A novel T cell–based ZIKV vaccine, encoding NS1 protein, confers protection against systemic infection.
Abstract
The causal association of Zika virus (ZIKV) with microcephaly, congenital malformations in infants, and Guillain-Barré syndrome in adults highlights the need for effective vaccines. Thus far, efforts to develop ZIKV vaccines have focused on the viral envelope. ZIKV NS1 as a vaccine immunogen has not been fully explored, although it can circumvent the risk of antibody-dependent enhancement of ZIKV infection, associated with envelope antibodies. Here, we describe a novel DNA vaccine encoding a secreted ZIKV NS1, that confers rapid protection from systemic ZIKV infection in immunocompetent mice. We identify novel NS1 T cell epitopes in vivo and show that functional NS1-specific T cell responses are critical for protection against ZIKV infection. We demonstrate that vaccine-induced anti-NS1 antibodies fail to confer protection in the absence of a functional T cell response. This highlights the importance of using NS1 as a target for T cell–based ZIKV vaccines.
INTRODUCTION
Zika virus (ZIKV) is a flavivirus transmitted via the bite of infected Aedes aegypti mosquitoes. Historically, ZIKV infections were considered asymptomatic and self-limiting and were associated with the development of Guillain-Barré syndrome in adults, a polyneuropathy that can result in paralysis (1). The explosive spread of ZIKV in the Americas in 2015 to 2016 was causally associated with serious birth defects in infants born to mothers infected during pregnancy, including microcephaly and a range of neurological abnormalities and birth defects termed congenital Zika syndrome (2). Human-to-human transmission of ZIKV has been established, with ZIKV being the only known arbovirus that is transmitted sexually with persistence in the reproductive tissues of both males and females for prolonged periods of time (3, 4). Currently, there is no licensed vaccine available to protect against ZIKV infection.
ZIKV has biological similarities to other flaviviruses, such as dengue virus (DENV), West Nile virus (WNV), and Japanese encephalitis virus (JEV). ZIKV contains a positive-sense RNA genome encoding one polyprotein, which is co- and posttranslationally cleaved into structural proteins [capsid (C), premembrane/membrane (prM/M), and envelope (E)] and nonstructural proteins (NS1–NS5). ZIKV virions are comprised of prM/M and E proteins, with E presented on the outer surface of a mature virion representing the primary antigenic target of neutralizing antibodies (nAb) (5). Hence, ZIKV prM and E have been the focus of most experimental ZIKV vaccines (6–12). All prM/E-based vaccines were able to induce nAb and provide protection in mouse models of ZIKV infection, whereas DNA, adenoviral, and inactivated virus vaccines have shown efficacy in nonhuman primate models of ZIKV disease (7, 8). DNA and inactivated virus vaccines have progressed to phase 1 clinical trials and exhibit immunogenicity and safety (13, 14).
Sequence and structural homology between some flavivirus E proteins can result in high degree of antibody cross-reactivity and resultant antibody-dependent enhancement (ADE) of infection, through the engagement of immunoglobulin G (IgG) antibodies with cell surface Fcγ receptors (15). Although there are no clinical data to support ZIKV ADE in humans, studies have shown cross-reactivity between human DENV and ZIKV antibodies, resulting in enhancement of ZIKV infection in vitro (15–17). Most recently, DENV-specific antibodies have been shown to enhance vertical ZIKV transmission in ZIKV-infected pregnant mice, resulting in a severe microcephaly-like syndrome (18). Therefore, NS1 is a promising vaccine target that eliminates the risk of ADE, because NS1 is not expressed on the surface of ZIKV virions, and NS1-specific antibodies are thus unlikely to enhance the infection.
NS1 is essential for viral replication; although it exists primarily as a membrane-associated homodimer in infected cells, it has both intracellular and extracellular functions (19–21). Intracellular dimeric NS1 plays a key role in viral replication and localizes to sites of viral RNA synthesis, where it is incorporated in the viral replication complex and associated vesicle packets (22). NS1 protein is also trafficked to the plasma membrane, where it binds the surface of infected cells and is secreted into the extracellular space as a hexametric lipoprotein particle (23). Secreted- and membrane-associated NS1 homodimers are highly immunogenic, and NS1 has been found to contribute to the pathogenesis of DENV infection (20, 24). Soluble NS1 has also been found to facilitate ZIKV acquisition by mosquitoes and to contribute to evasion of host interferon induction (25, 26). The molecular mechanisms of NS1 are relatively well established for DENV and WNV (23, 27); however, ZIKV disease is very different from DENV, and a greater understanding of the distinctive role of ZIKV NS1 in disease pathogenesis is emerging (28).
Previous studies have shown that passive immunization with DENV NS1–specific antibodies confers protection against DENV, while WNV NS1–specific monoclonal antibodies prevent lethal infection in mice (29, 30). Furthermore, vaccination with DENV-1, DENV-3, or DENV-4 NS1 provided cross-protection against a heterologous DENV-2 lethal challenge (24). Recently, it has been proposed that ZIKV NS1 antibodies mediate antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent pathways, and a modified vaccinia ankara (MVA)–based NS1 vaccine showed protection in an intracranial model of ZIKV infection (31, 32). Passive transfer of human monoclonal NS1 antibodies displayed partial protective efficacy against lethal challenge in Stat2−/− mice (32). Inclusion of NS1 in an adenoviral vaccine (Ad2) that also encoded prM/E enhanced vaccine efficacy in ZIKV-challenged neonatal mice born to maternally immunized Balb/c dams (33). Immunization of Balb/c mice with a recombinant vesicular stomatitis virus encoding prM-E-NS1 showed that NS1 can confer partial protection from ZIKV disease in Balb/c mice that were administered anti–IFNAR1 (interferon-α/β receptor subunit 1) blocking antibody (34). However, to date, no ZIKV NS1–based vaccine has evaluated the role of NS1-specific cell-mediated immune (CMI) responses in protection against systemic ZIKV infection in an immunocompetent mouse model.
Here, we constructed three different ZIKV NS1 DNA vaccines and assessed the ability of each to induce NS1-specific humoral and cell-mediated immune responses and protective efficacy against ZIKV challenge in Balb/c mice. We show that immunogenicity against ZIKV NS1 is critically dependent on efficient secretion of NS1. We demonstrate that while NS1-specific antibodies contribute to the control of ZIKV infection, they do not, however, confer protection. In contrast, protection is achieved only in the presence of fully functional CD8+ and CD4+ T cell responses, most likely directed by the recognition of novel NS1-specific cytotoxic CD8+ T lymphocyte (CTL) and T helper (TH) cell epitopes present in the NS1 C-terminal β-ladder domain. Our data demonstrate that NS1 alone can confer protection against ZIKV and highlight the importance of NS1 and T cell responses in ZIKV vaccine development.
RESULTS
Antibody and CMI responses induced by ZIKV NS1 DNA vaccines
We constructed three different ZIKV NS1 DNA vaccines encoding (i) wild-type NS1 (pVAX-NS1), (ii) secreted NS1 with a tissue plasminogen activator (TPA) leader sequence introduced upstream of the NS1 to ensure efficient secretion (pVAX-tpaNS1), and (iii) NS1 secreted as a heptamer by fusion to a chimeric version of the oligomerization domain from the chicken complement inhibitor C4b-binding protein termed IMX313P (pVAX-tpaNS1-IMX313P) (fig. S1). Fusion of vaccine antigens to IMX313 results in self-assembly into soluble heptametric structures after expression, resulting in increased magnitude of antibody and T cell responses, as well as protective efficacy when compared with the same dose of monomeric antigen (35–38).
NS1 expression from all DNA vaccines was validated by Western immunoblotting and indirect immunofluorescence of 293T cells transfected with the NS1 DNA vaccines (fig. S1). We detected secreted NS1 in the supernatants of human embryonic kidney (HEK) 293T cells transfected with pVAX-tpaNS1 and pVAX-tpaNS1-IMX313P, with the NS1 protein in the latter being secreted as an oligomerized heptamer (fig. S1C).
To assess the humoral immunogenicity of the NS1 DNA vaccines, groups of Balb/c mice (n = 7) received three immunizations of 50 μg of each of the NS1 DNA vaccines or control pVAX intradermally (i.d.) into the ear pinnae (Fig. 1B). Serum NS1-specific antibody responses following vaccination with the different DNA vaccines were assessed by enzyme-linked immunosorbent assay (ELISA) using immobilized recombinant NS1 as the capture antigen.
Fig. 1. Antibody responses induced by NS1 DNA vaccination in Balb/c mice.
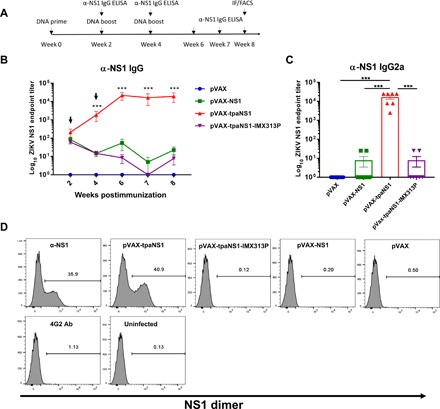
Six to 8-week-old Balb/c mice were immunized with different NS1 DNA vaccine candidates. (A) Timeline of vaccination and antibody assays. FACS, fluorescence-activated cell sorting. (B) Kinetics of NS1-specific endpoint IgG ELISA titers. Arrows indicate time points when DNA vaccine boosts were given. Titers are expressed as the reciprocal of the serum dilution and plotted as log10. The data represent mean responses in each group (n = 7) ± SEM. ***P < 0.001 (Kruskal-Wallis H test). (C) Endpoint IgG2a titers against ZIKV NS1 measured at week 8 after immunization using rabbit anti-mouse immunoglobulin isotype-specific antibodies recognizing IgG2a (***P < 0.001; Kruskal-Wallis H test). (D) Flow cytometric analysis of the efficacy of hyperimmune mouse sera in binding the ZIKV NS1 dimer expressed on the surface of ZIKV-infected Vero cells. Vero cells were infected with ZIKVPRVABC59 at multiplicity of infection (MOI) of 0.1 and 48 hours and later stained with pooled sera from immunized mice. Flaviviral 4G2 antibody was used as a negative control, while mouse monoclonal anti-ZIKV NS1 was used as a positive control.
