Abstract
Seed dormancy (SD) and longevity (SL) may share developmental and genetic mechanisms, as both traits are developed in the same maternal environment and evolved to coordinate the timing of germination and the life span of seedbanks. To test the hypothesis, allelic variants at the SD1-2, 7-1, 7-2, and 12 loci from weedy and cultivated rice (Oryza sativa) were assembled into the same genetic background, and 16 homozygous lines selected as a tetragenic system. These lines were evaluated for SD measured by germination at 7, 21, 35, and 150 days of after-ripening (DAR), and for SL measured by the seed decay rate and survivability in the soil of a rice field for 7 months. Pyramiding the alleles from weedy rice lengthened the dormancy duration, and seeds survived in the soil remained dormant at the excavation. Germination levels at 7 to 150 DAR were correlated positively with the seed decay rate (r = 0.41–0.53) and negatively with the survivability (r = −0.45 to −0.28) in the tetragenic system. All four loci contributed to genotypic variation for each of the SD and SL measurements through main and/or epistatic (two- to four-order interactions) effects. SD7-1 (identical to the pericarp color gene Rc) played a major role in regulating seedbank longevity when interacted with the other SD gene(s). This research provided evidence that natural genes controlling SD are involved in regulation of soil seedbank longevity. Thus, accumulation of SD genes in a population could result in persistence of wild plants and weeds in conventional tillage systems.
Subject terms: Plant genetics, Epistasis
Introduction
Seed dormancy (SD) and longevity (SL) are adaptive traits evolved to regulate the timing of germination and the life span of soil seedbanks in local ecosystems. Both traits vary in degree in many species, including conspecific/congeneric crop-weed complexes. For example, weedy forms of rice (Oryza spp.), oats (Avena spp.), sorghum (Sorghum bicolor), and sunflowers (Helianthus annuus) usually yield dormant seeds, which could stay alive in the soil for years resulting in persistent seedbanks (Goss and Brown 1939; Miller et al. 1985; Noldin et al. 2006; Brunick 2008; Abeysekara 2010; Baek and Chung 2012; Adugna 2013). In contrast, crop cultivars have weak dormancy, and their seeds left in the field after harvesting could barely survive over the next season. It is generally believed that primary dormancy is required for seeds to enter soil seedbanks after dispersal from the mother plant (Roberts 1972; Simpson 1990). However, it is uncertain to what extent the primary dormancy could affect seedbank longevity, if natural genes controlling SD are also involved in regulation of SL, and if SD and soil seedbank persistence are related (Thompson et al. 2003; Saatkamp et al. 2009; Sano et al. 2016; Finch-Savage and Footitt 2017). Answers to these questions could help to define the biological factors influencing seedbank dynamics, weed persistence, and consequences of gene flow from genetically engineered crops into wild/weedy relatives (Buhler et al. 1997; Ellstrand et al. 2010).
The SD and SL traits may share developmental and genetic mechanisms, as they are developed in the same maternal environment before maturation (Bewley et al. 2013). The shared mechanisms could be inferred by quantitative trait loci (QTL) associated with SD and SL. In Arabidopsis thaliana, some SD QTL were co-localized with the QTL for SL in storage (storability) on short chromosome segments, and the storability was negatively correlated with the dormancy duration (Nguyen et al. 2012). SL is influenced by more factors in a field than in storage, such as secondary dormancy induced to respond to seasonal changes in temperature or moisture (Hilhorst 2007). Thus, further research could be extended from lab to field environments to identify shared and specific genes for the traits using an ecological genetic approach (Anderson et al. 2011).
Weedy rice refers to various forms of the Oryza plants adapted to rice-growing areas from tropical to temperate regions. The most popular form of weedy rice is ‘red’ rice, which is characterized by red pericarp color controlled by the Rc gene (Oka 1988; Delouche et al. 2007). Seeds of red rice usually have strong dormancy and could survive in the soil for up to a decade (Goss and Brown 1939; Cohn and Hughes 1981; Noldin et al. 2006; Abeysekara et al. 2010; Baek and Chung 2012; Zhang et al. 2013). A majority of QTL associated with SD in weedy rice are common in map position between tropical and temperate ecotypes (Gu et al. 2004; Jing et al. 2008; Subudhi et al. 2012; Mispan et al. 2013; Zhang et al. 2017). Some of the common QTL, including SD1-2, 7-1, 7-2, and 12, were isolated as single Mendelian factors or cloned, which is a reductionistic approach used to characterize single genes for a complex trait. For example, SD7-1 is identical to Rc, a transcription factor gene (Sweeney et al. 2006), with the functional allele activating the flavonoid biosynthesis in the lower epidermal cell layer of the pericarp tissue to produce red pigments and also promoting biosynthesis and accumulation of the abscisic acid (ABA) hormone to induce the primary dormancy (Gu et al. 2011). SD1-2 is the gibberellin (GA) synthesis enzyme gene OsGA20ox2, with the loss-of-function mutant delaying seed development and maturation programs resulting in enhanced primary dormancy (Ye et al. 2015). SD12, 1-2, and 7-1 were associated specifically with types of dormancy imposed by the embryo, endosperm, and maternal tissues, respectively, while SD7-2 is likely responsible for the dormancy release (Ye et al. 2013; Gu et al. 2015). Allelic variants at each of the SD1-2, 7-1, 7-2, and 12 loci from weedy and cultivated rice were introduced into the same genetic background as isogenic lines (ILs) in a series of the previous research. These ILs were used to develop a tetragenic system to address the above-stated questions in this research.
Two and higher orders of epistasis were involved in the regulation of SD (Gu et al. 2004) and may also contribute to the genotypic variation in SL. Assembling allelic variants at isolated loci into the same recipient, a holistic approach, is required to precisely model their epistatic interactions. This is particularly important for the present research, as a single gene could not provide a sufficient range of genotypic variation in SD to evaluate its relationship with SL in the soil over a winter season. Thus, the initial objective of this research was to develop a set of 16 ILs for all tetragenic homozygotes of the SD1-2, 7-1, 7-2, and 12 loci, each having two alleles, to enlarge the genotypic variation. Then, the 16 lines were grown under the same conditions, and evaluated for SD in a controlled environment and for SL in a rice field. Data sets from different environments or experiments were used to estimate the strength and direction of correlation between SD and seedbank longevity, and to partition genotypic variations for each of the trait measurements into main and epistatic components of the four loci.
