Abstract
Alzheimer’s disease is a devastating neurodegenerative disorder with no cure. Countless promising therapeutics have shown efficacy in rodent Alzheimer’s disease models yet failed to benefit human patients. While hope remains that earlier intervention with existing therapeutics will improve outcomes, it is becoming increasingly clear that new approaches to understand and combat the pathophysiology of Alzheimer’s disease are needed. Human induced pluripotent stem cell (iPSC) technologies have changed the face of preclinical research and iPSC-derived cell types are being utilized to study an array of human conditions, including neurodegenerative disease. All major brain cell types can now be differentiated from iPSCs, while increasingly complex co-culture systems are being developed to facilitate neuroscience research. Many cellular functions perturbed in Alzheimer’s disease can be recapitulated using iPSC-derived cells in vitro, and co-culture platforms are beginning to yield insights into the complex interactions that occur between brain cell types during neurodegeneration. Further, iPSC-based systems and genome editing tools will be critical in understanding the roles of the numerous new genes and mutations found to modify Alzheimer’s disease risk in the past decade. While still in their relative infancy, these developing iPSC-based technologies hold considerable promise to push forward efforts to combat Alzheimer’s disease and other neurodegenerative disorders.
Subject terms: Neuroscience, Molecular biology, Cell biology, Diseases, Genetics
Introduction
Alzheimer’s disease (AD) is a devastating and ultimately fatal form of neurodegeneration characterized by progressive loss of cognition and disruption of basic functions, such as swallowing, walking, attention, and memory [1]. As the sixth leading cause of death in the USA, the disease places a tremendous emotional and financial burden on families, caregivers, and the health care system [2]. Projections show that this burden will grow as domestic and world populations continue to age [3]. AD has been the subject of intense research efforts over the past 40 years, with human genetic studies identifying numerous mutations that either cause, or alter risk for the disease. In parallel, cellular and rodent models have been workhorses in deepening our understanding of numerous pathophysiological mechanisms associated with disease progression. Despite these efforts, there remains no cure for AD. The only current FDA approved treatments for AD target cholinergic and/or glutamatergic neuronal function, providing modest and transient cognitive benefit, but do not alter disease course or underlying neurodegeneration [4, 5]. The continued failure of promising therapeutics to provide benefit in human patients points to the need for improved model systems that better mimic the pathophysiology of AD patients. The advent in the past decade of techniques to generate human induced pluripotent stem cells (iPSCs) and to differentiate them into the various cell types of the body, including brain cells, has ushered in a new era of neurodegenerative disease research that offers renewed hope in tackling an old problem. Here we provide an overview of iPSC-based models of AD that have been developed, or are being developed, as well as highlighting important research directions going forward.
Alzheimer’s disease
First described by Alois Alzheimer in 1907, AD is characterized by progressive cognitive decline, initially and particularly, affecting short-term memory, but later also language, mood, movement, and physiological functions [1]. Early onset familial AD (FAD) strikes in the fourth or fifth decade of life, while sporadic late onset AD (SAD) typically develops after the age of 70. The pathological hallmarks of AD—shared by FAD and SAD—are senile plaques and neurofibrillary tangles (NFTs), accompanied by progressive neurodegeneration [6]. Plaques are extracellular aggregates composed largely of amyloid-β (Aβ) peptides, while NFTs are intracellular inclusions rich in hyperphosphorylated tau protein. Various forms of Aβ induce cellular dysfunction and toxicity in vitro and in vivo. Likewise, mutated or hyperphosphorylated tau species are prone to aggregation and can cause neuronal dysfunction and cell death. Intriguingly, while both Aβ and tau can exhibit toxicity independently, their pathologies are linked; numerous studies show that Aβ buildup can induce tau hyperphosphorylation [7]. Other common pathological signs of AD include gliosis, inflammation, blood–brain barrier (BBB) disruptions, metabolic disturbances, altered endocytic and cellular degradation pathways, and elevated DNA damage [8]. The relative importance and interconnections between these phenomena remain subjects of intense research.
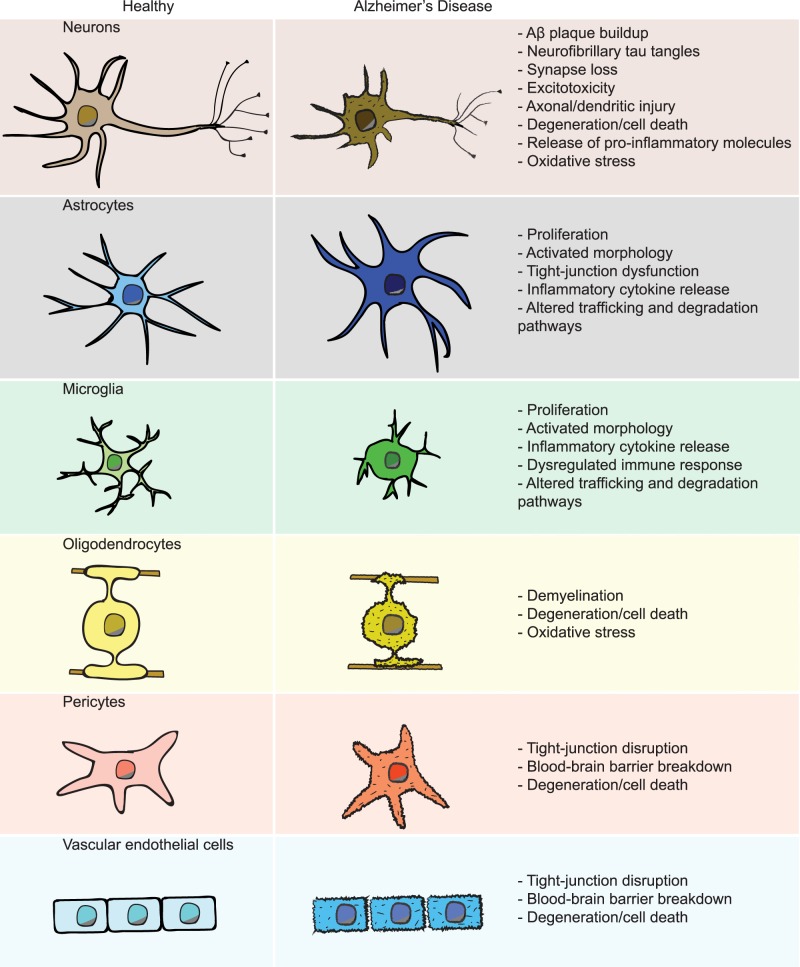
Following many years of focus on neuron-based mechanisms of neurodegeneration in AD, recent genetic studies have shifted attention to a more holistic view incorporating the functions of multiple different cell types of the brain [8, 9] (Fig. 1). Neuron dysfunction and degeneration undoubtedly underlies a large part of the characteristic cognitive decline during AD, but the brain also consists of many other non-neuronal cell types [8, 10, 11]. These cells are increasingly being recognized for helping to maintain proper brain function as well as ensuring the long-term health and survival of neurons. Oligodendrocytes insulate neuronal axons and promote fast synaptic transmission [12]. Astrocytes and microglia closely associate with synapses, the sites of communication between neurons, to form what is known as the “tripartite synapse” [13, 14]. Astrocytes, in addition to playing critical roles in neurotransmitter recycling, also perform important metabolic functions in the brain [15]. Microglia, the resident innate immune cells of the brain, are involved in the pruning of synapses, in particular during development but also in the context of adult synaptic plasticity [16]. Throughout life, microglia also act as sentinels that detect and remove exogenous and endogenous invaders and debris, including pathogens, dying brain cells, cancerous cells and protein aggregates such as Aβ plaques [17, 18]. All of the mentioned glial cell types can also contribute trophic support for other brain cells, secrete pro- or anti-inflammatory factors, and participate in clearance of toxic substances from the brain milieu [8]. Aside from glia, the cells of the BBB form a physical impediment to the passage of most cells and molecules between the central nervous system and peripheral blood circulation. These specialized blood vessels are composed of vascular endothelial cells (VECs), pericytes, and astrocytes that can break down during AD and exacerbate its progression [19]. The functions and contribution of each of these non-neuronal brain cell types to AD-associated neurodegeneration remains incompletely understood.
Fig. 1.
Brain cell types in Alzheimer’s disease. A summary of the major human brain cell types and the alterations they exhibit in AD
FAD exhibits an early onset of symptoms and is dominantly inherited, which facilitated the identification in the early 1990’s of disease-causing mutations in three different genes, encoding the amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) [20–22]. It was already known that the Aβ peptide is a major constituent of amyloid plaques, and is derived from sequential proteolytic cleavage of APP [23]. Subsequent studies revealed that PSEN1 and PSEN2 are components of the gamma secretase complex, which carries out amyloidogenic cleavage of APP to produce Aβ peptides ranging from 36 to 43 amino acids in length. Aβ40 and Aβ42 are the most common isoforms, with longer forms such as Aβ42 and Aβ43 being more aggregation prone [24]. AD-linked mutations in APP, PSEN1, and PSEN2, as well as APP duplications, all increase either the total amount of Aβ42, or the ratio of Aβ42/Aβ40 produced by neurons [25, 26].
Together with the observation that amyloid buildup is a prominent event in disease pathology, these genetic findings laid the foundation for the “amyloid cascade hypothesis” of AD, placing neuron-derived Aβ at the top of a cascade ultimately leading to neurodegeneration and cognitive decline [27]. The identification of specific mutations that can cause AD also allowed for the development of cellular and animal models to study the pathophysiological alterations that link these mutations to neurodegeneration.
Despite a less aggressive progression, SAD shares the major characteristics of FAD, including Aβ plaque deposition and NFT pathology, and accounts for the vast majority (>95%) of all AD cases [28]. While no SAD-causative mutations have been found, a considerable number of genetic loci that increase or decrease the risk for developing SAD have now been reported [29–34]. The first identified was the APOE locus, encoding apolipoprotein E (APOE) [35]. The APOE2, APOE3 and APOE4 alleles correspond to APOE with cysteine at amino acid positions 112 and 158, cysteine at 112 and arginine and 158, or arginine at 112 and 158, respectively. Compared to the major APOE3 allele, APOE2 has been reported to be protective, while APOE4 increases late-onset AD risk by ~three-fold for heterozygous carriers and 15-fold for homozygous carriers [36]. Despite its partial penetrance, the relatively high frequency of APOE4 in the general population (~13%) makes it the single largest cause of AD [37]. APOE is most studied as a lipid carrier secreted from astrocytes that facilitates Aβ clearance from the brain, however, recent studies have revealed potentially detrimental roles of APOE4 also in neurons and microglial cells [38, 39]. Genome-wide association studies (GWAS) in the last decade have identified numerous additional SAD risk genes, many of which are expressed primarily in non-neuronal cells of the brain [29–34] (Table 1). Combined with the persistent failure of AD clinical trials, largely aimed at reducing Aβ production by neurons, these recent genetic findings have begun to shift the focus of AD research toward better understanding the roles and functions of non-neuronal cells during neurodegeneration in AD. In addition, while FAD appears to be caused primarily by overproduction of Aβ, it has become clear that other mechanisms, including defective clearance or aberrant degradation of Aβ, are likely to be important drivers of many SAD cases.
Table 1.
