Abstract
CYP2C19 and CYP2D6 are important drug-metabolizing enzymes that are involved in the metabolism of around 30% of all medications. Importantly, the corresponding genes are highly polymorphic and these genetic differences contribute to interindividual and interethnic differences in drug pharmacokinetics, response, and toxicity. In this study we systematically analyzed the frequency distribution of clinically relevant CYP2C19 and CYP2D6 alleles across Europe based on data from 82,791 healthy individuals extracted from 79 original publications and, for the first time, provide allele confidence intervals for the general population. We found that frequencies of CYP2D6 gene duplications showed a clear South-East to North-West gradient ranging from <1% in Sweden and Denmark to 6% in Greece and Turkey. In contrast, an inverse distribution was observed for the loss-of-function alleles CYP2D6*4 and CYP2D6*5. Similarly, frequencies of the inactive CYP2C19*2 allele were graded from North-West to South-East Europe. In important contrast to previous work we found that the increased activity allele CYP2C19*17 was most prevalent in Central Europe (25–33%) with lower prevalence in Mediterranean-South Europeans (11–24%). In summary, we provide a detailed European map of common CYP2C19 and CYP2D6 variants and find that frequencies of the most clinically relevant alleles are geographically graded reflective of Europe’s migratory history. These findings emphasize the importance of generating pharmacogenomic data sets with high spatial resolution to improve precision public health across Europe.
Subject terms: Genetic variation, Genetics research, Epidemiology
Introduction
Interindividual variability in therapeutic drug response can result in adverse drug reactions (ADRs) or lack of efficacy and constitutes a key challenge for health care systems. Notably, 40–70% of patients experience insufficient drug response or drug toxicity and ADRs account for 6.5% of all hospital admissions of which up to 30% are life threatening in at-risk subpopulations [1–4]. Genetic polymorphisms in drug-metabolizing enzymes, transporters, or drug targets explain around 20–30% to these interindividual differences [5].
Cytochrome P450 (CYP) enzymes constitute a polymorphic superfamily, consisting of 57 functional members in humans [6], that metabolize >80% of all clinically used medications [7]. Among those, CYP2C19 and CYP2D6 are of particular clinical relevance, as they are highly polymorphic and implicated in the metabolism of numerous widely prescribed drugs. CYP2C19 substrates include the tricyclic antidepressants amitriptyline, clomipramine, doxepin and imipramine, the selective serotonin reuptake inhibitors citalopram and sertraline, the antifungal voriconazole, as well as the antiplatelet agent clopidogrel. CYP2C19*2 (rs4244285) is the most common allelic variant in Caucasians and results in aberrant splicing and loss-of-enzyme activity [8]. In contrast, the regulatory polymorphism rs12248560 defining CYP2C19*17 increases transcriptional activity and causes the ultrarapid CYP2C19 metabolism [9].
CYP2D6 metabolizes around 25% of currently prescribed drugs, including various antidepressants, neuroleptics, beta-blockers, opioids, antiemetics, and antiarrhythmics. Of the more than 100 allelic variants for CYP2D6 that have been described so far, CYP2D6*4 (rs3892097) is the most prevalent loss-of-function allele in Caucasian individuals. Furthermore, CYP2D6 harbors functionally relevant copy number variations (CNVs) in which the whole open reading frame is duplicated (e.g., CYP2D6*1×N and CYP2D6*2×N) or deleted (CYP2D6*5), resulting in increased or decreased metabolism of CYP2D6 substrates, respectively.
While frequencies of CYP2C19 and CYP2D6 variations have been extensively studied, these studies were either focused on selected geographical regions or analyzed data aggregated by ethnicity or ancestry [10–12]. Therefore, in the present study, we systematically analyzed 79 original publications covering 82,791 healthy volunteers throughout Europe for CYP2C19 and CYP2D6 variants to provide a high-resolution map of pharmacogenetically relevant variability across European populations. Analysis of this consolidated data set revealed that the loss-of-function variants CYP2C19*2, CYP2D6*4, and CYP2D6*5 were graded from Northern Europe to the Mediterranean, whereas CYP2D6 duplications showed an inverse pattern. Furthermore, in contrast to previous reports we find clear evidence that CYP2C19*17 is most common in Central Europe, whereas prevalence is lower in South Europeans. Combined, these data reveal the extent of intra-European pharmacogenetic variability and underscore the importance of using local genomic information for conducting pharmacogenetic analyzes, clinical trials, and precision public health.
