Abstract
Oropouche virus (OROV) causes an acute, systemic febrile illness, and in certain regions of South America, this represents the second most common human arboviral infection after dengue virus. A new real-time RT-PCR was developed for OROV and reassortant species. The new OROV rRT-PCR proved linear across 6–7 orders of magnitude with a lower limit of 95% detection of 5.6–10.8 copies/µL. Upon testing dilutions of OROV and Iquitos virus reference genomic RNA, all dilutions with >10 copies/µL were detected in both the OROV rRT-PCR and a comparator molecular assay, but the OROV rRT-PCR detected more samples with ≤10 copies/µL (8/14 vs 0/13, respectively, p = 0.002). In a set of 100 acute-phase clinical samples from Paraguay patients with a suspected arboviral illness, no patients tested positive for OROV RNA using either assay. The OROV rRT-PCR provides a sensitive molecular assay for the study of this important yet neglected tropical arboviral infection.
Keywords: Oropouche virus, Orthobunyavirus, Reverse Transcriptase PCR, Quantitative Real-Time PCR
1. Introduction
Oropouche virus (OROV) is a member of the genus Orthobunyavirus (family Peribunyaviridae) that is transmitted predominantly by the biting midge, Culicoides paraenesis (Pinheiro, et al., 1981; Pinheiro, et al., 1982; Romero-Alvarez & Escobar, 2018; Travassos da Rosa, et al., 2017). Human infections with OROV have been documented in Brazil, Peru, Ecuador, Panama, and Trinidad and Tobago, where the virus was first identified (Alva-Urcia, et al., 2017; Romero-Alvarez & Escobar, 2018; Wise, et al., 2018). In certain regions of the Amazon basin, OROV represents the second most-common human arboviral infection, following dengue virus (DENV) (Travassos da Rosa, et al., 2017). A recent single-season study from eastern Peru documented more acute OROV infections than DENV, and these were second only to chikungunya virus (CHIKV) among patients with a confirmed arboviral infection (Alva-Urcia, et al., 2017).
Human infections with OROV and certain reassortant species may result in Oropouche fever, with one estimate that 63% of infections are symptomatic (Aguilar, et al., 2011; Ladner, et al., 2014; Romero-Alvarez & Escobar, 2018). Patients typically present with fever, headache, myalgias and arthralgias (Romero-Alvarez & Escobar, 2018; Silva-Caso, et al., 2019; Wise, et al., 2018). This non-specific syndrome cannot be clinically differentiated from other common febrile illnesses in regions of endemicity, such as dengue, chikungunya, Zika, and Mayaro, among others. Therefore, the diagnosis of OROV infections requires a high index of suspicion and access to accurate diagnostic methods. Confirmation of the diagnosis is important as a different management approach may be warranted, given the relatively mild and self-limited course of Oropouche fever, and misdiagnosis could result in the inefficient use of public health resources aimed at controlling mosquito-borne viruses.
Orthobunyaviruses are negative-sense, segmented RNA viruses, and each viral genome consists of a small (S), medium (M), and large (L) segment. As a result, reassortant viral species can emerge containing new combinations of genome segments. Three OROV reassortants have been identified to date: Madre de Dios virus (MDDV), Iquitos virus (IQTV), and Perdões virus (PERDV) (Aguilar, et al., 2011; Ladner, et al., 2014; Tilston-Lunel, et al., 2015). These viruses share the OROV S and L segments but contain different M segments. MDDV and IQTV have been associated with human disease (Aguilar, et al., 2011; Ladner, et al., 2014). Infections with OROV and these reassortant species are most commonly diagnosed using molecular methods (Bastos, et al., 2014; Cardoso, et al., 2015; Moreli, et al., 2002; Naveca, et al., 2017; Weidmann, et al., 2003), though serological testing and viral culture can also confirm the diagnosis (Figueiredo & Da Rosa, 1988; Navarro, et al., 2016; Saeed, et al., 2001a; Wise, et al., 2018). A small number of molecular assays have been published, but method comparisons between the diagnostics have not been performed (Moreli, et al., 2002; Naveca, et al., 2017). Assays most commonly target the S segment, though a set of hemi-nested RT-PCRs targeting the M segment have been published for the differentiation of reassortants (Nunes, et al., 2019).