Alzheimer’s disease risk genes
| Human Brain FPKM | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Mutation type | Molecular function | % identity | Neurons | Astrocytes | Microglia | Oligodendrocytes | Endothelial |
| APP | Coding | Integral membrane protein | 97.3 | 160.9 | 16.6 | 6.9 | 109.5 | 41.0 |
| PSEN1 | Coding | Protease, γ-secretase complex | 92.7 | 5 | 7.4 | 5.7 | 16.5 | 2.7 |
| PSEN2 | Coding | Protease, γ-secretase complex | 96 | 1.1 | 1 | 0.3 | 0.3 | 1.3 |
| ABCA7 | Both | ATP-binding cassette transporter | 76.4 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| ACE | Non-coding | Metalloprotease | 82.8 | 0.1 | 0.1 | 0.1 | 0.1 | 0.4 |
| ADAM10 | Non-coding | Metalloprotease | 95.9 | 9.1 | 7.4 | 22.6 | 15.6 | 6.3 |
| ADAMTS1 | Non-coding | Metalloprotease | 80.8 | 3.5 | 0.9 | 0.1 | 9.7 | 3.1 |
| APOE | Coding | Lipoprotein | 71.7 | 0.5 | 3.3 | 0.5 | 0.2 | 0.1 |
| BIN1 | Non-coding | Endocytic adaptor | 95.6 | 1.2 | 0.9 | 6.9 | 6.7 | 1.7 |
| CASS4 | Non-coding | Tyrosine kinase docking | 63.1 | 0.3 | 0.2 | 8.5 | 0.3 | 1.5 |
| CD2AP | Non-coding | Scaffolding, actin cytoskeleton | 86.5 | 1.4 | 1.7 | 7.9 | 1.4 | 4 |
| CD33 | Non-coding | Surface receptor | 39.0a | 0.1 | 0.2 | 9.5 | 1 | 0.1 |
| CELF1 | Non-coding | RNA-binding protein | 99.6 | 7.9 | 6.3 | 9.5 | 4.9 | 3.1 |
| CLU | Non-coding | Extracellular chaperone | 76.6 | 19.3 | 384.2 | 0.5 | 9.6 | 15.6 |
| CR1 | Non-coding | Surface receptor | 48.9 | 0.1 | 0.1 | 0.2 | 0.1 | 0.1 |
| EPHA1 | Non-coding | Receptor tyrosine kinase | 87.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| FERMT2 | Non-coding | Extracellular matrix scaffolding | 98.2 | 3.3 | 43.2 | 2.9 | 6.9 | 6.8 |
| HLA-DRB1 | Non-coding | Antigen presentation | 58.9a | 0.6 | 1.1 | 27.1 | 3 | 0.7 |
| INPP5D | Non-coding | Phosphatidylinositol phosphatase | 87.5 | 0.1 | 0.5 | 18.9 | 1.4 | 2.3 |
| IQCK | Non-coding | Calmodulin-binding domain | 71.8 | 4.7 | 20.8 | 0.2 | 6.4 | 0.6 |
| MEF2C | Non-coding | Transcription factor | 93.5 | 51.9 | 3.8 | 44 | 3.9 | 4.1 |
| MS4A6A | Non-coding | Transmembrane protein | 53.7a | 0.1 | 0.7 | 24 | 5.2 | 0.1 |
| PICALM | Non-coding | Endocytosis, clathrin assembly | 96.5 | 12.3 | 14.6 | 65.6 | 37.3 | 20.5 |
| PTK2B | Non-coding | Tyrosine kinase | 95.3 | 2 | 0.9 | 1.3 | 0.4 | 0.8 |
| SLC24A4 | Non-coding | Na+/K+/Ca2+ exchanger | 94.4 | 1.9 | 0.2 | 0.5 | 0.2 | 0.1 |
| SORL1 | Non-coding | Endocytic receptor/sorting | 93.2 | 9.9 | 17.9 | 78.9 | 6.7 | 0.8 |
| SPI1 | Non-coding | Transcription factor | 87.9 | 0.1 | 0.1 | 0.3 | 0.1 | 0.1 |
| TREM2 | Coding | Surface receptor | 50.6 | 0.1 | 0.4 | 27.1 | 1.4 | 0.4 |
| TXNDC3 | Non-coding | Thioredoxin domain | 63.5 | 0.1 | 0.1 | 0.2 | 0.2 | 0.1 |
| WWOX | Non-coding | Oxidoreductase | 93.7 | 4.6 | 5.1 | 1.1 | 1.8 | 0.8 |
| ZCWPW1 | Non-coding | Zinc finger domain | 60 | 0.2 | 0.5 | 0.5 | 0.7 | 0.1 |
FAD causative genes are shown in bold, SAD risk genes in normal font [20–22, 29–32, 34, 35]. Percent amino acid identity of mouse to human orthologue is indicated (ensembl.org [46])
aIndicates multiple orthologues. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) of AD-linked genes from purified human brain cell types are also indicated (brainrnaseq.org [226])
Alzheimer’s disease models
As the most common neurodegenerative disorder, AD has been studied intensively. Valuable contributions to our understanding of this disease have been made in numerous different systems, including post-mortem patient brain samples, human and animal cell lines, as well as invertebrate, zebrafish and rodent models of disease. Post-mortem brain samples are vital for identifying the cellular and molecular changes associated with neurodegeneration, but provide no ability to alter or intervene in the course of disease. Cultured human and rodent cells have been useful for examining the effects of Aβ and other disease-related molecules on cellular function and health, however as dividing cells they do a poor job of modeling neurodegeneration and many other age-related aspects of AD. Further, while primary brain cells can be cultured from rodents, studies using human cells have utilized mostly non-neuronal cell lines. Non-mammalian models overexpressing disease-related proteins can also serve as in vivo screening systems to identify conserved mechanisms of toxicity or protection, however, the considerable evolutionary distance between these models and humans limits their utility.
Mice have formed the backbone of AD research for a quarter century due to the powerful genetic toolkit available and comparatively close evolutionary relationship with humans [40]. Mice possess most of the same major brain regions and neurotransmitter systems as humans, and are uniquely amenable to rapid assessment of neuronal and circuit function, as well as cognitive performance.
Mouse models that successfully mimic age-dependent neurodegeneration rely mostly on neuronal overexpression of human proteins carrying FAD-causing mutations [41, 42]. For instance, 5xFAD mice use the mouse Thy1 promoter to overexpress human APP harboring three disease-associated mutations (the Florida (I716V), London (V717I), and Swedish (K670N/M671L) mutations) together with PSEN1 harboring two FAD-linked mutations (the L286V and M146L mutations) [43]. However, this need to express mutated human proteins, typically at considerably higher levels than their endogenous counterparts, in order to achieve a neurodegenerative phenotype hints at some of the limitations of mice as an AD model. In fact, across neurodegenerative diseases, mouse mutations corresponding to human disease-linked mutations rarely result in neurodegenerative phenotypes. This may be due in part to the fact that age is the single greatest risk factor for neurodegeneration and mice have much shorter lifespans than humans. There may also be intrinsic differences in the resilience of mouse and human neurons in the face of oxidative stress, pathologic protein aggregates or other perturbations, though the mechanisms are not fully understood.
In addition to the well-established FAD models, mouse models of numerous SAD genes have also been developed. At many disease-associated loci, the exact mutation(s) linked to AD risk have not been identified, necessitating examination of SAD risk gene knockout mutations [31]. As these mutations alone generally do not cause neurodegeneration, examining their effects on pathology has typically been done in the context of established FAD-mutation mouse models. This approach has identified numerous mechanisms by which SAD risk genes impact AD pathology and has broadened our appreciation of the contributions of non-neuronal cell types to brain health and neurodegeneration. Despite this progress, our understanding of the effects of SAD risk mutations, and of the exact roles that each cell type plays during neurodegeneration, remains far from complete.
In the face of the immense impact that mouse models have made to our understanding of AD mechanisms and pathophysiology, it is also important to acknowledge the potential caveats of mouse studies. While mice are much closer evolutionarily to humans than most other genetic model systems, there remain considerable differences in the functions of proteins, signaling pathways, cellular processes and the interactions between different cell types when comparing the two species [44, 45]. At the protein level, this is highlighted by examining the amino acid sequence identity of different proteins that either cause, or alter the risk for, AD (Table 1). The largely neuronally expressed FAD proteins are nearly identical between human and mouse, exhibiting greater than 90% amino acid identity. In stark contrast, the proteins encoded by a number of SAD risk genes, including the microglial cell surface proteins TREM2, CD33, CR1 and MS4A6A, are only about 50% identical between human and mouse, comparable to the difference between human and insect presenilin proteins [46]. These cross-species differences may also extend more broadly to cell types, such that mice are generally a better model for understanding neuronal phenomena than they are for processes occurring in innate immune cells such as microglia.
While the exact reasons remain uncertain, the sobering reality is that hundreds of clinical trials over more than 30 years have failed to provide an effective treatment that alters the course of neurodegeneration in AD [47]. Hope remains that earlier detection of at-risk individuals, enabling earlier intervention with existing therapeutics, will eventually yield benefit in AD [9]. However, the field has already begun to embrace new ideas—in particular a more balanced view of brain function and dysfunction, acknowledging the importance of multiple brain cell types and their complex, interconnected functions [8]. Another avenue of potentially critical importance is the emerging ability to model human disease using human cells that is becoming increasingly feasible with developing iPSC technologies.
Induced pluripotent stem cells
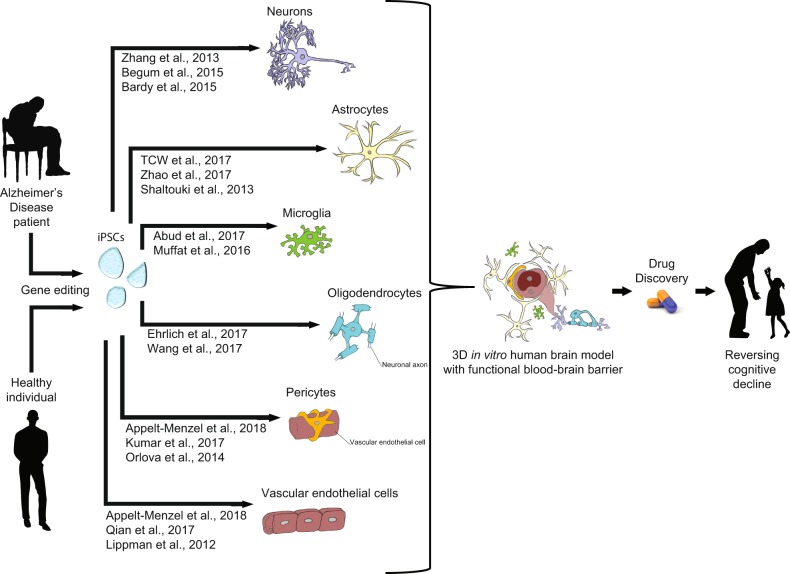
Since the first publication describing how to generate iPSCs from human somatic cells in 2007, there has followed an explosion of methods to direct differentiation into the varied cell types of the body, including brain cells [48–52]. Methods of inducing neural progenitor cells (NPCs), numerous neuron subtypes, as well as astrocytes, microglia, oligodendrocytes, endothelial cells, and pericytes have all been established (Fig. 2). Further, co-culture models incorporating multiple brain cell types have been developed as first steps towards better modeling the complex interactions that occur between these cells in vivo. In all cases, the research community is continuing to refine these methods. Differentiation protocols continue to increase the yield, purity, and maturity of brain cell types. Three dimensional (3D) and co-culture models are still developing, but even now can be utilized to generate AD hallmark pathologies that do not arise in 2D monocultures. Further improving these models to facilitate more meaningful neurodegenerative disease research will be a major goal.
Fig. 2.
Human iPSC differentiation to brain cell types. Somatic cells from patients or healthy individuals can be reprogrammed to iPSCs and subsequently differentiated into all major brain cell types for in vitro studies. Such studies can examine cellular functions as well as how they are impacted by AD hallmark pathologies or AD-linked mutations. Genome editing techniques can be used to introduce or correct AD-linked mutations to examine phenotypes in isogenic backgrounds. 3D and co-culture models allow for examination of interactions occurring between cell types and sub-types to better model processes occurring in vivo. These and developing techniques hold promise for better understanding the relevant pathomechanisms underlying AD, and will hopefully facilitate development of effective therapeutics to combat dementia
As age is the primary risk factor for AD and other neurodegenerative disorders, it may seem counterintuitive to study AD using stem cells. It is worth noting though that even in the early stages following differentiation, neurons derived from AD patient iPSCs, and from iPSCs carrying FAD mutations, generally exhibit AD-related phenotypes such as elevated Aβ production [53–55]. These early alterations presumably parallel the understudied early stages of disease progression that occur in vivo. 3D cultures derived from the same sources allow for the age-dependent accumulation of Aβ and tau aggregates on a timescale of months [56, 57]. While still far short of the timeline for development of pathology in the human brain, such cultures can be maintained for years in vitro if desired, in line with the lifespans of laboratory mouse strains [58, 59]. Importantly, unlike most other AD model systems, such cultures do not require exogenous overexpression of mutant proteins for the development of disease-relevant pathologies. Thus, while iPSC models are far from perfect, they provide a number of advantages over other systems that are likely to facilitate novel insights into AD pathomechanisms.
Initially, iPSC lines derived from healthy individuals and from patients exhibiting a given disease were isolated, differentiated into the cell type(s) of interest, and compared. A plethora of iPSC lines have been generated from early and late-onset AD patients—as well as healthy age- and sex-matched individual—and numerous repositories have been established around the world to make such lines available to researchers. Comparing cells derived from healthy vs. diseased individuals can identify cellular phenotypes associated with a disease state that may transcend individual genetic differences. Indeed, as GWAS have shown, many different genetic alterations can contribute to the development of AD, so in some cases shared cellular phenotypes can unify disparate genetic changes [8]. Generating iPSC-derived brain cells from a specific individual also has potential applications for personalized medicine and could eventually allow for identification and treatment of patient-specific alterations underlying disease [60, 61].
Genetic diversity, however, can also be a major hinderance to experimental analysis. When trying to understand the function of a specific gene or mutation for instance, additional genetic differences may mask or exacerbate a phenotype. Thus, tools for manipulating gene expression and generating mutations in iPSCs and their derivatives have been critical developments in the field, allowing for introduction or correction of specific mutations without altering the remaining genetic background. The CRISPR/Cas9 system has been revolutionary in this regard, allowing for targeted mutagenesis and base pair resolution editing of eukaryotic genomes [62–64]. While care needs to be taken to rule out off target mutations generated by CRIPSR/Cas9 mutagenesis, this technique allows for examination of the effects of targeted mutations in an otherwise identical (isogenic) genetic background. Genome editing can be used to introduce disease-associated mutations into iPSC lines from healthy individuals, or to correct mutations in cell lines from diseased patients. The nuclease-dead Cas9 (dCas9) system further allows for targeted repression or activation of gene expression in a temporally controlled and reversible manner without the need of gene editing [65, 66]. Complementing examinations of patient-derived iPSC lines, these techniques have begun to yield novel findings about the pathological mechanisms underlying AD, and promise to push forward research in the field of neurodegeneration and beyond.