Materials and methods
Development of ILs for tetragenic genotypes of four loci
SD1-2 (A/a), 7-1 (B/b), 7-2 (C/c), and 12 (D/d) were identified as QTL from a cross between ‘EM93-1’ and ‘SS18-2’ in different generations (Gu et al. 2004; Ye et al. 2010). EM93-1 is an early maturation line of cultivated rice (O. sativa subs. indica), while SS18-2 is an accession of weedy rice from Thailand. Genotypes for the four loci can be written as AAbbccdd for EM93-1 or aaBBCCDD for SS18-2, with the upper- and lower-case letters indicating dormancy-enhancing and -reducing alleles, respectively. Each of the QTL alleles from SS18-2 was introduced into the EM93-1 background by recurrent backcrossing and genome-wide marker-assisted selection (MAS) to develop the ILs for SD1-2 (ILabcd), SD7-1 (ILABcd), SD7-2 (ILAbCd), and SD12 (ILAbcD) in a series of the previous research (Gu et al. 2010, 2011; Ye et al. 2013).
The ILs for the four loci were used as parents in three crosses to pyramid two or more alleles from SS18-2 in the background of EM93-1 (ILAbcd) in this research (Fig. S1). The first two crosses were made between ILAbcD and ILABcd to select ILABcD, or between ILabcd and ILAbCd to select ILabCd from the F2 populations. The third cross was made between ILABcD and ILabCd to select ILs for the remaining nine genotypes for the four loci (Fig. S1).
Plant cultivation, genotyping and seed harvesting
Germinated seeds from the 16 ILs were transferred to 200-cell plug trays (Bootstrap Farmers), with one seed per cell, and cultured with a nutrient solution (Yoshida et al. 1976) for 3 weeks. Seedlings were genotyped with the SD1-2, 7-1, 7-2, and 12 markers to verify the identity and purity of the ILs. The markers were selected from the QTL underlying genes or peak positions (Gu et al. 2010; Gu et al. 2011; Ye et al. 2013). DNA samples were prepared from the leaf tissue and genotyped using methods described in the previous research.
Seedlings were transplanted into pots, which were filled with clay mixed with Sunshine® Mix #1 medium, with one plant per pot (12 × 12 × 15-cm dimensions), in a greenhouse. Each line of 24 plants was placed in a container (90 × 60 × 18-cm dimensions). Both SD1-2 and 7-1 have a pleiotropic effect on plant height (Ye et al. 2013). Thus, the 16 lines were distributed based on plant height (tall or semidwarf) to eliminate a light-blocking effect on photosynthesis. Day/night temperatures were set at 30/23° and day-lengths were natural in the greenhouse. Flowering date was tagged when the first panicle of a plant emerged from the leaf sheath. Seeds were harvested at 40 days after flowering, air dried in the greenhouse for 3 days, cleaned and stored in a freezer (−20°) to maintain the status of primary dormancy.
SD assessment
SD is characterized by delay of germination or reduction in germination percentage, which varies in degree with genotypes and durations of after-ripening (AR). Thus, the 16 ILs were evaluated for the dormancy degrees by germination at 7, 21, 35, and 150 days of AR (DAR). Seed samples from 12 plants for each line were moved from the freezer to the room conditions (24–25 °C) for given DAR prior to germination. To prepare a germination test, a sample of about 50 seeds from each plant was distributed on a filter paper in a 9-cm Petri dish, and soaked with 8 ml water. Germination was conducted in incubators at 30 °C, 100% relative humidity (RH), and dark conditions for 7 days. Germinated seeds (radicle protrusion > 3 mm) were counted for individual samples to calculate germination percentages. Thus, for an AR time point, each line had 12 replicates to estimate its genotypic mean and environmental variance.
Seed burial trial and survivability assessment
A burial trial was conducted in a rice field on the Missouri Rice Research Farm in Glennonville (36.58° N, 90.15° W), MO, USA. About 1500 well-developed seeds from each of the 16 ILs were evenly allocated to five samples. A sample of about 300 seeds was mixed with river sand and packed in a 50-ml centrifuge tube, which was predrilled with 20 holes (2-mm diameter) around the wall to circulate air and water. After 7 days of storage at ambient temperatures, the seed samples were buried in the soil at 20 cm deep in the end of September. The field was planted to the rice crop before the trial and the burial area kept undisturbed over the winter to spring seasons for 7 months. The buried samples were excavated in the end of the next April and air dried to evaluate the seed decay rate and survivability.
The seed decay rate measured a proportion of seeds that lost the morphological integrity in the burial trial. A morphologically intact seed was a spikelet with a solid endosperm enclosed in the hull (lemma and palea), while a decayed seed was the spikelet either empty (only the hull left) or having a rotten endosperm tissue. Thus, each of the buried samples was counted for numbers of intact (n1) and decayed (n2) seeds, and the sample size N (N = n1 + n2). The seed decay rate was calculated as the percentage of n2/N.
Seed survivability measured a proportion of seeds that were alive at the excavation and capable to germination after given days of air drying. At the 5th day after the excavation, the intact seeds left in a sample were sterilized with a bleach solution (one part 5.25% hypochlorite and four parts water) for 15 min. and rinsed with running water for 10 min. The clean seeds were germinated in a buffer containing 0.12% Fungicide VITAVAX® T-L (Gustafson McKinney, TX) at 30 °C, 100% RH, and dark conditions for 7 days. Germinated seeds (n3) were counted, but the germination percentages were <5%, based on the number of intact seeds in a sample. The extremely low germination suggests that some of the intact seeds could remain dormant.
The nongerminated seeds in the first test were air dried in a greenhouse for 15 days to break dormancy. The dried seeds were germinated again in new Petri dishes using the above-stated methods. Germinated seeds (n4) in the second test were counted daily for 7 days. Seed survivability for a sample was calculate as (n3 + n4)/N, where the summation (n3 + n4) was the total number of seeds germinated in the first and second tests.