OROV epidemiology has not been well studied outside of Brazil and portions of eastern Peru (Romero-Alvarez & Escobar, 2018). Paraguay reports high numbers of dengue cases annually (Pan American Health Organization, 2018), though even in dedicated arboviral studies, 50% of acute suspected dengue cases test negative for DENV (Cardozo, et al., 2017). OROV may account for a portion of these cases, but to date this has not been studied.
The objectives of the current study, therefore, were to develop and evaluate a new rRT-PCR for the detection of OROV and reassortant species that would work in tandem with current laboratory protocols. This new assay, termed the OROV rRT-PCR, was then compared to a reference rRT-PCR, and using both assays, we tested acute-phase clinical samples from Paraguayan patients with a suspected arboviral illness.
2. Materials and Methods
2.1. Ethics statement
The study of acute arboviral illnesses in Paraguay was reviewed and approved by the Science and Ethical Committees at the Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción (IICS-UNA) and the Emory University IRB. Written informed consent was obtained for all patients, and assent was obtained for children ≥6 years old. Consent forms included language regarding the testing of samples with new methods such as for the current study.
2.2. Assay design and optimization
Primers and probes were initially designed from an alignment of all OROV S-segment or nucleocapsid sequences available in GenBank (n=140 sequences, access 29 August 2017). The design was then refined with a second alignment that included all S-segment or nucleocapsid sequences for OROV, MDDV, IQTV, PERDV and Jatobal virus (JATV; n=149 total sequences, access 02 April 2019). Alignments were performed by ClustalX (DNAStar, Madison, WI). A region at the 5’ end of the S-segment with sequences conserved across ≥95% strains was identified as a potential target for assay development. Primers and probes were designed using Primer3 software (http://bioinfo.ut.ee/primer3). Oligonucleotide sequences were then checked manually against the alignment, and degenerate bases or multiple oligonucleotides were designed to account for common variants (Table 1).
Table 1.
Primer and probe sequences for OROV assays.
| Name | Sequence (5’ → 3’) | Conc.a | Locationb |
|---|---|---|---|
| OROV rRT-PCR | |||
| OROV Forward-S | GACAAGTSCTCAATGCTGGTGT | 200nM | 92–113 |
| OROV Forward-K | GACAAGTGCTCAATGCTKGTGT | 200nM | |
| OROV Reverse | CGTTGTCCGGSACTGGATT | 200nM | 247–265 |
| OROV Probe-Yc | TGGTTGACCTYACTTTTGGTGGGGT | 200nM | 179–203 |
| OROV Probe-Rc | TGGTTGACCTTACTTTTRGTGGGGT | 200nM | |
| Comparator rRT-PCR | |||
| Forward Primer | CATTTGAAGCTAGATACGGACAA | 1,000nM | 74–96 |
| Reverse Primerd | GGCACTGGATTCGACTGGA | 1,000nM | 239–257 |
| Probec | CAATGCTGGTGTTGTTAGAGTCTTCTTCCT | 600nM | 102–230 |
Concentration of each oligonucleotide in the final reaction mixture
Genomic locations for viral primers and probes are provided based on the following reference sequences: Oropouche virus isolate H759483 (GenBank: HQ830492.1).
5’ fluor and 3’ quencher pairs were FAM and BHQ-1
Reverse primer was redesigned from the assay by Weidmann, et al., 2003
Quantified, synthesized ssDNA oligonucleotides containing the target region from two reference strains [accession numbers H759483 (H75), and BeH29090 (BeH)] were used for the analytical evaluation of the OROV rRT-PCR. Strains were selected that matched the alignment consensus sequence (H75) and contained a common variant sequence (BeH). Using these synthesized target sequences, primer and probe concentrations were optimized by varying the concentration of each oligonucleotide in the final reaction mixture from 100 to 400nM.