Human neural progenitor cell and neuron models
As non-dividing cells, neurons face considerable challenges in maintaining their health and proper function over a lifetime that can span many decades. Indeed, the numerous neurodegenerative diseases that afflict humanity attest to these challenges and speak to the importance of better understanding both the changes that occur during aging, as well as the mechanisms that normally help to ensure neuron health and survival. iPSCs can be differentiated into NPCs, which can subsequently be patterned to different neuronal lineages [67–69]. Both passive and directed differentiation protocols having been developed for numerous different neuron subtypes, including glutamatergic, GABAergic, cholinergic and dopaminergic neurons, though existing protocols are biased strongly towards excitatory neurons [53, 68–75]. Early studies served primarily to validate the iPSC-derived models themselves and to test whether these models recapitulated findings from the large body of AD literature based on human post-mortem brain samples, rodent and other studies (Table 2 highlights select studies of AD using iPSC-derived cells). Consistently, human neuronal function and survival are compromised by treatment with exogenous Aβ [76–82]. Similarly, NPCs and neurons derived from SAD and FAD patients exhibit elevated Aβ42 production and/or Aβ42/Aβ40 ratio as well as increased tau phosphorylation [38, 53–55, 83–93]. Studies using iPSC-derived neurons also suggest neuron sub-type differences in Aβ secretion and susceptibility to Aβ-induced toxicity that vary between glutamatergic and GABAergic neurons, as well as between glutamatergic neurons expressing markers of different brain regions [79, 83, 94].
Table 2.
Select important AD studies utilizing iPSC-derived brain cells
| Model | Reference | Mutation(s) | Significance |
|---|---|---|---|
| Neurons | Israel et al. [89] | sAD, APPDp | Early study of iPSC modeling; elevated Aβ, p-tau, endosome accumulation in AD neurons |
| Shi et al. [98] | Down syndrome | Elevated Aβ secretion, Aβ aggregation, tau phosphorylation in DS neurons | |
| Wang et al. [39] | Isogenic APOE3, APOE4 and APOE null | Elevated Aβ and p-tau levels, GABAergic neuron degeneration in APOE4 neurons; identification of small molecule APOE4 structure corrector | |
| Astrocytes | Oksanen et al. [128] | Isogenic PSEN1ΔE9 | Increased Aβ production and oxidative stress, altered cytokine release and Ca2+ homeostasis, reduced neuronal support function in PSEN1 astrocytes |
| Lin et al. [38] | Isogenic APOE3 and APOE4 | Impaired Aβ clearance and increased cholesterol content of APOE4 astrocytes | |
| Microglia | Lin et al. [38] | Isogenic APOE3 and APOE4 | Reduced Aβ uptake from media and fAD organoids, reduced morphological complexity of APOE4 microglia |
| 3D cultures | Choi et al. [56] | Overexpression of APPK670N/M671L, APPV717I, PSEN1ΔE9 | Robust deposition of Aβ and filamentous tau in vitro; demonstrates that Aβ can cause tau deposition |
| Park et al. [170] | Overexpression of APPK670N/M671L, APPV717I, PSEN1ΔE9 | Triculture model system incorporating iPSC-derived neurons, astrocytes, and immortalized human microglia; recapitulates AD phenotypes, microglial recruitment, and neuroinflammation |
Numerous studies have focused on characterizing additional effects of FAD-linked mutations on induced neurons. Glutamatergic neurons expressing mutated APPV717I were used to demonstrate that this amino acid substitution alters APP subcellular distribution and cleavage by both β- and γ-secretases, underlying the perturbed Aβ production and elevated tau phosphorylation observed in these cells [92]. In contrast, β-secretase-mediated cleavage of APP, and Aβ production, were reduced by the protective APPA673T mutation [95]. Neurons carrying APP duplications exhibited more and larger early endosomes, while those harboring the APPV717I or APPK670N/M671L mutations exhibited reduced mitophagy and defects in low density lipoprotein endocytosis, indicative of functional impairment in cellular uptake, trafficking and degradation pathways [89, 90, 96]. APPE693Δ neurons further exhibit endoplasmic reticulum and oxidative stress [78]. iPSC lines from Down’s syndrome (DS) patients have also been studied as a model of AD. DS patients develop early onset AD, thought to result from triplication of the APP gene as part of trisomy 21 [97]. DS iPSC-derived neurons show elevations in Aβ secretion and phosphorylated tau similar to that caused by FAD-linked mutations [98–101]. Intriguingly, deletion of the supernumerary copy of APP from DS cells was able to restore Aβ production to control levels and correct many of the gene expression alterations caused by trisomy 21, but was not able to restore altered tau phosphorylation, indicating Aβ-dependent and independent phenotypes in DS [101].
AD-linked mutations in PSEN1 and PSEN2 have also been modeled in iPSC-derived glutamatergic neurons, revealing a wide range of phenotypes. Isogenic PSEN1ΔE9 and PSEN1null iPSC-derived neurons were used to test whether this AD-linked PSEN1 mutation acts via loss- or gain-of-function, demonstrating in fact, a gain of γ-secretase function without loss of other functions [90]. iPSC-derived neurons carrying PSEN1A246E, PSEN1V89L, and PSEN1L150P mutations, like neurons derived from SAD-patient cells, were also more sensitive to Aβ-induced toxicity and oxidative stress than cells from healthy individuals [54, 102]. Furthermore, multiple defects in cellular trafficking and degradation pathways have been documented in AD patients and animal models [103–105]. Consistently, fibroblasts and iPSC-derived neurons from multiple PSEN1-mutation carriers show defects in autophagic, ubiquitin proteasome and endo-lysosomal degradation pathways, as well as in mitophagy [55, 87, 106, 107]. Potentially arising from defective mitophagy, mitochondrial dysfunction, elevated oxidative stress and oxidative damage have also been described in PSEN1P117L- and PSEN1A246E-carrying human cells [107, 108]. Elevated apoptotic cell death and DNA damage response pathway activation was additionally observed in neurons carrying AD-linked PSEN1 mutations [55, 84]. Furthermore, NPCs carrying PSEN1S169del and PSEN1A246E mutations exhibited accelerated differentiation into glutamatergic neurons, though it remains unknown whether this affects the functional properties of mature neurons [84]. Interestingly, iPSC-derived cholinergic neurons carrying the PSEN2N141I mutation exhibited reduced excitability and impaired insulin-induced Ca2+ influx compared to isogenic controls, suggesting a functional defect consistent with that found in AD patients [53, 85, 109].
Patient-derived SAD cells often exhibit many of the same phenotypes as cells bearing FAD mutations when differentiated into neurons. In addition to elevated Aβ levels and tau phosphorylation, iPSC-derived neurons from SAD patients can exhibit enlarged endosomes, mitochondrial dysfunction, activation of ER, and oxidative stress pathways, elevated DNA damage as well as increased sensitivity to Aβ toxicity and oxidative stress [54, 78, 89, 110, 111]. Altered gene expression, early neuronal differentiation and maturation, as well as perturbed activity of the transcription factor REST have also been observed [112]. Importantly, these phenotypes are not observed in cells derived from all SAD patients, reinforcing the genetic heterogeneity of SAD [78, 89, 102, 110, 111, 113].
In some cases, the effects of SAD risk genes have also been modeled. The first identified SAD risk factor, APOE4 has been the most studied risk mutation in iPSC-derived cells, as it has been in rodent and other models. iPSC-derived neurons carrying APOE4 produce more Aβ and have higher levels of tau phosphorylation compared to APOE3 cells [35, 38, 39, 110, 114]. Surprisingly, elevated p-tau levels were not dependent on Aβ in APOE4 cells, indicating perturbation of additional pathways regulating tau phosphorylation [39]. APOE4 also induced endosome abnormalities, defects in autophagy and mitophagy, and widespread gene expression alterations in neurons [38, 96, 106, 112]. Interestingly, synaptic structure and function were also altered by the presence of APOE4, with glutamatergic neurons exhibiting more synaptic sites and increased frequency of spontaneous miniature synaptic transmission compared to isogenic APOE3 controls [38]. APOE4 GABAergic interneurons experienced degeneration in culture (though glutamatergic and dopaminergic neurons did not), while cholinergic neurons exhibited elevated sensitivity and altered Ca2+ signaling in response to glutamate toxicity [39, 110]. Intriguingly, iPSC-derived excitatory neurons lacking the SAD risk factor CLU were less sensitive to Aβ-induced toxicity, consistent with CLU single nucleotide polymorphisms being associated with reduced AD risk [31, 115]. Sullivan and colleagues took a broader approach, performing a shRNA knockdown screen of more than 50 AD candidate genes in iPSC-derived neurons, examining effects on Aβ secretion and tau phosphorylation [116]. Identifying the SAD risk factor FERMT2 as a modifier of both cellular phenotypes, the authors targeted FERMT2 with CRISPR/Cas9 mutagenesis, confirming that reducing its levels in neurons can decrease Aβ secretion and tau phosphorylation [116]. Future studies using iPSC-derived neurons will examine phenotypes associated with additional AD-risk genes, and promise to identify mechanisms underlying their roles in AD risk.
Astrocytes
The most abundant cell type in the human brain, astrocytes, provide physical, energetic, metabolic, and trophic support to neurons and other brain cells [117, 118]. They play important roles in vasomodulation, inflammation and wound healing. Astrocytes also interact closely with neurons and their synapses; astrocytic calcium signalling is regulated by various neurotransmitters and calcium waves can propagate over considerable distances via gap junctions. Astrocytes can regulate neuronal excitability and synaptic transmission via modulation of ion concentrations and by regulating the uptake and recycling of neurotransmitters such as glutamate and GABA [119, 120]. In a neurodegenerative milieu, astrocytes can proliferate and enter a reactive state that is potentially toxic to neurons [121]. Further, astrocytes are a major source of cholesterol and other lipids that are critical for many cellular functions, as well as lipoproteins such as APOE, which are thought to be important regulators of brain Aβ clearance and degradation [122].
Multiple different protocols for differentiation of iPSCs into astrocytes have been developed [123–127]. Astrocytes generated from patients carrying both FAD-linked PSEN1M146L and SAD-linked APOE4 mutations exhibited reduced morphological complexity and altered localization of marker proteins, indicating similar effects of FAD and SAD mutations [126]. PSEN1ΔE9 astrocytes showed elevated release and reduced uptake of Aβ42, altered Ca2+ homeostasis, increased reactive oxygen species production, altered cytokine release and impaired fatty acid oxidation [128, 129]. iPSC-derived astrocytes have also been shown to promote the survival, maturation and function of co-cultured human neurons, effects that can be impaired by PSEN1ΔE9 and APOE4 mutations [128, 130, 131]. Further, compared to isogenic APOE3 astrocytes, APOE4 cells showed extensive gene expression alterations, cholesterol accumulation, and reduced ability to internalize Aβ42 [38]. APOE4 astrocytes were also found to express and secrete lower levels of APOE protein, which additionally exhibited reduced lipidation, confirming earlier findings from mouse studies [38, 131]. While additional FAD and SAD-linked mutations remain to be examined, these findings indicate considerable overlap in the effects of PSEN1 and APOE mutations on human astrocyte function.
Microglia
Microglia are the resident innate immune cells of the brain. They play numerous support roles in the developing, adult and aging brain, ranging from synaptic pruning, to clearance of dying cells and other debris, to the regulation of neuroinflammation [132]. Their involvement in neurodegenerative processes has long been appreciated, but until recently microglia were generally thought to respond to disease-associated pathologies rather than to play a causative role [133]. Improved sequencing technologies in the past 10 years have allowed GWAS to identify a surprising number of microglial genes as risk factors for late onset AD, including TREM2, CD33, HLA-DRB1, INPP5D, MS4A6A, CASS4, and SPI1 [29–32]. These findings clearly implicate microglial dysfunction as a driver of neurodegeneration under certain circumstances.
