Sir,
We read with great interest the recently published article by Lee and colleagues reporting variants in ARSA and their association with Parkinson’s disease (Lee et al., 2019). Deficiency of arylsulfatase A is a known cause of metachromatic leukodystrophy (MLD), an autosomal recessive lysosomal storage disease. The study describes a patient with MLD and a family history of Parkinson’s disease. The patient was a compound heterozygous carrier of two rare missense ARSA mutations, p.L300S (c.899T>C, rs199476389) and p.C174Y (c.521G>A, rs199476381). Screening of ARSA in two family members with Parkinson’s disease and two unaffected members found that the p.L300S mutation segregated with Parkinson’s disease, but not the p.C174Y mutation. Next, a candidate gene analysis of ARSA was conducted in 92 familial and 92 sporadic Parkinson’s disease patients, and the results were compared to the allele frequencies within the Integrative Japanese Genome Variation Database. This screening identified a common missense variant, p.N352S (c.1055A>G, rs2071421), that was more frequent in healthy Japanese individuals than in familial and sporadic Parkinson’s disease cases (P = 0.026 and P = 0.0349, respectively). The authors concluded that the p.N352S variant may be protective against the development of Parkinson’s disease. They also found that ARSA deficiency increases α-synuclein aggregation and secretion, suggesting a potential link between ARSA mutations and α-synuclein pathology.
α-Synucleinopathies are a heterogeneous group of neurodegenerative disorders characterized by fibrillar aggregates of insoluble α-synuclein protein in the cytoplasm of specific neurons and glial cells. These disorders include Parkinson’s disease, Lewy body dementia (LBD), multiple system atrophy (MSA), and REM-sleep behaviour disorder (RBD), a prodromal α-synucleinopathy (Goedert et al., 2017; Postuma et al., 2019). Advances in genetics have implicated lysosomal dysfunction in the pathogenesis of several α-synucleinopathies. For example, variants within the lysosomal genes GBA (Sidransky et al., 2009) and SMPD1 (Alcalay et al., 2019) have been associated with an increased risk of Parkinson’s disease. Accumulation of α-synuclein has been observed in some lysosomal storage disorders suggesting a pathobiological link between these two disease groups (Shachar et al., 2011). Similarly, GBA variants have been associated with LBD (Nalls et al., 2013; Geiger et al., 2016), MSA (Mitsui et al., 2015; Sklerov et al., 2017) and RBD (Gan-Or et al., 2015). These intriguing observations prompted us to investigate ARSA variants in cohorts of α-synucleinopathies.
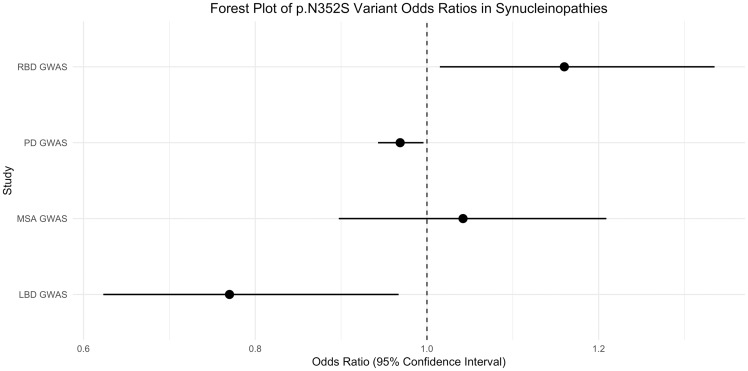
First, we sought to examine the association between the ARSA p.N325S variant and α-synucleinopathies by analysing genome-wide association study (GWAS) data from cohorts of Parkinson’s disease cases and proxy cases (n = 56 306 cases, n = 1 417 791 controls), LBD (n = 556 cases, n = 1418 controls), MSA (n = 896 cases, n = 3881 controls) and RBD (n = 1046 cases, recruited as isolated, polysomnography-confirmed RBD before conversion to α-synucleinopathy, n = 11 961 controls). All participants were of European ancestry and underwent similar genotyping, and standardized quality control procedures are described in detail elsewhere (Sailer et al., 2016; Nalls et al., 2019). The common p.N352S variant was reliably imputed in all cohorts (R2 > 0.9; allele frequency distributions are shown in Table 1). In the Parkinson’s disease cohort, the allele frequencies of the p.N325S variant were very similar in patients (0.1334) and controls (0.1354). In other α-synucleinopathy cohorts, the direction of effect was not consistent (Table 1). After correction for multiple testing, our analyses found no significant association of the p.N352S ARSA variant with α-synucleinopathies.