2.3. OROV rRT-PCR performance
All rRT-PCR reactions in this study were performed on a Rotor-Gene Q 5plex HRM instrument (Qiagen) using 25µL reactions of the SuperScript III Platinum One-Step qRT-PCR Kit (Thermo Fisher) and 5µL of nucleic acid eluate. Cycling conditions for the OROV rRT-PCR were the following: 52°C x 15min, 94°C x 2min, and 45 cycles of 94°C x 15sec, 55°C x 40sec (acquire in green), and 68°C x 20sec. The crossing threshold was set manually, as described (Waggoner, et al., 2016a), and any exponential curve that crossed this threshold at ≤ cycle 40 was considered positive.
2.4. Analytical evaluation
The linear range of the assay was determined by testing synthesized targets from each reference strain in quadruplicate at 8.0, 6.0, 4.0, 2.0, and 1.0 log10 copies/µL. The linear range was defined as the range of concentrations at which all replicates were detected and the R2 values of the linear regression was ≥0.99. The lower limit of 95% detection (95% LLOD) was determined by testing 10 replicates of 2-fold serial dilutions from 20–2.5 copies/µL (H75) and 40–5 copies/µL (BeH).
Assay exclusivity was evaluated by testing genomic RNA from the following viruses (strain in parentheses): Rift Valley fever virus (h85/09); La Crosse virus (3 clinical isolates); ZIKV (MR766); DENV serotype-1 (Hawaii 1944), −2 (NGC), −3 (Sleman/78) and −4 (H241); CHIKV (R80422); Mayaro virus (TRVL 4675); yellow fever virus (17D and Asibi strains); and West Nile virus (NAL strain).
2.5. Comparator assay
A modified version of the rRT-PCR for OROV described by Weidmann, et al, served as the molecular comparator for the current study (hereinafter referred to as the comparator assay) (Weidmann, et al., 2003). Modifications included a new reverse primer which had been previously designed at the University of Texas Medical Branch and the use of a different reaction kit compared to the original publication. Cycling conditions and interpretation were otherwise maintained as described previously (Weidmann, et al., 2003).
2.6. Testing of reference strains
Genomic RNA extracted from four viral strains was provided by the World Reference Center for Emerging Viruses and Arboviruses: TRVL 9760 and Oropouche 7767 (OROV strains); IQT 9924 (an IQTV strain); and BeAn 423380 (JATV). Serial dilutions (10−2, 10−4, 10−5 and 10−6) of TRVL 9760, Oropouche 7767, and IQT 9924 were tested sequentially in the comparator assay followed by the OROV rRT-PCR (single freeze-thaw cycle, stored at 4°C between runs). Genome copy numbers in each dilution were calculated from a 4-point standard curve of H75, performed on each run of the OROV rRT-PCR.
2.7. Clinical samples
Acute-phase sera from 100 Paraguayan patients with a suspected arboviral illness were tested sequentially in the comparator assay and the OROV rRT-PCR (during a single freeze-thaw cycle). Patients presented with an acute febrile illness of ≤7 day’s duration to one of two outpatient clinical sites in metro Asunción between January and May 2018. Serum was collected and stored at −80°C. Total nucleic acids were extracted from 200µL of serum using an eMAG instrument (bioMérieux) and eluted into 50µL of elution buffer (Buffer 3). All patients had previously been tested for Zika virus (ZIKV), CHIKV, and DENV by rRT-PCR (Waggoner, et al., 2016a). We selected samples from all patients that tested negative for DENV (n=73) and an additional 27 samples from DENV-positive patients to further evaluate specificity. No patients tested positive for ZIKV or CHIKV.
In order to evaluate for potential rRT-PCR inhibitors in extracted serum, dilutions of TRVL 9760, Oropouche 7767, and IQT 9924 were tested side-by-side in reactions of the OROV rRT-PCR that had been 1) spiked with eluted total nucleic acids from serum samples collected in Paraguay (5µL/reaction) or 2) brought to the final volume with nuclease-free water. All serum samples included in this experiment had previously tested negative for DENV, ZIKV, CHIKV and OROV, using both the OROV rRT-PCR and comparator assay.
