Abstract
Sudden cardiac death (SCD) is often associated with structural abnormalities of the heart during autopsy. This study sought to compare the diagnostic yield of postmortem genetic testing in (1) cases with structural findings of uncertain significance at autopsy to (2) cases with autopsy findings diagnostic of cardiomyopathy. We evaluated 57 SCD cases with structural findings at cardiac autopsy. Next-generation sequencing using a panel of 77 primary electrical disorder and cardiomyopathy genes was performed. Pathogenic and likely pathogenic variants were classified using American College of Medical Genetics (ACMG) consensus guidelines. In 29 cases (51%) autopsy findings of uncertain significance were identified whereas in 28 cases (49%) a diagnosis of cardiomyopathy was established. We identified a pathogenic or likely pathogenic variant in 10 cases (18%); in 1 (3%) case with non-specific autopsy findings compared with 9 (32%) cases with autopsy findings diagnostic of cardiomyopathy (p = 0.0054). The yield of genetic testing in SCD cases with autopsy findings consistent with cardiomyopathy is comparable with the yield in cardiomyopathy patients that are alive. Genetic testing in cases with findings of uncertain significance offers lower clinical utility than in cardiomyopathy, with lower yields than detected previously. This highlights the need for stringent evaluation of variant pathogenicity.
Subject terms: Disease genetics, Cardiovascular diseases
Introduction
Sudden cardiac death (SCD) is an important cause of death in the young. Predominant causes vary by age and include coronary artery disease, cardiomyopathy and primary electrical disorders [1]. SCD and cardiac arrest in the young due to cardiomyopathies are, however, uncommon both in the general population [2] and during participation in competitive sport [3]. They remain important because of their inherited nature and hence potential value in family screening. In 30–40% no diagnosis can be established at autopsy despite toxicological and histopathological analysis [4, 5]. These cases are classified as sudden arrhythmic death syndrome (SADS) and we have recently reported a 13% yield of pathogenic and likely pathogenic variants from postmortem genetic testing (i.e. molecular autopsy) in a large set of 302 SADS cases [5]. In the current study, we aimed to determine the diagnostic yield of genetic testing in cases with autopsy findings of uncertain significance and compare them to cases with autopsy findings diagnostic of cardiomyopathy.
Materials and methods
Study population
Suitable cases were included from two cohorts of SCD cases as previously described [5]. Cohort 1 consisted of a population-based coronial series (Cardiac Inherited Disease Registry, Auckland, New Zealand 2000–2009, [n = 11] [6, 7]; consecutive referrals for autopsy (Royal Brompton Hospital, UK 2007–2011 [n = 6], Sheffield Children’s Hospital, UK 1985–2001 [n = 1]); and consecutive referrals at the time of familial cardiac evaluation following a SCD (St George’s and Lewisham Hospitals, UK 2009–2011 [n = 3]; Academic Medical Centre, Netherlands 1995–2011 [n = 2]). Cohort 2 consisted of 34 consecutive referrals for autopsy to St George’s Hospital, UK 2012–2015. The study was approved by Research Ethics Service Wandsworth (Ref: 10H/0803/72). SCD was defined as death within 1 h of onset of symptoms, or an unwitnessed death with the individual being seen in good health within 24 h of death. Clinical history, prior symptoms, circumstance of death and family history were collected by direct contact with next of kin and from coroners’ and pathologists’ reports.
Autopsy evaluation
Cardiac autopsy was performed by locally recognised expert pathologists and included histological analysis. Cases where than classified into those (1) with autopsy findings of uncertain significance and (2) cases with autopsy findings diagnostic of cardiomyopathy. Findings of uncertain significance were defined as cardiac structural changes identified at autopsy of uncertain relevance to the cause of death. Adjudication of autopsy findings was performed by two of the authors using recent guidelines for autopsy investigation of SCD [8] as previously described [9] and then confirmed by an expert pathologist (MNS).
