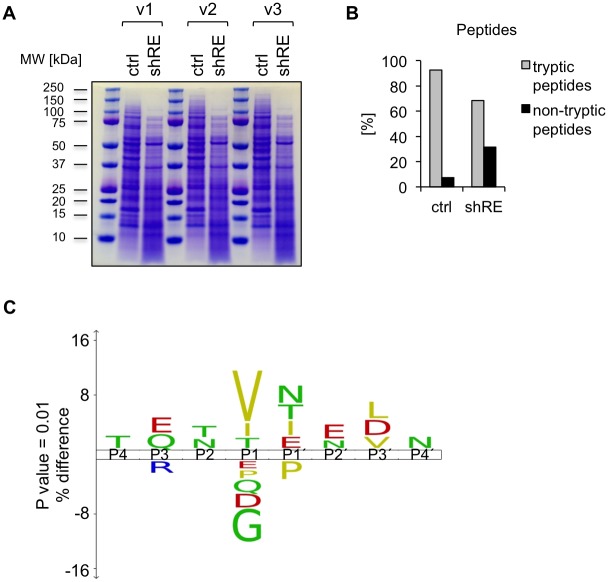
To analyze the incidence of proteolytic cleavage and to identify the contributing proteases on a proteome-wide scale, we employed LC-MS proteomics, referring to the approach, described by Gupta et al. 2010 [53] which allows the mapping of proteolytic cleavage sites in any MS dataset. Kasumi-1/shRE and Kasumi-1/ctrl cells were lysed in RIPA buffer, supplemented with commercially available Protease Inhibitor Cocktail, and separated by SDS-PAGE. The pattern of the protein bands after Coomassie staining indicates increased proteolytic activity in the Kasumi-1/shRE lysates (Fig 2A) in line with our previous observation of RUNX1-ETO knockdown resulting in protein degradation in Kasumi-1 cells (Fig 1I). The proteins were excised from the gel, trypsinized and subjected to LC-MS analysis. Trypsin is a highly specific protease, cleaving only after Lysine (K) or Arginine (R). Thus, we were able to differentiate between peptides, which can be ascribed to trypsin digestion as a part of the procedure for MS sample preparation (tryptic peptides), and peptides resulting from proteolysis by other proteases, either due to endogenous proteolytic activity, or during cell lysis (non-tryptic peptides). Peptides with K or R at position P1 of their N- and C-terminal ends were considered tryptic, while all peptides with at least one end being not K or R and the other end being not the first or the last amino acid of the protein, were considered to be products of proteolytic cleavage during cell lysis. Based on this analysis, we identified a total of 17,490 peptides, with 5767 and 3370 peptides being unique to Kasumi-1/ctrl and Kasumi-1/shRE, respectively. We found 7.4% of the peptides from Kasumi-1/ctrl but 31.5% of the peptides from Kasumi-1/shRE to be of non-tryptic origin, which reflects a more than 4-fold increase of proteolytic activity in Kasumi-1/shRE compared to control cells (Fig 2B).
Fig 2. Mapping of protease cleavage sites by mass spectrometry.
A) Whole cell lysates of Kasumi-1/ctrl and Kasumi-1/shRE cells were prepared in RIPA buffer supplemented with cOmplete Protease Inhibitor Cocktail (Roche) on day twelve after transduction. Proteins were separated by SDS PAGE, stained with Coomassie blue (n = 3) and subjected to LC-MS analysis. B) The identified peptides were categorized as tryptic peptides, resulting from trypsin digestion during the MS sample preparation procedure, or non-tryptic peptides, resulting from proteolytic cleavage during cell lysis. C) IceLogo showing amino acids at positions P4-P4´, which are enriched or depleted in peptides from Kasumi-1/shRE compared to Kasumi-1/ctrl with p<0.01. Red, acidic; blue, basic; yellow, nonpolar hydrophobic; green, polar neutral amino acids. Results shown in B) and C) are representative for one of three independent measurements.