Methods
For the present study we performed a systematic literature survey of the PubMed database covering articles published before December 2018. The search query criteria were (CYP2C19 or CYP2D6) AND (allele OR genotype OR frequency OR prevalence OR polymorphism) AND (European). All studies reporting genotype or allele frequencies of CYP2C19*2 (rs4244285; NC_000010.11:g.94781859 G > A), CYP2C19*17 (rs12248560; NC_000010.11:g.94761900C > T), CYP2D6*3 (rs35742686; NC_000022.11:g.42128242delT), CYP2D6*4 (rs3892097; NC_000022.11:g.42128945C > T), CYP2D6*5 (CYP2D6 gene deletion), or of functional gene duplications (CYP2D6*1×N or CYP2D6*2×N) in healthy individuals of clear geographic origin within a European country were included. Variant positions are provided based on GRCh38. Only original research articles available in English were considered. In addition, we included data from the Genome Aggregation Database [13], the 1000 Genomes Project [14], the SweGen project [15], and the Estonian biobank [16]. As a result, we identified 79 original articles and 82,791 individuals were included in the analysis (Supplementary Tables 1 and 2). For countries for which multiple studies were available, data were aggregated using a weighted average approach using the studies’ cohort sizes as weighting factor. For additional information about the haplotypes in question we refer the interested reader to the website of the Pharmacogene Variation Consortium (https://www.pharmvar.org).
Results
Frequencies of important CYP2C19 alleles exhibit large intra-European differences
For CYP2C19 we assessed the prevalence of the loss-of-function allele CYP2C19*2 and the increased function variant CYP2C19*17. In Europe, the frequency of CYP2C19*2 was the highest in Cyprus (21%) and Malta (20%), whereas the lowest prevalence was reported in Czech Republic (8%; Fig. 1; Table 1). Furthermore, frequencies were high in Romani individuals (20.8%). Overall, CYP2C19*2 was slightly more prevalent in Northern and Western European countries, such as Finland (17.5%), the Faroe Islands (18.8%), and France (17.7%), compared with countries on the Mediterranean coast, including Italy (11.8%) and Turkey (11.3%).
Fig. 1.
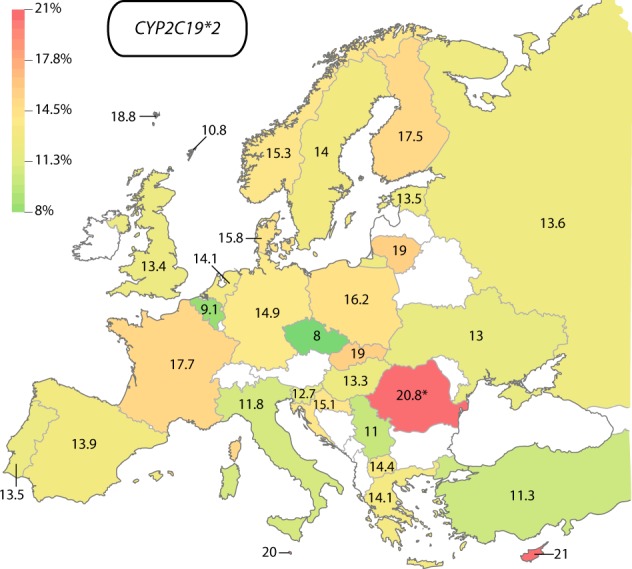
European map of CYP2C19*2 allele frequencies. The lowest frequencies were found in the Czech republic (8%, green), whereas highest frequencies were described in Cyprus (21%). Frequency in Romania (indicated by asterisk) refers exclusively to the Romani population
Table 1.
Frequencies of important CYP2C19 and CYP2D6 alleles in Europe
| Functional consequence | Frequency in % | |||||||
|---|---|---|---|---|---|---|---|---|
| Country/Geographic region | Studies | Individuals | CYP2C19 | CYP2D6 | ||||
| *2 | *17 | *3 | *4 | *5 | Dupl | |||
| Inactive | Increased function | Inactive | Inactive | Inactive | Increased function | |||
| Austria | 1 | 93 | / | / | 0.5 (1.2) | 14 (5.9) | 1.6 (2.1) | 1.6 (2.1) |
| Belgium | 1 | 121 | 9.1 (4.3) | / | / | / | / | / |
| Croatia | 3 | 1119–1247 | 15.2 (1.7) | 23.5 (2) | 2.3 (0.7) | 16.7 (1.7) | 1 (0.5) | 3.4 (0.8) |
| Cyprus | 1 | 40 | 21 (10.6) | 11 (8.1) | 4 (5.1) | 21 (10.6) | / | / |