2.8. Statistical analyses
Linear regression and basic statistics (mean, standard deviation) were performed using Excel software (IBM). 95% LLOD was calculated by probit analysis (SPSS, IBM), and the qualitative detection of reference-strain dilutions was compared by Fisher’s exact test (GraphPad QuickCalcs, accessed May 2019).
3. Results
3.1. Analytical evaluation
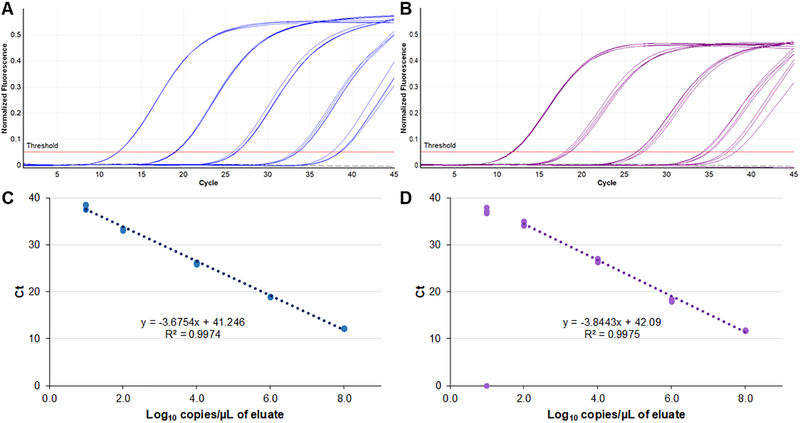
The optimized concentration of primers and probes in the final reaction mixture are shown in Table 1. The dynamic range of the assay extended from 8.0 to 1.0 log10 copies/µL for the H75 target sequence (Figure 1A). For the BeH target, the linear range extended from 8.0 to 2.0 log10 copies/µL. Ct values remained linear to 1.0 log10 copies/µL, however, only 3 of 4 replicates tested positive at that concentration (Figure 1B). The 95% LLOD for was 5.6 and 10.8 copies/µL for the H75 and BeH targets, respectively. No curves were observed crossing the threshold after cycle 40 (Figures 1 and 2), which was used as the cut-off for positive results in this assay.
Figure 1.
Linear range of the OROV rRT-PCR for two type strains: H759483 (blue, A and C) and BeH29090 (purple, B and D). Five concentrations (8.0, 6.0, 4.0, 2.0 and 1.0 log10 copies/µL) were run in quadruplicate on a single run of the assay. All curves and data points are displayed. Gray dashed line (A and B), no template control.
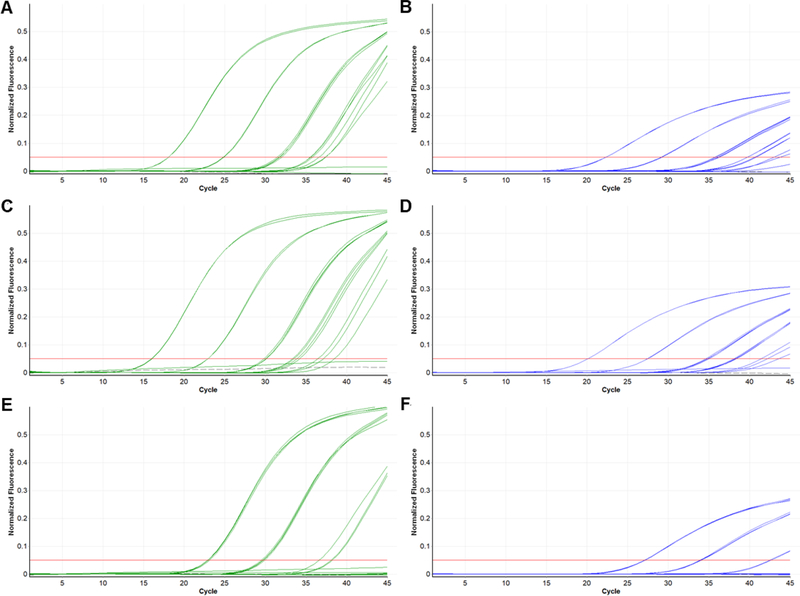
Figure 2.