Next-generation sequencing and quality control
Next-generation sequencing was performed as previously described [5]. In summary, target regions were captured by in-solution hybridisation target capture, using the Agilent SureSelect system (cohort 1) or the Illumina TruSight Cardio system (cohort 2). For SureSelect capture, custom hybridisation-capture probes were designed using the Agilent eArray platform to target 201 genes implicated in cardiovascular disease. RNA baits targeted all exons of all Ensembl (v54) transcripts, including untranslated regions, with a 100-bp extension into adjacent introns. The TruSight Cardio Kit (Illumina) consists of comprehensive coverage of 174 genes (coding sequence region only) with known associations to 17 inherited cardiac conditions [10]. Libraries were prepared according to the manufacturer’s instructions and sequenced on the Illumina HiSeq (cohort 1) or NextSeq (cohort 2). Variant were annotated using ANNOVAR [11]. We generated alignment summary metrics, coverage and callability metrics. We considered a base “callable” if it was sequenced with minimum read depth = 10×, base quality ≥ 20 and mapping quality ≥ 10. Target base callability of >90% was achieved in 90% and 91% of samples from cohort 1, 2, respectively. Gene level callability is shown in Supplementary Table 1. We analysed 77 genes (Supplementary Table 2) present in both capture systems that have been previously associated with primary electrical diseases or cardiomyopathies.
Classification of genetic variants
Variants with a minor allele frequency (MAF) >1/10,000 in the Genome Aggregation Database (gnomAD), synonymous variants not located at splice sites and non-truncating variants in TTN were excluded. We applied American College of Medical Genetics (ACMG) consensus guidelines [12] to classify variants as (1) pathogenic, (2) likely pathogenic or as (3) variant of unknown significance (VUS). This included evidence from population data; identification of the variant in clinically affected patients in public and in-house disease databases; functional evidence; and concordance between the gene, type of variant and disease mechanism. Variants were adjudicated by two investigators as previously described [5]. Pathogenic and likely pathogenic variants according to ACMG guidelines were considered likely to cause disease in the patient. Genetic variants were submitted to ClinVar (www.ncbi.nlm.nih.gov/clinvar/) and the following accession number was obtained UB5428733.
Statistical analyses
Comparison was performed using the Mann–Whitney U test for non-normally distributed continuous variables and the chi-square or Fisher’s exact test, as suitable, for categorical variables. All statistical analyses were performed using R (version 3.2.1.), and a p < 0.05 was used to indicate statistical significance. Percentages reported for clinical parameters are based on non-missing data (n = 50).
Results
In total, 28 SCD cases with a diagnosis of cardiomyopathy and 29 cases with findings of uncertain significance during autopsy were included (median age of 29 and 32 years, respectively, p-value = 0.7659, Table 1). The most prevalent circumstances of death in the overall cohort were during sleep (n = 15, 30%) or rest (n = 14, 28%) with death occurring during exercise in 10 cases (20%). No statistically significant difference was observed in demographic and clinical characteristic between cases with autopsy findings of uncertain significance and cases with autopsy findings diagnostic of cardiomyopathy. Out of 28 SCD cases diagnosed as cardiomyopathy 6 (21%) had a premorbid diagnosis of cardiomyopathy and 3 (11%) had a family history of cardiomyopathy.
Table 1.
