To the Editor
Bronchiolitis is the leading cause of infant hospitalizations in the US (1). Two distinct viruses – respiratory syncytial virus (RSV) and rhinovirus (RV) – account for most cases of severe bronchiolitis (i.e, bronchiolitis requiring hospitalization) (1). Although bronchiolitis is traditionally discussed as a single entity and currently-available treatment does not vary by viral etiology (2), emerging evidence suggests heterogeneity in the pathobiology of bronchiolitis by infecting virus. For instance, epidemiological investigations have shown that children with RSV bronchiolitis have different demographics, higher acute severity, and lower risk of developing childhood asthma compared to RV bronchiolitis (3,4). Studies have further demonstrated multiple clinical phenotypes of bronchiolitis (5), with between-virus (RSV vs. RV) differences in upper airway metabolomic profiles and bacterial colonization (6). However, it remains unclear whether the host systemic (i.e., serum) response differs by infecting virus. To address this knowledge gap, we examined the difference between RSV and RV infections using metabolomic profiling (i.e., comprehensive characterization of small-molecule metabolites that represent the functional activity of the host) to serum samples of infants with severe bronchiolitis.
We analyzed data from the 35th Multicenter Airway Research Collaboration (MARC-35), an ongoing, multicenter, prospective cohort study of infants with severe bronchiolitis (7). The study design, setting, participants, and methods of data collection have been reported previously (8). Briefly, MARC-35 enrolled infants (age <12 months) hospitalized with attending physician diagnosis of bronchiolitis at 17 sites across 14 US states during 2011–2014 winter seasons. Bronchiolitis was defined by the American Academy of Pediatrics guidelines – acute respiratory illness with some combination of rhinitis, cough, tachypnea, wheezing, crackles, and retractions (2). We excluded infants with previous enrolment, those who were transferred to a participating hospital >24 hours after the original hospitalization, those who were consented >24 hours after hospitalization, or those with known heart-lung disease, immunodeficiency, immunosuppression, or gestational age <32 weeks. The institutional review board at each of the participating hospitals approved the study. Written informed consent was obtained from the parent or guardian.
Metabolomic profiling of serum samples collected within 24 hours of hospitalization was performed with ultra-high performance liquid chromatography-tandem mass spectrometry, as detailed previously (9). To examine the between-virus difference in the overall metabolomic profile, orthogonal partial least squares-discriminatory analysis (OPLS-DA) with 2000 permutations was performed. To determine individual discriminatory metabolites, linear random-effects models adjusting for age, sex, race/ethnicity, prematurity, history of systemic corticosteroid use, and potential patient clustering at hospital-level were constructed, with the use of Benjamini-Hochberg false discovery rate (FDR). We did not adjust for acute severity markers (e.g., mechanical ventilation use) as these were considered intermediates in the association of interest. The analysis was performed using R version 3.5 (10) and MetaboAnalyst 3.0 (11).
The current investigation analyzed 116 infants with bronchiolitis who had, by design, over-representation of rhinovirus infection – 79 with RSV-only (68%) and 37 with RV-only (32%). Overall, the median age was 3.0 months (IQR 1.2–5.7 months), 62% were male, and 38% were non-Hispanic white. Most patient characteristic did not differ (P>0.20), including sex, race, maternal smoking status, smoke exposure, eczema, gestational age, receipt of breast milk, and antibiotic use. Infants with RSV-only bronchiolitis were younger than those with RV-only (P=0.002).
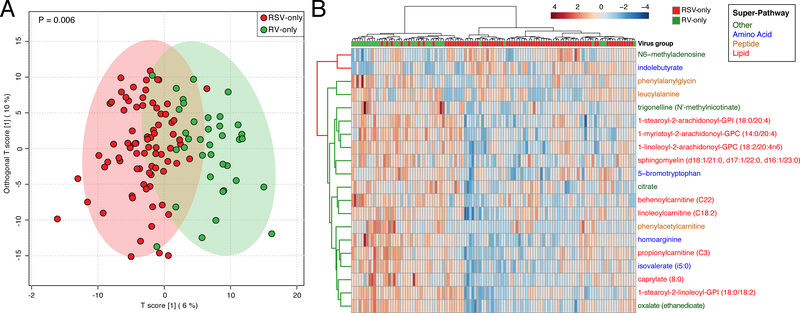
Serum metabolomic testing identified a total of 708 metabolites from 91 sub-pathways within 8 super-pathways. In OPLS-DA, the overall metabolomic profile of infants with RSV-only and that of RV-only clustered distinctly (Ppermutation=0.006; Figure 1A). Of the 708 identified metabolites, 151 metabolites had significantly different intensities between groups after adjusting for potential confounders and patient clustering: 21 metabolites were higher in the RSV-only group, while 130 metabolites were higher in the RV-only group (FDR<0.05).
Figure 1. Association of respiratory syncytial virus (RSV)-only and rhinovirus (RV)-only bronchiolitis with serum metabolomic profile.
Data based on serum metabolites among infants with RSV-only (n=79) compared to those with RV-only (n=37) infection.