Protocols for the differentiation of iPSCs into microglia have only recently become available, and will undoubtedly be utilized more heavily to examine the effects of AD-linked mutations in the coming years [134–140]. Induced microglia derived from healthy patients are capable of synaptic pruning, phagocytosis and Aβ uptake, they secrete diverse cytokines and exhibit altered gene expression in response to treatment with exogenous Aβ [134, 135]. Microglia induced from SAD-patient iPSCs were found to have altered phagocytosis and elevated release of certain cytokines following treatment with lipopolysaccharide [139]. Lin et al. compared APOE4 microglia to isogenic APOE3 controls, finding extensive gene expression alterations suggestive of a pro-inflammatory phenotype, reduced morphological complexity, as well as an impaired ability to internalize Aβ from culture media and to clear Aβ aggregates from 3D cerebral organoids [38]. Recent studies have also examined microglia derived from patients carrying non-AD-linked TREM2W50C and TREM2T66M mutations, finding reduced viability under stress conditions and impaired phagocytosis of certain substrates [138, 141]. AD-linked TREM2 mutations, as well as those of other FAD and SAD-linked genes, await examination in iPSC-derived microglia. The findings of such studies could be of particular interest given the significant differences in sequence similarity, and potentially protein function, between the human and mouse orthologues of many microglial disease genes.
Oligodendrocytes
The primary role of oligodendrocytes is generation of the myelin sheath that wraps the axons of many nerve cells, together forming the white matter of the central nervous system [12, 142]. The myelin sheath, composed primarily of lipids, electrically insulates axons to prevent ion leak and promote rapid propagation of electrical signals over distance. This insulator property contributes to fast excitatory and inhibitory synaptic transmission that underlies cognitive function and coordinated movement. Additionally, oligodendrocytes provide trophic support to neurons, can mediate inflammation and contribute to the regulation of metabolic waste in the brain [12, 142]. AD patients and mouse models exhibit loss of white matter and oligodendrocyte dysfunction early in disease progression, likely contributing to neuronal dysfunction and degeneration [143, 144]. Consistently, multiple AD risk genes play important functions in oligodendrocyte biology. Protocols for differentiation and induction of oligodendrocytes from iPSCs exist, and induction of oligodendrocytes and myelination in 3D organoid models has also recently been reported [136, 145–150]. However, these human systems have not yet been utilized to examine the effects of AD-related pathologies on myelination and oligodendrocyte function.
The blood–brain barrier
The human brain is a highly vascularized and energetically demanding organ, containing more than 600 km of blood vessels that supply oxygen, glucose, and nutrients to support brain functions, while facilitating removal of carbon dioxide and other metabolic wastes [151]. At the same time, the cerebrospinal fluid (CSF) that bathes the brain has a distinct chemical composition essential for proper neuronal function, and molecules, cells and pathogens carried by the blood can cause brain inflammation and toxicity [19]. Thus, systemic blood is separated from the brain and CSF by the BBB, a specialized structure formed of VECs, pericytes and astrocyte end-feet; this barrier restricts transit of many molecules between the two compartments [19]. BBB dysfunction and breakdown are observed in AD and multiple other neurodegenerative diseases, exacerbating not only brain inflammatory responses, but also impairing blood flow, the delivery of oxygen, glucose and nutrients, as well as the clearance of Aβ and other neurotoxic substances from the brain [152]. Further, the BBB can act as an impediment to the transit of many therapeutics into the brain, so the development of models to test the passage of potential treatments preclinically is of interest [152].
While BBB dysfunction is involved in AD pathophysiology, the exact mechanisms by which it occurs, as well as the potential roles of AD-linked mutations, remain largely unknown. In vitro BBB models have been established using rodent and human primary cells, and in recent years have begun to incorporate human iPSC-derived cells [153, 154]. iPSC-derived brain endothelial cells have been used either alone, in combination with iPSC-derived neural cells and astrocytes, or together with rodent primary pericytes and astrocytes for such models [155–159]. However, while pericyte differentiation protocols exist, no BBB model incorporating all of iPSC-derived endothelial cells, pericytes and astrocytes has yet been reported [160, 161]. The development of a functional BBB model will be of interest not only for screening potential therapeutics to better predict their entry into to the brain, but also to understand whether mutations linked to neurodegenerative disease predispose to BBB dysfunction.
Co-culture, 3D culture, and in vivo systems
It has become increasingly clear in recent years that multiple different brain cell types can contribute to AD progression [8]. Thus, examining their interactions and impacts on each other is of critical importance; indeed, the ability to do this may be the single greatest strength of AD animal models, and the greatest weakness of in vitro systems. As such, considerable efforts have been made to build models incorporating multiple iPSC-derived brain cell types. In addition, 3D neuronal models, either alone or together with other brain cell types, have allowed for a more faithful recapitulation of Aβ plaques and NFTs, AD hallmark pathologies that are rarely observed in 2D cultures [56, 57, 162]. Together, these advances are allowing the field to more and more accurately model AD using human cells in vitro.
3D tissue culture has a long history, but a landmark for studies of the nervous system was the first description of “cerebral organoids” derived from human iPSCs in 2013 [163, 164]. This approach utilizes the self-organizing properties of iPSCs and their progenitors after embedding in Matrigel, a complex mixture of extracellular matrix and other secreted proteins. The resulting organoids can exhibit regionalization and organized expression of layer- and brain region-specific markers [163]. Organoids grown from iPSCs derived from SAD, FAD or DS patients exhibit elevated Aβ production, the formation of Aβ aggregates, increased tau phosphorylation and altered early endosome markers [57, 165]. Similar 3D culture models—made from NPCs overexpressing FAD proteins with AD-linked mutations—demonstrate bonafide Aβ plaques and NFTs, and demonstrate in human cells that Aβ pathology can drive tau phosphorylation and aggregation [56]. Neurospheroids, made either from NPCs overexpressing mutant FAD proteins, or iPSCs derived from SAD patients, can also recapitulate AD phenotypes and exhibit similar proteomic changes to AD patients [166, 167].
Such neuron-based 3D culture models often also develop astrocytes during the course of development, though they exhibit a paucity of other brain cell types [38, 58, 168, 169]. To study both neuron and astrocyte function, organoids derived from isogenic APOE3 and APOE4 iPSCs were compared; age- and APOE4-dependent elevations of Aβ and phosphorylated tau were found to corresponded temporally to the appearance of astrocytes, thus indicating the critical importance of APOE to astrocytic function [38]. The same study further incorporated iPSC-derived APOE3 and APOE4 microglia into organoids derived from FAD patient cells to monitor microglial effects on amyloid pathology, finding that after 30 days of co-culture organoids with APOE3 microglia exhibited fewer Aβ puncta than those containing APOE4 microglia [38].
A human triculture model incorporating NPC-derived neurons and astrocytes, together with immortalized human microglia, has also been developed to study how interactions between these three cell types affect hallmark AD pathologies and neuroinflammation [170]. Similarly, a recent study reported an organoid model that exhibits spontaneous development of microglial cells [171]. Additional recent studies demonstrate methods to induce oligodendrocytes and myelination in organoid systems [148, 149]. Another intriguing development involves the fusion of organoids initially directed toward excitatory or inhibitory neuronal fates to model the developmental migration of inhibitory neurons and functional integration of these two key neuronal sub-types [172–174]. 3D co-culture models such as these provide an opportunity to examine the effects of AD-linked mutations present in one cell type or sub-type on phenotypes of other brain cells, as well as potential interactions that may occur between different AD-linked mutations. Ultimately such models could incorporate multiple different neuronal sub-types, together with oligodendrocytes, astrocytes and microglia, as well BBB components and vasculature to create a simplified “brain in a dish”.
An additional strategy to examine iPSC-derived brain cell phenotypes and cell-cell interactions is their incorporation into the nervous systems of living rodents. In order to avoid immunogenic rejection of the graft, host strains must lack an adaptive immune system, as in the case of the SCID and Rag2−/− mouse strains [175, 176]. Xenografting of iPSCs—transplanting human iPSCs into mice—has been performed for iPSC-derived neurons and microglia [135, 177, 178], as well as for whole 3D organoids that exhibited functional integration with in vivo neural circuits and vasculature [179]. Such iPSC “xenocultures” open a new window to study in vivo cell type specific interactions using human cells during aging and neurodegeneration.
Challenges and future directions
The development of iPSC technologies provides the attractive possibility of using differentiated human cells as platforms for drug and mutagenesis screening, reviewed by Elitt and colleagues [180]. Multiple small-scale compound screens using differentiated neuronal subtypes have already targeted disease-related pathways in AD [39, 181–184], Parkinson’s Disease [185], Huntington’s Disease [186], and frontotemporal dementia [187]. A recent effort screened over 1600 compounds for reductions of tau phosphorylation in FAD neurons, identifying numerous hits and ultimately moving forward our understanding of the biology underlying p-tau accumulation [188]. shRNA screening has also been used to test the effects of knocking down more than 50 different AD candidate genes on Aβ and p-tau levels in iPSC-derived neurons [116]. Thus far such screens have targeted primarily neuronal cells, however, human glial cells will also be utilized as screening platforms going forward. Refining high-yield 3D culture techniques to facilitate screening of multiple co-cultured brain cell types holds strong potential to become the gold-standard for future central nervous system drug development. Proof-of-principle has already been demonstrated in a landmark paper that recapitulated the pathology of Zika virus infection in 3D organoids, including a small-scale screen to alleviate its symptoms [189].
Human stem cell technologies also raise intriguing possibilities for regenerative and personalized medicines, topics that have been reviewed elsewhere [60, 61, 190, 191]. A serious safety concern for regenerative medicine, however, is genomic instability exhibited by iPSCs, which can also be an issue for experimental studies using these cells [192–197]. Uncertainty remains about whether iPSCs are actually intrinsically more unstable than other cultured cells, but regardless, care should be taken to limit passage numbers and regularly check iPSC lines used for research for the presence of genomic alterations [198]. While genomic alterations in iPSCs have been reduced by replacing retroviral expression of somatic cell-to-iPSC reprogramming factors with integration-free delivery systems, additional avenues to reduce genomic instability remain a topic of active research [192, 193, 199].
Continuing to improve iPSC differentiation protocols, expanding the repertoire of brain cell sub-types that can be generated, as well as developing more complex 3D co-culture systems to model brain development and disease remain major goals of iPSC researchers. These efforts strive to improve the ease and speed of differentiation protocols, but importantly also the quality, purity and maturity of differentiated cells. Most iPSC differentiation protocols generate heterogeneous populations of the target cell type and various precursors, either necessitating analysis of impure cultures or use of additional methods to purify cell samples such as fluorescence activated cell sorting. This can be particularly problematic for neuron sub-types as even partially mature neurons are extremely sensitive to dissociation and sorting methods. Transcription factor-induced differentiation protocols hold promise to improve both the speed and conversion rate of iPSC-derived cells and have emerged for a number of brain cell types [71, 74, 75, 145, 200].
As a new and still developing field, improving the consistency and reproducibility of iPSC-derived cell types also remain major challenges. A wide range of protocols for differentiation to brain cell types are in use by different laboratories around the world, and even within a given laboratory. Indeed, even using the same differentiation protocol, considerable variability in cellular morphology and gene expression signature can occur between sites [201], with clear potential to affect experimental findings. Furthermore, variability between different clones derived from the same parental iPSC line have also been reported [202]. As such, better standardization of differentiation techniques and growth conditions, more thorough reporting of methodologies and adoption of stringent statistical analyses should help to minimize such variability. To this end, minimal standards for quality control have been suggested, in particular for large repositories of iPSC lines, in order to reduce experimental variability [203, 204]. Such measures include regular testing for donor identity and genetic integrity, testing for microbial contaminations and a standardized nomenclature for genetic modification of iPSC lines.
The ability to generate mature iPSC-derived brain cells that properly mimic their in vivo counterparts is another significant challenge. The various cell types of the brain grow and mature together, often relying on signals from other cell types to help shape their identity. Indeed, significant differences in the transcriptomes of human and mouse glial cells arise following purification from the brain and culture in vitro [205–208]. Consistently, iPSC-derived brain cells and 3D cultures often exhibit similarity to immature cells from the human nervous system [58, 209]. A better understanding of the identity and timing of key signals will enable researchers to better recapitulate the brain milieu in monocultures in vitro. Further, co-cultures incorporating multiple brain cell types, as well as utilization of extended culture times, will help to facilitate the maturation of each cell type so they more closely resemble their counterparts in the adult human brain [58].
3D co-culture systems will be increasingly important iPSC-derived models for neurodegeneration research. In addition to aiding the maturation of brain cell types, these systems promote development of AD hallmark pathologies that are generally not found in 2D cultures [56]. They further provide a platform to better understand the complex interactions and interrelated functions of brain cell types that have been both difficult to model in vitro and difficult to disentangle in vivo. The ability to introduce a specific brain cell type, carrying a mutation of interest, into an otherwise functioning co-culture system provides a powerful tool to isolate cell type- and mutation-specific effects that could be masked otherwise. Indeed, even an analysis of cell-type specific transcriptomic alterations in the presence of an AD-linked mutation in one cell type could be highly informative. Importantly, such a system would also allow for examination of potentially synergistic interactions between different AD risk mutations, an area that remains very poorly studied, but that is likely an important factor in the development of SAD in human patients.