Quantitative genetic analysis
Data collected from individual plants or burial samples were used to estimate three groups of parameters: heritability, genetic component effects, and linear correlation. Heritability (H2) was calculated using Eq. 1:
| 1 |
where Vp, Vg, and Ve are the phenotypic, genotypic, and environmental variances, respectively, for a measurement. One-way analysis of variance (ANOVA) was used to compute mean squares for the error (MSe) and genotype (MSg) components. Ve was estimated by MSe, Vg estimated as (MSg − MSe)/n, and n is the number of plants or samples for each of the ILs.
Genotypic variation for each of the measurements was partitioned into the main (additive, ‘a’s) and interaction (‘i’s, epistasis) components using the linear repression model:
| 2 |
where ym is the phenotypic value for sample m (m = 1 – N, the sample size for a line); μ is the intercept or mean of the model; xi, xj, xk, and xl are variables for genotypes at SD1-2, 7-1, 7-2, and 12, respectively, with i, j, k, or l = −1 for the genotype homozygous for the dormancy-reducing allele or 1 for the genotype homozygous for the dormancy-enhancing allele; a1, a2, a3, and a4 are partial regression coefficients of the variables and also estimates of additive effects for the loci i, j, k, and l, respectively; the ‘z’s are variables for two, three or four orders of epistatic interactions between/among the loci indicated in the subscripts, with values being products of the indicated variables; the ‘i’s are partial regression coefficients for the epistasis variables, or estimates of interactional effects; and em is the residue effect, including the random error and the effect unexplained by the model.
Germination percentages at different DAR, which are reversely proportional to the degree of primary dormancy, were correlated with the seed decay rate or survivability. The correlation strength was estimated with Pearson correlation coefficients. The ANOVA, regression and correlation analyses were performed using the SAS GLM, REG and CORR programs, respectively (SAS Institute Inc. 2013). A stepwise selection at a significance level of 5% was set to retain variables in the regression model (Eq. 2).
Results
Pyramiding SD genes lengthened the dormancy duration
The 16 lines are homozygous recombinants among the SD1-2, 7-1, 7-2, and 12 loci, each having two alleles, as confirmed by the marker genotypes (Fig. S2A). All the 16 ILs were similar to the recipient parent EM93-1 in flowering time and seed (spikelet and caryopsis) morphologies, except the pericarp color (Fig. S2B). Based on phenotypes for the pericarp color, the 16 genotypes were divided equally into the red and white groups, which are alike the donor (SS18-2) and recipient parents, respectively, and correspond exactly to the marker genotypes for SD7-1 (Fig. S2).
A wide range of variation in primary dormancy among the 16 ILs was detected by levels and distribution patterns of germination at/from 7 to 150 DAR. Heritability for germination percentage varied from about 0.6 at 7 and 150 DAR to about 0.9 at 21 to 35 DAR (Table 1). Thus, the genotypic differences for the SD loci accounted for most of the phenotypic variation in germination dynamics during the tested period.
Table 1.
List of correlation and heritability estimates for seed dormancy and longevity in the isogenic lines
| Variablea | Correlation coefficients (r) withb | Heritabilityc | |
|---|---|---|---|
| Seed decay rate | Seed survivability | ||
| Germination at 7 DAR | 0.449 (<0.0001) | −0.285 (0.0117) | 0.59 |
| Germination at 21 DAR | 0.535 (<0.0001) | −0.322 (0.0043) | 0.89 |
| Germination at 35 DAR | 0.513 (<0.0001) | −0.357 (0.0018) | 0.92 |
| Germination at 150 DAR | 0.414 (<0.0001) | −0.471 (<0.0001) | 0.60 |
| Seed decay rate | −0.541 (<0.0001) | 0.62 | |
| Seed survivability | 0.53 | ||
aGermination percentages at 7–150 days of after-ripening (DAR) were used to evaluate seed dormancy, and seed decay rate and survivability used to evaluate seed longevity in a 7-month field burial trial
bThe r values are significant at the probability levels listed in the parentheses
cEstimated based on Eq. (1)
Various patterns of germination distribution from 7 to 150 DAR were observed among the 16 ILs (Fig. 1). The AR time to 70% germination varied from 15 days for ILabcd to >150 days for ILaBCD. ILabcd had the shortest dormancy duration, as it carries dormancy-reducing (germination-promoting) alleles at the four loci. Pyramiding dormancy-enhancing (germination-inhibiting) alleles from two or more loci in the ILabcd background generally delayed germination, or lengthened the dormancy duration. However, the line with the longest dormancy duration was not ILABCD, the carrier of dormancy-enhancing alleles at the four loci. In fact, ILABCD and ILaBcD were similar in the distribution pattern, although they differ in genotype for SD1-2 and 7-2 (Fig. 1). The non-linear relationship between genotypes and phenotypes suggests the presence of epistatic interactions in the tetragenic system.
Fig. 1.
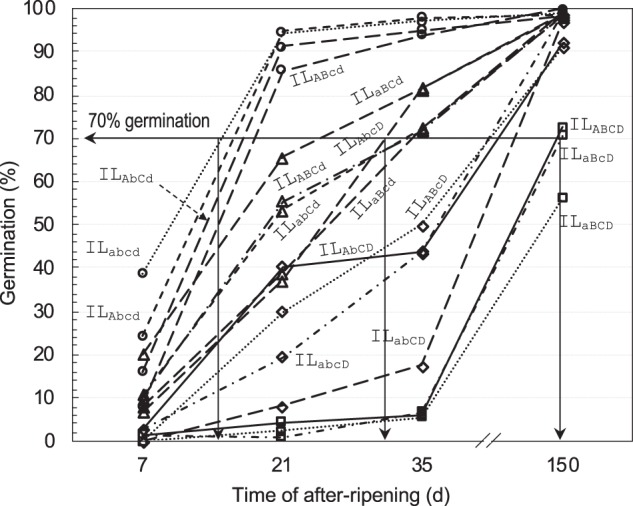
Genotypic differences in the degree or duration of seed dormancy among the 16 isogenic lines (ILs). The ILs are homozygous for the dormancy-enhancing/-reducing alleles at SD1-2 (A/a), 7-1 (B/b), 7-2 (C/c), and 12 (D/d), and tetragenic genotypes indicated by nonallelic combinations of the four loci in subscripts to individual ILs. Seed dormancy was evaluated by germination percentages at 7, 21, 35, and 150 days of after-ripening (DAR). Data shown are genotypic means of 12 plants. The horizontal line indicates the time required to reach 70% germination
Two to four orders of epistasis were involved in regulation of germination dynamics
Genotypic variations among the 16 ILs for germination percentages at different DAR were partitioned into additive (a) and epistatic (i) components of the four loci using the full model (Eq. 2). The additive effects were detected for SD7-1 (a2) and 12 (a4) at 7 to 150 DAR, and for SD1-2 (a1) and 7-2 (a3) after 7 DAR, with a4 being the greatest in magnitude (Table 2). The a2, a3, and a4 estimates were all negative, while the a1 values were positive, indicating that substitution of the dormancy-enhancing with the dormancy-reducing allele at a locus increased germination for SD7-1, 7-2, and 12, but reduced germination for SD1-2 in the tetragenic system.