Dilution series of strains IQT 9924 (A and B), TRVL 9760 (C and D), and OROV 7767 (E and F) tested in the OROV rRT-PCR (green; A, C and E) and comparator rRT-PCR (blue; B, D and F). Each strain was tested at the following dilutions (in duplicate or quadruplicate): undiluted stock; 10−2, 10−4, 10−5 and 10-6. Gray dashed line, no template control.
Exclusivity was evaluated by testing the OROV rRT-PCR against extracted genomic RNA from type strains of other bunyaviruses as well as flaviviruses and alphaviruses that may present in a similar fashion. No signal was detected in the OROV rRT-PCR from any of these viral strains.
3.2. Testing of reference strains
Serial dilutions of genomic RNA from three reference strains of OROV (TRVL 9760 and Oropouche 7767) and IQTV (IQT 9924) were prepared and tested sequentially in the comparator assay followed by the OROV rRT-PCR (Figure 2). Based on a standard curve of the H75 target sequence, the highest estimated viral loads tested for these samples were 6.86, 6.27 and 4.91 log10 copies/µL for the TRVL 9760, IQT 9924 and Oropouche 7767 strains, respectively. The lowest concentrations tested were <10 copies/µL for each strain. Although Ct values in the comparator assay were 2.2–6.0 cycles later (mean 3.9, standard deviation 1.2) than in the OROV rRT-PCR, qualitative results were concordant between the two assays for all samples with >10 copies/µL of RNA. The OROV rRT-PCR detected significantly more samples with ≤10 copies/µL (8/14 vs 0/13, p = 0.002). Higher MgSO4 concentrations were evaluated to maximize sensitivity of the comparator assay, but this did not result in changes to performance (data not shown). The JATV strain (BeAn 423308) was detected in both assays. This produced a consistent but weak signal in the OROV rRT-PCR but gave a similar signal to the other strains in the comparator assay (Figure S1).
3.3. Clinical samples
Acute phase serum samples from 100 patients with a suspected arboviral illness were tested for OROV using the comparator assay and the OROV rRT-PCR. Characteristics of the population are shown in Table 2. Patients presented with either an undifferentiated fever or two or more of the following symptoms: fever, rash, red eyes, arthralgia, or myalgia. All 73 patients without a known etiology for their fever tested negative for OROV RNA, and 27 dengue cases, tested to further evaluate clinical specificity, all tested negative.
Table 2.
Characteristics of 100 patients tested for OROV.
| Characteristic | All Patients |
|---|---|
| Gender, Female, n | 59 |
| Age, mean (SD) | 32.6 (13.3) |
| Day of symptoms, mean (SD) | 3.5 (1.7) |
| Hospitalized, n | 19 |
| DENV-positive, n | 27 |
| DENV-1 | 20 |
| DENV-2 | 3 |
| DENV-4 | 2 |
| No serotypea | 2 |
Abbreviations: DENV, dengue virus; SD, standard deviation
Both patients tested positive for DENV NS1 antigen and anti-DENV IgM
To mimic assay performance with total nucleic acid eluates from clinical samples, OROV (TRVL 9760 and Oropouche 7767) and IQTV (IQT 9924) strains were tested across a range of concentrations (0.2 – 6.8 log10 copies/µL) in reactions of the OROV rRT-PCR prepared with and without eluates from Paraguayan serum samples. Qualitative results were concordant in 18/18 replicates at or above the 95% LLOD, and Ct values for these replicates were a mean of 0.15 cycles later (SD 0.12) in samples with eluted nucleic acids compared to those prepared with water. Two of six replicates with concentrations at or below the 95% LLOD (2–6 copies/µL) tested negative when reactions were prepared with eluted nucleic acids, whereas all were detected in reactions prepared with water.
