Inflammatory activation of microglia in the brains of prematurely born infants can lead to permanent neurological sequelae. Van Steenwinckel et al. show that a reduction in microglial Wnt signalling is necessary and sufficient to drive a microglial phenotype causing hypomyelination, and establish the Wnt pathway as a viable therapeutic target.
Keywords: neuroprotection, 3DNA, neuroinflammation, neonatal encephalopathy, innate immunity
Abstract
Microglia of the developing brain have unique functional properties but how their activation states are regulated is poorly understood. Inflammatory activation of microglia in the still-developing brain of preterm-born infants is associated with permanent neurological sequelae in 9 million infants every year. Investigating the regulators of microglial activation in the developing brain across models of neuroinflammation-mediated injury (mouse, zebrafish) and primary human and mouse microglia we found using analysis of genes and proteins that a reduction in Wnt/β-catenin signalling is necessary and sufficient to drive a microglial phenotype causing hypomyelination. We validated in a cohort of preterm-born infants that genomic variation in the Wnt pathway is associated with the levels of connectivity found in their brains. Using a Wnt agonist delivered by a blood–brain barrier penetrant microglia-specific targeting nanocarrier we prevented in our animal model the pro-inflammatory microglial activation, white matter injury and behavioural deficits. Collectively, these data validate that the Wnt pathway regulates microglial activation, is critical in the evolution of an important form of human brain injury and is a viable therapeutic target.
Introduction
Neuroinflammation is a key pathological mechanism in almost all acute and degenerative diseases in both the adult and in the developing brain (Perry et al., 2010; Hagberg et al., 2012; Hickman et al., 2018). Microglia and infiltrating macrophages (MG/Mϕ) are major contributors to neuroinflammation (Perry et al., 2010). These cells are capable of acquiring numerous complex functional states dependent on the specific nature of an insult or injury, including cytotoxic responses, immune regulation, or injury resolution (Michell-Robinson et al., 2015; Wolf et al., 2017). Limiting the cytotoxic activation of MG/Mϕ while promoting injury resolution represents a rational neuroprotective strategy (Rivest, 2009; Miron et al., 2013) that requires an in-depth understanding of the molecular mechanisms controlling their phenotypes across forms of injury. Understanding of these regulatory mechanisms is increasing at an extraordinary rate for adult disorders. However, microglia do not reach maturity until around the time of birth in humans, or the equivalent developmental time point of approximately postnatal Day 14 in rodents (Butovsky et al., 2014; Bennett et al., 2016; Miyamoto et al., 2016; Krasemann et al., 2017). These microglia, termed pre-microglia (Matcovitch-Natan et al., 2016), have specialized functions in brain development (Thion et al., 2018), but there is a striking paucity of knowledge on the regulators of the microglia phenotype that is relevant to injury/insult to the developing brain.
Preterm birth is the commonest cause of death and disability in children under 5 years of age, exceeding deaths from malaria or pneumonia (Lim et al., 2012). Preterm birth, birth before 37 of 40 weeks of gestation, occurs in 15 million infants yearly; rates are increasing in developed countries (e.g. 7% of all births in the UK; 13% in the US) and we are limited in our predictive and therapeutic options for these vulnerable infants. Up to 60% of infants born preterm will be left with persistent cognitive and neuropsychiatric deficits including autism spectrum, attention-deficit disorders and epilepsy (Wood et al., 2000; Delobel-Ayoub et al., 2009). Insights into the previously poorly understood pathophysiology of preterm birth (Back et al., 2005; Billiards et al., 2008; Delobel-Ayoub et al., 2009; Moore et al., 2012; Verney et al., 2012) have enabled us to design animal models of improved relevance to the constellation of injuries seen in contemporary cohort of infants, collectively called encephalopathy of the premature infant. Encephalopathy of the premature infant involves cerebral white matter injury due to oligodendrocyte maturation blockade and the most severe of injuries widespread neuronal/axonal disease, both of which are linked to the activation of MG/Mϕ (Volpe, 2009b). The strongest predictor for outcomes we currently have in encephalopathy of the premature infant is the severity of white matter/connectivity changes (Woodward et al., 2006; Nosarti et al., 2008, 2014; Spittle et al., 2009). MG/Mϕ activation driving neuroinflammation in preterm-born infants is often caused by exposure to maternal foetal infections or inflammation, such as chorioamnionitis, or post-natal sepsis (Hagberg and Mallard, 2005; Hagberg et al., 2015). Endemic but often clinically silent maternal inflammatory events are considered the chief cause of preterm birth (Wu et al., 2009). As such, exposure to inflammation is both a driver of preterm birth and of brain injury, and further highlighting its importance, increased indices of exposure to inflammation are the strongest predictor of poor neurological outcome (Hillier et al., 1993; Dammann and Leviton, 2004; Wu et al., 2009).
The Wnt [wingless-type MMTV (mouse mammary tumour virus) integration site] pathways are divided into the canonical Wnt/β-catenin, the non-canonical Wnt/planar cell polarity (PCP) and the Wnt/calcium pathways. The Wnt pathways have well-studied critical roles in the early developmental events of body axis patterning, and brain cell fate specification, proliferation and migration (Sokol, 2015). Aberrant regulation of canonical WNT/β-catenin signalling is strongly implicated in the onset and progression of numerous cancers (Zhan et al., 2017). In development, the canonical Wnt pathway has been described as playing a repressive role in the maturation of oligodendrocytes (Shimizu et al., 2005; Feigenson et al., 2009, 2011; Yuen et al., 2014). However, our knowledge has increased and we appreciate that elaborate spatial and temporal regulation of Wnt signalling nudges oligodendrocytes through multiple stages of maturation (Zhao et al., 2016; and reviewed in Guo et al., 2015). Interestingly, Wnt signalling in endothelial cells has also been shown to have important roles in the transmigration of immune cells in the context of multiple sclerosis (Shimizu et al., 2016; Lengfeld et al., 2017). As such, it is clear that the effects of Wnt pathway activation are highly cell and context specific. However, a key regulatory role of the Wnt pathways in the response of MG/Mϕ to injury or insult has not been demonstrated, especially in the developing brain. However, activation of the Frizzled receptors (a family of G protein-coupled receptors) with Wnt ligands has complex effects on some facets of microglia activity in vitro (Halleskog et al., 2012; Halleskog and Schulte, 2013). Experimental evidence in other cell types also supports that the Wnt/β-catenin pathway is involved in aspects of inflammatory signalling, namely, there are links between expression of the inflammatory mediator COX2 (cyclooxygenase-2) and Wnt/β-catenin in cancer (Buchanan and DuBois, 2006), and evidence that β-catenin regulates at numerous levels the cytokine/pathogen induced activation of NF-κB (Ma and Hottiger, 2016). As such, in the hunt for a regulator of immature microglia activation state with a view to therapy design the Wnt pathway is an interesting and rationale candidate.
In the present study, we used our mouse model of encephalopathy of prematurity in which we clearly demonstrate a causal link between MG/Mϕ activation and a myelin deficit. This mouse model of encephalopathy of prematurity recapitulates the key neuropathological findings from preterm-born infants, including a myelination deficit, which is the focus of this study (Haynes et al., 2008; Volpe, 2009a; Verney et al., 2012). Using a combination of approaches across species, including human, we reveal for the first time that inhibition of the Wnt/β-catenin pathway is necessary and sufficient to drive this MG/Mϕ pro-inflammatory phenotypic transformation. We also verified the clinical relevance of the Wnt pathway to anatomical white matter structure in a cohort of human preterm-born infant brains using an integrated imaging genomics analysis. Finally, we used a novel 3DNA® nanocarrier that carries cargo across the blood–brain barrier following non-invasive intraperitoneal administration that then delivers cargo specifically to MG/Mϕ. Using this 3DNA nanocarrier we show that preventing Wnt pathway downregulation specifically in MG/Mϕ reduces the pro-inflammatory MG/Mϕ phenotype, white matter damage and long-term memory deficit in our model of encephalopathy of prematurity.
Altogether, these findings identify the Wnt/β-catenin pathway as a key regulator of MG/Mϕ activation state across species and a potential target for the treatment of encephalopathy of prematurity and other neuroinflammatory conditions.
Materials and methods
Study approval
Experimental protocols were approved by the institutional guidelines of the Institut National de la Santé et de la Recherché Scientifique (Inserm, France) (Approval 2012-15/676-0079), the Ethics committee, the services of the French ministry in charge of higher education and research according the directive 2010/63/EU of the European Parliament (Approval #9285-2016090611282348 and #9286-2016090617132750), and met the guidelines for the United States Public Health Service’s Policy on Humane Care and Use of Laboratory Animals (NIH, Bethesda, Maryland, USA).
The EPRIME study was reviewed and approved by the National Research Ethics Service, and all infants were studied following written consent from their parents.
For human microglia cell culture, all procedures had ethical approval from Agence de Biomédicine (Approval PFS12-0011). Written informed consent was received from the tissue donor.
Study design
A summary of the independent and total replicates for all experiments, described relative to the figures, is available in Supplementary Table 1.
Nomenclature of microglia phenotype
We adopted nomenclature consistent with our previous work in primary microglia (Chhor et al., 2013) and the work of others (Michell-Robinson et al., 2015) acknowledging that this is a simplification necessary to facilitate description of the data. To simplify, we distinguished three phenotypes according to the mRNA expression levels of markers listed in brackets: pro-inflammatory (Nos2, Ptgs2, Cd32, Cd86 and Tnfa), anti-inflammatory (Arg1, Cd206, Lgals3, Igf1, Il4 and Il10) and immunoregulatory (Il1rn, Il4ra, Socs3 and Sphk1).
Mice
Experiments were performed with male OF1 strain mice pups from adult females purchased from Charles River or transgenic male mice born in our animal facility. See the justification below in animal model section regarding use of male mice only. Transgenic mice B6.129P-Cx3cr1tm1Litt/J (CX3CR1-GFP) mice, B6.129-Ctnnb1tm2Kem/KnwJ (β-cateninlfox) mice and B6.129P2-Lyz2tm1(cre)Ifo/J (LysMCre) mice were purchased from The Jackson Laboratory. CX3CR1-GFP mice express EGFP in monocytes, dendritic cells, natural killer (NK) cells, and brain microglia under control of the endogenous Cx3cr1 locus. Only heterozygous (Cx3cr1GFP/+ and β-cateninΔ/+) mice were used in this study. Cx3cr1GFP/+ are obtained by crossing Cx3cr1GFP/GFP with C57bl6J (wild-type) mice. LysMCre mice are useful for Cre-lox studies of the myeloid cell lineage. Here, we used it to analyse β-catenin deficit in brain microglia by crossing β-cateninflox with LysMCre mice. Control mice were obtained by crossing β-cateninflox with C57bl6J (wild-type) mice.
Transgenic fish lines and fish maintenance
Transgenic (pu1::Gal4-UAS-TagRFP) (Sieger et al., 2012) fish were kept at 26.5°C in a 14-h light/10-h dark cycle. Embryos were raised at 28.5°C in E3 solution until 120 h post-fertilization (hpf) for analyses.
Drugs
Mouse cytokines (IL-1β, IL-4) were purchased from R&D systems. LPS-EB Ultrapure was purchased from InvivoGen. To inhibit the Wnt/β-catenin pathway, we used XAV939 (Sigma-Aldrich); and to activate it, we used CT99021 (Selleckchem), lithium chloride (Sigma-Aldrich) and L803mts (Tocris). Chelerythrine was purchased from Sigma-Aldrich. To decide on the choice of modulators of Wnt, including considerations for doses and consideration of off target effects we referred to reviews on the subject and previously published works (Fancy et al., 2011, 2014; Keats et al., 2014; Tran and Zheng, 2017). The L803mts peptide and a scrambled peptide (SCR) were purchased conjugated to short oligonucleotide with disulphide bond linker from BioSynthesis then hybridized to Cy3-labelled 3DNA at Genisphere. L803mts was chosen because of its structural properties allowing it to be conjugated to 3DNA and high tolerability have been demonstrated in mice (Kaidanovich-Beilin and Eldar-Finkelman, 2006).
Model of encephalopathy of prematurity induced by systemically driven neuroinflammation
Mice were housed under a 12-h light-dark cycle, had access to food and water ad libitum and were weaned into same sex groups at postnatal Day (P)21. Our animal facility has no known pathogens. On P1 pups were sexed and where necessary litters were culled to six to eight (transgenic mice) or 12 (OF1 mice) pups. Assessments of injury and outcomes were made only in male animals and all pups within a litter received identical treatment to reduce any effects of differing maternal care. All experiments include pups from at least three separate litters. Allocation to phosphate-buffered saline (PBS) or IL-1β exposure was made in a cage by cage alternating manner. Systemic inflammation was induced by intraperitoneal IL-1β exposure as described previously (Favrais et al., 2011). Only male mice were used, as only male pups display a myelin deficit following the induction of systemic inflammation to drive neuroinflammation in this paradigm. Specifically, female mice do not have a significant loss of the MBP protein or APC-positive cell number in adulthood like the male mice (P. Gressens, unpublished results). This higher severity of injury in male animals is in accordance with observations of a greater burden of neurological injury in male infants following perinatal insult (Johnston and Hagberg, 2007; Twilhaar et al., 2018). In brief, a 5 μl volume of PBS containing 10 μg/kg/injection of recombinant mouse IL-1β (R&D Systems) or of PBS alone (control) was injected intraperitoneal in male pups twice a day (morning and evening) on P1 to P4 and once in the morning at P5. Animal health was monitored via weight and general visual health assessment and there were no adverse events.
