Abstract
Mycobacterium tuberculosis (Mtb) is a major causal pathogen of human tuberculosis (TB), which is a serious health burden worldwide. The demand for the development of an innovative therapeutic strategy to treat TB is high due to drug-resistant forms of TB. Autophagy is a cell-autonomous host defense mechanism by which intracytoplasmic cargos can be delivered and then destroyed in lysosomes. Previous studies have reported that autophagy-activating agents and small molecules may be beneficial in restricting intracellular Mtb infection, even with multidrug-resistant Mtb strains. Recent studies have revealed the essential roles of host nuclear receptors (NRs) in the activation of the host defense through antibacterial autophagy against Mtb infection. In particular, we discuss the function of estrogen-related receptor (ERR) α and peroxisome proliferator-activated receptor (PPAR) α in autophagy regulation to improve host defenses against Mtb infection. Despite promising findings relating to the antitubercular effects of various agents, our understanding of the molecular mechanism by which autophagy-activating agents suppress intracellular Mtb in vitro and in vivo is lacking. An improved understanding of the antibacterial autophagic mechanisms in the innate host defense will eventually lead to the development of new therapeutic strategies for human TB.
Subject terms: Autophagy, Medical research
Tuberculosis: Helping infected cells take out the trash
Therapies that promote intracellular digestion of microbes could prove a valuable addition to antibiotic weapons against tuberculosis. Mycobacterium tuberculosis (Mtb) establishes itself within immune cells, and employs a variety of tricks to protect itself as it sickens its host. Researchers led by Eun-Kyeong Jo at Chungnam National University, Daejeon, South Korea, have reviewed efforts to defeat this pathogen by jump-starting a cellular ‘recycling’ pathway called autophagy. Autophagy helps cells break down both biomolecules aggregates and potential invaders, but Mtb can elude such digestion. Jo and colleagues highlight antimycobacterial agents that can potentially render Mtb vulnerable to autophagy, as well as promising cellular targets that may allow researchers to access this process. For example, evidence suggests that agents that activate a regulatory protein such as ERRα or PPARα could stimulate cellular degradation of Mtb.
Introduction
There remains a high demand for the development of new drugs against human tuberculosis (TB), which accounts for an estimated 1.3 million deaths globally1. TB is mainly caused by Mycobacterium tuberculosis (Mtb), a human pathogen that successfully resides in host macrophages and phagocytic cells2–4. Macrophages and phagocytes can trigger numerous innate immune signaling pathways, resulting in the activation of effector molecules to combat intracellular parasites, which can exploit host defense strategies through multiple escape mechanisms, leading to the arrest of phagosomal maturation2,4,5. Mtb and the host immune system are involved in complicated crosstalk, which requires further investigation. The development of new vaccines and therapeutics against TB requires a comprehensive understanding of the molecular mechanisms underlying the host–pathogen interactions during mycobacterial infection6,7.
Autophagy is an intracellular process involved in the housekeeping function and maintenance of cellular homeostasis in response to diverse stress conditions8,9. It is becoming clear that the autophagy pathway is vital in the host defense against infection by various intracellular pathogens, including Mtb, Salmonella enterica serovar Typhimurium, and Listeria monocytogenes through the enhancement of phagolysosome formation10–15. This pathway functions as a cell-autonomous defense system that delivers cytoplasmic cargos and bacterial phagosomes for lysosomal degradation10. Accumulating evidence has shown that autophagy contributes to innate and adaptive immune pathways in a variety of settings12,14,16,17. However, Mtb has evolved numerous strategies to manipulate host innate immune pathways and evade phagosomal acidification2,18–20. Furthermore, recent studies have reported that several autophagy genes do not play a critical role in antimycobacterial defense in murine systems in vivo21. Nevertheless, numerous drugs/agents are able to induce autophagy activation to promote the restriction and eradication of Mtb in vitro and in vivo22. Although there are no specific drugs targeting autophagy, the identification of autophagy-activating small molecules/agents is a promising and new therapeutic target based on host-directed therapy against TB22–24. In this review, we present a brief overview of autophagy/xenophagy during Mtb infection and highlight the autophagy-activating agents/molecules that promote host defense against Mtb. We subsequently focus on important recent studies concerning the discovery of new functions of NRs that promote host autophagy and antimicrobial responses against Mtb infection.
Overview of autophagy in mycobacterial infection
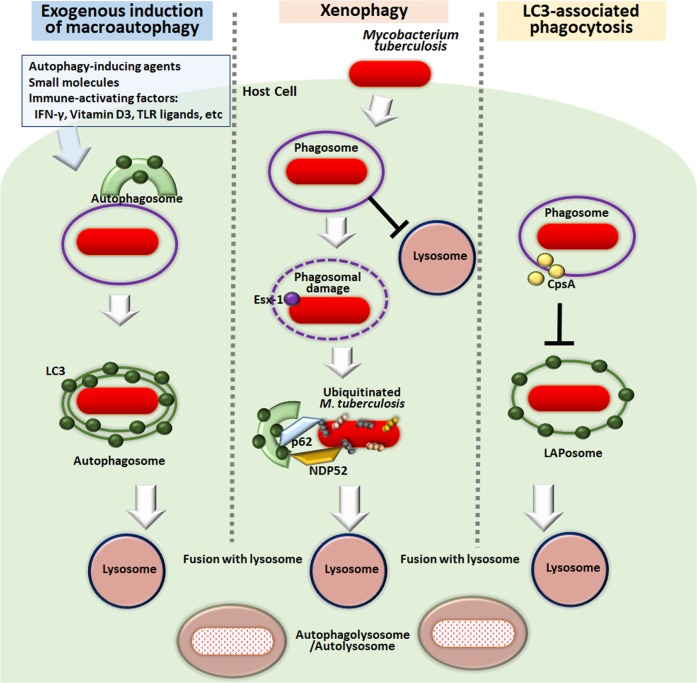
Autophagy (herein, “macroautophagy”) is a multistep process characterized by (1) the initiation of a double-membrane vesicle phagophore; (2) closure as an autophagosome; and (3) fusion with a lysosome to form an autolysosome capable of degrading intracytoplasmic cargo (Fig. 1)25. During this process, numerous autophagy-related genes (ATGs), first identified by Dr. Yoshinori Ohsumi26, were shown to play essential roles as part of the cellular machinery underlying autophagy27,28. In particular, the core machinery of the autophagy process is essential for autophagosome formation. Two ubiquitin-like protein conjugation systems (ATG12 and ATG8/LC3) play critical roles in the formation and ultimate closure of the double-membrane structures of autophagosomes29.