| Czech Republic | 2 | 42–265 | 8 (2.7) | 29 (4.6) | 1.6 (1.3) | 21.6 (4.2) | 3.1 (1.8) | / |
| Denmark | 3 | 579–634 | 15.8 (2.4) | 20.1 (2.6) | 2.2 (1) | 20.5 (2.6) | 5.9 (1.5) | 0.8 (0.6) |
| Estonia | 1 | 35,506–44,448 | 13.5 (0.3) | 26.4 (0.3) | 1.8 (0.1) | 16.7 (0.3) | 1.5 (0.1) | 0.3 (0.04) |
| Faroe Islands | 2 | 309–311 | 18.8 (3.6) | 15.4 (3.4) | 0.2 (0.4) | 33.4 (4.4) | / | / |
| Finland | 6 | 12,589–13,956 | 17.5 (0.5) | 19.6 (0.6) | 3.5 (0.3) | 10 (0.4) | 2.2 (0.2) | 4.3 (0.3) |
| France | 3 | 607 | 17.7 (2.6) | / | / | / | / | / |
| Germany | 8 | 923–1758 | 14.9 (1.4) | 24.9 (1.7) | 1.1 (0.4) | 19.6 (1.6) | 3.2 (0.7) | 1.3 (0.4) |
| Greece | 3 | 327 | 14.1 (3.2) | 18.2 (3.5) | 2.1 (1.3) | 17.7 (3.5) | / | 6 (2.2) |
| Hungary | 4 | 530–591 | 13.3 (2.3) | 23 (2.9) | 1.6 (0.9) | 19 (2.7) | 1.8 (0.9) | 1.8 (0.9) |
| Italy | 8 | 914–917 | 11.8 (1.8) | 22.1 (2.3) | 1 (0.5) | 16.4 (2) | 2.4 (0.8) | 3 (0.9) |
| Lithuania | 1 | 20 | 19 (14.4) | 25 (15.9) | 2 (5.2) | 24 (15.7) | / | / |
| Malta | 1 | 41 | 20 (10.3) | 26 (11.3) | 0 | 18 (9.9) | / | / |
| Netherlands | 5 | 1114–1158 | 14.1 (1.7) | 19 (1.9) | 1.5 (0.6) | 18.9 (1.9) | / | / |
| Norway | 3 | 83–403 | 15.3 (3) | 22 (3.4) | 0 | 21.1 (3.3) | 6 (2) | / |
| Orkney Islands | 1 | 88 | 10.8 (5.4) | / | / | / | / | / |
| Poland | 5 | 166–791 | 16.3 (2.2) | 29.8 (2.7) | 1.6 (0.7) | 20.8 (2.4) | / | / |
| Portugal | 4 | 279–400 | 13.4 (2.8) | / | 0.7 (0.7) | 17 (3.1) | 2.6 (1.3) | 3 (1.4) |
| Republic of North Macedonia | 2 | 100–184 | 14.4 (4.3) | 20.1 (4.9) | 2 (1.7) | 17 (4.6) | 1.5 (1.5) | 2.5 (1.9) |
| Romania (Romani) | 3 | 426–562 | 20.8 (2.8) | / | / | 22.5 (2.9) | / | / |
| Russia | 4 | 391–1663 | 13.6 (1.4) | 15 (1.4) | 1.2 (0.4) | 17.6 (1.5) | 1.6 (0.5) | 2.4 (0.6) |
| Sardinia | 2 | 76 | / | / | 2.6 (3) | 15.8 (6.9) | 1.3 (2.1) | 2 (2.6) |
| Serbia | 1 | 46 | 11 (7.6) | 18 (9.3) | 0 | 16 (8.9) | / | / |
| Slovakia | 1 | 26 | 19 (12.7) | 33 (15.2) | 2 (4.5) | 28 (14.5) | / | / |
| Slovenia | 3 | 1952–2081 | 12.7 (1.2) | 23 (1.5) | 1.8 (0.5) | 16.8 (1.3) | / | / |
| Spain | 14 | 1215–2328 | 14 (1.2) | 17.1 (1.3) | 1.2 (0.4) | 18.6 (1.3) | 2.3 (0.5) | 3.5 (0.6) |
| Sweden | 6 | 1816–2020 | 14 (1.3) | 19.2 (1.4) | 1.6 (0.5) | 20.7 (1.5) | 4.1 (0.7) | 0.5 (0.3) |
| Turkey | 6 | 689–785 | 11.3 (1.9) | 24 (2.5) | 0.7 (0.5) | 13.2 (2) | 1.8 (0.8) | 5.6 (1.4) |
| Ukraine | 2 | 52–689 | 13 (2.1) | 25 (2.7) | 2 (0.9) | 18.9 (2.5) | / | / |
| United Kingdom | 2 | 91–168 | 13.4 (4.3) | 24.2 (5.4) | 3.3 (2.3) | 24.2 (5.4) | / | / |
Note that the number of individuals in a given country or geographic region for whom genotype data are available can differ between alleles. Values in brackets indicate the 90% confidence intervals
On the contrary, CYP2C19*17 was most common in Central Europe with highest frequencies in Slovakia (33%), Poland (29.8%), and the Czech Republic (29%; Fig. 2); Table 1. However, the CYP2C19 genotyping data reported for Slovakia included only 26 subjects and should thus be interpreted with caution [17]. In contrast, frequencies were lower in Southern European countries, such as Spain (17.1%), Greece (18.2%), and Cyprus (11%), as well as Scandinavia (19–22%) and Russia (15%).