In a next step, we looked at the distribution of the amino acids surrounding the cleavage sites of the non-tryptic peptides to assign the responsible proteases. The amino acid P4-P4´ of peptides derived from Kasumi-1/ctrl and Kasumi-1/shRE have been integrated into an IceLogo [54] to identify conserved sequence patterns (Fig 2C). The IceLogo shows a prominent enrichment of the hydrophobic amino acids Valine (V), Isoleucine (I) and Threonine (T) in position P1, representing the amino acid before the cleavage site. In addition, a noticeable over-representation of the acidic amino acids Glutamate (E) and Aspartate (D) in the positions P1´-P3´, downstream of the cleavage site has been observed. To draw conclusions from the proteolytic signature in our dataset to the corresponding proteases, we searched the literature and the Peptidase Database Merops (http://www.ebi.ac.uk/merops/) [55] for established protease cleavage patterns. As the lysosomal protease CTSG is a well-known target of RUNX1-ETO and strongly induced by stable anti-RUNX1-ETO RNAi in our experiments (Fig 1H+1I) we focused on lysosomal enzymes. According to O´Donoghue et al. [56], CTSG has a preference for the aromatic amino acids Phenylalanine (F) and Tyrosine (Y) in position P1 and does not match the observed cleavage patterns in our dataset. However, we did find overlaps between the proteolytic signatures of Kasumi-1/shRE peptides and the signature for Neutrophil Elastase (ELANE), which shows enrichment for Isoleucine (I), Valine (V) and Threonine (T) at position P1 of its substrates, as shown by substrate specificity profiling [56]. In addition, Vizovisek et al. [57] have demonstrated an over-representation of Glutamate (E) and Aspartate (D) at positions P1´-P4´ of cleavage sites detected by Cathepsins K, L and S (CTSK/L/S), which also fits the cleavage site pattern observed in our study.
ELANE and CTSG contribute to cell lysis-induced protein degradation in response to long-term RUNX1-ETO silencing in Kasumi-1 cells
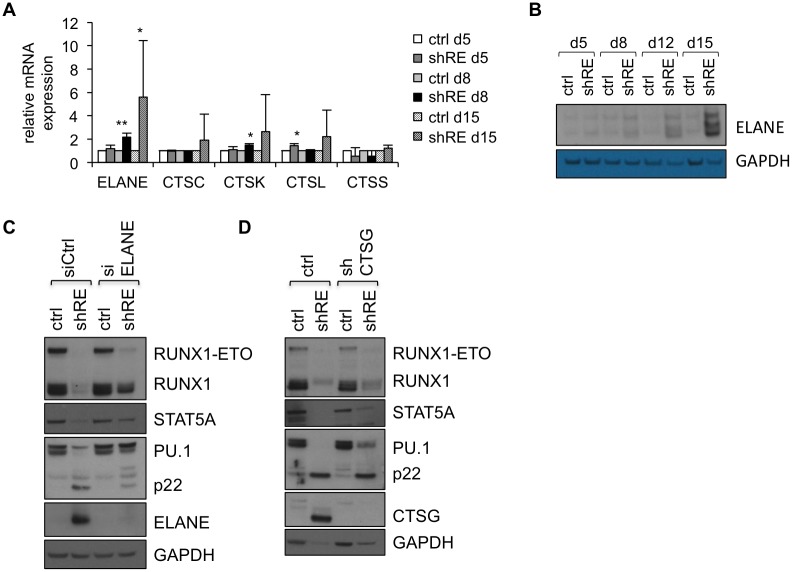
To narrow down the spectrum of proteases involved in protein degradation in RUNX1-ETO-silenced Kasumi-1 cells, we examined RUNX1-ETO-dependent expression of ELANE and CTSK/L/S in Kasumi-1 by qRT-PCR (Fig 3A). Cathepsin C (CTSC, Dipeptidyl-peptidase I) has been included in our analysis, as it is a common activator of CTSG and ELANE [58]. Only a weak induction was observed for CTSC and CTSK/L/S in response to RUNX1-ETO silencing in Kasumi-1 cells, while the expression of ELANE mRNA was clearly enhanced at the later time points after RUNX1-ETO knockdown. In addition, upregulation of ELANE protein expression in Kasumi-1/shRE has been confirmed by Western Blot (Fig 3B). A possible direct regulation of ELANE by RUNX1-ETO has already been excluded by Lausen et al. 2006 [59], but ELANE expression is controlled by hematopoietic transcription factors, such as the well-known RUNX1-ETO targets C/EBPα and PU.1 [60,61]. This suggests an indirect induction of ELANE expression in response to the release of C/EBPα and PU.1 suppression in Kasumi-1/shRE.
Fig 3. ELANE and CTSG contribute to cell lysis-induced protein degradation in RUNX1-ETO-depleted Kasumi-1 cells.
