Abstract
Given the global outbreak of breast cancer and its debilitating effect on women's health, it is not surprising that tremendous efforts have been made with an aim of shedding more light on the mechanisms involved in the pathogenesis of this type of cancer. Among the long list of risk factors associated with this malignancy, recently, the role of microRNAs (miRNAs or miRs) has turned into a hotspot for breast cancer investigations. miRNAs approximately 20 nucleotides in length and are located in either an exon or an intron, playing a role in the regulation of gene expression. In the present study, we extracted RNA from both the serum and cancerous tissue of breast cancer patients and after synthesizing the cDNA, we performed quantitative PCR to determine the expression levels of miR-9 and miR-192. The resulting data revealed that while the mRNA expression level of miR-9 was significantly decreased in the breast cancer tissues, there was no noticeable change in the expression level of this miRNA in the serum samples. Likewise, we found that the marked downregulation of miR-192 was only restricted to the cancerous tissues, but was not found in the serum of patients. Based on the meaningful downregulation of the expression of miR-9 and miR-192, this study provides a plausible framework for these miRNAs as effective biomarkers for breast cancer patients.
Keywords: miR-9, miR-192, qPCR, breast cancer
Introduction
Breast cancer is a serious life-threatening disease, which with its different stages, ranging from benign to invasive malignant tumors, has a debilitating effect on the health and quality of life of women (1). Despite considerable progress being made in the early detection and diagnosis, the complex nature of breast cancer renders this malignancy an incurable disease due to the acquisition of the chemoresistant phenotype and the metastatic spread of cancer cells to distant organs (2). Of note, it has been demonstrated that women who are younger than 45 years are at a higher risk of developing breast cancer in modern countries (3,4). Marked improvements in the laboratory and etiological investigations have revolutionized the major understanding of the relevant mechanisms involved in the pathogenesis of breast cancer, providing new perspectives for the evolution of this type of cancer. Recent studies have suggested that there is a tight association between the incidence of breast cancer and numerous risk factors, such as sex, age, smoking, alcohol addiction, high-fat diet and salt consumption, infection diseases, pollution and radiation (5-8).
Among the extensive list of risk factors associated with breast cancer, the significant role of microRNAs (miRNAs or miRs), which are crucial small molecules responsible for regulation, has been noted (9). The pivotal role of miRNAs in several types of human cancer has been well-established in a mounting body of recent investigations. Although some miRNAs serve as oncogenes and activate cancer-related signaling pathways, others play the roles of tumor suppressors, preventing numerous biomolecules that are important for the formation of malignancies (10-13). It has been suggested that miR-9, an important miRNA which participates in cancer progression, can downregulate E-cadherin expression in breast cancer and thereby increase the risk of cancer metastasis in patients by reducing the ability of malignant cells to adhere to the cancer microenvironment (10-12). According to bioinformatics tools (miRWalk), there are several gene targets for miR-9, such as claudin 14, cyclin G1 and leptin (14). The importance of miRNAs in the regulation of cancer signaling pathways has attracted increasing attention to the mechanisms involved in the regulation of these small non-coding RNAs. It has been revealed that epigenetic alterations, such as DNA methylation, can alter the expression levels of miRNAs (15). In a study conducted by Botla et al, it was reported that the addition of a methyl group into the miR-192 promoter decreased the expression of this miRNA in cancer tissue, leading to the progression of pancreatic cancer (16). Bioanformatic tools (miRWalk) suggest that miR-192 is able to target several genes, for example, cyclin T2, epiregulin and plexin B2(14). Although multiple lines of evidence emphasize the significant role of miRNAs in different human cancers, to the best of our knowledge, there are no studies available to date on the role of miR-9 and miR-192 in breast cancer. In the present study, we aimed to examine the expression levels of these miRNAs in breast cancer tissues and serum.
Subjects and methods
Tissue samples
The present study was approved by the Shahid Beheshti University of Medical Sciences (IR.SBMU. MSP.REC.1397.505, grant no. 13756). To evaluate and compare the expression levels of miR-9 and miR-192 in serum and cancerous tissue, 38 breast cancer tissues and their adjacent normal tissues were collected from Taleghani Hospital between 2017 and 2019 (Table I). Informed consent was obtained from the patients prior to enrolment in this study. All tissues were stored in RNAlater (Qiagen GmbH) at -20˚C and all serum samples were stored in -70˚C. An expert pathologist confirmed the stage of breast cancer and all patients’ information was recorded. Of note, the samples of those patients who had been treated with chemotherapy were excluded from the study. The inclusion criteria in this study were no treatment with chemotherapy or radiotherapy.