Fig. 2.
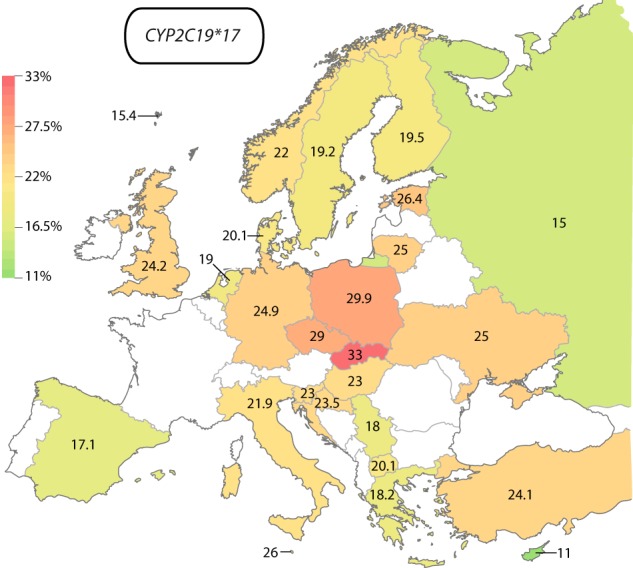
European map of CYP2C19*17 allele frequencies. The lowest frequencies were found in Cyprus (11%, green), whereas highest frequencies were described in Slovakia (33%)
CYP2D6 gene duplications are graded from South-East to North-West Europe
Functional duplications of CYP2D6 (CYP2D6*1×N and CYP2D6*2×N) were most prevalent in the South-East European countries Greece (6%) and Turkey (5.6%), while lower frequencies were found in South-Western Europe, including Spain (3.5%), Italy (3%), and Portugal (3%; Fig. 3; Table 1). In contrast, frequencies in Northern and Central Europe, including Austria (1.6%), Germany (1.3%), Denmark (0.8%), and Sweden (0.5%), were substantially lower. Surprisingly, CYP2D6 duplications were common in Finland (4.3%) at levels similar to Southern Europe.
Fig. 3.
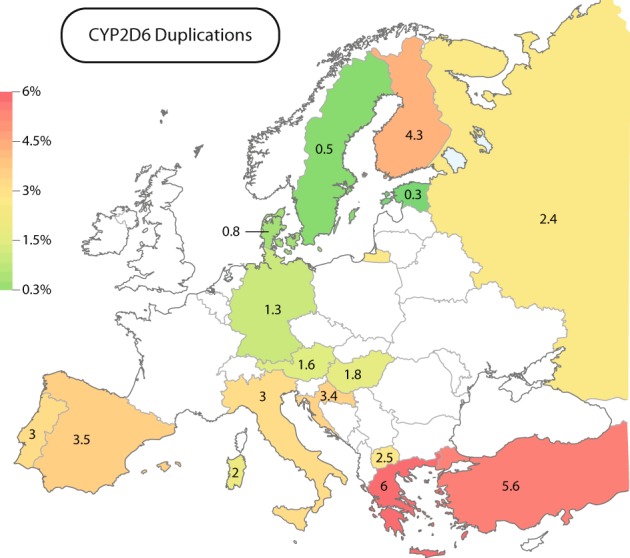
European map of CYP2D6 allele duplications (CYP2D6*1×N and CYP2D6*2×N). The lowest frequencies were found in Northern European countries, such as Estonia (0.3%) and Sweden (0, 5%, green), whereas highest frequencies were described in South-Eastern Europe (Greece; 6% and Turkey; 5.6%, red)
CYP2D6 loss-of-function alleles are distributed along a North-to-South gradient
Importantly, the CYP2D6 loss-of-function alleles CYP2D6*4 and CYP2D6*5 showed an inverse profile (Fig. 4; Table 1). CYP2D6*4 was most prevalent throughout Northern and Central Europe with frequencies pivoting around 20–25%. The highest CYP2D6*4 frequency in Europe was observed on the Faroe Islands (33.4%). In contrast, frequencies were substantially lower in most Southern European countries, such as Turkey (13.2%), Italy (16.4%), and Greece (17.7%). Notably, Finns contradict this trend with a population frequency of 10%, which is substantially lower than in neighboring Sweden (19.2%), Norway (22%), and Estonia (16.7%).
Fig. 4.