The titers induced by pVAX-tpaNS1 vaccination were significantly higher than those induced by pVAX-NS1 or pVAX-tpaNS1-IMX313P (***P < 0.001) (Fig. 1B). pVAX-tpaNS1 immunization resulted in 4 log titers of ZIKV NS1–specific antibodies as detected by endpoint ELISA. NS1 antibody titers increased 1 log each following the second (week 2) and third (week 4) vaccine boosts and remained steady (4 log) for at least 4 weeks following the last vaccination. Immunization with either pVAX-NS1 or pVAX-tpaNS1-IMX313P DNA vaccines induced ~2 log antibody titers following prime, however failing to induce a significant increase in titers following boost. In addition, we determined the extent to which IgG2a contributed to the anti-NS1 antibody response induced by DNA immunization (Fig. 1C), as previous work has shown an association between anti-NS1 IgG2a and protective effects of flavivirus anti-NS1 antibodies via complement and ADCC activation (30, 31, 39). Endpoint IgG2a anti-NS1 titers measured 4 weeks after the last vaccine dose showed that high IgG2a anti-NS1 antibody titers were induced only by immunization with pVAX-tpaNS1 (***P < 0.001) (Fig. 1C). Endpoint titers of anti-NS1 IgG2a were comparable to the titers of total anti-NS1 IgG (Fig. 1, B and C), suggesting that IgG2a response was predominant.
Flaviviral anti-NS1 IgG2a has been shown to target NS1 dimers expressed on infected Vero cells and to mediate ADCC via engagement of IgG2a antibodies with cell surface FcγRIII receptors (30, 31). Therefore, we tested the ability of sera from mice immunized with three different NS1 DNA vaccines to detect cell surface NS1 in ZIKV-infected Vero cells by flow cytometry (Fig. 1D). Only pooled sera from pVAX-tpaNS1–vaccinated mice bound to NS1 on ZIKV-infected Vero cells at a frequency similar to that observed with the positive control (polyclonal commercial antibody, mean = 40.9% versus 35.9%, respectively). sera from pVAX-NS1 and pVAX-tpaNS1-IMX313P failed to recognize NS1 on infected Vero cells (0.2 and 0.12%, respectively). As expected, sera from control pVAX mice failed to bind to surface NS1 (0.5%). The specificity of NS1 recognition by sera from pVAX-tpaNS1 mice was confirmed by the inability of sera to bind to uninfected Vero cells. Overall, the data suggest superior immunogenicity of secreted native NS1 as obtained with the pVAX-tpaNS1 vaccine in eliciting anti-NS1 humoral responses.
To assess the T cell immunogenicity of the different NS1 DNA vaccines, we immunized Balb/c mice (n = 7) as before (Fig. 2A). Two weeks after the last immunization, we quantified NS1-specific T cell responses by IFN-γ enzyme-linked immunospot (ELISpot). Splenocytes were stimulated with four peptide pools derived from panels of overlapping 13- or 15-mer peptides, spanning the entire ZIKVPRVABC59 NS1, with each pool containing 27 to 29 individual overlapping peptides. Significant levels of NS1-specific IFN-γ responses were only detected in mice vaccinated with pVAX-tpaNS1 in response to stimulation with NS1 pools 3 and 4, corresponding to amino acids 172 to 352 of the ZIKVPRVABC59 NS1 protein [mean spot-forming units (SFU) = 472 and 920, respectively] (Fig. 2B). No significant T cell responses were detected following vaccination of mice with pVAX-NS1, pVAX-tpaNS1-IMX313P, or pVAX (Fig. 2B). Summary analysis of the total NS1-specific T cell responses to all the pools highlights the superior immunogenicity of pVAX-tpaNS1 in eliciting NS1-specific T cell responses (Fig. 2C).
Fig. 2. Characterization of cell-mediated responses induced by ZIKV NS1 DNA vaccination in Balb/c mice.
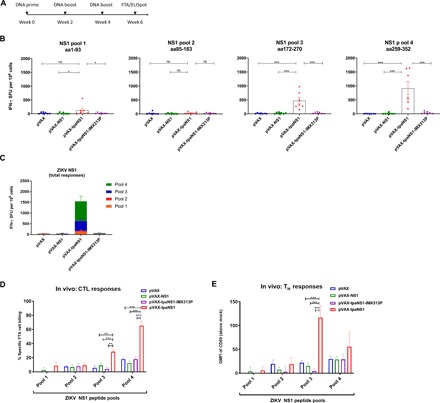
(A) Timeline of vaccination and T cell assays. (B) ELISpot analysis of ZIKV-NS1–specific IFN-γ secretion in splenocytes in response to immunization with different ZIKV-NS1 DNA vaccines. aa, amino acid; ns, not significant. The data are presented as mean (n = 7) ± SEM spot-forming units (SFU) per 106 splenocytes. (C) Total ZIKV-NS1–specific IFN-γ responses representing the contribution of each NS1 peptide pool to the total NS1-specific response. Data represent the average numbers of SFU per 106 splenocytes with values representing the mean responses in each ±SEM. (D) In vivo NS1-specific CTL responses induced by NS1 DNA vaccinations. Female Balb/c mice (n = 7 per group) were vaccinated with pVAX-NS1, pVAX-tpaNS1, pVAX-tpaNS1-IMX313P, and control pVAX DNA. Thirteen days later, FTA was performed where 1.5 × 106 peptide-pulsed cells (for each target cell cluster) or mock target cells were injected intravenously All the peptide-pulsed cell targets were gated and analyzed for the percentage recovery relative to mock targets to determine the specific CTL cell loss using the equation [(percent mock targets − percent peptides-pulsed targets)/percent mock targets] × 100. The bar graphs show the mean ± SEM of ZIKV NS1–specific in vivo CTL responses in pVAX, pVAX-NS1, pVAX-tpaNS1, and pVAX-tpaNS1-IMX313P pulsed with NS1 peptides (10 μg/ml) in NS1 pools 1 to 4. (E) In vivo TH cell responses to ZIKV NS1 in different DNA vaccine groups. Analysis of expression [geometric mean fluorescent intensity (GMFI)] of the activation marker CD69 on FTA B220+ target cells pulsed with ZIKV NS1 peptide pools 1 to 4 (10 μg/ml). Mean ± SEM representing the GMFI of CD69 (above mock) on FTA B220 + cells (*P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis H test).
Monitoring T cell responses in vivo after immunization is of critical importance for developing efficacious vaccines. Fluorescent target array (FTA) technology simultaneously measures in vivo CTL (CD8) and TH (CD4) T cell responses in real time against numerous autologous target cells (naïve splenocytes) pulsed with different peptides corresponding to vaccine antigens. This technique involves labeling of autologous naïve splenocytes (target cells) with different concentrations and combinations of carboxyfluorescein succinimidyl ester (CFSE), CellTrace Violet (CTV), and Cell Proliferation Dye eFluor670 (CPD) to generate target cell clusters with unique florescent signatures that can be easily distinguishable using flow cytometry following recovery of the injected targets from FTA-challenged animals (40, 41). CFSE, CTV, and CPD label live cells with multiple, stable fluorescence intensities at discrete emission wavelengths. We have shown that CFSE, CTV, and CPD can be used in combination to generate an array of >250 discernible target cell clusters, allowing for comprehensive analysis of magnitude, breadth, epitope-variant cross-reactivity, and avidity of T cell responses in vivo (41).
FTA is based on the recovery of fluorescently labeled naïve autologous target cells loaded with major histocompatibility complex class I (MHC-I)–binding peptides and, hence, measures CTL-mediated target cell killing events in vivo and defines the contribution of antigen-specific CD8+ T cells to immunogenicity of a vaccine. In addition, FTA can measure the CD4+ TH cell activity based on the ability of TH cells to help activate cognate B cells directly. This is achieved by measuring the up-regulation of activation marker CD69 on FTA B220+ target cells pulsed with MHC-II–binding peptides. Thus, the FTA allows clear delineation of the individual contributions of CD8+ and CD4+ T cells to vaccine efficacy and immunogenicity in vivo following vaccination. Therefore, to directly measure CTL and TH T responses in vivo 14 days after immunization with different NS1 DNA vaccines, we conducted an FTA simultaneously measuring in vivo CTL and TH cell responses against autologous target cells (naïve splenocytes) pulsed with peptides spanning ZIKV NS1 (40–43).
We generated FTA using splenocytes from naïve mice pulsed with peptide pools 1 to 4 (10 μg/ml) of ZIKV NS1 and injected the target cells into vaccinated mice (Fig. 2A). T cell responses were assessed 18 hours later by flow cytometry analysis of FTA target cells recovered from the spleen of vaccinated mice (fig. S2). The highest in vivo NS1-specific CTL responses were detected against NS1 pool 4–pulsed target cells in mice vaccinated with pVAX-tpaNS1 (mean ± SEM, 70% killing; ***P < 0.001) (Fig. 2D). Significant CTL responses were also detected in pVAX-tpaNS1–vaccinated mice against NS1 pool 3–pulsed target cells (mean ± SEM, 30% killing) (**P < 0.01; ***P < 0.001) but not against pool 1–pulsed or pool 2–pulsed target cells. Mice immunized with pVAX-NS1, pVAX-tpaNS1-IMX313P, or pVAX showed no significant killing.
Upon recognition of cognate antigens, TH cells present costimulation to B cells resulting in the up-regulation of CD69 on B cells, and monitoring the expression levels of CD69 on naïve B cells correlates with the magnitude of TH cell responses in vivo in the FTA analysis. Examination of in vivo TH cell responses showed that mice vaccinated with pVAX-tpaNS1 elicited the greatest and significantly higher responses to NS1 pool 3 (mean ± SEM, 120) when compared with the other vaccines (mean ± SEM, 14; P < 0.01; ***P < 0.001) (Fig. 2E). Significant TH responses were not detected against other NS1 peptide pools for any other vaccine groups relative to the pVAX control. Overall, the in vivo FTA data showed that immunization with pVAX-tpaNS1 induced strong CD8+ and CD4+ T cell responses and that these responses are likely driven by the recognition of T cell epitopes present in the C terminus (amino acids 172 to 352) of the ZIKVPRVABC59 NS1 protein.
In vivo identification of novel ZIKV NS1 CTL and TH T cell epitopes
We then set out to identify the T cell epitopes driving the ZIKV NS1–specific CTL and TH cell responses within the newly identified immunodominant (ID) NS1 region (amino acids 172 to 352) induced by pVAX-tpaNS1 DNA vaccination following our standard prime boost protocol in Balb/c mice (Fig. 3A). To achieve this, we conducted an FTA using 57 individual overlapping ZIKV NS1 peptides present in pools 3 and 4 of ZIKVPRVABC59 NS1 protein (amino acids 172 to 352) (Fig. 3B), while total pools of peptides for NS1, pool 3, and pool 4 were used as controls.
Fig. 3. In vivo mapping of the ZIKV NS1–specific CTL and TH epitopes.