(A) Orthogonal partial least squares–discriminatory analysis (OPLS-DA) score scatter plot. Each dot is the representation of the serum metabolomic profile of a single infant in a two-dimensional space. The ellipses represent the 95% confidence interval. The overall serum metabolomic profiles of infants with RSV-only and that of RV-only clustered distinctly (P=0.006 with 2000 random permutations).
(B) Unsupervised hierarchical clustering of the 20 most significant metabolites using Euclidean distance with Ward linkage. Infants are shown in columns. Metabolites are shown in rows with the color representing the log-transformed intensity of metabolites, where blue is low and red is high. The row dendrogram is colored according to the metabolite being increased in RSV-only (red) or RV-only (green) and metabolite names are colored according to their super-pathway.
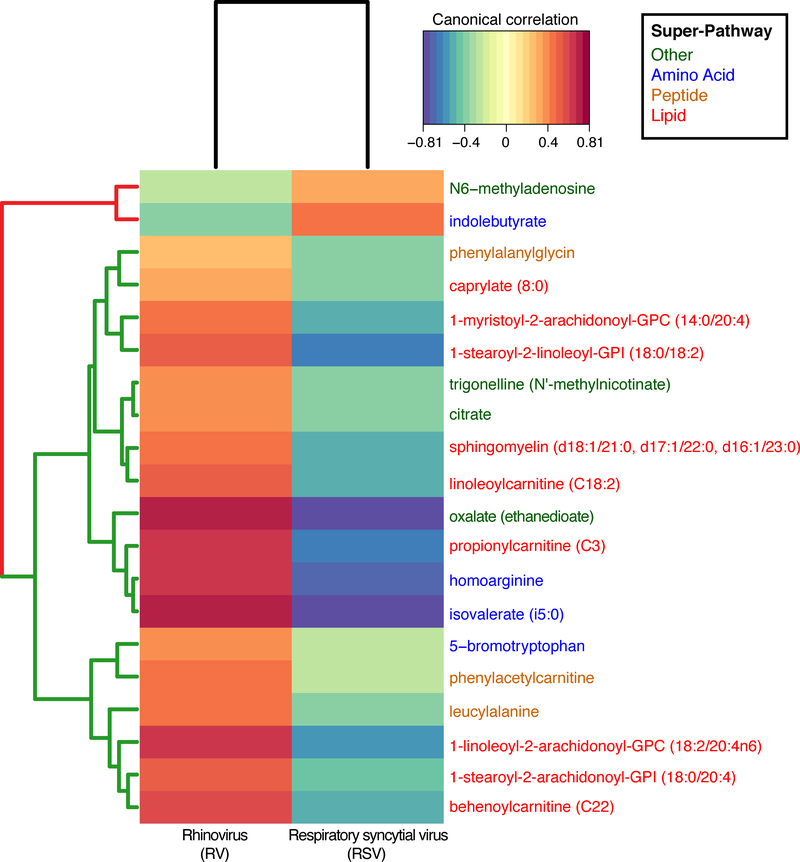
Lipids accounted for 76% (16/21) of metabolites that were significantly enriched in RSV-only, but 35% (45/130) of metabolites significantly enriched in RV-only (P<0.001). In the RSV-only group, palmitoleate (16:1n7) (long-chain fatty acid sub-pathway) was the most significantly enriched lipid. In the RV-only group, 1-stearoyl-2-arachidonoyl-GPI (18:0/20:4) (phosphatidylinositol sub-pathway) was the most significantly enriched lipid. By contrast, amino acid metabolites accounted for only 10% (2/21) of metabolites significantly enriched in RSV-only, while they accounted for 34% (44/130) of metabolites enriched in RV-only (P=0.04). For example, within the amino acid super-pathway, the RV-only group had significantly enriched N-acetyl metabolites – N-acetylarginine, N-acetylglutamate, N-acetyltaurine, N-acetylvaline. and N-acetylleucine (all FDR<0.05). Similar to the OPLS-DA, the unsupervised clustering of 20 most significant metabolites separated infants with RSV-only bronchiolitis from those with RV-only bronchiolitis. (Figure 1B). These differences between the qualitative virus status were also supported by the correlations between the quantitative viral differences (i.e., virus genomic load based on inverse cycle threshold values) and significant metabolites (Figure 2).
Figure 2. Correlations of the viral genomic load (measured as the inverse cycle threshold value) with 20 most significant metabolites.
Data based on serum metabolomics among infants with respiratory syncytial virus (RSV)-only (n=79) compared to those with rhinovirus (RV)-only (n=37) bronchiolitis. Only the 20 most significant metabolites are shown. Canonical correlations are based on sparse partial least squares (sPLS) analysis of the metabolite intensity with the inverse viral genomic load. The row dendrogram is colored according to the metabolite being increased in RSV-only (red) or RV-only (green); the metabolite names are colored according to their super-pathway.