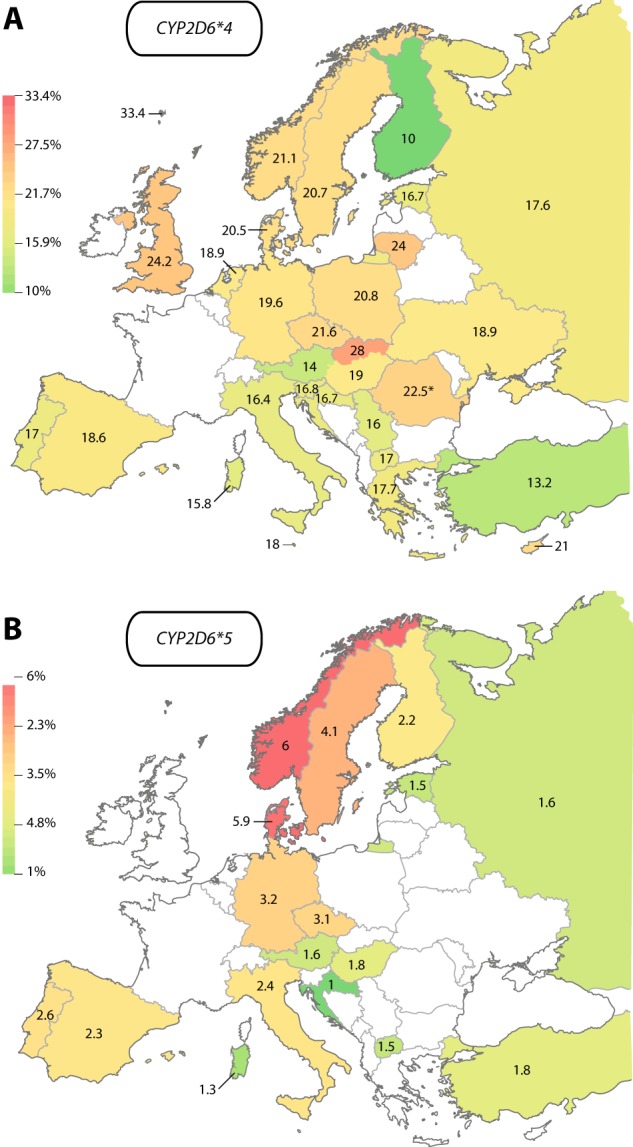
European maps of the CYP2D6 loss-of-function alleles CYP2D6*4 and CYP2D6*5. a CYP2D6*4 frequencies differed between 10% in Finland (green) and 33.4% on the Faroe Islands (red). Frequency in Romania (indicated by asterisk) refers exclusively to the Romani population. b CYP2D6*5 was most common in Norway (6%, red) and Denmark (5.9%), whereas it was most rare in Croatia (1%)
Similar trends were observed for the CYP2D6 deletion variant CYP2D6*5, which was most frequent in Norway (6%), Denmark (5.9%), and Sweden (4.1%), whereas prevalence in Central Europe pivoted around 3% and lowest CYP2D6*5 frequencies were observed in Southern European countries, such as Croatia (1%), Sardinia (1.3%), North Macedonia (1.5%), and Turkey (1.8%). Again, population frequency of CYP2D6*5 in Finland (2.2%) contrasted surrounding Scandinavian countries and was more similar to prevalence rates in Central Europe.
In contrast to CYP2D6*4 and CYP2D6*5, no clear gradients were detected for CYP2D6*3, whose frequencies pivoted around 0–2% throughout Europe. Notable exceptions are the relatively high, geographically disperse frequencies in Cyprus (4%), Finland (3.5%), and the UK (3.3%; Supplementary Fig. 1).
Discussion
Interethnic differences in drug pharmacokinetics or dynamics constitute important factors to consider for increasingly multinational drug development programs and genetic variability in drug-metabolizing enzymes constitutes an important factor underlying these differences. As a result, the labels of multiple marketed drugs, including rosuvastatin, carbamazepine, and tacrolimus, contain recommendations to adjust starting doses based on ethnicity [18]. CYP2C19 and CYP2D6 harbor multiple genetic polymorphisms, which differ substantially between ethnic groups and geographic regions and can entail clinically important differences in drug response. To date, numerous studies have analyzed the frequencies of these polymorphisms; yet, the available allele frequency data have, to our knowledge, not yet been systematically consolidated into high-resolution maps of CYP2C19 and CYP2D6 variability within Europe. We therefore compiled data from 79 original publications resulting in aggregated genotypes for the most relevant CYP2C19 and CYP2D6 alleles from 82,791 healthy individuals. Notably, while most studies provided data from unrelated individuals, we cannot exclude relatedness across studies. However, we do not expect this fraction to significantly impact the accuraccy of the reported frequencies.
