Abstract
Seed mass is a key component of adaptation in plants and a determinant of yield in crops. The climatic drivers and genomic basis of seed mass variation remain poorly understood. In the cereal crop Sorghum bicolor, globally-distributed landraces harbor abundant variation in seed mass, which is associated with precipitation in their agroclimatic zones of origin. This study aimed to test the hypothesis that diversifying selection across precipitation gradients, acting on ancestral cereal grain size regulators, underlies seed mass variation in global sorghum germplasm. We tested this hypothesis in a set of 1901 georeferenced and genotyped sorghum landraces, 100-seed mass from common gardens, and bioclimatic precipitation variables. As predicted, 100-seed mass in global germplasm varies significantly among botanical races and is correlated to proxies of the precipitation gradients. With general and mixed linear model genome-wide associations, we identified 29 and 56 of 100 a priori candidate seed size genes with polymorphisms in the top 1% of seed mass association, respectively. Eleven of these genes harbor polymorphisms associated with the precipitation gradient, including orthologs of genes that regulate seed size in other cereals. With FarmCPU, 13 significant SNPs were identified, including one at an a priori candidate gene. Finally, we identified eleven colocalized outlier SNPs associated with seed mass and precipitation that also carry signatures of selection based on FST scans and PCAdapt, which represents a significant enrichment. Our findings suggest that seed mass in sorghum was shaped by diversifying selection on drought stress, and can inform genomics-enabled breeding for climate-resilient cereals.
Subject terms: Agricultural genetics, Evolutionary biology
Introduction
Local adaptation exists when the average fitness of organisms in their local environment is higher than conspecifics from other environments (Des Marais et al. 2013; Blanquart et al. 2013). Long-term selection acting on phenotypic traits can lead to adaptive divergence, characterized by the divergent morphological characteristics and allele frequencies among local populations (Hoban et al. 2016). Climate is a major factor shaping local adaptation in plants, and the genomic basis of climate adaptation is beginning to be revealed (Eckert et al. 2010; Fournier-Level et al. 2011; Lasky et al. 2012; Kort et al. 2014; Siepielski et al. 2017).
Life-history theory suggests a trade-off between seed mass and seed number, given a constant amount of available resources to produce the seeds (Smith and Fretwell 1974). Increasing seed size provides more reserves for seedling growth, but greater number of seeds provides more propagules and bet-hedging (Olofsson et al. 2009). In semi-arid and arid regions, precipitation-mediated water availability may act as a selective pressure on seed size (Hallett et al. 2011), but findings on seed size adaptation to drought risk have been contradictory (Leishman et al. 2000). In crops, which evolve under both natural and artificial selection, grain size is a main component determining grain yield, subject to similar ecophysiological trade-offs (Sadras 2007).
Sorghum [Sorghum bicolor (L.) Moench] is a cereal crop grown in smallholder and commercial agricultural systems in drought-prone regions worldwide (Monk et al. 2014). Sorghum was domesticated in Africa (Wendorf et al. 1992), and diffused widely across diverse agroclimatic zones, including arid, semi-arid, and sub-humid regions, in Africa, Asia, and the Americas (Deu et al. 2006; Morris et al. 2013). Botanists have noted that seed size in common-garden experiments is typically inversely correlated to the abundance of precipitation in the environment of origin, both among and within the botanical races of sorghum (Harlan et al. 1972). Consistent with models of ecophysiological trade-offs, a negative correlation between grain mass and grain number has been observed in sorghum (Yang et al. 2010; Burow et al. 2014; Tao et al. 2018).
The molecular genetic basis of seed size variation has been a major target study due to its potential to facilitate yield improvements (Li et al. 2011; Sosso et al. 2015; Ma et al. 2016). In sorghum, several studies have reported quantitative trait loci (QTL) for seed mass using biparental linkage mapping and genome-wide association study (GWAS) (Paterson et al. 1995; Brown et al. 2006; Feltus et al. 2006; Srinivas et al. 2009; Upadhyaya et al. 2012; Zhang et al. 2015; Han et al. 2015; Boyles et al. 2016; Tao et al. 2018), but the quantitative trait nucleotides have not been identified. By contrast, in the model cereal rice (Oryza sativa) molecular networks underlying grain size variation have been characterized in detail (Zuo and Li 2014). Comparative studies suggest ancestral regulatory networks may underlie grain size variation (including mass variation) in cereals and other plants (Zhang et al. 2015).
Ecological and population genomic approaches for detecting signatures of local adaptation have progressed with advances in genotyping (Mita et al. 2013; Tiffin and Ross-Ibarra 2014; Forester et al. 2016). Common garden experiments, which bring germplasm from multiple environments into a common environment, can reveal phenotypic and genomic signatures of local adaptation by isolating heritable genotype effects (Savolainen et al. 2013; de Villemereuil et al. 2016). GWAS of phenotypes or environmental factors can be used to locate genomic regions controlling the putative adaptive traits (Bergelson and Roux 2010; Savolainen et al. 2013). Genome scans for differentiation among populations from different environments can identify adaptive loci without explicitly modeling environmental variation (Beaumont and Balding 2004; Martins et al. 2016).
Genomic signatures of climate adaptation have been reported in many widely-distributed crops including maize, barley, soybean, pearl millet, and sorghum (Vigouroux et al. 2011; Abebe et al., 2015; Lasky et al. 2015; Swarts et al. 2017; Bandillo et al. 2017). Seed size variation in sorghum and other crops has been subject to selection during domestication and diversification (Tao et al. 2017). However, whether sorghum seed mass variation is locally adaptive to global precipitation gradient remains unknown. In sorghum, about 20% of global nucleotide variation can be explained by environmental variables (Lasky et al. 2015), but it is not known how much of this variation affects trait variation, and what trait variation is environmentally adaptive. In this study, we tested the hypothesis that sorghum seed mass variation reflects adaptation to precipitation gradients due to genetic variation in ancestral seed size regulators, and found multiple lines of evidence (phenotypic, geographic, and genomic) consistent with this hypothesis.
Materials and methods
Genotypic and environmental data
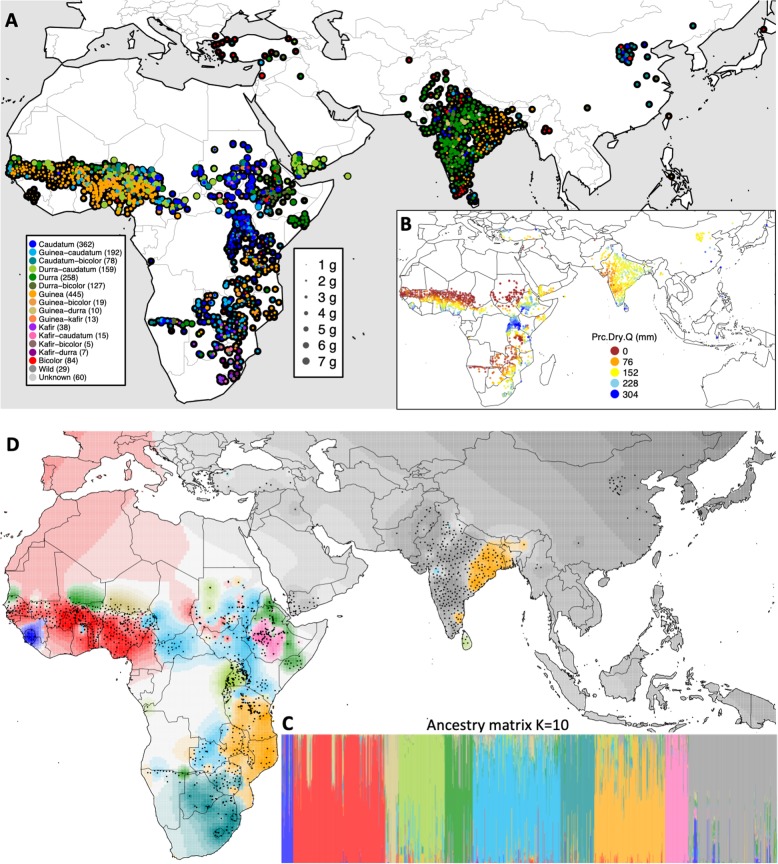
Public genotyping-by-sequencing (GBS) data for 404,627 SNPs and WorldClim bioclimatic variables (Hijmans et al. 2005) for 1901 georeferenced sorghum accessions were obtained from Dryad Digital Repository at 10.5061/dryad.jc3ht (Lasky et al. 2015) (Table S1). The diverse accessions include landraces of all five major botanical races of sorghum and ten intermediate races. In addition, 29 accessions of sorghum wild relatives and 60 accessions with unknown race designations were included. These accessions originated across the majority of the agroclimatic zones in Africa and Asia where sorghum has traditionally been cultivated (Fig. 1a and Table S1).