Morphological analysis of microglia
P1 and P3 brains from Cx3Cr1-GFP mice administered PBS or IL-1β were fixed for 24 h in 4% buffered formalin (QPath, Labonord SAS). Cerebral tissue was sliced along a sagittal plane on a calibrated vibratome (VT1000 S, Leica) into 100-µm thick free-floating slices. Morphological analysis of microglia was assessed as described previously (Verdonk et al., 2016) using a spinning disk confocal system (Cell Voyager - CV1000) with a UPLSAPO 40×/NA 0.9 objective and the use of a 488 nm laser. A mosaic of >100 pictures covering ∼3.17 mm² of tissue (cortex and white matter) was generated per brain. An automatic image analysis using a custom designed script developed with the Acapella™ image analysis software (version 2.7, Perkin Elmer Technologies) was carried out to obtain density, cell body area, number of primary, secondary and tertiary processes and the area covered by the microglia processes in 2D. A complexity index was calculated to analyse the morphological modifications induced by IL-1β.
Neural tissue dissociation and CD11B+ microglia or O4+ oligodendrocyte magnetic-activated cell sorting
At P1, P2, P3, P5 and P10, brains were collected for cell dissociation and CD11B+ cell separation using a magnetic coupled antibody anti-CD11B (MACS Technology), according to the manufacturer’s protocol (Miltenyi Biotec) and as described previously (Schang et al., 2014). In brief, mice were intracardially perfused with NaCl 0.9%. After removing the cerebellum and olfactory bulbs, the brains until were pooled (n = 2–3 per sample except at P10, n = 1) and dissociated using the Neural Tissue Dissociation Kit containing papain and the gentleMACS Octo Dissociator with Heaters. Microglia were enriched using the anti-CD11B (MG) MicroBeads, pelleted and conserved at −80°C. The purity of the eluted CD11B+ fraction was verified as described previously (Krishnan et al., 2017). At P5 and P10, brains were collected for, O4+ oligodendrocytes cell separation using a magnetic coupled antibody anti-O4 (MACS Technology), according to the manufacturer’s protocol (Miltenyi Biotec) and as described previously (Schang et al., 2014).
RNA extraction and quantification of gene expression by real-time qPCR
Total RNA was extracted with the RNeasy® mini kit according to the manufacturer’s instructions (Qiagen). RNA quality and concentration were assessed by spectrophotometry with the NanodropTM apparatus (ThermoFisher Scientific). Total RNA (500 ng) was subjected to reverse transcription using the iScriptTM cDNA synthesis kit (Bio-Rad). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed in triplicate for each sample using SYBR® Green Supermix (Bio-Rad) for 40 cycles with a 2-step program (5 s of denaturation at 95°C and 10 s of annealing at 60°C). Amplification specificity was assessed with a melting curve analysis. Primers were designed using Primer3 plus software (sequences are provided in Supplementary Table 2). Specific mRNA levels were calculated after normalization of the results for each sample with those for Rpl13a mRNA (reference gene). The data are presented as relative mRNA units with respect to control group (expressed as fold change over control value).
Immunohistofluorescence
For frozen sections, P1 or P5 mice were intracardially perfused with 4% paraformaldehyde in 0.12 M phosphate buffer solution under isoflurane anaesthesia. Brains were then post-fixed in 4% paraformaldehyde (Sigma-Aldrich) overnight at 4°C After 2 days in 30% sucrose 0.12 M phosphate buffer solution, brains were frozen at −45°C in isopentane (Sigma-Aldrich) before storage at −80°C until sectioning on a cryostat (14 μm thickness). Immunohistofluorescence was performed as described previously (Miron et al., 2013). Slides were permeabilized and blocked for 1 h and primary antibody was applied overnight at 4°C in a humid chamber. To detect microglia, rabbit antibody to IBA1 (Wako, 1:500) was used. To detect proinflammatory microglia we used mouse antibody to iNOS (BD Biosciences, 610329, 1:100), rat antibody to CD16/CD32 (BD Pharmingen, 553141, 1:500), and rabbit antibody to COX2 (Abcam, Ab15191, 1:500). To detect anti-inflammatory microglia, we used goat antibody to Arginase-1 (ARG1) (Santa Cruz Biotechnology, sc-18355, 1:50), and rabbit antibody to mannose receptor (MR) (Abcam, ab64693, 1:600). To detect microglia and infiltrating macrophage (Mϕ) rat antibody to CD68 (Abcam, ab53444, 1:100) was used. Fluorescently conjugated secondary antibody to goat IgG (A21432), antibodies to rabbit IgG (A11034, A10042), antibodies to rat IgG (A21434, A11006, A21247) and antibody to mouse IgG (A21042), were applied for 2 h at 20–25°C in a humid chamber (1:500, Invitrogen). Following counterstaining with Hoechst, slides were coverslipped with Fluoromount-G (Southern Biotech, Cliniscience). Antibody isotype controls (Sigma-Aldrich) added to sections at the same final concentration as the respective primary antibodies showed little or no nonspecific staining. All manual cell counts were performed by an investigator who had been blinded to the group allocation of the sample.
In vivo treatments with XAV939, 3DNA L803mts Cy3, and gadolinium
XAV939 (250 μM, 0.5 nmol/injection) or PBS/dimethyl sulphoxide (DMSO) (vehicle) and 3DNA L803mts Cy3 (200 ng/injection) or 3DNA SCR Cy3 (200 ng/injection) (control peptide) were injected into the right ventricle of mice pups at P1 using a 26-G needle linked to a 10 μl Hamilton syringe mounted on a micromanipulator and coupled to a microinjector (Harvard Apparatus; outflow 2 μl/min), 3DNA SCR or L803mts Cy3 were injected 1 h before intraperitoneal injection of PBS or IL-1β. Three hours after PBS or IL-1β injection or 4 h after XAV939 or vehicle injection, pups were sacrificed for CD11B+ cell sorting. Chronic treatment with 3DNA L803mts Cy3 were performed by intraperitoneal co-injections of PBS or IL-1β with 3DNA L803mts Cy3 (500 ng/injection) or 3DNA L803mts Cy3 (500 ng/injection) twice a day between P1 and P3 and once a day at P4 and P5. The effect of this treatment on myelination was analysed at P10, P15 and P30, and behavioural tests were realized in adulthood (2–3 months). To analyse uptake of 3DNA in liver and spleen, one intraperitoneal injection of 3DNA Cy3 (500 ng/injection) with PBS or IL-1β was performed and animals sacrificed 4 h later. To analyse uptake of 3DNA in brain, animals were sacrificed 4 h after intracerebroventricular injection at P1, and at the end of the intraperitoneal treatment at P5.
Selective depletion of pro-inflammatory microglia in white matter injury was performed using gadolinium chloride (GdCl3) as described previously (Miron et al., 2013) with slight modifications. Briefly, PBS or gadolinium (200 nmol/injection, Sigma) was injected into the corpus callosum of mouse pups at P1, 1 h before intraperitoneal injections of PBS or IL-1β using a 26-G needle linked to a 10 μl Hamilton syringe mounted on a micromanipulator and coupled to a microinjector (Harvard apparatus, outflow 2 μl/min). Effect of pro-inflammatory microglia depletion on microglia cell phenotype and myelination was analysed at P15.
Western blotting
Frozen right anterior cortex from P15 mice was homogenized in RIPA Buffer (Sigma-Aldrich) containing protease inhibitors (cOmplete Tablets, Roche) in gentleMACS M tubes using a gentleMACS dissociator (Miltenyi Biotec) as per the manufacturer’s instructions. Samples were centrifuged (10 000g, 10 min, 4°C) and the pellets were stored for later use. Equal amounts of protein (25 μg) as determined by BCA protein assays (Sigma-Aldrich), were diluted with Laemmli sample buffer (Bio-Rad) containing 2-mercaptoethanol (Sigma-Aldrich) and then separated in Mini-PROTEAN® TGX gels (Any kD™, Bio-Rad, 80 V for 1 h 50 min). Proteins were then electrotransferred (Trans-Blot® Turbo™, Bio-Rad) onto a 0.2 μm nitrocellulose membrane (Trans-Blot® Turbo™ Transfer Pack, mini, Bio-Rad). The membrane was cut into an upper and lower portion and both were incubated in blocking solution [5% bovine serum albumin, 0.1% Tween 20 in Tris-buffered saline (TBS)] for 1 h. The upper and lower parts were incubated, respectively, with mouse anti-β-actin (Sigma-Aldrich AC-74, 1:20 000) and rat anti-MBP (Millipore MAB386 1:500) overnight at 4°C in blocking solution. Blots were rinsed with 0.1% Tween 20 in TBS and incubated for 1 h with a HRP-conjugate goat anti-mouse IgG (1:2000; Sigma-Aldrich) or HRP-conjugate goat anti-rat IgG (1:10 000; Invitrogen) in blocking solution. The blots were washed three times with 0.1% Tween 20 in TBS for 5 min. Membranes were processed with the Clarity™ Western ECL substrate (Bio-Rad), and the proteins of interest were investigated with Syngene PXi (Syngene) coupled to acquisition software. The immunoreactivity of four isoforms of MBP was compared with that of actin controls using NIH ImageJ software (http://rsb.info.nih.gov/ij/).
Electron microscopy
At P30 mice were transaortically perfused with 20 ml saline followed by 100 ml of ice-cold 2% paraformaldehyde with 2% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4 (PB). Brains were post-fixed overnight in 2% paraformaldehyde at 4°C. Sagittal sections were cut on a vibratome at 70-μm thickness, post-fixed in 1% glutaraldehyde for 10 min, treated with 1% osmium for 10 min, dehydrated in an ascending series of ethanol, which included 1% uranyl acetate in 70% ethanol. Sections were then treated with propylene oxide, equilibrated overnight in Durcupan™ ACM (Fluka), and flat-embedded on glass slides for 48 h at 60°C. Blocks of the trunk of the corpus callosum (+0.1 mm to −0.1 mm from bregma) close to the midline were cut out from the sections and glued to blank cylinders of resin. Ultrathin sections were cut on a Reichert Ultracut S microtome and collected on Pioloform®-coated single-slot grids. Sections were stained with lead citrate and examined with a Hitachi HT7700 electron microscope (Hitachi High-Technologies) equipped with an AMT XR-41B 4 Megapixel camera (Advanced Microscopy Techniques).
The myelinated axon diameter was measured on cross-sectional axons. For each animal, images were acquired at three dorsoventral positions of three rostrocaudal levels. The thickness of the myelin sheath was assessed by determining the G-ratio (axon diameter/total fibre diameter). An average of 1600 measurements of myelinated axons per animal were performed, using the Fiji version of ImageJ (Schindelin et al., 2012). The axons were pooled by size according to their small (0.2–0.4 μm), medium (0.4–0.8 μm) or large (>0.8 μm) diameter. Results were compared by one-way ANOVA followed by Bonferroni’s multiple comparison test, using GraphPad Prism 5.0 (Graph-Pad Software, San Diego, CA, USA). The value of P < 0.05 was considered statistically significant.
Behavioural assessments
Actimetry
The horizontal (spontaneous locomotion) and vertical (rearing) activities were individually assessed in transparent activity cages (20 × 10 × 12 cm) with automatic monitoring of photocell beam breaks (Imetronic). An actimetry test was performed at ≈ 2 months (Day 65), with recording every 30 min over 23 h, in order to evaluate the effect of IL-1β and 3DNA treatment intraperitoneal injections on spontaneous locomotor activity, as modified locomotor activity could lead to biased results from other behavioural tests requiring locomotion.
Open-field test
The open-field test was performed at ≈2.5 months (Day 80) using a square white open-field apparatus (100 × 100 × 50 cm) made of plastic permeable to infrared light. Distance travelled and time spent in inactivity were recorded by the VideoTrack system (Viewpoint®) during the 9-min test.
Barnes maze test
The Barnes maze test was performed at 3 months (Days 90–105). The maze is a wet, white and circular platform (80-cm diameter), brightly illuminated (400 lx), raised 50 cm above the floor, with 18 holes (4.5 cm) equally spaced around the perimeter. A white hidden escape box (4 × 13 × 7 cm), representing the target, was located under one of the holes. Prior to the test, each mouse was subjected to a habituation trial where the mouse was directly put in the escape box for 30 s.