A) RUNX1-ETO dependent mRNA expression of ELANE and CTSC/K/L/S in Kasumi-1 cells was determined by qRT-PCR and normalized to housekeeping control. Data are shown as mean 2-ΔΔCT +/- SD (n = 3) and p-values were calculated by two-sided student´s t-test (*p<0.05, **p<0.01). B) Whole cell lysates of Kasumi-1/shRE and control cells have been prepared in SDS sample buffer and analyzed for ELANE protein expression by Western Blot. C) Kasumi-1/ctrl and Kasumi-1/shRE cells were consecutively electroporated with ELANE-specific or control siRNA on days five and eight after lentiviral transduction with control virus or shRE. Whole cell lysates were prepared in high-salt lysis buffer on day eleven after RUNX1-ETO depletion and protein expression was analyzed by Western Blot. D) Kasumi-1 cells were transduced with CTSG-specific shRNA (Kasumi-1/shCTSG) or control virus (Kasumi-1/ctrl), followed by transduction with RUNX1-ETO-specific shRNA or control virus, two days after the first lentiviral transduction. Whole cell lysates were prepared in high-salt lysis buffer on day twelve after RUNX1-ETO knockdown (respectively day 14 after CTSG knockdown) and analyzed by Western Blot. Western Blots are representative for three independent experiments.
The combined results of the gene expression analysis and the consultation of available protease cleavage patterns suggest a contribution of ELANE to the proteolytic activity observed in Kasumi-1/shRE. As a consequence, depletion of ELANE in Kasumi-1 should rescue protein expression after RUNX1-ETO knockdown. Indeed, repeated electroporation of Kasumi-1/ctrl and Kasumi-1/shRE with ELANE-specific siRNA partially reversed cell lysis-triggered degradation of RUNX1, STAT5A and PU.1, and diminished detection of PU.1 p22 in Kasumi-1/shRE (Fig 3C). Although the proteolytic cleavage patterns described for CTSG did not resemble the proteolytic signature generated from our dataset, we still considered a contribution of CTSG to the degradation of proteins in RUNX1-ETO-silenced Kasumi-1 cells. Expression of CTSG was strongly induced in response to RUNX1-ETO depletion in our experiments (Fig 1H+1I). Furthermore, CTSG has been described to cleave STAT5 in a cell lysis-dependent mode of action [38], and we found the protein levels of STAT5A to be decreased in Kasumi-1/shRE (Fig 1I), dependent on the cell lysis procedure (S2 Fig). Consequently, we investigated a possible contribution of CTSG to the cell lysis-induced protein degradation in Kasumi-1/shRE by shRNA-mediated depletion of CTSG in Kasumi-1 cells, followed by knockdown of RUNX1-ETO. Indeed, silencing of CTSG expression diminished protein degradation in Kasumi-1/shRE during the preparation of whole cell lysates, as shown for RUNX1, STAT5A, PU.1 and GAPDH (Fig 3D). In contrast to ELANE, CTSG does not seem to contribute to the generation of PU.1 p22. Knockdown of ELANE and CTSG was confirmed by Western Blot (Fig 3C+3D) and qRT-PCR (S3 Fig). Finally, the most right lane in Fig 3C and 3D demonstrates specific silencing of ELANE and CTSG by RNAi, respectively.
Taken together, we describe a mechanism by which the high abundance of lysosomal proteases in combination with high-salt cell lysis conditions leads to the degradation of cellular proteins during the preparation of whole cell lysates. This phenomenon has led to false interpretation of protein expression data before as in case of the intensely discussed STAT5γ isoform which has been shown to be generated by CTSG during the preparation of nuclear extracts [38]. This might present a major pit fall for the investigation of protein expression under conditions which promote proteolytic activity such as the increased expression of lysosomal proteases in response to RUNX1-ETO depletion.
Silencing of RUNX1-ETO in t(8;21) positive cells has been shown to promote granulocytic differentiation accompanied by the expression of lysosomal proteins [26,28]. As a consequence, the upregulation of lysosomal proteases may also compromise proteomic studies concerning leukemic differentiation.
Using LC-MS data and sequence analysis of peptide cleavage sites, we propose the lysosomal proteases ELANE and CTSK/L/S as possible candidates for the degradation of cellular proteins in Kasumi-1/shRE triggered by cell lysis. The contribution of ELANE and CTSG was confirmed by si/shRNA knockdown although the LC-MS analysis did not show any footprints of CTSG activity on Kasumi-1/shRE derived peptides. This may be due to limited sensitivity of this screening approach. The contribution of CTSK/L/S has not been further evaluated, as we observed only a weak induction in response to RUNX1-ETO silencing in Kasumi-1 cells. However, the regulation of lysosomal proteases is not limited to gene expression, but relies on a complex network of activation cascades including the proteolytic processing of inactive zymogens [62–64] and the regulation of endogenous protease inhibitors. For instance, CTSG and ELANE are able to activate Cathepsin B [65], which in turn activates CTSL via CTSD [66]. The endogenous inhibitor of CTSK and CTSL, Serpin B13 [67], is induced by RUNX1 [68] and, hypothetically, could be a possible target for RUNX1-ETO suppression, leading to an increase in CTSK and CTSL activity in Kasumi-1/shRE cells.