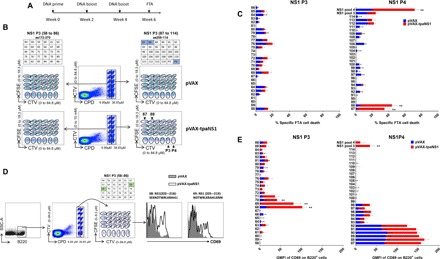
(A) Timeline of vaccination and FTA assay. (B) Epitope mapping of ZIKV NS1 CTL epitopes using the FTA assay. Flow diagram of the target cell matrices for individual peptides present in pools 3 and 4. Double-discriminated lymphocytes were gated before analysis of ZIKV NS1 pools 3 and 4 individual peptide targets and analysis of total responses to all peptides present in NS1 peptide pool 3 or 4 based on the CPD and CTV emission. Individual peptide targets were then gated on the basis of the CFSE and CTV emission. Tables depict which peptides correspond to cell targets presented in the flow dot plots. The dot plots are the targets gated from a representative pVAX-tpaNS1– and a representative pVAX-vaccinated mouse. (C) The bar graphs show the mean ± SEM of ZIKV NS1–specific CTL responses in pVAX-tpaNS1– and pVAX-vaccinated mice against target cells pulsed with individual NS1 peptides (10 μg/ml) in NS1 pools 3 (58 to 86) and 4 (87 to 114) and CTL responses against NS1 peptide pools 3 or 4. (D) Analysis of FTA B cell expression of the activation marker CD69 in pVAX-tpaNS1–vaccinated animals relative to control pVAX-vaccinated animals as an in vivo measure of ZIKV NS1–specific TH cell responses. B220+ cells from all targets were delineated on the basis of the CPD and CTV emission, and the individual peptide targets were then gated on the basis of the CFSE and CTV emission. The dot plot depicts the targets gated from a representative pVAX-tpaNS1–vaccinated mouse pulsed with peptides present in ZIKV NS1 pool 3, as shown in the table above the dot plot. The GMFI of CD69 was determined by flow cytometry. The representative histogram plots show up-regulation of CD69 expression on B220+ cell targets pulsed with peptide 68 or 69 (10 μg/ml) of NS1 pool 3 in representative pVAX-tpaNS1–vaccinated animal compared with control pVAX. (E) In vivo ZIKV NS1 TH epitope mapping. The bar graphs show the mean CD69 GMFI ± SEM of each B220+ target pulsed with peptide (10 μg/ml) (**P < 0.01, nonparametric Mann-Whitney U test).**
As expected, the greatest CTL killing was observed against target cells pulsed with NS1 peptide pool 4 (mean ± SEM, 60% killing) and for targets pulsed with peptides 87 or 88 (**P < 0.001) (Fig. 3C). We identified an overlapping sequence “MKGPWHSEELEI” (amino acids 262 to 273) present in both peptides 87 and 88 (**P < 0.001) (Fig. 4C). CTL responses for pool 3 were modest compared with previous observations. No significant CTL responses above control (pVAX) were detected against targets pulsed with any of the individual peptides in NS1 pool 3 (amino acids 172 to 270).
Fig. 4. Functional confirmation of ZIKV NS1–specific CTL and TH epitopes.
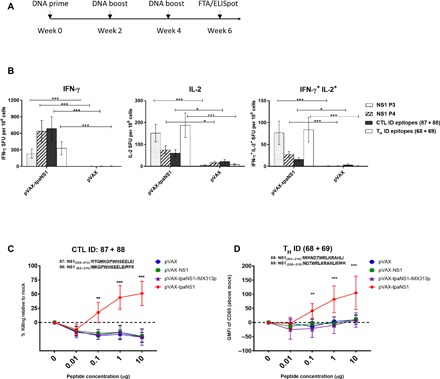
(A) Timeline of vaccination and FTA assay. Groups of Balb/c mice (n = 7 per group) were vaccinated with NS1 DNA vaccine constructs, and 13 days after the last vaccine dose, IFN-γ ELISpot and FTA assays were performed. (B) Assessment of NS1-specific polyfunctional (IL-2/IFN-γ) responses induced by pVAX-tpaNS1 DNA immunization. FluoroSpot analysis of ZIKV NS1–specific IFN-γ and IL-2 production in pVAX-tpaNS1– and pVAX-vaccinated mice. Splenocytes were harvested 2 weeks after the third immunization and were stimulated with ZIKV NS1 pool 3, NS1 pool 4, NS1 CTL ID peptides (87 + 88), and NS1 TH ID peptides (68 + 69). NS1-specific in vitro secretion of IFN-γ and IL-2, and dual secretion of IFN-γ/IL-2 were measured. The data are presented as mean (n = 7) ± SEM SFU per 106 splenocytes, and the nonparametric Mann-Whitney U test was used to analyze the statistical significance. *P < 0.05; ***P < 0.001. (C and D) FTA assessment of the magnitude and functional avidity of the NS1 CTL or TH ID epitopes in vivo following vaccination with different NS1 DNA vaccines. Naïve splenocytes were pulsed with titrated concentrations of ZIKV NS1 CTL ID peptides (87 and 88) or TH ID peptides (68 and 69). Peptide-pulsed and mock target cells (1.5 × 106 for each target cell cluster) were injected intravenously into vaccinated mice, and splenocytes were harvested 15 hours later and analyzed by flow cytometry. (C) % specific killing was calculated for the ZIKV NS1 CTL epitopes above mock at each peptide concentration. Data show mean ± SEM of NS1-specific CTL killing. (D) Mean ± SEM representing the GMFI of CD69 (above mock) on FTA B220+ target cells pulsed with titrated concentrations of the identified ZIKV NS1 TH peptides 68 and 69. Kruskal-Wallis H test was used to analyze the statistical significance. **P < 0.005; ***P < 0.001.
Next, the same FTA was used to evaluate NS1-specific TH epitopes for their ability to up-regulate CD69 expression on target B220+ cells in mice vaccinated with pVAX-tpaNS1 (**P < 0.01) (Fig. 3D). Statistically significant TH responses were only detected against target cells pulsed with peptides spanning NS1 pool 3 (**P < 0.01) of pVAX-tpsNS1–vaccinated mice with the most dominant responses detected against targets pulsed with peptides 68 (amino acids 202 to 216) and 69 (amino acids 205 to 2019) with an overlapping sequence “NDTWRLKRAHLI” (amino acids 205 to 216) (Fig. 3E). No TH responses were detected in pVAX-tpaNS1–vaccinated mice against targets pulsed with any peptides within NS1 pool 4. Together, detailed in vivo FTA-based epitope mapping analysis revealed novel dominant NS1-specific CTL and TH cell epitopes.
Functional confirmation of the novel ZIKV NS1 CTL and TH cell epitopes
We analyzed the specificity, magnitude, and functional avidity of the NS1-specific T cell responses induced by the newly identified NS1 CTL and TH ID epitopes in vitro and in vivo following NS1 DNA prime boost vaccination (Fig. 4A). Fluorospot analysis of splenocytes from pVAX-tpaNS1–vaccinated mice showed that CTL epitopes primarily induced IFN-γ secretion and TH epitopes induced interleukin-2 (IL-2) secretion (Fig. 4B). Although robust polyfunctional IFN-γ+/IL-2+ CTL responses were observed following stimulation with the identified ID CTL epitopes, we observed the most significant increase in polyfunctional responses in NS1-specific CD4+ T cells (Fig. 4B). These data not only validate the identified ID CTL and TH cell epitopes as being ID relative to IFN-γ and IL-2 secretion but also highlight that pVAX-tpaNS1 vaccination can induce polyfunctional antiviral T cell responses.
Functional avidity of T cell receptors for their cognate antigen is a crucial determinant of T cell functionality, as high-avidity T cells can sense and respond to low levels of cognate antigen, a characteristic associated with more potent responses against many infections. Therefore, the ability of all NS1 DNA vaccine constructs to induce NS1-specific CTL or TH responses with high functional avidity was tested in vivo with a FTA comprising targets pulsed with titrated concentrations of the ID CTL or TH cell epitopes of NS1. We performed flow cytometric analysis of the FTA cells isolated from vaccinated mice as described above.
FTA analysis of the CTL responses against the two CTL ID ZIKV NS1 peptides revealed that only the pVAX-tpaNS1 prime-boost immunization generated CTL responses with high avidity and ability to recognize target cells pulsed with low concentrations (0.1 μg/ml) of peptide (**P < 0.01, ***P < 0.001) (Fig. 4C). Similarly, pVAX-tpaNS1 vaccination induced TH responses of greatest avidity when pulsed with varying concentrations of the NS1 TH epitope (**P < 0.01, ***P < 0.001) compared with all other NS1 DNA vaccines (Fig. 4D). Furthermore, when FTA targets were pulsed with ZIKV NS1 pool 4 lacking CTL ID peptides 87 and 88, in vivo killing was completely abrogated in pVAX-tpaNS1–vaccinated mice, thus confirming that the identified peptides contain functional NS1 CTL epitopes that are recognized in vivo (fig. S3).
Our findings confirm that the novel ZIKV NS1 CTL and TH epitopes are responsible for strong cytotoxic and TH activity induced by pVAX-tpaNS1 immunization inducing NS1-specific CD8+ and CD4+ T cells with high functional avidity coupled with robust polyfunctional responses. Collectively, our investigations of the immunogenicity of ZIKV NS1 as a vaccine antigen showed that only the pVAX-tpaNS1 DNA vaccine induced significant adaptive responses, reaffirming the notion that TPA-driven NS1 secretion determines the immunogenicity of ZIKV NS1 in a DNA vaccine.
Immunization with pVAX-tpaNS1 DNA vaccine protects against ZIKV challenge
To assess the protective efficacy of pVAXtpaNS1 DNA vaccine against ZIKV challenge in vivo, we immunized Balb/c mice (n = 10) intradermally three times with either 50 or 100 μg of pVAXtpaNS1 DNA or with 100 μg of control pVAX (Fig. 5A). A recent study demonstrated that a high concentration of NS1 monoclonal antibodies limited the ZIKV disease severity in Stat2 −/− mice, and in line with this finding, we wanted to determine whether immunization with a higher dose (100 μg) would result in higher titers of vaccine-induced anti-NS1 antibodies that may limit ZIKV viremia after challenge (32). Immunization with 100 μg of pVAX-tpaNS1 DNA did not significantly increase anti-NS1 antibody titers compared to the 50 μg dose (Fig. 5B).
Fig. 5. Protective efficacy of the pVAX-tpaNS1 vaccine.
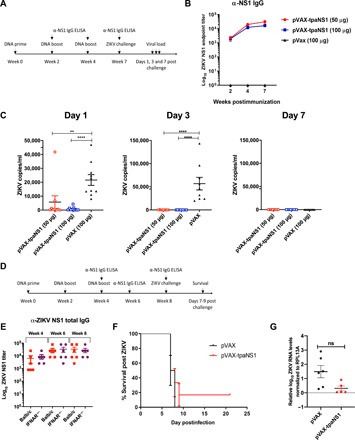
(A) Timeline of vaccination, challenge, and viral load determinations. Female Balb/c mice (n = 10) were immunized intradermally three times with either 50 or 100 μg of the pVAX-tpaNS1 DNA vaccine. Control pVAX mice received three 100-μg doses. (B) anti-NS1 antibody titer as measured by ELISA. (C) Mice were challenged 3 weeks after the last dose by the intravenous route with 200 PFU of ZIKVPRVABC59. Serum ZIKV VL on days 1, 3, and 7 after challenge are shown (**P < 0.005, ****P < 0.0001, Kruskal-Wallis H test). (D) Timeline of vaccination, determination of NS1 antibody responses and lethal challenge of female IFNAR−/− mice. (E) Female IFNAR−/− (n = 6) and Balb/c (n = 7) mice were immunized intradermally three times with 50 μg of pVAX-tpaNS1 or pVAX control DNA vaccine. Anti-ZIKV NS1 antibody titers were determined at weeks 4, 6, and 8 of the immunization schedule by NS1 ELISA and compared with antibody titers in Balb/c mice. (F) Survival of pVAX-tpaNS1– or pVAX-vaccinated IFNAR−/− mice after subcutaneous challenge with 103 CCID50 of ZIKVMR766. Mice were euthanized when ethically defined endpoints had been reached. (G) Viremia in pVAX-tpaNS1– or pVAX-vaccinated IFNAR−/− mice after challenge with ZIKVMR766. Limit of detection 2 log10CCID50/ml. Error bars represent SEM.