For the learning phase, the mouse was placed in the centre of the circular maze and allowed to explore the platform and holes for a maximum of 3 min. The distance travelled was recorded using the VideoTrack system (Viewpoint®). Latency to reach the escape box, and number of errors (number of empty holes visited) were manually noted. When the mouse found and entered the escape box, the VideoTrack recording was stopped, and the mouse remained in the escape box for an additional 10 s before returning to its home cage. If the mouse did not enter spontaneously, it was gently put toward the escape box, before returning to its home cage. On the first day of training, mice underwent two trials after the habituation trial; thereafter, three trials were given per day, with a 2-h interval.
On the fifth day of learning the 90-s probe trial was carried out to assess spatial memory performances. The escape box was removed and time spent and distance travelled in each sextant (defined by one of the six parts of the maze, which includes three holes) were recorded. The target zone was defined as the part that contains the target hole and two adjacent holes.
In the long-term trial, 15 days after learning, a new trial, with the escape box, was used to assess long-term retention memory. Escape latency and distance travelled to reach the target were recorded.
Immunohistochemistry
Brains were collected at P15 or P30 and immersed immediately after sacrifice in 4% formaldehyde for 4 days at room temperature, prior to dehydration and paraffin embedding. Section was realized using a microtome. Immunostaining was performed as previously described (Favrais et al., 2011) using mouse antibody to MBP (MAB382, Millipore, France 1:500) or rabbit antibody to OLIG2 (JP18953 IBL 1:200). The intensity of the MBP immunostaining in the anterior corpus callosum and the number of OLIG2+ cells were assessed by a densitometry analysis through NIH ImageJ Software (http://imagej.nih.gov/ij/). Optical density was deduced from grey scale standardized to the photomicrograph background. Corpus callosum and/or cingulum were defined as region of interest. One measurement per section (on 40 000 μm2 area) and four sections were analysed in each brain.
Primary mouse microglia culture
Primary mixed glial cell cultures were prepared from the cortices of P1 OF1 mice, as described previously (Chhor et al., 2013). After 14 days, microglia were purified and pelleted via centrifugation and resuspended in Dulbecco’s modified Eagle medium/penicillin-streptomycin/10% foetal bovine serum (DMEM/PS/10% FBS) at a concentration of 4 × 105 cells/ml. One or two millilitres per well of cell suspension was plated in 12-well (for qRT-PCR) or 6-well (for ELISA) culture plates, respectively. For immunofluorescence, 250 μl of cell suspension was plated in µ-Slide 8 Well Glass Bottom chamber slides (Ibidi, BioValley). Post-plating, media was replaced after 1 h. Based on previous work from our lab (Chhor et al., 2013), the day after plating, microglia were exposed for 4 h to DMEM (control) or IL-1β at 50 ng/ml, IL-4 at 20 ng/ml, diluted in DMEM. For RT-qPCR or ELISA analysis, media were removed and plates frozen at −80°C. For immunofluorescence experiments, cells were fixed at room temperature with 4% paraformaldehyde for 20 min.
Mixed mouse glial and oligodendrocyte progenitor cell cultures
Cortical mixed glial cultures and purified oligodendrocyte progenitor cells (OPCs) were obtained from the cortices of P1 OF1 mice as previously described with slight modification (Shiow et al., 2017). Mixed glial cells were resuspended in MEM-C, which consisted of minimal essential medium (MEM) supplemented with 10% FBS (Gibco), GlutaMAX™ (Gibco), and 1% PS, and plated at 2 × 105 cells/cm2 onto T75 flasks for subsequent OPC purification or Ibidi 8-well chamber slides (BioValley) for mixed glial culture treatment. For this purpose, mixed glial cultures were proliferated in MEM-C for 7 days, then maintained for 10–12 days in differentiation medium, which consisted of DMEM-F12 with glutamine (Gibco), N2 (Gibco) and 1% PS. Half of the wells were exposed to IL-1β (50 ng/ml) and medium was changed every 2–3 days. Alternatively, mixed glial cultures plated onto T75 flasks were maintained for 9–12 days in MEM-C and purified OPC cultures were prepared by a differential shake method (McCarthy and de Vellis, 1980). OPCs were seeded onto Ibidi 8-well chamber slides at a density of 3 × 104 cells/cm2 in proliferation medium, which consisted of MACS Neuro Medium with NeuroBrew® 21 (Miltenyi Biotec), GlutaMAX™, 1% PS, FGFb and PDGFA (10 ng/ml each, Sigma-Aldrich). After 72 h, differentiation was induced by FGFb and PDGFA withdrawal and by adding T3 (40 ng/ml, Sigma). Half of the wells were exposed to IL-1β (50 ng/ml). After 10–12 days (for mixed glial cultures) or 72 h (for OPCs) of IL-1β, cells were fixed for 20 min with 4% paraformaldehyde.
Primary human microglia culture
Within 1 h of scheduled termination (age 19 and 21 weeks of amenorrhea) brain tissues were collected from two human foetuses without any neuropathological alterations. Brain tissue (4 g) was minced and further mechanically dissociated using a 1 ml micropipettor in Hank’s Balanced Salt Solution with Ca2+ and Mg2+. Using a 70-μM strainer, a single cell suspension was obtained. MG/Mϕ isolation were obtained using anti-CD11B (MG) microbeads (MACS Technology), according to the manufacturer’s protocol (Miltenyi Biotec), as above. CD11B+ MG/Mϕ were pelleted via centrifugation and resuspended in DMEM/PS/10% FBS at a concentration of 5 × 106 cells/ml. Cells were plated in 12-well plates (1 ml/well) for qRT-PCR analysis. For immunofluorescence analysis, cells were plated in µ-Slide 8 Well Glass Bottom (Ibidi, BioValley). Forty-eight hours after plating, microglia were exposed for 4 h to DMEM (control) or lipopolysaccharide (LPS) 10 ng/ml diluted in DMEM. For RT-qPCR media were removed and plates frozen at −80°C. For immunofluorescence experiments, cells were fixed at room temperature with 4% paraformaldehyde for 20 min.
Immunocytofluorescence
Immunofluorescence staining was performed as described previously (Favrais et al., 2011), using mouse antibody to MBP (MAB382, 1:500; Millipore), rabbit antibody to OLIG2 (18953, 1:500 IBL-America), rabbit antibody to beta-catenin (E247, 1:100; Millipore), rabbit antibody to COX2 (Ab15191, 1:400; Abcam) goat antibody to ARG1 (sc-18354, 1:400; Santa-Cruz) and IL1RA (sc-8482, 1:200, Santa-Cruz). Fluorescently conjugated secondary antibody (all ThermoFisher Scientific) to mouse IgG (A21236), to rabbit IgG (A21206) and to goat IgG (A11055), were applied for 2 h at 20–25°C in a humid chamber (1:500, Invitrogen). Slides were coverslipped with Fluoromount-G (Southern Biotech). To analyse MBP labelling, the MBP area was assessed as the percentage of total area above threshold using NIH ImageJ software (http://imagej.nih.gov/ij/) and was normalized to the number of Olig2+ cells. Means were expressed as fold change over control condition.
Microarrays and data preprocessing
Gene expression quantification, including RNA extraction and quality assurance, was performed by Miltenyi Biotec on a total of 24 samples of CD11B+ cells and 20 samples of O4+ cells from the brains of mice exposed to IL-1β or PBS between P1 and P5, and sacrificed at P5 or P10. RNA was extracted and hybridized to Agilent Whole Mouse Genome Oligo Microarrays (8x60K).
Bioinformatic gene co-expression network reconstruction
Expression data
Each cell type and experimental condition was analysed separately, to investigate changes in gene expression over time in response to exposure to IL-1β or PBS. Expression values for microglia (CD11B+) and oligodendrocytes (O4+) at P5 and P10 were quantile normalized.
Differential expression
Probes showing high variability across time points within each cell type were retained for further analysis (coefficient of variation above 50th centile). Probes showing differential expression between time points were identified using unpaired t-tests with Benjamini-Hochberg correction for multiple testing in the Bioconductor multitest package (Gentleman et al., 2004; Schafer and Strimmer, 2005; Opgen-Rhein and Strimmer, 2007), with a false discovery rate threshold (FDR) of 10%.
Network reconstruction
The co-expression network was inferred using Graphical Gaussian Models (GGM), implemented within the software package ‘GeneNet’ (Schafer and Strimmer, 2005) in R (http://www.r-project.org/). This analysis computes partial correlations, which are a measure of conditional independence between two genes i.e. the correlation between the expression profiles of two genes after the common effects of all other genes are removed. Correction was made for multiple comparisons by setting the local FDR at ≤1%. Individual networks were reconstructed for temporally differentially expressed genes for each cell type in each experimental condition (i.e. two cell types × two conditions). The input for each GGM was a matrix of differentially expressed normalized mRNA transcript levels. The genes in each of these four co-expression networks (Supplementary Fig. 3E) were functionally annotated by calculating significant enrichment of KEGG pathways using the Broad Institute MSigDB database (http://software.broadinstitute.org/gsea/msigdb/index.jsp) (Subramanian et al., 2005) (Supplementary Table 3).
Differential expression analysis was performed using the limma package (Smyth, 2004). The mouse datasets were annotated with human Ensembl gene ID using the biomaRt Bioconductor R package (Durinck et al., 2009) and selecting human genes that were ‘one-to-one’ orthologues with mouse genes. We multiplied the adjusted P-value (the −log10 of P-value) by the log-transformed fold change to generate a gene-level score, which was used as a metric to ‘rank’ genes. Gene set enrichment analysis GSEA (Subramanian et al., 2005) was applied genome-wide to the ranked list of gene scores (reflecting both the significance and the magnitude of expression changes in IL-1β exposed MG/Mϕ) to test if a group of genes (MSigDB gene sets) occupy higher (or lower) positions in the ranked gene list than what it would be expected by chance. Gene set enrichment scores and significance level of the enrichment (normalized enrichment score, P-value, FDR) and enrichment plots were provided in the GSEA output format developed by Broad Institute of MIT and Harvard (permutations = 10 000). The GSEA was carried out accounting for the direction of IL-1β effect, i.e. considering whether a gene is up- or downregulated under IL-1β exposure at P5 and P10.
Wnt/β-catenin pathway modulation in primary microglia
Microglia transfection was realized using negative control siRNA or siRNA Axin2 (ON TARGET Plus Control Pool and ON TARGET Plus mouse Axin2-06, Thermo Scientific, Dharmacon) at a final concentration of 30 μM. Transfection was realized using the MACSfectin™ transfection reagent (Miltenyi Biotec) in a mixture of Opti-MEM™ and DMEM mediums for 48 h prior to IL-1β stimulation. Axin2 mRNA knockdown was evaluated by qRT-PCR. XAV939 and CT99021 (1 μM) or PBS/DMSO (vehicle) were added to media 30 min before DMEM (control) or IL-1β (50 ng/ml) treatments.
PSer45 β-catenin and β-catenin quantification by ELISA
Quantification of PSer45 β-catenin and β-catenin by enzyme-linked immunosorbent assay (ELISA) was realized using β-catenin pSer45 + Total PhosphoTracer ELISA Kit (ab119656, Abcam) for primary microglia or β-catenin ELISA Kit (ab1205704, Abcam) for magnetically-activated cell sorted (MACS) cells. Briefly, cell lysates were obtained using lysis buffer of the kit. Total protein level was quantified using BCA method [Bicinchoninic Acid Solution and Copper (II) Sulfate Solution from Sigma]. The ELISA was carried out according to the manufacturer’s instruction. Quantification of PSer45 β-catenin is a ratio PSer45 β-catenin/total β-catenin. Total β-catenin was normalized using total protein level. Total β-catenin data are expressed as fold change over control values.
Wnt/β-catenin pathway modulation in Zebrafish
Microglia were visualized using pu1::Gal4-UAS::TagRFP. Zebrafish embryos aged 72 hpf were used for LPS microinjection (5 ng/injection) in hindbrain. The Wnt/β-catenin pathway was modulated by adding LiCl (80 mM), XAV939 (5 µM), CT99021 (3 µM) in E3 solution. Fluorescently-labelled embryos were imaged using a microscope equipped with an ApoTome system (Zeiss) fitted with an AxioCam MRm camera (Zeiss) controlled by the Axiovision software. The thickness of the z-stacks was always between 2.5 and 3 µm. Fluorescence intensity was quantified by NIH ImageJ software (http://imagej.nih.gov/ij/) on greyscale images.
Preterm infant cohort
Preterm infants were recruited as part of the EPRIME [Evaluation of Magnetic Resonance (MR) Imaging to Predict Neurodevelopmental Impairment in Preterm Infants] study and were imaged at term-equivalent age over a 3-year period (2010–13) at a single centre. The EPRIME study was reviewed and approved by the National Research Ethics Service, and all infants were studied following written consent from their parents. A total of 290 infants (gestational age 23.57 to 32.86 weeks, median 30 weeks) were scanned at term-equivalent age (38.29 to 58.28 weeks); additional cohort details are available in Supplementary Table 4. Pulse oximetry, temperature, and heart rate were monitored throughout the period of image acquisition; ear protection in the form of silicone-based putty placed in the external ear (President Putty, Coltene) and Mini-muffs (Natus Medical Inc.) were used for each infant.