Fig. 1.
Global diversity panel of sorghum landraces with seed mass variation. a Origin of global diverse sorghum landrace accessions and their 100-seed mass in common-garden (BLUPs). Accessions are colored by botanical races (morphological type) with the size of each colored circle proportional to 100-seed mass (with each colored circle plotted on a solid black circle for clarity). b The precipitation (precipitation in the driest quarter, in mm) where each sorghum landrace accession originated. c The bar plot of sorghum landraces ancestry coefficients based on ADMIXTURE analysis and d corresponding spatial population structure map, colored by ancestry coefficients for ten putative ancestral populations (K = 10)
Phenotypic data
Seed size was estimated for 1901 sorghum landraces using 100-seed mass obtained from the US National Plant Germplasm System’s Germplasm Resources Information Network (GRIN) or the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) genebanks (Table S1). Two sets of 100-seed mass data from GRIN (United States) and ICRISAT (India) represent two common-garden experiments. If multiple observations were recorded in GRIN, the mean of all observations for each accession was calculated. To account for the effect of location, best linear unbiased predictors (BLUPs) of 100-seed mass from the two genebanks were estimated for all accessions using a random linear model implemented in R package lme4 (Bates et al. 2015) as follows:
where yij is the 100-seed mass of the ith accession in the jth location, μ is the overall mean, Gi is the random genotypic effect for the ith accession, Ej is the random environmental effect of the jth location, and εij is the residual. To test the differences of 100-seed mass among botanical races, one-way analysis of variance (ANOVA) and post hoc Tukey HSD test (TukeyHSD in R) were conducted using 100-seed mass BLUPs.
Phenotype and precipitation correlations
To characterize the relationship between 100-seed mass BLUPs and each of the five precipitation variables (annual precipitation: Ann.Prc; precipitation in the wettest quarter: Prc.Wet.Q; precipitation in the driest quarter: Prc.Dry.Q; precipitation in the warmest quarter: Prc.Wrm.Q; and precipitation in the coldest quarter: Prc.Cld.Q. The distribution of precipitation variables is skewed, so data were transformed as log10(precipitation variable + 1), with the one added to avoid undefined values. Five linear regression analyses were run with 100-seed mass BLUP as dependent variable and each of the five transformed precipitation variables as independent variables using the lm function in R package stats v3.5.0. A partial correlation analysis using partial.cor in R package RcmdrMisc was used to estimate the independent effect of each of the five precipitation factors on 100-seed mass variation, while controlling for the remaining four precipitation factors.
Population genomic analyses
Population structure was characterized with principal component analysis (PCA) using 258,777 SNPs in TASSEL 5.0 (Bradbury et al. 2007), after filtering for SNP with a minor allele frequency >0.02. The genomic ancestry estimates were inferred by a model-based program ADMIXTURE (Alexander et al. 2009), when the pre-defined number range of genetic clusters (K) is from 2 to 17. Since the model in ADMIXTURE does not explicitly take (linkage disequilibrium (LD) into account, a LD-pruned data set of 317,836 SNPs was prepared for ADMIXTURE using --indep-pairwise 50 5 0.5 parameters in PLINK 1.07 (Purcell et al. 2007). The geographic structure was characterized through spatial mapping of the ancestry coefficients inferred by ADMIXTURE, along with the longitude and latitude coordinates of each accession, using R package tess3r (Caye et al. 2016; Martins et al. 2016). LD was calculated using PopLDdecay 3.30 with -MAF 0.05 for minimum minor allele frequency filtering and -MaxDist 500 for maximum distance (500 kb) between two SNPs (Zhang et al. 2018). The squared correlation coefficients (r2) were used to estimate LD across the whole population and for each major botanical race. Smoothed LD decay curves were fitted by the spline function in R.
Genome-wide association study of 100-seed mass and precipitation variables
A total of 146,130 biallelic SNPs with minor allele frequency (MAF) >0.05 were used for phenotypic GWAS and environmental GWAS, as a genome scan for local adaptation. A general linear model (GLM; no kinship or structure term), a mixed linear model (MLM) (Yu et al. 2006), and a fixed and random model circulating probability unification (FarmCPU) (Liu et al. 2016) in the R package GAPIT (Lipka et al. 2012) were performed to scan for genome-wide associations between SNPs and BLUPs of sorghum 100-seed mass. For MLM, model.selection = TRUE was selected to determine the optimal number of PCs, which was zero. For FarmCPU, PCA.total = 5 was used. The individual genetic relatedness matrix (K), which was calculated based on VanRaden method (VanRaden 2008) using the default setting for the kinship.algorithm option in GAPIT. For GLM, a threshold at the top 1% smallest P-values was set to identify the outlier SNPs for 100-seed mass and precipitation analyses. Given that the nominal P-values for GLM are expected to be inflated, we reported the percentiles for the outlier SNPs. For MLM, Bonferroni correction of 3.4 × 10−7 (α/number of markers, where α = 0.05) and 1% outlier thresholds were used. Due to the nature of the model, Bonferroni correction of 3.4 × 10−7 threshold was used but the top 1% smallest P-values threshold was not used for FarmCPU. To examine the degree of model fitness, quantile-quantile (Q–Q) plots of the GWAS models were constructed via plotting quantile distribution of observed −log10 (p) on the y-axis versus the quantile distribution of expected −log10(p) on x-axis.
Genome-wide scans for local adaptation
Two approaches were applied to screen the genomic signatures of adaptation based on allele frequencies. First, an ancestral allele frequency differentiation test was employed to detect the genome-wide outlier loci that could be under selection using the snmf function (Martins et al. 2016) in R package LEA (Frichot and François 2015). This method can compute fixation indices (FST) and cope with the ambiguous subpopulations simultaneously. Because the assignments of genetic clusters captured most of the genetic variations among different genetic clusters when the number of principal components (K) = 10, the number of genetic clusters was specified as 10, and then we identified outlier loci at this level. SNPs with the top 1% smallest P-values were considered outliers. Second, a PCA-based statistic was utilized to detect outlier SNPs that may be involved in local adaptation using R package pcadapt (Luu et al. 2017). The PCAdapt method assumes that SNPs excessively related with population structure are candidates for local adaptation, and the PCAdapt method can also deal with admixed individuals without any pre-defined populations. As with SNMF-based FST, K was specified as 10.
A priori candidate genes for seed mass variation
A total of 100 a priori candidate genes of sorghum seed mass on Sorghum bicolor were considered in this study, including a published set of 99 orthologs of known cereal seed mass/size genes (Tao et al. 2017) and one sorghum gene Sb04g015420 mapped from a seed size mutant (Jiao et al. 2016). The extent of LD decay (150 kb) was applied to identify the a priori candidate genes of sorghum seed mass, which are nearby the outlier SNPs derived from the genome-wide association studies of 100-seed mass and precipitation, and the selection scans. All genes mentioned in the study were a priori candidates, unless they are noted as post hoc candidates.