Three weeks after the last dose, vaccinated mice were challenged with ZIKVZKV2015 and viral loads (VL) were quantitated by reverse transcription polymerase chain reaction (RT-PCR), as previously described (6). In this model, pVAX-vaccinated Balb/c mice challenged intravenously with ZIKVZKV2015 develop 6 days of detectable viremia, with VL detectable on day 1 (mean, 22,000 copies/ml) that peak on day 3 (mean, 56,114 copies/ml) and are cleared by day 7 after challenge (Fig. 5C).
In contrast, immunization with either 50 or 100 μg of pVAX-tpaNS1 provided protection against ZIKVZKV2015 challenge with no detectable viremia (****P < 0.0001) on day 3 after challenge (Fig. 5C), while significantly lower VL were detected on day 1 (**P < 0.01; ****P < 0.0001). There was no significant difference in the VL between the two vaccine doses (Fig. 5B). Immunization with the pVAX-tpaNS1 DNA vaccine conferred protection after challenge.
We next immunized type I IFN receptor–deficient IFNAR−/− mice with 50 μg of pVAX tpaNS1 or pVAX DNA to assess whether pVAX-tpaNS1 immunization can confer protection in this model of ZIKV disease (Fig. 5D). There were no significant differences in the anti-NS1 antibody titers induced by vaccination of IFNAR−/− mice compared with the titers in Balb/c mice as measured by ELISA (Fig. 5E). Four weeks after immunization, the IFNAR−/− mice were challenged subcutaneously with a lethal dose of 103 CCID50 (cell culture infectious dose 50%) MR766 strain of ZIKV using an established protocol (9). Immunization with pVAX-tpaNS1 DNA did not protect IFNAR−/− mice against a lethal ZIKV challenge (Fig. 5E), and VL were not significantly different between the pVAX- and pVAX-tpaNS1–vaccinated groups (Fig. 5G). Thus, inferring that protection against ZIKV following pVAX-tpaNS1 DNA vaccination, even in the presence of high anti-NS1 antibody titer, requires intact type I IFN signaling.
Immune correlates of protection against ZIKV challenge
To investigate the immunological mechanism required for protection against ZIKV challenge after pVAX-tpaNS1 vaccination, we purified IgG from the serum of Balb/c mice vaccinated with pVAX-tpaNS1. Passive transfer of varying concentrations of purified IgG via the intravenous route resulted in mean NS1-specific antibody titers of 2.9 log10 (high) and 2.2 (low) in recipient mice as measured by ELISA (Fig. 6A). Recipient and control naïve mice (n = 4) were challenged intravenously with 200 plaque-forming units (PFU) of ZIKVZKV2015, and the VL were assayed on days 1 and 3 after challenge. All animals were viremic with no significant differences between groups (Fig. 6B). There was a trend toward lower VL in mice that received high anti-NS1 IgG, indicating that anti-NS1 IgG likely contributes to viral control but is not protective in the absence of NS1-specific T cell immunity.
Fig. 6. Mechanism of protection.
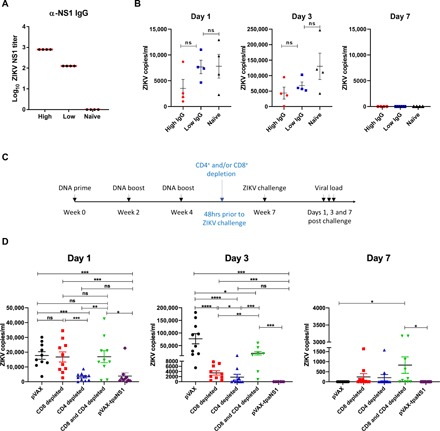
(A) NS1-specific serum antibody titers in recipient Balb/c mice following adoptive transfer of high or low amounts of IgG purified from the serum of Balb/c mice vaccinated with pVAX-tpaNS1 DNA vaccine. (B) Serum VL on days 1, 3, and 7 after challenge in Balb/c mice that received adoptive transfer of purified IgG from vaccinated mice. Mice were challenged intravenously with 200 PFU of ZIKVZKV2015. Naïve mice were challenged and used as controls. (C) Timeline of vaccination, CD4+ and/or CD8+ T cell depletion, challenge with ZIKVZKV2015, and VL determination. (D) Serum VL in pVAX-tpaNS1 DNA–vaccinated mice that were depleted of CD4+ and/or CTLs before challenge with ZIKVZKV2015. Serum ZIKV VL on days 1, 3, and 7 after challenge are shown. pVAX immunized mice were used as controls (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Kruskal-Wallis H test).
Next, we depleted CD8+ and/or CD4+ T lymphocytes (>99% efficiency) in pVAX-tpaNS1–vaccinated mice (n = 10) 2 days before ZIKVZKV2015 challenge (Fig. 6C). Depletion of CD8+ T cells completely abrogated viral control on day 1 after challenge (Fig. 6D). CD4+ T cell–depleted mice had significantly lower VL than pVAX control or CD8+-depleted mice (***P < 0.001) with VL levels similar to those detected in undepleted pVAX-tpaNS1–vaccinated mice (Fig. 6D). In addition, mice depleted of both CD8+ and CD4+ T cells were also unable to control ZIKV infection on day 1 despite the presence of high-titer anti-NS1 antibodies. Collectively, data indicate that NS1-specific CD8+ T cells play an important role in the early control of ZIKV infection in mice immunized with pVAX-tpaNS1.
By day 3, pVAX-tpaNS1–vaccinated mice have cleared the ZIKV infection, whereas T cell–depleted groups showed varying degrees of viral control when compared with pVAX or pVAX-tpaNS1. This was expected as anti-NS1 antibodies have been shown to restrict ZIKV infection via complement activation and ADCC (31, 32). CD8+ T cell–depleted mice remained viremic with a significantly higher VL than CD4+ T cell–depleted mice (*P < 0.05) or pVAX-tpaNS1 (*P < 0.001)–vaccinated mice, indicating the need for a sustained CD8+ T cell response in the rapid and complete clearance of ZIKV infection. This was further confirmed as CD8+/CD4+ T cell–depleted mice had a significantly higher VL than CD8+ (**P = 0.001) or CD4+-depleted mice (***P < 0.0001), suggesting that CTL and TH cell responses are necessary at the peak of viremia to restrict ZIKV replication but that early viral clearance is critically dependent on the action of CD8+ T cells. At day 7, only 20% of CD4+-depleted mice remained viremic, whereas 50% of CD8+- and CD4+/CD8+-depleted animals had a detectable load compared with pVAX or pVAX-tpaNS1. Together, our data indicate that T cell–mediated immunity, rather than anti-NS1 antibodies, is crucial for the protection and elimination of ZIKV infection following pVAX-tpaNS1 immunization.
DISCUSSION
Here, we describe a novel DNA vaccine encoding a secreted form of ZIKV NS1, demonstrating that NS1 immunization alone can confer protection against systemic ZIKV infection in immunocompetent Balb/c mice. Immunization with pVAX-tpaNS1 DNA induced strong CTL and polyfunctional TH responses driven by the recognition of novel ID NS1 T cell epitopes, present in the highly conserved C-terminal β-ladder of ZIKV NS1 (44). We show that T cell responses are crucial for protection as vaccine-induced anti-NS1 antibodies may aid in the control of viral replication but are insufficient to confer protection in the absence of a functional NS1-specific T cell response.
We demonstrate that when NS1 is used in a DNA vaccine, its immunogenicity was dependent on TPA leader sequence–driven expression, likely allowing for antigen processing and presentation to occur similar to events during a natural ZIKV infection (45). If NS1 is not efficiently secreted or if the secreted NS1 is oligomerized by fusion to IMX313P, then its immunogenicity is abrogated. Fusion of IMX313P to the C terminus of NS1 may induce an “inside-out” heptamer, forcing the hydrophobic N-terminal β-roll domain to be exposed to the outer regions of the heptamer. This may alter NS1 uptake, processing and/or presentation, and effective adaptive immunity.
Prior studies have focused on elucidating anti-NS1 antibody responses and examined protection in immunodeficient mice, namely, A129, STAT2−/−, and IFNAR1 antibody-depleted mice (32–34). Our data support the findings that NS1 vaccination induces robust humoral response and that anti-NS1 antibodies, although nonneutralizing, recognize cell surface–associated NS1 and contribute to immunity against ZIKV, most likely via FcγR-mediated complement and ADCC, as others have shown (31, 32). However, these immunodeficient models do not allow for comprehensive examination of T cell responses or their role in providing vaccine-induced protective immunity against ZIKV infection.
