Metagenomic analysis identifies microbial signatures preceding necrotizing enterocolitis development in premature infants.
Abstract
Necrotizing enterocolitis (NEC) is a devastating intestinal disease that occurs primarily in premature infants. We performed genome-resolved metagenomic analysis of 1163 fecal samples from premature infants to identify microbial features predictive of NEC. Features considered include genes, bacterial strain types, eukaryotes, bacteriophages, plasmids, and growth rates. A machine learning classifier found that samples collected before NEC diagnosis harbored significantly more Klebsiella, bacteria encoding fimbriae, and bacteria encoding secondary metabolite gene clusters related to quorum sensing and bacteriocin production. Notably, replication rates of all bacteria, especially Enterobacteriaceae, were significantly higher 2 days before NEC diagnosis. The findings uncover biomarkers that could lead to early detection of NEC and targets for microbiome-based therapeutics.
INTRODUCTION
Necrotizing enterocolitis (NEC) is widely studied yet poorly understood. First described in the early 1800s (1), NEC is a disorder of intestinal inflammation that can progress to bowel necrosis, sepsis, and death (2). NEC affects 7% of very low birth weight infants born in the United States each year, and mortality rates have remained around 20 to 30% for several decades (2). The direct cause or causes of NEC remain unknown.
The primary risk factor for NEC is preterm birth (2). Immature enterocytes exhibit hyperactive immune responses through the Toll-like receptor 4 (TLR4) pathway in response to bacterial lipopolysaccharide (LPS), which can lead to bowel damage (3). Experimental NEC occurs in conventionally raised animals but not those reared in a germ-free environment (4, 5). These observations suggest that the intestinal microbiome plays a role in the disease and lead to the prevailing hypothesis that an excessive immune response to aberrations in the composition and function of gut microbial communities is the most likely basis for the pathogenesis of NEC. Although no single microbe has been consistently identified as a biomarker for NEC, increased abundance of bacteria in the phylum Proteobacteria is a frequently reported microbial pattern in NEC infants (6). Most fecal microbiome-based profiling studies of NEC use 16S ribosomal RNA (rRNA) amplicon sequencing, which provides a general overview of the bacteria present but does not reveal metabolic features that could contribute to NEC pathogenesis.
Genome-resolved methods may provide new insights into NEC development. The approach has several advantages over 16S rRNA amplicon sequencing. As the method is not reliant on polymerase chain reaction amplification nor specific probes, all DNA can be sequenced, allowing detection of bacteriophages, plasmids, eukaryotes, and viruses. Bioinformatic techniques can also infer in situ bacterial replication rates directly from metagenomic data (7), an important metric, as some microbiome-related diseases have a signal related to bacterial replication but not relative abundance (8). Genome assembly and annotation can provide functional information about organisms present and possibly reveal genes associated with NEC. Further, whole-genome comparisons provide strain discrimination and thus detailed testing of Koch’s postulates. Last, mapping to reference genomes is not required for genome detection, allowing for the discovery of novel bacterial clades (9). While identification of a single causative strain, virus, or toxin would be the most actionable result for clinicians, any associations could potentially be used as biomarkers to identify early warning signs of NEC, and microbial communities associated with NEC could be targeted with microbiome-altering techniques such as probiotics, prebiotics, or other approaches (10).
RESULTS
Metagenomic characterization of premature infant fecal samples
We analyzed 1163 fecal metagenomes from 34 preterm infants who developed NEC and 126 preterm infants without NEC (Fig. 1). Premature infant participants were matched for gestational age and calendar date and recruited from the University of Pittsburgh Medical Center Magee-Womens Hospital (Pittsburgh, PA) over a 5-year period. Fecal samples were banked, and specific samples were later chosen for DNA extraction and sequencing to preferentially study samples immediately before NEC onset. An average of 7.2 samples per infant, mostly from the first month of life, was sequenced, and a total of 4.6 tera–base pairs of shotgun metagenomic sequencing were generated (table S1). Detailed sequencing information (table S1) and patient metadata (table S2) are provided.
Fig. 1. Metagenomic characterization of 1163 samples from 160 premature infants.
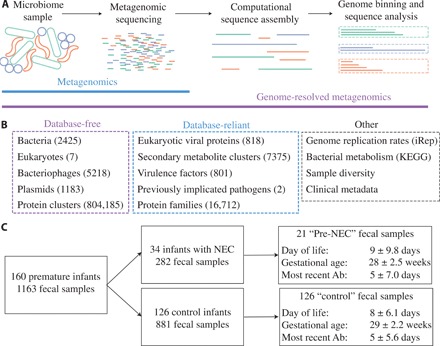
(A) Schematic of metagenomics versus genome-resolved metagenomics. Metagenomics involves DNA extraction from a microbiome sample, followed by library preparation and sequencing. In genome-resolved metagenomics, this is followed by sequence assembly and binning to generate draft-quality microbial genomes. (B) Metagenomes were characterized using database-free and database-reliant methods. The number of features in each category is listed in parentheses. See Materials and Methods for details. (C) Flow chart of the 160 premature infants recruited for inclusion in this study from the same neonatal intensive care unit over a 5-year period. Pre-NEC and control samples are a subset of the total fecal samples that are matched for DOL, gestational age, and recent antibiotic administration (Ab), and for NEC infants, samples are within 2 days before NEC diagnosis. The median and SD of matched metrics are reported.
We performed extensive computational analyses on all samples to recover genomes de novo and determine their phylogeny, metabolic potential, and replication rates [index of replication (iRep) (7)]. We also searched samples for eukaryotic viruses, virulence factors, secondary metabolite gene clusters, and previously implicated pathogens (Fig. 1) (11, 12). This analysis resulted in 36 giga–base pairs of assembled sequence, 2425 dereplicated bacterial genomes (average of 92% completeness and 1.1% contamination), 5218 bacteriophage genomes, 1183 plasmid genomes, 7 eukaryotic genomes, and 804,185 de novo protein clusters (Fig. 1B and table S3). As NEC can be a rapidly progressive disorder, for most statistical tests, we defined NEC samples as those taken within 2 days before NEC diagnosis (“pre-NEC” samples). For infants who did not develop NEC, only one sample from the period associated with NEC onset was used (“control” samples). Pre-NEC and control samples were matched for day of life (DOL), gestational age, and recent antibiotic administration (Fig. 1C and figs. S1 and S2). For other analyses, when explicitly stated, we used all samples.
Klebsiella pneumoniae is enriched in samples from infants with NEC
The gut microbiomes of all infants were dominated by Proteobacteria, regardless of NEC development (Fig. 2, A and B). As compared to previous studies of full-term infants (13, 14), the premature infants in this study had increased Enterobacteriaceae (a family of Proteobacteria to which many nosocomial pathogens belong) and notably low abundances of Actinobacteria and Bacteroidetes. Factors that could select for these organisms include prophylactic antibiotics given to all premature infants at birth, high rates of birth by cesarean section, predominance of formula feeding, and immaturity of the intestine and immune system. Compared to control infants, the NEC infant microbiomes exhibited lower abundances of Firmicutes (P = 3.7 × 10−7, Wilcoxon rank sum test) and higher abundances of Enterobacteriaceae (P = 8.9 ×10−7, Wilcoxon rank sum test) than the microbiomes of control infants (Fig. 2A). The general association of Enterobacteriaceae and infants who go on to develop NEC has been described previously (15), but this prior analysis was not restricted to the period immediately before NEC detection. In our study, the gut microbiomes of infants who developed NEC were not significantly enriched in Enterobacteriaceae in pre-NEC versus control samples (P = 0.15, Wilcoxon rank sum test), so the association of Enterobacteriaceae and NEC infants overall may be due to the proliferation of these bacteria after the administration of antibiotics to treat NEC (fig. S2B).