Table I.
Nucleotide sequences of primers used for RT-qPCR.
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| U6 | GAGAAGATTAGCATGGCCCCT | ATATGGAACGCTTCACGAATTTGC |
| miR-9 | CTTTGGTTATCTAGCTGTATGAGTCGT | ATCCAGTGCAGGGTCCGA |
| miR-192 | CTGACCTATGAATTGACAGCCGT | ATCCAGTGCAGGGTCCGA |
RNA extraction and cDNA synthesis
To extract RNA from breast tissues and serum, all samples were digested with RNX-plus solution (Cinnagen). Following the addition of chloroform, propanol was used to precipitate the RNA samples. The purity and quality of the extracted RNA were confirmed by electrophoresis and using a Nanodrop spectrophotometer (Eppendorf). Subsequently, 10 µl of RNA from each sample were added to the reaction containing 0.5 µl primer of RT miRNA-9, 0.5 µl primer of RT miRNA-192, 0.5 µl of the U6 reverse primer and 9 µl of reverse transcriptase (BioFACT) to synthesize the cDNA. The PCR (Bio Intellectica PCR) schedule was as follows: 5 min at 95˚C and 40 min at 50˚C. In order to ensure a suitable concentration, the cDNA was diluted twice sterile water.
Quantitative PCR (qPCR)
To evaluate the expression level of miR-9 and miR-192 in the breast cancer tissue and serum, qPCR was performed using Rotor-Gene 6000 (Corbett Life Science) in 36-well Gene Discs, using a final volume of 20 µl. We then combined 10 µl of BIOFACT™ 2X real-time PCR master mix (for SYBR-Green I; BioFACT), 1 µl (10 pmol) of forward primer, 1 µl of (10 pmol) reverse primer, 2 µl of 1/2 diluted cDNA and 6 μl of sterile water to evaluate the expression of miR-9 and miR-192. All reactions were performed in triplicate, simultaneously, to confirm the results. The sequences of the primers used in this study are presented in Table I. The thermal cycling conditions were 95˚C for 10 min followed by 40 cycles of 95˚C for 20 sec, 55˚C for 20 sec and 72˚C for 20 sec. The values for the relative quantification were calculated using the 2-ΔΔCq expression formula (17).
Statistical analysis
To analysis the results of miRNA expression, Graph-PadPrism software (version 5.1) was used. The experimental data are expressed as the means ± standard deviation of 3 independent assays. The unpaired, two-tailed Student's t-test was used to analyze the statistical differences between groups using Graph-Pad Prism software. P<0.05 was considered to indicate a statistically significant difference.
Results
Characteristics of patients with breast cancer
In the present study, 38 breast cancer tissues and their adjacent normal tissues coupled with serum samples were collected from breast cancer patients hospitalized at Taleghani Hospital. Following the extraction of RNA from both tissue and serum samples, the expression levels of miR-9 and miR-192 were examined by RT-qPCR analysis. It is worth noting that 52% of the patients presented with stage III-IV of the disease. The characteristics of each patient, including their tumor size, tumor location, family history, and the genetic status of the tumors, are summarized in Table II.
Table II.
Clinical characteristics of the 38 patients with breast cancer.