Our data demonstrate that the protein degradation in response to long-term RUNX1-ETO RNAi does not occur in vivo, but depends on cell lysis conditions which allow the release of proteases from the lysosomes. Yet, it cannot be excluded, that the strong induction of proteolytic activity by RUNX1-ETO depletion has any impact on the cells and their environment in vivo, as the functions of CTSG and ELANE are not restricted to the lysosomal compartment, but can be secreted [69–71] to modulate the activity of cytokines and their receptors [72].
Supporting information
Oligonucleotides used for cloning of CTSG-specific shRNA with underlined target sequence.
(TIF)
A) Transduction efficacy of lentivirally transduced Kasumi-1 cells was determined by flow cytometry based on GFP-reporter fluorescence. B) Whole cell lysates were prepared in high-salt lysis buffer on day 5 after lentiviral transduction and expression of RUNX1-ETO target genes was determined by Western Blot (n = 4).
(TIF)
Kasumi-1/ctrl and Kasumi-1/shRE cells were treated with Bortezomib (A) or Chloroquine (B) on day 13 after lentiviral transduction, as indicated. Whole cell lysates were analyzed for the expression of RUNX1-ETO target genes by Western Blot. Proteasomal and lysosomal inhibition was confirmed by detecting accumulation of ubiquitinylated proteins or processing of the autophagy marker LC3A/B (microtubule-associated proteins 1A/1B light chain 3B), respectively. PI staining was performed to evaluate cytotoxicity of the different inhibitors by flow cytometry and the percentage of PI-positive cells is shown at the bottom of each Western Blot. Data are representative for three independent experiments. C) Lysates of Kasumi-1/ctrl and Kasumi-1/shRE cells were prepared at day 14 after shRNA-mediated RUNX1-ETO knockdown and analyzed by Western Blot. The application of different lysis conditions demonstrates the impact of the cell lysis procedure on protein stability in RUNX1-ETO-silenced Kasumi-1 cells. Data are representative for one of three independent experiments.
(TIF)
ELANE and CTSG mRNA levels were measured by qRT-PCR and normalized to housekeeping control. Data are shown as log2 of mean 2-ΔΔCT +/- SD and p-values were determined by two-sided student´s t-test. *p<0.05, ***p<0.001.
(TIF)
Acknowledgments
We thank Karin Battmer and Hanna Kirchhoff for excellent technical help and reading of the manuscript.
Data Availability
Data underlying the results of this study have been uploaded as Supporting Information files and to figshare at the following links: https://doi.org/10.6084/m9.figshare.9784493.v1 and https://doi.org/10.6084/m9.figshare.9724856.v1.
Funding Statement
This work was supported by Deutsche Forschungsgemeinschaft (https://www.dfg.de) ED 34/4 to M.E. and SCHE 550/6-1 to M.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998. October 1;92(7):2322–33. [PubMed] [Google Scholar]
- 2.Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002. December 15;100(13):4325–36. [DOI] [PubMed] [Google Scholar]
- 3.Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992. October 1;80(7):1825–31. [PubMed] [Google Scholar]
- 4.Nisson PE, Watkins PC, Sacchi N. Transcriptionally active chimeric gene derived from the fusion of the AML1 gene and a novel gene on chromosome 8 in t(8;21) leukemic cells. Cancer Genet Cytogenet. 1992. October 15;63(2):81–8. 10.1016/0165-4608(92)90384-k [DOI] [PubMed] [Google Scholar]
- 5.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993. July;12(7):2715–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998. December;18(12):7185–91. 10.1128/mcb.18.12.7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998. December;18(12):7176–84. 10.1128/mcb.18.12.7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A. 1998. September 1;95(18):10860–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata H, Spencer TE, Oñate SA, Jenster G, Tsai SY, Tsai MJ, et al. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–64; discussion 164–165. [PubMed] [Google Scholar]
- 10.Chen JD, Li H. Coactivation and corepression in transcriptional regulation by steroid/nuclear hormone receptors. Crit Rev Eukaryot Gene Expr. 1998;8(2):169–90. 10.1615/critreveukargeneexpr.v8.i2.40 [DOI] [PubMed] [Google Scholar]