4. Discussion
OROV and OROV-reassortant species represent important human arboviral pathogens in Central and South America (Romero-Alvarez & Escobar, 2018; Travassos da Rosa, et al., 2017). In many regions of the Amazon basin, such viruses represent the second most common arboviral infection, following only DENV (Travassos da Rosa, et al., 2017). However, the incidence of OROV infection in many parts of South America, including Paraguay, remains unknown (Romero-Alvarez & Escobar, 2018; Travassos da Rosa, et al., 2017). This stems from the non-specific, and frequently mild, manifestations that result from infection (Romero-Alvarez & Escobar, 2018) as well as the limited availability of specific diagnostic tests for OROV. The comparator assay used in our study has previously been employed to confirm an OROV infection in Ecuador (Weidmann, et al., 2003; Wise, et al., 2018), but it has not been published in the current, modified form. A more recent rRT-PCR has also been described, which utilizes a minor-groove binding hydrolysis probe (Naveca, et al., 2017), but overall, there have been few diagnostics published for OROV.
The OROV rRT-PCR developed in the current study provides analytically sensitive and specific detection of pathogens in the OROV-species complex using standard hydrolysis probe chemistry. We have demonstrated the sensitive detection of OROV type strains, IQTV, and target sequences from representative virus strains in GenBank. Available S-segment sequences from MDDV and PERDV were included in the alignment used for assay development, and based on in silico analysis, these viruses would amplify with similar efficiency to the IQTV 9924 strain. Importantly for our laboratory, this assay utilizes a standard cycling protocol and is often performed in combination with assays for other arboviruses such as yellow fever virus and Mayaro virus. This simplifies testing workflow and reduces turn-around-time.
In addition to the aforementioned reference strains, we tested a characterized strain of JATV, which is a related Simbu-group orthobunyavirus that was isolated from a ring-tailed coati in Brazil in 1985 (Figueiredo & Da Rosa, 1988). Although originally reported to be another OROV reassortant (Saeed, et al., 2001b), JATV is now considered to be a phylogenetic outgroup from the OROV-species complex (Ladner, et al., 2014; Tilston-Lunel, et al., 2015). This strain was detectable in the OROV rRT-PCR, but the amplification curves were flat and distinct from the OROV strains and reassortants, consistent with more recent sequencing and phylogenetic analysis of this virus (Ladner, et al., 2014). The importance of JATV detection remains unclear, as this has only been isolated once and it has not been associated with human disease. Notably, should this virus emerge to cause an outbreak, it would be detectable in the OROV rRT-PCR at viral loads observed for other systemic arboviral infections (Waggoner, et al., 2016b; Weidmann, et al., 2003).
No acute Oropouche cases were detected in the current study. Clinically, patients presented with an illness that was compatible with acute Oropouche fever at an average of 3.5 days after symptom onset, when viremia would be expected to be detectable. Viral load kinetics in OROV infections have not been characterized, but viral loads are expected to be relatively high in the acute setting to facilitate transmission to biting midges (Pinheiro, et al., 1982). Although Paraguay reports high annual incidence rates of dengue, the epidemiology of OROV in the country is unknown and may differ significantly from neighboring regions of Brazil. Ours was primarily an urban patient population, and sporadic OROV infections may not occur in this setting without proximity to amplifying vertebrate hosts (Romero-Alvarez & Escobar, 2018). As such, surveillance for OROV among DENV-negative cases in Paraguay, including patients from outlying areas, is warranted to determine the contribution of OROV to arboviral illness in the country and monitor for emergence in naïve populations.
5. Conclusions
The OROV rRT-PCR provides a new, sensitive and specific molecular assay for viruses in the OROV-species complex, which can now be implemented to study this important yet neglected tropical arboviral infection.
Supplementary Material
Figure S1. Amplification of two dilutions of JATV strain BeAn 423380 in the OROV rRT-PCR (A) and comparator rRT-PCR (B). JATV curves (black) are displayed in relation to two dilutions of TRVL 9760 (green and blue curves). Gray dashed line, no template control.
Acknowledgements
Reference bunyavirus strains were kindly provided by the World Reference Center for Emerging Viruses and Arboviruses (OROV, IQTV, and JATV) and Amy Leber (La Crosse virus). We thank the members of the study team based at the Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción, and Hospital Villa Elisa in Paraguay for their dedication and excellent work, and we are grateful to the study participants and their families.