Fig. 1. Autophagy pathway activation during Mtb infection.
After phagocytosis, Mtb can reside in phagosomes to escape phagosomal acidification. Numerous immunological and pharmacological autophagy activators (box on the left) can enhance the restriction of intracellular Mtb growth by overcoming Mtb escape from phagosomal maturation. Mtb phagosome damage through the ESX-1 system is able to trigger ubiquitination of Mtb and its DNA to recruit autophagic adaptors, thereby linking this system to the autophagic machinery. Although much less is known about LAP during Mtb infection, Mtb CpsA has been reported as an inhibitory component in resistance to LAP during infection. IFN interferon, LC3 Microtubule-associated proteins 1A/1B light chain 3B, NDP52 Nuclear domain 10 protein 52, TLR Toll-like receptor
Previous studies have shown that the Th1 cytokine Interferon (IFN)-γ activates autophagy in macrophages, leading to an increase in antimicrobial host defense against Mtb infection12. Numerous additional studies have reported that activation of macroautophagy can promote phagosomal acidification and antimicrobial responses in murine and human macrophages, suggesting that autophagy may represent a promising host-targeting therapeutic strategy against Mtb infection22,24,30. Notably, a recent study by Kimmey et al. showed that ATG5, but no other autophagy genes, plays a unique role in host protection during Mtb infection in mouse models21. Interestingly, this protective effect was not mediated through autophagy activation but through the amelioration of excessive inflammatory responses caused by polymorphonuclear neutrophils21. These observations suggest that the contribution of individual autophagy genes alone is not sufficient to control the growth of intracellular Mtb. Overall, further investigation is warranted to understand whether activation of autophagy by small molecules and/or compounds could enhance the inhibition of intracellular Mtb replication in vivo.
Once regarded as a simple, nonspecific catabolic process, autophagy has proven far more sophisticated than originally thought and is capable of targeting and degrading specific cellular components, including mitochondria, endoplasmic reticulum, lysosomes, and even invading bacteria31,32. Xenophagy is a form of selective autophagy in which cells are able to target and selectively capture bacteria, including Mtb or Salmonella Typhimurium, for autophagic degradation14,28. Specific processes capable of triggering xenophagy include Mtb phagosomal permeabilization through the ESX-1 secretion system, which can trigger xenophagy activation through ubiquitin-mediated-dependent pathways13. Two examples of these pathways are the ubiquitin ligases Parkin and Smurf1, which are involved in the ubiquitination of cytosolic Mtb, followed by its delivery to autophagic machinery13,33. The recognition of cytosolic Mtb DNA by the DNA sensor cGAS is required to target Mtb to the ubiquitin-mediated xenophagy pathway34. The cGAS-STING pathway is required for type I IFN production, which can compromise host protective immunity against Mtb infection, though the activation of these processes can vary depending on the particular Mtb strain35,36. Under most circumstances, the elimination of intracellular Mtb by xenophagy is considered beneficial to the host cells; however, the excessive activation of xenophagy by an Mtb eis-deletion mutant induced host cell death and failed to elicit any protective effects in vivo37. Taken together, these data suggest that xenophagy activation should be coordinated in conjunction with the appropriate immune responses to promote a more rapid resolution of harmful inflammation, increase cell death and limit the spread of infection.
Another type of noncanonical autophagy pathway is LC3-associated phagocytosis (LAP), which has mainly been studied in fungal infections38,39. LAP is an essential link between pattern receptor receptors and phagosomal maturation, helping to enhance the effect of antimicrobial peptides on intracellular pathogens and regulate a variety of physiological functions, including the clearance of apoptotic cells, antigen presentation and type I IFN signaling40,41. A recent study found that the Mtb CpsA protein contributes to Mtb escape from the LAP pathway by inhibiting the recruitment of NADPH oxidase 2 (NOX2) to the mycobacterial phagosome42. This discovery of the Mtb CpsA protein as a key player in the escape from the LAP pathway has highlighted the need to explore mycobacterial effectors and investigate their ability to modulate canonical and noncanonical autophagic processes during infection42. The host autophagy protein Rubicon activates LAP, while inhibiting canonical autophagy43. It is necessary to clarify the exact role of Rubicon in autophagy and/or LAP activation during Mtb infection. A schematic overview of autophagy activation during Mtb infection is shown in Fig. 1. In addition, future studies are needed to elucidate the relationship between canonical autophagy and LAP in shaping host protective immune responses during Mtb infection.
Promotion of antimycobacterial host defense by autophagy-activating drugs/reagents
Mtb and many other pathogens employ numerous strategies to inhibit autophagy2,19,44,45. Here, we discuss how the treatment of autophagy-activating agents promotes antimicrobial host defenses in vitro and in vivo by overcoming the ability of bacteria to block xenophagy and dampening excessive inflammation during infection (Table 1).
Table 1.
