Abstract
The gut microbiota has been proposed to be an important environmental factor in the development of rheumatoid arthritis (RA). Here, we review a growing body of evidence from human and animal studies that supports the hypothesis that intestinal microbiota play a role in RA. Previous studies from we and others showed an altered composition of the microbiota in early RA patients. A recent study demonstrated that Prevotella species are dominant in the intestine of patients in the preclinical stages of RA. In addition, Prevotella-dominated microbiota isolated from RA patients contributes to the development of Th17 cell-dependent arthritis in SKG mice. Moreover, it was reported that periodontal bacteria correlates with the pathogenesis of RA. In this review, we discuss the link between oral bacteria and the development of arthritis. However, many questions remain to be elucidated in terms of molecular mechanisms for the involvement of intestinal and oral microbiota in RA pathogenesis.
Subject terms: Translational research, Acute inflammatory arthritis, Rheumatoid arthritis
Rheumatoid arthritis: gut and mouth microbes linked to autoimmune disease
Microbes living in the gut and mouth have been implicated in the development of rheumatoid arthritis (RA) and treatments that promote the growth of healthier bacterial communities may help weaken this autoimmune disease. Yuichi Maeda and Kiyoshi Takeda from Osaka University, Japan, review data from mice and humans linking RA to altered microbial compositions in the gut. They focus on a particular bacterium called Prevotella copri, which is found at much higher numbers in the gastrointestinal tracts of people with newly diagnosed RA than in those without the disease. Certain mouth-dwelling bacteria may also help exacerbate RA through the induction of antibodies directed against the host. The exact molecular mechanism by which gut and oral microbes contribute to RA remains unclear.
Introduction
Rheumatoid arthritis (RA) is a systemic inflammatory disease characterized by polyarthritis that leads to joint destruction. Despite rapid progress being made in the treatment of RA1,2, the etiology of RA is not fully understood. It has been reported that combinations of genetic and environmental factors are involved in RA development3,4. The concordance rates for RA in monozygotic twins are ~15%, suggesting that environmental factors are important for RA development5. Exposure to many environmental factors, including smoking, hormones, microbiota, and infections, may be involved in the induction of the disease6–11. Among these environmental factors, the gut microbiota plays an important role in the development of arthritis in mice12–15. Recent studies showed that immunoglobulin A (IgA) anti-citrullinated protein (CCP) antibodies are detectable for several years before the onset of arthritis in humans16,17. These findings suggest that RA originates at mucosal sites, such as the gut and oral cavity. Considering that antimicrobial drugs such as minocycline or salazosulfapyridine are effective in some RA patients, the gut and oral microbiota appear to be correlated with the disease18,19.
Here, we review recent works showing the altered composition of the gut microbiota observed in RA patients. Moreover, we describe the correlations between the gut microbiota and human or murine arthritis in previous studies. We also discuss recent evidence that Prevotella species directly contribute to the development of arthritis in mice.
Dysbiosis triggers arthritis in animal models
Several studies on experimental murine arthritis have clearly demonstrated the importance of the intestinal microbiota in the pathogenesis of arthritis (Table 1). When mice are reared in germ-free (GF) conditions or treated with antibiotics, they do not develop arthritis12,20. However, the inoculation of specific microbes is sufficient to induce arthritis in GF-conditioned mice12,21,22, suggesting that the gut microbiota plays an important role in the development of arthritis.
Table 1.