Table 2.
Summary of genetic component effects of the four loci on germination at different days of after-ripening (DAR)
| Parametera | Germination (%) at DARb | |||||||
|---|---|---|---|---|---|---|---|---|
| 7 | 21 | 35 | 50 | |||||
| Est.b | Prob. | Est. | Prob. | Est. | Prob. | Est. | Prob. | |
| u | 10.1 | <0.0001 | 46.5 | <0.0001 | 57.9 | <0.0001 | 91.2 | <0.0001 |
| a1 | ns | 10.7 | <0.0001 | 9.1 | <0.0001 | 2.6 | 0.009 | |
| a2 | −2.8 | 0.0057 | −9.4 | <0.0001 | −8.6 | <0.0001 | −6.0 | <0.0001 |
| a3 | ns | −4.4 | 0.0009 | −9.3 | <0.0001 | −3.3 | 0.0011 | |
| a4 | −8.1 | <0.0001 | −27.4 | <0.0001 | −27.6 | <0.0001 | −7.4 | <0.0001 |
| i12 | ns | −3.1 | 0.0148 | −3.7 | 0.0018 | 3.1 | 0.0020 | |
| i13 | ns | ns | −3.6 | 0.0028 | −2.0 | 0.0434 | ||
| i14 | ns | ns | 5.5 | <0.0001 | 2.4 | 0.0148 | ||
| i23 | 4.2 | <0.0001 | ns | 3.4 | 0.0042 | ns | ||
| i24 | ns | ns | −5.3 | <0.0001 | −5.8 | <0.0001 | ||
| i34 | ns | ns | −5.2 | <0.0001 | −3.0 | 0.0026 | ||
| i123 | ns | 9.9 | <0.0001 | 6.9 | <0.0001 | ns | ||
| i124 | ns | ns | ns | −3.3 | 0.0012 | |||
| i134 | ns | ns | 2.6 | 0.0241 | ns | |||
| i234 | 3.2 | 0.0018 | 5.2 | 0.0001 | ns | ns | ||
| i1234 | 2.0 | 0.0430 | 3.0 | 0.0177 | ns | ns | ||
aThe model mean (u), and gene additive (a) and international (i) effects were estimated based on Eq (2), and the subscripts 1–4 indicate the SD1-2, 7-1, 7-2, and 12 loci, respectively
bA positive (negative) estimate (Est.) indicates that substituting a dormancy-enhancing with a dormancy-reducing allele promoted (reduced) germination. Effects not significant (ns) are not listed
The epistatic effects were resulted from six two-, four three- and one four-order interactions (Table 2). The six two-order interactions were all detected at 35 DAR, and were also significant at 7 (i23), both 21 and 150 (i12), or 150 (i13, i14, i24, and i34) DAR. The three- or four-order interactions were detected at 7–21 (i234 and i1234), 21–35 (i123), 35 (i134), or 150 (i124) DAR. Of the four loci, only SD7-1 was involved in one or more of the two- or three-order interactions at 7–150 DAR, although it (a2 = −2.8% to −9.6%) had a smaller main effect on germination inhibition than SD12 (a4 = −7.4% to −27.6%) during the period. The two orders of epistasis effected to increase or decrease germination, depending on types or DAR, while the higher orders of epistasis acted to increase germination, except for i124 that reduced germination Table 2).
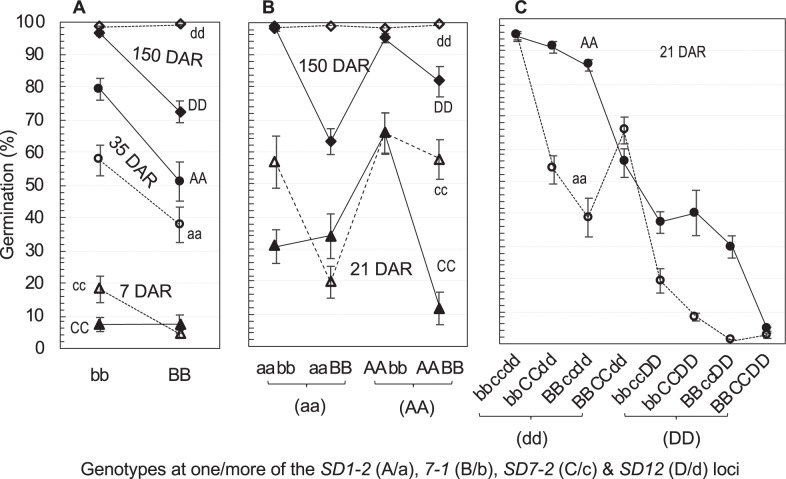
The epistatic interactions varied in the nonparallel relationship between genotypes and phenotypes (i.e., genotypic means for germination percentage at given DAR), as shown by distinct patterns (Fig. 2). The interaction patterns are presented as nonparallel distributions of two genotypes for a locus along with genotypes for the other one, two or three loci, and the distributions increased in diversity with the number of loci involved in an epistasis (Fig. 2). For example, the genotypic difference of SD1-2 (AA-aa) in the germination percentage at 21 DAR varied from positive, null, or negative with genotypes for the other three loci in the tetragenic epistasis (Fig. 2c), while the null or negative effect was absent in the digenic epistasis with SD7-1 (Fig. 2a). Similarly, the genotypic difference of SD7-2 was more diverse in the trigenic epistasis with the SD1-2 and 7-1 loci than in the digenic epistasis with SD7-1 (Fig. 2a, b). The diversity in the interaction pattern suggests that pyramiding a dormancy gene in some genetic backgrounds may not cause a phenotypic change as expected based on its additive effect.