Autophagy-activating agents in antimicrobial host defense during mycobacterial infection
| Reagent/drug | Class | Mycobacterial species | Experimental model | Mechanism of action | Ref |
|---|---|---|---|---|---|
| Rapamycin | mTORC1 complex inhibitor | M. bovis BCG, Mtb | RAW264.7 cells, BMDM, and human MDM | Enhancement of mycobacterial phagosome colocalization with LC3, and increases acidification of mycobacterial phagosomes | 12 |
| Small molecule enhancers of rapamycin (SMER) | mTORC1 complex inhibitor | M. bovis BCG | Human PBMC | Induction of autophagy through inhibition of mTOR pathway | 46 |
| Vitamin D | Vitamin | Mtb | Human monocytes, MDM, THP-1, and RAW 264.7 cells | Increased transcriptional activation of ATG5 and ATG6 through cathelicidin-dependent MAPK and C/EBPβ signaling. Recruitment of cathelicidin to autophagosomes through the Ca2+ and AMPK-dependent pathways. | 47 |
| Mtb | Human MDM | Cathelicidin LL-37 and autophagic flux activation | 48 | ||
| IFN-γ | Cytokine | Mtb | Human T cells, monocytes, MDM, and BMDM | Induction of autophagy and production of cathelicidin via vitamin D-dependent pathway | 49 |
| M. bovis BCG | RAW264.7, human U937, 293T, and HeLa cells | Induction of autophagy via Irgm1 | 50 | ||
| Metformin | Antidiabetic drug | M. bovis BCG, Mtb | THP-1 cells, human MDM, and mice | Enhancement of mROS production, phagosome-lysosome fusion, and upregulation of lipidated LC3 form | 51 |
| 4-phenylbutyrate (PBA) | Histone deacetylase inhibitor | Mtb | Human MDM and THP-1 cells | LL-37-dependent activation of autophagy by PBA and/or vitamin D | 52 |
| Nitazoxanide | Antiprotozoal drug | Mtb | Human PBMC, THP-1, MCF-7, MEF, and HEK 293T cells | Inhibition of mTORC1, a negative regulator of autophagy via NQO1 | 53 |
| Fluoxetine | Selective serotonin reuptake inhibitor | Mtb | J774 cells and BMDM | Increased TNF-α production and autophagy Induction | 54 |
| Gefitinib | EGFR inhibitor | Mtb | J774 cells, BMDM, human MDM, and mice | Autophagy induction and Inhibition of EGFR-mediated p38 activation | 54 |
| Carbamazepine | Anticonvulsant | M. bovis BCG, Mtb, M. marinum | RAW264.7 cells, human MDM, alveolar macrophages, zebrafish RAW264.7 cells and mice | mTOR-independent autophagy through IP3 depletion and AMPK activation | 55 |
| Valproic acid | Anticonvulsant | M. bovis BCG, Mtb | RAW264.7 cells, human MDM, and alveolar macrophages | mTOR-independent autophagosome formation through ATG12 and inhibition of intracellular bacterial growth | 55 |
| AICAR | AMPK activator | Mtb, BCG, M. marinum | RAW264.7 cells, THP-1 cells, BMDM, mice, and flies | Activation of autophagy through AMPK-PGC1α pathway via C/EBPβ signaling | 56 |
| M. bovis BCG, Mtb | BMDM, RAW264.7 cells, HEK 293T cells, and mice | ERRα-mediated transcriptional activation of autophagy genes | 57 | ||
| Resveratrol | SIRT1 activator | Mtb | BMDM, RAW264.7 cells, HEK 293T cells, and mice | SIRT1-ERRα interaction to activate ATG gene transcription | 57 |
| M. bovis BCG, Mtb | THP-1 cells and mice | Induction of autophagolysosome in a SIRT1-dependent manner | 58 | ||
| SRT1720 | SIRT1 activator | M. bovis BCG, Mtb | Human MDM, THP-1 cells, and mice | Induction of autophagolysosome in a SIRT1-dependent manner | 58 |
| Honokiol | SIRT3 activator | Mtb | BMDM, human MDM, and mice | Induction of autophagosome and autophagic flux in a SIRT3-dependent manner | 59 |
| Isoniazid, Pyrazinamide | Antibiotics | Mtb | BMDM, human MDM, and mice | Autophagy activation by ROS, Ca2+, and AMPK-dependent pathway (in Mtb-infected macrophages) | 60 |
| Loperamide | Anticonvulsant | Mtb | BMDM, murine avleolar macrophages, human avleolar macrophages, MDM, and mice | Increased induction of ATG16L1, LC3 mRNA expression, colocolization of LC3 with Mtb, and reduction of TNF-α production | 61 |
| Thiostrepton (TSR) | Thiopeptide antibiotic drug | M. marinum | RAW264.7 cells and zebrafish | Autophagy activation by endoplasmic reticulum stress pathways | 62 |
| Statin | Cholesterol-inhibiting drugs | Mtb | Human PBMC, MDM, BMDM, and mice | Reduction of cholesterol levels within phagosomal membranes, promotion of phagosomal maturation and autophagy | 63 |
| Dehydroepiandrosterone (DHEA) | Steroid hormone | Mtb | THP-1 cells | Induction of autophagosome formation | 64 |
| Nortriptyline | Anti-depressant | M. bovis BCG, Mtb | Human MDM, HeLa cells | Induction of autophagosome formation and autophagy flux | 65 |
| GW7647, Wy14643 | PPARα agonist | M. bovis BCG, Mtb | BMDM and mice | Autophagy induction via TFEB, and enhanced lipid catabolism | 66 |
| GSK4112 | NR1D1 agonist | Mtb | THP-1 cells | Increased autophagic flux and TFEB signaling | 67 |
| Gamma-aminobutyric acid (GABA) | Neurotransmitter | M. bovis BCG, Mtb, M. marinum | BMDM, RAW 264.7 cells, human MDM, mice, zebrafish, and files | Induces autophagic flux via GABAAR, intracellular calcium release, GABARAPL1 induction | 68 |
BMDM bone marrow-derived macrophages, MDM monocyte-derived macrophages, PBMC peripheral blood mononuclear cells, TFEB transcription factor EB
Previous studies have shown that rapamycin, small molecule enhancers of rapamycin (SMER), vitamin D, interferon-γ, metformin, and 4-phenylbutyrate (PBA) displayed antimicrobial activity against Mtb in human or murine macrophages by enhancing the activation of the autophagy pathway12,46–52. In human macrophages, a link between vitamin D-induced autophagy and human cathelicidin microbial peptide (LL-37) has been demonstrated47,52. Interestingly, PBA and the active form of vitamin D3 (1,25[OH]2D3) were shown to improve intracellular killing of Mtb in human macrophages through LL-37 expression and autophagy52.
