Humans are characterized by vast interindividual variability, as a result of the ~3.5–5 million genomic variants observed per genome, a significant proportion of which are rare and can potentially be of clinical relevance. The same rule applies for those genes that encode the various enzymes involved in absorption, distribution, metabolism, excretion, and toxicity of medicines, also referred to as ADMET genes or pharmacogenes. In particular, it has been previously shown that every individual bears, on average, ~18,000 variants in their 231 pharmacogenes, the majority of which are rare variants, some of which may be deleterious regarding gene function [1]. This vast interindividual genetic variability reciprocally leads to remarkable interethnic differences in the allele frequencies of genomic biomarkers in those pharmacogenes, with significant consequences not only in public health but also in drug development [2].
In a recent study, Petrović et al. have systematically assessed the interethnic distribution of clinically relevant genomic biomarkers in the CYP2C19 and CYP2D6 pharmacogenes in 33 European populations [3]. These authors have extracted relevant genotyping data from 79 original studies referring to 82,791 healthy individuals across Europe and concluded that the prevalence of certain pharmacogenomic biomarkers present with significant differences among populations. In particular, frequencies of CYP2D6 gene duplications showed a clear Southeast to Northwest gradient ranging from <1% in Sweden and Denmark to 6% in Greece in Turkey, while an inverse distribution was obvious for the CYP2D6*4, CYP2D6*5, and CYP2C19*2 loss-of-function alleles. These data indicate that some of the clinically actionable pharmacogenomic biomarkers present with a distinct geographical gradient, which is consistent with European populations’ migratory history.
In recent years, the concept of population pharmacogenomics (PGx) has gained momentum. In a similar multicenter study (the Euro-PGx project), coordinated by the Golden Helix Foundation (www.goldenhelix.org) and conducted from 2010 until 2015, Mizzi et al. analysed over 1800 individuals from 22 different populations, mostly European but also of Middle Eastern and South African origin, using the DMET+ microarray-based and conventional-genotyping approaches. These authors demonstrated significant differences in several pharmacogenomic biomarker allele frequencies among different European populations [4], particularly in a number of clinically actionable pharmacogenomic biomarkers, affecting drug efficacy and/or toxicity of 51 drug treatment modalities. Interestingly, these differences are also reflected not only on the prevalence of high-risk genotypes in these populations, as far as common pharmacogenomic biomarkers in the CYP2C9, CYP2C19, CYP3A5, VKORC1, SLCO1B1, and TPMT pharmacogenes are concerned but also on the expected differences in the predicted genotype-based warfarin dosing among these populations [4]. Similarly, Lakiotaki et al. demonstrated significant differences of pharmacogenomic biomarker allele frequencies that directly reflect on the genomic structure of the 28 populations comprising the 1000 genome project [5].
There are important implications of population PGx, both in terms of public health policies and also, and most importantly, on drug development. In the first case, prior knowledge of pharmacogenomic biomarker allele frequencies in a certain population, may help towards establishing medication prioritization guidelines, which could contribute towards improving the quality of life of the patients and also minimizing the national healthcare expenditure by reducing possible adverse drug reactions. This is of utmost importance as far as pharmacogenomic biomarkers with high prevalence are concerned. Two important examples is the screening for the HLA*1502 variant in Southeast Asian populations, leading to Steven–Johnson syndrome/toxic epidermolysis necrosa, where, contrary to the European populations, is highly prevalent [6], and reciprocally, CYP2D6 variants, related to tamoxifen and several other medications, in which case, ~6–10% of Europeans are considered to be poor CYP2D6 metabolizers, as compared with <1% of East Asians [7]. As such, once incorporating PGx into national healthcare policies, taking the prevalence of (the clinically actionable) pharmacogenomic variant alleles into serious consideration, provision of healthcare can become more cost-effective by ensuring better treatment, proper medication prescription and, above all, identification of subpopulations at certain disease and adverse drug reactions risk [2]. The latter has broad implications, not only for developing but also for developed countries, which constitutes a big mosaic of ethnically diverse subpopulations.
Population PGx may also play a decisive role in drug development. In prospective genome-guided randomized clinical trials, participants’ selection is based on their pharmacogenomic biomarker status [8]. It is anticipated that this, so-called genome-enrich, clinical study design (see also https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-good-pharmacogenomic-practice-first-version_en.pdf) may identify those participants’ subgroups that are more likely to respond better to a given drug, or be more susceptible to a potential adverse drug reaction, which could significantly expedite the entire drug development process, while also allowing participation from a variety of racial/ethnic, ancestral, and geographic backgrounds, even from the developing world, addressing the issue of limited ethnic representation in several clinical trials. The above may have a very positive impact on the drug development and/or repositioning costs.
In their study, Petrović et al. stress the need for refining pharmacogenomic biomarker mapping in different (sub)populations, which is particularly important even among populations of the same racial group [3], allowing for cost-effective and more precise provision of healthcare services. This can be only achieved by encouraging well-designed projects that address this topic. Such a project, closely related to the Euro-PGx project, is currently ongoing in Southeast Asia (the 100 pharmacogenes project), aiming to analyze—by targeted resequencing—100 pharmacogenes in ~1400 individuals, mostly of Southeast Asian origin, and also of Greek and Emirati descent. Findings from these studies should then be made available through well curated online resources, documenting clinically relevant genomic variants allele frequencies, such as the FINDbase database (www.findbase.org), that has been active in the field since 2006 [9]. Ultimately, development of companion diagnostics solutions, e.g., ethnic-specific panels for pharmacogenomic testing, bearing those pharmacogenomic biomarkers that are relevant for specific population(s) based on findings from relevant projects, may represent a cost-effective choice not only for pharmacogenomic testing, particularly for low- and medium-income countries, but also for genome-enriched clinical studies (Fig. 1).