Fig. 2. Comparison of microbes in premature infants who do and do not develop NEC.
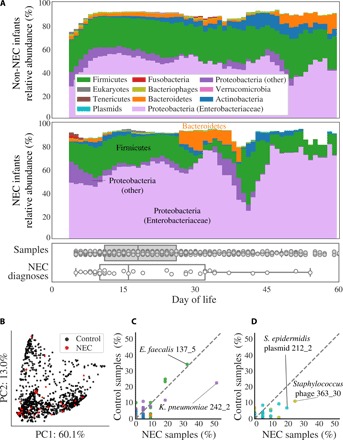
(A) The compositional profile of microbes colonizing infants who were and were not diagnosed with NEC. Bacteria were classified on the basis of their phyla, and other microbes were classified on the basis of their domain. Each color represents the percentage of reads mapping to all organisms belonging to a taxon, and the stacked boxes for each sample show the fraction of reads in that dataset accounted for by the genomes assembled from the sample. Proteobacteria were subdivided into the family Enterobacteriaceae and other. All relative abundance values were averaged over a 5-day sliding window. Boxplots show the DOL in which samples were collected (top) and in which infants were diagnosed with NEC (bottom). (B) Principal components analysis (PCA) based on weighted UniFrac distance for all samples from NEC infants (red) and control infants (black). (C and D) Percentage of NEC infants versus the percentage of non-NEC infants colonized by strains of (C) bacteria or (D) bacteriophage (gold) and plasmids (blue). The taxonomies of four strains with extreme values are provided, of which only K. pneumoniae strain 242_2 is significantly enriched in NEC samples (P < 0.05, Fisher’s exact test). Colonization by bacteria is defined as the presence of a strain at ≥0.1% relative abundance. Plasmid and bacteriophage detection required a read-based genome breadth of coverage of ≥50%. Each dot represents a strain, and dashed lines show a 1:1 colonization rate.
We performed a principal components analysis (PCA) based on weighted UniFrac distance to compare the microbiomes of all samples from all time points (Fig. 2B). The first two principal components explained 73% of the overall variance, but samples collected from NEC infants (red) did not cluster separately from control infants (black dots). Consideration of higher principal components (up to the fifth principal component) did not separate pre-NEC and control samples, and samples coded by clinical metadata also did not cluster together (fig. S3).
To identify strains enriched in pre-NEC samples, we calculated the percentage of pre-NEC versus control samples carrying each assembled bacterial, bacteriophage, and plasmid genome (Fig. 2, C and D). K. pneumoniae strain 242_2 was the most associated with NEC and was present above the threshold of detection in 52% of pre-NEC samples versus 23% of control samples (P = 0.008, Fisher’s exact test) (table S4). Closely related bacteria [>99% average nucleotide identity (ANI)] colonized up to 35% of all infants (Fig. 2C). This is likely the result of colonization by the same hospital-associated bacteria (16) in multiple infants. No organisms in this study satisfied Koch’s postulate that a disease causing organism should be found in all NEC infants and no healthy patients.
Bacterial replication rates are higher before NEC development
Bacterial replication rates are measured from metagenomic data by determining the difference in DNA sequencing coverage at the origin versus terminus of replication, yielding an iRep value that correlates with traditional doubling time measurements (7, 8). iRep values of bacteria overall were significantly higher in pre-NEC versus control samples (P = 0.0003, Wilcoxon rank sum test) in a cohort balanced for DOL, gestational age, and recent antibiotic administration (Fig. 3). Further, iRep values followed a notable pattern in relation to NEC diagnosis: Bacterial replication was stable four or more days before NEC diagnosis, increased daily in 3 days before diagnosis, and crashed following diagnosis (probably due to subsequent antibiotic administration) (Fig. 3A). Individual species did not have enough data points to be plotted confidently (minimum of five measurements per DOL), but genomes of the family Enterobacteriaceae displayed even higher pre-NEC iRep values than bacteria overall (Fig. 3, A and B). Increased bacterial replication before NEC could promote disease onset or merely be a reaction to changing conditions in the gut that led to NEC.
Fig. 3. Bacterial replication rates are significantly higher before NEC development.
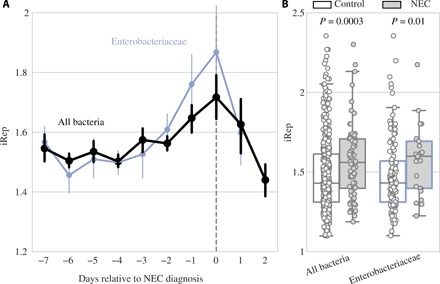
(A) Replication rates for bacterial groups relative to day of NEC diagnosis. Dots represent the mean value for each group on each day, and error bars represent SEM. DOL in which growth rates were calculated from at least five infants are shown. (B) Growth rates in control (white) versus pre-NEC (gray) samples. P values shown from Wilcoxon rank sum test.
Machine learning identifies additional differences between NEC and control cases
We measured 2119 features for each of the 1163 metagenomic samples (Fig. 1 and table S5). To evaluate which features are most different between pre-NEC and control samples, we developed a machine learning (ML) classifier. Multiple ML algorithms were evaluated, and although all performed with similar accuracy (table S4), the boosted gradient classifier was ultimately chosen because of its known ability to handle class imbalance. The classifier was trained on all 2119 features to predict whether samples were pre-NEC or control, and accuracy was measured through cross-validation over 100 iterations. The classifier achieved a median accuracy of 64% on balanced sets; 14% better than random chance. While a classifier with this accuracy may have limited utility in a clinical setting, it allowed us to interrogate which features were most informative for differentiating pre-NEC and control samples.
The most important individual features used by the ML classifier were replication rates (iRep values), KEGG (Kyoto Encyclopedia of Genes and Genomes) modules, secondary metabolite gene clusters, and overall plasmid abundance (Fig. 4). iRep values of both specific bacterial taxa and median iRep values overall were some of the most important features (Fig. 4B), while KEGG modules accounted for more than 50% of the total feature importance (Fig. 4A and table S5). A similar number of KEGG modules were associated and anti-associated with NEC (Fig. 4C), but the descriptions of the modules associated with NEC (e.g., erythritol and galactitol transport systems) and anti-associated with NEC (e.g., sodium and capsular polysaccharide transport systems) bear no obvious relationship to the disease (table S5). Secondary metabolite gene clusters were the second most important category overall (Fig. 4A), but unlike KEGG modules, very few were anti-associated with NEC (Fig. 4C). The most significant secondary metabolite gene cluster encodes an unusual operon of biosynthetic genes found in Klebsiella (cluster 416). In other species, similar operons are implicated in the biosynthesis of quorum-sensing butyrolactones (17). The second most significant cluster of genes occurs in Enterococcus and is involved in biosynthesis of a sactipeptide resembling subtilosin A1, an antimicrobial agent with known hemolytic activity (cluster 438) (table S3) (18). Another cryptic secondary metabolite gene cluster with a high feature importance (cluster 432) is closely related to a previously characterized cluster on a plasmid of enterotoxin-producing Clostridium perfringens adjacent to the enterotoxin gene (cpe) and β2 toxin gene (cpb2) (19). Overall, high plasmid abundance was correlated with pre-NEC samples (Fig. 4B), and K. pneumoniae plasmids in particular were significantly more abundant in pre-NEC samples (P = 0.03) (fig. S2E). The prevalence of K. pneumoniae in pre-NEC samples (Fig. 2C) may explain the high abundance of K. pneumoniae plasmids in these samples.