Fig. 2.
Patterns for two- (a), three- (b) or four- (c) order interactions between/among the seed dormancy loci. The upper/lower-case letters indicate the dormancy-enhancing/-reducing alleles at the SD1-2 (A/a), 7-1 (B/b), 7-2 (C/c) or 12 (D/d) loci. Data shown are genotypic means (circles, triangles, or diamonds) and standard errors (bars) for germination percentages at 7, 21, 35, or 150 days of after-ripening (DAR)
Dormancy and red pericarp tended to maintain seed morphological integrity in the soil
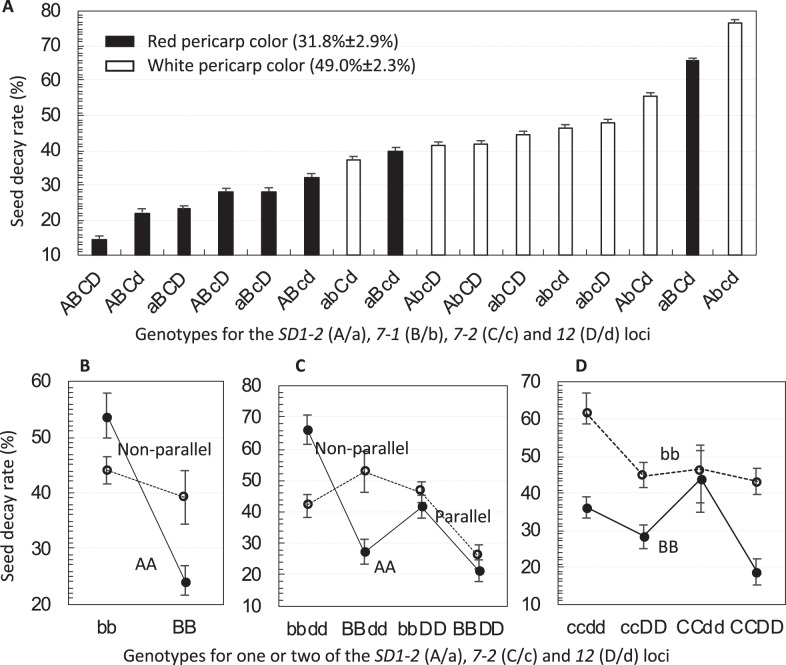
The 16 lines varied in the seed decay rate from 15% for ILADCD to 77% for ILAbcd after 7 months of the burial (Fig. 3a). Genotypic variation accounted for most of the variation in the seed decay rate (H2 = 0.62), which was correlated positively (r = 0.41–0.53) with the germination levels at 7–150 DAR (Table 1) or negatively with the degree of primary dormancy. Interestingly, red pericarp-colored lines distributed mainly in the low end of the percentage scale, and the mean seed decay rate was 17% lower for the eight red (RcRc) than for the eight white (rcrc) ILs (Fig. 3a). Thus, both dormancy and pigmented pericarp appeared to help maintain the morphological integrity of seeds in the soil.
Fig. 3.
Genotypic variation in the seed decay rate among the 16 lines. a Sorted genotypic differences. The 16 lines are genotypes homozygous for SD1-2 (A/a), 7-1 (B/b), 7-2 (C/c) and 12 (D/d), and indicated by combinations of the dormancy-enhancing/-reducing (upper/lower case) alleles at the four loci. Data shown are genotypic means (columns) and standard errors (bars) of five seed samples buried in a rice field for 7 months. b–d Two- or three-order interaction patterns
The genotypic variation was partitioned into six additive or epistatic components of the four loci (Table 3). Additive effect was significant for SD7-1 (a2 = 9.4%), 7-2 (a3 = 3.1%), and 12 (a4 = 5.9%), but not for SD1-2. The positive a-values indicate that substitution of a dormancy-enhancing with a dormancy-reducing allele accelerated seed decay in the soil.
Table 3.
Summary of genic effects of the four loci on seed decay rate and viability in the field burial trial
| Parametera | Seed decay rate | Seed viability | ||
|---|---|---|---|---|
| Estimate (%)b | Probability | Estimate (%) | Probability | |
| u | 39.6 | <0.0001 | 9.2 | <0.0001 |
| a2 | −9.4 | <0.0001 | 7.7 | <0.0001 |
| a3 | −3.1 | 0.03 | 3.1 | 0.0030 |
| a4 | −5.9 | <0.0001 | ns | |
| i12 | −4.8 | 0.0007 | ns | |
| i14 | ns | −2.6 | 0.0126 | |
| i23 | ns | 2.6 | 0.0131 | |
| i124 | −4.9 | 0.0006 | 2.5 | 0.0147 |
| i234 | 3.1 | 0.0289 | ns | |
aThe model mean (u), and gene additive (a) and international (i) effects were estimated based on Eq. (2), and the subscripts 1 to 4 indicate the SD1-2, 7-1, 7-2. and 12 loci, respectively
bA positive (negative) estimate indicates that substituting a dormancy-enhancing with a dormancy-reducing allele increased (decreased) the effect. Effects not significant (ns) are not listed
Epistatic effects arose from three two- or three-order interactions involving SD7-1 (Table 3). Although SD1-2 had no main effect, it interacted with SD7-1 (i12 = −5%), or SD7-1 and 12 (i124 = −5%), to reduce seed decay. The SD1-2 (A/a) × 7-1 (B/b) interaction was strongly regulated by SD12 (D/d), as shown by the presence of a nonparallel pattern in the dd, but not in the DD, background (Fig. 3b, d). The other interaction occurred among the SD7-1, 7-2 (C/c) and 12 loci, and acted to increase the seed decay rate (i234 = 3.1%). In this trigenic system, SD7-1 had little effect on the seed decay rate in the CCdd background (Fig. 3d).