Murine models of arthritis known to be correlated with the gut microbiota
| Mice strain | Environmental condition | Mechanism of involvement of arthritis | Intestinal bacteria correlated with induction of arthritis | Ref. |
|---|---|---|---|---|
| SKG | GF, SPF: no arthritis conventional: arthritis |
Production of auto-reactive T cells Activation of innate immunity by fungi |
Prevotella-dominated microbiota | 20, 22, 23 |
| IL-1ra−/− | GF: no arthritis conventional: arthritis |
Activation of TLR2 and TLR4 Th17 cells ↑ Treg cells ↓ |
Lactobacillus Bifidus Helicobacter |
21, 24 |
| K/BxN |
GF: no arthritis SPF: arthritis |
Production of GPI-antibody Th17 cell expansion in the intestine |
SFB | 12 |
| CIA |
ABX: reduced severity of arthritis SPF: arthritis |
Production of anti-type II collagen antibody and serum inflammatory cytokines | – | 27 |
GF germ-free, SPF specific pathogen free, GPI glucose-6-phosphate isomerase, SFB segmented filamentous bacteria, TLR Toll-like receptor, Treg cells regulatory T cells, CIA collagen-induced arthritis, ABX antibiotics, Ref references
Previous studies from we and others demonstrated that SKG mice, which spontaneously develop chronic T cell-mediated arthritis under conventional conditions, do not develop the disease under GF conditions20,23. However, a limited bacterial consortium, altered Schaedler flora, is sufficient to induce arthritis with a curdlan injection. We also showed that monocolonization of GF-SKG mice with Prevotella copri is sufficient to induce arthritis with a fungal injection20. These results indicate that a particular commensal bacterium is sufficient to induce arthritis in SKG mice.
As another model of arthritis, interleukin (IL)-1 receptor antagonist knockout (IL1rn−/−) mice spontaneously develop T cell-mediated arthritis under specific-pathogen-free conditions21. These mice do not develop arthritis under GF conditions. However, monocolonization of the mice with Lactobacillus bifidus induces arthritis. Recently, Rogier et al. revealed the importance of IL-1 receptor antagonists in maintaining the diversity and composition of the commensal microbiota. IL1rn−/− mice display decreased bacterial richness and diversity, and their altered microbiota is characterized by a high abundance of Helicobacter species and a low abundance of Ruminococcus species. The Th17 cell population is increased in the intestinal lamina propria of IL1rn−/− mice, and the phenotype is transferable to wild-type mice. Tobramycin treatment decreases the abundance of the commensal microbiota, such as Helicobacter species, and suppresses arthritis in IL1rn−/− mice. Furthermore, by using IL1-rn and TLR4 double-knockout mice, the dysbiosis in IL1rn−/− mice was shown to be TLR4-dependent24.
K/BxN T cell receptor transgenic mice develop inflammatory arthritis with high titers of autoantibodies directed against glucose-6-phosphate isomerase25,26. When the mice are reared under GF conditions, they do not develop the disease and display reduced numbers of Th17 cells in the small intestine and spleen12. Monocolonization with segmented filamentous bacteria is sufficient to cause Th17 cell-dependent arthritis in these mice.
Recently, Widian et al. reported that intestinal dysbiosis triggers collagen-induced arthritis (CIA) via mucosal immune responses. Dysbiosis and mucosal inflammation precede the development of CIA27. Treatment with antibiotics was found to reduce the disease severity, as well as the levels of anti-type II collagen antibodies and serum inflammatory cytokines. Therefore, certain gut commensal microbiota is sufficient to induce arthritis in mice. However, more intensive analyses are needed to analyze which bacterium shows a strong effect on the development of arthritis.
Dysbiosis in human RA patients
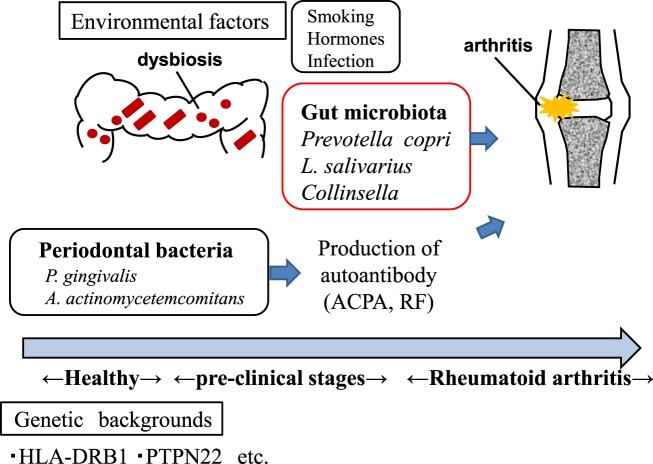
Recent accumulating evidence supports the hypothesis that the gut microbiota plays a pivotal role in the development of human arthritis (Fig. 1). Several case–control studies have shown that the composition of the intestinal microbiota is altered in RA patients (Table 2).