| Characteristics | Percentage of patients |
|---|---|
| Age of >60 years | 69% |
| Tumor localization | Left, 45%; right, 55% |
| Family history of breast cancer | Absent, 88.1%; present, 11.9% |
| Lymph node metastasis | Negative, 48%; positive, 52% |
| Tumor size | >2 cm, 62.8%; <2 cm, 37.2% |
| Tumor stage | I-II, 48%; III-IV, 52% |
| Estrogen receptor (ER) status | Negative, 54.3%; positive, 45.7% |
| Progesterone receptor (PR) status | Negative, 46.6%; positive, 53.4% |
Expression level of miR-9 in breast cancer tissues and serum samples
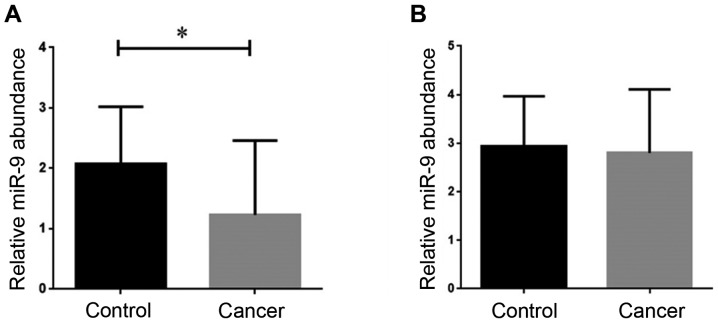
To evaluate the expression level of miR-9 in both breast cancer tissues and serum, we performed RT-qPCR. As presented in Fig. 1A, the results of RT-qPCR revealed in the breast cancer tissue samples, the expression of miR-9 was significantly downregulated compared with the normal tissue samples (P<0.05). However, the results regarding miRNA-9 expression in serum from the breast cancer patients revealed that while the expression level of this miRNA was slightly decreased in the breast cancer samples, this alteration in gene expression was not considered meaningful according to the statistical analysis (Fig. 1B).
Figure 1.
Evaluation of the expression level of miR-9 in the tissue and serum of both breast cancer patients and healthy counterparts. (A) The expression level of miR-9 was significantly decreased in the breast cancer tissues (*P<0.05) in comparison with normal tissues. (B) There was no significant difference in the expression level of miR-9 in the serum samples collected from either breast cancer patients or the healthy volunteers. Values are presented as the means ± SD of 3 independent experiments.
Evaluation of the expression level of miR-192 in breast cancer tissues and serum samples
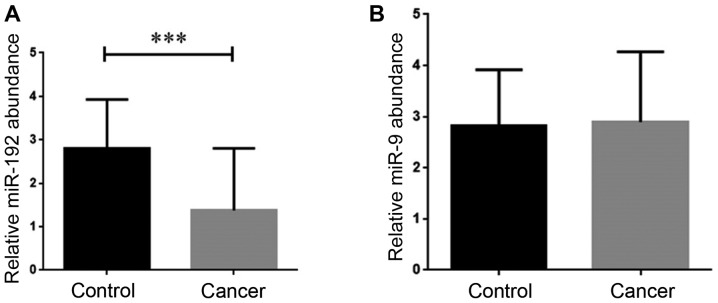
Given the critical participation of miR-192 in the progression of different types of human cancer, it was of particular interest to evaluate whether there is an alteration in the expression of this miRNA in the breast cancer patients tissues and serum samples. We found that the mRNA expression level of miR-192 was significantly decreased in the breast cancer tissues (P<0.001; Fig. 2A). After analyzing the expression of miRNA-192 in the serum from the breast cancer patients, we failed to find any noticeable changes in its expression in the cancerous serum samples in comparison with the healthy counterparts (Fig. 2B).
Figure 2.
Investigation of the expression level of miR-192 in the tissue and serum of breast cancer patients and healthy volunteers. (A) There was a significant decrease in the level of miR-192 in cancerous tissues (***P<0.001). (B) The expression level of this miRNA remained unaltered in the serum samples. Values are presented as the means ± SD of 3 independent experiments.
Discussion
Since miRNAs are strongly considered to be responsible for the regulation of the main mechanisms of in cancer pathogenesis, exploiting this small non-coding RNAs has been currently under intense investigation for future cancer management strategies. By bifurcating at several signaling pathways, miRNAs have a noticeable impact on the diverse intracellular process, and it has been suggested that epigenetic alterations, such as methylation are the main mechanisms through which miRNA expression is regulated (16,18). Among the panoply of miRNAs involved in tumor progression, metastasis and drug resistance, miR-9 and miR-192 are the most important ones.