Several pharmacologic agents have been identified for their ability to induce autophagy to promote antimicrobial effects against Mtb infection. For example, the antiprotozoal drug nitazoxanide and its analogs activate autophagosome formation and mTORC1 inhibition, thus restricting Mtb proliferation in vitro53. In addition, a chemical screening study using a high-content microscopic assay identified small molecules that inhibit mycobacterial growth in macrophages by targeting host autophagy activation. It was noted that both fluoxetine (a selective serotonin reuptake inhibitor) and gefitinib (an inhibitor of the epidermal growth factor receptor) activate autophagy and reduce Mtb growth in macrophages and in vivo54. Another study with cell-based screening of FDA-approved drugs ascertained that the anticonvulsant carbamazepine and valproic acid enhanced mTOR-independent autophagic killing of Mtb in human macrophages55. Recent studies have revealed that AMPK activator (5-Aminoimidazole-4-carboxamide 1-β-D-ribofuranoside, AICAR), sirtuin (SIRT) 1 activator (resveratrol, RSV or SRT1720) or a SIRT3 activator (Honokiol) were beneficial for promoting host defenses against mycobacterial infection through autophagy induction, AMPK activation or reduced inflammation56–59.
While host-directed therapy has recently emerged as a new therapeutic strategy for the treatment of human TB, accumulating evidence strongly suggests that antimycobacterial antibiotics exert activities through dual modes, acting on both intracellular bacteria and host autophagy activation60. The induction of autophagy by treating macrophages with isoniazid and pyrazinamide was required for successful chemotherapeutic effects against intracellular Mtb. The mechanisms of autophagy activation involved the antibiotic-mediated triggering of hydroxyl radicals and cellular reactive oxygen species in Mtb-infected macrophages60. Accumulating evidence suggests that drug repurposing, based on autophagy activation, shows promise in the development of new host-directed therapeutics against Mtb infection. Carbamazepine, loperamide, and valproic acid induce ATG expression and autophagy, which are associated with the control of the intracellular growth of Mtb in murine alveolar cells and alveolar macrophages61. Recently, thiostrepton (TSR), a thiopeptide antibiotic possessing a quinaldic acid moiety, has been shown to have a dual action on direct targeting to the bacterial ribosome and the induction of ER stress-mediated autophagy to promote the elimination of intracellular mycobacteria62. The cholesterol-lowering drugs, statins showed beneficial effects against intracellular Mtb growth through the promotion of phagosomal maturation and autophagy activation63. In addition, the immunomodulatory drug, dehydroepiandrosterone (DHEA) was beneficial in controlling Mtb load through an autophagy mechanism, which contributes to the clearance of Mtb and the prevention of tissue damage64. Moreover, the FDA-approved antidepressant drug, nortriptyline can increase autophagosome formation and xenophagic flux against mycobacteria through the synergistic activation of autophagy with IFN-γ65. Peroxisome proliferator-activated receptor (PPAR) α agonists (GW7647 and Wy14643) and NR subfamily 1, group D, member 1 (NR1D1) agonist (GSK4112) enhance xenophagic flux via transcription factor EB (TFEB) signaling66,67. In our recent report, the major inhibitory neurotransmitter, gamma-aminobutyric acid (GABA) promotes antimicrobial responses and autophagy activation through macrophage type A GABA receptor (GABAAR), intracellular calcium release, and the GABA type A receptor-associated protein-like 168. Together, these drugs or agents may act as new therapeutics of host-induced autophagy, thereby enhancing host protection against TB.
Nuclear receptors and autophagy in mycobacterial infection
NRs are important for innate immune responses to control inflammatory responses and infection69. In recent reports, emerging evidence suggests that several NRs play critical roles in autophagy activation to promote the innate host defense against mycobacterial infection. The vitamin D-mediated beneficial effects on the restriction of intracellular Mtb growth in macrophages have been studied; however, additional clinical trials of vitamin D-adjunctive therapies for TB are needed to consider all genetic variants23,70,71. NR1D1, an orphan NR, also exerts antimycobacterial effects through the reinforcement of autophagic flux and lysosome biogenesis in human macrophages67. We recently showed that orphan NR, estrogen-related receptor α (ERRα; NR3B1, ERR1, ESRRA), promotes macrophage autophagy in response to various autophagy stimulators, including AICAR and RSV57. In addition, other studies have reported a role for PPARα in the activation of host defenses in macrophages through autophagy and lysosomal biogenesis66. In a recent study of the expression profile of NRs in Mtb-infected macrophages or dendritic cells72, several NRs, such as N4a3 and Rora, were identified. Given the findings that numerous NRs appear to be involved in the regulation of autophagy in host cells, future studies are needed to investigate the novel functions of new NRs and their complex interplay with Mtb in the context of autophagy. In this review, we focus on recent studies of the functions of two NRs, ERRα and PPARα.
ERRα and autophagy
ERRα is the first orphan family member of NRs in which the physiological ligands have not been identified. ERRα, along with other members of the ERRs, does not bind estrogens and preferentially binds to an estrogen-related response element (ERRE) to regulate target genes containing these binding elements in their promoter/enhancer regions73. Previous functional studies have shown that ERRα plays a transcriptional activating role through an interaction with the transcriptional coactivator PPARγ coactivator-1α (PGC-1α)74,75. ERRα function has been widely studied in the regulation of mitochondrial and metabolic gene transcription, particularly in muscle differentiation, thermogenesis, and in heart and bone functions76. Previous works have shown that ERRα is a central regulator of innate immune function, including the regulation of toll-like receptor-induced inflammatory responses and antimicrobial responses against intracellular bacterial infection77,78. Recently, a new function of ERRα was revealed in the negative regulation of antiviral responses through the inhibition of type-I interferon signaling79.
The involvement of ERRα, in cooperation with PGC-1α, in the mitochondrial quality control and regulation of autophagy has been shown80. ERRα deficiency was associated with incomplete autophagy and necrotic cell death in adrenocortical cancer through the control of bioenergetic metabolism81. Thyroid hormone induces ERRα, which is essential in the regulation of DRP1-mediated mitochondrial fission and mitophagy through the expression of autophagy-initiating kinase ULK182.
