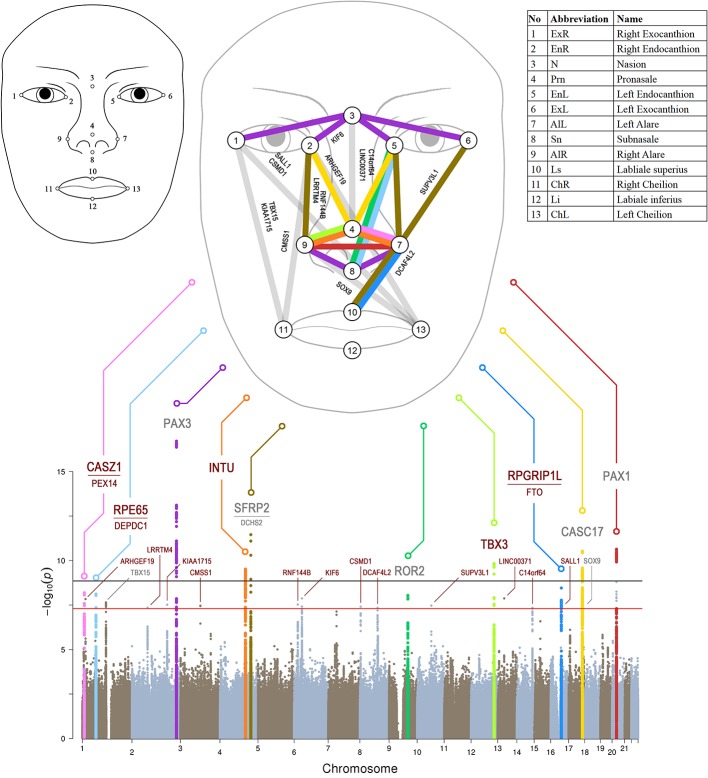
Figure 1. Manhattan plot from meta-analysis discovery GWAS (N = 10,115 Europeans) for 78 Euclidean distances between 13 facial landmarks.
Results from meta-analysis of four GWASs in Europeans (N = 10,115), which were separately conducted in RS, TwinsUK, ALSPAC, and PITT for 78 facial shape phenotypes that are displayed in a signal ‘composite’ Manhattan plot at the bottom of the figure. All loci consisting of SNPs reaching study-wide suggestive significance (p<5 × 10−8, red line) were nominated according to the nearby candidate genes in the associated loci, and the top 10 loci were highlighted in colors. The black line indicates study-wide significance after multiple trait correction (p<1.2×10−9). Novel and previously established face-associated genetic loci were differentiated using colored and gray text, respectively. Annotation of the 13 facial landmarks used to derive the 78 facial phenotypes and the associated facial phenotypes are illustrated in the upper part of the figure.