Image acquisition
MRI was performed on a Philips 3-T system using an 8-channel phased array head coil. The 3D-MPRAGE and high-resolution T2-weighted fast spin echo images were obtained before diffusion tensor imaging (DTI). Single-shot EPI DTI was acquired in the transverse plane in 32 non-collinear directions using the following parameters: repetition time: 8000 ms; echo time: 49 ms; slice thickness: 2 mm; field of view: 224 mm; matrix: 128 × 128 (voxel size: 1.75 × 1.75 × 2 mm3); b-value: 750 s/mm2. Data were acquired with a SENSE factor of 2.
Data selection and quality control
The T2-weighted MRI anatomical scans were reviewed to exclude subjects with extensive brain abnormalities, major focal destructive parenchymal lesions, multiple punctate white matter lesions or white matter cysts, as these infants represent a heterogeneous minority (1–3%) with different underlying biology and clinical features to the general preterm population (Hamrick et al., 2004; van Haastert et al., 2011). All MRIs were assessed for the presence of image artefacts (inferior-temporal signal dropout, aliasing, field inhomogeneity, etc.) and severe motion (for head-motion criteria see below). All exclusion criteria were designed so as not to bias the study but preserve the full spectrum of clinical heterogeneity typical of a preterm born population. Two hundred and ninety infants had images suitable for tractography and associated genetic information.
DTI analysis was performed using FMRIB's Diffusion Toolbox (FDT v2.0). Images were registered to their non-diffusion weighted (b0) image and corrected for differences in spatial distortion due to eddy currents. Non-brain tissue was removed using the brain extraction tool (BET) (Smith, 2002). Diffusion tensors were calculated voxel-wise, using a simple least-squares fit of the tensor model to the diffusion data. From this, the tensor eigenvalues and fractional anisotropy maps were calculated.
Probabilistic tractography
Regions of interest for seeding tracts were obtained by segmentation of the brain based on a 90-node anatomical neonatal atlas (Shi et al., 2011), and the resulting segmentations were registered to the diffusion space using a custom neonatal pipeline (Ball et al., 2010). Tractography was performed on DTI data using a modified probabilistic tractography that estimates diffusive transfer between voxels (Robinson et al., 2008) using cortico-cortical connections only. A weighted adjacency matrix of brain regions was produced for each infant: self-connections along the diagonal were removed; symmetry was enforced; and the redundant lower triangle removed. The edges of these connectivity graphs were vectorized by concatenating the rows for each individual connectivity matrix, and appending them to form a single matrix of n individuals by p edges. Each phenotype matrix was adjusted for major covariates (post-menstrual age at scan and at birth) and reduced to its first principal component.
Genome-wide genotyping
Saliva samples from 290 preterm infants with imaging data were collected using Oragene DNA OG-250 kits (DNAGenotek Inc.) and genotyped on Illumina HumanOmniExpress-24 v1.1 chip. Filtering was carried out using PLINK (https://www.cog-genomics.org/plink2). All individuals had missing call frequency <0.1. Single nucleotide polymorphisms (SNPs) with Hardy-Weinberg equilibrium exact test P ≥ 1 × 10−6, minor allele frequency ≥ 0.01 and genotyping rate > 0.99 were retained for analysis. After these filtering steps, 659 674 SNPs remained. Additional details of these analyses and the STREGA details are found in the original description of the cohort (Boardman et al., 2014).
Preterm infant imaging genomics analysis of the Wnt pathway
A list of all genes in the WNT signalling pathway (entry hsa04310) in the Kyoto Encyclopaedia of Genes and Genomes (KEGG) was used as a gene-set of interest (n = 141 genes). Gene-set analysis was carried out with the Joint Association of Genetic Variants (JAG) tool (Lips et al., 2012, 2015) to test the joint effect of all SNPs located in the Wnt pathway. This procedure consists of three parts: (i) SNP to gene annotation; (ii) self-contained testing (i.e. association); and (iii) competitive testing (i.e. enrichment). An enriched pathway can be defined as one whose genes are more strongly associated with the phenotype than those genes outside the pathway. Phenotype permutation is used to evaluate gene-set significance, implicitly controlling for linkage disequilibrium, sample size, gene size, number of SNPs per gene, and number of genes in a gene-set. This will result in a P-value (with a traditional significance threshold <0.05) if there is an enrichment for variants associated with the phenotype within the gene-set of interest compared to random matched gene-sets.
Following SNP-gene mapping, genes in the WNT-set were tested for association with the original phenotype (query data), and this was repeated with 1000 permutations of the phenotype (self-contained/association test). This association testing tests the null hypothesis that no pathway genes are associated with the phenotype and combines the individual gene association P-values into a single P-value for the entire pathway. The competitive/enrichment test was then done to test the null hypothesis that the pathway genes are no more associated with the phenotype than 300 randomly drawn non-pathway gene-sets, with 1000 phenotype permutations. The association analysis was repeated gene-by-gene within the WNT gene-set to investigate which members of the gene-set might be predominantly contributing to the collective signal.
Statistical analysis
No statistical methods were used to predetermine sample sizes, but these were similar to those generally used in the field. Data are expressed as the mean values ± standard error of the mean (SEM). Data were first tested for normality using the Kolmogorov-Smirnov normality test for n = 5–7 and D'Agostino and Pearson omnibus normality test for n > 7. F-test (single comparisons) or Bartlett's test (multiple comparison) for ANOVA was used. Multiple comparisons in the same dataset were analysed by one-way ANOVA with Newman-Keuls post hoc test, two-way ANOVA with Bonferroni post hoc or Kruskal-Wallis test with Dunns post hoc. Single comparisons to control were made using two-tailed Student's t-test or Mann-Whitney test. P < 0.05 was considered to be statistically significant. For the Barnes maze probe trial, univariate t-test was performed to compare the per cent time spent in the target sextant to the theoretical value 16.67% (i.e. when the mouse spent equal time within each sextant). Data handling and statistical processing was performed using Microsoft Excel and GraphPad Prism Software.
Data availability
All new data are available from the authors on request, subject to ethical restrictions related to the human studies.
Results
Microglial activation drives hypomyelination in our model of encephalopathy of the premature infant
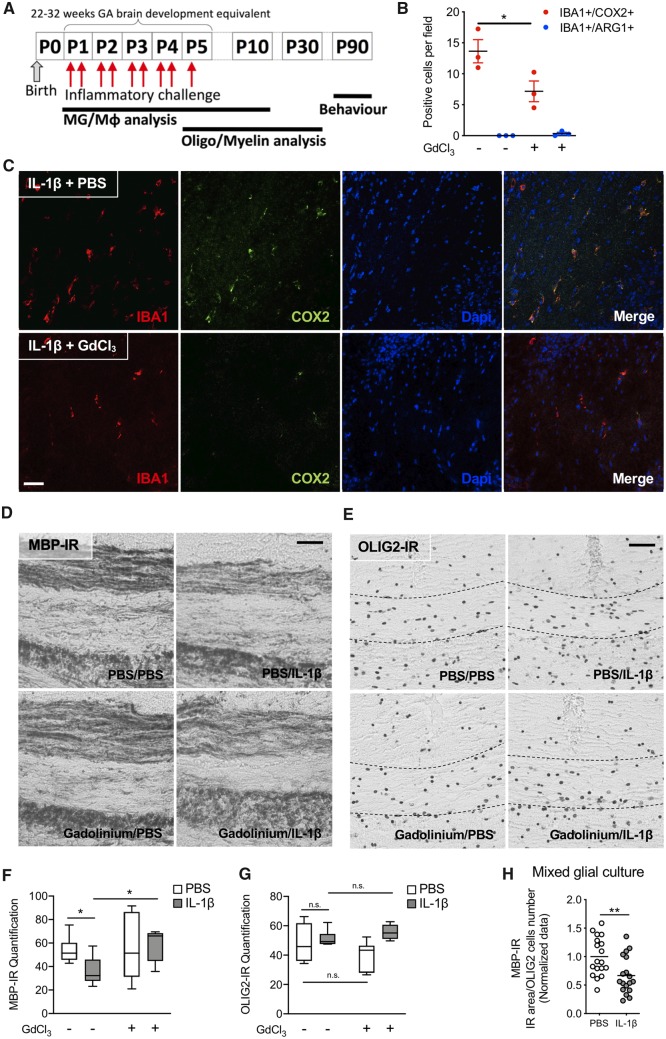
To understand the mechanisms underpinning encephalopathy of the premature infant in contemporaneous cohorts we previously set up a model mimicking its neuropathological, behavioural and imaging phenotypes (Favrais et al., 2011; Krishnan et al., 2017). Of note, the neuropathological similarities between our model and clinical studies include MG/Mϕ activation and hypomyelination due to the maturation blockade of oligodendrocytes but limited cell death (Billiards et al., 2008; Verney et al., 2010, 2012). Specifically, intraperitoneal injections of IL-1β (10 μg/kg) were administered to mouse pups twice daily on P1–4 and once at P5 (Fig. 1A). P1–P5 in the mouse is a developmental period roughly corresponding to 22–32 weeks of human pregnancy (Marret et al., 1995) and this is the greatest period of vulnerability for white matter damage in preterm-born infants. Exposure to systemic IL-1β recapitulates the systemic inflammatory insult of maternal/foetal infections. Specifically, intraperitoneal IL-1β injection causes a complex systemic response including increased blood levels of IL-6, TNFα and IL-1β (Favrais et al., 2011) that lead to neuroinflammatory responses including microglial activation with increased cytokines and chemokines (Krishnan et al., 2017; Shiow et al., 2017). The eventual outcome is an oligodendrocyte maturation delay that can be observed specifically as an increase in the numbers of NG2+ and PDGFRa+ OPC/pre-oligodendrocyte populations, decreased expression of MBP, MAG and MOG, and altered axonal myelination into adulthood (Favrais et al., 2011; Schang et al., 2014; Shiow et al., 2017) associated with cognitive dysfunction (Favrais et al., 2011). In this study, we verified a direct causal link between MG/Mϕ activation and neuropathology in our model by selectively killing pro-inflammatory MG/Mϕ and observing a reduction in myelin injury. Specifically, we used an intracerebral injection of GdCl3, which kills pro-inflammatory MG/Mϕ via competitive inhibition of calcium mobilization and damage to the plasma membrane. We validated in vivo that this approach is effective at reducing the numbers of pro-inflammatory microglia in the developing brain, and it has previously been validated in vivo in adult models of injury (Fulci et al., 2007; Miron et al., 2013; Du et al., 2014). We chose this approach and not ablation of microglia with a tamoxifen-driven transgenic or other pharmacological approach (i.e. PLX3397, ganciclovir) as our studies target maturing oligodendrocytes (as found in the preterm infant brain) that are present from P1. These conditional and pharmacological depletion methods cannot practically be made effective at P1 and this window of development is key to this model. Also, embryonic depletion of microglia itself alters brain development (Squarzoni et al., 2014) in ways that would alter our experimental paradigm. GdCl3 (200 nmol) was injected into the corpus callosum of P1 mice prior to intraperitoneal injection of IL-1β or PBS. In the corpus callosum at P3, after 72 h of exposure to systemic inflammation, immunohistological analysis revealed that the majority of MG/Mϕ were IBA1+/COX2+ and the numbers of these cells was reduced by ∼50% by GdCl3 (Fig. 1B and C). This GdCl3-mediated reduction in MG/Mϕ prevented the typical loss of MBP in the corpus callosum at P30 (Fig. 1D and E), but OLIG2 was not altered by exposure to systemic inflammation, as reported previously (Favrais et al., 2011) or GdCl3 (Fig. 1F and G). A link between microglial activation and hypomyelination can also be recapitulated with inflammatory activation of mixed glial cultures containing OPCs, microglia and astrocytes. In these in vitro conditions, there is reduced MBP+ cell density, without affecting the total number of oligodendrocytes (Fig. 1H). Exposure of pure OPC cultures (in the absence of microglia or astrocytes) to an inflammatory stimulant, IL-1β, did not cause hypomyelination (data not shown). Altogether, these results validate in our model a causal link between MG/Mϕ activation and the hypomyelination that is a hallmark of encephalopathy of prematurity.
Figure 1.