Enrichment analysis
The colocalized outlier SNPs detected through GLM of 100-seed mass, GLM of Prc.Dry.Q, SNMF-based FST, or PCAdapt were compared to the set of a priori 100 seed mass candidate genes. First, enrichment analysis for the colocalized top 1% outlier SNPs detected from GLM of 100-seed mass BLUPs and GLM of Prc.Dry.Q was performed. Enrichment of colocalized SNPs was compared to random SNPs derived from the genome-wide 146,130 biallelic SNPs. Second, enrichment analysis for the colocalized top 1% outlier SNPs detected from GLM of 100-seed mass BLUPs, GLM of Prc.Dry.Q and genome-wide scan of selection (SNMF-based FST or PCAdapt) were carried out. Significance of enrichment was tested for the 1% outlier SNPs of GLM of 100-seed mass BLUPs versus GLM of Prc.Dry.Q and the top 1% outlier SNPs detected from the scan of selections (SNMF-based FST or PCAdapt), relative to random SNPs. Both enrichment analyses were calculated using permTest function in R package regioneR with the randomization strategy of “resampleRegions” and 1000 permutations (Gel et al. 2016).
RESULTS
Characterization of seed mass among different sorghum botanical races
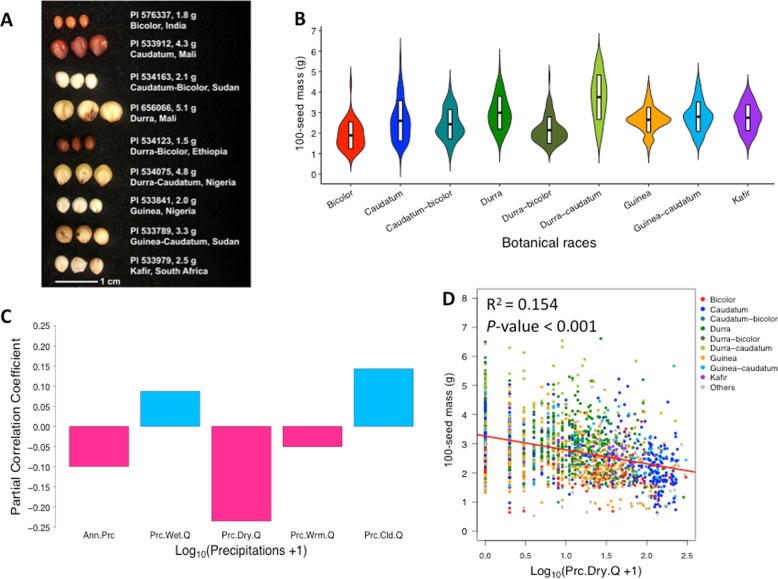
Across the global diversity of georeferenced sorghum accessions, we observed a large variation in 100-seed mass (Fig. 1a; Table 1). The sorghum accessions are classified into five botanical races (bicolor, caudatum, durra, guinea, and kafir) and ten intermediate races based on inflorescence and seed morphology (Harlan et al. 1972). To characterize variations of seed mass among different botanical races and wild relatives of sorghum, we calculated the means of each botanical race and wild species. The georeferenced GRIN accessions used in this study have an average 100-seed mass of 2.7 g, and a range in mass from 0.62 g to 5.8 g (Table 1). Among the nine botanical races with more than 20 accessions, durra-caudatum (3.8 ± 1.08 g) and durra (3.0 ± 0.8 g) sorghums have the heaviest 100-seed mass, while bicolor (1.9 ± 0.65 g) and durra-bicolor (2.1 ± 0.7 g) sorghums have the lightest 100-seed mass (Table 1; Fig. 2a, b). Notably, durra and durra-caudatum sorghums predominate in the most arid portion of the traditional sorghum range (the Sahel, the Arabian peninsula, and the western plateau of India), while durra-bicolor sorghum predominates in the humid highlands of Ethiopia (Fig. 1a, b). Bicolor sorghums, which are hypothesized to be an ancestral form (Harlan et al. 1972), have no notable geographic distribution. There is a significant difference of 100-seed mass among botanical races (F[16, 1884], P < 10−15). Post hoc comparisons based on the Tukey HSD test indicated that 41 of 136 pairwise comparisons among botanical race are significant when adjusted P < 0.01 (Table S2).
Table 1.
Summary of variation for 100-seed mass in global sorghum germplasm
| Botanical race | Number of accessions | Seed mass (g)a | ||
|---|---|---|---|---|
| GRIN | ICRISAT | BLUP | ||
| Bicolor | 84 | 1.9 ± 0.65 [0.9, 4.4] | 1.9 ± 0.60 [0.64, 3.8] | −0.73 ± 0.47 [−1.7, 0.54] |
| Caudatum | 362 | 2.6 ± 0.99 [0.7, 5.8] | 2.7 ± 1.0 [0.63, 6.5] | −0.07 ± 0.84 [−1.7, 2.9] |
| Caudatum-bicolor | 78 | 2.4 ± 0.72 [1.0, 4.2] | 2.8 ± 1.0 [0.99, 6.0] | −0.12 ± 0.67 [−1.5, 1.7] |
| Durra | 258 | 3.0 ± 0.80 [1.5, 5.2] | 3.2 ± 0.87 [1.2, 6.6] | 0.31 ± 0.67 [−1.3, 3.0] |
| Durra-bicolor | 127 | 2.1 ± 0.65 [1.2, 4.7] | 2.5 ± 0.71 [1.4, 5.4] | −0.36 ± 0.57 [−1.2, 2.1] |
| Durra-caudatum | 159 | 3.8 ± 1.1 [1.7, 5.7] | 3.7 ± 1.2 [1.5, 6.5] | 0.79 ± 0.95 [−1.0, 2.9] |
| Guinea | 445 | 2.7 ± 0.60 [0.94, 4.4] | 2.6 ± 0.76 [0.8, 5.0] | −0.11 ± 0.60 [−1.6, 1.7] |
| Guinea-caudatum | 192 | 2.8 ± 0.72 [1.4, 4.8] | 2.9 ± 0.74 [1.2, 5.7] | 0.07 ± 0.59 [−1.2, 1.6] |
| Kafir | 38 | 2.8 ± 0.63 [1.7, 3.9] | 2.8 ± 0.43 [1.8, 4.4] | −0.01 ± 0.37 [−0.9, 1.0] |
| Wild | 29 | 1.8 ± 0.71 [0.6, 3.3] | 1.3 ± 0.56 [0.53, 2.5] | −1.1 ± 0.48 [−1.8, 0.11] |
| Overall | 1901 | 2.7 ± 0.89 [0.6, 5.8] | 2.8 ± 0.97 [0.53, 6.6] | 0.0 ± 0.77 [−1.8, 3.0] |
aGiven are the mean ± standard deviation, and [minimum, maximum] across accessions for each botanical race
Fig. 2.
Global variation of common-garden seed mass in sorghum accessions with respect to botanical races and precipitation at the origin. a Representative differences of seed mass among sorghum accessions, with accession ID, 100-seed mass, botanical race, and country of origin. b Violin plot of 100-seed mass of different sorghum botanical races. Data from nine botanical races in GRIN seed bank and including more than 20 accessions are shown. The means and the standard deviations of 100-seed mass of each botanical races were represented with the black crossbars. c Partial correlations between five precipitation variables and 100-seed mass of sorghum accessions. The positive partial correlation coefficients are colored by blue and the negative partial correlation coefficients are colored by red. d Correlation of 100-seed mass and log10(Prc.Dry.Q + 1) for sorghum accessions. The accessions are colored by botanical race, with botanical races including less than 20 accessions colored in gray
Relationship between seed mass and precipitation variables
To characterize the relationship between seed mass and precipitation, we correlated 100-seed mass and each of five transformed bioclimatic precipitation variables (naccession = 1897). The correlation of Prc.Dry.Q accounts for 15.4% of the variance of 100-seed mass (R2 = 0.15, P < 0.001), more than the correlation of the other four precipitation variables: Prc.Wrm.Q (R2 = 0.082, P < 0.001), Ann.Prc (R2 = 0.051, P < 0.001), Prc.Cld.Q (R2 = 0.047, P < 0.001), Prc.Wet.Q (R2 = 0.008, P < 0.001) (Fig. 2d; Fig. S1). To further explore the independent effect of each of the five precipitation variables on the 100-seed mass, we conducted a partial correlation analysis. The precipitation variables Prc.Wet.Q (r = 0.087, adj. P = 0.0004) and Prc.Cld.Q (r = 0.14, adj. P < 0.0001) had individually positive partial correlation to 100-seed mass BLUPs. By contrast, Ann.Prc (r = −0.10, adj. P < 0.0001), Prc.Dry.Q (r = −0.23, adj. P < 0.0001), and Prc.Wrm.Q (r = −0.05, adj. P = 0.03) had individually negative partial correlation to 100-seed mass BLUPs. Among the five partial correlation tests, the strongest partial correlation was observed between 100-seed mass BLUPs and the Prc.Dry.Q (r = −0.23, adj. P < 0.0001) (Fig. 2c), so Prc.Dry.Q was targeted in further analyses.