SD1-2, 7-1, 7-2, and 12 interacted as systems in regulating seed survival in the soil
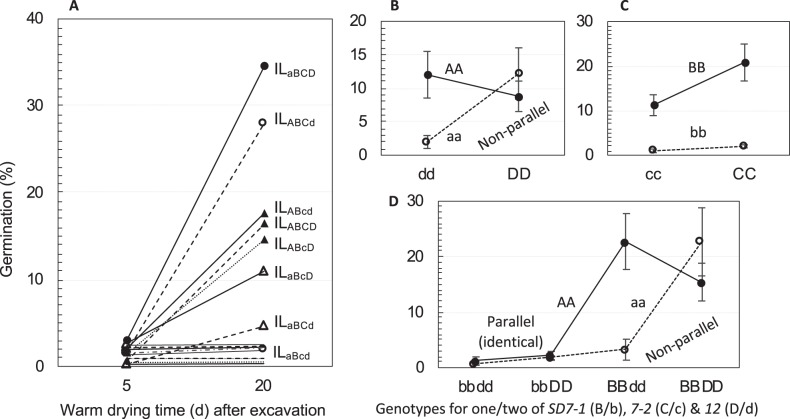
The germination level of the intact seeds at 5 days after the excavation was too low (0.2–2.9%) to discern genotypic variation among the 16 ILs. With an additional period (15 days) of air drying, seven of the ILs had 3–30% increase and the other lines had little change in germination (Fig. 4a). Thus, seeds that survived the burial were largely dormant at the excavation. The seed survivability had a moderate level of heritability (0.53), and was correlated negatively with the seed decay rate (r = −0.54) and the germination levels at 7 to 150 DAR (r = −0.47 to −0.28; Table 1), or positively with the degree of primary dormancy.
Fig. 4.
Genotypic variations in germination ability of buried seeds among the 16 lines. a. Genotypic differences in germination percentage at 5 and 20 days after the excavation. The isogenic lines (ILs) are genotypes homozygous for the SD1-2 (A/a), 7-1 (B/b), 7-2 (C/c) and 12 (D/d) loci. Data shown are genotypic means of five seed samples buried in a rice field for 7 months. Only the eight red pericarp-colored ILs are indicated by combinations of the dormancy-enhancing/-reducing alleles at the four loci in the subscripts, and they all contain the dormancy-enhancing alleles at SD7-1/Rc. Note that the buried seeds were dead or dormant at 5 days, or dead or germinated at 20 days. b–d. Two- or three-order interaction patterns
Genetic analysis of the survivability data identified additive effects for SD7-1 (a2) and 7-2 (a3), and epistatic effects from two-(ia2a3 and ia1a4) or three-(ia1a2a3) order interactions (Table 3). The two-order interactions differed in the nonparallel pattern (Fig. 4b, c) and effected to increase (ia2a3 = 2.6%) or reduce (ia1a4 = −2.6%) seed survival. SD7-1 (B/b) was involved in both of the di- and tri-genic epistases, and the latter was characterized by the SD1-2 × SD12 interaction in the BB, but not in the bb, background (Fig. 4b and d).
SD7-1/Rc had a major effect on maintaining seed morphological integrity and survivability in the burial trial. The seven lines with increased germination at 20 days after excavation (Fig. 4a) all have red pericarp or the SD7-1/Rc gene, and they also contain the dormancy-enhancing alleles at one, two or three of the SD1-2, 7-2, and 12 loci. The red line ILaBcd was identical to the eight white lines in seed survivability. These results suggest that the pleiotropic gene SD7-1/Rc alone had little effect on seed survival in this field experiment.
Discussion
Usefulness of a synthesized multigenic system for research on a complex trait
QTL mapping followed by map-based cloning, a reductionistic approach, has been frequently used to isolate and characterize individual genes for complex traits for about 20 years (Frary et al. 2000). Thus, allelic variants at single loci for a trait could be isolated from different donors into the same genetic background as ILs. For example, ILabcd is isogenic to ILAbcd, ILaBcd, ILabCd, and ILabcD for SD1-2, 7-1, 7-2, and 12, respectively. Such ILs are ideal to assess genetic effects, expression profiles, or regulatory mechanisms precisely for single genes, but not for two or more genes in a system. In this research, we synthesized a tetragenic system for SD using the previously isolated alleles from weedy and cultivated rice.
With the tetragenic system and the full model (Eq. 2), we confirmed that two to four orders of epistatic interactions contribute to the genotypic variation in SD or germination dynamics, which were first detected in a mapping population segregating for traits that influence seed development or harvesting, such as shattering and flowering time (Gu et al. 2004). In the tetragenic system, pyramiding dormancy genes lengthened the dormancy duration (Fig. 1), but epistatic interactions modified the phenotypes in positive or negative directions, resulting deviations of genotypic means from expectations based on additive effects (Table 2). In agriculture, SD too strong could cause nonuniform germination of cultivars or feral weeds, while the dormancy too weak may cause the pre-harvest sprouting (PHS) problem. Thus, it is necessary to include epistatic components in modeling marker-assisted/genomic selection for SD genes to manipulate germination ability of crop varieties.
This tetragenic system was developed not only for the present but also for future research to elucidate how two or more genes interact to regulate SD, SL, and germination at physiological or molecular levels. The SD1-2, 7-1, 7-2, and 12 loci were identified as a transcription factor/GA synthesis gene or associated with specific seed tissues or the ABA/GA signaling pathway (Gu et al. 2011, 2015, Ye et al. 2013, 2015). These lines of isolated information can be verified and synthesized in the multigenic system using systems biology approach and omics tools.
Implications of genetic correlation between SD and SL
This research provided unambiguous evidence, likely for the first time, that natural genes controlling SD are also involved in regulation of soil seedbank longevity. It was multiple genes that worked together to promote seed survival in the soil for 7 months (Fig. 4). The functional allele of SD7-1/Rc from weedy red rice played a major role in maintaining seed morphological integrity and viability in the field. However, a single gene, including SD7-1/Rc, appeared not to provide a sufficient degree of dormancy and longevity for seeds to survive in the soil over the winter to spring seasons. In cropping systems managed by a conventional tillage practice, seeds survived in the soil for months are possible to contribute to the next plant population of weeds. Thus, agronomic and biotechnical techniques that reduce the strength of SD could help manage weed problems or mitigate the risk of transgene flow into wild/weed relatives.
Additional to the genetic mechanisms shared with SD, there must be other genes that naturally occurred during evolution to regulate SL in wild or weedy rice. The correlation strengths evaluated in this research (Table 1) accounted for only a small proportion (R2 < 30%) of variances in the seed decay rate or survivability. This suggests that the specific genes may contribute more to genotypic variation in seedbank longevity than the SD genes. Research is being conducted to scan for QTL associated specifically with SL in the soil along the genome of weedy rice.