Frequency of functional CYP2D6 gene duplications was highest in Greece and Turkey and lowest in Scandinavian countries, which is in accordance with decreasing frequencies of ultrarapid metabolizers in a direction from Southern to Northern European populations [19]. Globally, CYP2D6 duplication is most common in North-East Africa and the Middle East with frequencies of 7–16% [20–22]. It has been speculated that the evolutionary basis for this gradient is the role of CYP2D6 in the detoxification of plant alkaloids, which allowed carriers of duplicated alleles to tap food sources during times of starvation that would have been toxic for normal CYP2D6 metabolizers [23]. Inversely, frequencies of the loss-of-function alleles CYP2D6*4 and CYP2D6*5 were highest in Scandinavia and lowest on the Mediterranean with further decreasing frequencies in Ethiopia and the Arabian peninsula [20–22]. These data thus corroborate the hypothesis that CYP2D6 metabolic capacity might have been under selective pressure specifically in North-East Africa and subsequent migration events resulted in the high frequencies of ultrarapid CYP2D6 metabolizers in Southern Europe.
We observed that CYP2C19*2 was graded from North-West to South-East Europe. Interestingly, we observed a high frequency of CYP2C19*2 in Romani (20.8%) that was significantly different from the hosting Hungarian population (13.3%; p < 0.01; [24]). The Roma minority originates from North-West India, and due to a series of population bottlenecks with multiple founder events and low number of interethnic marriages constitutes a relatively homogeneous ethnic group [25]. As a consequence of this complex population history, CYP2C19*2 frequencies in Roma were similar to those reported in North Indian populations [26]. Thus, pharmacogenetic variability in Roma is distinctly different from European populations and affiliation to a Roma group might be a factor of consideration for treatment decisions of CYP2C19 substrates.
The distribution of CYP2C19*17 was highest in Central Europe and lower in Southern European countries. Our findings are in drastic contrast to a meta-analysis performed by Fricke-Galindo et al. who reported that CYP2C19*17 is predominantly found in Mediterranean countries with frequencies of 42% [11]. However, we find that frequencies are substantially lower throughout Southern Europe, pivoting around 20–25%. Careful revisiting of the original data revealed that instead of the frequency (14.9%), Fricke-Galindo et al. erroneously used the number of individuals (n = 42) for the Spanish population [27] as population frequency. Our findings of moderate CYP2C19*17 frequencies in Southern Europe align with data from Northern African and Middle Eastern populations in which CYP2C19*17 allele frequencies between 17.9% and 26.9% have been reported for Ethiopians, Saudi Arabians, Kurds, and Turks [9, 28–30]. Furthermore, low frequencies (15.9%) have been found in Sephardic Jews [31] who originated from Jews on the Iberian peninsula in the 15th century, which are in close agreement with the aggregated prevalence we found in contemporary Spanish individuals (17.1%). The distribution of CYP2C19 alleles thus reflects the migratory history of European populations.
These findings have potentially important implications, as CYP2C19 genotype is included as a pharmacogenomic biomarker in the drug labels of 22 medications. Furthermore, guidelines issued by pharmacogenetics expert workgroups (CPIC and DPWG) provide recommendations to optimize genotype-guided prescription for 14 drugs [32]. For instance, CYP2C19 genotype affects treatment efficacy and risk of adverse events when treated with the antidepressant escitalopram [33], and for ultrarapid CYP2C19 metabolizers it is recommended to select an alternative drug not predominantly metabolized by CYP2C19. As the cost effectiveness of pharmacogenetic implementation is dependent on carrier frequencies, falsely high population frequencies might erroneously incentivize pre-emptive CYP2C19 genotyping.
Notably, while genotype data for CYP2C19 and CYP2D6 were available for more than 80,000 individuals from 31 European countries, cohort coverage was geographically highly unequal (Table 1). For eight countries less than 100 individuals were genotyped and, as a result, population frequencies in these countries could only be estimated with wide confidence intervals. Thus, these analyzes incentivize the country-specific expansion of genotype data to further refine estimates of intra-European CYP allele frequencies. Furthermore, while CYP genotype-derived activity scores constitute important proxies for the prediction of metabolic capacity, they can only explain a fraction of the observed functional variability [34]. One underlying reason could be rare variants beyond the tested polymorphisms that contribute to gene function. In this regard CYP2C19 and CYP2D6 have indeed been found to harbor a plethora of rare genetic single nucleotide variants (SNVs) with putative functional importance [35–37]. Furthermore, rare population-specific CNVs can contribute to functional variability. For instance, CYP2C19 has recently been found to be deleted specifically in Finns with frequencies of 0.8% [38]. However, information regarding the prevalence of these rare SNVs and CNVs is currently not available with high geographic resolution and the generation of such sequencing-based pharmacogenomic data sets constitutes an interesting avenue for future research that will help to refine genotype-guided drug response predictions [39, 40].
In conclusion, we provide refined maps of clinically important CYP2C19 and CYP2D6 genetic variability across European populations. Our findings support the need for refined pharmacogenomic mapping to guide precision public health.