Genome-wide SNP variation in global georeferenced germplasm
We performed two approaches to characterize the population structure and spatial genetic differentiation of the global sorghum data set. PCA and ancestry analyses showed similar complex hierarchical population structure reflecting geographic origin and botanical race (Fig. 1c, d; Fig. S2A). LD dropped to half its maximum value at ~25 kb and to background level (r2 < 0.1) at ~150 kb (Fig. S2B). LD substantially varied among the five major botanical races and sorghum wild relatives, and the LD of wild sorghum decayed much faster than the domesticated botanical races. Among domesticated sorghum, bicolor sorghum had the fasted LD decay followed by caudatum, durra, and guinea sorghum (Fig. S2B).
Genome-wide association studies of seed mass
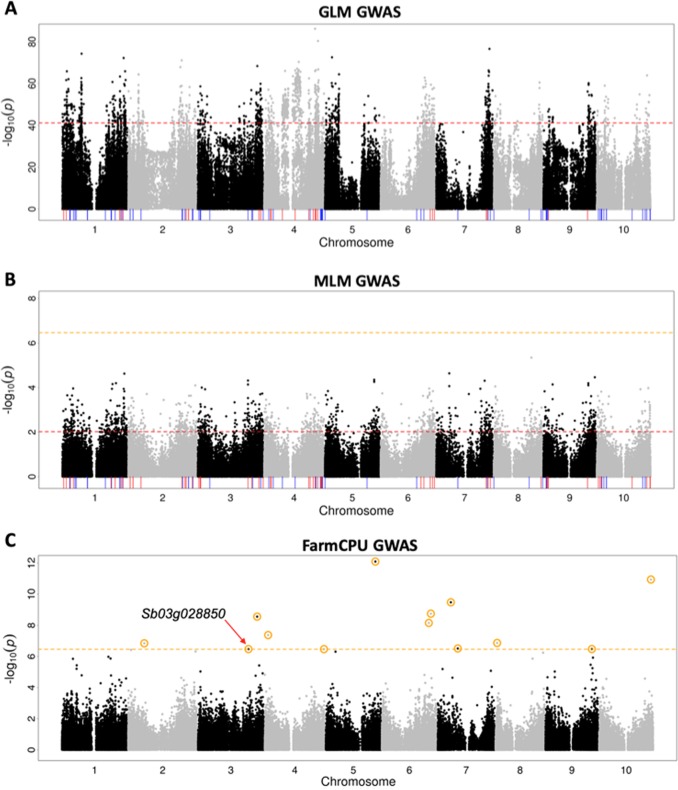
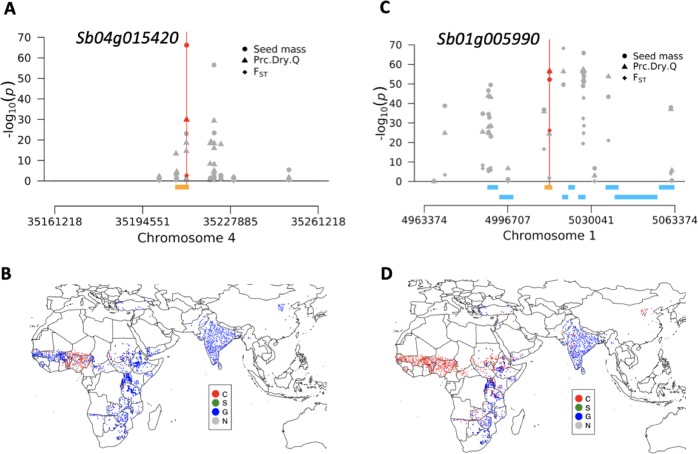
To identify genomic regions associated with seed mass in sorghum, we scanned for associations between SNPs and BLUPs of 100-seed mass using GLM, MLM, and FarmCPU. The Q–Q plots were shown in Fig. S3 A-C. The 1% outlier SNPs for 100-seed mass associations (n = 1461) were distributed on all ten chromosomes. Among these SNPs, 96 outlier SNPs were colocalized with 29 a priori candidate genes of seed mass (within 150 kb) on chromosomes 1, 2, 3, 4, 6, 7, and 9 (Fig. 3a; Table S3). The most significant SNP of the 96 outlier SNPs was on chromosome 4 (S4_57856768, MAF = 0.50, 99.9th percentile), ~9 kb from the ortholog of OsPGL2 (Sb04g027930) (Table S3). Additionally, one of the 29 most significant SNPs (S4_35211218, MAF = 0.14, 98.8th percentile) was in the sorghum mutant seed size gene Sb04g015420 (Table S3; Fig. 6a).
Fig. 3.
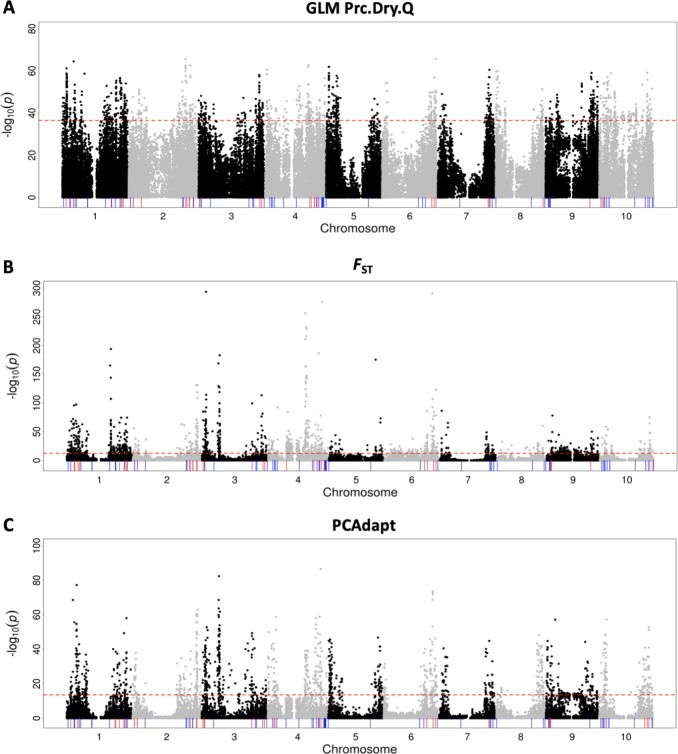
Genome-wide association studies of seed mass. Negative log10 P-values are plotted against genome position for a GLM, b MLM, and c FarmCPU GWAS of 100-seed mass BLUPs. Red horizontal dashed line indicates the genome-wide thresholds of the top 1% outlier SNPs with for GLM (−log10(p) = 41.4) and MLM (−log10(p) = 2.0). Orange horizontal dashed line indicates the genome-wide thresholds of Bonferroni correction (P < 0.05) for MLM and FarmCPU (−log10(p) = 6.5). Red vertical lines indicate a priori candidate genes, which are within 150 kb from a top 1% outlier SNPs of seed mass for GLM (n = 29) or MLM (n = 56). Blue lines indicate all other a priori candidate genes. Orange circles indicate 13 outlier SNPs of seed mass and the a priori candidate gene Sb03g028850 (ZmSh2) colocalized with the outlier SNP S3_57075248 labeled for FarmCPU
Fig. 6.