Demographic and clinical characteristics in the sudden death cohort
| Autopsy findings | ||
|---|---|---|
| Variable | uncertain significance (n = 29) | diagnostic of cardiomyopathy (n = 28) |
| Age at death, median age [IQR], years | 32 [21–38] | 29 [24–38] |
| Male gender, no. (%) | 86% [25/29] | 68% [19/28] |
| Circumstances of death, no./total no. (%)a | ||
| Exercise and extreme emotion | 25% [6/24] | 19% [5/26] |
| Sleep and rest | 63% [15/24] | 54% [14/26] |
| Light activity | 12% [3/24] | 27% [7/26] |
| Symptoms prior to death, no./total no. (%)a | ||
| Syncope and/or seizures | 19% [5/26] | 7% [2/27] |
| Palpitations | 8% [2/26] | 11% [3/27] |
| None | 73% [19/26] | 81% [22/27] |
| Family history of cardiomyopathy, no./total no. (%)a | 7% [2/29] | 11% [3/28] |
aPlease note missing data for these parameters and that percentages are based on non-missing data
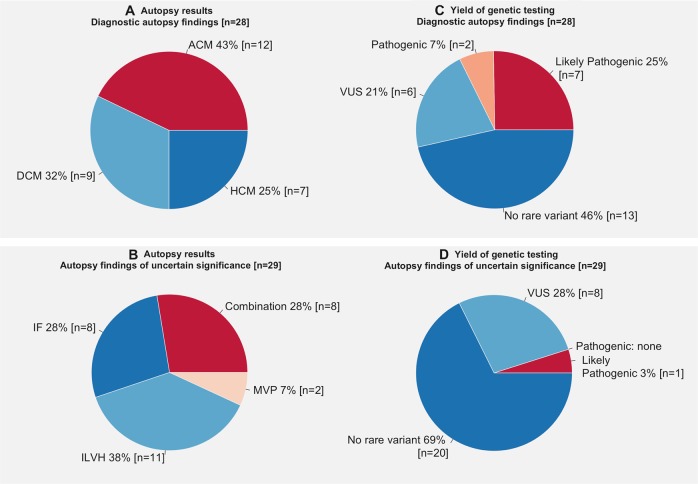
The main diagnoses established at autopsy were hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM) and arrhythmogenic cardiomyopathy (ACM) in 7, 9 and 12 cases, respectively (Fig. 1a). Autopsy findings of uncertain significance were identified in 29 cases (Fig. 1b), namely: idiopathic fibrosis (n = 8), idiopathic left ventricular hypertrophy (i.e. without disarray) (n = 11), or a combination thereof (n = 8) and isolated mitral valve prolapse without associated myocardial fibrosis and extensive myxomatous degeneration (n = 2).
Fig. 1.
Overview of structural finding during autopsy and yield of genetic testing. Figure summarising autopsy findings and yield of genetic testing in cases with autopsy findings diagnostic of cardiomyopathy (a, c) and cases with autopsy findings of uncertain significance (b, d). ACM arrhythmogenic cardiomyopathy, CAD coronary artery disease, DCM dilated cardiomyopathy, HCM hypertrophic cardiomyopathy, IF idiopathic fibrosis, ILVH idiopathic left ventricular hypertrophy, LP likely pathogenic, MVP mitral valve prolapse, P pathogenic
Genetic testing uncovered a total of 40 rare protein-altering variants (MAF < 1 in 10,000) in 23 different genes after variant filtering (Supplementary Table 3). In 33 patients (58%) no rare variant was identified: 20 cases with autopsy findings of uncertain significance and 13 cases diagnosed with cardiomyopathy. Manual curation against ACMG guidelines yielded two pathogenic and eight likely pathogenic variants in 10 cases in the overall cohort, with an overall yield of 18% (10 of 57 patients, Table 2). The majority of the yield was from cases diagnosed with cardiomyopathy (9 of 28 [32%], 95% confidence interval: 18–51% Fig. 1c) compared with non-specific findings (1 of 29 [3%], 95% confidence interval: 0.006–17%, p = 0.0054, Fig. 1d). The two pathogenic variants were in PKP2 and TMEM43, whereas the likely pathogenic variants were identified in MYBPC3 (n = 4), DSP (n = 2), TNNI3 (n = 1) and TTN (n = 1). In one patient who died at the age of 26 with ACM one likely pathogenic variant and one rare VUS were identified in DSP (Table 2, Supplementary Table 3 case P1). One case with non-specific findings (idiopathic fibrosis) hosted the likely pathogenic TTN variant suggesting that this may be an early presentation of DCM as suggested by recent data from Finland [13]. No pathogenic or likely pathogenic variants were identified in genes associated with primarily electrical disorders.
Table 2.