Fig. 1. Both genetic and environmental factors are involved in the pathogenesis of arthritis.
The gut and oral microbiota may contribute to the development of arthritis. P. gingivalis Porphyromonas gingivalis, A. actinomycetemcomitans Aggregatibacter actinomycetemcomitans, L. salivarius Lactobacillus salivarius, ACPA anti-citrullinated protein antibodies, RF rheumatoid factor
Table 2.
Altered composition of the gut microbiota in human RA patients
| Country | Increased bacteria | Reduced bacteria | Method | Ref. |
|---|---|---|---|---|
| USA | Prevotella (Prevotella copri) | Bacteroides | 16S rRNA sequencing | 30 |
| Japan | Prevotella (Prevotella copri) | Bacteroides | 16S rRNA sequencing | 20 |
| USA | Collinsella | Faecalibacterium | 16S rRNA sequencing | 34 |
| China | Lactobacillus salivarius etc. | Veillonella, Haemophilus etc. | Metagenomic shotgun sequence | 32 |
Vaahtovuo et al.28 analyzed the composition of the microbiota in patients with untreated early RA or fibromyalgia using a technique based on flow cytometry, 16S rRNA hybridization, and DNA staining. In the Bacteroides fragilis subgroup, the genera Bifidobacterium and Eubacterium rectale–Clostridium coccoides were decreased in RA patients. These results are comparable to previous results in patients with Crohn’s disease29.
Scher et al.30 found using 16S rRNA gene sequencing that patients with untreated new-onset RA in American populations harbored an increased abundance of P. copri and a reduced abundance of Bacteroides species in the intestine. Interestingly, the relative abundance of P. copri was inversely correlated with the presence of shared epitope risk alleles. We further found that some Japanese patients with recent-onset RA carry an increased abundance of the genus Prevotella, especially P. copri, and a decreased abundance of Bacteroides species in the intestine20. Very recently, preclinical phase RA patients in European countries were shown to harbor a high abundance of Prevotella species, including P. copri, in the intestine, suggesting that dysbiosis precedes the development of arthritis31.
A study in China demonstrated that RA patients had an increased abundance of Lactobacillus salivarius in the gut, on the teeth, and in the saliva, based on metagenomic shotgun sequencing32. In contrast, Haemophilus species were found to be depleted at all three sites in RA patients. The abundance of P. copri in the gut was elevated in the first year after disease onset. Interestingly, the dysbiosis observed in RA patients was partially restored after treatment with disease-modifying drugs. Furthermore, in China, Liu et al.33 found that fecal Lactobacillus species were enriched in RA patients compared with healthy controls (HCs).
Chen et al. reported that compared with HCs, patients with RA show decreased gut microbial diversity, which correlates with autoantibody levels and disease duration. Interestingly, methotrexate induces an increase in species richness and diversity. The relative abundance of Collinsella was found to be increased in RA patients. In contrast, Faecalibacterium, which is generally recognized as a beneficial microbe, is decreased in RA patients. Inoculation of Collinsella into CIA-susceptible mice induces severe arthritis. In vitro experiments showed that Collinsella increases gut permeability and induces IL-17A expression, suggesting that Collinsella is a candidate arthritogenic bacterium in the human intestine34. In summary, P. copri, L. salivarius, and Collinsella are the dominant gut microbiota in patients with early RA and may be involved in its pathogenesis. The reason for the different candidate arthritogenic intestinal bacteria is possibly due to the host genetic background and environmental exposures, such as diet.