Fig. 4. ML identifies differences between pre-NEC and control samples.
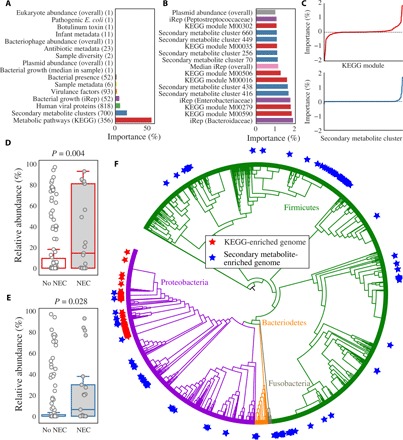
(A) Sum of all individual importances for each feature category. The number of features in each category is listed in parentheses. (B) Importance of all individual features associated with NEC with classifier importances over 1%. (C) Signed importances of all individual KEGG modules (top, red) and secondary metabolite clusters (bottom, blue). Negative values are negatively associated with pre-NEC samples, and positive values are positively associated with pre-NEC samples. (D and E) Relative abundance of genomes enriched in important KEGG modules (D) and important secondary metabolite-enriched genomes (E) in pre-NEC versus control samples. P values shown from Wilcoxon rank sum test. (F) Distribution of genomes enriched in important KEGG modules (red stars) and important secondary metabolite clusters (blue stars) around a phylogenetic tree of all recovered bacterial genomes. Genomes enriched in important KEGG modules are more clustered on the tree than those enriched in important secondary metabolite clusters.
Feature importances were also analyzed in combination. Each bacterial strain was assigned an importance value based on the sum of the importance scores for the KEGG modules encoded by its genome. A histogram of all genome importance scores was generated (fig. S4), and 150 genomes were visually determined to have KEGG importance values higher than the typical distribution (hereinafter referred to as “organisms of interest”) (table S3). The organisms of interest were significantly more abundant in pre-NEC samples as compared to control samples (P = 0.004) (Fig. 4D), and they cluster phylogenetically (Fig. 4F). A total of 97% were in the family Enterobacteriaceae, and of those, 90% were in the genus Klebsiella.
Secondary metabolite biosynthetic gene clusters identified as important by the ML classifier occur in 218 organisms that are significantly associated with pre-NEC samples (Fig. 4E). Several types of secondary metabolite gene clusters were enriched in these genomes (P < 0.01, Fisher’s exact test), including sactipeptides, bacteriocins, and butyrolactones (encoded by 382, 286, and 11 genomes, respectively) (table S3). As opposed to organisms of interest, these bacteria were spread around the phylogenetic tree (Fig. 4F). This may indicate that the clusters themselves are associated with pre-NEC samples. Overall, the results point to an association of quorum sensing and antimicrobial peptide production with NEC onset.
Bacteria associated with NEC encode specific types of fimbriae
We leveraged the gene content information provided by genome-resolved metagenomics to search for proteins associated with (i) pre-NEC samples and (ii) organisms of interest. Three clustering algorithms were evaluated for their ability to reconstruct known clusters of ribosomal proteins (table S4), and a hybrid Markov Cluster algorithm approach (20) performed best. Application of the algorithm to the 36,701,491 proteins reconstructed in this study yielded 804,277 protein clusters, none of which was statistically associated with NEC (Fisher’s exact test with false discovery rate correction) (Fig. 5A). However, 85 protein clusters were associated with organisms of interest with high precision and recall (>0.7) (Fig. 5B). The most common protein family (pFam) annotations for these clusters were fimbriae and adenosine triphosphate–binding cassette (ABC) transport proteins (table S6). However, only genomes encoding fimbrial proteins also had a significant association with NEC (P = 0.02, Wilcoxon rank sum test with Benjamini-Hochberg false discovery rate correction; table S6).
Fig. 5. Genomes encoding fimbriae are associated with NEC development.
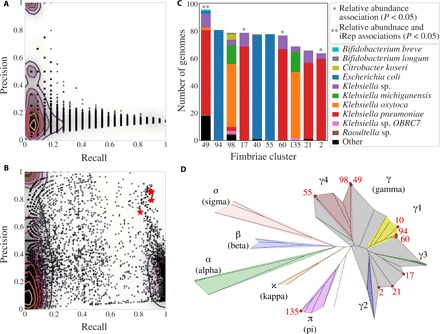
(A and B) Association of protein clusters with pre-NEC samples (A) and organisms of interest (B). Each dot represents a protein cluster. Recall is (A) the number of pre-NEC samples the cluster is in/the total number of pre-NEC samples and (B) the number of organisms of interest the cluster is in/the total number of organisms of interest. Precision is (A) the number of pre-NEC samples the cluster is in/the number of total pre-NEC samples and (B) the number of organisms of interest the cluster is in/the total number of genomes the cluster is in. Clusters annotated as fimbriae are marked with red stars. Contour lines are drawn to indicate density. (C) The number of bacterial genomes encoding each fimbriae cluster, the species-level phylogenetic profile of genomes encoded by each fimbriae cluster, and each cluster’s association with NEC. (D) Phylogenetic tree of CU proteins built using IQtree. Three amino acid sequences from each de novo CU cluster and three reference amino acid sequences from each defined CU clade were included in the tree. Colors mark the phylogenetic breadth spanned by reference sequences, and stars represent de novo CU clades. For all de novo clusters, the three randomly chosen sequences fell extremely close to each other on the tree.
Comparison of fimbrial operons against public databases revealed that the majority encode chaperone-usher (CU)–type fimbriae. A classification scheme exists for CU fimbriae based on usher protein pFam [PF00577.19 (21)]. The 32,646 usher proteins identified in our sequencing data (table S6) were clustered into groups based on amino acid sequence identity, and the 10 most prevalent groups were placed in a phylogenetic tree with reference sequences from each subtype of CU fimbriae (Fig. 5D). All 10 fimbriae clusters fit into the established CU fimbriae taxonomy, with 9 of 10 falling in the γ superclade and 1 into the π clade (Fig. 5D). Four fimbriae clusters identified in this study were significantly more abundant in pre-NEC samples, and genomes encoding cluster 49 (γ4 clade) also had significantly higher iRep values in pre-NEC samples (Fig. 5C). Twenty-seven genomes that encode fimbrial cluster 49 were not identified as genomes of interest, yet they were at significantly higher abundance and have significantly higher iRep values when considering all samples from NEC versus control infants (P < 0.01, Wilcoxon rank sum test) (fig. S4). This suggests that fimbrial cluster 49 itself may be associated with NEC and not incidentally associated with metabolically important genomes.
Biomarkers of NEC are most informative closer to NEC diagnosis
Statistical tests uncovered four factors significantly associated with pre-NEC samples (samples taken within 2 days before NEC diagnosis): iRep values overall (Fig. 3B), genomes encoding specific types of secondary metabolite gene clusters (sactipeptides, bacteriocins, and butyrolactones) (table S3), Klebsiella (Fig. 2C), and fimbriae cluster 49 (Fig. 5C). We performed a similar analysis each day up to 8 days before NEC diagnosis (Fig. 6). Genomes encoding specific types of secondary metabolite gene clusters and Klebsiella genomes were always significantly more abundant in NEC samples, although the effect size of the difference became slightly higher closer to NEC diagnosis. iRep values and the abundance of genomes encoding fimbriae cluster 49, on the other hand, were only significantly higher 3 days and 1 day before diagnosis, respectively.