Despite their obvious utility, iPSC-derived 3D co-culture systems remain very much works in progress. An ideal system would include multiple neuronal sub-types, each type of glial cell as well as BBB components, however, in practice reduced systems modelling only some aspects of brain function are likely to be more tractable in many cases. Regardless, continued improvement of brain cell-type differentiation protocols, definition of optimal media and conditions for co-culture experiments as well as identification of better substrates and scaffolding matrices for 3D brain cultures all hold promise for improving these models. The introduction of vasculature, either by self-organizing properties of BBB cells, or by other means, is also of great interest. Organoids often exhibit some degree of cell death and dysfunction in their deeper layers; a functional vasculature to facilitate nutrient, oxygen and waste exchange would likely improve the health of cells in 3D culture. The use of miniature spinning “bioreactors” to provide constant movement of culture media can also circumvent some of these issues and improve the homogeneity of organoid cultures [210, 211].
The ability of iPSC-based models to mimic diseases of aging such as AD has also been questioned, in part due to the loss of age- and environment-dependent cellular and epigenetic signatures that occurs during iPSC reprogramming [212–215]. Such epigenetic de-differentiation is undesirable when attempting to recapitulate disease-related phenotypes in vitro and also limits the application of iPSCs to personalized medicine. One way to overcome this loss of aging signatures is to bypass the iPSC stage with direct reprogramming, whereby fibroblasts and certain other somatic cells can be differentiated directly into neurons or other cell types to study neurodegenerative disease [216–222]. Directly reprogrammed cells retain many age-related cellular and transcriptomic alterations, and in at least some cases are better able to model age-related disease than iPSC-derived neurons [223]. Thus far methods for direct differentiation of neurons, neural precursor cells, astrocytes and oligodendrocytes have been described [217, 218, 221, 222]. Direct reprogramming also has limitations, in particular a relatively poor reprogramming efficiency and low yield of reprogrammed cells [224], however, continued refinement of direct induction methods should allow access to additional brain cell types and better facilitate the study of brain cells with intact aging signatures.
A final frontier of in vitro brain modeling is the generation of coordinated neuronal activity. While iPSC-derived neurons and 3D models exhibit synaptic activity, and connectivity between inhibitory and excitatory neurons has been reported, they do not possess coordinated circuit functions [172–174]. AD patients and rodent models demonstrate widespread defects in circuit properties involving multiple neuronal subtypes [9]. Indeed, recent work demonstrates the potential for therapeutic benefit that can be derived by modulating higher order brain rhythms, controlled by GABAergic interneurons, in AD mouse models [225]. Examination of such circuit-based mechanisms in human iPSC-derived systems would be of great interest, but will require continued efforts to optimise differentiation protocols to generate neuronal sub-types, improve available tools to modulate and monitor neuronal cells in culture, as well as to devise strategies to integrate different neuronal sub-types together to form functional circuits.
Conclusions
In little more than a decade since the advent of human iPSC technologies we have developed the ability to generate all of the main brain cell types from pluripotent cells. Increasingly complex 3D co-culture systems are also emerging that allow us to reconstitute many of the key interactions between brain cells. These technologies have already contributed greatly to our understanding of human development and human disease, including neurodegenerative disorders such as AD. As these techniques continue to be refined to better mimic in vivo conditions, our ability to model AD using human cells will improve. The functions of SAD risk genes in iPSC-derived brain cell types remains almost completely unexplored, and even the effects of FAD-linked mutations outside of neurons are mostly uncertain. Further, our understanding of how such mutations, either alone or in in combination, affect the interactions between the different cell types of the brain is in its infancy. iPSC-derived brain cell types hold considerable promise for allowing us to answer these questions, and ultimately to identify and implement effective treatments for AD.
Acknowledgements
We would like to thank Joel Blanchard, Vishnu Dileep, Hiruy Meharena and Matheus Victor for their thoughtful comments on the manuscript. This work was supported by Swiss National Science Foundation grant P300PB_177920 to W.T.R and NIH grants RF1 AG054012, RF1 054321, R37 NS051874, Cure Alzheimer’s Fund, the Robert and Renee Belfer Family Foundation and the JBP Foundation grants to L.H.T.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, et al. National institute on aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 2012;123:1–11. doi: 10.1007/s00401-011-0910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kochanek KD, Murphy SL, Xu J, Arias E. Deaths: final data for 2017. Natl Vital Stat Rep. 2019;68. https://www.cdc.gov/nchs/data/nvsr/nvsr68/nvsr68_09-508.pdf. [PubMed]
- 3.Hurd MD, Martorell P, Delavande A, Mullen KJ, Langa KM. Monetary costs of dementia in the United States. N Engl J Med. 2013;368:1326–34. doi: 10.1056/NEJMsa1204629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qaseem A, Snow V, Cross JT, Forciea MA, Hopkins R, Shekelle P, et al. Current pharmacologic treatment of dementia: a clinical practice guideline from the American college of physicians and the American academy of family physicians. Ann Intern Med. 2008;148:370–8. doi: 10.7326/0003-4819-148-5-200803040-00008. [DOI] [PubMed] [Google Scholar]
- 5.Tan L. Efficacy and safety of donepezil, galantamine, rivastigmine, and memantine for the treatment of Alzheimer’s disease: a systematic review and meta-analysis. J Alzheimers Dis. 2014;41:615–31. doi: 10.3233/JAD-132690. [DOI] [PubMed] [Google Scholar]
- 6.Jeong S. Molecular and cellular basis of neurodegeneration in Alzheimer’s disease. Mol Cells 2017. 10.14348/molcells.2017.0096. [DOI] [PMC free article] [PubMed]
- 7.Wray S, Noble W. Linking amyloid and tau pathology in alzheimer’s disease: the role of membrane cholesterol in a -mediated tau toxicity. J Neurosci. 2009;29:9665–7. doi: 10.1523/JNEUROSCI.2234-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Strooper B, Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164:603–15. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 9.Canter RG, Penney J, Tsai LH. The road to restoring neural circuits for the treatment of Alzheimer’s disease. Nature. 2016;539:187–96. doi: 10.1038/nature20412. [DOI] [PubMed] [Google Scholar]
- 10.Palop JJ, Mucke L. Amyloid-Β-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci. 2010;13:812–8. doi: 10.1038/nn.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Bartheld CS, Bahney J, Herculano-Houzel S. The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J Comp Neurol. 2016;524:3865–95. doi: 10.1002/cne.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimou L, Simons M. Diversity of oligodendrocytes and their progenitors. Curr Opin Neurobiol. 2017;47:73–9. doi: 10.1016/j.conb.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Allen NJ, Eroglu C. Cell biology of astrocyte-synapse interactions. Neuron. 2017;96:697–708. doi: 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eroglu C, Barres BA. Regulation of synaptic connectivity by glia. Nature. 2010;468:223–31. doi: 10.1038/nature09612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bélanger M, Allaman I, Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14:724–38. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Hong S, Dissing-Olesen L, Stevens B. New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol. 2016;36:128–34. doi: 10.1016/j.conb.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–12. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 18.Hickman S, Izzy S, Sen P, Morsett L, Khoury J. Microglia in neurodegeneration. Nat Neurosci. 2018;21:1359–69. doi: 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV. Establishment and dysfunction of the blood-brain barrier. Cell. 2015;163:1064–78. doi: 10.1016/j.cell.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–6. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 21.Schellenberg GD, Bird TD, Wijsman EM, Orr HT, Anderson L, Nemens E, et al. Genetic linkage evidence for a familial Alzheimer’s disease locus on chromosome 14. Science. 1992;258:668–71. doi: 10.1126/science.1411576. [DOI] [PubMed] [Google Scholar]
- 22.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science (80-) 1995;269:973–7. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 23.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, et al. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–6. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 24.Pauwels K, Williams TL, Morris KL, Jonckheere W, Vandersteen A, Kelly G, et al. Structural basis for increased toxicity of pathological Aβ42:Aβ40 ratios in alzheimer disease. J Biol Chem. 2012;287:5650–60. doi: 10.1074/jbc.M111.264473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takami M, Nagashima Y, Sano Y, Ishihara S, Morishima-Kawashima M, Funamoto S, et al. gamma-secretase: successive tripeptide and tetrapeptide release from the transmembrane domain of beta-carboxyl terminal fragment. J Neurosci. 2009;29:13042–52. doi: 10.1523/JNEUROSCI.2362-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez MA, Klutkowski JA, Freret T, Wolfe MS. Alzheimer presenilin-1 mutations dramatically reduce trimming of long amyloid β-peptides (Aβ) by γ-secretase to increase 42-to-40-residue Aβ. J Biol Chem. 2014;289:31043–52. doi: 10.1074/jbc.M114.581165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–5. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 28.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011;7:263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2012;368:117–27. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–16. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang KL, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci. 2017;20:1052–61. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel D, Mez J, Vardarajan BN, Staley L, Chung J, Zhang X, et al. Association of rare coding mutations with alzheimer disease and other dementias among adults of european ancestry. JAMA Netw Open. 2019;2:e191350. doi: 10.1001/jamanetworkopen.2019.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science (80-) 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 36.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein e and Alzheimer disease: Risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–18. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ridge Perry G., Hoyt Kaitlyn B., Boehme Kevin, Mukherjee Shubhabrata, Crane Paul K., Haines Jonathan L., Mayeux Richard, Farrer Lindsay A., Pericak-Vance Margaret A., Schellenberg Gerard D., Kauwe John S.K., Adams Perrie M., Albert Marilyn S., Albin Roger L., Apostolova Liana G., Arnold Steven E., Asthana Sanjay, Atwood Craig S., Baldwin Clinton T., Barber Robert C., Barmada Michael M., Barnes Lisa L., Barral Sandra, Beach Thomas G., Becker James T., Beecham Gary W., Beekly Duane, Bennett David A., Bigio Eileen H., Bird Thomas D., Blacker Deborah, Boeve Bradley F., Bowen James D., Boxer Adam, Burke James R., Burns Jeffrey M., Buxbaum Joseph D., Cairns Nigel J., Cantwell Laura B., Cao Chuanhai, Carlson Chris S., Carlsson Cynthia M., Carney Regina M., Carrasquillo Minerva M., Carroll Steven L., Chui Helena C., Clark David G., Corneveaux Jason, Crane Paul K., Cribbs David H., Crocco Elizabeth A., Cruchaga Carlos, De Jager Philip L., DeCarli Charles, Demirci F. Yesim, Dick Malcolm, Dickson Dennis W., Doody Rachelle S., Duara Ranjan, Ertekin-Taner Nilufer, Evans Denis A., Faber Kelley M., Fairchild Thomas J., Fallon Kenneth B., Fardo David W., Farlow Martin R., Ferris Steven, Foroud Tatiana M., Frosch Matthew P., Galasko Douglas R., Gearing Marla, Geschwind Daniel H., Ghetti Bernardino, Gilbert John R., Goate Alison M., Graff-Radford Neill R., Green Robert C., Growdon John H., Hakonarson Hakon, Hamilton Ronald L., Hamilton-Nelson Kara L., Hardy John, Harrell Lindy E., Honig Lawrence S., Huebinger Ryan M., Huentelman Matthew J., Hulette Christine M., Hyman Bradley T., Jarvik Gail P., Jicha Gregory A., Jin Lee-Way, Jun Gyungah, Kamboh M. Ilyas, Karydas Anna, Katz Mindy J., Kauwe John S.K., Kaye Jeffrey A., Kim Ronald, Kowall Neil W., Kramer Joel H., Kukull Walter A., Kunkle Brian W., LaFerla Frank M., Lah James J., Larson Eric B., Leverenz James B., Levey Allan I., Li Ge, Lieberman Andrew P., Lin Chiao-Feng, Lipton Richard B., Lopez Oscar L., Lunetta Kathryn L., Lyketsos Constantine G., Mack Wendy J., Marson Daniel C., Martin Eden R., Martiniuk Frank, Mash Deborah C., Masliah Eliezer, McCormick Wayne C., McCurry Susan M., McDavid Andrew N., McKee Ann C., Mesulam Marsel, Miller Bruce L., Miller Carol A., Miller Joshua W., Montine Thomas J., Morris John C., Mukherjee Shubhabrata, Murrell Jill R., Myers Amanda J., Naj Adam C., O'Bryant Sid, Olichney John M., Pankratz Vernon S., Parisi Joseph E., Partch Amanda, Paulson Henry L., Perry William, Peskind Elaine, Petersen Ronald C., Pierce Aimee, Poon Wayne W., Potter Huntington, Quinn Joseph F., Raj Ashok, Raskind Murray, Reiman Eric M., Reisberg Barry, Reisch Joan S., Reitz Christiane, Ringman John M., Roberson Erik D., Rogaeva Ekaterina, Rosen Howard J., Rosenberg Roger N., Royall Donald R., Sager Mark A., Sano Mary, Saykin Andrew J., Schneider Julie A., Schneider Lon S., Seeley William W., Smith Amanda G., Sonnen Joshua A., Spina Salvatore, St George-Hyslop Peter, Stern Robert A., Swerdlow Russell H., Tanzi Rudolph E., Thornton-Wells Tricia A., Trojanowski John Q., Troncoso Juan C., Tsuang Debby W., Valladares Otto, Van Deerlin Vivianna M., Van Eldik Linda J., Vardarajan Badri N., Vinters Harry V., Vonsattel Jean Paul, Wang Li-San, Weintraub Sandra, Welsh-Bohmer Kathleen A., Wendland Jens R., Wilhelmsen Kirk C., Williamson Jennifer, Wingo Thomas S., Winslow Ashley R., Wishnek Sarah, Woltjer Randall L., Wright Clinton B., Wu Chuang-Kuo, Younkin Steven G., Yu Chang-En, Yu Lei. Assessment of the genetic variance of late-onset Alzheimer's disease. Neurobiology of Aging. 2016;41:200.e13-200.e20. doi: 10.1016/j.neurobiolaging.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron. 2018;98:1141–1154.e7. doi: 10.1016/j.neuron.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Najm R, Xu Q, Jeong DE, Walker D, Balestra ME, et al. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector article. Nat Med. 2018;24:647–57. doi: 10.1038/s41591-018-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perlman RL. Mouse models of human disease: an evolutionary perspective. Evol Med Public Heal. 2016;2016:170–6. doi: 10.1093/emph/eow014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esquerda-Canals G, Montoliu-Gaya L, Güell-Bosch J, Villegas S. Mouse models of Alzheimer’s disease. J Alzheimer’s Dis. 2017;57:1171–83. doi: 10.3233/JAD-170045. [DOI] [PubMed] [Google Scholar]