Prevotella copri: a possible trigger of RA
Several reports have described the altered composition of the microbiota in RA patients. Among these studies, we and others found that Prevotella species, especially P. copri, are the dominant fecal microbiota in early RA patients20,30. Scher et al.30 have shown that P. copri exacerbates murine colitis in mice administered dextran sulfate sodium in their drinking water. However, it remains unclear whether the dysbiosis observed in RA patients triggers the development of arthritis.
To answer this question, a novel approach was taken to generate the intestinal microbiota: humanized mice20. Fecal samples were anaerobically obtained from early RA patients and HCs, diluted, and orally inoculated into GF-SKG mice. SKG mice develop T cell-mediated arthritis by the activation of innate immunity, resembling human RA23. Both Prevotella-dominated RA microbiota and HC microbiota successfully colonized GF-SKG mice. The SKG mice colonized with a Prevotella-dominated microbiota from RA patients (RA-SKG mice) showed severe arthritis and increased numbers of Th17 cells in the large intestine. Lymphocytes isolated from popliteal lymph nodes and the large intestine secreted IL-17 in response to the arthritis-related autoantigen RPL23A. In vitro analyses revealed that P. copri had the ability to induce the production of Th17-related cytokines such as IL-6 and IL-2320. Thus, these data strongly indicate that Prevotella species, especially P. copri, trigger the development of arthritis. Further analyses are needed to investigate whether intestinal barrier or immune cell populations are altered in patients during the initial stages of RA.
Recently, using liquid chromatography–tandem mass spectrometry, Pianta et al.35 identified a novel HLA-DR-presented peptide in a 27-kDa P. copri protein (Pc-p27) from peripheral blood mononuclear cells of RA patients. Pc-p27 stimulated Th1 responses in 42% of RA patients, although the authors did not show correlations between these proteins and P. copri abundance in the intestine. A subgroup of RA patients showed IgA responses to Pc-p27 or whole P. copri cells. Interestingly, a subgroup of RA patients had P. copri 16S rDNA in their synovial fluid. The authors further identified two novel HLA-DR-presented peptide autoantigens, N-acetylglucosamine-6-sulfatase (GNS) and filamin A (FLNA)36. T cell and B cell responses to GNS and FLNA were observed in 52% and 56% of RA patients, respectively. Interestingly, the GNS and FLNA HLA-DR-presented T cell epitopes have sequence homology with Prevotella epitopes. Moreover, GNS and FLNA autoantibodies positively correlated with P. copri antibody levels. Thus, the authors clearly demonstrated a relationship between microbial peptides from gut commensal bacteria and autoimmune responses affecting joints.
By contrast, several studies have demonstrated that the genus Prevotella is one of the major commensal bacteria in healthy subjects and plays beneficial roles in the host. In Africa and tropical Asia, healthy individuals were found to harbor a high abundance of Prevotella in the intestine37. It has been reported that the human gut microbiota can be divided into three enterotypes38, characterized by high levels of Bacteroides, Prevotella, and Ruminococcus. Therefore, Prevotella species are detectable in the intestine of RA patients and also some healthy individuals. It would be an interesting future issue to clarify which Prevotella species component leads to the development of arthritis.
A recent study demonstrated that Prevotella histicola from the intestine of healthy humans decreased the severity of CIA in HLA-DQ8 mice39. HLA-DQ8 mice were immunized with collagen and orally inoculated with P. histicola. The mice treated with P. histicola showed ameliorated arthritis through a reduction in intestinal permeability. P. histicola increased regulatory T cell numbers in the gut and reduced antigen-specific Th17 responses. The sequences of P. histicola were completely different from those of P. copri. These results indicate that some Prevotella species, such as P. histicola, can suppress the induction of arthritis. In summary, P. copri and P. histicola show different effects on arthritis. These differences might derive from the genetic diversity among Prevotella species.