Fig. 6. Biomarkers of NEC are most informative closer to NEC diagnosis.
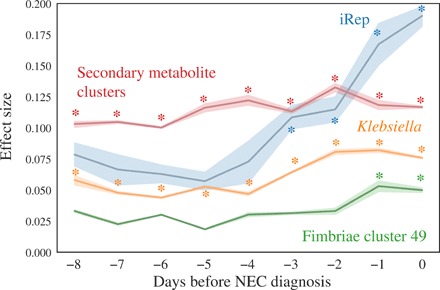
The effect size for difference of each feature in pre-NEC versus control samples is shown based on a Wilcoxon rank sum test over a 2-day sliding window (e.g., −5 compares samples collected from −6 to −4 days relative to NEC diagnosis to control samples). Comparisons with P < 0.05 are marked with asterisks.
DISCUSSION
Given that we found no single predictor of NEC and identified several factors as important by ML, our results support prior indications that NEC is a complex and likely multifactorial disease (2, 22). Of the four aspects of the gut microbiome that differ in pre-NEC compared to control samples (Fig. 6), the iRep values of all organisms in each sample had the highest effect size. Given that iRep is a measure of bacterial replication rather than relative abundance, the result highlights that reliance on relative abundance alone could be misleading. This is largely due to the fact that relative abundance metrics are themselves misleading because an organism can increase in relative abundance simply due to the decline in relative abundances of other organisms. For this reason, it is also unclear whether increased replication leads to increased bacterial biomass, as there could be concomitantly higher death rates from heightened inflammatory response during NEC, production of antimicrobials, etc. The higher bacterial replication rate before NEC diagnosis could be sustained by nutrient release from the breakdown of gut tissue. Alternatively, increased bacterial replication may trigger the onset of NEC, possibly because high activity of a specific organism leads to imbalance in concentrations of compounds in the gut environment.
Secondary metabolite gene clusters of specific types (bacteriocins, sactipeptides, and butyrolactones) were significantly enriched in pre-NEC compared to control samples (table S3). Bacteriocins are small peptides that kill closely related bacteria, and when produced, cell lysis could contribute to onset NEC via release of immunostimulatory compounds such as LPS. Sactipeptides are a class of posttranslationally modified peptides with diverse bioactivities (23). The sactipeptide with the highest overall importance is related to a subtilosin (antimicrobial agent) with known hemolytic activity. All sactipeptides identified in this study were encoded by Firmicutes, including C. perfringens and Clostridium difficile (fig. S2 and table S3). Production of sactipeptides by these species could trigger NEC through direct toxicity to human cells or via release of immunostimulatory bacterial compounds following bacterial cell lysis. This phenomenon could explain previous reports that implicate Clostridium in development of NEC (24–26). Follow-up studies involving proteomics and/or transcriptomics are needed to establish whether these gene clusters are expressed in situ in the infant gut.
Butyrolactones are generally involved in quorum sensing in Actinobacteria (17) but, in this study, were mostly found encoded in genomes of Proteobacteria, and more than half were identified in Klebsiella genomes. Whereas known quorum-sensing systems in Proteobacteria are responsible for the production of virulence factors, including fimbriae (27, 28), the functions of butyrolactones in Proteobacteria remain unstudied. Higher proportions of Klebsiella were found in infants who went on to develop NEC, and their capacity to produce secondary metabolites and fimbriae could explain this association.
Organisms with genomes encoding for fimbriae cluster 49 were at significantly higher abundances on both the day of and the day before NEC diagnosis. Fimbriae are known stimulants of TLR4 receptors (29), immune receptors that are overexpressed in premature infants and previously linked to NEC in animal studies (30, 31). Fimbriae are the hallmark pathogenicity factors of uropathogenic Escherichia coli (32), a group of organisms that have been previously implicated as a causative agent of NEC (11). Uropathogenic E. coli was specifically evaluated in this study and not found to be significantly enriched in pre-NEC compared to control samples (table S5 and Fig. 2C). The associations in prior work and the current study may instead reflect a general link between fimbriae and TLR4 receptor stimulation.
An advantage of genome-resolved metagenomics is that it provides whole community information, going far beyond what can be deduced from 16S rRNA gene surveys that are the hallmark of most prior and much current human microbiome research. Here, we applied this approach to a sufficiently large dataset to achieve statistical power unprecedented in a genome-resolved metagenomic study and found that there is likely no single bacteriophage, plasmid, eukaryote, virus, or even gene that is responsible for NEC. However, we identify several promising associations through ML, many of which have previously been proposed to explain NEC onset, but none of which alone can explain all cases. Bacteria of the genus Klebsiella emerged from our analyses as organisms of potential importance, with secondary metabolite, LPS, and fimbriae production all being possible contributors. The association of these bacteria, as well as bacteria of the Clostridium genera, with NEC and their presence in the neonatal intensive care unit (16) supports prior reports proposing that colonization by nosocomial microbes in premature infants may be clinically significant. Overall, we provide insight into how previously proposed but distinct explanations for the development of NEC are interconnected and identify bacterial growth rates as the strongest predictor of disease onset.
MATERIALS AND METHODS
Subject recruitment, sample collection, and metagenomic sequencing
This study was reviewed and approved by the University of Pittsburgh Institutional Review Board (IRB PRO12100487 and PRO10090089). This study made use of many different previously analyzed infant datasets. These datasets have previously published descriptions of the study design, patient selection, and sample collection and are referred to as NIH1 (33), NIH2 (16), NIH3 (34), NIH4 (35), NIH5 (36), and Sloan2 (16). Stool samples were collected from infants and stored at −80°C. DNA was extracted from frozen fecal samples using a MoBio PowerSoil DNA isolation kit with modifications (33). DNA libraries were prepared using the Illumina Nextera kit (NIH1, NIH2, and NIH3), KAPA Biosciences Hyper Plus Illumina library preparation reagents (NIH5), or PrepX DNA library preparation kits in conjunction with the Apollo 324 robot following factory recommendations (NIH4 and Sloan2). Libraries were sequenced on an Illumina HiSeq 2500 (NIH1, NIH2, NIH3, and Sloan2), Illumina HiSeq 3000 (NIH4), or Illumina HiSeq 4000 (NIH5). All samples were collected with parental consent. Collated sequencing and health information for all infants and samples are provided in the Supplementary Materials of this manuscript (tables S1 and S2).
Metagenomic profiling
Read processing and assembly
Reads from all samples were trimmed using Sickle (www.github.com/najoshi/sickle), and reads that were mapped to the human genome with Bowtie 2 (37) under default settings were discarded. Reads from all samples were assembled independently using IDBA-UD (38) under default settings. Coassemblies were performed for each infant as well, where reads from all samples from that infant were combined and assembled together. Scaffolds of <1 kb in length were discarded, and remaining scaffolds were annotated using Prodigal (39) to predict open reading frames using default metagenomic settings.
Recovery of de novo bacterial genomes
DasTool (40) was used to select the best bacterial bins from the combination of three programs for automatic binning—abawaca (https://github.com/CK7/abawaca), concoct (41), and maxbin2 (42). Cross-mapping was performed between samples for each infant to generate differential abundance signals, and each sample was binned independently. For each infant, dRep v1.4.2 (43) was then used on all bins created from all samples from that infant to generate an infant-specific genome set, using a minimum completeness of 50%, maximum contamination of 15%, the ANImf algorithm, 99% secondary clustering threshold, and 25% minimum coverage overlap.