Functions of SD1-2 in regulation of SD
The dormancy-enhancing and -reducing alleles of a functionally differentiated gene are expected to reduce and promote germination, respectively, before the dormancy is fully broken. This was true for SD7-1, 7-2, and 12, but not completely for SD1-2, in the tetragenic system. For example, the four lines that have the dormancy-enhancing alleles at one of the four loci (ILAbcd, ILaBcd, ILabCd, and ILabcD) all had lower (15–36%) germination at 7 DAR than the nondormant control ILabcd (open columns on Fig. 5). However, of the four lines that have the dormancy-reducing alleles at one of the four loci, three (ILAbCD, ILABcD, and ILABCd) had higher (19–26%), and ILaBCD had lower (16%) germination at 150 DAR than the dormant control ILABCD (filled columns on Fig. 5). Thus, the dormancy-reducing allele of SD1-2 inhibited, rather than promoted, germination when combined with the other SD genes. This discovery raises a concern about the definition for functionally differentiated alleles at an SD locus.
Fig. 5.
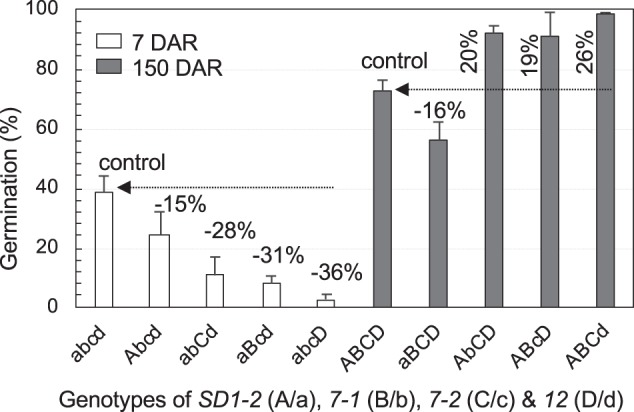
Allelic variation of SD1-2 (A/a) in effect on germination with genotypes of SD7-1 (B/b), 7-2 (C/c), and 12 (D/d) loci. The upper- and lower-case letters indicate dormancy-enhancing (germination-reducing) and -reducing (germination-promoting) alleles at the four loci, respectively, in the background of the ‘abcd’ genotype (non-dormant control). Note that SD1-2 differed from the other three loci in that the dormancy-reducing and -enhancing alleles reduced and promoted germination, respectively, in the genotypes where the other loci fixed for the dormancy-enhancing alleles. The allelic variation of SD1-2 was exemplified by germination at 7 and 150 days of after-ripening (DAR)
SD1-2 is a GA synthesis enzyme gene (OsGA20ox2) at the semidwarf 1 (sd1) locus in rice (Sasaki et al. 2002). SD1-2’s loss-of-function mutants, either naturally occurring or induced, reduced the GA content in early developing seeds, which in turn delayed the tissue (particularly the endosperm) morphogenesis, ABA accumulation, and maturation programs (e.g., dehydration and physiological maturity), resulting in the enhancement of primary dormancy (Ye et al. 2015). Although sd1 has been widely used for rice breeding since the green revolution in 1960s (Khush 2001), this gene alone appeared insufficient to provide the resistance to PHS for many cultivars (Lee et al. 2018). Thus, further research is needed to understand physiological and molecular mechanisms underlying interactions of SD1-2 with the other SD gene(s), particularly those for embryo dormancy.
Supplementary information
Assembling Seed Dormancy Genes into a System Identified Their Effects on Seedbank Longevity in Weedy Rice
Acknowledgements
We thank Dr. Michael E. Foley for initiating the seed dormancy project, Chad Michel for the field management. This research was supported by grants from the United States Department of Agriculture—National Institute of Food and Agriculture (2013-33522-21097, 2018-33522-28795, and 2019-67014-29238) and National Research Initiative (2008-35301-19058), Natural Science Foundation (IOS 1021382 and 0641376), and the American Seed Research Foundation, and by South Dakota Agricultural Extension Station.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41437-019-0253-8) contains supplementary material, which is available to authorized users.
References
- Abeysekara ASK, Herath HMS, Wickrame UB, Nugaliyadde L, Johnson DE (2010) Germinability, viability, and longevity of weedy and cultivated rice in Sri Lanka. In: International rice research conference. Hanoi, Vietnam
- Adugna A. Ecotypic variation for seed dormancy, longevity and germination requirements in wild/weedy Sorghum bicolor in Ethiopia: implications for seed mediated transgene dispersal and persistence. SpringerPlus. 2013;2:248. doi: 10.1186/2193-1801-2-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Willis JH, Mitchell-Olds T. Evolutionary genetics of plant adaptation. Trends Genet. 2011;27:258–266. doi: 10.1016/j.tig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek JS, Chung NJ. Seed wintering and deterioration characteristics between weedy and cultivated rice. Rice. 2012;5:21. doi: 10.1186/1939-8433-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Bradford K, Hilhorst H, Nonogaki H (2013) Seeds—physiology of development, germination and dormancy. 3rd edn. Spinger, New York
- Brunick RL (2008) Seed dormancy in domesticated and wild sunflowers (Helianthus annuus L.): types, longevity and QTL discovery. Dissertation, Oregon State University Libraries & Press, Corvallis, USA
- Cohn MA, Hughes JA. Seed dormancy in red rice (Oryza sativa L.) I. Effect of temperature on dry after-ripening red rice. Weed Sci. 1981;29:402–404. doi: 10.1017/S0043174500039898. [DOI] [Google Scholar]
- Delouche JC, Burgos NR, Gealy DR, De San Martin GZ, Labrada R, Larinde M, Rosell C. Weedy rices—origin, biology, ecology, and control. FAO plant production & protection paper 188. Rome: FAO; 2007. [Google Scholar]
- Buhler DD, Hartzler RG, Forcella F. Weed seed bank dynamics. J Crop Prod. 1997;1:145–168. doi: 10.1300/J144v01n01_07. [DOI] [Google Scholar]
- Ellstrand NC, Heredia SM, Leak-Garcia JA, Heraty JM, Burger JC, Yao L, Nohzadeh-Malakshah S, Ridley CE. Crops gone wild: evolution of weeds and invasives from domesticated ancestors. Evol Appl. 2010;3:494–504. doi: 10.1111/j.1752-4571.2010.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Footitt S. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J Exp Bot. 2017;68:843–856. doi: 10.1093/jxb/erw477. [DOI] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Goss WL, Brown E. Buried red rice. J Am Soc Agron. 1939;31:633–637. doi: 10.2134/agronj1939.00021962003100070007x. [DOI] [Google Scholar]