- 11.Hwang ES, Hong JH, Bae SC, Ito Y, Lee SK. Regulation of c-fos gene transcription and myeloid cell differentiation by acute myeloid leukemia 1 and acute myeloid leukemia-MTG8, a chimeric leukemogenic derivative of acute myeloid leukemia 1. FEBS Lett. 1999. March 5;446(1):86–90. 10.1016/s0014-5793(99)00190-8 [DOI] [PubMed] [Google Scholar]
- 12.Linggi B, Müller-Tidow C, van de Locht L, Hu M, Nip J, Serve H, et al. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002. July;8(7):743–50. 10.1038/nm726 [DOI] [PubMed] [Google Scholar]
- 13.Zhuang W-Y, Cen J-N, Zhao Y, Chen Z-X. Epigenetic silencing of Bcl-2, CEBPA and p14(ARF) by the AML1-ETO oncoprotein contributing to growth arrest and differentiation block in the U937 cell line. Oncol Rep. 2013. July;30(1):185–92. 10.3892/or.2013.2459 [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Gural A, Sun X-J, Zhao X, Perna F, Huang G, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science. 2011. August 5;333(6043):765–9. 10.1126/science.1201662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Soria N, McKenzie L, Draper J, Ptasinska A, Issa H, Potluri S, et al. The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation. Cancer Cell. 2018. 08;34(4):626–642.e8. 10.1016/j.ccell.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vangala RK, Heiss-Neumann MS, Rangatia JS, Singh SM, Schoch C, Tenen DG, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003. January 1;101(1):270–7. 10.1182/blood-2002-04-1288 [DOI] [PubMed] [Google Scholar]
- 17.Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001. April;7(4):444–51. 10.1038/86515 [DOI] [PubMed] [Google Scholar]
- 18.Choi Y, Elagib KE, Delehanty LL, Goldfarb AN. Erythroid inhibition by the leukemic fusion AML1-ETO is associated with impaired acetylation of the major erythroid transcription factor GATA-1. Cancer Res. 2006. March 15;66(6):2990–6. 10.1158/0008-5472.CAN-05-2944 [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004. August 27;305(5688):1286–9. 10.1126/science.1097937 [DOI] [PubMed] [Google Scholar]
- 20.Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, et al. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol. 2001. October;21(19):6470–83. 10.1128/MCB.21.19.6470-6483.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klisovic MI, Maghraby EA, Parthun MR, Guimond M, Sklenar AR, Whitman SP, et al. Depsipeptide (FR 901228) promotes histone acetylation, gene transcription, apoptosis and its activity is enhanced by DNA methyltransferase inhibitors in AML1/ETO-positive leukemic cells. Leukemia. 2003. February;17(2):350–8. 10.1038/sj.leu.2402776 [DOI] [PubMed] [Google Scholar]
- 22.Shia W-J, Okumura AJ, Yan M, Sarkeshik A, Lo M-C, Matsuura S, et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood. 2012. May 24;119(21):4953–62. 10.1182/blood-2011-04-347476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ptasinska A, Assi SA, Mannari D, James SR, Williamson D, Dunne J, et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012. August;26(8):1829–41. 10.1038/leu.2012.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhoades KL, Hetherington CJ, Harakawa N, Yergeau DA, Zhou L, Liu LQ, et al. Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood. 2000. September 15;96(6):2108–15. [PubMed] [Google Scholar]
- 25.Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci U S A. 2001. August 28;98(18):10398–403. 10.1073/pnas.171321298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidenreich O, Krauter J, Riehle H, Hadwiger P, John M, Heil G, et al. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003. April 15;101(8):3157–63. 10.1182/blood-2002-05-1589 [DOI] [PubMed] [Google Scholar]
- 27.Martinez N, Drescher B, Riehle H, Cullmann C, Vornlocher H-P, Ganser A, et al. The oncogenic fusion protein RUNX1-CBFA2T1 supports proliferation and inhibits senescence in t(8;21)-positive leukaemic cells. BMC Cancer. 2004. August 6;4:44 10.1186/1471-2407-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunne J, Cullmann C, Ritter M, Soria NM, Drescher B, Debernardi S, et al. siRNA-mediated AML1/MTG8 depletion affects differentiation and proliferation-associated gene expression in t(8;21)-positive cell lines and primary AML blasts. Oncogene. 2006. October 5;25(45):6067–78. 10.1038/sj.onc.1209638 [DOI] [PubMed] [Google Scholar]