Supplementary information
Acknowledgements
The work in the authors’ laboratories is supported by the Swedish Research Council [grant agreement numbers: 2016–01153 and 2016–01154], by the European Union’s Horizon 2020 research and innovation program U-PGx [grant agreement number 668353], by the Strategic Research Programme in Diabetes at Karolinska Institutet, by the Lennart Philipson Foundation, and by the Harald and Greta Jeansson Foundation. No writing assistance was utilized in the production of this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0480-8) contains supplementary material, which is available to authorized users.
References
- 1.Impicciatore P, Choonara I, Clarkson A, Provasi D, Pandolfini C, Bonati M. Incidence of adverse drug reactions in paediatric in/out-patients: a systematic review and meta-analysis of prospective studies. Br J Clin Pharmacol. 2001;52:77–83. doi: 10.1046/j.0306-5251.2001.01407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pirmohamed M, James S, Meakin S, Green S, Scott AK, Walley TJ, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. Brit Med J. 2004;329:15–9. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budnitz DS, Shehab N, Kegler SR, Richards CL. Medication use leading to emergency department visits for adverse drug events in older adults. Ann Intern Med. 2007;147:755–65. doi: 10.7326/0003-4819-147-11-200712040-00006. [DOI] [PubMed] [Google Scholar]
- 4.Lauschke Volker, Ingelman-Sundberg Magnus. The Importance of Patient-Specific Factors for Hepatic Drug Response and Toxicity. International Journal of Molecular Sciences. 2016;17(10):1714. doi: 10.3390/ijms17101714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauschke VM, Ingelman-Sundberg M. Prediction of drug response and adverse drug reactions: from twin studies to next generation sequencing. Eur J Pharm Sci. 2019;130:65–77. doi: 10.1016/j.ejps.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 6.Fujikura K, Ingelman-Sundberg M, Lauschke VM. Genetic variation in the human cytochrome P450 supergene family. Pharm Genom. 2015;25:584–94. doi: 10.1097/FPC.0000000000000172. [DOI] [PubMed] [Google Scholar]
- 7.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 8.de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419–22. [PubMed] [Google Scholar]
- 9.Sim SC, Risinger C, Dahl M-L, Aklillu E, Christensen M, Bertilsson L, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–13. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther. 2017;102:688–700. doi: 10.1002/cpt.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fricke-Galindo I, Céspedes-Garro C, Rodrigues-Soares F, Naranjo ME, Delgado A, de Andres F, et al. Interethnic variation of CYP2C19 alleles, ‘predicted’ phenotypes and ‘measured’ metabolic phenotypes across world populations. Pharm J. 2016;16:113–23. doi: 10.1038/tpj.2015.70. [DOI] [PubMed] [Google Scholar]
- 12.Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017;19:69–76. doi: 10.1038/gim.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ameur A, Dahlberg J, Olason P, Vezzi F, Karlsson R, Martin M, et al. SweGen: a whole-genome data resource of genetic variability in a cross-section of the Swedish population. Eur J Hum Genet. 2017;25:1253–60. doi: 10.1038/ejhg.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reisberg S, Krebs K, Lepamets M, Kals M, Mägi R, Metsalu K, et al. Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: challenges and solutions. Genet Med. 2018;20:4. doi: 10.1038/s41436-018-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizzi C, Dalabira E, Kumuthini J, Dzimiri N, Balogh I, Basak N, et al. A European spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PLoS ONE. 2016;11:e0162866. doi: 10.1371/journal.pone.0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasuda SU, Zhang L, Huang S-M. The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin Pharmacol Ther. 2008;84:417–23. doi: 10.1038/clpt.2008.141. [DOI] [PubMed] [Google Scholar]
- 19.Arvanitidis K, Ragia G, Iordanidou M, Kyriaki S, Xanthi A, Tavridou A, et al. Genetic polymorphisms of drug-metabolizing enzymes CYP2D6, CYP2C9, CYP2C19 and CYP3A5 in the Greek population. Fundam Clin Pharmacol. 2007;21:419–26. doi: 10.1111/j.1472-8206.2007.00510.x. [DOI] [PubMed] [Google Scholar]
- 20.Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharm Exp Ther. 1996;278:441–6. [PubMed] [Google Scholar]