- Gu X-Y, Foley ME, Horvath DP, Anderson JV, Feng J, Zhang L, Mowry CR, Ye H, Suttle JC, Kadowaki K, Chen Z. Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates ABA and flavonoid synthesis in weedy red rice. Genetics. 2011;189:1515–1524. doi: 10.1534/genetics.111.131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X-Y, Kianian SF, Foley ME. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa) Genetics. 2004;166:1503–1516. doi: 10.1534/genetics.166.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X-Y, Liu T, Feng J, Suttle JC, Gibbons J. The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice. Plant Mol Biol. 2010;73:97–104. doi: 10.1007/s11103-009-9555-1. [DOI] [PubMed] [Google Scholar]
- Gu X-Y, Zhang J, Ye H, Zhang L, Feng J. Genotyping endosperms to determine genes regulating seed dormancy through the embryo, endosperm or maternal tissues in rice. G3 (Bethesda) 2015;5:183–193. doi: 10.1534/g3.114.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst HWM (2007) Definition and hypothesis of seed dormancy. In: Bradford KJ, Nonogaki H (eds) Seed development, dormancy and germination, Blackwell Publishing, Oxford, UK, p 50–71
- Jing W, Jiang L, Zhang W, Zhai H, Wan J. Mapping QTL for seed dormancy in weedy rice. Acta Agron Sin. 2008;34:737–742. [Google Scholar]
- Khush GS. Green revolution: the way forward. Nat Rev Genet. 2001;2:815–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- Lee GA, Jeon YA, Lee HS, Hyun DY, Lee JR, Lee MC, Lee SY, Ma KH, Koh HJ. Variation in pre-harvest sprouting resistance, seed germination and changes in abscisic acid levels during grain development in diverse rice genetic resources. Plant Genet Resour. 2018;16:18–27. doi: 10.1017/S1479262116000319. [DOI] [Google Scholar]
- Miller SD, Nalewaja JD, Gillespie G (1985) Wild oats seed longevity and production Farm Res 43:1. http://hdl.handle.net/10365/5656
- Mispan MS, Zhang L, Feng J, Gu X-Y. Quantitative trait locus and haplotype analyses of wild and crop-mimic traits in U.S. weedy rice. G3. 2013;3:1049–1059. doi: 10.1534/g3.113.006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TP, Keizer P, van Eeuwijk F, Smeekens S, Bentsink L. Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis. Plant Physiol. 2012;160:2083–2092. doi: 10.1104/pp.112.206649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noldin JA, Chandler JM, McCauley GN. Seed longevity of red rice ecotypes buried in soil. Planta Daninha. 2006;24:611–620. doi: 10.1590/S0100-83582006000400001. [DOI] [Google Scholar]
- Oka HI. Origin of cultivated rice. Tokyo, Japan: Japan Scientific Society Press; 1988. [Google Scholar]
- Roberts E. H. Viability of Seeds. Dordrecht: Springer Netherlands; 1972. Dormancy: a Factor Affecting Seed Survival in the Soil; pp. 321–359. [Google Scholar]
- Saatkamp A, Affre L, Dutoit T, Poschlod P. The seed bank longevity index revisited: limited reliability evident from a burial experiment and database analyses. Ann Bot. 2009;104:715–724. doi: 10.1093/aob/mcp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano N, Rajjou L, North HM, Debeaujon I, Marion-Poll A, Seo M. Staying alive: molecular aspects of seed longevity. Plant Cell Physiol. 2016;57:660–674. doi: 10.1093/pcp/pcv186. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. (2013) Base SAS® 9.4 procedures guide: statistical procedures. 2nd edn. Cary, NC: SAS Institute Inc.
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Simpson GM (1990) Seed dormancy in grasses. Cambridge University Press, Cambridge, New York
- Subudhi PK, Parco A, Singh PK, Deleon T, Karan R, Biradar H, Cohn MA, Brar DS, Sasaki T. Genetic architecture of seed dormancy in US weedy rice in different genetic backgrounds. Crop Sci. 2012;52:2564–2575. doi: 10.2135/cropsci2012.04.0228. [DOI] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson K, Ceriani RM, Bakker JP, Bekker RM. Are seed dormancy and persistence in soil related? Seed Sci Res. 2003;13:97–100. doi: 10.1079/SSR2003128. [DOI] [Google Scholar]
- Ye H, Beighley DH, Feng J, Gu X-Y. Genetic and physiological characterization of two clusters of quantitative trait loci associated with seed dormancy and plant height in rice. G3. 2013;3:323–331. doi: 10.1534/g3.112.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Feng J, Zhang L, Zhang J, Mispan MS, Cao Z, Beighley DH, Yang J, Gu X-Y. Map-based cloning of qSD1-2 identified a gibberellin synthase gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 2015;169:2152–2165. doi: 10.1104/pp.15.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Foley ME, Gu X-Y. New seed dormancy loci detected from weedy rice-derived advanced populations with major QTL alleles removed from the background. Plant Sci. 2010;179:612–619. doi: 10.1016/j.plantsci.2010.04.003. [DOI] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. Laboratory Manual for Physiological Studies of Rice. 3rd edn. Manila, Philippines: International Rice Research Institute; 1976. [Google Scholar]
- Zhang L, Lou J, Foley ME, Gu XY. Comparative mapping of seed dormancy loci between tropical and temperate ecotypes of weedy rice (Oryza sativa L.) G3. 2017;7:2605–2614. doi: 10.1534/g3.117.040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dai W, Song X, Qiang S. A model of the relationship between weedy rice seed-bank dynamics and rice-crop infestation and damage in Jiangsu Province, China. Pest Manag Sci. 2013;70:716–724. doi: 10.1002/ps.3649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Assembling Seed Dormancy Genes into a System Identified Their Effects on Seedbank Longevity in Weedy Rice