MG/Mϕ are required to induce hypomyelination in our model of encephalopathy of prematurity. (A) Schematic of the experimental paradigm for modelling systemic inflammation-associated encephalopathy of prematurity and the timing of analysis; MG/Mϕ and oligodendrocytes (Oligo). Of note, experiments needed to be performed between P1 and P5 as this is when oligodendrocyte maturation in the mouse matches that found in vulnerable preterm-born infants, those born between 22 and 32 weeks gestation (GA). (B–H) Demonstration that oligodendrocyte injury in our model is dependent on activated MG/Mϕ by inducing cell death in these cells. Specifically, PBS or IL-1β exposed mice were injected in the corpus callosum with vehicle (PBS) or GdCl3; 200 nmol at P1. (B) Scatter plot showing the reduced numbers of pro-inflammatory (IBA1+/COX2+) and stable numbers of anti-inflammatory (IBA1+/ARG1+) MG/Mϕ in corpus callosum at P3 (mean ± SEM, Mann-Whitney test, *P < 0.05, n = 3/group) as illustrated by representative images in C of IBA1-immunoreactivity (IR), COX2-IR and DAPI (scale bar = 100 μm). Following GdCl3 treatment, representative images showing MBP-IR (D) and OLIG2-IR (E) (scale bars = 20 μm) in the corpus callosum at P15, and min to max box and whisker plots of the quantification of MBP-IR (F) and OLIG2-IR (G) (Mann-Whitney test, *P < 0.05, n = 5–6/group). The requirement for the presence of MG/Mϕ to illicit demyelination was also verified in vitro in mixed glial cultures in H, scatter plots of MBP-IR normalized by OLIG2+ cell number in mixed cell culture (mean, Mann-Whitney test **P < 0.01 and n = 18/group).
Microglial activation is persistent with specific temporal patterns in our model of encephalopathy of the preterm infant
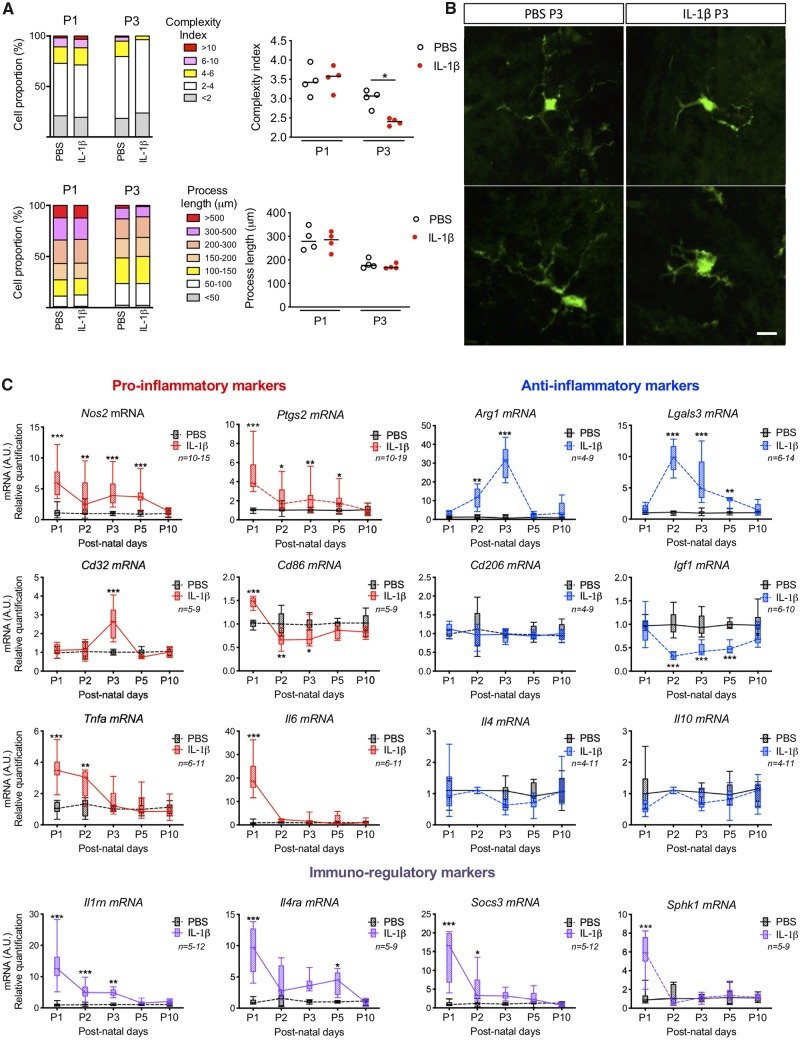
We comprehensively characterized the morphology, gene and protein expression of MG/Mϕ over time in our model to assess the relationship between activation states and injury. Our morphological analysis of GFP+ MG/Mϕ from CX3CR1GFP/+ mice revealed a moderate but significant reduction of complexity (arborization) at P3 when comparing MG/Mϕ from IL-1β versus PBS-injected animals, but no difference in process length or density, cell body area, numbers of primary, secondary and tertiary processes, or the area covered by the microglia processes in 2D (Fig. 2A and B). Cell morphology was measured at 3 h after the first inflammatory challenge (at P1) and at P3 when animals had been exposed to >48 h of systemic inflammation driven neuroinflammation in vibratome-cut sections using a custom designed script developed with the Acapella™ image analysis (Verdonk et al., 2016).
Figure 2.
MG/Mϕ have a subtly altered morphology and bi-phase alterations to phenotype in our model of encephalopathy of prematurity. (A) Quantification of the complexity of MG/Mϕ ramifications and process length in CX3CR1GFP/+ mice exposed by intraperitoneal injection of PBS or IL-1β for 3 h (P1) or for 48 h (P3). The panel shows the proportion (%) and the scatter plots of the complexity index and the process length MG/Mϕ (mean, Mann-Whitney test, *P < 0.05, n = 4/group). (B) Representative images of IL-1β induced decrease in the complexity index in GFP+ MG/Mϕ at P3 (scale bar = 25 μm). (C) MG/Mϕ phenotype over time is represented by min to max box and whiskers plots of pro-inflammatory, anti-inflammatory and immuno-regulatory markers levels by RT-qPCR in CD11B+ MG/Mϕ from brain in PBS or IL-1β injected mice. See also Supplementary Figs 1, 2 and Supplementary Table 5. mRNA levels are presented as a fold-change relative to PBS group. Two-way ANOVA with post hoc Bonferroni’s test ,*P < 0.05, **P < 0.01, ***P < 0.001, n/group are indicated under the legend.
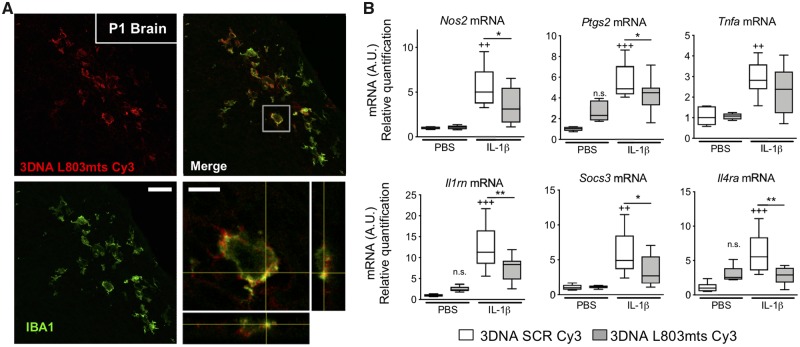
We further studied MG/Mϕ activation using expression analysis of 16 genes and six protein markers previously associated with polarized expression states (Chhor et al., 2013; Miron et al., 2013) at five time points. Gene expression analysis was performed in CD11B+ cells isolated by MACS (Miltenyi Biotec) from PBS- or IL-1β-exposed mice as per our previous studies (Chhor et al., 2017; Krishnan et al., 2017; Shiow et al., 2017). Prior FACS analysis has demonstrated that the ratio of microglia to other CD11B+ cells in this isolate is greater than 50 to 1 (Krishnan et al., 2017). Nevertheless, because we cannot exclude the presence of Mϕ completely we use MG/Mϕ as the descriptor for these cell isolates. Also, the purity of the MACS isolation was confirmed as >95% CD11B+ MG/Mϕ by fluorescence-activated cell sorting (FACS) and qRT-PCR, also as previously (Schang et al., 2014; Krishnan et al., 2017). Quantitative RT-PCR-based gene expression analysis (Fig. 2C) and immunofluorescence (Supplementary Fig. 1A–D) showed that systemic exposure to IL-1β induced a robust pro-inflammatory MG/Mϕ activation state at P1, only 3 h following the first injection of IL-1β. Specifically, five of six markers of a pro-inflammatory state were significantly increased including Il6 increased by 20-fold, and a 5-fold increases in Ptgs2 and Nos2, all P < 0.001. Of note, Ptgs2 and Nos2 remained elevated for 5 days, but Il6 and Tnfa were returned to normal by P3. There was a similar robust increase in markers of immune-regulation at P1 as all four markers examined were increased including an 8-fold increase in Socs3 and >10-fold increase for Sphk1 and Il1rn, all P < 0.001. Expression of markers associated with an anti-inflammatory activation state were the most effected in animals exposed to systemic IL-1β exposure at P2 and P3, which is 48 or 72 h after the start of IL-1β injections. These findings of a temporal change in MG/Mϕ activation profiles were corroborated with immunohistological analysis of protein levels (Supplementary Fig. 1A–D).
By combining our observations of morphology, gene and protein changes in MG/Mϕ after systemic IL-1β exposure we have characterized a persistent state of activation responsible for injury to the developing white matter.
The Wnt pathway is comprehensively downregulated in activated microglia in vivo and in vitro
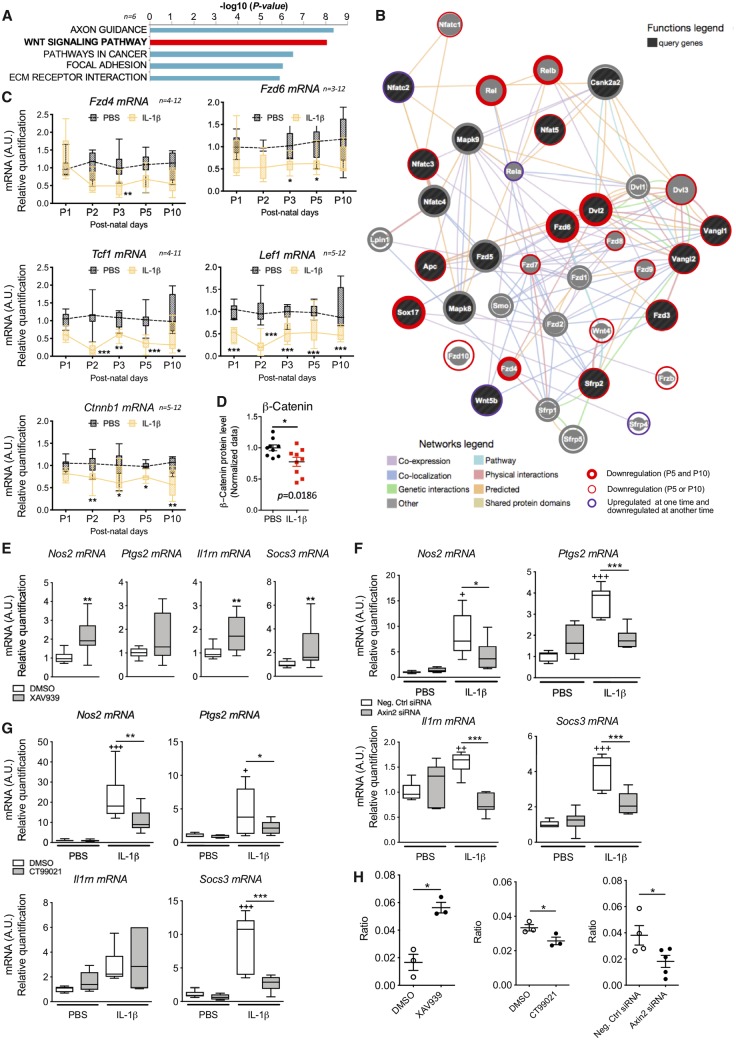
In mouse pups exposed to systemic IL-1β we have characterized a persistent activation of microglia between P1 and until at least P5, which induces a partial blockade of OPC maturation and subsequently hypomyelination (Favrais et al., 2011). Using gene and predicted protein network-based analysis, we sought to identify molecular pathways regulating this MG/Mϕ activation driving OPC injury. We hypothesized that the best time to pick-up the potential disrupting regulator pathways specific to developing microglia was P5, when microglia are activated and when the first effects of OPCs have been observed, and P10, when microglia activation is apparently resolved but injury to OPCs is well established. As such, we undertook genome-wide transcriptomics analysis of MG/Mϕ (CD11B+) and OPCs (O4+) MACSed at P5 and P10 from the brains of mice exposed to IL-1β or PBS. The purity of the O4+ MACS isolation was verified with qPCR (Schang et al., 2014) as outlined for CD11B fractions above. Genes showing differential expression between time points, P5 and P10, were identified for each cell-type, MG/Mϕ (CD11B+) or OPCs (O4+), within each condition, IL-1β or PBS. Gene co-expression networks were inferred for each of the four sets of differentially expressed genes using GGMs (Opgen-Rhein and Strimmer, 2007). GGMs use partial correlations to infer significant co-expression relationships. We applied an FDR of <1% between any microarray probe pair in each of the four sets of differentially expressed genes sets previously identified: PBS-OPC, IL-1β-OPC, PBS-MG/Mϕ and IL-1β-MG/Mϕ. These four co-expression networks were found to exhibit unique topologies and functional annotations (Supplementary Fig. 2A, B and Supplementary Tables 3 and 5). Compared to the three other conditions networks, the IL-1β exposed MG/Mϕ co-expression network was the most highly interconnected and the densest indicating a strong co-regulation of gene expression under this condition (Supplementary Fig. 2A and B). Biological pathway analysis using the Broad Institute MsigDB database (Subramanian et al., 2005) indicated enrichment for Wnt signalling pathway genes within the IL-1β exposed MG/Mϕ network (FDR = 8.5 × 10−7; Fig. 3A and Supplementary Table 3). Because of the proposed importance of the Wnt pathway in OPC maturation (Fancy et al., 2009) we undertook a specific analysis of data from the O4+ OPCs from mice subjected to systemic exposure to IL-1β. Analysis of microarray data from the O4+ OPCs did not show any significant enrichment for Wnt signalling genes in the gene co-expression response (Supplementary Table 3). These data highlight two important facts about Wnt dysregulation: (i) that Wnt dysregulation is likely not a mechanism that directly effects oligodendrocytes in this model; and (ii) that changes in Wnt are not simply ubiquitous in the developing CNS in response to inflammation. Returning to the analysis of MG/Mϕ, to highlight the directionality of the data, we looked for enrichments for Wnt signalling pathway sets among the genes significantly downregulated by IL-1β exposure at P5 and P10 (Supplementary Table 6, see ‘Materials and methods’ section). The known interactions among the Wnt pathway genes plus predicted interactors were then retrieved from an extensive set of functional association data by the GeneMania tool (Warde-Farley et al., 2010) and we present this sub-network in Fig. 3B. We annotated this figure to highlight that the vast majority of targets predicted to interact in this model have downregulated expression. Finally, qualitative gene profiling with qRT-PCR in CD11B+ MG/Mϕ confirmed the downregulation of mRNA expression for numerous Wnt pathway members including multiple Frizzled receptors, Lef1 and Ctnnb1 in MG/Mϕ from IL-1β-exposed mice (Fig. 3C). The downregulation of Ctnnb1 mRNA was also accompanied by a significant decrease in the expression of β-catenin protein in CD11B+ MG/Mϕ at P3 corresponding to the time point when the greatest morphological changes and pro-inflammatory gene activation is observed (Fig. 3D).