- 42.Sasaguri H, Nilsson P, Hashimoto S, Nagata K, Saito T, De Strooper B, et al. APP mouse models for Alzheimer’s disease preclinical studies. EMBO J. 2017;36:2473–87. doi: 10.15252/embj.201797397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, et al. Intraneuronal beta-Amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–40. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tompkins RG. SP0136 genomic responses in mouse models poorly mimic human inflammatory diseases. Ann Rheum Dis. 2014;73:37.3–37. [Google Scholar]
- 45.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2014;112:1167–72. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cummings J. Lessons learned from Alzheimer disease: clinical trials with negative outcomes. Clin Transl Sci. 2018;11:147–52. doi: 10.1111/cts.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaffe RB. Induction of pluripotent stem cells from adult human fibroblasts by defined factors: commentary. Obstet Gynecol Surv. 2008;63:153. [Google Scholar]
- 49.Chuah JKC, Zink D. Stem cell-derived kidney cells and organoids: recent breakthroughs and emerging applications. Biotechnol Adv. 2017;35:150–67. doi: 10.1016/j.biotechadv.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Pournasr B, Duncan SA. Modeling inborn errors of hepatic metabolism using induced pluripotent stem cells. Arterioscler Thromb Vasc Biol. 2017;37:1994–9. doi: 10.1161/ATVBAHA.117.309199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Devalla Harsha D., Passier Robert. Cardiac differentiation of pluripotent stem cells and implications for modeling the heart in health and disease. Science Translational Medicine. 2018;10(435):eaah5457. doi: 10.1126/scitranslmed.aah5457. [DOI] [PubMed] [Google Scholar]
- 52.Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18:573–84. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortiz-Virumbrales M, Moreno CL, Kruglikov I, Marazuela P, Sproul A, Jacob S et al. CRISPR/Cas9-Correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2N141I neurons. Acta Neuropathol Commun 2017;5. 10.1186/s40478-017-0475-z. [DOI] [PMC free article] [PubMed]
- 54.Ochalek A, Mihalik B, Avci HX, Chandrasekaran A, Téglási A, Bock I et al. Neurons derived from sporadic Alzheimer’s disease iPSCs reveal elevated TAU hyperphosphorylation, increased amyloid levels, and GSK3B activation. Alzheimer’s Res Ther 2017;9. 10.1186/s13195-017-0317-z. [DOI] [PMC free article] [PubMed]
- 55.Wezyk M, Szybinska A, Wojsiat J, Szczerba M, Day K, Ronnholm H, et al. Overactive BRCA1 affects Presenilin 1 in induced pluripotent stem cell-derived neurons in Alzheimer’s disease. J Alzheimer’s Dis. 2018;62:175–202. doi: 10.3233/JAD-170830. [DOI] [PubMed] [Google Scholar]
- 56.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–8. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez C, Armijo E, Bravo-Alegria J, Becerra-Calixto A, Mays CE, Soto C. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol Psychiatry. 2018;23:2363–74. doi: 10.1038/s41380-018-0229-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron. 2017;95:779–790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Speakman JR, Mitchell SE, Mazidi M. Calories or protein? The effect of dietary restriction on lifespan in rodents is explained by calories alone. Exp Gerontol. 2016;86:28–38. doi: 10.1016/j.exger.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 60.Chen IY, Matsa E, Wu JC. Induced pluripotent stem cells: at the heart of cardiovascular precision medicine. Nat Rev Cardiol. 2016;13:333–49. doi: 10.1038/nrcardio.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cota-Coronado A, Ramírez-Rodríguez PB, Padilla-Camberos E, Díaz éNstor F, Flores-Fernández JM, Ávila-Gónzalez D, et al. Implications of human induced pluripotent stem cells in metabolic disorders: from drug discovery toward precision medicine. Drug Disco Today. 2019;24:334–41. doi: 10.1016/j.drudis.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 62.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–9. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 65.Gilbert La, Larson MH, Morsut L, Liu Z, Gloria A, Torres SE, et al. NIH public access. Cell. 2014;154:442–51. [Google Scholar]
- 66.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–9. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–72. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–86. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, et al. Parkinson’s Disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–77. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Pak CH, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–98. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Begum AN, Guoynes C, Cho J, Hao J, Lutfy K, Hong Y. Rapid generation of sub-type, region-specific neurons and neural networks from human pluripotent stem cell-derived neurospheres. Stem Cell Res. 2015;15:731–41. doi: 10.1016/j.scr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9:575–8. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- 74.Sun AX, Yuan Q, Tan S, Xiao Y, Wang D, Khoo ATT, et al. Direct induction and functional maturation of forebrain GABAergic neurons from human pluripotent stem cells. Cell Rep. 2016;16:1942–53. doi: 10.1016/j.celrep.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 75.Yuan F, Chen X, Fang K-H, Wang Y, Lin M, Xu S-B et al. Induction of human somatostatin and parvalbumin neurons by expressing a single transcription factor LIM homeobox 6. Elife 2018;7. 10.7554/elife.37382. [DOI] [PMC free article] [PubMed]
- 76.Wicklund Linn, Leão Richardson N., Strömberg Anne-Marie, Mousavi Malahat, Hovatta Outi, Nordberg Agneta, Marutle Amelia. β-Amyloid 1-42 Oligomers Impair Function of Human Embryonic Stem Cell-Derived Forebrain Cholinergic Neurons. PLoS ONE. 2010;5(12):e15600. doi: 10.1371/journal.pone.0015600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu X, Lei Y, Luo J, Wang J, Zhang S, Yang XJ, et al. Prevention of β-amyloid induced toxicity in human iPS cell-derived neurons by inhibition of Cyclin-dependent kinases and associated cell cycle events. Stem Cell Res. 2013;10:213–27. doi: 10.1016/j.scr.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 78.Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013;12:487–96. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 79.Vazin T, Ball KA, Lu H, Park H, Ataeijannati Y, Head-Gordon T, et al. Efficient derivation of cortical glutamatergic neurons from human pluripotent stem cells: a model system to study neurotoxicity in Alzheimer’s disease. Neurobiol Dis. 2014;62:62–72. doi: 10.1016/j.nbd.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nieweg K, Andreyeva A, van Stegen B, Tanriöver G, Gottmann K. Alzheimer's disease-related amyloid-β induces synaptotoxicity in human iPS cell-derived neurons. Cell Death & Disease. 2015;6(4):e1709–e1709. doi: 10.1038/cddis.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hong W, Wang Z, Liu W, O’Malley TT, Jin M, Willem M, et al. Diffusible, highly bioactive oligomers represent a critical minority of soluble Aβ in Alzheimer’s disease brain. Acta Neuropathol. 2018;136:19–40. doi: 10.1007/s00401-018-1846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berry BJ, Smith AST, Long CJ, Martin CC, Hickman JJ. Physiological Aβ concentrations produce a more biomimetic representation of the Alzheimer’s Disease phenotype in iPSC derived human neurons. ACS Chem Neurosci. 2018;9:1693–701. doi: 10.1021/acschemneuro.8b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muratore CR, Zhou C, Liao M, Fernandez MA, Taylor WM, Lagomarsino VN, et al. Cell-type dependent Alzheimer’s disease phenotypes: probing the biology of selective neuronal vulnerability. Stem Cell Rep. 2017;9:1868–84. doi: 10.1016/j.stemcr.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J, Zhao H, Ma Y, Shi G, Song J, Tang Y et al. Early pathogenic event of Alzheimer's disease documented in iPSCs from patients with PSEN1 mutations. Oncotarget 2016;8. 10.18632/oncotarget.13776. [DOI] [PMC free article] [PubMed]
- 85.Moreno CL, Guardia L Della, Shnyder V, Ortiz-Virumbrales M, Kruglikov I, Zhang B et al. IPSC-derived familial Alzheimer’s PSEN2 N141I cholinergic neurons exhibit mutation-dependent molecular pathology corrected by insulin signaling. Mol Neurodegener 2018;13. 10.1186/s13024-018-0265-5. [DOI] [PMC free article] [PubMed]
- 86.Chang Kuo-Hsuan, Lee-Chen Guey-Jen, Huang Ching-Chang, Lin Jia-Li, Chen Yi-Jing, Wei Pei-Chi, Lo Yen-Shi, Yao Chin-Fa, Kuo Ming-Wei, Chen Chiung-Mei. Modeling Alzheimer’s Disease by Induced Pluripotent Stem Cells Carrying APP D678H Mutation. Molecular Neurobiology. 2018;56(6):3972–3983. doi: 10.1007/s12035-018-1336-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li L, Roh JH, Chang EH, Lee Y, Lee S, Kim M, et al. iPSC modeling of Presenilin1 mutation in Alzheimer’ s disease with cerebellar ataxia. Exp Neurobiol. 2018;27:350–64. doi: 10.5607/en.2018.27.5.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, et al. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20:4530–9. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 89.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–20. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Woodruff G, Young JE, Martinez FJ, Buen F, Gore A, Kinaga J, et al. The Presenilin-1 δE9 mutation results in reduced γ-secretase activity, but not total loss of PS1 function, in isogenic human stem cells. Cell Rep. 2013;5:974–85. doi: 10.1016/j.celrep.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sproul Andrew A., Jacob Samson, Pre Deborah, Kim Soong Ho, Nestor Michael W., Navarro-Sobrino Miriam, Santa-Maria Ismael, Zimmer Matthew, Aubry Soline, Steele John W., Kahler David J., Dranovsky Alex, Arancio Ottavio, Crary John F., Gandy Sam, Noggle Scott A. Characterization and Molecular Profiling of PSEN1 Familial Alzheimer's Disease iPSC-Derived Neural Progenitors. PLoS ONE. 2014;9(1):e84547. doi: 10.1371/journal.pone.0084547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LNP, et al. The familial alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–36. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Q, Waltz S, Woodruff G, Ouyang J, Israel MA, Herrera C, et al. Effect of potent γ-secretase modulator in human neurons derived from multiple presenilin 1-induced pluripotent stem cell mutant carriers. JAMA Neurol. 2014;71:1481–9. doi: 10.1001/jamaneurol.2014.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao M-C, Muratore CR, Gierahn TM, Sullivan SE, Srikanth P, De Jager PL, et al. Single-cell detection of secreted Aβ and sAPPα from human IPSC-derived neurons and astrocytes. J Neurosci. 2016;36:1730–46. doi: 10.1523/JNEUROSCI.2735-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maloney JA, Bainbridge T, Gustafson A, Zhang S, Kyauk R, Steiner P, et al. Molecular mechanisms of Alzheimer disease protection by the A673T allele of amyloid precursor protein. J Biol Chem. 2014;289:30990–1000. doi: 10.1074/jbc.M114.589069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat Neurosci. 2019;22:401–12. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Antonarakis SE. Down syndrome and the complexity of genome dosage imbalance. Nat Rev Genet. 2017;18:147–63. doi: 10.1038/nrg.2016.154. [DOI] [PubMed] [Google Scholar]
- 98.Shi Y., Kirwan P., Smith J., MacLean G., Orkin S. H., Livesey F. J. A Human Stem Cell Model of Early Alzheimer's Disease Pathology in Down Syndrome. Science Translational Medicine. 2012;4(124):124ra29–124ra29. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang CY, Chen SM, Lu HE, Lai SM, Lai PS, Shen PW et al. N-butylidenephthalide attenuates Alzheimer’s disease-like cytopathy in down syndrome induced pluripotent stem cell-derived neurons. Sci Rep 2015;5. 10.1038/srep08744. [DOI] [PMC free article] [PubMed]