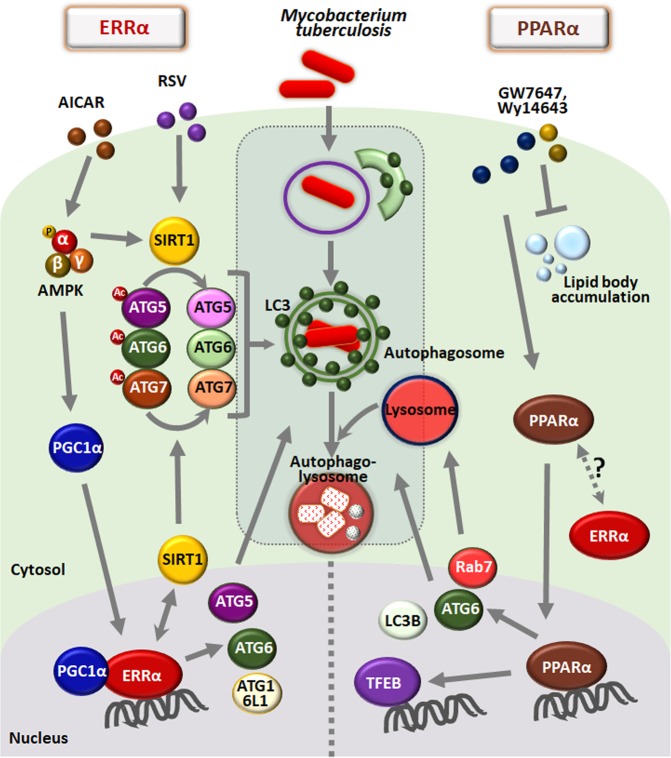
Notably, ERRα was found to be a key transcriptional regulator of numerous ATGs, including ATG5, ATG6, and ATG16L1, which contain ERR response elements in their promoter/enhancer regions57. Although ERRα has no physiological ligands, AMPK and SIRT1 activation enhances the induction of ERRα mRNA and proteins, thereby enhancing the formation of autophagosomes and autophagic flux in macrophages. In addition, ERRα plays a posttranslational regulatory role through the deacetylation of several autophagy proteins, including ATG5, ATG6, and ATG7, all of which are regulated through interactions with SIRT1. Furthermore, ERRα-deficient mice show defective antimicrobial and excessive inflammatory responses against mycobacterial infection, indicating that ERRα is a possible target of antimicrobial innate defenses during Mtb infection57. The transcriptional and posttranslational mechanisms by which ERRα regulates the autophagy pathway are shown in Fig. 2.
Fig. 2. The roles of ERRα and PPARα in autophagy and host defense against Mtb infection.
(Left) ERRα, which is induced by either AMPK or SIRT1 activation, contributes to the induction of autophagosome formation in BMDMs. ERRα is required for the transcriptional activation of several ATGs containing ERR response elements in the promoters. In addition, the cooperation of ERRα with SIRT1 promotes the deacetylation of ATG5, ATG6, and ATG7, thereby activating autophagy at the posttranslational level. ERRα-mediated autophagy activation results in increased phagosomal maturation and antimicrobial responses during Mtb infection. (Right) PPARα, which is activated by PPARα ligands (GW7647 and Wy14643), contributes to enhanced autophagosomal formation and maturation in BMDMs. PPARα is essential for the transcriptional activation of several ATGs, TFEB and lipid catabolism. PPARα reinforces antimicrobial responses to mycobacterial infection by inducing autophagic maturation, TFEB, and lipid catabolism. AICAR, 5-Aminoimidazole-4-carboxamide 1-β-D-ribofuranoside; RSV resveratrol
PPARα and Autophagy
The NR PPARs include three isoforms (α, δ, and γ)83, which form heterodimers with retinoid X receptor and bind to AGGTCANAGGTCA, the peroxisome proliferator response element (PPRE), to induce or repress the transcription of target genes84,85. The PPAR target genes are mostly involved in metabolic homeostasis in various tissues, including the liver, adipose tissues, heart and muscle85–87. Of the three isoforms of PPARs, PPARα is an important coordinator of lipid metabolism and vascular and inflammatory responses86,87. Since PPARα is critically involved in fatty acid oxidation (FAO), lipid and glucose metabolism, and inflammation, the dysregulation of PPARα leads to various defects, such as metabolic, cardiovascular and inflammatory diseases88–90. In terms of immunological control, PPARα acts as a critical regulator in immune homeostasis against various inflammatory and infectious stimuli91–95. A novel connection between autophagy and PPARα to influence lipid metabolism and innate immunity has been proposed, where autophagy activation by PPARα was shown to promote autophagic lipid degradation and innate host defenses66. PPARα activation elevates autophagy, particularly in the transcriptional activation of ATGs66,96, which is essential for the regulation of the autophagy process in various tissues and cells97. Importantly, there exists a great deal of evidence for crosstalk between PPARα and TFEB66,98,99, which is a master regulator of autophagy, lysosomal function and biogenesis, and lipid catabolism98–100. Indeed, TFEB is recognized as an important transcriptional factor for the regulation of immune and inflammatory responses100,101. Combined with our recent study showing that SIRT3 induces antibacterial autophagy against Mtb infection through PPARα59, the function of PPARα in the host defense against intracellular Mtb infection might be primarily mediated through its activation of autophagy59,66.