Table 1.
Analysis of the p.N352S ARSA variant in α-synucleinopathies
| Cohort | Cases, n | Controls, n | Frequency (affected) | Frequency (unaffected) | P-valuea | OR (CI) |
|---|---|---|---|---|---|---|
| LBD GWASb | 556 | 1418 | 0.1061 | 0.1326 | 0.024 | 0.77 (0.623–0.967) |
| MSA GWASb | 896 | 3881 | 0.1384 | 0.1336 | 0.592 | 1.042 (0.897–1.209) |
| RBD GWASb | 1046 | 11 961 | 0.1456 | 0.1292 | 0.030 | 1.16 (1.015–1.335) |
| PD GWASb | 56 306c | 1 417 791 | 0.1334d | 0.1354d | 0.022 | 0.969 (0.943–0.996) |
CI = confidence interval; GWAS = genome-wide association study; LBD = Lewy body dementia; MSA = multiple system atrophy; OR = odds ratio; PD = Parkinson’s disease; RBD = REM sleep behaviour disorder.
Uncorrected P-value, all results were not significant after correction for multiple comparisons.
p.N352S was found in imputed genotyping files with an R2 value of 0.975 for dementia with Lewy bodies GWAS, 0.953 for multiple system atrophy GWAS, 0.997 for RBD GWAS, and >0.96 for Parkinson’s disease GWAS.
Including proxy cases.
Frequency estimates were based on a subset of the data including 21 478 cases and 24 388 controls.
Next, we aimed to examine whether rare, potentially pathogenic variants in ARSA are associated with α-synucleinopathies. For this purpose, we performed burden analysis of these ARSA variants (annotated as stop-gain, frameshift, or marked as ‘pathogenic’ by ClinVar) in European-ancestry exome datasets from 1311 Parkinson’s disease patients and 571 matched control subjects, demonstrating lack of association, with higher frequency of potentially pathogenic variants in controls (frequency in patients/controls = 0.0015/0.004, P = 0.226). We further performed burden analysis in 264 definite MSA patients and 462 neuropathologically healthy control subjects (Pihlstrom et al., 2018) (including non-synonymous variants only, no frameshift, stop-gain or ClinVar ‘pathogenic’ variants were identified in this cohort), and here too, no association was found (frequency in patients/controls = 0.0076/0.0043, P = 0.517). The p.L300S variant was not observed in any of these datasets.
We are concerned about several conclusions regarding the p.N352S variant and the role of pathogenic ARSA variants that have been drawn in the Lee et al. (2019) article. First, as p.N352S is a common polymorphism (Table 1), a GWAS would be able to determine with certainty if this locus is significant on a genome-wide level, and this is not seen in well-powered cohorts, including a Japanese Parkinson’s disease GWAS of 2011 patients and 18 381 controls, which did not identify an association in this locus (Satake et al., 2009). Second, the hypothesis arguing that p.N352S is protective in autosomal dominant Parkinson’s disease would ideally be investigated by assessing penetrance or age at onset in carriers of known autosomal dominant variants. Third, regarding p.N352S being a coding and reportedly functional variant, population-specific effects are unlikely. The variant shows particularly variable frequencies across populations in gnomAD, with frequencies between 0.06 and 0.33 in different populations (0.1243 and 0.1733 in European and East Asian populations, respectively, https://gnomad.broadinstitute.org/). Given the high frequency of the variant, it is unlikely that it has a large effect size. The authors nominated their protective variant based on a comparison between a small cohort of sporadic Parkinson’s disease patients (n = 92) and a Japanese database, seemingly without adjustment for covariates, such as age, sex, or ancestry. In addition, the same healthy individuals were used for comparing both the familial and sporadic Parkinson’s disease cohorts. Consequently, bias within this small control cohort may have affected the results. Lastly, our burden analyses, as well as a previous burden analysis (Robak et al., 2017), did not identify an association between rare, potentially pathogenic ARSA variants and α-synucleinopathies.
Figure 1.
Forest plot of the p.N352S variant odds ratios and 95% confidence intervals in α-synucleinopathies..
In conclusion, our analyses do not support a significant association between common and rare ARSA variants and α-synucleinopathies despite adequate power. Our cohorts were of European ancestry and it is possible that with a larger Asian cohort, the reported association of the ARSA p.N352S variant with Parkinson’s disease would be lost and mimic our findings. However, our results do not completely rule out a potential role for ARSA in Parkinson’s disease, and additional large-scale familial and case-control studies are necessary to determine whether ARSA is associated with α-synucleinopathies.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Acknowledgements
We would like to thank all of the subjects who donated their time and biological samples to be part of this study. We thank the members of the International RBD Study Group. We would like to thank the NIH NeuroBioBank for contributing tissue samples. We would also like to thank all members of the International Parkinson Disease Genomics Consortium (IPDGC). For a complete overview of members, acknowledgements and funding, please see http://pdgenetics.org/partners.