These observations lend additional support to the emerging concept that there is heterogeneity in bronchiolitis pathogenesis (3–7), at least, by two major respiratory viruses (RSV and RV). We found that the overall serum metabolomic profiles and a large proportion of individual metabolites differed significantly between the viral groups. Consistent with the current study, we previously demonstrated relationships between respiratory viruses, different metabolic pathways in the nasopharyngeal airway, and disease severity in infants with severe bronchiolitis (6,8). These investigations of airway metabolomics demonstrated not only the associations of lipid metabolites with higher disease severity, but also the associations of RV infection with N-acetyl amino acid metabolites. While serum data most likely reflects the host responding to environmental stimuli systemically, whether local and/or circulating metabolites can alter virus infectivity merits further investigation. Our data corroborate earlier studies that showed the between-virus differences in host local response and molecular mediators in nasopharyngeal aspirates (12,13), and extend them by demonstrating the virus-specific metabolomic signatures reflecting the differences in host systemic response.
different metabolomic profiles are an important determinant of the susceptibility of infection and/or different respiratory virus. Alternatively, a metabolomic profile could simply be a marker of an infant who is prone to specific virus infection. Additionally, reverse causation – i.e., infection to a specific virus and subsequent host response perturbate metabolic states – is also possible. This study has potential limitations. First, the analytic cohort was relatively small and these novel findings will need to be validated in independent datasets. Second, the cross-sectional nature of the study design precludes temporal analysis and additional experimentation in model systems will be necessary to determine cause or effect. Third, we examined the relationships of respiratory virus with serum metabolome data while bronchiolitis involves a complex interplay among viral, microbiome, and host. Additional datasets (e.g., metagenomics, host genomics, transcriptomics) would gain a more holistic understanding of disease mechanism. Lastly, although our study sample was racially/ethnically- and geographically-diverse, all were hospitalized for bronchiolitis. Therefore, inferences should be made cautiously to children with mild-to- moderate bronchiolitis. Regardless, our findings remain directly relevant to 130,000 hospitalized American children each year (1).
In summary, the current multicenter analysis demonstrated, for the first time, that the virus infecting the airways of infants with severe bronchiolitis is significantly associated with different serum metabolomic signatures. In conjunction with prior studies, our findings suggest that the pathobiology of bronchiolitis differs between RSV and RV infection, which has direct implications for the development of targeted treatment strategies. Our data should advance research into the complex interplay between respiratory viruses and host response and their contributions to bronchiolitis morbidities.
Acknowledgments
Financial support: This work was supported by the grants U01 AI-087881, R01 AI-114552, R01 AI-108588, R01 AI-134940, and UG3/UH3 OD-023253 from the National Institutes of Health (Bethesda, MD). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest: The authors have no financial relationships relevant to this article to disclose.
REFERENCES
- 1.Hasegawa K, Mansbach JM, Camargo CA. Infectious pathogens and bronchiolitis outcomes. Expert Rev Anti Infect Ther 2014: 12:817–828. [DOI] [PubMed] [Google Scholar]
- 2.Ralston SL, Lieberthal AS, Meissner HC et al. Clinical Practice Guideline: The Diagnosis, Management, and Prevention of Bronchiolitis. Pediatrics 2014: 134:e1474–e1502. [DOI] [PubMed] [Google Scholar]
- 3.Jackson DJ, Gangnon RE, Evans MD et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med 2008: 178:667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubner FJ, Jackson DJ, Evans MD et al. Early life rhinovirus wheezing, allergic sensitization, and asthma risk at adolescence. J Allergy Clin Immunol 2017: 139:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumas O, Mansbach JM, Jartti T et al. A clustering approach to identify severe bronchiolitis profiles in children. Thorax 2016: 71:712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart CJ, Hasegawa K, Wong MC et al. Respiratory Syncytial Virus and Rhinovirus Bronchiolitis Are Associated with Distinct Metabolic Pathways. J Infect Dis 2018: 217:1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansbach JM, Hasegawa K, Henke DM et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol 2016: 137:1909–1913e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart CJ, Mansbach JM, Wong MC et al. Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis: A multiomic analysis. Am J Respir Crit Care Med 2017: 196:882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart CJ, Ajami NJ, Mansbach JM et al. Serum Metabolome Is Associated With the Nasopharyngeal Microbiota and Disease Severity Among Infants With Bronchiolitis. J Infect Dis Published Online First: 2019. doi: 10.1093/infdis/jiz021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Team RC. R: A language and environment for statistical computing. 2014http://www.r-project.org/
- 11.Xia J, Sinelnikov IV, Han B et al. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res 2015: 43:W251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Steenhuijsen Piters WAA, Heinonen S, Hasrat R et al. Nasopharyngeal Microbiota, Host Transcriptome, and Disease Severity in Children with Respiratory Syncytial Virus Infection. Am J Respir Crit Care Med 2016: 194:1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assumpção Neto E, Durigon TS, Oliveira-Junior MC et al. New serum and nasal immunological biomarkers in pediatric infection caused by respiratory syncytial virus In: 7.4 Paediatric Respiratory Infection and Immunology. European Respiratory Society; 2016: PA1277. [Google Scholar]