Seed mass and precipitation associations of alleles in two candidate genes. a A regional Manhattan plot around confirmed sorghum seed mass gene Sb04g015420 (orange bar). The GLM of 100-seed mass identified the outlier SNP S4_35211218 (red circle) inside the gene. However, the outlier SNP S4_35211218 was not identified as a significant SNP by GLM of precipitation (red triangle point) and SNMF-based FST (red diamond). c A regional Manhattan plot around post hoc candidate gene Sb01g005990 (orange bar), which putatively encodes a glutathione S-transferase. GLM of 100-seed mass, GLM of precipitation and SNMF-based FST all identified the outlier SNP S1_5013374 (represented by red circle, triangle point, and diamond, respectively) in the gene. Global allele distribution for b S4_35211218 and d S1_5013374, with color code indicating the genotype for each given accession
No significant SNPs were identified by MLM with a threshold −log10(p) = 6.5 after Bonferroni correction (Fig. 3b). With MLM and top 1% associated SNPs as threshold, we identified associated genomic regions on all ten chromosomes. Among the outlier SNPs, 126 SNPs on the nine chromosomes were colocalized within 150 kb flanking regions of 56 a priori candidate genes of seed mass (Fig. 3b; Table S4). The most significant of the 126 outlier SNPs was on chromosome 3 (S3_57075248, MAF = 0.38, P < 10−4), 58 kb from the putative ortholog of ZmSh2 (Sb03g028850) (Table S4). Similarly, one of the 56 most significant SNPs was in a priori gene Sb04g022620, which is the ortholog of OsSDG725 (Table S4).
Thirteen significant SNPs were identified by FarmCPU with a threshold −log10(p) = 6.5 after Bonferroni correction (Fig. 3c; Table S5). The thirteen SNPs were distributed on nine chromosomes, except Chromosome 1. These SNPs were colocalized within 150 kb flanking regions of 293 sorghum genes (Table S6). Among the thirteen SNPs, the twelfth most significant SNP, S3_57075248 (MAF = 0.38, P < 10−6), is 58 kb from the putative ortholog of ZmSh2 (Sb03g028850). Sb03g028850 is the only a priori candidate gene of sorghum seed mass included in the detected 293 sorghum genes via FarmCPU.
Genome-wide association studies of precipitations
To identify candidate genes of seed mass that are associated with precipitation gradients, we explicitly tested the genome-wide associations between SNPs and precipitation gradients. Because precipitation in the driest quarter (Prc.Dry.Q) exhibited the strongest correlation to the 100-seed mass BLUPs based on both the linear regression analysis and partial correlation analysis, the Prc.Dry.Q was used as the proxy for the precipitation gradient in further analysis. The 1% outlier SNPs for Prc.Dry.Q associations (n = 1461) were identified on ten chromosomes. Of these SNPs, 112 were colocalized with the 36 a priori candidate genes on nine chromosomes, excluding chromosome 5 (Fig. 4a; Table S7). The most highly associated SNP that was colocalized with an a priori candidate gene was S2_64282627 (MAF = 0.37, 99.9th percentile). This SNP is 146 kb from candidate seed mass gene Sb02g029300, the ortholog of OsGW8 (Table S7). No outlier SNPs fell inside an a priori candidate gene. The Q–Q plot was shown in Fig. S3D.
Fig. 4.
Genome-wide scans for adaptation. Negative log10-transformed P-values from the genome-wide SNP scans are plotted against the genome position for a GLM association with precipitation in the driest quarter (Prc.Dry.Q), b SNMF-based FST, and c PCAdapt using K = 10. Red horizontal dashed line indicates the genome-wide threshold of the top 1% outlier SNPs for association with Prc.Dry.Q (−log10(p) = 36.5), SNMF-based FST (−log10(p) = 12.5) and PCAdapt (−log10(p) = 13.5). Red vertical lines represent a priori candidate genes of seed mass above the given thresholds for association with Prc.Dry.Q (n = 36), SNMF-based FST (n = 35), and PCAdapt (n = 37). Blue vertical segments indicate the undetected 64, 65, and 63 a priori candidate genes of seed mass below the thresholds for association with Prc.Dry.Q, SNMF-based FST, and PCAdapt, respectively
Genome-wide scans for local adaptation
Genomic regions displaying signatures of selection among populations could suggest adaptation to diverse environmental gradients. We conducted SNMF-based FST and PCAdapt analyses to detect outlier SNPs involving allele frequency differentiation. The top 1% outlier SNPs identified from both methods were distributed on all ten chromosomes. From SNMF-based FST, 133 outlier SNPs identified on chromosomes 1, 2, 3, 4, 6, 9, and 10 were colocalized with the 150 kb flanking regions of 35 a priori candidate seed mass genes (Fig. 4b; Table S8). The most significant SNP among the 133 outliers was S6_58925412 (MAF = 0.082, P < 10−23), which is ~20 kb from the seed mass candidate gene Sb06g030550, an ortholog of OsFLO2 (Table S8). Additionally, two outlier SNPs fell inside two a priori candidate genes of seed mass on chromosome 3. First, S3_57017386 (MAF = 0.09, P < 10−99) was located inside Sb03g028850, the ortholog of ZmSh2 (Table S8). Second, S3_71459932 (MAF = 0.12, P < 10−42) was in Sb03g044160, the ortholog of OsGW8 (Table S8).
As a complement to the SNMF-based FST analysis, PCAdapt identified 120 outlier SNPs on eight chromosomes, which were colocalized within 150 kb of 37 a priori candidate genes of seed mass (Fig. 4c; Table S9). The most significant SNP of the 120 outlier SNPs was S2_72877577 (MAF = 0.15, P < 10−60), ~5 kb away from seed mass candidate gene Sb02g038670, an ortholog of OsBG2 (Table S9). In accordance with the SNMF-based FST results, S3_71459932 (MAF = 0.12, P < 10−15) was also detected in PCAdapt, located inside the ortholog of OsGW8 (Sb03g044160; Table S9).
Colocalized SNPs and enrichment analysis
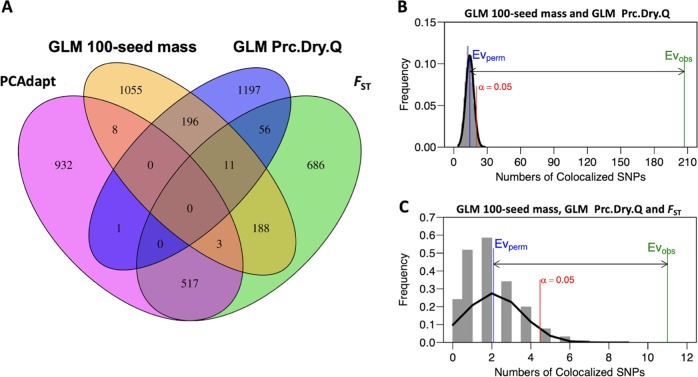
Enrichment of a priori seed mass genes with GWAS and genome scan outliers may provide additional evidence of seed mass adaptation. A total of 207 colocalized outlier SNPs diffusing on the ten chromosomes were identified based on the top 1% outlier SNPs detected by GLM of 100-seed mass BLUPs and GLM of precipitation (Fig. 5a; Table S10). There was a significant enrichment for the colocalized 207 outlier SNPs (P < 0.001; Fig. 5b). A total of 11 a priori sorghum seed mass candidate genes on chromosome 1, 2, 3, 4, 6, 7, and 9, which fell into 150 kb flanking the colocalized 207 outlier SNPs, were identified (Table 2). A different set of 11 colocalized outlier SNPs at four loci (chromosomes 1, 4, 7, and 8) were identified based on the top 1% outlier SNPs detected by GLM of 100-seed mass, GLM of precipitation and SNMF-based FST method (Table S11). Seven sorghum genes (not a priori candidates) nearest (<2 kb) to the eleven colocalized outlier SNPs were identified (Table S11). However, no colocalized SNPs were identified based on the top 1% outlier SNPs detected by GLM of 100-seed mass, GLM of precipitation, and PCAdapt (Fig. 5a). There was a significant enrichment for the colocalized eleven outlier SNPs (P < 0.001; Fig. 5c). However, no a priori sorghum 100-seed mass candidate genes were identified, because none of them fell into 150 kb flanking the eleven outlier SNPs.