Correlation between periodontal bacteria and arthritis
Recent studies have revealed that periodontal disease is correlated with an increased risk of RA in humans and in mouse models of arthritis40–42. The presence of periodontitis in patients with RA is associated with anti-CCP antibody levels43. Moreover, periodontitis is correlated with the disease activity of RA44. In addition, treatment of periodontitis ameliorates the disease activity of RA45,46. These results suggest that periodontal bacteria are correlated with RA pathogenesis.
Porphyromonas gingivalis, one of the major periodontal bacteria, is the only known pathogen that expresses a bacterial peptidylarginine deiminase47–49. Reports have shown that P. gingivalis infection positively correlates with the production of anti-CCP antibody responses in RA patients50,51. In a CIA model, oral inoculation of P. gingivalis was found to exacerbate arthritis through the increased production of IL-1742. The arthritogenic effects of P. gingivalis are dependent on the bacterial strain, presence of fimbriae, and time of infection52. Recently, Sato et al.53 showed that P. gingivalis, but not Prevotella intermedia, exacerbate arthritis by modulating the gut microbiota and increasing the proportion of Th17 cells in mesenteric lymph nodes. However, the roles of other pathogens in the development of arthritis have not been fully investigated.
Aggregatibacter actinomycetemcomitans, a periodontal bacterium, was recently proposed to connect periodontitis to RA because of its ability to induce citrullinated autoantigens. Konig et al. reported that the pore-forming toxin leukotoxin-A produced by A. actinomycetemcomitans, but not by other periodontal pathogens, drives hypercitrullination in neutrophils54. Moreover, antibodies against A. actinomycetemcomitans and leukotoxin-A were found to be highly detectable in human RA patients. In summary, periodontal bacteria such as P. gingivalis and A. actinomycetemcomitans may contribute to autoantibody production and autoimmunity in RA. Further analyses are needed to elucidate whether A. actinomycetemcomitans induces anti-CCP antibody production in vivo.
Conclusion
In this review, we have summarized the role of the intestinal microbiota in human RA and in murine models of arthritis. Several studies have demonstrated that P. copri is present in early RA patients and contributes to the induction of arthritis. However, the precise molecular mechanisms by which P. copri exacerbates human arthritis are still unknown. L. salivarius and Collinsella were found to be the dominant gut microbiota in other cohorts. Moreover, it was reported that periodontal bacteria such as P. gingivalis and A. actinomycetemcomitans may induce the production of anti-CCP antibodies, leading to the development of arthritis. Further studies are needed to clarify the mechanistic links between these specific bacteria and RA development in humans. The manipulation of dysbiosis would be a novel preventative strategy in RA patients.
Author Contributions
Y.M. and K.T. wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishimoto N, et al. Drug free REmission/low disease activity after cessation of tocilizumab (Actemra) Monotherapy (DREAM) study. Mod. Rheumatol. 2014;24:17–25. doi: 10.3109/14397595.2013.854079. [DOI] [PubMed] [Google Scholar]
- 3.Klareskog L, Padyukov L, Ronnelid J, Alfredsson L. Genes, environment and immunity in the development of rheumatoid arthritis. Curr. Opin. Immunol. 2006;18:650–655. doi: 10.1016/j.coi.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Mankia K, Emery P. Preclinical rheumatoid arthritis: progress toward prevention. Arthritis Rheumatol. 2016;68:779–788. doi: 10.1002/art.39603. [DOI] [PubMed] [Google Scholar]
- 5.Silman AJ, et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br. J. Rheumatol. 1993;32:903–907. doi: 10.1093/rheumatology/32.10.903. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe F. The effect of smoking on clinical, laboratory, and radiographic status in rheumatoid arthritis. J. Rheumatol. 2000;27:630–637. [PubMed] [Google Scholar]
- 7.Masdottir B, et al. Smoking, rheumatoid factor isotypes and severity of rheumatoid arthritis. Rheumatology. 2000;39:1202–1205. doi: 10.1093/rheumatology/39.11.1202. [DOI] [PubMed] [Google Scholar]