Figure 3.
The Wnt pathway is downregulated in pro-inflammatory microglia and modulates microglia activation in vitro. (A) Transcriptomic data reveal that the Wnt signalling pathway is strongly associated with MG/Mϕ activation in our model and (B) builds a cohesive network of multi-modal interactions between Wnt pathway genes in the MG/Mϕ co-expression network. (C) Validation of a selection of Wnt targets from the array data showing min to max box and whiskers plots of mRNA level presented as a fold-change relative to PBS group by RT-qPCR in CD11B+ MG/Mϕ from PBS or IL-1β-exposed mice. See also Supplementary Fig. 3. Two-way ANOVA, post hoc Bonferroni’s test, *P < 0.05, **P < 0.01 ***P < 0.001, n/group is indicated on the graph. (D) Scatter plot of β-catenin protein level from ELISA in CD11B+ MG/Mϕ from PBS or IL-1β-exposed mice at P3. Data are expressed as fold-change relative to PBS group (mean ± SEM, Student’s t-test, *P < 0.05, n = 9/group). (E) β-Catenin pathway inhibition, with XAV939, induced a pro-inflammatory like activation in primary microglia. Min to max box and whiskers plots of mRNA levels are presented as a fold-change relative to vehicle group (Student’s t-test, *P < 0.05, **P < 0.01, n = 12/group). (F) β-Catenin pathway activation induced by blocking Axin2 with siRNA (see also Supplementary Fig. 3I, ∼45% Axin2 mRNA decrease). (G) Inhibition of GSK3β, a β-catenin inhibitor, with CT99021 reduced primary microglia activation induced by IL-1β. Min to max box and whiskers plots of mRNA levels presented as a fold-change relative to vehicle group. One-way ANOVA with post hoc Newman-Keuls’s test. Effects of IL-1β on gene expression are shown with +P < 0.05, ++P < 0.01, +++P < 0.001; effects of Axin2 siRNA or CT99021 to alter the IL-1β effects are shown with *P < 0.05, **P < 0.01, ***P < 0.001, n = 6/group). (H) Effects of modulating Wnt with XAV939, Axin2 siRNA and CT99021 on phosphorylation of β-catenin (PSer45 β-catenin/β-catenin ratio) in primary microglia. Scatter plots, mean ± SEM, Mann-Whitney test *P < 0.05, n = 3/group.
To extend this ex vivo CD11B+ cell data and set up an in vitro model for further study, qRT-PCR and immunofluorescence analysis of mouse primary cultured microglia were undertaken. The cultures are >97% pure for microglia as validated with immunohistochemistry and qRT-PCR, as previously reported (Chhor et al., 2013). Exposure of microglia to IL-1β in vitro induces a similar pro-inflammatory phenotype as is found in MG/Mϕ from mice exposed to systemic IL-1β (Supplementary Fig. 3A and B, cf. data in Fig. 2C). Furthermore, we observed a similar downregulation of Wnt pathway members including mRNA encoding Fzd receptors, Ctnnb1 and Tcf1 (Supplementary Fig. 3C). Inducing a pro-inflammatory activation in microglia in vitro also induced an upregulation of mRNA encoding Axin1 and Axin2, two specific inhibitors of β-catenin nuclear translocation (Supplementary Fig. 3C) and reduced the protein levels for β-catenin as assessed with immunofluorescence (Supplementary Fig. 3D), and ELISA (Supplementary Fig. 3E). There was no significant change in the ratio of phosphorylated (Pser45) β-catenin to β-catenin also assessed with ELISA (Supplementary Fig. 3E). In contrast, IL-4, a classic anti-inflammatory stimulus, increased Wnt/β-catenin activation as indicated by strongly increased Lef1 mRNA and decreased the Pser45 β-catenin/β-catenin ratio as measured by ELISA (Supplementary Fig. 3F and G).
Altogether these data demonstrate that a pro-inflammatory MG/Mϕ phenotype is associated with a robust and comprehensive downregulation of the gene and protein expression of members of the Wnt/β-catenin signalling pathway in vivo and in vitro.
Wnt/β-catenin pathway activity negatively correlates with pro-inflammatory microglia activation in vitro
We next sought to determine if Wnt/β-catenin pathway modulation alone is sufficient to drive phenotypic changes in microglia in vitro. Pharmacological inhibition of the Wnt/β-catenin pathway in vitro with XAV939, a tankyrase inhibitor that stabilizes the Wnt/β-catenin pathway inhibitor AXIN2 (Fancy et al., 2011), was sufficient to promote a pro-inflammatory phenotype, mimicking the effects of IL-1β (Fig. 3E). Conversely, the IL-1β-induced pro-inflammatory microglia phenotype was blocked by activating the Wnt pathway using siRNAs against AXIN2 (Fig. 3F; validation of siRNA efficacy is presented in Supplementary Fig. 3H) or a pharmacological inhibitor of GSK3β that phosphorylates and inactivates β-catenin, CT99021 (Ring et al., 2003) (Fig. 3G). As expected, we verified with ELISA that the Pser45 β-catenin/β-catenin ratio was increased with XAV939, and decreased with Axin2 siRNA and CT99021 (Fig. 3H). In contrast, we tested in PBS and IL-1β-exposed primary microglia a non-isoform specific block of PKC signalling with chelerythrine. Chelerythrine targets the non-canonical Wnt pathways and exposure of doses at 1–3 µM had no effect on microglia activation via qRT-PCR (Supplementary Fig. 4). It is worth noting that all pharmacological approaches are liable to off target effects, but altogether the multiple compounds, and genetic targeting create a cohesive link to the Wnt canonical pathway as being key to microglia activation.
Altogether, these data verify in pure cultures of primary microglia that modulating the Wnt/β-catenin pathway is sufficient and necessary to effect microglia activation state.
Wnt/β-catenin pathway activity negatively correlates with pro-inflammatory microglia activation in vivo
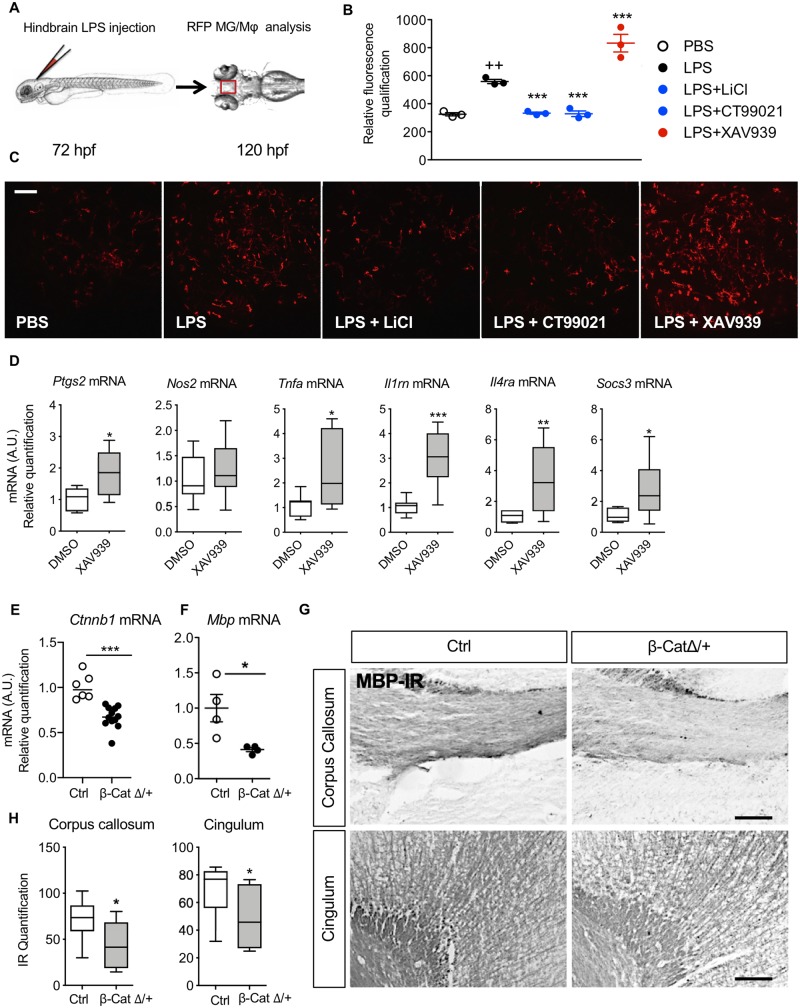
We further tested our working hypothesis that Wnt/β-catenin signalling drives a pro-inflammatory phenotype in MG/Mϕ in vivo using non-cell specific approaches in zebrafish and mice and then transgenic mice with MG/Mϕ specific β-catenin knock-down. In zebrafish larvae at 72 hpf that express RFP in MG/Mϕ (Tg[pU1::Gal4-UAS-TagRFP]) we injected into the hindbrain the prototypical pro-inflammatory agent LPS (Fig. 4A). LPS was used because of the unavailability of recombinant zebrafish IL-1β and prior work demonstrating the expression by zebrafish of receptors for LPS, the Toll-like 4 receptor (TLR4), and the requisite downstream signalling cascade (van der Sar et al., 2006). Although LPS and IL-1β use different signalling pathways, the end result of exposure to both is a complex neuroinflammatory milieu and it is this complex ‘soup’ and not the IL-1β alone that is effective. We compared the nature of the IL-1β and LPS responses in primary microglia using qRT-PCR for 12 genes to highlight the similarities in the overall inflammatory milieu induced by these inflammatory agents (Supplementary Fig. 3A), supporting our previous data comparing inflammatory protocols (Chhor et al., 2013). LPS-induced neuroinflammation in the zebrafish was confirmed after 48 h by a significant increase in fluorescent MG/Mϕ in the optic tectum (Fig. 4B and C). We observe a similar increase in MG/Mϕ immunofluorescence in our mouse model with IBA1 and MAC1 immunofluorescence (Favrais et al., 2011). As expected, exposure of zebrafish larvae to the Wnt/β-catenin antagonist XAV939 exacerbated the LPS-induced MG/Mϕ activation, while conversely, exposure to Wnt agonists acting via the canonical GSK3β dependent pathway with CT99021 or LiCl prevented the LPS-induced MG/Mϕ activation (Fig. 4B and C).
Figure 4.