To determine the taxonomy of bins, the amino acid sequences of all predicted genes were searched against the UniProt database using the usearch ublast command with a maximum e value of 0.0001. tRep (https://github.com/MrOlm/tRep/tree/master/bin) was used to convert the list of identified taxIDs into taxonomic levels. Briefly, this assigns a call to each taxonomy level when at least 50% of protein hits reach that taxonomic level.
Bacterial growth rates
iRep values (7) were calculated by first mapping reads from all samples in each infant to the dereplicated genome set from that infant using Bowtie 2. iRep values resulting from genomes with less than 0.9 breadth of coverage were discarded. To visualize growth rates over time (Fig. 3A), all iRep values from all bacteria were averaged together for each DOL relative to NEC and plotted using seaborn (https://seaborn.pydata.org/) with a confidence interval of 68% and discarding outliers. DOL in which less than five infants were profiled were manually removed.
Bacteriophages, plasmids, and eukaryotes
For all assemblies, circular contigs were identified using VICA (44), and bacteriophages were identified using VirSorter (45) and VirFinder (46). Bacteriophages were defined as scaffolds that were considered “level 2” or “level 1” by VirSorter or P < 0.01 by VirFinder. Plasmids were defined as scaffolds, which were circular, but not identified as bacteriophage according to the above definition. Bacteriophages and plasmids more than 10 kb in length were then each dereplicated separately on a per-infant basis using dRep version 2.0.5 with a primary clustering threshold of 0.9, the ANImf genomic comparison algorithm, a minimum coverage threshold of 0.5, a minimum length of 10 kilo–base pairs, an N50 weight score of 0, a contig length weight score of 1, no quality filtering, and the nearest point algorithm for genome clustering. All plasmid and bacteriophage genomes were then compared to each other using the same dRep command. Eukaryotes were assembled and binned from the gut samples of premature infants as previously reported (36).
Eukaryotic viruses
Eukaryotic viruses were analyzed using the 2014 vFam A HMM (Hidden Markov Model) collection (47), a set of HMMs designed for the identification of eukaryotic viruses within metagenomic sequence data. All hits with e values less than 1 × 10−5 were considered significant and retained. Reads were also mapped to a previously curated list of human viruses (48). This led to the identification of no viruses when individual samples were used and a very small number of viruses when combined sets of reads from each infant were used (Torque teno midi virus 2, Torque teno virus 14, and Macaca mulatta polyomavirus 1). This line of work was not followed up on due to lack of signal.
Diversity
Shannon diversity and overall bacterial richness were calculated for each sample. Shannon diversity was calculated using skbio.diversity.alpha.shannon (http://scikit-bio.org/). Richness was calculated as the number of bacteria with relative abundances over 0.1%.
KEGG modules
KEGG modules were annotated by using HMMER against an in-house HMM database built from the KEGG orthology groups (KOs) (www.genome.jp/kegg/). Briefly, all KEGG database proteins with KOs were compared with all-v-all global similarity search using USEARCH (49). MCL (Markov Cluster Algorithm) was then used to subcluster KOs (inflation_value = 1.1). Each subcluster was aligned using MAFFT (multiple alignment using fast Fourier transform) (50), and HMMs were constructed from subcluster alignments. HMMs were then scored against all KEGG sequences with KOs, and a score threshold was set for each HMM at the score of the highest-scoring hit outside of that HMM subcluster. KEGG modules were considered present in a genome if all necessary KOs were present in that genome. The abundance of each KEGG module in a sample was calculated as the summed relative abundance of all bacterial genomes containing that module.
Secondary metabolite gene clusters
To identify secondary metabolites, antismash-4.0.2 was run on each infant coassembly (51). The results were parsed using the custom script parse_antismash.py (https://github.com/MrOlm/Public-Scripts), and resulting key proteins were clustered using diamond with default settings (52). Alignments were filtered to only retain those with >75% amino acid identity and 50% alignment coverage. Hierarchical clustering was then performed using average amino acid identity and resolved using a distance threshold of 0.5 to assign each secondary metabolite gene cluster to a gene cluster family. Next, for each infant, the nucleotide sequences of all genes in a representative for each gene cluster family were concatenated together. The reads from each sample from that infant were mapped to this concatenation of genes to determine the dynamics of these genes in all samples from that infant. The breadth of each cluster was calculated as the weighted breadth (considering gene length) for all genes in that cluster.
Virulence factors
Virulence Factors Database (VFDB) was used to search for virulence factors (53). The database used was from 17 March 2017, containing 2597 sequences. Abricate was used to search all predicted protein sequences against the VFDB (https://github.com/tseemann/abricate). Metadata from the VFDB website (www.mgc.ac.cn/VFs/) were used to get additional information about the virulence factors. Approximately 15% of virulence factors were not included in this metadata file and were excluded from additional analysis.
Botulinum toxin
A blast database of all subtypes of botulinum neurotoxin was downloaded from https://bontbase.org/ (as accessed on 15 February 2018). Blastp was used to search the predicted amino acid sequences of all genes against the database. Hits with an e value less than 1 × 10−5 were considered valid.
Pathogenic E. coli
It was previously reported that pathogenic E. coli may be associated with NEC development, specifically the clades 73, 95, 127, 131, 144, 998, and 69 (11). To identify E. coli genomes of these sequencing types in our dataset, all genomes were multilocus sequence typing (MLST)–profiled using PubMLST (54) and the program “mlst” (https://github.com/tseemann/mlst). The MLST definition requires having seven genes; in cases where only six genes could be identified, if only one sequence type (ST) existed with those six gene types, the sequence type was inferred. Each sample with an E. coli genome of the above STs at more than 1% relative abundance was considered to have a “pathogenic” E. coli, in accordance with the previous study implicating pathogenic E. coli in NEC development (11).
Proteins
Three protein clustering methods were evaluated for use in this study—MMseqs2 (55) (run using default settings), CD-HIT (0.9 global sequence identity threshold and 200,000 MB memory limit), and a previously described hybrid Markov Cluster approach (20). Algorithms were evaluated on the basis of their ability to reconstruct known protein clusters, and the hybrid Markov Cluster approach performed best (table S4). This method was used to cluster the amino acid sequences of all predicted genes from all assembled scaffolds.
The average microbiome of NEC and control infants
To calculate the relative abundance of all microbes in each infant, a full “genome inventory” was generated for each infant by resolving the overlap between the recovered bacteria, eukaryote, bacteriophage, and plasmid genomes. Bacteriophage and plasmid genomes were first aligned using MUMmer (56), and in all cases where scaffolds were aligned with more than 95% ANI on more than 50% of the scaffold, the scaffold was removed from the plasmid list. The resulting scaffolds were next aligned to bacterial genomes, and all phage/plasmid scaffolds that aligned to bacterial genomes with the same thresholds were removed. Last, eukaryotic genomes were aligned to the remaining scaffolds, and in cases where similar scaffolds were detected, the scaffold was removed from the eukaryotic genome. Reads from all samples were then mapped to that infant’s genome inventory using Bowtie 2, and the relative abundance of each organism was calculated as the percentage of total sample reads that map to that genome (table S4).
To compare the microbiome between NEC and control infants, the microbiome of each cohort was averaged across all infants in that cohort (Fig. 2A) using the relative abundance values described in the previous paragraph. For each DOL, the average relative abundance of each taxon was first calculated. A 5-day sliding window was next applied, and values from samples in each window were averaged. For example, DOL 10 represents the average abundances from DOL 8 to 12.
Strain-level differences between NEC and control infants
To calculate the relative abundance of each bacterium in each sample, each sample was mapped to the infant-specific bacterial genome set for that infant using Bowtie 2. Relative abundances of all bacteria were calculated as the percentage of total sample reads mapping to each genome. Bacteria assembled from all infants were then compared to each other using dRep, and bacterial genomes with at least 99% ANI were considered to be the same “strain.” A bacterium was considered present in a sample if it had more than 0.1% relative abundance, and the fraction of pre-NEC and control samples in which each strain was present was calculated and plotted in Fig. 2C.