Importantly, a recent report showed that PPARα activation contributes to the enhancement of FAO and lipid catabolism in macrophages during Mtb infection66. It would be attractive to examine whether autophagy activation is linked to lipid body inhibition in terms of host defense against Mtb infection. A previous study showed that lipid droplets are delivered to lysosomes via the autophagy pathway, thereby hydrolyzing lipid droplets by the action of lysosomal acid lipase102. Thus, autophagy may be required for the regulation of lipid metabolism in macrophages during Mtb infection. PPARα-mediated host defense is summarized in Fig. 2. Gemfibrozil (lipid-lowering drug), an FDA-approved PPARα agonist, has been reported to inhibit the intracellular growth of wild-type and multidrug-resistant Mtb and suppress the activity of enoyl-CoA reductases103. For this reason, gemfibrozil may be a potential anti-TB drug candidate; however, it is unclear whether gemfibrozil-mediated antimicrobial responses depend on autophagy activation. It is an open question whether there is crosstalk between PPARα and ERRα in terms of antimycobacterial host defense. Defining the unique immunological features of autophagy-activating agents based on NR function may represent a rational path for designing improved therapeutics or protective vaccines against TB.
Concluding remarks
Autophagy activation by diverse exogenous stimuli has now been recognized for its role in antimicrobial host defense and in regulating immune and inflammatory responses during Mtb infection. However, the mechanisms controlling these antimicrobial responses are not completely understood. Accumulating evidence shows that autophagy-activating agents are crucial for innate host defense and for controlling excessive inflammatory responses against Mtb infection. Future studies are warranted to examine the effects of autophagy-modulating agents, used either alone or together with chemotherapeutic drugs, for their antimicrobial effects against Mtb infection in vivo and in clinical trials. Given the recent reports showing that both ERRα and PPARα modulate antibacterial autophagy, progress is expected in the development of new therapeutic approaches to treat other infectious diseases beyond tuberculosis. An improved understanding of the molecular mechanisms of autophagy-activating agents will eventually lead to the development of novel therapeutic strategies for human TB.
Acknowledgements
We are indebted to current and past members of our laboratory for discussions and investigations that contributed to this article. This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (Ministry of Science and ICT) (2017R1A5A2015385) and by the framework of international cooperation program managed by NRF of Korea (Grant Number: 2015K2A2A6002008). This research was supported by Chungnam National University Hospital Research Fund, 2017–2018. The authors have no financial conflicts of interest.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization Global tuberculosis report 2018. (World Health Organization, Geneva, 2018).
- 2.Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc. Natl Acad. Sci. USA. 2003;100:5437–5442. doi: 10.1073/pnas.0737613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V, Via LE, Fratti RA, Deretic D. Mycobacterial phagosome maturation, rab proteins, and intracellular trafficking. Electrophoresis. 1997;18:2542–2547. doi: 10.1002/elps.1150181409. [DOI] [PubMed] [Google Scholar]
- 4.Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 2009;11:1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Via LE, et al. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 1997;272:13326–13331. doi: 10.1074/jbc.272.20.13326. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann E, Machelart A, Song OR, Brodin P. Proteomics of mycobacterium infection: moving towards a better understanding of pathogen-driven immunomodulation. Front. Immunol. 2018;9:86. doi: 10.3389/fimmu.2018.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015;264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura S, Yoshimori T. New insights into autophagosome-lysosome fusion. J. Cell Sci. 2017;130:1209–1216. doi: 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- 9.Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Micro. Cell. 2016;3:588–596. doi: 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshimori T, Amano A. Group a Streptococcus: a loser in the battle with autophagy. Curr. Top. Microbiol. Immunol. 2009;335:217–226. doi: 10.1007/978-3-642-00302-8_10. [DOI] [PubMed] [Google Scholar]
- 11.Yuk JM, Yoshimori T, Jo EK. Autophagy and bacterial infectious diseases. Exp. Mol. Med. 2012;44:99–108. doi: 10.3858/emm.2012.44.2.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 13.Manzanillo PS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol. Cell. 2014;54:224–233. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Siqueira MDS, Ribeiro RM, Travassos LH. Autophagy and its interaction with intracellular bacterial pathogens. Front Immunol. 2018;9:935. doi: 10.3389/fimmu.2018.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang, Y. J., Kim, J. H. & Byun, S. Modulation of autophagy for controlling immunity. Cells8, E138 (2019). [DOI] [PMC free article] [PubMed]
- 17.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu CH, Liu H, Ge B. Innate immunity in tuberculosis: host defense vs pathogen evasion. Cell. Mol. Immunol. 2017;14:963–975. doi: 10.1038/cmi.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romagnoli A, et al. ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells. Autophagy. 2012;8:1357–1370. doi: 10.4161/auto.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandra P, et al. Mycobacterium tuberculosis Inhibits RAB7 recruitment to selectively modulate autophagy flux in macrophages. Sci. Rep. 2015;5:16320. doi: 10.1038/srep16320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimmey JM, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528:565–569. doi: 10.1038/nature16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta A, Misra A, Deretic V. Targeted pulmonary delivery of inducers of host macrophage autophagy as a potential host-directed chemotherapy of tuberculosis. Adv. Drug Deliv. Rev. 2016;102:10–20. doi: 10.1016/j.addr.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paik S, Kim JK, Chung C, Jo EK. Autophagy: a new strategy for host-directed therapy of tuberculosis. Virulence. 2018;10:488–459. doi: 10.1080/21505594.2018.1536598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann SHE, Dorhoi A, Hotchkiss RS, Bartenschlager R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Disco. 2018;17:35–56. doi: 10.1038/nrd.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 27.Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24:58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deretic V, et al. Immunologic manifestations of autophagy. J. Clin. Invest. 2015;125:75–84. doi: 10.1172/JCI73945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jo EK. Autophagy as an innate defense against mycobacteria. Pathog. Dis. 2013;67:108–118. doi: 10.1111/2049-632X.12023. [DOI] [PubMed] [Google Scholar]