Funding: Research was supported by NIH grants K08 AI110528 (JJW). In addition, the development of this collaboration was supported by funding from the Consejo Nacional de Ciencia y Tecnología (CONACYT) in Paraguay (ARS and JJW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar PV, Barrett AD, Saeed MF, Watts DM, Russell K, Guevara C, Ampuero JS, Suarez L, Cespedes M, Montgomery JM, Halsey ES, & Kochel TJ (2011) Iquitos virus: a novel reassortant Orthobunyavirus associated with human illness in Peru. PLoS neglected tropical diseases, 5: e1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva-Urcia C, Aguilar-Luis MA, Palomares-Reyes C, Silva-Caso W, Suarez-Ognio L, Weilg P, Manrique C, Vasquez-Achaya F, Del Valle LJ, & Del Valle-Mendoza J (2017) Emerging and reemerging arboviruses: A new threat in Eastern Peru. PloS one, 12: e0187897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos MS, Lessa N, Naveca FG, Monte RL, Braga WS, Figueiredo LT, Ramasawmy R, & Mourao MP (2014) Detection of Herpesvirus, Enterovirus, and Arbovirus infection in patients with suspected central nervous system viral infection in the Western Brazilian Amazon. Journal of medical virology, 86: 1522–1527. [DOI] [PubMed] [Google Scholar]
- Cardoso BF, Serra OP, Heinen LB, Zuchi N, Souza VC, Naveca FG, Santos MA, & Slhessarenko RD (2015) Detection of Oropouche virus segment S in patients and inCulex quinquefasciatus in the state of Mato Grosso, Brazil. Memorias do Instituto Oswaldo Cruz, 110: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo F, Rojas A, Franco L, Herebia L, Vallejos MA, Páez GM, Guillén Y, & Mendoza LM (2017) Búsqueda de flavivirus en individuos con sospecha clínica de dengue y con resultado negativo para el antígeno NS1 en Paraguay. Mem Inst Investig Cienc Salud, 15: 7–15. [Google Scholar]
- Figueiredo LT, & Da Rosa AP (1988) Jatobal virus antigenic characterization by ELISA and neutralization test using EIA as indicator, on tissue culture. Memorias do Instituto Oswaldo Cruz, 83: 161–164. [DOI] [PubMed] [Google Scholar]
- Ladner JT, Savji N, Lofts L, Travassos da Rosa A, Wiley MR, Gestole MC, Rosen GE, Guzman H, Vasconcelos PF, Nunes MR, T, JK, Lipkin WI, Tesh RB, & Palacios G (2014) Genomic and phylogenetic characterization of viruses included in the Manzanilla and Oropouche species complexes of the genus Orthobunyavirus, family Bunyaviridae. The Journal of general virology, 95: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreli ML, Aquino VH, Cruz AC, & Figueiredo LT (2002) Diagnosis of Oropouche virus infection by RT-nested-PCR. Journal of medical virology, 66: 139–142. [DOI] [PubMed] [Google Scholar]
- Navarro JC, Giambalvo D, Hernandez R, Auguste AJ, Tesh RB, Weaver SC, Montanez H, Liria J, Lima A, Travassos da Rosa JF, da Silva SP, Vasconcelos JM, Oliveira R, Vianez JL Jr., & Nunes MR (2016) Isolation of Madre de Dios Virus (Orthobunyavirus; Bunyaviridae), an Oropouche Virus Species Reassortant, from a Monkey in Venezuela. The American journal of tropical medicine and hygiene, 95: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca FG, Nascimento VAD, Souza VC, Nunes BTD, Rodrigues DSG, & Vasconcelos P (2017) Multiplexed reverse transcription real-time polymerase chain reaction for simultaneous detection of Mayaro, Oropouche, and Oropouche-like viruses. Memorias do Instituto Oswaldo Cruz, 112: 510–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MRT, de Souza WM, Savji N, Figueiredo ML, Cardoso JF, da Silva SP, da Silva de Lima CP, Vasconcelos HB, Rodrigues SG, Ian Lipkin W, Vasconcelos PFC, & Palacios G (2019) Oropouche orthobunyavirus: Genetic characterization of full-length genomes and development of molecular methods to discriminate natural reassortments. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases, 68: 16–22. [DOI] [PubMed] [Google Scholar]