- 100.Dashinimaev EB, Artyuhov AS, Bolshakov AP, Vorotelyak EA, Vasiliev AV. Neurons derived from induced pluripotent stem cells of patients with down syndrome reproduce early stages of Alzheimer’s disease type pathology in vitro. J Alzheimer’s Dis. 2017;56:835–47. doi: 10.3233/JAD-160945. [DOI] [PubMed] [Google Scholar]
- 101.Ovchinnikov DA, Korn O, Virshup I, Wells CA, Wolvetang EJ. The impact of APP on Alzheimer-like pathogenesis and gene expression in Down syndrome iPSC-Derived Neurons. Stem Cell Rep. 2018;11:32–42. doi: 10.1016/j.stemcr.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Armijo E, Gonzalez C, Shahnawaz M, Flores A, Davis B, Soto C. Increased susceptibility to Aβ toxicity in neuronal cultures derived from familial Alzheimer’s disease (PSEN1-A246E) induced pluripotent stem cells. Neurosci Lett. 2017;639:74–81. doi: 10.1016/j.neulet.2016.12.060. [DOI] [PubMed] [Google Scholar]
- 103.Nixon RA, Mathews PM, Cataldo AM. The neuronal endosomal-lysosomal system in Alzheimer’s disease. J Alzheimer’s Dis. 2001;3:97–107. doi: 10.3233/jad-2001-3114. [DOI] [PubMed] [Google Scholar]
- 104.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wolfe DM, Lee Jhyun, Kumar A, Lee S, Orenstein SJ, Nixon RA. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci. 2013;37:1949–61. doi: 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee JK, Jin HK, Park MH, Kim B, Lee PH, Nakauchi H, et al. Acid sphingomyelinase modulates the autophagic process by controlling lysosomal biogenesis in Alzheimer’s disease. J Exp Med. 2014;211:1551–70. doi: 10.1084/jem.20132451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martín-Maestro P, Gargini R, A. Sproul A, García E, Antón LC, Noggle S et al. Mitophagy Failure In Fibroblasts And iPSC-Derived Neurons of Alzheimer’s Disease-associated Presenilin 1 Mutation. Front Mol Neurosci 2017;10. 10.3389/fnmol.2017.00291. [DOI] [PMC free article] [PubMed]
- 108.Oka S, Leon J, Sakumi K, Ide T, Kang D, LaFerla FM et al. Human mitochondrial transcriptional factor A Breaks the mitochondria-mediated vicious cycle in Alzheimer’s disease. Sci Rep 2016;6. 10.1038/srep37889. [DOI] [PMC free article] [PubMed]
- 109.Du X, Wang X, Geng M Alzheimer’s disease hypothesis and related therapies. Transl Neurodegener 2018;7. 10.1186/s40035-018-0107-y. [DOI] [PMC free article] [PubMed]
- 110.Duan Lishu, Bhattacharyya Bula J, Belmadani Abdelhak, Pan Liuliu, Miller Richard J, Kessler John A. Stem cell derived basal forebrain cholinergic neurons from Alzheimer’s disease patients are more susceptible to cell death. Molecular Neurodegeneration. 2014;9(1):3. doi: 10.1186/1750-1326-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Birnbaum JH, Wanner D, Gietl AF, Saake A, Kündig TM, Hock C, et al. Oxidative stress and altered mitochondrial protein expression in the absence of amyloid-β and tau pathology in iPSC-derived neurons from sporadic Alzheimer’s disease patients. Stem Cell Res. 2018;27:121–30. doi: 10.1016/j.scr.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 112.Meyer K, Feldman HM, Lu T, Church GM, Tsai L, Yankner BA, et al. Article REST and neural gene network dysregulation in iPSC models of Alzheimer’s disease article REST and neural gene network dysregulation in iPSC Models of Alzheimer’s disease. Cell Rep. 2019;26:1112–1127.e9. doi: 10.1016/j.celrep.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Foveau B, Correia AS, Hébert SS, Rainone S, Potvin O, Kergoat MJ, et al. Stem cell-derived neurons as cellular models of sporadic Alzheimer’s disease. J Alzheimer’s Dis. 2019;67:893–910. doi: 10.3233/JAD-180833. [DOI] [PubMed] [Google Scholar]
- 114.Wadhwani Anil R., Affaneh Amira, Van Gulden Stephanie, Kessler John A. Neuronal apolipoprotein E4 increases cell death and phosphorylated tau release in alzheimer disease. Annals of Neurology. 2019;85(5):726–739. doi: 10.1002/ana.25455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Robbins JP, Perfect L, Ribe EM, Maresca M, Dangla-Valls A, Foster EM et al. Clusterin is required for β-amyloid toxicity in human iPSC-derived neurons. Front Neurosci 2018;12. 10.3389/fnins.2018.00504. [DOI] [PMC free article] [PubMed]
- 116.Sullivan SE, Liao M, Smith RV, White C, Lagomarsino VN, Xu J, et al. Candidate-based screening via gene modulation in human neurons and astrocytes implicates FERMT2 in A β and TAU proteostasis. Hum Mol Genet. 2019;28:718–35. doi: 10.1093/hmg/ddy376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weber B, Barros LF The astrocyte: Powerhouse and recycling center. Cold Spring Harb Perspect Biol 2015;7. 10.1101/cshperspect.a020396. [DOI] [PMC free article] [PubMed]
- 118.Molofsky AV, Deneen B. Astrocyte development: a guide for the perplexed. Glia. 2015;63:1320–9. doi: 10.1002/glia.22836. [DOI] [PubMed] [Google Scholar]
- 119.Bosworth AP, Allen NJ. The diverse actions of astrocytes during synaptic development. Curr Opin Neurobiol. 2017;47:38–43. doi: 10.1016/j.conb.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 120.Ben Haim L, Rowitch DH. Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci. 2016;18:31–41. doi: 10.1038/nrn.2016.159. [DOI] [PubMed] [Google Scholar]
- 121.Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–7. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mahley RW. Central nervous system lipoproteins. Arterioscler Thromb Vasc Biol. 2016;36:1305–15. doi: 10.1161/ATVBAHA.116.307023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Krencik R, Weick JP, Liu Y, Zhang ZJ, Zhang SC. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol. 2011;29:528–34. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Emdad L, D’Souza SL, Kothari HP, Qadeer ZA, Germano IM. Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells Dev. 2011;21:404–10. doi: 10.1089/scd.2010.0560. [DOI] [PubMed] [Google Scholar]
- 125.Shaltouki A, Peng J, Liu Q, Rao MS, Zeng X. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells. 2013;31:941–52. doi: 10.1002/stem.1334. [DOI] [PubMed] [Google Scholar]
- 126.Jones V C, Atkinson-Dell R, Verkhratsky A, Mohamet L. Aberrant iPSC-derived human astrocytes in Alzheimer's disease. Cell Death & Disease. 2017;8(3):e2696–e2696. doi: 10.1038/cddis.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.TCW J, Wang M, Pimenova AA, Bowles KR, Hartley BJ, Lacin E, et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Rep. 2017;9:600–14. doi: 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oksanen M, Petersen AJ, Naumenko N, Puttonen K, Lehtonen Š, Gubert Olivé M, et al. PSEN1 mutant iPSC-derived model reveals severe astrocyte pathology in Alzheimer’s disease. Stem Cell Rep. 2017;9:1885–97. doi: 10.1016/j.stemcr.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Konttinen H, Gureviciene I, Oksanen M, Grubman A, Loppi S, Huuskonen MT, et al. PPARβ/δ-agonist GW0742 ameliorates dysfunction in fatty acid oxidation in PSEN1ΔE9 astrocytes. Glia. 2019;67:146–59. doi: 10.1002/glia.23534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kuijlaars J, Oyelami T, Diels A, Rohrbacher J, Versweyveld S, Meneghello G et al. Sustained synchronized neuronal network activity in a human astrocyte co-culture system. Sci Rep 2016;6. 10.1038/srep36529. [DOI] [PMC free article] [PubMed]
- 131.Zhao J, Davis MD, Martens YA, Shinohara M, Graff-Radford NR, Younkin SG, et al. APOE ε4/ε4 diminishes neurotrophic function of human iPSC-derived astrocytes. Hum Mol Genet. 2017;26:2690–700. doi: 10.1093/hmg/ddx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Salter MW, Beggs S. Sublime microglia: expanding roles for the guardians of the CNS. Cell. 2014;158:15–24. doi: 10.1016/j.cell.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 133.Heppner FL, Ransohoff RM, Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci. 2015;16:358–72. doi: 10.1038/nrn3880. [DOI] [PubMed] [Google Scholar]
- 134.Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–67. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Rep. 2017;8:1516–24. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, et al. A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays A Neuronal-co-culture-specific Expression Profile And Inflammatory Response. Stem Cell Rep. 2017;8:1727–42. doi: 10.1016/j.stemcr.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brownjohn PW, Smith J, Solanki R, Lohmann E, Houlden H, Hardy J, et al. Functional Studies Of Missense TREM2 mutations in human stem cell-derived microglia. Stem Cell Rep. 2018;10:1294–307. doi: 10.1016/j.stemcr.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu M, Zhang L, Liu G, Jiang N, Zhou W, Zhang Y. Pathological changes in Alzheimer’s disease analyzed using induced pluripotent stem cell-derived human microglia-like cells. J Alzheimer’s Dis. 2019;67:357–68. doi: 10.3233/JAD-180722. [DOI] [PubMed] [Google Scholar]
- 140.McQuade A, Coburn M, Tu CH, Hasselmann J, Davtyan H, Blurton-Jones M, et al. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol Neurodegener. 2018;13:67. doi: 10.1186/s13024-018-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Garcia-Reitboeck P, Phillips A, Piers TM, Villegas-Llerena C, Butler M, Mallach A, et al. Human induced pluripotent stem cell-derived microglia-like cells harboring TREM2 missense mutations show specific deficits in phagocytosis. Cell Rep. 2018;24:2300–11. doi: 10.1016/j.celrep.2018.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Simons Mikael, Nave Klaus-Armin. Oligodendrocytes: Myelination and Axonal Support. Cold Spring Harbor Perspectives in Biology. 2015;8(1):a020479. doi: 10.1101/cshperspect.a020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bartzokis G. Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol Aging. 2011;32:1341–71. doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Desai MK, Mastrangelo MA, Ryan DA, Sudol KL, Narrow WC, Bowers WJ. Early oligodendrocyte/myelin pathology in Alzheimer’s disease mice constitutes a novel therapeutic target. Am J Pathol. 2010;177:1422–35. doi: 10.2353/ajpath.2010.100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ehrlich M, Mozafari S, Glatza M, Starost L, Velychko S, Hallmann A-L, et al. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc Natl Acad Sci USA. 2017;114:E2243–E2252. doi: 10.1073/pnas.1614412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu BY, Du ZW, Zhang SC. Differentiation of human oligodendrocytes from pluripotent stem cells. Nat Protoc. 2009;4:1614–22. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang S, Bates J, Li X, Schanz S, Chandler-Militello D, Levine C, et al. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12:252–64. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hubler Z, Allimuthu D, Bederman I, Elitt MS, Madhavan M, Allan KC, et al. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature. 2018;560:372–6. doi: 10.1038/s41586-018-0360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Madhavan M, Nevin ZS, Shick HE, Garrison E, Clarkson-Paredes C, Karl M, et al. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat Methods. 2018;15:700–6. doi: 10.1038/s41592-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci. 2019;22:484–91. doi: 10.1038/s41593-018-0316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zlokovic BV. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 152.Sweeney MD, Sagare AP, Zlokovic BV. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol. 2018;14:133–50. doi: 10.1038/nrneurol.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Osipova ED, Komleva YK, Morgun AV, Lopatina OL, Panina YA, Olovyannikova RY et al. Designing in vitro blood-brain barrier models reproducing alterations in brain aging. Front Aging Neurosci 2018;10. 10.3389/fnagi.2018.00234. [DOI] [PMC free article] [PubMed]
- 154.Miranda-Azpiazu P, Panagiotou S, Jose G, Saha S. A novel dynamic multicellular co-culture system for studying individual blood-brain barrier cell types in brain diseases and cytotoxicity testing. Sci Rep 2018;8. 10.1038/s41598-018-26480-8. [DOI] [PMC free article] [PubMed]
- 155.Mantle JL, Lee KH. A differentiating neural stem cell-derived astrocytic population mitigates the inflammatory effects of TNF-α and IL-6 in an iPSC-based blood-brain barrier model. Neurobiol Dis. 2018;119:113–20. doi: 10.1016/j.nbd.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 156.Canfield SG, Stebbins MJ, Morales BS, Asai SW, Vatine GD, Svendsen CN, et al. An isogenic blood–brain barrier model comprising brain endothelial cells, astrocytes, and neurons derived from human induced pluripotent stem cells. J Neurochem. 2017;140:874–88. doi: 10.1111/jnc.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Appelt-Menzel A, Cubukova A, Günther K, Edenhofer F, Piontek J, Krause G, et al. Establishment of a human blood-brain barrier co-culture model mimicking the neurovascular unit using induced pluri- and multipotent stem cells. Stem Cell Rep. 2017;8:894–906. doi: 10.1016/j.stemcr.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Qian Tongcheng, Maguire Shaenah E., Canfield Scott G., Bao Xiaoping, Olson William R., Shusta Eric V., Palecek Sean P. Directed differentiation of human pluripotent stem cells to blood-brain barrier endothelial cells. Science Advances. 2017;3(11):e1701679. doi: 10.1126/sciadv.1701679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lippmann ES, Azarin SM, Kay JE, Nessler RA, Wilson HK, Al-Ahmad A, et al. Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nat Biotechnol. 2012;30:783–91. doi: 10.1038/nbt.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Orlova VV, Drabsch Y, Freund C, Petrus-Reurer S, Van Den Hil FE, Muenthaisong S, et al. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler Thromb Vasc Biol. 2014;34:177–86. doi: 10.1161/ATVBAHA.113.302598. [DOI] [PubMed] [Google Scholar]