- 8.Caminer AC, Haberman R, Scher JU. Human microbiome, infections, and rheumatic disease. Clin. Rheumatol. 2017;36:2645–2653. doi: 10.1007/s10067-017-3875-3. [DOI] [PubMed] [Google Scholar]
- 9.de Oliveira GLV, Leite AZ, Higuchi BS, Gonzaga MI, Mariano VS. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology. 2017;152:1–12. doi: 10.1111/imm.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerreiro CS, Calado A, Sousa J, Fonseca JE. Diet, microbiota, and gut permeability—the unknown triad in rheumatoid arthritis. Front. Med. 2018;5:349. doi: 10.3389/fmed.2018.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tracy A, Buckley CD, Raza K. Pre-symptomatic autoimmunity in rheumatoid arthritis: when does the disease start? Semin. Immunopathol. 2017;39:423–435. doi: 10.1007/s00281-017-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans-Marin H, et al. Microbiota-dependent involvement of Th17 cells in murine models of inflammatory arthritis. Arthritis Rheumatol. 2018;70:1971–1983. doi: 10.1002/art.40657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horta-Baas G, Romero-Figueroa MDS. Intestinal dysbiosis and rheumatoid arthritis: a link between gut microbiota and the pathogenesis of rheumatoid arthritis. J. Immunol. Res. 2017;2017:4835189. doi: 10.1155/2017/4835189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D, Zeng MY, Nunez G. The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp. Mol. Med. 2017;49:e339. doi: 10.1038/emm.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielen MM, et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 2004;50:380–386. doi: 10.1002/art.20018. [DOI] [PubMed] [Google Scholar]
- 17.Rantapaa-Dahlqvist S, et al. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 2003;48:2741–2749. doi: 10.1002/art.11223. [DOI] [PubMed] [Google Scholar]
- 18.Tilley BC, et al. Minocycline in rheumatoid arthritis. A 48-week, double-blind, placebo-controlled trial. MIRA Trial Group. Ann. Intern. Med. 1995;122:81–89. doi: 10.7326/0003-4819-122-2-199501150-00001. [DOI] [PubMed] [Google Scholar]
- 19.O’Dell JR, et al. Treatment of early seropositive rheumatoid arthritis: a two-year, double-blind comparison of minocycline and hydroxychloroquine. Arthritis Rheum. 2001;44:2235–2241. doi: 10.1002/1529-0131(200110)44:10<2235::AID-ART385>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 20.Maeda Y, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016;68:2646–2661. doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- 21.Abdollahi-Roodsaz S, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Investig. 2008;118:205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehaume LM, et al. ZAP-70 genotype disrupts the relationship between microbiota and host, leading to spondyloarthritis and ileitis in SKG mice. Arthritis Rheumatol. 2014;66:2780–2792. doi: 10.1002/art.38773. [DOI] [PubMed] [Google Scholar]
- 23.Sakaguchi N, et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature. 2003;426:454–460. doi: 10.1038/nature02119. [DOI] [PubMed] [Google Scholar]
- 24.Rogier R, et al. Aberrant intestinal microbiota due to IL-1 receptor antagonist deficiency promotes IL-17- and TLR4-dependent arthritis. Microbiome. 2017;5:63. doi: 10.1186/s40168-017-0278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouskoff V, et al. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/S0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto I, Staub A, Benoist C, Mathis D. Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science. 1999;286:1732–1735. doi: 10.1126/science.286.5445.1732. [DOI] [PubMed] [Google Scholar]
- 27.Jubair WK, et al. Modulation of inflammatory arthritis in mice by gut microbiota through mucosal inflammation and autoantibody generation. Arthritis Rheumatol. 2018;70:1220–1233. doi: 10.1002/art.40490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 2008;35:1500–1505. [PubMed] [Google Scholar]