Previous work has shown that monoclonal anti-NS1 antibodies elicit protection against ZIKV infection in immunodeficient STAT2−/− mice and that this action is mediated by ADCC (32). This contrasts to our data demonstrating a crucial role for T cells in control of ZIKV infection. In the context of NS1 vaccine immunity, the involvement of CMI and nonneutralizing antibody responses (complement and ADCC) is most likely not mutually exclusive. Our data suggest that T cell responses are critical, showing that without a functional CD8+ and CD4+ T cell response, even in the presence of high-antibody titer, immunized mice were unable to control or clear ZIKV infection. This was supported by passive antibody transfer studies, as administration of anti-NS1 antibodies in the absence of vaccine-induced NS1-specific T cell responses was unable to confer protection from systemic ZIKV infection in Balb/c mice. Nevertheless, CD8+ and CD4+ T cell–depleted mice do show lower VL than control mice at the peak of viremia, indicating that vaccine-induced antibody-mediated responses contribute to the control of viral replication but are not protective per se. This is further supported by lethal challenge studies in vaccinated IFNAR−/− mice, where high anti-NS1 antibody titer failed to prevent mortality, although there was a trend for lower VL in pVAX-tpaNS1–vaccinated mice. It is worth noting that the degree of protection in the STAT2−/− study was achieved by passive transfer of a high concentration of purified monoclonal antibodies (mAbs), but protection was not sterilizing nor complete, most likely due to the lack of functional T cell responses in the STAT2 −/− model, which lacks STAT2-driven signaling pathways (32). STAT2 not only participates in the canonical type I IFN–driven JAK/STAT signaling pathway to induce type I IFN–stimulated gene expression but can also act in noncanonical pathways to influence type I IFN (α/β), type II IFN (IFN-γ), and type III IFN (IFN-λ) signaling (46). The model of ZIKV infection in STAT2−/− mice would favor a TH2-biased antibody response by precluding the roles of STAT2 in cross-presentation to CD8+ T cells and IFN-γ function, including the generation of CTL responses, thus affecting subsequent CTL activity (47, 48). In addition, we demonstrate that vaccination of IFNAR−/− mice induced high levels of anti-NS1 antibodies, comparable to those observed in Balb/c mice, but these mice, deficient in IFN α/β receptor signaling responses, were unable to control viral replication after challenge and eventually succumbed to infection. It is well known that IFN α/β plays a key role in priming adaptive T cell responses and directly influences the fate of both CD4+ and CD8+ T cells during the initial phases of antigen presentation, thus shaping the effector and memory T cell pool (49). Therefore, the absence of IFNAR signaling is predicted to result in a dysfunctional T cell response in the vaccinated IFNAR−/− mice, ablating the essential NS1-specific T cell–mediated protection. In contrast, in the immunocompetent Balb/c mouse model, T cell responses are fully intact, and the magnitude and duration of ZIKV viremia are comparable with that in humans (50). Thus, our study represents the first comprehensive evaluation of vaccine-induced NS1-specific T cell responses in vivo and their contribution to protection against ZIKV infection.
Notably, an MVA-NS1 vaccine was able to induce protection against ZIKV infection after an intracranial challenge, which is not the natural route of ZIKV infection (31). This study supports the involvement of humoral and cell-mediated immunity in NS1 control of ZIKV infection, but it did not investigate the mechanism(s) responsible for the control of ZIKV in the brain. Given that flavivirus infection in the brain is associated with CD8+ T cell infiltration and our data demonstrate that early elimination of ZIKV-infected cells is CD8+ T cell driven, it is likely that the protective immunity in the study of Brault et al. was also CD8+ T cell mediated.
Induction of strong NS1-specific T cell responses via ZIKV NS1 vaccination may induce favorable priming for enhanced immunological memory during a secondary heterologous flavivirus challenge, as it has been shown that flaviviruses can induce serotype cross-reactive T cell responses (51–53). Cross-reactive B and T cells have been shown to provide protection against a secondary heterotypic DENV infection, while skewing of CD8+ T cell cross-reactive responses toward primary infecting viruses during heterologous DENV infection did not impair immune responses (54, 55). Sequential immunizations with flaviviruses sharing CD4+ T cell epitopes have also been found to promote protection during subsequent heterologous infection (53). Recently, it has also been shown that T cell responses elicited by prior infection with DENV enhance the timing, magnitude, and quality of T cell responses to ZIKV during in vitro stimulation (56).
A recent study in naïve IFNAR−/− mice challenged with ZIKV demonstrated that CD4+ T cells are required for protection against a lethal ZIKV challenge in this model (57). This was not a vaccine study, but rather the focus was on ZIKV-reactive CD4+ T cell responses in an infection model, and CD4+ T cell epitopes were identified in structural and nonstructural ZIKV proteins. A different study reported that the CD4+ T cell response to primary infection was predominantly TH1 and was directed against a narrow range of ID ZIKV epitopes in E, NS3, NS4B, and NS5 proteins in the myeloid type I IFN receptor–deficient mice (LysMCre+Ifnar1fl/fl) (58). However, human CD4+ T cell response to ZIKV was shown to target epitopes in both structural and nonstructural proteins, with the most ID epitopes located in E, NS1, and NS5 (59, 60).
Furthermore, broad CD8+ T cell responses were detected during infection against ZIKV MR766 and Cambodian FSS13025 ZIKV strains in LysMCre+Ifnar1fl/fl mice (61). Notably, in the H-2b LysMCre+Ifnar1fl/fl model, E protein appeared to be the main target of the anti-ZIKV CD8+ T cell response, and the overall response against the MR766 ZIKV strain was broader than that to the FSS13025 strain, especially for NS1, NS3, and NS5 epitopes (61).
It is likely that T cell responses and epitope recognition induced by NS1 DNA vaccination in Balb/c mice may not mirror the epitopes recognized during primary ZIKV infection in LysMCre+Ifnar1fl/fl mice as the mouse strains were genetically different, and different strains of ZIKV and infection routes were used. Furthermore, ZIKV-specific CD4+ T cells were shown to be necessary for local control of viral infection in the lower female reproductive tract in LysMCre+Ifnar1fl/fl mice infected intravaginally, but not protective during intravenous infection such as ours (58). Collectively, all studies demonstrate the importance of T cells in the control of ZIKV infection and support ZIKV vaccine strategies that induce a protective T cell response to ZIKV.
The NS1 DNA ZIKV vaccine offers an attractive alternative to envelope-based ZIKV vaccines. Vaccines that target the NS1 protein do not risk inducing ADE in individuals living in areas endemic for DENV, and other flaviviruses and anti-NS1 mAbs do not enhance viral uptake in vitro (32). Sequence alignment analysis showed that NS1 ID T cell epitopes are highly conserved among ZIKV strains; therefore, a vaccine-induced T cell response targeting NS1 is likely to be cross-protective against all circulating strains.
In addition, DNA vaccines are relatively easy and cost-effective to manufacture on a large scale. They have excellent safety profiles for women of childbearing age and children, both critical target populations to prevent and mitigate ZIKV outbreaks (62) .
In summary, we evaluated three different ZIKV NS1 DNA vaccine candidates using an immunocompetent mouse model. We demonstrated that effective secretion of NS1 is critical for immunogenicity, showing that NS1 alone can confer protection against systemic ZIKV infection and that this protection is T cell mediated. This study bridges a major gap in the understanding of how ZIKV NS1 vaccine regulates CMI responses and protection in an immunocompetent host and highlights the importance of NS1 as a target for protective T cell–based ZIKV vaccines.
METHODS
DNA vaccines
ZIKV virus isolate Brazil-ZKV2015 (accession number KU497555) was used to design NS1 transgenes, which were produced synthetically and codon optimized for enhanced mammalian expression by GeneArt (Germany). ZIKV NS1 was cloned into pVax (Invitrogen) downstream of the CMV promoter, and a Kozak translation initiation sequence was included. Three plasmids were generated encoding either wild-type NS1 viz. (i) pVAX-NS1, or secreted NS1 generated by the upstream introduction of the human TPA leader sequence, viz. (ii) pVAX-tpaNS1 (63). To generate secreted NS1 fused to the oligomerization domain of the C4b-p (pVAX-tpaNS1-IMX313), TPA and the oligomerization domain of C4b-p (IMX313) were introduced at the N and C termini, respectively, of the ZIKV NS1 gene as we described (35, 64). Plasmids were produced with QIAGEN endotoxin-free gigaprep kits. Sequences were confirmed by double-stranded sequencing.
Animals
All mouse work was conducted in accordance with the Australian Code for the Care and Use of Animals for Scientific Purposes as defined by the National Health and Medical Research Council of Australia, and studies were approved by the University of Adelaide, the University of South Australia, and the South Australian Pathology Animal Ethics Committees (Adelaide, Australia) and by the Institutional Animal Care and Use Committees (IACUCs) at Beth Israel Deaconess Medical Center (Boston, USA). Balb/c and IFNAR−/− female mice 6 to 8 weeks of age were vaccinated with 50 μg of DNA vaccine in saline by the intradermal route in the ear pinnae as we described previously (43, 65). Balb/c mice were challenged intravenously 3 weeks after the last vaccine dose with 200 PFU of ZIKV-PRVABC59 or ZKV2015 as we described previously (6). IFNAR−/− mice were challenged 4 weeks after the last vaccine dose with 103 CCID50 MR766 as we described previously (9). ZIKVMR766–infected female IFNAR−/− mice were euthanized when ethically defined clinical end points were reached (primarily hind-limb paralysis). Animals were randomly allocated to groups. Immunologic and virologic assays were performed blinded. Sample size was determined to achieve 80% power to detect significant differences in protective efficacy.
Western blot
To detect NS1 expression, cell lysates and supernatant fluids were harvested from HEK293T cells transiently transfected (48 hours) with NS1 DNA vaccines using Lipofectamine LTX reagent (Life Technologies), and 50 μg of protein was analyzed in 10 to 12% (v/v) SDS–polyacrylamide gel electrophoresis under reducing [with β-mercaptoethanol (β-Me)] or nonreducing (without β-Me) conditions as we described (35). Mouse monoclonal anti-NS1 antibody (BioFront Technologies, BF-1225-16) and goat anti-mouse IgG horseradish peroxidase (HRP)–conjugated secondary antibody (Invitrogen) were used to detect NS1 expression essentially as we described previously (35, 66).
Immunofluorescence assay
HEK293T cells were cultured in 96-well plates in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in 5% CO2 and transfected with 200 ng of DNA by Lipofectamine LTX (Life Technologies) according to the manufacturer’s protocol. Antigen expression was detected as we described previously (43, 67). Briefly, at 48 hours after transfection, transfected cells were fixed with 4% formalin (Sigma), permeabilized with methanol at −20°C, and then blocked in 2.5% bovine serum albumin (Sigma) in phosphate-buffered saline (PBS) before the addition of anti-NS1 antibody (BioFront Technologies) at 37°C for 2 hours. Finally, cells were stained with DAPI (4′,6′-diamidino-2-phenylindole; Life Technologies) and visualized by fluorescence microscopy (Zeiss LSM-700), and the data were digitized using the Zen software (Zeiss).
Flow cytometry
Vero cells were infected with ZIKV PRVABC59 at multiplicity of infection (MOI) of 0.1, and 48 hours later, the cells were stained for 1 hour at 4°C with either pooled mouse sera, polyclonal rabbit anti-ZIKV NS1 antibody (GeneTex, GTX133307), or 4G2 mouse anti-flavivirus envelope antibody (Clonegene, NR-50327). Alexa Fluor 488–conjugated goat anti-mouse IgG (Invitrogen) secondary antibody was then added for 1 hour at 4°C in the dark, and cells were then fixed in 0.5% paraformaldehyde and analyzed on BD FACSCanto flow cytometer.
Enzyme-linked immunosorbent assay
To determine endpoint anti-NS1 antibody titers, Nunc MaxiSorp flat-bottom 96-well plates (Thermo Fisher Scientific) were coated overnight at 4°C with ZIKV NS1 protein (Sino Biological) at 1 μg/ml in PBS as described previously. The plates were washed four times with PBS +0.05% Tween 20 (PBST) and then blocked with StartingBlock Block Buffer (Thermo Fisher Scientific) for 5 min at room temperature. Serially diluted mouse serum samples were added to wells and incubated at 37°C for 1 hour, and the plates were washed four times. Bound antibodies were detected using HRP-conjugated goat anti-mouse IgG (GE Healthcare Life Sciences), and the optical density (OD) was read at 492 nm. To confirm that the IgG2a antibody isotype predominates the anti-NS1 humoral response, antibodies were captured as described above, and bound antibodies were detected using HRP-conjugated anti-mouse IgG2a (GE Healthcare Life Sciences). Endpoint titers were determined as the reciprocal of the highest serum sample dilution with an OD reading above the cutoff, set as 2SD above the mean OD of serum samples from prevaccinated or naïve mice.