Similar procedures were performed for the bacteriophage and plasmid genome sets of each infant. Mapping was performed to each infant set separately, and genomes were considered to be the same strain if they had 99% ANI over at least 50% of their genomes. Organisms were considered present in a sample if they were present with more than 50% genome breadth of coverage.
Principal components analysis
PCA was performed on the basis of the relative abundance of bacteria in each sample as assessed using weighted UniFrac distance. A phylogenetic tree was created by comparing all assembled bacterial genomes to each other using dRep primary clustering with a mash sketch size of 100,000, the weighted UniFrac distance between all samples was calculated using scikit-bio (http://scikit-bio.org/), and PCA was performed using scikit-learn.
Machine learning
Preparation of metagenomic data for ML
Many individual features were summarized before inclusion in the ML training dataset (table S5). For each sample, BTtoxin_abund describes the summed relative abundance of all botulinum toxins detected, BacterialNCBIGrowth describes the average iRep value of each identified bacterial taxonomic family, BacterialNCBITax describes the summed relative abundance of each identified bacterial taxonomic family, Bacteriophage_overall describes the summed relative abundance of all bacteriophage genomes with a breadth of at least 0.75, CatInfSampleMetadata describes clinical metadata about the infant (e.g., breastfeeding versus formula feeding, gender, and birth mode), CatSampleMetadata describes clinical metadata about the sample (e.g., current antibiotic administration, antibiotic administration in the last 5 days), Diversity describes the bacterial richness and shannon diversity, Eukaryotes_overall describes the summed relative abundance of all eukaryotic genomes with a breadth of at least 0.1, HumanViralProteins describes whether each vFam is detected, KEGG_modules describes the summed relative abundance of each KEGG module, Plasmid_overall describes the total summed relative abundance of all plasmid genomes with a breadth of at least 0.75, SampleMetadata describes continuous clinical variables (e.g., gestational age, weight, and days since antibiotic administration), SecMetabolites_cluster_mapping describes the coverage of each gene cluster family with a breadth of at least 0.5, VirFactor_cat_abund describes the summed relative abundance of each detected virulence factor category, Ward_ecoliPathogen_PE describes whether pathogenic E. coli was detected, and median_irep describes the median measured iRep value. In total, this leads to the calculation of 2119 features for each sample (table S5). See above methods for details on how individual features were calculated.
Algorithm development
Three ML methods were evaluated for their ability to classify pre-NEC versus control samples—a random forest classifier (sklearn.ensemble.RandomForestClassifier with 460 estimators and 10 max features) balanced using SMOTE (synthetic minority over-sampling technique; imblearn.combine.SMOTEENN), a gradient boosting classifier (sklearn.ensemble.GradientBoostingClassifier with 0.1 learning_rate, 10 max_depth, 46 max_features, 1483 minimum samples to split an internal node, and 200 estimators) balanced using SMOTE, and the same gradient boosting classifier without balancing. Hyperparameters were empirically determined using sklearn.model_selection.RandomizedSearchCV, and in general, many different combinations of hyperparameters gave similar results. Models were trained and evaluated using cross-validation for five iterations each (using sklearn.model_selection.StratifiedKFold with 10 splits, sklearn.model_selection.cross_val_predict, and sklearn.metrics.accuracy_score) and all achieved similar prediction ability (table S4).
To determine the accuracy of the gradient boosting classifier, 100 iterations were performed where each iteration consisted of (i) randomly balancing the input to include 21 pre-NEC samples and 21 control samples, (ii) classifying each sample in the input using 10-fold cross-validation (same methods as above), and (iii) calculating the percentage of samples that were correctly classified. The median accuracy value was reported.
Feature importance analysis
Feature importances were determined by 100 iterations of training the gradient boosted classifier on the full dataset of pre-NEC and control samples. Importance values were scaled for each iteration such that the overall sum equals 1. The median importance value for each feature is reported (table S5).
KEGG and secondary metabolite enriched genomes
Each bacterial genome was assigned a metabolic importance value by summing the median feature importances of each KEGG module encoded by that genome (see above methods for how KEGG modules were determined). A distribution of KEGG genomes importances was generated (fig. S4A), and on the basis of this distribution, genomes with importance values of more than 15 were considered organisms of interest. Each bacterial genome was also assigned an importance value equivalent to the highest importance value of all secondary metabolite clusters encoded by that genome. A distribution was generated (fig. S4B), and genomes with importances of more than 0.5 were considered enriched in important secondary metabolite clusters.
Phylogenetic tree
A phylogenetic tree was made to visualize the distributions of organisms of interest and organisms enriched in important secondary metabolite clusters (Fig. 4F). Ribosomal protein S3 was identified in bacterial genomes using pFam PF00189.19 and HMMER with a score cutoff of 50 (57). An archaeal outgroup was added, and all sequences were aligned using MAFFT (50) under default parameters. All positions with gaps in more than 50% of sequences were trimmed from the alignment, and FastTree was used with default parameters to generate a phylogenetic tree (58). The tree was visualized and annotated using iTOL (59).
Protein clustering
Protein association with NEC
Each protein cluster was considered present in a sample if a protein from that cluster had been assembled from the sample. Fisher’s exact test was run on each protein cluster to determine whether it was enriched in pre-NEC or control samples, and after Benjamini-Hochberg correction, no P values were statistically significant.
Protein association with organisms of interest
Each protein cluster was considered present in an organism of interest if a protein from that cluster was encoded in the organism’s genome. The recall and precision of each cluster with organisms of interest were calculated as follows: recall = the number of organisms of interest the cluster is in/the total number of organisms of interest; precision = the number of organisms of interest the cluster is in/the total number of genomes the cluster is in. The recall and precision of each protein cluster were plotted (Fig. 5B), and a threshold of 0.7 recall and 0.7 precision was visually established. Protein clusters with recall and precision of more than 0.7 were considered enriched in organisms of interest.
The 85 protein clusters enriched in organisms of interest were profiled using the pFam database (57) with provided noise cutoffs, and the two most common pFams were PF00419.19 (Fimbrial) and PF00005.26 (ABC transporter) with four proteins each. We next determined whether organisms encoding these proteins were enriched in pre-NEC samples. For each pFam with at least three proteins enriched in organisms of interest, we compared the total relative abundance of all bacteria encoding that pFam in pre-NEC versus control samples, as well as all iRep values of bacteria encoding that pFam in pre-NEC versus control samples using the Wilcoxon rank sum test with Benjamini-Hochberg P value correction (table S6).
Fimbriae
CU fimbriae were identified in our dataset using pFam PF00577.19 (usher protein) and clustered using usearch (49) with an identity threshold of 0.9. The taxonomic profile of each fimbriae cluster was determined on the basis of the taxonomy of organisms encoded by that cluster, and relative abundance and iRep associations with pre-NEC versus control samples were calculated using the Wilcoxon rank sum test applied to all bacterial genomes encoding each cluster. A similar procedure was performed using genomes that were not classified as organisms of interest but did encode fimbriae cluster 49, comparing between pre-NEC and control samples and between all samples from NEC infants and all samples from control infants (fig. S4).
A phylogenetic tree was made to establish the type of usher proteins identified in our study. Three reference sequences from each previously established type (21) were aligned with three representatives of each of our clusters using MAFFT. All columns with gaps in more than 50% of sequences were trimmed from the alignment, IQ-TREE was used with default parameters to generate a phylogenetic tree (60), and tree annotation was performed using iTOL (59).