- 29.Seksik P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scher JU, et al. Expansion of intestinal Prevotellacopri correlates with enhanced susceptibility to arthritis. ELife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alpizar-Rodriguez D, et al. Prevotellacopri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019;78:590–593. doi: 10.1136/annrheumdis-2018-214514. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Zhang D, Jia H. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr. Microbiol. 2013;67:170–176. doi: 10.1007/s00284-013-0338-1. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pianta A, et al. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017;69:964–975. doi: 10.1002/art.40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pianta A, et al. Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. J. Clin. Investig. 2017;127:2946–2956. doi: 10.1172/JCI93450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marietta EV, et al. Suppression of inflammatory arthritis by human gut-derived Prevotellahisticola in humanized mice. Arthritis Rheumatol. 2016;68:2878–2888. doi: 10.1002/art.39785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scher JU, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012;64:3083–3094. doi: 10.1002/art.34539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arvikar SL, et al. Clinical correlations with Porphyromonasgingivalis antibody responses in patients with early rheumatoid arthritis. Arthritis Res. Ther. 2013;15:R109. doi: 10.1186/ar4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Aquino SG, et al. Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-driven Th17 response. J. Immunol. 2014;192:4103–4111. doi: 10.4049/jimmunol.1301970. [DOI] [PubMed] [Google Scholar]
- 43.Dissick A, et al. Association of periodontitis with rheumatoid arthritis: a pilot study. J. Periodontol. 2010;81:223–230. doi: 10.1902/jop.2009.090309. [DOI] [PubMed] [Google Scholar]
- 44.de Smit M, et al. Periodontitis in established rheumatoid arthritis patients: a cross-sectional clinical, microbiological and serological study. Arthritis Res. Ther. 2012;14:R222. doi: 10.1186/ar4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortiz P, et al. Periodontal therapy reduces the severity of active rheumatoid arthritis in patients treated with or without tumor necrosis factor inhibitors. J. Periodontol. 2009;80:535–540. doi: 10.1902/jop.2009.080447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Katma MK, Bissada NF, Bordeaux JM, Sue J, Askari AD. Control of periodontal infection reduces the severity of active rheumatoid arthritis. J. Clin. Rheumatol. 2007;13:134–137. doi: 10.1097/RHU.0b013e3180690616. [DOI] [PubMed] [Google Scholar]
- 47.Lappin DF, et al. Influence of periodontal disease, Porphyromonas gingivalis and cigarette smoking on systemic anti-citrullinated peptide antibody titres. J. Clin. Periodontol. 2013;40:907–915. doi: 10.1111/jcpe.12138. [DOI] [PubMed] [Google Scholar]
- 48.Hendler A, et al. Involvement of autoimmunity in the pathogenesis of aggressive periodontitis. J. Dent. Res. 2010;89:1389–1394. doi: 10.1177/0022034510381903. [DOI] [PubMed] [Google Scholar]
- 49.Wegner N, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and alpha-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010;62:2662–2672. doi: 10.1002/art.27552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mikuls TR, et al. Periodontitis and Porphyromonas gingivalis in patients with rheumatoid arthritis. Arthritis Rheumatol. 2014;66:1090–1100. doi: 10.1002/art.38348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quirke AM, et al. Heightened immune response to autocitrullinated Porphyromonas gingivalis peptidylarginine deiminase: a potential mechanism for breaching immunologic tolerance in rheumatoid arthritis. Ann. Rheum. Dis. 2014;73:263–269. doi: 10.1136/annrheumdis-2012-202726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung H, et al. Arthritic role of Porphyromonas gingivalis in collagen-induced arthritis mice. PLoS ONE. 2017;12:e0188698. doi: 10.1371/journal.pone.0188698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato K, et al. Aggravation of collagen-induced arthritis by orally administered Porphyromonas gingivalis through modulation of the gut microbiota and gut immune system. Sci. Rep. 2017;7:6955. doi: 10.1038/s41598-017-07196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konig MF, et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016;8:369ra176. doi: 10.1126/scitranslmed.aaj1921. [DOI] [PMC free article] [PubMed] [Google Scholar]