Funding
This research was supported in part by the Intramural Research Program of the NIH National Institute of Neurological Disorders and Stroke and the National Institute on Aging (project numbers: ZIA-NS003154, Z01-AG000949). The MSA study was supported by The Multiple System Atrophy Trust, The MSA Coalition, The Wellcome Trust (the Synaptopathies strategic award (104033) and The Medical Research Council (MRC UK MR/J004758/1, G0802760, G1001253). The RBD study was funded by grants from the Michael J. Fox Foundation, the Canadian Consortium on Neurodegeneration in Aging (CCNA), the Canada First Research Excellence Fund (CFREF), awarded to McGill University for the Healthy Brains for Healthy Lives (HBHL) program. Z.G-O. is supported by the Fonds de recherche du Québec - Santé (FRQS) and Parkinson Quebec Chercheurs-boursiers award, and by the Young Investigator Award by Parkinson Canada.
Competing interests
The authors report no competing interests.
References
- Alcalay RN, Mallett V, Vanderperre B, Tavassoly O, Dauvilliers Y, Wu RYJ, et al. SMPD1 mutations, activity, and alpha-synuclein accumulation in Parkinson's disease. Mov Disord 2019; 34: 526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan-Or Z, Mirelman A, Postuma RB, Arnulf I, Bar-Shira A, Dauvilliers Y, et al. GBA mutations are associated with rapid eye movement sleep behavior disorder. Ann Clin Transl Neurol 2015; 2: 941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger JT, Ding J, Crain B, Pletnikova O, Letson C, Dawson TM, et al. Next-generation sequencing reveals substantial genetic contribution to dementia with Lewy bodies. Neurobiol Dis 2016; 94: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Jakes R, Spillantini MG. The synucleinopathies: twenty years on. J Parkinsons Dis 2017; 7: S51–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kanai K, Suzuki M, Kim WS, Yoo HS, Fu Y, et al. Arylsulfatase A, a genetic modifier of Parkinson's disease, is an alpha-synuclein chaperone. Brain 2019; 142: 2845–59. [DOI] [PubMed] [Google Scholar]
- Mitsui J, Matsukawa T, Sasaki H, Yabe I, Matsushima M, Durr A, et al. Variants associated with Gaucher disease in multiple system atrophy. Ann Clin Transl Neurol 2015; 2: 417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Duran R, Lopez G, Kurzawa-Akanbi M, McKeith IG, Chinnery PF, et al. A multicenter study of glucocerebrosidase mutations in dementia with Lewy bodies. JAMA Neurol 2013; 70: 727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Expanding Parkinson’s disease genetics: novel risk loci, genomic context, causal insights and heritable risk. bioRxiv 2019, 388165. doi: 10.1101/388165. [Google Scholar]
- Pihlstrom L, Schottlaender L, Chelban V, Consortium MSAE, Meissner WG, Federoff M, et al. Lysosomal storage disorder gene variants in multiple system atrophy. Brain 2018; 141: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postuma RB, Iranzo A, Hu M, Hogl B, Boeve BF, Manni R, et al. Risk and predictors of dementia and parkinsonism in idiopathic REM sleep behaviour disorder: a multicentre study. Brain 2019; 142: 744–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robak LA, Jansen IE, van Rooij J, Uitterlinden AG, Kraaij R, Jankovic J, et al. Excessive burden of lysosomal storage disorder gene variants in Parkinson's disease. Brain 2017; 140: 3191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer A, Scholz SW, Nalls MA, Schulte C, Federoff M, Price TR, et al. A genome-wide association study in multiple system atrophy. Neurology 2016; 87: 1591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet 2009; 41: 1303–7. [DOI] [PubMed] [Google Scholar]
- Shachar T, Lo Bianco C, Recchia A, Wiessner C, Raas-Rothschild A, Futerman AH. Lysosomal storage disorders and Parkinson's disease: Gaucher disease and beyond. Mov Disord 2011; 26: 1593–604. [DOI] [PubMed] [Google Scholar]
- Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med 2009; 361: 1651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklerov M, Kang UJ, Liong C, Clark L, Marder K, Pauciulo M, et al. Frequency of GBA variants in autopsy-proven multiple system atrophy. Mov Disord Clin Pract 2017; 4: 574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.