Fig. 5.
Colocalized outlier SNPs among phenotypic association, environmental association, and selection scans. a Venn diagrams of outlier SNPs identified jointly by GLM of 100-seed mass, GLM of precipitation in the driest quarter (Prc.Dry.Q), SNMF-based FST, and PCAdapt. Enrichment analysis for b seed mass association and precipitation associations, or c seed mass associations, precipitation associations, and FST. In panel b, c, the blue line (Evperm) represents the mean of colocalized SNPs from the null distribution with 1000 permutations, the red line represents α = 0.05 for the null distribution, and the green line (Evobs) represents the number of the observed colocalized SNPs
Table 2.
Eleven a priori sorghum seed mass candidate genes that colocalized with outlier SNPs for both GLM of 100-seed mass and GLM of Prc.Dry.Q
| SNP IDa | Geneb | Gene Startb | Gene Endb | Genec | Orthologd |
|---|---|---|---|---|---|
| S1_4315232 | Sb01g005170 | 4275513 | 4279485 | Sobic.001G056700 | ZmO2 |
| S1_65251930 | Sb01g041900 | 65135319 | 65141117 | Sobic.001G445900 | OsCYP90B1 |
| S2_65532747 | Sb02g030640 | 65662195 | 65664496 | Sobic.002G272700 | AtEOD3 |
| S2_68333169 | Sb02g033830 | 68370266 | 68371629 | Sobic.002G308400 | MYB |
| S2_68333169 | Sb02g033940 | 68469057 | 68478741 | Sobic.002G309600 | AtUPF1 |
| S3_69399186 | Sb03g042060 | 69485282 | 69485858 | Sobic.003G380900 | OsSERF1 |
| S4_50682434 | Sb04g021540 | 50589773 | 50601467 | Sobic.004G163700 | ZmSbeIIb |
| S4_58788981 | Sb04g028690 | 58829498 | 58833664 | Sobic.004G247000 | ZmGln-4 |
| S6_56374341 | Sb06g027465 | 56363893 | 56367340 | Sobic.006G203400 | OsGS2 |
| S7_57962621 | Sb07g023040 | 57901670 | 57902598 | Sobic.007G156800 | OsSGL1 |
| S9_5395395 | Sb09g004510 | 5453946 | 5459014 | Sobic.009G053600 | OsGS5 |
aSNP ID includes the chromosome number (before the underscore) and position (after the underscore) in v1.4 coordinates
bGene name and position in v1.4 sorghum genome assembly
cGene name in v3.0 sorghum genome assembly
dDetails on orthologs in ref. (Tao et al. 2017)
Discussion
Evidence for clinal adaptation of seed mass to precipitation gradients
Sorghum has wide variation in seed mass, over 7-fold among accessions of domesticated sorghum (Fig. 1a; Fig. 2a, b; Table 1). This observation runs counter to the literature on wild plants, which suggests that within-species variation in seed mass is small (<4-fold) relative to among-species and within-plant variation (Silvertown 1989; Leishman et al. 2000). While the genebank 100-seed mass data do not capture within-plant or among-plant variation for a given genotype, we do not expect these to be major contributors to the observed variance, since genebank seed is produced under near-optimal conditions where stress and microenvironment variation is minimized (Tao et al. 2018).
Ecological studies of wild plants suggest that plants adapted to drought-prone environments produce larger seeds (Baker 1972; Hallett et al. 2011), but whether this is a general phenomenon remains contentious (Leishman et al. 2000). In the present study, the significant differences of seed mass among sorghum botanical races from precipitation zones (Fig. 2a-d; Fig. S1 & S2) support the hypothesis that the precipitation gradient has shaped genetic variation of seed mass within and among botanical races of sorghum. For instance, durra sorghum, adapted to arid climates of Sahelian West Africa and western India, produce larger seeds than guinea sorghums, adapted to subtropical high rainfall climate zone in the Guinean West Africa and eastern India (Fig. 1a, b).
Based on our findings and previous studies, we hypothesize that large seeds are favored in arid agroclimatic zones due to a “seedling size effect”, where additional reserves in larger seeds help seedlings survive drought stress (Leishman et al. 2000). Given that wild sorghums have small seeds (Table 1), we hypothesize, parsimoniously, that small-seeded domesticated sorghum in humid agroclimatic zones reflect the ancestral state, rather than diversifying selection for reduced seed size in domesticated sorghum. We note that other environmental variables are correlated with precipitation across the global distribution of sorghum (Lasky et al. 2015). Therefore, further population and quantitative trait analyses will be required to confirm these hypotheses, and disentangle adaptation to precipitation from adaptation to temperature, soil, or other factors.
Genomic signatures of seed mass regulation and precipitation adaptation
To our knowledge, no natural variants controlling seed mass or size in sorghum have been identified (Tao et al. 2017, 2018). A top association from GLM GWAS of seed mass (S4_35211218) fell within a confirmed sorghum seed size and mass gene Sb04g015420 (Jiao et al. 2016), suggesting that this gene identified via mutant analysis also underlies natural variation in seed mass (Fig. 6a, b). Both the maize ortholog ZmSWEET4c and rice ortholog OsSWEET4 of Sb04g015420 regulate the grain filling via SWEET-mediated hexose transport (Sosso et al. 2015). Among the candidate genes identified by the seed mass GWAS, several (15/29 from GLM, 16/56, from MLM) have further evidence from colocalization with linkage mapping QTL intervals (Table S12). For example, Sb09g004510, an ortholog of GS5, colocalized with the interval of the sorghum grain weight QTL SDWT.LG9-2 (Brown et al. 2006). In rice and wheat, GS5 regulates seed size by promoting cell division (Li et al. 2011; Ma et al. 2016), so Sb09g004510 may play a similar role in sorghum. FarmCPU GWAS identified a promising a priori candidate gene of seed mass, Sb03g028850, which is a putative ortholog of ZmSh2 involving starch biosynthesis. Sb03g028850 was also identified by the MLM GWAS and with top 1% most significant association as the threshold.
Several lines of evidence in the present study supported the hypothesis that genetic variation around sorghum loci regulating seed mass was shaped by adaptation to precipitation gradients. Eleven a priori seed mass candidate genes were identified simultaneously from genomic signals of seed mass association and Prc.Dry.Q association (Fig. 5a, b; Table S11; Table 1), consistent with precipitation gradients shaping seed mass variation. Sb02g033830 (MYB67) is an ortholog of a MYB transcription factor in rice that was identified from a grain yield GWAS (Huang et al. 2012). Sb01g041900 is an ortholog of OsCYP90B1 and OsCYP90B2, brassinosteroid biosynthesis genes that regulate grain filling in rice (Wu et al. 2008). The seven nearest sorghum genes (not a priori candidates) to the eleven colocalized outlier SNPs may be considered post hoc candidate genes for seed mass adaptation (Table S12). Among the seven genes, Sb01g005990 (Fig. 6c, d) and Sb01g006000 putatively encode glutathione S-transferase (GST). GSTs have been implicated in drought tolerance in Arabidopsis (Chen et al. 2012), but to our knowledge no studies have suggested a role for GSTs in seed mass.
Identification of the overlapping genomic regions between positional and association genetic studies can further support the results of each method (Zhang et al. 2015). The GLM and MLM of seed mass identified 29 and 56 a priori candidate genes of seed mass (Table S3 & S4), respectively, suggesting the diverse mechanisms could regulate sorghum seed mass. For example, OsPGL2, the ortholog of Sb04g027930, is a bHLH transcription factor that positively regulates the rice grain length (Heang and Sassa 2012). Similarly, OsSDG725, the ortholog of Sb04g022620, is H3K36 methyltransferase that regulates brassinosteroid-dependent grain development in rice (Sui et al. 2012). Further functional investigations of these candidate genes will be required to confirm effects on seed mass and test for any role in drought adaptation.