- 29.Scherr M, Battmer K, Ganser A, Eder M. Modulation of gene expression by lentiviral-mediated delivery of small interfering RNA. Cell Cycle Georget Tex. 2003. June;2(3):251–7. [PubMed] [Google Scholar]
- 30.Dawodu D, Patecki M, Hegermann J, Dumler I, Haller H, Kiyan Y. oxLDL inhibits differentiation and functional activity of osteoclasts via scavenger receptor-A mediated autophagy and cathepsin K secretion. Sci Rep. 2018. August 2;8(1):11604 10.1038/s41598-018-29963-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A, et al. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood. 2011. April 21;117(16):4338–48. 10.1182/blood-2010-06-289058 [DOI] [PubMed] [Google Scholar]
- 32.Jochim N, Gerhard R, Just I, Pich A. Impact of clostridial glucosylating toxins on the proteome of colonic cells determined by isotope-coded protein labeling and LC-MALDI. Proteome Sci. 2011. August 17;9:48 10.1186/1477-5956-9-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Takeuchi T, Tanaka S, Kubo SK, Kayo T, Lu D, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. J Clin Invest. 1999. January;103(1):27–37. 10.1172/JCI4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, Welling C, et al. Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia. 2005. November;48(11):2313–21. 10.1007/s00125-005-1947-4 [DOI] [PubMed] [Google Scholar]
- 35.Akiyama M, Hatanaka M, Ohta Y, Ueda K, Yanai A, Uehara Y, et al. Increased insulin demand promotes while pioglitazone prevents pancreatic beta cell apoptosis in Wfs1 knockout mice. Diabetologia. 2009. April;52(4):653–63. 10.1007/s00125-009-1270-6 [DOI] [PubMed] [Google Scholar]
- 36.Masciarelli S, Capuano E, Ottone T, Divona M, De Panfilis S, Banella C, et al. Retinoic acid and arsenic trioxide sensitize acute promyelocytic leukemia cells to ER stress. Leukemia. 2018;32(2):285–94. 10.1038/leu.2017.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin W, Wu K, Li Y-Z, Yang W-T, Zou B, Zhang F, et al. AML1-ETO targets and suppresses cathepsin G, a serine protease, which is able to degrade AML1-ETO in t(8;21) acute myeloid leukemia. Oncogene. 2013. April 11;32(15):1978–87. 10.1038/onc.2012.204 [DOI] [PubMed] [Google Scholar]
- 38.Schuster B, Hendry L, Byers H, Lynham SF, Ward MA, John S. Purification and identification of the STAT5 protease in myeloid cells. Biochem J. 2007. May 15;404(1):81–7. 10.1042/BJ20061877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azam M, Erdjument-Bromage H, Kreider BL, Xia M, Quelle F, Basu R, et al. Interleukin-3 signals through multiple isoforms of Stat5. EMBO J. 1995. April 3;14(7):1402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azam M, Lee C, Strehlow I, Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997. June;6(6):691–701. 10.1016/s1074-7613(00)80445-8 [DOI] [PubMed] [Google Scholar]
- 41.Meyer J, Jucker M, Ostertag W, Stocking C. Carboxyl-truncated STAT5beta is generated by a nucleus-associated serine protease in early hematopoietic progenitors. Blood. 1998. March 15;91(6):1901–8. [PubMed] [Google Scholar]
- 42.Chakraborty A, Tweardy DJ. Granulocyte colony-stimulating factor activates a 72-kDa isoform of STAT3 in human neutrophils. J Leukoc Biol. 1998. November;64(5):675–80. 10.1002/jlb.64.5.675 [DOI] [PubMed] [Google Scholar]
- 43.Xia Z, Salzler RR, Kunz DP, Baer MR, Kazim L, Baumann H, et al. A novel serine-dependent proteolytic activity is responsible for truncated signal transducer and activator of transcription proteins in acute myeloid leukemia blasts. Cancer Res. 2001. February 15;61(4):1747–53. [PubMed] [Google Scholar]
- 44.Kato T, Sakamoto E, Kutsuna H, Kimura-Eto A, Hato F, Kitagawa S. Proteolytic conversion of STAT3alpha to STAT3gamma in human neutrophils: role of granule-derived serine proteases. J Biol Chem. 2004. July 23;279(30):31076–80. 10.1074/jbc.M400637200 [DOI] [PubMed] [Google Scholar]
- 45.Sherman MA, Secor VH, Brown MA. IL-4 preferentially activates a novel STAT6 isoform in mast cells. J Immunol Baltim Md 1950. 1999. March 1;162(5):2703–8. [PubMed] [Google Scholar]