In the present study, we compared the expression levels of miR-9 and miR-192 in both tissue and serum samples collected from breast cancer patients and healthy counterparts. In this study, there were some limitations of collecting serum and tissue samples. The expression levels of miRNA-9 and -192 were assessed by RT-qPCR method; however, northern blotting could have been performed to validate the results. The results of this study revealed that while the expression level of miR-9 was significantly diminished in the cancerous tissue, there was no distinct difference between the transcriptional activity of this miRNA in the serum of breast cancer patients and the healthy individuals. In accordance with this finding, Orangi and Motovali-Bashi claimed that the downregulation in the expression level of miR-9 in breast cancer patients was only restricted to the cancerous tissues and not to the patient plasma (19). Moreover, in another study, it was demonstrated that the miR-9 expression pattern was dependent on the stage of breast cancer. In spite of the fact that the expression of miR-9 was decreased in benign breast tumors, its expression level displayed an elevated profile in more advanced stages of the disease. In addition, Hasanzadeh et al demonstrated that in benign and malignant breast tumors, there was a downregulation compared to healthy breast tissues (20). This association between the expression of miR-9 and cancer stage has also been reported in gastric cancer, at least partially, due to the varied hypermethylation status in different phases of the malignancy (21). The reduction in the expression level of miR-9 has also been reported in other types of human cancer, including gastric and renal cell carcinoma (21,22). In the study conducted by Cheng et al, it was demonstrated that miR-9 and MicroRNA-221enhance the generation of breast cancer stem cells (23). Ma et al demonstrated that miRNA-9 was able to downregulate E-cadherin; therefore, metastasis in breast tumors occurred (24). However, another study delineated that the up-regulation of miR-9 provided an opportunity for breast cancer cells to invade to distant organs through the down-regulation of FOXO1(25). In line with these oncogenic properties of miR-9, several investigations have suggested that the expression of miR-9 can be used as a prognostic biomarker in triple-negative breast cancer (TNBC) and estrogen receptor (ER)-positive breast cancer (26,27).
Another miRNA, whose participation in different types of human tumors has been well-established in numerous studies is miR-192 (28-30). Notably, in the present study, we found that the expression of miR-192 was significantly decreased only in the breast cancer tissues, but not in the serum. Consistently, Hu et al introduced miR-192 as a tumor suppressor miRNA which, coupled with bone morphogenetic protein-6 (BMP-6), were downregulated in breast cancer, proposing these two molecules as novel therapeutic targets for breast cancer treatment (31). Likewise, the results of another study demonstrated that both the expression of miRNA-192-3p and miRNA-192-5p were decreased in stage IIIB colon cancer as compared to healthy tissues (32). The reduction in the expression level of this tumor suppressor miRNA has been reported in osteosarcoma, lung cancer and pancreatic cancer, where it has been reported that miR-192 exhibits a tight association with the regulation of cell proliferation and apoptosis (33-34). Moreover, it has been reported that the downregulation of miR-192 in hepatocellular carcinoma (HCC) may provide a signal that upregulates SLC39A6/SNAIL, a molecule involved in cell invasion, which in turn deteriorates patient outcome (35).
In conclusion, the findings of the present study suggested that the expression of both miR-9 and miR-192 was downregulated in breast cancer tissues, suggesting that these miRNAs could serve as effective biomarkers for the diagnosis of breast cancer.
Acknowledgements
The present study was conducted at the School of Medicine, Shahid Beheshti University of Medical Sciences.