Wnt/β-catenin pathway regulates MG/Mϕ activation and hypomyelination in vivo. Wnt signalling was modulated in vivo with pharmacological techniques in zebrafish (A–C) and mice (D) and using gene knockout in mice (E–G). (A) Location of LPS microinjection (5 ng/injection) into the zebrafish hindbrain at 72 hpf in pu1::Gal4-UAS::TagRFP morpholinos followed by analysis at 120 hpf. (B) Scatter plot of quantification of RFP labelled microglia and (C) representative images of RFP microglia in these zebrafish brains after LPS injection in the presence or not in their growth solution of Wnt/β-catenin pathway activators LiCl (80 mM) and CT99021 (3 μM) or Wnt inhibitor XAV939 (5 μM) (mean ± SEM, one-way ANOVA with post hoc Newman-Keuls’s test, **P < 0.01, ***P < 0.001). Scale bar = 40 μm. (D) Analysis of the phenotype of CD11B+ cells isolated from P1 mice injected intracerebroventricularly with XAV939 (0.5 nmol) alone (no intraperitoneal IL-1β), demonstrating increased MG/Mϕ activation with Wnt agonism. Min to max box and whiskers plots of quantification of Nos2, Ptgs2, Tnfa, Il1rn, Socs3 and Il4ra mRNA by RT-qPCR. mRNA levels are presented as a fold-change relative to PBS/DMSO group (Student’s t-test, *P < 0.05, **P < 0.01 and ***P < 0.001 n = 7–10/group. (E and F) β-Catenin deficit in microglia driven by β-cateninflox/+/LysMCre/+ (β-cateninΔ/+) induces hypomyelination. Scatter plots of quantification by RT-qPCR of (E) Ctnnb1 mRNA deficit in CD11B+ MG/Mϕ from P10 β-cateninΔ/+ mice (n = 6–12/group) and (F) Mbp mRNA deficit in the anterior brain at P10 in β-cateninΔ/+ mice. mRNA levels are presented as a fold-change relative to control (Ctrl) mice (β-catenin+/+/LysMCre/+) (n = 4/group) (mean, Mann-Whitney test, *P < 0.05, ***P < 0.01). (H) Min to max box and whiskers plot of grey density level of MBP immunoreactivity (MBP-IR) in the corpus callosum and cingulum at P30 relative to control mice. (Mann-Whitney test, *P < 0.05, n = 6/group and illustrated by representative images of MBP-IR in G. Scale bar = 40 μm.
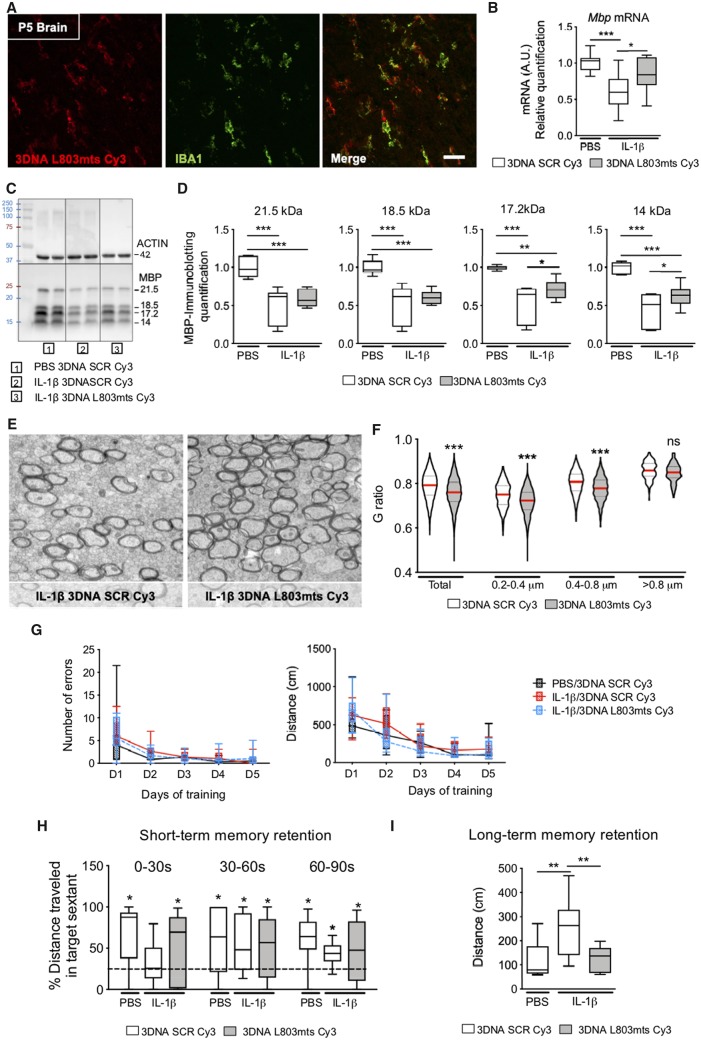
Subsequently, we studied the specific relationship between Wnt/β-catenin pathway activity and MG/Mϕ activation in mice using pharmacological and genetic approaches. First, we inhibited the Wnt/β-catenin pathway by intracerebroventricular injection of XAV939 in P1 pups and isolated CD11B+ MG/Mϕ 3 h later. In agreement with our hypothesis, in vivo Wnt inhibition with XAV939 mimicked the effects of systemic IL-1β on gene expression in MG/Mϕ (Fig. 4D). Furthermore, to target the Wnt pathway in MG/Mϕ specifically, we bred LysMCre/Cre with β-cateninFlox/Flox mice to generate β-CatΔ/+ transgenic mice. Recombination, and as such knockout of β-catenin occurs in ≥20% of MG/Mϕ in this Cre line (Cho et al., 2008; Derecki et al., 2012; Degos et al., 2013) and we verified that there was a 40% decrease in Ctnnb1 mRNA encoding β-catenin (P < 0.001, Fig. 4E). In the absence of external inflammatory stimuli, β-CatΔ/+ mice with microglia expressing reduced levels of β-catenin developed a myelin deficiency that mimicked the damaging effects of systemic IL-1β exposure in our encephalopathy of prematurity model. Specifically, these mice had reduced Mbp mRNA in the anterior cortex at P10 (Fig. 4F) and reduced MBP immunoreactivity in the corpus callosum and cingulum at P30 (Fig. 4G and H).
Collectively, these data show that in fish and mice that downregulation of the Wnt pathway is sufficient and necessary to drive a pro-inflammatory MG/Mϕ activation state. Of note, even a partial MG/Mϕ-specific β-catenin deletion in vivo in the absence of any external stimuli is sufficient to induce MG/Mϕ activation leading to a myelination defect.
The Wnt pathway regulates activation state in primary human microglia in vitro
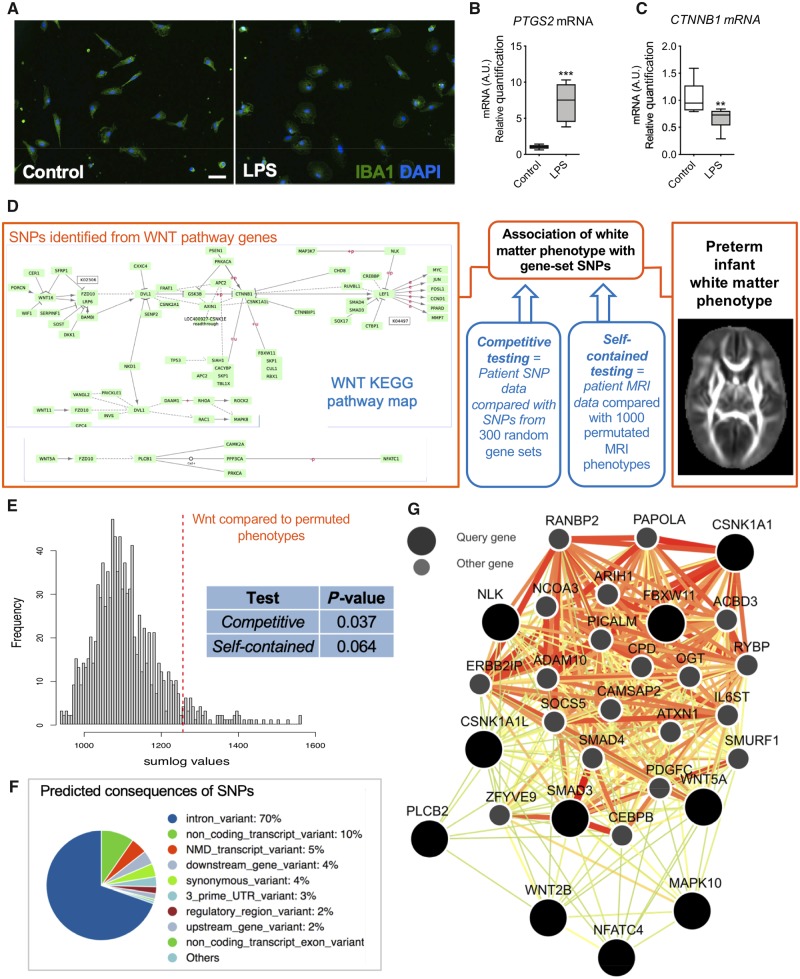
We then verified a similar relationship between decreased Wnt and increased pro-inflammatory activation in primary human MACSed CD11B+ MG/Mϕ isolated from cerebral tissues collected from scheduled terminations at gestational age 19 and 20 weeks. Based on the low numbers of cells available we aimed to ensure we induced a robust response so we chose to use LPS to induce a pro-inflammatory microglia response based on previous studies (Melief et al., 2012). Human primary MG/Mϕ were activated to a pro-inflammatory state as indicated by modified morphology (Fig. 5A) and increased PTGS2 (COX2) gene expression (Fig. 5B). In agreement with observations in our mouse and zebrafish experiments these pro-inflammatory activated human primary microglia had a reduction in their expression of mRNA for the β-catenin gene, CTNNB1 (Fig. 5C).
Figure 5.
Human microglia activation leads to Wnt/β-catenin downregulation and genetic variation in the Wnt pathway associates with preterm infant white matter phenotype. (A) Human primary microglia exposed to LPS showing immunoreactive changes with IBA1 (scale bar = 50 μm); (B) upregulated mRNA expression for COX2 (PTGS2); and (C) the associated decrease in the expression of the gene for β-catenin (CTNNB1). Min to max box and whiskers plots, Student’s t-test, **P < 0.01, ***P < 0.0001, n = 6–9/group. (D) Schematic of the analysis for any association between SNPs in the Wnt pathway and the preterm infant white matter phenotype. Common genetic variation (SNPs) in the WNT gene-set were enriched for variants associated with tractography features when compared to random matched gene-sets, i.e. competitive test, P = 0.037, 1000 permutations, (E, blue inset) and is also associated with white matter probabilistic tractography in preterm infants, compared with the null background, i.e. self-contained test, P = 0.064, 1000 permutations (E, blue inset). (F) The predicted consequences of all of the SNPs found in the top 10 ranked WNT genes shown in Table 1, a total of 42 SNPs. (G) The relationships between these 10 genes significantly associated with the tractography phenotype is reconstructed in a high confidence interaction network specific to the human brain, retrieved from known tissue-specific expression and regulatory data (Greene et al., 2015).
Genomic variance in Wnt pathway genes is relevant to human preterm infant white matter structure
Our experimental data show that genetic knockdown of Wnt/β-catenin signalling is sufficient in itself to drive brain injury in transgenic mice. In a conceptually similar manner, but with an expected much smaller effect size, we reasoned that genetic variation in Wnt pathway genes would be associated with surrogates of white matter-mediated connectivity in infants born preterm. We hypothesized that, at least in part, any link between WNT variation and connectivity would be mediated by differences in the innate response to inflammatory challenge in these preterm infants. Although our genetic analysis is agnostic with respect to cellular mechanism or type, a link between WNT variations and brain structure would support further research into WNTs role in injury, and using WNT variants for preterm-born infant injury/outcome stratification. To test our prediction of a link between WNT and preterm brain connectivity we performed an analysis where we jointly analysed connectivity data derived from MRI and genomics data from 290 preterm-born infants. This approach has previously uncovered novel genetic variants associated with brain connectivity phenotype (Krishnan et al., 2016, 2017). The preterm-born infants included in this study are described in more detail in Supplementary Table 4 and the inclusion/exclusion criteria are outlined in the ‘Materials and methods’ section. Imaging of the white matter in preterm-born infants associates with neurodevelopmental outcomes in preterm-born infants (Woodward et al., 2006; Spittle et al., 2009; Pandit et al., 2013), making an imaging-genomics approach relevant for generating hypotheses about the relevance of the Wnt pathway to functional outcomes for preterm-born infants.