Fig. 1.
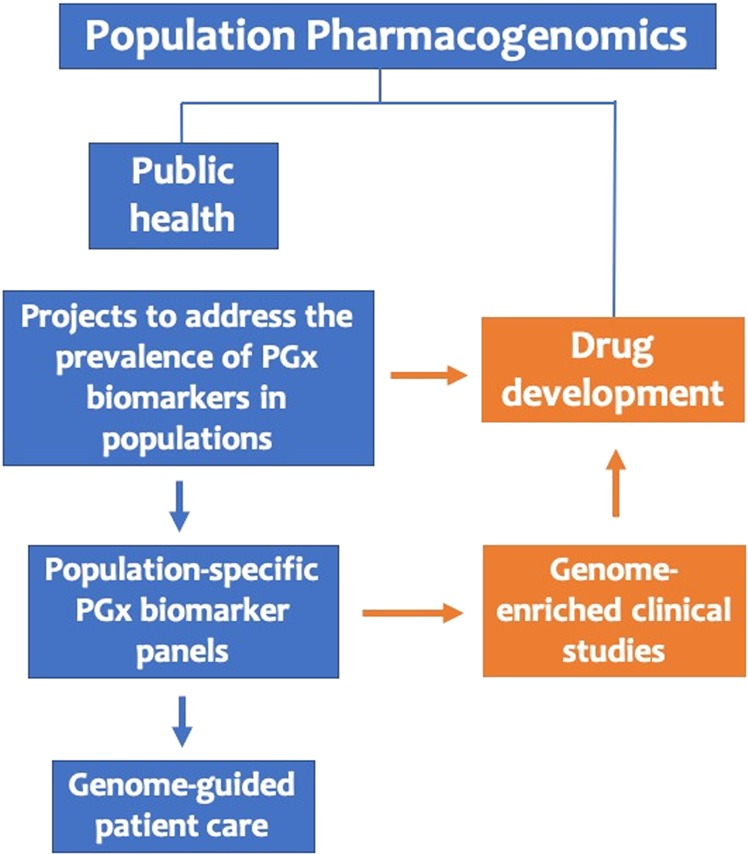
Possible applications of population PGx, in public health and drug discovery
Overall, differences in the prevalence of pharmacogenomic biomarkers in different populations can positively impact both on public health as well as in drug development and coupled with population-based cost-effectiveness analyses [10] may further expedite the integration of genome-guided drug treatment interventions into routine clinical care.
Acknowledgements
This work was partly funded by grants from the European Commission [(H2020-668353; U-PGx (www.upgx.eu) and H2020-860895; TranSYS)] to GPP.
Compliance with ethical standards
Conflict of interest
The author declares no conflict of interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mizzi C, Peters B, Mitropoulou C, Mitropoulos K, Katsila T, Agarwal MR, et al. Personalized pharmacogenomics profiling using whole-genome sequencing. Pharmacogenomics. 2014;15:1223–34. doi: 10.2217/pgs.14.102. [DOI] [PubMed] [Google Scholar]
- 2.Mette L, Mitropoulos K, Vozikis A, Patrinos GP. Pharmacogenomics and public health: implementing ‘populationalized’ medicine. Pharmacogenomics. 2012;13:803–13. doi: 10.2217/pgs.12.52. [DOI] [PubMed] [Google Scholar]
- 3.Petrović J, Pešić V, Lauschke VM. Frequencies of clinically important CYP2C19 and CYP2D6 alleles are graded across Europe. Eur J Hum Genet. 2019. (In press.) [DOI] [PMC free article] [PubMed]
- 4.Mizzi C, Dalabira E, Kumuthini J, Dzimiri N, Balogh I, Başak N, et al. A european spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PLoS One. 2016;11:e0162866. doi: 10.1371/journal.pone.0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakiotaki K, Kanterakis A, Kartsaki E, Katsila T, Patrinos GP, Potamias G. Exploring public genomics data for population pharmacogenomics. PLoS One. 2017;12:e0182138. doi: 10.1371/journal.pone.0182138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee MT, Mahasirimongkol S, Zhang Y, Suwankesawong W, Chaikledkaew U, Pavlidis C, et al. Clinical application of pharmacogenomics: the example of HLA-based drug-induced toxicity. Public Health Genom. 2015;17:248–55. doi: 10.1159/000366253. [DOI] [PubMed] [Google Scholar]
- 7.Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer. 2009;9:576–86. doi: 10.1038/nrc2683. [DOI] [PubMed] [Google Scholar]
- 8.Patrinos GP. Population pharmacogenomics: impact on public health and drug development. Pharmacogenomics. 2018;19:3–6. doi: 10.2217/pgs-2017-0166. [DOI] [PubMed] [Google Scholar]
- 9.Viennas E, Komianou A, Mizzi C, Stojiljkovic M, Mitropoulou C, Muilu J, et al. Expanded national database collection and data coverage in the FINDbase worldwide database for clinically relevant genomic variation allele frequencies. Nucleic Acids Res. 2017;45(D1):D846–D853. doi: 10.1093/nar/gkw949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder SR, Mitropoulou C, Patrinos GP, Williams MS. Economic evaluation of pharmacogenomics: a value-based approach to pragmatic decision making in the face of complexity. Public Health Genom. 2014;17:256–64. doi: 10.1159/000366177. [DOI] [PubMed] [Google Scholar]