Prospects for dissection and improvement of climate-adaptive seed mass variation
Identifying the most effective genomic approaches to dissect adaptive trait variation is an ongoing challenge (Mita et al. 2013; Tiffin and Ross-Ibarra 2014; Lipka et al. 2015; Forester et al. 2016; Bouchet et al. 2017). In this study, few outlier SNPs were colocalized among all methods (Fig. 5a). Similar findings were reported in a study of local adaptation in the wild progenitor of soybean Glycine soja (Bandillo et al. 2017). Different assumptions of each method may be one reason for the small number of colocalized outlier SNPs (Akey 2009).
Population structure observed in our study reflected highly structured global sorghum diversity, which can confound association approaches (Fig. 1c, d; Fig. S2A) (Morris et al. 2013). The geographic patterns and GWAS findings indicate seed mass is a trait that is strongly confounded with population structure (Fig. 3a). Although the MLM controlled inflation efficiently (Fig. 3b), no significant association outliers were identified by MLM at the Bonferroni threshold, suggesting false negatives due to a collinear polygenic and oligogenic components of variance (Myles et al. 2009; Lipka et al. 2015). As shown in the Q–Q plots (Fig. S3 A-C), FarmCPU model is best fitted our 100-seed mass data. The FarmCPU in our 100-seed mass GWAS study can relieve the confounding effect efficiently, such as populations structure, and strengthen the statistical power. The performance of FarmCPU in our study supported its developer’s statement on improved power (Liu et al. 2016). To break up population structure and gain mapping power, multi-parental mapping with multi-advanced generation intercross (MAGIC) or nested association mapping (NAM) populations should be an effective approach (Bouchet et al. 2017; Ongom and Ejeta 2018).
The phenotypic, geographic, and genomic signatures we characterized here provide new evidence that seed mass variation can be adaptive to precipitation gradients (Leishman et al. 2000). In addition to basic insights on ecology and evolution, these findings may help crop improvement programs to identify germplasm harboring adaptive alleles for seed mass and drought tolerance. Genomics knowledge on adaptive and agronomic traits can ultimately be leveraged into genomics-enabled breeding of climate-resilience in sorghum and other crops (Zuo and Li 2014; Tao et al. 2017).
Supplementary information
Acknowledgements
The work was supported by Bill & Melinda Gates Foundation through the “TERRA-REF partnership: Sorghum Genomics Toolbox”. We thank Oliver François for advice on implementing LEA, and Narinder Singh for preliminary analyses that motivated the study. We thank the editor and anonymous peer reviewers for the comments to improve our manuscript. This study is contribution 20-006-J from the Kansas Agricultural Experiment Station.
Data availability
All data are publicly available. SNP genotype, geographic, and precipitation data are available from Dryad Digital Repository at 10.5061/dryad.jc3ht and provided in Table S1. The 100-seed mass data from GRIN, ICRISAT, and BLUPs data can be accessed in Table S1.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41437-019-0249-4) contains supplementary material, which is available to authorized users.
References
- Abebe TD, Naz AA, Léon J. Landscape genomics reveal signatures of local adaptation in barley (Hordeum vulgare L.) Front Plant Sci. 2015;6:813. doi: 10.3389/fpls.2015.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 2009;19:711–722. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker HG. Seed weight in relation to environmental conditions in California. Ecology. 1972;53:997–1010. [Google Scholar]
- Bandillo NB, Anderson JE, Kantar MB, Stupar RM, Specht JE, Graef GL, et al. Dissecting the genetic basis of local adaptation in soybean. Sci Rep. 2017;7:17195. doi: 10.1038/s41598-017-17342-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet. 2010;11:867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- Blanquart F, Kaltz O, Nuismer SL, Gandon S. A practical guide to measuring local adaptation. Ecol Lett. 2013;16:1195–1205. doi: 10.1111/ele.12150. [DOI] [PubMed] [Google Scholar]
- Bouchet S, Olatoye MO, Marla SR, Perumal R, Tesso T, Yu J, et al. Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics. 2017;206:573–585. doi: 10.1534/genetics.116.198499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles Richard E., Cooper Elizabeth A., Myers Matthew T., Brenton Zachary, Rauh Bradley L., Morris Geoffrey P., Kresovich Stephen. Genome-Wide Association Studies of Grain Yield Components in Diverse Sorghum Germplasm. The Plant Genome. 2016;9(2):0. doi: 10.3835/plantgenome2015.09.0091. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Brown P, Klein P, Bortiri E, Acharya C, Rooney W, Kresovich S. Inheritance of inflorescence architecture in sorghum. Theor Appl Genet. 2006;113:931–942. doi: 10.1007/s00122-006-0352-9. [DOI] [PubMed] [Google Scholar]
- Burow G, Xin Z, Hayes C, Burke J. Characterization of a multiseeded (msd1) mutant of sorghum for increasing grain yield. Crop Sci. 2014;54:2030–2037. [Google Scholar]
- Caye K, Deist TM, Martins H, Michel O, François O. TESS3: fast inference of spatial population structure and genome scans for selection. Mol Ecol Resour. 2016;16:540–548. doi: 10.1111/1755-0998.12471. [DOI] [PubMed] [Google Scholar]
- Chen J-H, Jiang H-W, Hsieh E-J, Chen H-Y, Chien C-T, Hsieh H-L, et al. Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 2012;158:340–351. doi: 10.1104/pp.111.181875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, Hernandez KM, Juenger TE. Genotype-by-environment interaction and plasticity: exploring genomic responses of plants to the abiotic environment. Annu Rev Ecol Evol Syst. 2013;44:5–29. [Google Scholar]
- Deu M, Rattunde F, Chantereau J. A global view of genetic diversity in cultivated sorghums using a core collection. Genome. 2006;49:168–180. doi: 10.1139/g05-092. [DOI] [PubMed] [Google Scholar]
- Eckert AJ, Van Heerwaarden J, Wegrzyn JL, Nelson CD, Ross-Ibarra J, González-Martínez SC, et al. Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae) Genetics. 2010;185:969–982. doi: 10.1534/genetics.110.115543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltus F, Hart G, Schertz K, Casa A, Kresovich S, Abraham S, et al. Alignment of genetic maps and QTLs between inter- and intra-specific sorghum populations. Theor Appl Genet. 2006;112:1295–1305. doi: 10.1007/s00122-006-0232-3. [DOI] [PubMed] [Google Scholar]
- Forester BR, Jones MR, Joost S, Landguth EL, Lasky JR. Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol Ecol. 2016;25:104–120. doi: 10.1111/mec.13476. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- Frichot E, François O. LEA: an R package for landscape and ecological association studies. Methods Ecol Evol. 2015;6:925–929. [Google Scholar]
- Gel B, Díez-Villanueva A, Serra E, Buschbeck M, Peinado MA, Malinverni R. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics. 2016;32:289–291. doi: 10.1093/bioinformatics/btv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett LM, Standish RJ, Hobbs RJ. Seed mass and summer drought survival in a Mediterranean-climate ecosystem. Plant Ecol. 2011;212:1479. [Google Scholar]
- Han L, Chen J, Mace ES, Liu Y, Zhu M, Yuyama N, et al. Fine mapping of qGW1, a major QTL for grain weight in sorghum. Theor Appl Genet. 2015;128:1813–1825. doi: 10.1007/s00122-015-2549-2. [DOI] [PubMed] [Google Scholar]
- Harlan JR, Wet D, J JM. A simplified classification of cultivated sorghum. Crop Sci. 1972;12:172–176. [Google Scholar]
- Heang D, Sassa H. Overexpression of a basic helix–loop–helix gene Antagonist of PGL1 (APG) decreases grain length of rice. Plant Biotechnol. 2012;29:65–69. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- Hoban S, Kelley JL, Lotterhos KE, Antolin MF, Bradburd G, Lowry DB, et al. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am Nat. 2016;188:379–397. doi: 10.1086/688018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Burke J, Chopra R, Burow G, Chen J, Wang B, et al. A sorghum mutant resource as an efficient platform for gene discovery in grasses. Plant Cell Online. 2016;28:1551–1562. doi: 10.1105/tpc.16.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort HD, Vandepitte K, Bruun HH, Closset-Kopp D, Honnay O, Mergeay J. Landscape genomics and a common garden trial reveal adaptive differentiation to temperature across Europe in the tree species Alnus glutinosa. Mol Ecol. 2014;23:4709–4721. doi: 10.1111/mec.12813. [DOI] [PubMed] [Google Scholar]