- 31.Mizumura K, Choi AM, Ryter SW. Emerging role of selective autophagy in human diseases. Front. Pharm. 2014;5:244. doi: 10.3389/fphar.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 33.Franco LH, et al. The ubiquitin ligase Smurf1 FUNCTIONS IN SELECTIVE AUTOPHagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe. 2017;21:59–72. doi: 10.1016/j.chom.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson RO, et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce Type I interferons and activate autophagy. Cell Host Microbe. 2015;17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNab FW, et al. Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-gamma for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages. J. Immunol. 2014;193:3600–3612. doi: 10.4049/jimmunol.1401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiens KE, Ernst JD. The mechanism for Type I interferon induction by Mycobacterium tuberculosis is bacterial strain-dependent. PLoS Pathog. 2016;12:e1005809. doi: 10.1371/journal.ppat.1005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin DM, et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010;6:e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sprenkeler EG, Gresnigt MS, van de Veerdonk FL. LC3-associated phagocytosis: a crucial mechanism for antifungal host defence against Aspergillus fumigatus. Cell. Microbiol. 2016;18:1208–1216. doi: 10.1111/cmi.12616. [DOI] [PubMed] [Google Scholar]
- 39.Martinez J, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Henault J, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–997. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green DR, Oguin TH, Martinez J. The clearance of dying cells: table for two. Cell Death Differ. 2016;23:915–926. doi: 10.1038/cdd.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koster S, et al. Mycobacterium tuberculosis is protected from NADPH oxidase and LC3-associated phagocytosis by the LCP protein CpsA. Proc. Natl Acad. Sci. USA. 2017;114:E8711–E8720. doi: 10.1073/pnas.1707792114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong SW, Sil P, Martinez J. Rubicon: LC3-associated phagocytosis and beyond. FEBS J. 2018;285:1379–1388. doi: 10.1111/febs.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorbara MT, Girardin SE. Emerging themes in bacterial autophagy. Curr. Opin. Microbiol. 2015;23:163–170. doi: 10.1016/j.mib.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 45.Yoshikawa Y, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 46.Floto RA, et al. Small molecule enhancers of rapamycin-induced TOR inhibition promote autophagy, reduce toxicity in Huntington’s disease models and enhance killing of mycobacteria by macrophages. Autophagy. 2007;3:620–622. doi: 10.4161/auto.4898. [DOI] [PubMed] [Google Scholar]
- 47.Yuk JM, et al. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 2009;6:231–243. doi: 10.1016/j.chom.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Campbell GR, Spector SA. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012;8:e1002689. doi: 10.1371/journal.ppat.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fabri M, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci. Transl. Med. 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 51.Singhal A, et al. Metformin as adjunct antituberculosis therapy. Sci. Transl. Med. 2014;6:263ra159. doi: 10.1126/scitranslmed.3009885. [DOI] [PubMed] [Google Scholar]
- 52.Rekha RS, et al. Phenylbutyrate induces LL-37-dependent autophagy and intracellular killing of Mycobacterium tuberculosis in human macrophages. Autophagy. 2015;11:1688–1699. doi: 10.1080/15548627.2015.1075110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam KK, et al. Nitazoxanide stimulates autophagy and inhibits mTORC1 signaling and intracellular proliferation of Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002691. doi: 10.1371/journal.ppat.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanley SA, et al. Identification of host-targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS Pathog. 2014;10:e1003946. doi: 10.1371/journal.ppat.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schiebler M, et al. Functional drug screening reveals anticonvulsants as enhancers of mTOR-independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO Mol. Med. 2015;7:127–139. doi: 10.15252/emmm.201404137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang CS, et al. The AMPK-PPARGC1A pathway is required for antimicrobial host defense through activation of autophagy. Autophagy. 2014;10:785–802. doi: 10.4161/auto.28072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim SY, et al. ESRRA (estrogen-related receptor alpha) is a key coordinator of transcriptional and post-translational activation of autophagy to promote innate host defense. Autophagy. 2018;14:152–168. doi: 10.1080/15548627.2017.1339001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng CY, et al. Host sirtuin 1 regulates mycobacterial immunopathogenesis and represents a therapeutic target against tuberculosis. Sci. Immunol. 2017;2:eaaj1789. doi: 10.1126/sciimmunol.aaj1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim TS, et al. SIRT3 promotes antimycobacterial defenses by coordinating mitochondrial and autophagic functions. Autophagy. 2019;15:1356–1375. doi: 10.1080/15548627.2019.1582743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JJ, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11:457–468. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 61.Juarez E, et al. Loperamide restricts intracellular growth of Mycobacterium tuberculosis in lung macrophages. Am. J. Respir. Cell Mol. Biol. 2016;55:837–847. doi: 10.1165/rcmb.2015-0383OC. [DOI] [PubMed] [Google Scholar]
- 62.Zheng Q, et al. Thiopeptide antibiotics exhibit a dual mode of action against intracellular pathogens by affecting both host and microbe. Chem. Biol. 2015;22:1002–1007. doi: 10.1016/j.chembiol.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 63.Parihar SP, et al. Statin therapy reduces the Mycobacterium tuberculosis burden in human macrophages and in mice by enhancing autophagy and phagosome maturation. J. Infect. Dis. 2014;209:754–763. doi: 10.1093/infdis/jit550. [DOI] [PubMed] [Google Scholar]
- 64.Bongiovanni B, et al. Effect of cortisol and/or DHEA on THP1-derived macrophages infected with Mycobacterium tuberculosis. Tuberculosis. 2015;95:562–569. doi: 10.1016/j.tube.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 65.Sundaramurthy V, et al. Integration of chemical and RNAi multiparametric profiles identifies triggers of intracellular mycobacterial killing. Cell Host Microbe. 2013;13:129–142. doi: 10.1016/j.chom.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 66.Kim YS, et al. PPAR-alpha activation mediates innate host defense through induction of TFEB and lipid catabolism. J. Immunol. 2017;198:3283–3295. doi: 10.4049/jimmunol.1601920. [DOI] [PubMed] [Google Scholar]