- 161.Kumar A, D’Souza SS, Moskvin OV, Toh H, Wang B, Zhang J, et al. Specification and diversification of pericytes and smooth muscle cells from mesenchymoangioblasts. Cell Rep. 2017;19:1902–16. doi: 10.1016/j.celrep.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kim YH, Choi SH, D’Avanzo C, Hebisch M, Sliwinski C, Bylykbashi E, et al. A 3D human neural cell culture system for modeling Alzheimer’s disease. Nat Protoc. 2015;10:985–1006. doi: 10.1038/nprot.2015.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hattori N. Cerebral organoids model human brain development and microcephaly. Mov Disord. 2014;29:185–185. doi: 10.1002/mds.25740. [DOI] [PubMed] [Google Scholar]
- 164.Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol. 2017;216:31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Raja Waseem K., Mungenast Alison E., Lin Yuan-Ta, Ko Tak, Abdurrob Fatema, Seo Jinsoo, Tsai Li-Huei. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLOS ONE. 2016;11(9):e0161969. doi: 10.1371/journal.pone.0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jorfi M, D’Avanzo C, Tanzi RE, Kim DY, Irimia D. Human neurospheroid arrays for in vitro studies of Alzheimer’s disease. Sci Rep. 2018;8:2450. doi: 10.1038/s41598-018-20436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen M, Lee HK, Moo L, Hanlon E, Stein T, Xia W. Common proteomic profiles of induced pluripotent stem cell-derived three-dimensional neurons and brain tissue from Alzheimer patients. J Proteom. 2018;182:21–33. doi: 10.1016/j.jprot.2018.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Duan L, Peng C-Y, Pan L, Kessler JA. Human pluripotent stem cell-derived radial glia recapitulate developmental events and provide real-time access to cortical neurons and astrocytes. Stem Cells Transl Med. 2015;4:437–47. doi: 10.5966/sctm.2014-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Yang SM, Berger DR, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Park J, Wetzel I, Marriott I, Dréau D, D’Avanzo C, Kim DY, et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci. 2018;21:941–51. doi: 10.1038/s41593-018-0175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM et al. Microglia Innately develop within cerebral organoids. Nat Commun. 2018;9:4167. [DOI] [PMC free article] [PubMed]
- 172.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–9. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–398.e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sloan SA, Andersen J, Pașca AM, Birey F, Pașca SP. Generation and assembly of human brain region–specific three-dimensional cultures. Nat Protoc. 2018;13:2062–85. doi: 10.1038/s41596-018-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bosma G, Custer R, Rosman M. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 176.Shinkai Y, Rathbun Gary, Lam KP, Oltz EM, Stewart V, Mendelsohn M, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–67. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 177.Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, et al. Rescue of fragile X Syndrome neurons by DNA methylation editing of the FMR1 gene. Cell. 2018;172:979–992.e6. doi: 10.1016/j.cell.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hasselmann J, Coburn MA, England W, Velez DXF, Shabestari SK, Tu CH, et al. Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo. Neuron. 2019;103:1–18. doi: 10.1016/j.neuron.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol. 2018;36:432–41. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Elitt MS, Barbar L, Tesar PJ. Drug screening for human genetic diseases using iPSC models. Hum Mol Genet. 2018;27:R89–R98. doi: 10.1093/hmg/ddy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Young JE, Fong LK, Frankowski H, Petsko GA, Small SA, Goldstein LSB. Stabilizing the Retromer Complex in a Human Stem Cell Model of Alzheimer’s Disease Reduces TAU Phosphorylation Independently of Amyloid Precursor Protein. Stem Cell Rep. 2018;10:1046–58. doi: 10.1016/j.stemcr.2018.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kondo T, Imamura K, Funayama M, Tsukita K, Miyake M, Ohta A, et al. iPSC-based compound screening and in vitro trials identify a synergistic anti-amyloid β combination for Alzheimer’s disease. Cell Rep. 2017;21:2304–12. doi: 10.1016/j.celrep.2017.10.109. [DOI] [PubMed] [Google Scholar]
- 183.Kimura J, Shimizu K, Kajima K, Yokosuka A, Mimaki Y, Oku N, et al. Nobiletin reduces intracellular and extracellular β-amyloid in iPS cell-derived Alzheimer’s disease model neurons. Biol Pharm Bull. 2018;41:451–7. doi: 10.1248/bpb.b17-00364. [DOI] [PubMed] [Google Scholar]
- 184.Brownjohn PW, Smith J, Portelius E, Serneels L, Kvartsberg H, De Strooper B, et al. Phenotypic screening identifies modulators of amyloid precursor protein processing in human stem cell models of Alzheimer’s disease. Stem Cell Rep. 2017;8:870–82. doi: 10.1016/j.stemcr.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Burbulla LF, Song P, Mazzulli JR, Zampese E, Wong YC, Jeon S, et al. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science (80-) 2017;357:1255–61. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dickey AS, Sanchez DN, Arreola M, Sampat KR, Fan W, Arbez N, et al. PPARδ activation by bexarotene promotes neuroprotection by restoring bioenergetic and quality control homeostasis. Sci Transl Med. 2017;9:eaal2332. doi: 10.1126/scitranslmed.aal2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Simone R, Balendra R, Moens TG, Preza E, Wilson KM, Heslegrave A, et al. G‐quadruplex‐binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol Med. 2017;10:22–31. doi: 10.15252/emmm.201707850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.van der Kant R, Langness VF, Herrera CM, Wagner SL, Bang AG, Goldstein LSB, et al. Cholesterol metabolism is a druggable axis that independently regulates tau and amyloid-B in iPSC-derived Alzheimer’ s disease neurons. Cell Stem Cell. 2019;24:363–375.e9. doi: 10.1016/j.stem.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Watanabe M, Buth JE, Vishlaghi N, de la Torre-Ubieta L, Taxidis J, Khakh BS, et al. Self-organized cerebral organoids with human-specific features predict effective drugs to combat zika virus infection. Cell Rep. 2017;21:517–32. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sproul AA. Being human: the role of pluripotent stem cells in regenerative medicine and humanizing Alzheimer’s disease models. Mol Asp Med. 2015;43–44:54–65. doi: 10.1016/j.mam.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 191.Chen YC, Tsai KL, Hung CW, Ding DC, Chen LH, Chang YL, et al. Induced pluripotent stem cells and regenerative medicine. J Clin Gerontol Geriatr. 2011;2:1–6. [Google Scholar]
- 192.Yoshihara M, Hayashizaki Y, Murakawa Y. Genomic instability of iPSCs: challenges towards their clinical applications. Stem Cell Rev Rep. 2017;13:7–16. doi: 10.1007/s12015-016-9680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rebuzzini P, Zuccotti M, Redi CA, Garagna S. Achilles’ heel of pluripotent stem cells: genetic, genomic and epigenetic variations during prolonged culture. Cell Mol Life Sci. 2016;73:2453–66. doi: 10.1007/s00018-016-2171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ji J, Ng S, Sharma V, Neculai D, Hussein S, Sam M, et al. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–40. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- 195.Sugiura M, Kasama Y, Araki R, Hoki Y, Sunayama M, Uda M, et al. Induced pluripotent stem cell generation-associated point mutations arise during the initial stages of the conversion of these cells. Stem Cell Rep. 2014;2:52–63. doi: 10.1016/j.stemcr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Taapken SM, Nisler BS, Newton MA, Sampsell-Barron TL, Leonhard KA, McIntire EM, et al. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–4. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- 197.Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–31. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 198.Kwon EM, Connelly JP, Hansen NF, Donovan FX, Winkler T, Davis BW, et al. iPSCs and fibroblast subclones from the same fibroblast population contain comparable levels of sequence variations. Proc Natl Acad Sci. 2017;114:1964–9. doi: 10.1073/pnas.1616035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhang M, Wang L, An K, Cai J, Li G, Yang C et al. Lower genomic stability of induced pluripotent stem cells reflects increased non-homologous end joining. Cancer Commun 2018;38. 10.1186/s40880-018-0313-0. [DOI] [PMC free article] [PubMed]
- 200.Canals I, Hidalgo I, Ginisty A, Miskinyte G, Timmerman R, Bryder D, et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods. 2018;16:134–134. doi: 10.1038/s41592-018-0264-z. [DOI] [PubMed] [Google Scholar]
- 201.Volpato V, Smith J, Sandor C, Ried JS, Baud A, Handel A, et al. Reproducibility of molecular phenotypes after long-term differentiation to human iPSC-derived neurons: A Multi-Site Omics Study. Stem Cell Rep. 2018;11:897–911. doi: 10.1016/j.stemcr.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mills JA, Wang K, Paluru P, Ying L, Lu L, Galvão AM, et al. Clonal genetic and hematopoietic heterogeneity among human-induced pluripotent stem cell lines. Blood. 2013;122:2047–51. doi: 10.1182/blood-2013-02-484444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Assou S, J. Bouckenheimer JDV. Concise review: deciphering the mechanism behind induced pluripotent stem cell generation. Stem Cells. 2018;36:814–21. doi: 10.1002/stem.2797. [DOI] [PubMed] [Google Scholar]
- 204.Sullivan S, Stacey GN, Akazawa C, Aoyama N, Baptista R, Bedford P, et al. Quality control guidelines for clinical-grade human induced pluripotent stem cell lines. Regen Med. 2018;13:859–66. doi: 10.2217/rme-2018-0095. [DOI] [PubMed] [Google Scholar]
- 205.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–40. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. An environment-dependent transcriptional network specifies human microglia identity. Science (80-) 2017;356:1248–59. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB, Barres BA. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron. 2017;94:759–773.e8. doi: 10.1016/j.neuron.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci USA. 2015;112:15672–7. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–54. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming GL. Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nat Protoc. 2018;13:565–80. doi: 10.1038/nprot.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Baxter M, Withey S, Harrison S, Segeritz CP, Zhang F, Atkinson-Dell R, et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J Hepatol. 2015;62:581–9. doi: 10.1016/j.jhep.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ravaioli Francesco, Bacalini Maria, Franceschi Claudio, Garagnani Paolo. Age-Related Epigenetic Derangement upon Reprogramming and Differentiation of Cells from the Elderly. Genes. 2018;9(1):39. doi: 10.3390/genes9010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Roessler R, Smallwood SA, Veenvliet JV, Pechlivanoglou P, Peng SP, Chakrabarty K, et al. Detailed analysis of the genetic and epigenetic signatures of iPSCs-derived mesodiencephalic dopaminergic neurons. Stem Cell Rep. 2014;2:520–33. doi: 10.1016/j.stemcr.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–54. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 216.Mertens J, Paquola ACM, Ku M, Hatch E, Böhnke L, Ladjevardi S, et al. Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell. 2015;17:705–18. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–7. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 218.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–3. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Huh CJ, Zhang B, Victor MB, Dahiya S, Batista LF, Horvath S, et al. Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts. eLife. 2016;5:1–14. doi: 10.7554/eLife.18648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng PY, et al. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84:311–23. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Corti S, Nizzardo M, Simone C, Falcone M, Donadoni C, Salani S, et al. Direct reprogramming of human astrocytes into neural stem cells and neurons. Exp Cell Res. 2012;318:1528–41. doi: 10.1016/j.yexcr.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, et al. Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol. 2013;31:434–40. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Victor MB, Richner M, Olsen HE, Lee SW, Monteys AM, Ma C, et al. Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat Neurosci. 2018;21:341–52. doi: 10.1038/s41593-018-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mertens J, Reid D, Lau S, Kim Y, Gage FH. Aging in a dish: iPSC-derived and directly induced neurons for studying brain aging and age-related neurodegenerative diseases. Annu Rev Genet. 2018;52:271–93. doi: 10.1146/annurev-genet-120417-031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Iaccarino HF, Singer AC, Martorell AJ, Rudenko A, Gao F, Gillingham TZ, et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature. 2016;540:230–5. doi: 10.1038/nature20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]