Effect size calculations
To determine when signals first become apparent relative to NEC diagnosis, control samples were compared to samples collected over different sliding 3-day windows (Fig. 6). To compare the signal at 5 days before NEC diagnosis, for example, a rarefied set of samples was chosen from 4 to 6 days before diagnosis where one sample from each infant that has a sample in that window was randomly chosen. This procedure was repeated 10 times, and the average effect size and 95% confidence intervals were plotted. The effect size was calculated on the basis of the Wilcoxon rank sum test statistic [as calculated by SciPy (scipy.stats.ranksums)] using the formula: effect size = {test statistic/square root [(observations in population 1) + (observations in population 2)]}. For iRep, all iRep values were compared between the two sets; for secondary metabolite gene clusters, the total relative abundance of genomes encoding secondary metabolite gene clusters classified as producing sactipeptides, bacteriocins, or butyrolactones was compared; for Klebsiella, the total relative abundances of all genomes classified as the genus Klebsiella were compared; and for Fimbriae cluster 49, the total relative abundances of all genomes encoding fimbriae cluster 49 were compared.
Supplementary Material
Acknowledgments
Funding: This research was supported, in part, by the NIH under awards RAI092531A and R01-GM109454, the Alfred P. Sloan Foundation under grant APSF-2012-10-05, and National Science Foundation Graduate Research Fellowships under grant no. DGE 1106400 (to M.O.). The study was approved by the University of Pittsburgh Institutional Review Board (Protocol PRO10090089). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. Author contributions: M.R.O., M.J.M., and J.F.B. designed the study. M.R.O. performed metagenomic analyses. N.B. and Y.S.S. guided statistical and ML analyses. A.C.-C. contributed to secondary metabolite analyses. R.B. recruited infants for the study, and B.A.F. performed all DNA extractions. M.R.O. and J.F.B. wrote the manuscript, and all authors contributed to manuscript revisions. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Reads are available under BioProject under BioProject PRJNA294605; SRA studies SRP052967, SRP114966, and SRP012558; and SRA accessions SRR5405607 to SRR5406014 (16, 33, 35, 36). De novo assembled genomes are available at https://doi.org/10.6084/m9.figshare.c.4740080.v1. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaax5727/DC1
Table S1. Metagenomic sequencing depth and read quality information.
Table S2. Patient metadata.
Table S3. Information about dereplicated secondary metabolite clusters, de novo–assembled genomes, and genome-wide importances of genomes.
Table S4. Accuracy of ML algorithms and protein clustering algorithms and mapping-based abundances of bacterial taxa.
Table S5. Full feature table provided to the ML classifier and importances of all features resulting from the ML classifier.
Table S6. Proteins enriched in genomes of interest and identified fimbrial genes.
Fig. S1. Metagenomic characterization of 1163 samples from 160 premature infants.
Fig. S2. Fecal samples taken before NEC diagnosis have a higher abundance of plasmids from specific bacterial taxa.
Fig. S3. PCA is unable to separate pre-NEC and control samples.
Fig. S4. ML feature importance values reveal organismal associations with NEC.
REFERENCES AND NOTES
- 1.Obladen M., Necrotizing enterocolitis—150 years of fruitless search for the cause. Neonatology 96, 203–210 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Neu J., Walker W. A., Necrotizing enterocolitis. N. Engl. J. Med. 364, 255–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claud E. C., Lu L., Anton P. M., Savidge T., Walker W. A., Cherayil B. J., Developmentally regulated IκB expression in intestinal epithelium and susceptibility to flagellin-induced inflammation. Proc. Natl. Acad. Sci. U.S.A. 101, 7404–7408 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afrazi A., Sodhi C. P., Richardson W., Neal M., Good M., Siggers R., Hackam D. J., New insights into the pathogenesis and treatment of necrotizing enterocolitis: Toll-like receptors and beyond. Pediatr. Res. 69, 183–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence G., Bates J., Gaul A., Pathogenesis of neonatal necrotising enterocolitis. Lancet 319, 137–139 (1982). [DOI] [PubMed] [Google Scholar]
- 6.Pammi M., Cope J., Tarr P. I., Warner B. B., Morrow A. L., Mai V., Gregory K. E., Kroll J. S., McMurtry V., Ferris M. J., Engstrand L., Lilja H. E., Hollister E. B., Versalovic J., Neu J., Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome 5, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown C. T., Olm M. R., Thomas B. C., Banfield J. F., Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256–1263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korem T., Zeevi D., Suez J., Weinberger A., Avnit-Sagi T., Pompan-Lotan M., Matot E., Jona G., Harmelin A., Cohen N., Sirota-Madi A., Thaiss C. A., Pevsner-Fischer M., Sorek R., Xavier R. J., Elinav E., Segal E., Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morowitz M. J., Denef V. J., Costello E. K., Thomas B. C., Poroyko V., Relman D. A., Banfield J. F., Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc. Natl. Acad. Sci. U.S.A. 108, 1128–1133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronda C., Chen S. P., Cabral V., Yaung S. J., Wang H. H., Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 16, 167–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward D. V., Scholz M., Zolfo M., Taft D. H., Schibler K. R., Tett A., Segata N., Morrow A. L., Metagenomic sequencing with strain-level resolution implicates uropathogenic E. coli in necrotizing enterocolitis and mortality in preterm infants. Cell Rep. 14, 2912–2924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S., Lebreton F., Mansfield M. J., Miyashita S. I., Zhang J., Schwartzman J. A., Tao L., Masuyer G., Martínez-Carranza M., Stenmark P., Gilmore M. S., Doxey A. C., Dong M., Identification of a botulinum neurotoxin-like toxin in a commensal strain of enterococcus faecium. Cell Host Microbe 23, 169–176.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penders J., Thijs C., Vink C., Stelma F. F., Snijders B., Kummeling I., van den Brandt P. A., Stobberingh E. E., Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Bokulich N. A., Chung J., Battaglia T., Henderson N., Jay M., Li H., Lieber A. D., Wu F., Perez-Perez G. I., Chen Y., Schweizer W., Zheng X., Contreras M., Dominguez-Bello M. G., Blaser M. J., Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8, 343ra82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrow A. L., Lagomarcino A. J., Schibler K. R., Taft D. H., Yu Z., Wang B., Altaye M., Wagner M., Gevers D., Ward D. V., Kennedy M. A., Huttenhower C., Newburg D. S., Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 1, 13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks B., Olm M. R., Firek B. A., Baker R., Thomas B. C., Morowitz M. J., Banfield J. F., Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat. Commun. 8, 1814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y.-L., Shen X.-L., Yu P., Bai L.-Q., Li Y.-Q., Gamma-butyrolactone regulatory system of Streptomyces chattanoogensis links nutrient utilization, metabolism, and development. Appl. Environ. Microbiol. 77, 8415–8426 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang T., Geng H., Miyyapuram V. R., Sit C. S., Vederas J. C., Nakano M. M., Isolation of a variant of subtilosin A with hemolytic activity. J. Bacteriol. 191, 5690–5696 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamoto K., Fisher D. J., Li J., Sayeed S., Akimoto S., McClane B. A., Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 188, 1585–1598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meheust R., Burstein D., Castelle C. J., Banfield J. F., Biological capacities clearly define a major subdivision in Domain Bacteria. bioRxiv 10.1101/335083 , (2018). [Google Scholar]