Overview of pathogenic and likely pathogenic variants in the sudden death cohort
| Case | Age at death (years) | Sex | Circumstances of death | Symptoms before death | Autopsy finding | Gene | Chr | Starta | End | Ref/Alt | MAFb dbSNP | cDNA changea | Amino acid changea | ACMG classification | ACMG criteriac |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 26 | F | Light activity | Seizures | ACM | DSP | 6 | 7580818 | 7580818 | T/G | None | c.4395T>G | p.(Y1465*) | Likely pathogenic | PVS1.PM2 |
| P2 | 28 | M | Light activity | None | ACM | DSP | 6 | 7581692 | 7581692 | C/T | None | c.5269C>T | p.(Q1757*) | Likely pathogenic | PVS1, PM2 |
| P3 | 20 | M | Exercise | None | ACM | TMEM43 | 3 | 14183165 | 14183165 | C/T | None | c.1073C>T | p.(S358L) | Pathogenic | PS3, PS4, PM2, PP3 |
| rs63750743 | |||||||||||||||
| P4 | 37 | M | Sleep | None | ACM | PKP2 | 12 | 32955433 | 32955438 | GGGTGT/G | 1.6 × 10−5 | c.2066_2070delACACC | p.(H689Pfs*8) | Pathogenic | PVS1, PS4 |
| P5 | 29 | M | Light activity | Seizures | DCM | TNNI3 | 19 | 55665462 | 55665462 | C/T | 4 × 10−5 | c.485G>A | p.(R162Q) | Likely pathogenic | PS4, PP5, PP3 |
| rs397516354 | |||||||||||||||
| P6 | 29 | M | Exercise | None | HCM | MYBPC3 | 11 | 47373030 | 47373030 | A/AC | None | c.51dupG | p.(S18Tfs*31) | Likely pathogenic | PVS1, PM2 |
| P7 | 19 | F | Sleep | Palpitations | HCM | MYBPC3 | 11 | 47359112 | 47359112 | T/C | None | c.2432A>G | p.(K811R) | Likely pathogenic | PM1, PM2, PP3, PP5 |
| rs375675796 | |||||||||||||||
| P8 | 19 | M | NA | None | HCM | MYBPC3 | 11 | 47372895 | 47372905 | CGTGTGCCCTCT/C | None/rs397515925 | c.177_187delAGAGGGCACAC | p.(E60Afs*49) | Likely pathogenic | PVS1 |
| P9 | 46 | F | Sleep | None | HCM | MYBPC3 | 11 | 47372960 | 47372960 | C/CG | None | c.121dupC | p.(R41Pfs*8) | Likely pathogenic | PVS1, PM2 |
| P10 | 40 | M | NA | NA | IF | TTN | 2 | 179440001 | 179440001 | T/A | None | c.70858A>T | p.(R23620*) | Likely pathogenic | PVS1, PM2 |
ACM arrhythmogenic cardiomyopathy, Alt alternate allele, cDNA coding DNA, Chr chromosome, DCM dilated cardiomyopathy, F female, HCM hypertrophic cardiomyopathy, IF idiopathic fibrosis, LP likely pathogenic, M male, MAF minor allele frequency, NA not available, P pathogenic, Ref reference allele
aGenomic position refers to GRCh37. The following transcripts were used: DSP: NM_004415.4; MYBPC3:NM_000256.3; PKP2:NM_001005242.2; TNNI3:NM_000363.4; TMEM43:NM_024334.2; TTN:NM_001267550.2
bMAF is based on total number of individuals in the Genome Aggregation Database (gnomAD)
cSee ACMG guidelines [12] for further information on classification criteria
Discussion
This study indicates that the yield of pathogenic and likely pathogenic variants following molecular autopsy in cases with cardiomyopathy (32%) is comparable with the yield in living cardiomyopathy patients [14] but substantially greater than in cases with non-specific findings (3%). However, the yield in cases with non-specific findings did not differ significantly from that in our SADS cases from the same original SCD cohorts (13%) (p = 0.2312) [5]. Across all groups we used a stringent MAF threshold of 1 in 10,000 and then applied ACMG variant classification guidelines [12]. Hertz et al. have previously studied cases with non-specific structural changes at autopsy [15] and reported a much higher prevalence (29%) of rare variants with likely functional effects. This had relied upon a higher MAF threshold (1 in 100) and computational approaches to classify variant pathogenicity which may have overestimated the yield. Interestingly, Papadakis et al. evaluated 41 families of SCD cases with non-specific findings at autopsy and diagnosed 45% with primary electrical diseases on clinical grounds alone without the availability of postmortem genetic testing [9]. This was similar to the results of clinical evaluation in families of SADS cases. The main clinical diagnosis was, however, the Brugada syndrome, where the diagnostic return of genetic testing is only ~20% [16]. It is therefore possible that by chance pathogenic or likely pathogenic variants for Brugada syndrome were not identified in the current cohort, regardless of whether Brugada syndrome may have been an important cause of death.