- 46.Sherman MA, Powell DR, Brown MA. IL-4 induces the proteolytic processing of mast cell STAT6. J Immunol Baltim Md 1950. 2002. October 1;169(7):3811–8. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki K, Nakajima H, Kagami S-I, Suto A, Ikeda K, Hirose K, et al. Proteolytic processing of Stat6 signaling in mast cells as a negative regulatory mechanism. J Exp Med. 2002. July 1;196(1):27–38. 10.1084/jem.20011682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bovolenta C, Testolin L, Benussi L, Lievens PM, Liboi E. Positive selection of apoptosis-resistant cells correlates with activation of dominant-negative STAT5. J Biol Chem. 1998. August 14;273(33):20779–84. 10.1074/jbc.273.33.20779 [DOI] [PubMed] [Google Scholar]
- 49.Lokuta MA, McDowell MA, Paulnock DM. Identification of an additional isoform of STAT5 expressed in immature macrophages. J Immunol Baltim Md 1950. 1998. August 15;161(4):1594–7. [PubMed] [Google Scholar]
- 50.Lee C, Piazza F, Brutsaert S, Valens J, Strehlow I, Jarosinski M, et al. Characterization of the Stat5 protease. J Biol Chem. 1999. September 17;274(38):26767–75. 10.1074/jbc.274.38.26767 [DOI] [PubMed] [Google Scholar]
- 51.Piazza F, Valens J, Lagasse E, Schindler C. Myeloid differentiation of FdCP1 cells is dependent on Stat5 processing. Blood. 2000. August 15;96(4):1358–65. [PubMed] [Google Scholar]
- 52.Xia Z, Sait SN, Baer MR, Barcos M, Donohue KA, Lawrence D, et al. Truncated STAT proteins are prevalent at relapse of acute myeloid leukemia. Leuk Res. 2001. June;25(6):473–82. 10.1016/s0145-2126(00)00158-2 [DOI] [PubMed] [Google Scholar]
- 53.Gupta N, Hixson KK, Culley DE, Smith RD, Pevzner PA. Analyzing protease specificity and detecting in vivo proteolytic events using tandem mass spectrometry. Proteomics. 2010. August;10(15):2833–44. 10.1002/pmic.200900821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009. November;6(11):786–7. 10.1038/nmeth1109-786 [DOI] [PubMed] [Google Scholar]
- 55.Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018. 04;46(D1):D624–32. 10.1093/nar/gkx1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Donoghue AJ, Jin Y, Knudsen GM, Perera NC, Jenne DE, Murphy JE, et al. Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PloS One. 2013;8(9):e75141 10.1371/journal.pone.0075141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vizovišek M, Vidmar R, Van Quickelberghe E, Impens F, Andjelković U, Sobotič B, et al. Fast profiling of protease specificity reveals similar substrate specificities for cathepsins K, L and S. Proteomics. 2015. July;15(14):2479–90. 10.1002/pmic.201400460 [DOI] [PubMed] [Google Scholar]
- 58.Adkison AM, Raptis SZ, Kelley DG, Pham CTN. Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J Clin Invest. 2002. February;109(3):363–71. 10.1172/JCI13462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lausen J, Liu S, Fliegauf M, Lübbert M, Werner MH. ELA2 is regulated by hematopoietic transcription factors, but not repressed by AML1-ETO. Oncogene. 2006. March 2;25(9):1349–57. 10.1038/sj.onc.1209181 [DOI] [PubMed] [Google Scholar]
- 60.Oelgeschläger M, Nuchprayoon I, Lüscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol. 1996. September;16(9):4717–25. 10.1128/mcb.16.9.4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nuchprayoon I, Shang J, Simkevich CP, Luo M, Rosmarin AG, Friedman AD. An enhancer located between the neutrophil elastase and proteinase 3 promoters is activated by Sp1 and an Ets factor. J Biol Chem. 1999. January 8;274(2):1085–91. 10.1074/jbc.274.2.1085 [DOI] [PubMed] [Google Scholar]
- 62.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012. January;1824(1):68–88. 10.1016/j.bbapap.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishimura Y, Kawabata T, Furuno K, Kato K. Evidence that aspartic proteinase is involved in the proteolytic processing event of procathepsin L in lysosomes. Arch Biochem Biophys. 1989. June;271(2):400–6. 10.1016/0003-9861(89)90289-0 [DOI] [PubMed] [Google Scholar]
- 64.Rowan AD, Mason P, Mach L, Mort JS. Rat procathepsin B. Proteolytic processing to the mature form in vitro. J Biol Chem. 1992. August 5;267(22):15993–9. [PubMed] [Google Scholar]