- Lasky JR, Des Marais DL, McKay JK, Richards JH, Juenger TE, Keitt TH. Characterizing genomic variation of Arabidopsis thaliana: the roles of geography and climate. Mol Ecol. 2012;21:5512–5529. doi: 10.1111/j.1365-294X.2012.05709.x. [DOI] [PubMed] [Google Scholar]
- Lasky JR, Upadhyaya HD, Ramu P, Deshpande S, Hash CT, Bonnette J, et al. Genome-environment associations in sorghum landraces predict adaptive traits. Sci Adv. 2015;1:e1400218. doi: 10.1126/sciadv.1400218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leishman MR, Wright IJ, Moles AT, Westoby M (2000) The evolutionary ecology of seed size. In: Fenner M (ed) Seeds: The Ecology of Regeneration in Plant Communities. CABI, Wallingford, p 31–57
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- Lipka AE, Kandianis CB, Hudson ME, Yu J, Drnevich J, Bradbury PJ, et al. From association to prediction: statistical methods for the dissection and selection of complex traits in plants. Curr Opin Plant Biol. 2015;24:110–118. doi: 10.1016/j.pbi.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, Buckler ES, Zhang Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016;12:e1005767. doi: 10.1371/journal.pgen.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu K, Bazin E, Blum MGB. pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol Ecol Resour. 2017;17:67–77. doi: 10.1111/1755-0998.12592. [DOI] [PubMed] [Google Scholar]
- Ma L, Li T, Hao C, Wang Y, Chen X, Zhang X. TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol J. 2016;14:1269–1280. doi: 10.1111/pbi.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins H, Caye K, Luu K, Blum MGB, François O. Identifying outlier loci in admixed and in continuous populations using ancestral population differentiation statistics. Mol Ecol. 2016;25:5029–5042. doi: 10.1111/mec.13822. [DOI] [PubMed] [Google Scholar]
- Mita SD, Thuillet A-C, Gay L, Ahmadi N, Manel S, Ronfort J, et al. Detecting selection along environmental gradients: analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol Ecol. 2013;22:1383–1399. doi: 10.1111/mec.12182. [DOI] [PubMed] [Google Scholar]
- Monk R, Franks C, Dahlberg J (2014) Sorghum. In: Smith S, Diers B, Specht J, Carver B (eds) Yield Gains in Major US Field Crops, Crop ScienceSociety of America, Madison, WI, USA. pp 293–310
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA. 2013;110:453–458. doi: 10.1073/pnas.1215985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, et al. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson H, Ripa J, Jonzén N. Bet-hedging as an evolutionary game: the trade-off between egg size and number. Proc R Soc Lond B Biol Sci. 2009;276:2963–2969. doi: 10.1098/rspb.2009.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongom PO, Ejeta G (2018) Mating design and genetic structure of a multi-parent advanced generation inter-cross (MAGIC) population of sorghum (Sorghum bicolor (L.) Moench). G3 Genes Genomes Genet 8:331–341 [DOI] [PMC free article] [PubMed]
- Paterson AH, Lin Y-R, Li Z, Schertz KF, Doebley JF, Shannon RMP, et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadras VO. Evolutionary aspects of the trade-off between seed size and number in crops. Field Crops Res. 2007;100:125–138. [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. Ecological genomics of local adaptation. Nat Rev Genet. 2013;14:807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- Siepielski AM, Morrissey MB, Buoro M, Carlson SM, Caruso CM, Clegg SM, et al. Precipitation drives global variation in natural selection. Science. 2017;355:959–962. doi: 10.1126/science.aag2773. [DOI] [PubMed] [Google Scholar]
- Silvertown J. The paradox of seed size and adaptation. Trends Ecol Evol. 1989;4:24–26. doi: 10.1016/0169-5347(89)90013-X. [DOI] [PubMed] [Google Scholar]
- Smith CC, Fretwell SD. The optimal balance between size and number of offspring. Am Nat. 1974;108:499–506. [Google Scholar]
- Sosso D, Luo D, Li Q-B, Sasse J, Yang J, Gendrot G, et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet. 2015;47:1489–1493. doi: 10.1038/ng.3422. [DOI] [PubMed] [Google Scholar]
- Srinivas G, Satish K, Madhusudhana R, Nagaraja Reddy R, Murali Mohan S, Seetharama N. Identification of quantitative trait loci for agronomically important traits and their association with genic-microsatellite markers in sorghum. Theor Appl Genet. 2009;118:1439–1454. doi: 10.1007/s00122-009-0993-6. [DOI] [PubMed] [Google Scholar]
- Sui P, Jin J, Ye S, Mu C, Gao J, Feng H, et al. H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 2012;70:340–347. doi: 10.1111/j.1365-313X.2011.04873.x. [DOI] [PubMed] [Google Scholar]
- Swarts K, Gutaker RM, Benz B, Blake M, Bukowski R, Holland J, et al. Genomic estimation of complex traits reveals ancient maize adaptation to temperate North America. Science. 2017;357:512–515. doi: 10.1126/science.aam9425. [DOI] [PubMed] [Google Scholar]
- Tao Yongfu, Mace Emma, George-Jaeggli Barbara, Hunt Colleen, Cruickshank Alan, Henzell Robert, Jordan David. Novel Grain Weight Loci Revealed in a Cross between Cultivated and Wild Sorghum. The Plant Genome. 2018;11(2):0. doi: 10.3835/plantgenome2017.10.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Mace ES, Tai S, Cruickshank A, Campbell BC, Zhao X, et al. Whole-genome analysis of candidate genes associated with seed size and weight in Sorghum bicolor reveals signatures of artificial selection and insights into parallel domestication in cereal crops. Front Plant Sci. 2017;8:1237. doi: 10.3389/fpls.2017.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Ross-Ibarra J. Advances and limits of using population genetics to understand local adaptation. Trends Ecol Evol. 2014;29:673–680. doi: 10.1016/j.tree.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Upadhyaya HD, Wang Y-H, Sharma S, Singh S, Hasenstein KH. SSR markers linked to kernel weight and tiller number in sorghum identified by association mapping. Euphytica. 2012;187:401–410. [Google Scholar]
- VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y, Mariac C, Mita SD, Pham J-L, Gérard B, Kapran I, et al. Selection for earlier flowering crop associated with climatic variations in the Sahel. PLoS ONE. 2011;6:e19563. doi: 10.1371/journal.pone.0019563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villemereuil P, Gaggiotti OE, Mouterde M, Till-Bottraud I. Common garden experiments in the genomic era: new perspectives and opportunities. Heredity. 2016;116:249–254. doi: 10.1038/hdy.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendorf F, Close AE, Schild R, Wasylikowa K, Housley RA, Harlan JR, et al. Saharan exploitation of plants 8000 years BP. Nature. 1992;359:721–724. [Google Scholar]
- Wu C, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, et al. Brassinosteroids regulate grain filling in rice. Plant Cell. 2008;20:2130–2145. doi: 10.1105/tpc.107.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, van Oosterom EJ, Jordan DR, Doherty A, Hammer GL. Genetic variation in potential kernel size affects kernel growth and yield of sorghum. Crop Sci. 2010;50:685. [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zhang C, Dong S-S, Xu J-Y, He W-M, Yang T-L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2018;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- Zhang D, Li J, Compton RO, Robertson J, Goff VH, Epps E, et al. Comparative genetics of seed size traits in divergent cereal lineages represented by sorghum (Panicoidae) and rice (Oryzoidae) G3 Genes Genomes Genet. 2015;5:1117–1128. doi: 10.1534/g3.115.017590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet. 2014;48:99–118. doi: 10.1146/annurev-genet-120213-092138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available. SNP genotype, geographic, and precipitation data are available from Dryad Digital Repository at 10.5061/dryad.jc3ht and provided in Table S1. The 100-seed mass data from GRIN, ICRISAT, and BLUPs data can be accessed in Table S1.