- 21.McLellan RA, Oscarson M, Seidegård J, Evans D, Ingelman-Sundberg M. Frequent occurrence of CYP2D6 gene duplication in Saudi Arabians. Pharmacogenetics. 1997;7:187–91. doi: 10.1097/00008571-199706000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Fuselli S, Dupanloup I, Frigato E, Cruciani F, Scozzari R, Moral P, et al. Molecular diversity at the CYP2D6 locus in the Mediterranean region. Eur J Hum Genet. 2004;12:916–24. doi: 10.1038/sj.ejhg.5201243. [DOI] [PubMed] [Google Scholar]
- 23.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharm J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 24.Sipeky C, Weber A, Szabo M, Melegh BI, Janicsek I, Tarlos G, et al. High prevalence of CYP2C19*2 allele in Roma samples: study on Roma and Hungarian population samples with review of the literature. Mol Biol Rep. 2013;40:4727–35. doi: 10.1007/s11033-013-2569-4. [DOI] [PubMed] [Google Scholar]
- 25.Martínez-Cruz B, Mendizabal I, Harmant C, de Pablo R, Ioana M, Angelicheva D, et al. Origins, admixture and founder lineages in European Roma. Eur J Hum Genet. 2016;24:937–43. doi: 10.1038/ejhg.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamba JK, Dhiman RK, Kohli KK. CYP2C19 genetic mutations in North Indians. Clin Pharmacol Ther. 2000;68:328–35. doi: 10.1067/mcp.2000.109365. [DOI] [PubMed] [Google Scholar]
- 27.Vicente J, González-Andrade F, Soriano A, Fanlo A, Martínez-Jarreta B, Sinués B. Genetic polymorphisms of CYP2C8, CYP2C9 and CYP2C19 in Ecuadorian Mestizo and Spaniard populations: a comparative study. Mol Biol Rep. 2014;41:1267–72. doi: 10.1007/s11033-013-2971-y. [DOI] [PubMed] [Google Scholar]
- 28.Al-Jenoobi FI, Alkharfy KM, Alghamdi AM, Bagulb KM, Al-Mohizea AM, Al-Muhsen S, et al. CYP2C19 genetic polymorphism in Saudi Arabians. Basic Clin Pharmacol Toxicol. 2013;112:50–54. doi: 10.1111/j.1742-7843.2012.00919.x. [DOI] [PubMed] [Google Scholar]
- 29.Saeed LH, Mayet AY. Genotype-phenotype analysis of CYP2C19 in healthy saudi individuals and its potential clinical implication in drug therapy. Int J Med Sci. 2013;10:1497–502. doi: 10.7150/ijms.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dehbozorgi M, Kamalidehghan B, Hosseini I, Dehghanfard Z, Sangtarash MH, Firoozi M, et al. Prevalence of the CYP2C19*2 (681 G>A), *3 (636 G>A) and *17 (‑806 C>T) alleles among an Iranian population of different ethnicities. Mol Med Rep. 2018;17:4195–202. doi: 10.3892/mmr.2018.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott SA, Martis S, Peter I, Kasai Y, Kornreich R, Desnick RJ. Identification of CYP2C19*4B: pharmacogenetic implications for drug metabolism including clopidogrel responsiveness. Pharm J. 2012;12:297–305. doi: 10.1038/tpj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bank PC, Caudle KE, Swen JJ, Gammal RS, Whirl-Carrillo M, Klein TE, et al. Comparison of the guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin Pharmacol Ther. 2018;103:599–618. doi: 10.1002/cpt.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jukić MM, Haslemo T, Molden E, Ingelman-Sundberg M. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: a retrospective study based on 2,087 patients. Am J Psychiatry. 2018;175:463–70. doi: 10.1176/appi.ajp.2017.17050550. [DOI] [PubMed] [Google Scholar]
- 34.Gaedigk A, Dinh J, Jeong H, Prasad B, Leeder J. Ten years’ experience with the CYP2D6 activity score: a perspective on future investigations to improve clinical predictions for precision therapeutics. J Pers Med. 2018;8:15. doi: 10.3390/jpm8020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozyra M, Ingelman-Sundberg M, Lauschke VM. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med. 2017;19:20–9. doi: 10.1038/gim.2016.33. [DOI] [PubMed] [Google Scholar]
- 36.Bush WS, Crosslin DR, Owusu-Obeng A, Wallace J, Almoguera B, Basford MA, et al. Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin Pharmacol Ther. 2016;100:160–9. doi: 10.1002/cpt.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingelman-Sundberg M, Mkrtchian S, Zhou Y, Lauschke VM. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum Genom. 2018;12:26. doi: 10.1186/s40246-018-0157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos M, Niemi M, Hiratsuka M, Kumondai M, Ingelman-Sundberg M, Lauschke VM, et al. Novel copy-number variations in pharmacogenes contribute to interindividual differences in drug pharmacokinetics. Genet Med. 2018;20:622–9. doi: 10.1038/gim.2017.156. [DOI] [PubMed] [Google Scholar]
- 39.Lauschke VM, Ingelman-Sundberg M. Precision medicine and rare genetic variants. Trends Pharm Sci. 2016;37:85–6. doi: 10.1016/j.tips.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Lauschke VM, Ingelman-Sundberg M. How to consider rare genetic variants in personalized drug therapy. Clin Pharmacol Ther. 2018;103:745–8. doi: 10.1002/cpt.976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