Enzyme-linked immunospot
ZIKV NS1–specific cellular immune responses were assessed by IFN-γ ELISpot assay. The 114-peptide array spanning the entire NS1 of the PRVABC59 strain of ZIKV was obtained through BEI Resources, the National Institute of Allergy and Infectious Diseases, and the National Institutes of Health: peptide array, ZIKV, and PRVABC59 (nonstructural protein 1, NR-50534). Individual peptides are 13- or 15-mers with 12–amino acid overlap, and detailed information on their length and sequence is provided by BEI Resources (www.beiresources.org/Catalog/BEIPeptideArrays/NR-50534.aspx).
The peptides were divided into four pools, each containing 27 to 29 individual peptides. Mouse IFN-γ ELISpot assay was performed on red blood cell–depleted splenocytes from immunized mice, which were stimulated with different NS1 peptide pools for 36 hours at 37°C, essentially as we described previously (43, 68). Briefly, multiscreen-IP HTS plates (Merck Millipore) were coated with anti-mouse IFN-γ (clone AN18, Mabtech), and secreted IFN-γ was detected with anti-mouse IFN-γ biotin (clone R4-6A2, Mabtech) followed by streptavidin–alkaline phosphatase (AP) (Sigma-Aldrich) and SigmaFast BCIP (5-bromo-4-chloro-3-indolyl phosphate)/NBT (nitroblue tetrazolium; Sigma-Aldrich).
The number of NS1-specific IL-2– and IFN-γ-producing cells in splenocytes from vaccinated mice was analyzed using FluoroSpot kit (Mabtech) according to the manufacturer’s instructions. In brief, splenocytes were stimulated with 4 μg of either NS1 peptide pool 3, pool 4, CTL, or TH pools. Samples were incubated for 36 hours at 37°C, after which spots were developed following the manufacturer’s instructions.
Developed spots were counted automatically by use of an ELISpot reader (Autoimmun Diagnostika GmbH, Germany), and the number of spots for unstimulated splenocytes (<50) was subtracted from the number of spots for the peptide pool–stimulated splenocytes to generate the number of specific spot-forming units per 106 cells. Data are presented as mean ± SEM.
FTA assay
The FTA assay was used to examine in vivo the magnitude and quality of T cell responses generated after vaccination. Fluorescent cell labeling and peptide pulsing were performed as per established protocols (40–43). Different concentrations and combinations of cell tracking dyes CFSE, CTV, and CPD were used to delineate several populations of target cells by fluorescence. Target cells from naïve mice were dye labeled, peptide pulsed, and injected intravenously into vaccinated mice. The percentage recovery of FTA targets loaded with MHC-binding peptides is a measure of cell killing by antigen-specific CTLs, while up-regulation of activation marker CD69 on FTA B220+ target cells pulsed with MHC-II–binding peptides represents a measure of antigen-specific CD4+ T cell help.
To generate the FTA used for NS1 epitope mapping (Fig. 4), splenocytes from naïve age-matched Balb/c mice were pooled, split evenly in six ways, and labeled with either 84.8, 22.95, 6.21, 1.67, 0.595, or 0 μM CTV. The cells were then washed thrice, and the cells from each aliquot were split evenly five ways and labeled with 18.3, 5.17, 1.46, 0.441 or 0 μM CFSE to result in 30 distinct populations of target cells. Following CFSE labeling, the cells were washed again and split into two to delineate individual peptides in pool 3 or pool 4 of ZIKV NS1. Each target cell population was then pulsed with each peptide (10 μg/ml) in NS1 pools 3 and 4 (peptides 58 to 114) and total pool 3 or 4 of NS1 for 4 hours at 37°C + 5% CO2. The peptide-pulsed targets were washed, and all NS1 pool 3– or pool 4–specific target cells were pooled and labeled with 9.89 or 38.65 μM CPD, respectively. The labeled cells were washed three times and pooled for intravenous injection into immunized mice (2.26 × 105 cells per target cell cluster in 200 μl PBS per mouse). Fifteen hours later, the splenocytes were harvested, depleted of red blood cells, stained with B220 and CD69, and analyzed by flow cytometry (BD FACSCanto II flow cytometer) as described for Fig. 3.
To generate the FTA used in the experiments for Figs. 3 and 5, splenocytes from naive aged-matched Balb/c mice were pooled; split evenly five ways; labeled with either 84.8, 22.95, 6.21, 1.67, or 0 μm CTV; and washed. The cells were then split into three aliquots and labeled with 5.17, 1.46, or 0.441 μM CFSE to result in 15 populations. Five 5.17 μM CFSE–labeled populations were pulsed with different concentrations (10, 1, 0.1, or 0.01 μg/ml) of identified CTL peptides (peptides 87 and 88) or mock pulsed. The 1.46 μM CFSE–labeled populations were pulsed with different concentrations (10, 1, 0.1, or 0.01 μg/ml) of identified TH peptides (peptides 68 and 69) or mock pulsed. The 0.441 μM CFSE populations were pulsed with NS1 pool 1, pool 2, pool 3, pool 4, or pool 4 (10 μg/ml) minus the CTL peptides (87 and 88). All labeled targets were pulsed for 4 hours at 37°C + 5% CO2 and upon completion of stimulation were washed. Target populations were then pooled and labeled with 38.65 μM CPD, washed, and injected (1.5 × 106 cells per target cell cluster) into immunized mice before analysis of splenocytes as described above.
FlowJo Tree Star (version 8.8.7) software was used to generate the flow cytometry plots. The percentage of specific FTA loss (as a measure of CTL activity) was calculated using the following formula: % loss = [(mock target value − peptide-pulsed target value)/mock target value] × 100. GraphPad Prism 6 software was used for statistical analysis and to construct the graphs presented in this study.
IgG purification and adoptive transfer
Serum was collected from pVAX-tpaNS1–vaccinated mice, and polyclonal IgG was purified using protein G purification kits (Thermo Fisher Scientific). Varying amounts of purified IgG was infused by the intravenous route into naive recipient mice before ZIKV challenge.
CD4+ and CD8+ T lymphocyte depletion
Anti-CD4 (GK1.5) and/or anti-CD8 β (Lyt3.2 clone 53-5.8; Bio X Cell) monoclonal antibodies were administered at doses of 0.1 mg per mouse to pVAX-tpaNS1 DNA–vaccinated mice by the intraperitoneal route on day −2 before ZIKV challenge. To confirm that CD4 + T cell depletion was maintained through the acute phase of infection, splenocytes were collected and analyzed on days 2 and 5 after treatment. Antibody depletions were >99.9% efficient as determined by flow cytometry using the following antibodies: α-CD4-APC-Cy7 (clone RM4-5), α-CD8-PerCP-Cy 5.5 (clone 53-6.7), and α-CD3-AF700 (clone 500A2).
Reverse transcription polymerase chain reaction
RT-PCR assays in Balb/c mice were used to monitor VL as previously described (6). RNA was extracted from plasma with QIAcube HT (Qiagen, Germany). The wild-type ZIKV BeH815744 Cap gene was used as a standard. RNA was purified (Zymo Research), and RNA quality and concentration were assessed by the BIDMC Molecular Core Facility. Log dilutions of the RNA standard were reverse transcribed and included with each RT-PCR assay. VL were calculated as virus particles per milliliter. RT-PCR assessment of VL in sera of IFNAR−/− mice was performed on day 2 (peak of viremia) using ZIKV M primers [as previously described (9)] with normalization to RPL13A.
Statistical analyses
Immunologic and virologic data analysis was performed using GraphPad Prism version 8 (GraphPad Software) and IBM SPSS Statistics software using Kruskal-Wallis H test or Mann-Whitney U tests, with *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 considered significant.
Supplementary Material
Acknowledgments
Funding: This work was supported by a grant from the National Foundation for Medical Research and Innovation (NFMRI), Australia (no. 274) and in part by a grant from The Hospital Research Foundation, Australia. D.K.W. and A.C.S. were supported by the Early Career Fellowships from The Hospital Research Foundation (THRF; Australia). M.M. was the recipient of the University of Adelaide Postgraduate Award (Australia), and Z.A.M. was the recipient of the International Postgraduate Research Scholarship (IPRS) and Australian Postgraduate Award (APA; Australia). Author contributions: E.J.G. and B.G.-B. designed the ZIKV NS1 DNA vaccines. B.G.-B. characterized the vaccines, performed animal experiments and immunological assays, directed the project, analyzed the results, and wrote the manuscript. D.K.W. performed animal experiments and FTA analysis and analyzed the results. M.M. characterized the vaccines, performed animal experiments, and analyzed the results. P.A., R.L.P., and R.A.L. performed viral load assessments and adoptive transfer studies. N.A.P., Z.A.M., and A.S. performed animal experiments. N.S.E. and M.R.B. prepared ZIKV for the challenge experiments. J.G. helped design the vaccines. J.C. provided vital reagents and assay support. E.J.G., S.A.R., J.D.H., D.H.B., and all other authors edited the manuscript. Competing interests: J.D.H. sits on the Scientific Advisory Board, is a paid consultant, receives research funding, and is a minor shareholder in the vaccines company Sementis Ltd. (Australia). B.G.-B., D.K.W., and E.J.G. are inventors on a patent related to this work filed by The University of Adelaide (AU2018902659, 23 July 2018; PCT/AU2019/050770, 23 July 2018). All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The BIDMC and vaccine construct materials can be shared with a standard material transfer agreement upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaax2388/DC1
Fig. S1. DNA vaccine design and antigen expression.
Fig. S2. FTA analysis.
Fig. S3. Epitope confirmation.
Fig. S4. ZIKV viral load over time in T cell–depleted mice.