- 21.Nuccio S.-P., Bäumler A. J., Evolution of the chaperone/usher assembly pathway: Fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballance W. A., Dahms B. B., Shenker N., Kliegman R. M., Pathology of neonatal necrotizing enterocolitis: A ten-year experience. J. Pediatr. 117, S6–S13 (1990). [DOI] [PubMed] [Google Scholar]
- 23.Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Cotter P. D., Craik D. J., Dawson M., Dittmann E., Donadio S., Dorrestein P. C., Entian K. D., Fischbach M. A., Garavelli J. S., Göransson U., Gruber C. W., Haft D. H., Hemscheidt T. K., Hertweck C., Hill C., Horswill A. R., Jaspars M., Kelly W. L., Klinman J. P., Kuipers O. P., Link A. J., Liu W., Marahiel M. A., Mitchell D. A., Moll G. N., Moore B. S., Müller R., Nair S. K., Nes I. F., Norris G. E., Olivera B. M., Onaka H., Patchett M. L., Piel J., Reaney M. J. T., Rebuffat S., Ross R. P., Sahl H. G., Schmidt E. W., Selsted M. E., Severinov K., Shen B., Sivonen K., Smith L., Stein T., Süssmuth R. D., Tagg J. R., Tang G. L., Truman A. W., Vederas J. C., Walsh C. T., Walton J. D., Wenzel S. C., Willey J. M., van der Donk W. A., Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morowitz M. J., Poroyko V., Caplan M., Alverdy J., Liu D. C., Redefining the role of intestinal microbes in the pathogenesis of necrotizing enterocolitis. Pediatrics 125, 777–785 (2010). [DOI] [PubMed] [Google Scholar]
- 25.de la Cochetiere M.-F., Piloquet H., des Robert C., Darmaun D., Galmiche J. P., Roze J. C., Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: The putative role of Clostridium. Pediatr. Res. 56, 366–370 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Dittmar E., Beyer P., Fischer D., Schäfer V., Schoepe H., Bauer K., Schlösser R., Necrotizing enterocolitis of the neonate with Clostridium perfringens: Diagnosis, clinical course, and role of alpha toxin. Eur. J. Pediatr. 167, 891–895 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Sturbelle R. T., de Avila L. F. d. C., Roos T. B., Borchardt J. L., de Cássia dos Santos da Conceição R., Dellagostin O. A., Leite F. P. L., The role of quorum sensing in Escherichia coli (ETEC) virulence factors. Vet. Microbiol. 180, 245–252 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Rutherford S. T., Bassler B. L., Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, a012427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer H., Yamamoto M., Akira S., Beutler B., Svanborg C., Mechanism of pathogen-specific TLR4 activation in the mucosa: Fimbriae, recognition receptors and adaptor protein selection. Eur. J. Immunol. 36, 267–277 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Leaphart C. L., Cavallo J., Gribar S. C., Cetin S., Li J., Branca M. F., Dubowski T. D., Sodhi C. P., Hackam D. J., A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. J. Immunol. 179, 4808–4820 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Jilling T., Simon D., Lu J., Meng F. J., Li D., Schy R., Thomson R. B., Soliman A., Arditi M., Caplan M. S., The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J. Immunol. 177, 3273–3282 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiles T. J., Kulesus R. R., Mulvey M. A., Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 85, 11–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raveh-Sadka T., Firek B., Sharon I., Baker R., Brown C. T., Thomas B. C., Morowitz M. J., Banfield J. F., Evidence for persistent and shared bacterial strains against a background of largely unique gut colonization in hospitalized premature infants. ISME J. 10, 2817–2830 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raveh-Sadka T., Firek B., Sharon I., Baker R., Brown C. T., Thomas B. C., Morowitz M. J., Banfield J. F., Gut bacteria are rarely shared by co-hospitalized premature infants, regardless of necrotizing enterocolitis development. eLife 4, e05477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman S. F., Olm M. R., Morowitz M. J., Banfield J. F., Machine learning leveraging genomes from metagenomes identifies influential antibiotic resistance genes in the infant gut microbiome. mSystems 3, e00123-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olm M. R., West P. T., Brooks B., Firek B. A., Baker R., Morowitz M. J., Banfield J. F., Genome-resolved metagenomics of eukaryotic populations during early colonization of premature infants and in hospital rooms. Microbiome 7, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng Y., Leung H. C. M., Yiu S. M., Chin F. Y. L., IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Hyatt D., Chen G.-L., LoCascio P. F., Land M. L., Larimer F. W., Hauser L. J., Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieber C. M. K., Probst A. J., Sharrar A., Thomas B. C., Hess M., Tringe S. G., Banfield J. F., Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alneberg J., Bjarnason B. S., de Bruijn I., Schirmer M., Quick J., Ijaz U. Z., Lahti L., Loman N. J., Andersson A. F., Quince C., Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Wu Y.-W., Simmons B. A., Singer S. W., MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Olm M. R., Brown C. T., Brooks B., Banfield J. F., dRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crits-Christoph A., Gelsinger D. R., Ma B., Wierzchos J., Ravel J., Davila A., Casero M. C., DiRuggiero J., Functional interactions of archaea, bacteria and viruses in a hypersaline endolithic community. Environ. Microbiol. 18, 2064–2077 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Roux S., Enault F., Hurwitz B. L., Sullivan M. B., VirSorter: Mining viral signal from microbial genomic data. PeerJ 3, e985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren J., Ahlgren N. A., Lu Y. Y., Fuhrman J. A., Sun F., VirFinder: A novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 5, 69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skewes-Cox P., Sharpton T. J., Pollard K. S., DeRisi J. L., Profile hidden Markov models for the detection of viruses within metagenomic sequence data. PLOS ONE 9, e105067 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rampelli S., Soverini M., Turroni S., Quercia S., Biagi E., Brigidi P., Candela M., ViromeScan: A new tool for metagenomic viral community profiling. BMC Genomics 17, 165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar R. C., Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber T., Blin K., Duddela S., Krug D., Kim H. U., Bruccoleri R., Lee S. Y., Fischbach M. A., Müller R., Wohlleben W., Breitling R., Takano E., Medema M. H., antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q., VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jolley K. A., Maiden M. C. J., BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinegger M., Söding J., MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Delcher A. L., Salzberg S. L., Phillippy A. M., Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics Chapter 10, Unit 10.3 (2003). [DOI] [PubMed] [Google Scholar]
- 57.El-Gebali S., Mistry J., Bateman A., Eddy S. R., Luciani A., Potter S. C., Qureshi M., Richardson L. J., Salazar G. A., Smart A., Sonnhammer E. L. L., Hirsh L., Paladin L., Piovesan D., Tosatto S. C. E., Finn R. D., The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price M. N., Dehal P. S., Arkin A. P., FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letunic I., Bork P., Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaax5727/DC1
Table S1. Metagenomic sequencing depth and read quality information.
Table S2. Patient metadata.
Table S3. Information about dereplicated secondary metabolite clusters, de novo–assembled genomes, and genome-wide importances of genomes.
Table S4. Accuracy of ML algorithms and protein clustering algorithms and mapping-based abundances of bacterial taxa.
Table S5. Full feature table provided to the ML classifier and importances of all features resulting from the ML classifier.
Table S6. Proteins enriched in genomes of interest and identified fimbrial genes.
Fig. S1. Metagenomic characterization of 1163 samples from 160 premature infants.
Fig. S2. Fecal samples taken before NEC diagnosis have a higher abundance of plasmids from specific bacterial taxa.
Fig. S3. PCA is unable to separate pre-NEC and control samples.
Fig. S4. ML feature importance values reveal organismal associations with NEC.