- 67.Chandra V, Bhagyaraj E, Nanduri R, Ahuja N, Gupta P. NR1D1 ameliorates Mycobacterium tuberculosis clearance through regulation of autophagy. Autophagy. 2015;11:1987–1997. doi: 10.1080/15548627.2015.1091140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim JK, et al. GABAergic signaling linked to autophagy enhances host protection against intracellular bacterial infections. Nat. Commun. 2018;9:4184. doi: 10.1038/s41467-018-06487-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin HS, Kim TS, Jo EK. Emerging roles of orphan nuclear receptors in regulation of innate immunity. Arch. Pharm. Res. 2016;39:1491–1502. doi: 10.1007/s12272-016-0841-6. [DOI] [PubMed] [Google Scholar]
- 70.Bekele A, et al. Daily adjunctive therapy with vitamin D3 and phenylbutyrate supports clinical recovery from pulmonary tuberculosis: a randomized controlled trial in Ethiopia. J. Intern. Med. 2018;284:292–306. doi: 10.1111/joim.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ganmaa D, et al. High-Dose Vitamin D3 during Tuberculosis Treatment in Mongolia. A Randomized Controlled Trial. Am. J. Respir. Crit. Care Med. 2017;196:628–637. doi: 10.1164/rccm.201705-0936OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saini A, et al. An accord of nuclear receptor expression in M. tuberculosis infected macrophages and dendritic cells. Sci. Rep. 2018;8:2296. doi: 10.1038/s41598-018-20769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Audet-Walsh E, Giguere V. The multiple universes of estrogen-related receptor alpha and gamma in metabolic control and related diseases. Acta Pharmacol. Sin. 2015;36:51–61. doi: 10.1038/aps.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J. Biol. Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 75.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 76.Huss JM, Garbacz WG, Xie W. Constitutive activities of estrogen-related receptors: Transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochim. Biophys. Acta. 1852;1912–1927:2015. doi: 10.1016/j.bbadis.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 77.Yuk JM, et al. Orphan nuclear receptor ERRalpha controls macrophage metabolic signaling and A20 expression to negatively regulate TLR-induced inflammation. Immunity. 2015;43:80–91. doi: 10.1016/j.immuni.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 78.Sonoda J, et al. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes Dev. 2007;21:1909–1920. doi: 10.1101/gad.1553007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He X, et al. ERRalpha negatively regulates type I interferon induction by inhibiting TBK1-IRF3 interaction. PLoS Pathog. 2017;13:e1006347. doi: 10.1371/journal.ppat.1006347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qi Z, Ding S. Transcriptional regulation by nuclear corepressors and PGC-1alpha: implications for mitochondrial quality control and insulin sensitivity. PPAR Res. 2012;2012:348245. doi: 10.1155/2012/348245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casaburi I, et al. Estrogen related receptor alpha (ERRalpha) a promising target for the therapy of adrenocortical carcinoma (ACC) Oncotarget. 2015;6:25135–25148. doi: 10.18632/oncotarget.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh BK, et al. Thyroid hormone receptor and ERRalpha coordinately regulate mitochondrial fission, mitophagy, biogenesis, and function. Sci. Signal. 2018;11:eaam5855. doi: 10.1126/scisignal.aam5855. [DOI] [PubMed] [Google Scholar]
- 83.Dreyer C, et al. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68:879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 84.Berger J, Moller DE. The mechanisms of action of PPARs. Annu. Rev. Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 85.Michalik L, et al. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol. Rev. 2006;58:726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- 86.Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015;62:720–733. doi: 10.1016/j.jhep.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 87.Lee WS, Kim J. Peroxisome proliferator-activated receptors and the heart: lessons from the past and future directions. PPAR Res. 2015;2015:271983. doi: 10.1155/2015/271983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Djouadi F, et al. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator- activated receptor alpha- deficient mice. J. Clin. Invest. 1998;102:1083–1091. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watanabe K, et al. Constitutive regulation of cardiac fatty acid metabolism through peroxisome proliferator-activated receptor alpha associated with age-dependent cardiac toxicity. J. Biol. Chem. 2000;275:22293–22299. doi: 10.1074/jbc.M000248200. [DOI] [PubMed] [Google Scholar]
- 90.Devchand PR, et al. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 91.Mansouri RM, Bauge E, Staels B, Gervois P. Systemic and distal repercussions of liver-specific peroxisome proliferator-activated receptor-alpha control of the acute-phase response. Endocrinology. 2008;149:3215–3223. doi: 10.1210/en.2007-1339. [DOI] [PubMed] [Google Scholar]
- 92.Huang W, Eum SY, Andras IE, Hennig B, Toborek M. PPARalpha and PPARgamma attenuate HIV-induced dysregulation of tight junction proteins by modulations of matrix metalloproteinase and proteasome activities. FASEB J. 2009;23:1596–1606. doi: 10.1096/fj.08-121624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Standage SW, Caldwell CC, Zingarelli B, Wong HR. Reduced peroxisome proliferator-activated receptor alpha expression is associated with decreased survival and increased tissue bacterial load in sepsis. Shock. 2012;37:164–169. doi: 10.1097/SHK.0b013e31823f1a00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Drosatos K, et al. Inhibition of c-Jun-N-terminal kinase increases cardiac peroxisome proliferator-activated receptor alpha expression and fatty acid oxidation and prevents lipopolysaccharide-induced heart dysfunction. J. Biol. Chem. 2011;286:36331–36339. doi: 10.1074/jbc.M111.272146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Penas F, et al. Treatment in vitro with PPARalpha and PPARgamma ligands drives M1-to-M2 polarization of macrophages from T. cruzi-infected mice. Biochim Biophys. Acta. 2015;1852:893–904. doi: 10.1016/j.bbadis.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 96.Lee JM, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13:1619–1628. doi: 10.1080/15548627.2017.1343770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ghosh A, et al. Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J. Biol. Chem. 2015;290:10309–10324. doi: 10.1074/jbc.M114.610659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brady OA, Martina JA, Puertollano R. Emerging roles for TFEB in the immune response and inflammation. Autophagy. 2018;14:181–189. doi: 10.1080/15548627.2017.1313943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Visvikis O, et al. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ouimet M, et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reich-Slotky R, et al. Gemfibrozil inhibits Legionella pneumophila and Mycobacterium tuberculosis enoyl coenzyme A reductases and blocks intracellular growth of these bacteria in macrophages. J. Bacteriol. 2009;191:5262–5271. doi: 10.1128/JB.00175-09. [DOI] [PMC free article] [PubMed] [Google Scholar]