REFERENCES AND NOTES
- 1.Cao-Lormeau V.-M., Blake A., Mons S., Lastѐre S., Roche C., Vanhomwegen J., Dub T., Baudouin L., Teissier A., Larre P., Vial A.-L., Decam C., Choumet V., Halstead S. K., Willison H. J., Musset L., Manuguerra J.-C., Despres P., Fournier E., Mallet H.-P., Musso D., Fontanet A., Neil J., Ghawché F., Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387, 1531–1539 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Araújo T. V. B., Ximenes R. A. d. A., Miranda-Filho D. d. B., Souza W. V., Montarroyos U. R., de Melo A. P. L., Valongueiro S., de Albuquerque M. d. F. P. M., Braga C., Filho S. P. B., Cordeiro M. T., Vazquez E., Cruz D. d. C. S., Henriques C. M. P., Bezerra L. C. A., Castanha P. M. d. S., Dhalia R., Marques-Júnior E. T. A., Martelli C. M. T., Rodrigues L. C.; investigators from the Microcephaly Epidemic Research Group; Brazilian Ministry of Health; Pan American Health Organization; Instituto de Medicina Integral Professor Fernando Figueira; State Health Department of Pernambuco , Association between microcephaly, Zika virus infection, and other risk factors in Brazil: Final report of a case-control study. Lancet Infect. Dis. 18, 328–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turmel J. M., Abgueguen P., Hubert B., Vandamme Y. M., Maquart M., Le Guillou-Guillemette H., Leparc-Goffart I., Late sexual transmission of Zika virus related to persistence in the semen. Lancet 387, 2501 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Murray K. O., Gorchakov R., Carlson A. R., Berry R., Lai L., Natrajan M., Garcia M. N., Correa A., Patel S. M., Aagaard K., Mulligan M. J., Prolonged detection of Zika virus in vaginal secretions and whole blood. Emerg. Infect. Dis. 23, 99–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stettler K., Beltramello M., Espinosa D. A., Graham V., Cassotta A., Bianchi S., Vanzetta F., Minola A., Jaconi S., Mele F., Foglierini M., Pedotti M., Simonelli L., Dowall S., Atkinson B., Percivalle E., Simmons C. P., Varani L., Blum J., Baldanti F., Cameroni E., Hewson R., Harris E., Lanzavecchia A., Sallusto F., Corti D., Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353, 823–826 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Larocca R. A., Abbink P., Peron J. P., Zanotto P. M. d. A., Iampietro M. J., Badamchi-Zadeh A., Boyd M., Ng'ang'a D., Kirilova M., Nityanandam R., Mercado N. B., Li Z., Moseley E. T., Bricault C. A., Borducchi E. N., Giglio P. B., Jetton D., Neubauer G., Nkolola J. P., Maxfield L. F., De La Barrera R. A., Jarman R. G., Eckels K. H., Michael N. L., Thomas S. J., Barouch D. H., Vaccine protection against Zika virus from Brazil. Nature 536, 474–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbink P., Larocca R. A., De La Barrera R. A., Bricault C. A., Moseley E. T., Boyd M., Kirilova M., Li Z., Ng'ang'a D., Nanayakkara O., Nityanandam R., Mercado N. B., Borducchi E. N., Agarwal A., Brinkman A. L., Cabral C., Chandrashekar A., Giglio P. B., Jetton D., Jimenez J., Lee B. C., Mojta S., Molloy K., Shetty M., Neubauer G. H., Stephenson K. E., Peron J. P. S., Zanotto P. M. d. A., Misamore J., Finneyfrock B., Lewis M. G., Alter G., Modjarrad K., Jarman R. G., Eckels K. H., Michael N. L., Thomas S. J., Barouch D. H., Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science 353, 1129–1132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbink P., Larocca R. A., Visitsunthorn K., Boyd M., De La Barrera R. A., Gromowski G. D., Kirilova M., Peterson R., Li Z., Nanayakkara O., Nityanandam R., Mercado N. B., Borducchi E. N., Chandrashekar A., Jetton D., Mojta S., Gandhi P., LeSuer J., Khatiwada S., Lewis M. G., Modjarrad K., Jarman R. G., Eckels K. H., Thomas S. J., Michael N. L., Barouch D. H., Durability and correlates of vaccine protection against Zika virus in rhesus monkeys. Sci. Transl. Med. 9, eaao4163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prow N. A., Liu L., Nakayama E., Cooper T. H., Yan K., Eldi P., Hazlewood J. E., Tang B., Le T. T., Setoh Y. X., Khromykh A. A., Hobson-Peters J., Diener K. R., Howley P. M., Hayball J. D., Suhrbier A., A vaccinia-based single vector construct multi-pathogen vaccine protects against both Zika and chikungunya viruses. Nat. Commun. 9, 1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan C., Muruato A. E., Nunes B. T. D., Luo H., Xie X., Medeiros D. B. A., Wakamiya M., Tesh R. B., Barrett A. D., Wang T., Weaver S. C., Vasconcelos P. F. C., Rossi S. L., Shi P.-Y., A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat. Med. 23, 763–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richner J. M., Himansu S., Dowd K. A., Butler S. L., Salazar V., Fox J. M., Julander J. G., Tang W. W., Shresta S., Pierson T. C., Ciaramella G., Diamond M. S., Modified mRNA vaccines protect against Zika virus infection. Cell 168, 1114–1125 e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardi N., Hogan M. J., Pelc R. S., Muramatsu H., Andersen H., DeMaso C. R., Dowd K. A., Sutherland L. L., Scearce R. M., Parks R., Wagner W., Granados A., Greenhouse J., Walker M., Willis E., Yu J.-S., McGee C. E., Sempowski G. D., Mui B. L., Tam Y. K., Huang Y.-J., Vanlandingham D., Holmes V. M., Balachandran H., Sahu S., Lifton M., Higgs S., Hensley S. E., Madden T. D., Hope M. J., Karikó K., Santra S., Graham B. S., Lewis M. G., Pierson T. C., Haynes B. F., Weissman D., Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 543, 248–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudinski M. R., Houser K. V., Morabito K. M., Hu Z., Yamshchikov G., Rothwell R. S., Berkowitz N., Mendoza F., Saunders J. G., Novik L., Hendel C. S., Holman L. A., Gordon I. J., Cox J. H., Edupuganti S., McArthur M. A., Rouphael N. G., Lyke K. E., Cummings G. E., Sitar S., Bailer R. T., Foreman B. M., Burgomaster K., Pelc R. S., Gordon D. N., DeMaso C. R., Dowd K. A., Laurencot C., Schwartz R. M., Mascola J. R., Graham B. S., Pierson T. C., Ledgerwood J. E., Chen G. L.; VRC 319; VRC 320 study teams , Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: Randomised, open-label, phase 1 clinical trials. Lancet 391, 552–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modjarrad K., Lin L., George S. L., Stephenson K. E., Eckels K. H., De La Barrera R. A., Jarman R. G., Sondergaard E., Tennant J., Ansel J. L., Mills K., Koren M., Robb M. L., Barrett J., Thompson J., Kosel A. E., Dawson P., Hale A., Tan C. S., Walsh S. R., Meyer K. E., Brien J., Crowell T. A., Blazevic A., Mosby K., Larocca R. A., Abbink P., Boyd M., Bricault C. A., Seaman M. S., Basil A., Walsh M., Tonwe V., Hoft D. F., Thomas S. J., Barouch D. H., Michael N. L., Preliminary aggregate safety and immunogenicity results from three trials of a purified inactivated Zika virus vaccine candidate: Phase 1, randomised, double-blind, placebo-controlled clinical trials. Lancet 391, 563–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul L. M., Carlin E. R., Jenkins M. M., Tan A. L., Barcellona C. M., Nicholson C. O., Michael S. F., Isern S., Dengue virus antibodies enhance Zika virus infection. Clin. Transl. Immunol. 5, e117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priyamvada L., Quicke K. M., Hudson W. H., Onlamoon N., Sewatanon J., Edupuganti S., Pattanapanyasat K., Chokephaibulkit K., Mulligan M. J., Wilson P. C., Ahmed R., Suthar M. S., Wrammert J., Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl. Acad. Sci. U.S.A. 113, 7852–7857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T., Sakuntabhai A., Cao-Lormeau V.-M., Malasit P., Rey F. A., Mongkolsapaya J., Screaton G. R., Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat. Immunol. 17, 1102–1108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathore A. P. S., Saron W. A. A., Lim T., Jahan N., St John A. L., Maternal immunity and antibodies to dengue virus promote infection and Zika virus–induced microcephaly in fetuses. Sci. Adv. 5, eaav3208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rastogi M., Sharma N., Singh S. K., Flavivirus NS1: A multifaceted enigmatic viral protein. Virol. J. 13, 131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller D. A., Young P. R., The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 98, 192–208 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Watterson D., Modhiran N., Young P. R., The many faces of the flavivirus NS1 protein offer a multitude of options for inhibitor design. Antivir. Res. 130, 7–18 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Mackenzie J. M., Jones M. K., Young P. R., Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220, 232–240 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Akey D. L., Brown W. C., Dutta S., Konwerski J., Jose J., Jurkiw T. J., DelProposto J., Ogata C. M., Skiniotis G., Kuhn R. J., Smith J. L., Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 343, 881–885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beatty P. R., Puerta-Guardo H., Killingbeck S. S., Glasner D. R., Hopkins K., Harris E., Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med. 7, 304ra141 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Song H., Qi J., Haywood J., Shi Y., Gao G. F., Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat. Struct. Mol. Biol. 23, 456–458 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Liu J., Liu Y., Nie K., Du S., Qiu J., Pang X., Wang P., Cheng G., Flavivirus NS1 protein in infected host sera enhances viral acquisition by mosquitoes. Nat. Microbiol. 1, 16087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edeling M. A., Diamond M. S., Fremont D. H., Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc. Natl. Acad. Sci. U.S.A. 111, 4285–4290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puerta-Guardo H., Glasner D. R., Espinosa D. A., Biering S. B., Patana M., Ratnasiri K., Wang C., Beatty P. R., Harris E., Flavivirus NS1 triggers tissue-specific vascular endothelial dysfunction reflecting disease tropism. Cell Rep. 26, 1598–1613.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan S.-W., Lu Y.-T., Huang C.-H., Lin C.-F., Anderson R., Liu H.-S., Yeh T.-M., Yen Y.-T., Wu-Hsieh B. A., Lin Y.-S., Protection against dengue virus infection in mice by administration of antibodies against modified nonstructural protein 1. PLOS ONE 9, e92495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung K. M., Nybakken G. E., Thompson B. S., Engle M. J., Marri A., Fremont D. H., Diamond M. S., Antibodies against West Nile Virus nonstructural protein NS1 prevent lethal infection through Fc gamma receptor-dependent and -independent mechanisms. J. Virol. 80, 1340–1351 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brault A. C., Domi A., McDonald E. M., Talmi-Frank D., McCurley N., Basu R., Robinson H. L., Hellerstein M., Duggal N. K., Bowen R. A., Guirakhoo F., A Zika vaccine targeting NS1 protein protects immunocompetent adult mice in a lethal challenge model. Sci. Rep. 7, 14769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey M. J., Duehr J., Dulin H., Broecker F., Brown J. A., Arumemi F. O., Bermúdez González M. C., Leyva-Grado V. H., Evans M. J., Simon V., Lim J. K., Krammer F., Hai R., Palese P., Tan G. S., Human antibodies targeting Zika virus NS1 provide protection against disease in a mouse model. Nat. Commun. 