Funding
The present article was financially supported by the Research Department of the School of Medicine Shahid Beheshti University of Medical Sciences. (IR. SBMU. MSP.REC.1397.505, grant no. 13756).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
EF and ST designed and performed the cell culture and molecular experiments. HG and FT performed the statistical analyses. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
This study was approved by the Shahid Beheshti University of Medical Sciences (IR.SBMU. MSP.REC.1397.505, grant no. 13756). Informed consent was obtained from the patients prior to enrolment in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Davies B, Miles DW, Happerfield LC, Naylor MS, Bobrow LG, Rubens RD, Balkwill FR. Activity of type IV collagenases in benign and malignant breast disease. Br J Cancer. 1993;67:1126–1131. doi: 10.1038/bjc.1993.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greaney ML, Sprunck-Harrild K, Ruddy KJ, Ligibel J, Barry WT, Baker E, Meyer M, Emmons KM, Partridge AH. Study protocol for Young & Strong: A cluster randomized design to increase attention to unique issues faced by young women with newly diagnosed breast cancer. BMC Public Health. 2015;15(37) doi: 10.1186/s12889-015-1346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villarreal-Garza C, Aguila C, Magallanes-Hoyos MC, Mohar A, Bargalló E, Meneses A, Cazap E, Gomez H, López-Carrillo L, et al. Breast cancer in young women in Latin America: An unmet, growing burden. Oncologist. 2013;18:26–34. doi: 10.1634/theoncologist.18-S2-26. [DOI] [PubMed] [Google Scholar]
- 4.Knaul F, Bustreo F, Ha E, Langer A. Breast cancer: why link early detection to reproductive health interventions in developing countries? Salud Publica Mex. 2009;51:220–227. doi: 10.1590/s0036-36342009000800012. [DOI] [PubMed] [Google Scholar]
- 5.Faghihloo E, Saremi MR, Mahabadi M, Akbari H, Saberfar E. Prevalence and characteristics of Epstein-Barr virus-associated gastric cancer in Iran. Arch Iran Med. 2014;17:767–770. [PubMed] [Google Scholar]
- 6.Mirzaei H, Goudarzi H, Eslami G, Faghihloo E. Role of viruses in gastrointestinal cancer. J Cell Physiol. 2018;233:4000–4014. doi: 10.1002/jcp.26194. [DOI] [PubMed] [Google Scholar]
- 7.Pormohammad A, Azimi T, Falah F, Faghihloo E. Relationship of human herpes virus 6 and multiple sclerosis: A systematic review and meta-analysis. J Cell Physiol. 2018;233:2850–2862. doi: 10.1002/jcp.26000. [DOI] [PubMed] [Google Scholar]
- 8.Vaezjalali M, Rashidpour S, Rezaee H, Hajibeigi B, Zeidi M, Gachkar L, Aghamohamad S, Najafi R, Goudarzi H. Hepatitis B viral DNA among HBs antigen negative healthy blood donors. Hepat Mon. 2013;13(e6590) doi: 10.5812/hepatmon.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai EC. microRNAs: Runts of the genome assert themselves. Curr Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 11.Tavakolian S, Goudarzi H, Eslami G, Faghihloo E. Transcriptional regulation of epithelial to mesenchymal transition related genes by lipopolysaccharide in human cervical cancer cell line HeLa. Asian Pac J Cancer Prev. 2019;20:2455–2461. doi: 10.31557/APJCP.2019.20.8.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 13.Pourbagheri-Sigaroodi A, Bashash D, Safaroghli-Azar A, Farshi-Paraasghari M, Momeny M, Mansoor FN, Ghaffari SH. Contributory role of microRNAs in anti-cancer effects of small molecule inhibitor of telomerase (BIBR1532) on acute promyelocytic leukemia cell line. Eur J Pharmacol. 2019;846:49–62. doi: 10.1016/j.ejphar.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 14.Dweep H, Gretz N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(697) doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 15.Ortiz IMDP, Barros-Filho MC, Dos Reis MB, Beltrami CM, Marchi FA, Kuasne H, do Canto LM, de Mello JBH, Abildgaard C, Pinto CAL, et al. Loss of DNA methylation is related to increased expression of miR-21 and miR-146b in papillary thyroid carcinoma. Clin Epigenetics. 2018;10(144) doi: 10.1186/s13148-018-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botla SK, Savant S, Jandaghi P, Bauer AS, Mücke O, Moskalev EA, Neoptolemos JP, Costello E, Greenhalf W, Scarpa A, et al. Early epigenetic downregulation of microRNA-192 expression promotes pancreatic cancer progression. Cancer Res. 2016;76:4149–4159. doi: 10.1158/0008-5472.CAN-15-0390. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Faghihloo E, Sadeghizadeh M, Shahmahmoodi S, Mokhtari-Azad T. Cdc6 expression is induced by HPV16 E6 and E7 oncogenes and represses E-cadherin expression. Cancer Gene Ther. 2016 doi: 10.1038/cgt.2016.51. Nov 11. doi.org/10.1038/cgt.2016.51. [DOI] [PubMed] [Google Scholar]