- Pan American Health Organization (2018) Dengue. In
- Pinheiro FP, Hoch AL, Gomes ML, & Roberts DR (1981) Oropouche virus. IV. Laboratory transmission by Culicoides paraensis. The American journal of tropical medicine and hygiene, 30: 172–176. [PubMed] [Google Scholar]
- Pinheiro FP, Travassos da Rosa AP, Gomes ML, LeDuc JW, & Hoch AL (1982) Transmission of Oropouche virus from man to hamster by the midge Culicoides paraensis. Science, 215: 1251–1253. [DOI] [PubMed] [Google Scholar]
- Romero-Alvarez D, & Escobar LE (2018) Oropouche fever, an emergent disease from the Americas. Microbes and infection / Institut Pasteur, 20: 135–146. [DOI] [PubMed] [Google Scholar]
- Saeed MF, Nunes M, Vasconcelos PF, Travassos Da Rosa AP, Watts DM, Russell K, Shope RE, Tesh RB, & Barrett AD (2001a) Diagnosis of Oropouche virus infection using a recombinant nucleocapsid protein-based enzyme immunoassay. Journal of clinical microbiology, 39: 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed MF, Wang H, Suderman M, Beasley DW, Travassos da Rosa A, Li L, Shope RE, Tesh RB, & Barrett AD (2001b) Jatobal virus is a reassortant containing the small RNA of Oropouche virus. Virus Res, 77: 25–30. [DOI] [PubMed] [Google Scholar]
- Silva-Caso W, Aguilar-Luis MA, Palomares-Reyes C, Mazulis F, Weilg C, Del Valle LJ, Espejo-Evaristo J, Soto-Febres F, Martins-Luna J, & Del Valle-Mendoza J (2019) First outbreak of Oropouche Fever reported in a non-endemic western region of the Peruvian Amazon. Molecular diagnosis and clinical characteristics. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases [DOI] [PubMed]
- Tilston-Lunel NL, Hughes J, Acrani GO, da Silva DE, Azevedo RS, Rodrigues SG, Vasconcelos PF, Nunes MR, & Elliott RM (2015) Genetic analysis of members of the species Oropouche virus and identification of a novel M segment sequence. The Journal of general virology, 96: 1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos da Rosa JF, de Souza WM, Pinheiro FP, Figueiredo ML, Cardoso JF, Acrani GO, & Nunes MRT (2017) Oropouche Virus: Clinical, Epidemiological, and Molecular Aspects of a Neglected Orthobunyavirus. The American journal of tropical medicine and hygiene, 96: 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Davila MJ, Tellez Y, Sahoo MK, Balmaseda A, Harris E, & Pinsky BA (2016a) Single-Reaction Multiplex Reverse Transcription PCR for Detection of Zika, Chikungunya, and Dengue Viruses. Emerging infectious diseases, 22: 1295–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner JJ, Gresh L, Vargas MJ, Ballesteros G, Tellez Y, Soda KJ, Sahoo MK, Nunez A, Balmaseda A, Harris E, & Pinsky BA (2016b) Viremia and Clinical Presentation in Nicaraguan Patients Infected With Zika Virus, Chikungunya Virus, and Dengue Virus. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 63: 1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann M, Rudaz V, Nunes MR, Vasconcelos PF, & Hufert FT (2003) Rapid detection of human pathogenic orthobunyaviruses. Journal of clinical microbiology, 41: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise EL, Pullan ST, Marquez S, Paz V, Mosquera JD, Zapata S, Jackson SK, Fejer G, Trueba G, & Logue CH (2018) Isolation of Oropouche Virus from Febrile Patient, Ecuador. Emerging infectious diseases, 24: 935–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Amplification of two dilutions of JATV strain BeAn 423380 in the OROV rRT-PCR (A) and comparator rRT-PCR (B). JATV curves (black) are displayed in relation to two dilutions of TRVL 9760 (green and blue curves). Gray dashed line, no template control.