- 65.Dalet-Fumeron V, Guinec N, Pagano M. In vitro activation of pro-cathepsin B by three serine proteinases: leucocyte elastase, cathepsin G, and the urokinase-type plasminogen activator. FEBS Lett. 1993. October 18;332(3):251–4. 10.1016/0014-5793(93)80643-9 [DOI] [PubMed] [Google Scholar]
- 66.Laurent-Matha V, Derocq D, Prébois C, Katunuma N, Liaudet-Coopman E. Processing of human cathepsin D is independent of its catalytic function and auto-activation: involvement of cathepsins L and B. J Biochem (Tokyo). 2006. March;139(3):363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jayakumar A, Kang Y, Frederick MJ, Pak SC, Henderson Y, Holton PR, et al. Inhibition of the cysteine proteinases cathepsins K and L by the serpin headpin (SERPINB13): a kinetic analysis. Arch Biochem Biophys. 2003. January 15;409(2):367–74. 10.1016/s0003-9861(02)00635-5 [DOI] [PubMed] [Google Scholar]
- 68.Boyapati A, Ren B, Zhang D-E. SERPINB13 is a novel RUNX1 target gene. Biochem Biophys Res Commun. 2011. July 22;411(1):115–20. 10.1016/j.bbrc.2011.06.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meier HL, Heck LW, Schulman ES, MacGlashan DW. Purified human mast cells and basophils release human elastase and cathepsin G by an IgE-mediated mechanism. Int Arch Allergy Appl Immunol. 1985;77(1–2):179–83. 10.1159/000233779 [DOI] [PubMed] [Google Scholar]
- 70.Bangalore N, Travis J. Comparison of properties of membrane bound versus soluble forms of human leukocytic elastase and cathepsin G. Biol Chem Hoppe Seyler. 1994. October;375(10):659–66. 10.1515/bchm3.1994.375.10.659 [DOI] [PubMed] [Google Scholar]
- 71.Owen CA, Campbell MA, Sannes PL, Boukedes SS, Campbell EJ. Cell surface-bound elastase and cathepsin G on human neutrophils: a novel, non-oxidative mechanism by which neutrophils focus and preserve catalytic activity of serine proteinases. J Cell Biol. 1995. November;131(3):775–89. 10.1083/jcb.131.3.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Döring G. The role of neutrophil elastase in chronic inflammation. Am J Respir Crit Care Med. 1994. December;150(6 Pt 2):S114–117. 10.1164/ajrccm/150.6_Pt_2.S114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides used for cloning of CTSG-specific shRNA with underlined target sequence.
(TIF)
A) Transduction efficacy of lentivirally transduced Kasumi-1 cells was determined by flow cytometry based on GFP-reporter fluorescence. B) Whole cell lysates were prepared in high-salt lysis buffer on day 5 after lentiviral transduction and expression of RUNX1-ETO target genes was determined by Western Blot (n = 4).
(TIF)
Kasumi-1/ctrl and Kasumi-1/shRE cells were treated with Bortezomib (A) or Chloroquine (B) on day 13 after lentiviral transduction, as indicated. Whole cell lysates were analyzed for the expression of RUNX1-ETO target genes by Western Blot. Proteasomal and lysosomal inhibition was confirmed by detecting accumulation of ubiquitinylated proteins or processing of the autophagy marker LC3A/B (microtubule-associated proteins 1A/1B light chain 3B), respectively. PI staining was performed to evaluate cytotoxicity of the different inhibitors by flow cytometry and the percentage of PI-positive cells is shown at the bottom of each Western Blot. Data are representative for three independent experiments. C) Lysates of Kasumi-1/ctrl and Kasumi-1/shRE cells were prepared at day 14 after shRNA-mediated RUNX1-ETO knockdown and analyzed by Western Blot. The application of different lysis conditions demonstrates the impact of the cell lysis procedure on protein stability in RUNX1-ETO-silenced Kasumi-1 cells. Data are representative for one of three independent experiments.
(TIF)
ELANE and CTSG mRNA levels were measured by qRT-PCR and normalized to housekeeping control. Data are shown as log2 of mean 2-ΔΔCT +/- SD and p-values were determined by two-sided student´s t-test. *p<0.05, ***p<0.001.
(TIF)
Data Availability Statement
Data underlying the results of this study have been uploaded as Supporting Information files and to figshare at the following links: https://doi.org/10.6084/m9.figshare.9784493.v1 and https://doi.org/10.6084/m9.figshare.9724856.v1.