9, 4560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Qu L., Ye X., Yi C., Zheng X., Hao M., Su W., Yao Z., Chen P., Zhang S., Feng Y., Wang Q., Yan Q., Li P., Li H., Li F., Pan W., Niu X., Xu R., Feng L., Chen L., Incorporation of NS1 and prM/M are important to confer effective protection of adenovirus-vectored Zika virus vaccine carrying E protein. NPJ Vaccines 3, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li A., Yu J., Lu M., Ma Y., Attia Z., Shan C., Xue M., Liang X., Craig K., Makadiya N., He J. J., Jennings R., Shi P.-Y., Peeples M. E., Liu S.-L., Boyaka P. N., Li J., A Zika virus vaccine expressing premembrane-envelope-NS1 polyprotein. Nat. Commun. 9, 3067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomusange K., Wijesundara D., Gummow J., Garrod T., Li Y., Gray L., Churchill M., Grubor-Bauk B., Gowans E. J., A HIV-Tat/C4-binding protein chimera encoded by a DNA vaccine is highly immunogenic and contains acute EcoHIV infection in mice. Sci. Rep. 6, 29131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Leneghan D. B., Miura K., Nikolaeva D., Brian I. J., Dicks M. D., Fyfe A. J., Zakutansky S. E., de Cassan S., Long C. A., Draper S. J., Hill A. V., Hill F., Biswas S., Enhancing immunogenicity and transmission-blocking activity of malaria vaccines by fusing Pfs25 to IMX313 multimerization technology. Sci. Rep. 6, 18848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masavuli M. G., Wijesundara D. K., Underwood A., Christiansen D., Earnest-Silveira L., Bull R., Torresi J., Gowans E. J., Grubor-Bauk B., A hepatitis C virus DNA vaccine encoding a secreted, oligomerized form of envelope proteins is highly immunogenic and elicits neutralizing antibodies in vaccinated mice. Front. Immunol. 10, 1145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spencer A. J., Hill F., Honeycutt J. D., Cottingham M. G., Bregu M., Rollier C. S., Furze J., Draper S. J., Søgaard K. C., Gilbert S. C., Wyllie D. H., Hill A. V., Fusion of the Mycobacterium tuberculosis antigen 85A to an oligomerization domain enhances its immunogenicity in both mice and non-human primates. PLOS ONE 7, e33555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H.-W., Huang H.-W., Hu H.-M., Chung H.-H., Wu S.-H., Chong P., Tao M.-H., Pan C.-H., A poorly neutralizing IgG2a/c response elicited by a DNA vaccine protects mice against Japanese encephalitis virus. J. Gen. Virol. 95, 1983–1990 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Quah B. J., Wijesundara D. K., Ranasinghe C., Parish C. R., Fluorescent target array killing assay: A multiplex cytotoxic T-cell assay to measure detailed T-cell antigen specificity and avidity in vivo. Cytometry A 81, 679–690 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Wijesundara D. K., Ranasinghe C., Jackson R. J., Lidbury B. A., Parish C. R., Quah B. J., Use of an in vivo FTA assay to assess the magnitude, functional avidity and epitope variant cross-reactivity of T cell responses following HIV-1 recombinant poxvirus vaccination. PLOS ONE 9, e105366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quah B. J., Wijesundara D. K., Ranasinghe C., Parish C. R., Fluorescent target array T helper assay: A multiplex flow cytometry assay to measure antigen-specific CD4+ T cell-mediated B cell help in vivo. J. Immunol. Methods 387, 181–190 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Wijesundara D. K., Gummow J., Li Y., Yu W., Quah B. J., Ranasinghe C., Torresi J., Gowans E. J., Grubor-Bauk B., Induction of genotype cross-reactive, hepatitis C virus-specific, cell-mediated immunity in DNA-vaccinated mice. J. Virol. 92, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown W. C., Akey D. L., Konwerski J. R., Tarrasch J. T., Skiniotis G., Kuhn R. J., Smith J. L., Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 23, 865–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilgenfeld R., Zika virus NS1, a pathogenicity factor with many faces. EMBO J. 35, 2631–2633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park C., Li S., Cha E., Schindler C., Immune response in Stat2 knockout mice. Immunity 13, 795–804 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Xu J., Lee M. H., Chakhtoura M., Green B. L., Kotredes K. P., Chain R. W., Sriram U., Gamero A. M., Gallucci S., STAT2 is required for TLR-induced murine dendritic cell activation and cross-presentation. J. Immunol. 197, 326–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tau G. Z., Cowan S. N., Weisburg J., Braunstein N. S., Rothman P. B., Regulation of IFN-γ signaling is essential for the cytotoxic activity of CD8+ T cells. J. Immunol. 167, 5574–5582 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huber J. P., Farrar J. D., Regulation of effector and memory T-cell functions by type I interferon. Immunology 132, 466–474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen L. R., Jamieson D. J., Honein M. A., Zika Virus. N. Engl. J. Med. 375, 294–295 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Singh R., Rothman A. L., Potts J., Guirakhoo F., Ennis F. A., Green S., Sequential immunization with heterologous chimeric flaviviruses induces broad-spectrum cross-reactive CD8+ T cell responses. J. Infect. Dis. 202, 223–233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trobaugh D. W., Yang L., Ennis F. A., Green S., Altered effector functions of virus-specific and virus cross-reactive CD8+ T cells in mice immunized with related flaviviruses. Eur. J. Immunol. 40, 1315–1327 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saron W. A. A., Rathore A. P. S., Ting L., Ooi E. E., Low J., Abraham S. N., St John A. L., Flavivirus serocomplex cross-reactive immunity is protective by activating heterologous memory CD4 T cells. Sci. Adv. 4, eaar4297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiskopf D., Angelo M. A., de Azeredo E. L., Sidney J., Greenbaum J. A., Fernando A. N., Broadwater A., Kolla R. V., De Silva A. D., de Silva A. M., Mattia K. A., Doranz B. J., Grey H. M., Shresta S., Peters B., Sette A., Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. U.S.A. 110, E2046–E2053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zompi S., Santich B. H., Beatty P. R., Harris E., Protection from secondary dengue virus infection in a mouse model reveals the role of serotype cross-reactive B and T cells. J. Immunol. 188, 404–416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grifoni A., Pham J., Sidney J., O’Rourke P. H., Paul S., Peters B., Martini S. R., de Silva A. D., Ricciardi M. J., Magnani D. M., Silveira C. G. T., Maestri A., Costa P. R., de-Oliveira-Pinto L. M., de Azeredo E. L., Damasco P. V., Phillips E., Mallal S., de Silva A. M., Collins M., Durbin A., Diehl S. A., Cerpas C., Balmaseda A., Kuan G., Coloma J., Harris E., Crowe J. E. Jr., Stone M., Norris P. J., Busch M., Vivanco-Cid H., Cox J., Graham B. S., Ledgerwood J. E., Turtle L., Solomon T., Kallas E. G., Watkins D. I., Weiskopf D., Sette A., Prior dengue virus exposure shapes t cell immunity to Zika virus in humans. J. Virol. 91, e01469-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hassert M., Wolf K. J., Schwetye K. E., DiPaolo R. J., Brien J. D., Pinto A. K., CD4+T cells mediate protection against Zika associated severe disease in a mouse model of infection. PLOS Pathog. 14, e1007237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elong Ngono A., Young M. P., Bunz M., Xu Z., Hattakam S., Vizcarra E., Regla-Nava J. A., Tang W. W., Yamabhai M., Wen J., Shresta S., CD4+ T cells promote humoral immunity and viral control during Zika virus infection. PLOS Pathog. 15, e1007474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lai L., Rouphael N., Xu Y., Natrajan M. S., Beck A., Hart M., Feldhammer M., Feldpausch A., Hill C., Wu H., Fairley J. K., Lankford-Turner P., Kasher N., Rago P., Hu Y. J., Edupuganti S., Patel S. M., Murray K. O., Mulligan M. J.; Emory Zika Patient Study Team , Innate, T-, and B-Cell responses in acute human Zika patients. Clin. Infect. Dis. 66, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Badolato-Corrêa J., Sánchez-Arcila J. C., Alves de Souza T. M., Santos Barbosa L., Conrado Guerra Nunes P., da Rocha Queiroz Lima M., Gandini M., Bispo de Filippis A. M., Venâncio da Cunha R., Leal de Azeredo E., de-Oliveira-Pinto L. M., Human T cell responses to dengue and Zika virus infection compared to Dengue/Zika coinfection. Immun. Inflamm. Dis. 6, 194–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elong Ngono A., Vizcarra E. A., Tang W. W., Sheets N., Joo Y., Kim K., Gorman M. J., Diamond M. S., Shresta S., Mapping and role of the CD8+ T cell response during primary Zika virus infection in mice. Cell Host Microbe 21, 35–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartsch S. M., Asti L., Cox S., Durham D. P., Randall S., Hotez P. J., Galvani A. P., Lee B. Y., What is the value of different Zika vaccination strategies to prevent and mitigate Zika outbreaks. J. Infect. Dis. 220, 920–931 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garrod T., Grubor-Bauk B., Yu S., Gargett T., Gowans E. J., Encoded novel forms of HSP70 or a cytolytic protein increase DNA vaccine potency. Hum. Vaccin. Immunother. 10, 2679–2683 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomusange K., Wijesundara D., Gummow J., Wesselingh S., Suhrbier A., Gowans E. J., Grubor-Bauk B., Mucosal vaccination with a live recombinant rhinovirus followed by intradermal DNA administration elicits potent and protective HIV-specific immune responses. Sci. Rep. 6, 36658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gargett T., Grubor-Bauk B., Garrod T. J., Yu W., Miller D., Major L., Wesselingh S., Suhrbier A., Gowans E. J., Induction of antigen-positive cell death by the expression of perforin, but not DTa, from a DNA vaccine enhances the immune response. Immunol. Cell Biol. 92, 359–367 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Garrod T. J., Grubor-Bauk B., Gargett T., Li Y., Miller D. S., Yu W., Major L., Burrell C. J., Wesselingh S., Suhrbier A., Gowans E. J., DNA vaccines encoding membrane-bound or secreted forms of heat shock protein 70 exhibit improved potency. Eur. J. Immunol. 44, 1992–2002 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Gummow J., Li Y., Yu W., Garrod T., Wijesundara D., Brennan A. J., Mullick R., Voskoboinik I., Grubor-Bauk B., Gowans E. J., A Multiantigenic DNA vaccine that induces broad hepatitis C virus-specific T-Cell responses in mice. J. Virol. 89, 7991–8002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grubor-Bauk B., Yu W., Wijesundara D., Gummow J., Garrod T., Brennan A. J., Voskoboinik I., Gowans E. J., Intradermal delivery of DNA encoding HCV NS3 and perforin elicits robust cell-mediated immunity in mice and pigs. Gene Ther. 23, 26–37 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaax2388/DC1
Fig. S1. DNA vaccine design and antigen expression.
Fig. S2. FTA analysis.
Fig. S3. Epitope confirmation.
Fig. S4. ZIKV viral load over time in T cell–depleted mice.