- 19.Orangi E, Motovali-Bashi M. Evaluation of miRNA-9 and miRNA-34a as potential biomarkers for diagnosis of breast cancer in Iranian women. Gene. 2019;687:272–279. doi: 10.1016/j.gene.2018.11.036. [DOI] [PubMed] [Google Scholar]
- 20.Hasanzadeh A, Mesrian Tanha H, Ghaedi K, Madani M. Aberrant expression of miR-9 in benign and malignant breast tumors. Mol Cell Probes. 2016;30:279–284. doi: 10.1016/j.mcp.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Luo H, Zhang H, Zhang Z, Zhang X, Ning B, Guo J, Nie N, Liu B, Wu X. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res. 2009;28(82) doi: 10.1186/1756-9966-28-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, Wood CG, Wu X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29:5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- 23.Cheng CW, Yu JC, Hsieh YH, Liao WL, Shieh JC, Yao CC, Lee HJ, Chen PM, Wu PE, Shen CY. Increased cellular levels of microRNA-9 and microRNA-221 correlate with cancer stemness and predict poor outcome in human breast cancer. Cell Physiol Biochem. 2018;48:2205–2218. doi: 10.1159/000492561. [DOI] [PubMed] [Google Scholar]
- 24.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu DZ, Chang B, Li XD, Zhang QH, Zou YH. MicroRNA-9 promotes the proliferation, migration, and invasion of breast cancer cells via down-regulating FOXO1. Clin Transl Oncol. 2017;19:1133–1140. doi: 10.1007/s12094-017-1650-1. [DOI] [PubMed] [Google Scholar]
- 26.Jang MH, Kim HJ, Gwak JM, Chung YR, Park SY. Prognostic value of microRNA-9 and microRNA-155 expression in triple-negative breast cancer. Hum Pathol. 2017;68:69–78. doi: 10.1016/j.humpath.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X, Marian C, Makambi KH, Kosti O, Kallakury BV, Loffredo CA, Zheng YL. MicroRNA-9 as potential biomarker for breast cancer local recurrence and tumor estrogen receptor status. PLoS One. 2012;7(e39011) doi: 10.1371/journal.pone.0039011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filipska M, Skrzypski M, Czetyrbok K, Stokowy T, Stasiłojć G, Supernat A, Jassem J, Żaczek AJ, Bigda J. MiR-192 and miR-662 enhance chemoresistance and invasiveness of squamous cell lung carcinoma. Lung Cancer. 2018;118:111–118. doi: 10.1016/j.lungcan.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Chen ZJ, Yan YJ, Shen H, Zhou JJ, Yang GH, Liao YX, Zeng JM, Yang T. miR-192 is overexpressed and promotes cell proliferation in prostate cancer. Med Princ Pract. 2019;28:124–132. doi: 10.1159/000496206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Jia LS, Yuan W, Wu Z, Wang HB, Xu T, Sun JC, Cheng KF, Shi JG. Low miR-34a and miR-192 are associated with unfavorable prognosis in patients suffering from osteosarcoma. Am J Transl Res. 2015;7:111–119. [PMC free article] [PubMed] [Google Scholar]
- 31.Hu F, Meng X, Tong Q, Liang L, Xiang R, Zhu T, Yang S. BMP-6 inhibits cell proliferation by targeting microRNA-192 in breast cancer. Biochim Biophys Acta. 2013;1832:2379–2390. doi: 10.1016/j.bbadis.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Li P, Ou Q, Braciak TA, Chen G, Oduncu FS. MicroRNA-192-5p is a predictive biomarker of survival for Stage IIIB colon cancer patients. Jpn J Clin Oncol. 2018;48:619–624. doi: 10.1093/jjco/hyy019. [DOI] [PubMed] [Google Scholar]
- 33.Shang G, Mi Y, Mei Y, Wang G, Wang Y, Li X, Wang Y, Li Y, Zhao G. MicroRNA-192 inhibits the proliferation, migration and invasion of osteosarcoma cells and promotes apoptosis by targeting matrix metalloproteinase-11. Oncol Lett. 2018;15:7265–7272. doi: 10.3892/ol.2018.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng S, Cong S, Zhang X, Bao X, Wang W, Li H, Wang Z, Wang G, Xu J, Du B, et al. MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells. Nucleic Acids Res. 2011;39:6669–6678. doi: 10.1093/nar/gkr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lian J, Jing Y, Dong Q, Huan L, Chen D, Bao C, Wang Q, Zhao F, Li J, Yao M, et al. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget. 2016;7:2672–2683. doi: 10.18632/oncotarget.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.