The most common uncertain findings in our study were idiopathic left ventricular hypertrophy (n = 11), idiopathic fibrosis of the myocardium (n = 8) or a combination thereof (n = 8). It remains unclear whether these uncertain findings during autopsy are innocent bystander or truly involved in sudden death. Both idiopathic left ventricular hypertrophy as well as myocardial fibrosis may present as a partial phenotypic expression of an underlying cardiomyopathy in association with sudden death. This has been suggested for myocardial fibrosis by a recent Finnish study, where the authors identified a higher yield than our study of pathogenic or likely pathogenic cardiomyopathy associated variants in 10% of sudden death cases in whom myocardial fibrosis was noted during autopsy (in genes associated to ACM [n = 2], HCM [n = 4] and DCM [n = 4]) [13]. In line with our findings, no variants were identified in genes associated to primary electrical disorders.
As we only screened our cohort for 77 genes, we cannot exclude the possibility of pathogenic variants in other genes that are associated to these subtle findings during autopsy and predispose to sudden death. Furthermore, a polygenic architecture with a combination of genetic variation, underlying these findings and sudden death cannot be excluded. In addition, rather than a genetic cause, sudden death could be the results of environmental factors (e.g. infection) with or without interaction with predisposing genetic factors.
Limitations
This study is limited by the small number of cases included (n = 57) and the lack of comprehensive clinical evaluation of relatives. The combined yield of genetic and clinical screening of relatives is therefore unknown. Although autopsies were performed at different centres by locally recognised experts using international guidelines, this may have resulted in non-uniformity of autopsy results. Unfortunately, quantitative criteria for autopsy diagnoses are still limited. Although included cases were either consecutive referrals or from population studies, we acknowledge that there may be an element of referral bias.
Conclusion
This study highlights the importance of applying stringent criteria to classify variants, resulting in a lower yield of actionable genetic results in cases with non-specific findings at autopsy. Currently, whilst molecular autopsy in these cases offers less clinical utility than in cardiomyopathy, its yield is not significantly less than in SADS cases based on these data. Larger studies coupled with family findings are necessary to elucidate the importance of monogenic diseases and the role for genetic testing in this group.
Supplementary information
Acknowledgements
This study was funded in part by Cardiac Risk in the Young (ERB, HR, MP, SS, MS, GM and SP); the British Heart Foundation (ERB, HR and SC) including BHF Clinical Research Training Fellowship FS/11/71/28918: Future diagnostic role and novel genetic loci in SADS; The Robert Lancaster Memorial Fund sponsored by McColl’s RG Ltd (ERB, YW and CM); NMRC Singapore (SC); Leducq Foundation (SC); NIHR Royal Brompton Cardiovascular BRU (support to JSW, RG, SAC and JAT); MRC UK (SC, JSW); Tanoto Foundation (SC); SingHealth/Duke-NUS Precision Medicine Institute (PRISM) (SC); Cure Kids (New Zealand) (JRS, JC and DRL); Wellcome Trust (JSW) and the Dutch Heart Foundation CVON-PREDICT project (CVON2012-10) (NL, EML, AAMW and CRB). All authors take responsibility for all aspects of the reliability and freedom from bias of the data presented and their discussed interpretation.
Members of the European Reference Network for rare and low prevalence diseases of the heart, Guard-Heart
Najim Lahrouchi, Elisabeth M. Lodder, Stathis Papatheodorou, Chris Miles, Michael Papadakis, Della Cole, Jacob Tfelt-Hansen, Christian van der Werf, Yanushi D. Wijeyeratne, Janice Till, Sanjay Sharma, Arthur A.M. Wilde, Mary N. Sheppard, Connie R. Bezzina, Elijah R. Behr.
Compliance with ethical standards
Conflict of interest
AAMW has undertaken consulting for Sorin in the past. ERB has received unrestricted research funds from Biotronik and St Jude Medical. The remaining authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mary N. Sheppard, Connie R. Bezzina, Elijah R. Behr
Change history
11/6/2019
The original article contained an incorrect symbol in Table 1. This has been corrected in the PDF and HTML.
Supplementary information
The online version of this article (10.1038/s41431-019-0500-8) contains supplementary material, which is available to authorised users.
References
- 1.Finocchiaro G, Papadakis M, Robertus J-L, Dhutia H, Steriotis AK, Tome M, et al. Etiology of Sudden Death in Sports. J Am Coll Cardiol. 2016;67:2108–15. doi: 10.1016/j.jacc.2016.02.062. [DOI] [PubMed] [Google Scholar]
- 2.Bagnall RD, Weintraub RG, Ingles J, Duflou J, Yeates L, Lam L, et al. A prospective study of sudden cardiac death among children and young adults. N Engl J Med. 2016;374:2441–52. doi: 10.1056/NEJMoa1510687. [DOI] [PubMed] [Google Scholar]
- 3.Landry CH, Allan KS, Connelly KA, Cunningham K, Morrison LJ, Dorian P. Sudden cardiac arrest during participation in competitive sports. N Engl J Med. 2017;377:1943–53. doi: 10.1056/NEJMoa1615710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahrouchi N, Behr ER, Bezzina CR. Next-generation sequencing in post-mortem genetic testing of young sudden cardiac death cases. Front Cardiovasc Med. 2016;3:13. doi: 10.3389/fcvm.2016.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lahrouchi N, Raju H, Lodder EM, Papatheodorou E, Ware JS, Papadakis M, et al. Utility of post-mortem genetic testing in cases of sudden arrhythmic death syndrome. J Am Coll Cardiol. 2017;69:2134–45. doi: 10.1016/j.jacc.2017.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skinner JR, Crawford J, Smith W, Aitken A, Heaven D, Evans C-A, et al. Prospective, population-based long QT molecular autopsy study of postmortem negative sudden death in 1–40 year olds. Heart Rhythm. 2011;8:412–9. doi: 10.1016/j.hrthm.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Gladding PA, Evans C-A, Crawford J, Chung SK, Vaughan A, Webster D, et al. Posthumous diagnosis of long QT syndrome from neonatal screening cards. Heart Rhythm. 2010;7:481–6. doi: 10.1016/j.hrthm.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 8.Basso C, Aguilera B, Banner J, Cohle S, d’Amati G, de Gouveia RH, et al. Guidelines for autopsy investigation of sudden cardiac death: 2017 update from the Association for European Cardiovascular Pathology. Virchows Arch. 2017;471:691–705. doi: 10.1007/s00428-017-2221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadakis M, Raju H, Behr ER, De Noronha SV, Spath N, Kouloubinis A, et al. Sudden cardiac death with autopsy findings of uncertain significance: potential for erroneous interpretation. Circ Arrhythmia Electro. 2013;6:588–96. doi: 10.1161/CIRCEP.113.000111. [DOI] [PubMed] [Google Scholar]
- 10.Pua CJ, Bhalshankar J, Miao K, Walsh R, John S, Lim SQ, et al. Development of a comprehensive sequencing assay for inherited cardiac condition genes. J Cardiovasc Transl Res. 2016;9:3–11. doi: 10.1007/s12265-016-9673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the association for molecular pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Junttila MJ, Holmström L, Pylkäs K, Mantere T, Kaikkonen K, Porvari K, et al. Primary myocardial fibrosis as an alternative phenotype pathway of inherited cardiac structural disorders. Circulation. 2018;137:2716–26. doi: 10.1161/CIRCULATIONAHA.117.032175. [DOI] [PubMed] [Google Scholar]
- 14.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8:1308–39. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Hertz CL, Christiansen SL, Ferrero-Miliani L, Dahl M, Weeke PE, Ottesen GL, et al. Next-generation sequencing of 100 candidate genes in young victims of suspected sudden cardiac death with structural abnormalities of the heart. Int J Leg Med. 2016;130:91–102. doi: 10.1007/s00414-015-1261-8. [DOI] [PubMed] [Google Scholar]
- 16.Wilde AAM, Behr ER. Genetic testing for inherited cardiac disease. Nat Rev Cardiol. 2013;10:571–83. doi: 10.1038/nrcardio.2013.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.