In this paired imaging-genomics analysis, we assessed SNPs in our preterm cohort. SNPs are DNA sequence variations where a single nucleotide varies between individual members of a species that can help pinpoint contributions of specific genes to disease states. To define the intracerebral anatomical connectivity phenotype in our preterm cohort, we used 3 T diffusion MRI and optimized probabilistic tractography as described previously (Robinson et al., 2008; Shi et al., 2011), adjusting the results for post-menstrual age at scan and at birth. Regions of interest for seeding tractography were obtained by segmentation of the brain based on a 90-node anatomical neonatal atlas (Shi et al., 2011) focusing on cortico-cortical connections. DNA extracted from saliva was genotyped and a gene-set of-interest defined using SNPs in the WNT signalling pathway (entry hsa04310) of the KEGG database (n = 141 genes).
SNPs were mapped to all of the genes in the KEGG Wnt pathway using the JAG tool and Genome Reference Consortium Human Build 37 (GRCh37; hg19), using 2 kb up/downstream, in line with NCBI practices (NCBI, 2005). Following SNP-gene mapping, we tested the null hypothesis that variation in Wnt pathway genes is not associated with the preterm tractography phenotype using JAG (Lips et al., 2015). Specifically, genes in the WNT-set were tested in two ways (Fig. 5D). First, we performed a ‘competitive/enrichment test’ to determine whether the Wnt pathway genes were no more associated with the connectivity phenotype than 300 randomly drawn non-WNT-pathway gene-sets. Second, we performed a ‘self-contained/association test’ to determine whether there is a comparable association between the original connectivity phenotype and WNT, versus 1000 random permutations of the phenotype and WNT. The ‘competitive test’, in which we compared to randomly generated sets of genes, showed a significant enrichment of genetic variants associated with the phenotype in the WNT gene-set compared to random gene sets (empirical P = 0.037, Fig. 5E inset). The conservative ‘self-contained’ test in which we compared to 1000 permutated phenotypes indicated that the WNT signalling gene-set was associated with our phenotype; i.e. white matter probabilistic tractography features (empirical P = 0.064; Fig. 5E histogram and inset). We performed a gene-by-gene analysis of SNPs within the WNT signalling gene-set to investigate which genes might be predominantly contributing to the collective pathway enrichment signal. This test highlighted a group of 10 genes with individual significant association with the tractography phenotype (empirical P ≤ 0.05, 1000 permutations). These 10 genes included eight associated with Wnt/β-catenin signalling NFATC4, CSNK1A1, MAPK10, WNT2B, SMAD3, FBXW11, NLK, CSNK1A1L, and two associated with the Wnt/Ca2+ or Wnt/PCP pathways, PLCB2 and WNT5A (Table 1, Fig. 5F and Supplementary Table 7). Although, we cannot determine specifically how these SNP variants would alter the brain phenotype we found using the Genome-scale Integrated Analysis of gene Networks in Tissues (GIANT) tool that these genes created a coherent interaction network within a human brain-specific gene interaction network (Fig. 5G and Supplementary Table 8). GIANT hosts searchable genome-scale functional maps of human tissues, integrating an extensive collection of experimental datasets from publications including expression, regulatory and protein data (Greene et al., 2015). We also performed an exploratory investigation of function for the 42 SNPs from the 10 genes predominantly contributing to the collective pathway enrichment signal to provide information related to function. We used the Gene Effect Predictor within the Consortium Human Build 37 (GRCh37; hg19) tool to study the 42 SNPs, whose full details are given in Supplementary Table 9. All 42 SNPs existed in the database and they were predicted to have 463 consequences on the genome (Supplementary Table 10). Specifically, these SNPs overlapped with 103 separate gene transcripts, seven of them overlapped regulatory sequences and overall 95% were synonymous variants and 5% were missense variants (Fig. 5F).
Table 1.
Functional descriptions of the 10 genes within the WNT gene-set
| Gene symbol | Description | Adjusted P-value | nSNP |
|---|---|---|---|
| NFATC4 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | 0.001 | 9 |
| CSNK1A1 | Casein kinase 1, alpha 1 | 0.002 | 52 |
| MAPK10 | Mitogen-activated protein kinase 10 | 0.002 | 4 |
| WNT2B | Wingless-type MMTV integration site family, member 2B | 0.007 | 6 |
| SMAD3 | SMAD family member 3 | 0.014 | 57 |
| FBXW11 | F-box and WD repeat domain containing 11 | 0.015 | 4 |
| WNT5A | Wingless-type MMTV integration site family, member 5A | 0.023 | 12 |
| NLK | Nemo-like kinase | 0.026 | 11 |
| CSNK1A1L | Casein kinase 1, alpha 1-like | 0.033 | 11 |
| PLCB2 | Phospholipase C, beta 2 | 0.038 | 5 |
These linked imaging-genomic analyses demonstrate a relationship between genetic variation in the Wnt pathway and an important clinical phenotype. They provide adjunct support for a causal relationship between the Wnt pathway and the brain phenotype that has been established with our experimental data.
The neuroprotective effects of a Wnt pathway agonist delivered via 3DNA nanocarrier
There is a paucity of neurotherapeutic options for damage to the preterm-born infant brain and many other neuroinflammatory-mediated neurological disorders. We have clearly demonstrated a link between decreased Wnt/β-catenin pathway activation and a pro-inflammatory microglia activation state. In addition, we have shown the relevance of this pathway in human microglia and preterm infant brain structure. As such, we complete this study by demonstrating that the Wnt pathway is a viable therapeutic target. To achieve this, we specifically increased Wnt pathway activity in MG/Mϕ using a peptide inhibitor of GSK3β, which has the effect of preventing the formation of the β-catenin degradation complex. We delivered this peptide to MG/Mϕ in vitro and in vivo by conjugating it to a 3DNA nanocarrier together with a fluorescent tag (Cy3) (Genisphere). 3DNA nanocarriers are interconnected monomeric subunits of DNA of ∼200 nM in diameter that are intrinsically capable of endosomal escape (Muro, 2014). In brief, we validated that our 3DNA-peptide conjugate had microglia specificity and immunomodulatory efficacy in vitro, that is crosses the blood–brain barrier, that it has cell specificity in vivo, and finally that it is neurotherapeutic in vivo in our systemic-inflammation (IL-1β) driven model of encephalopathy of the preterm-born infant.
First, we demonstrated in vitro that 3DNA nanocarrier Cy3 tagged conjugates are specifically internalized by microglia in mixed glial cultures (Supplementary Fig. 5A) and by microglia in primary cultures after 4 h of incubation (Supplementary Fig. 5B). When 3DNA was conjugated to both Cy3 and the peptide L803mts, an agonist of canonical Wnt signalling via inhibition of GSK3β, it was also specifically internalized by microglia in primary culture (Supplementary Fig. 6A). We also used the 3DNA nanocarrier tagged with Cy3 to deliver a cargo of L803mts to MG/Mϕ that had been activated with IL-1β in vitro. Treatment of primary microglia with L803mts delivered with 3DNA reversed the IL-1β-induced activation of the microglia but had no effect on PBS-treated microglia as assessed with gene expression analysis (Supplementary Fig. 6B).
Moving in vivo, we aimed to generate the proof-of-concept that the 3DNA nanocarrier delivering L803mts and tagged with Cy3 can be taken up by MG/Mϕ and influence MG/Mϕ phenotype. Specifically, the L803mts and Cy3 conjugated 3DNA nanocarrier or the scrambled (SCR) peptide control and Cy3 conjugated 3DNA nanocarrier was injected intracerebroventricular into P1 mice, 1 h prior to our standard intraperitoneal IL-1β injection. Four hours after intracerebroventricular injection, we observed Cy3 fluorescence exclusively co-localized with IBA-1+ staining in the periventricular white matter (Fig. 6A and Supplementary Video 1). In the same paradigm, using qRT-PCR analysis of isolated CD11B+ MG/Mϕ we demonstrated that intracerebroventricularly delivered 3DNA loaded with L803mts and tagged with Cy3 reversed the effect of IL-1β on the MG/Mϕ phenotype but that the 3DNA-L803mts-Cy3 had no significant effect on MG/Mϕ from PBS-treated mice (Fig. 6B). As intracerebroventricular injections cause aggravated tissue injury we subsequently undertook a translationally orientated trial using intraperitoneal delivery of Cy3-tagged 3DNA conjugated to L803mts. The 3DNA L803mts Cy3 or 3DNA SCR Cy3 control was injected intraperitoneal daily between P1 and P5, in parallel with PBS or IL-1β. At P5, immunofluorescence in the anterior periventricular white matter confirmed that 3DNA L803mts Cy3 was specifically taken up by IBA1+ MG/Mϕ (Fig. 7A). We also assessed uptake of our 3DNA conjugate by populations of macrophage in the body after intraperitoneal delivery at P5. Using the same immunohistochemistry techniques that clearly show uptake in MG/Mϕ we did not find Cy3 positive staining in the spleen or liver (Supplementary Fig. 5C and D).
Figure 6.
Nanocarrier delivery of a Wnt/β-catenin pathway activator L803mts regulates MG/Mϕ activation in our encephalopathy of prematurity model. (A) Representative images of the in vivo uptake of 3DNA by IBA1+ cells in the subventricular white matter 4 h after intracerebroventricular administration of 3DNA SCR Cy3 (control) or 3DNA L803mts Cy3 (200 ng); scale bar = 40 μm. Bottom right panel shows orthogonal view of white box inset showing co-localization in detail, scale bar = 10 μm. See also Supplementary Figs 4A, B, 5, and Supplementary Video 1. (B) Min to max box and whiskers plots of Nos2, Ptgs2, Tnfa, Il1rn, Socs3 and Il4ra mRNA by qRT-PCR in CD11B+ MG/Mϕ cells from intracerebroventricular injected PBS or IL-1β-treated mice with 3DNA SCR Cy3 or 3DNA L803mt at P1. mRNA levels are presented as a fold-change relative to vehicle group (one-way ANOVA with post hoc Newman-Keuls’s test, ++P < 0.01, +++P < 0.001 for statistically significant difference between the PBS and IL-1β groups also exposed to 3DNA SCR Cy3. *P < 0.05, **P < 0.01 for the effects of 3DNA Wnt modulator delivery, n = 5–6 for PBS groups and n = 9 for IL-1β groups).
Figure 7.
Nanocarrier delivery of a Wnt/β-catenin pathway activator L803mts in microglia reduces IL-1β-induced hypomyelination and memory deficit. (A) Representative images of in vivo uptake of 3DNA by IBA1+ cells in subventricular white matter at P5 after intraperitoneal injections of 3DNA L803mts Cy3 or 3DNA SCR Cy3 (500 ng/injection). Scale bar = 40 μm. (B) Min to max box and whiskers plot of Mbp mRNA by qRT-PCR in the anterior brain of mice at P10. mRNA levels are presented as a fold-change relative to the 3DNA SCR Cy3/PBS group (one way-ANOVA with Newman-Keuls’s test, *P < 0.05, **P < 0.01, n = 9–12/group). See also Supplementary Fig. 5C and D. (C) Representative image of actin and MBP immuno-blot; and (D) the min to max box and whiskers plots of quantification of the four isoforms of MBP in the anterior brain of mice at P15. Protein levels were normalized to actin and are presented as a fold-change relative to the 3DNA SCR Cy3/PBS group (one way-ANOVA with Newman-Keuls’s test, *P < 0.05, **P < 0.01, ***P < 0.001, n = 6–8/group). (E) Representative image of electron microscope (EM) image of the corpus callosum from mice at P30. (F) Violin plots of the G-ratios from the EM analysis showing improvements in the Wnt agonist group, 3DNA L803mts (Kruskall-Wallis test, ***P < 0001 n = 4–5/group). Microglial targeted Wnt agonism also induced functional improvements. (G) Data from trials of the Barnes maze for spatial learning. (H) Short-term memory retention and (I) long-term memory retention in 3-month-old mice. See also Supplementary Fig. 6. Memory retention present in the 30-s trial period on Day 5 of the test (probe trial, in H) was measured by recording of the distance travelled in the target sextant (min to max box and whiskers plot, univariate t-test *P < 0.05 n = 10/group). (I) The long-term memory deficits were measured via the distance travelled to reach the target on the 15th day after the start of testing (min to max box and whiskers plot, one-way ANOVA with Bonferroni post hoc was used **P < 0.01. n = 9/group).
To validate the in vivo neurotherapeutic efficacy of our novel nanocarrier-mediated WNT agonist therapy, we assessed the IL-1β-induced myelination deficit by measuring Mbp mRNA at P10, MBP protein via western blotting at P15 and axonal myelination via electron microscopy at P30. We have previously shown in this model that there is an accumulation of immature oligodendrocytes (NG2+ and PDGFRa+), reduced small calibre axonal myelination and reduced expression of myelin proteins (Favrais et al., 2011; Schang et al., 2014; Rangon et al., 2018).Treatment with intraperitoneal 3DNA L803mts Cy3 significantly prevented the IL-1β-induced reduction in Mbp mRNA (Fig. 7B) and alleviated the IL-1β-induced reduction in MBP proteins (Fig. 7C, D and Supplementary Fig. 7A). Finally, we observed that these effects of 3DNA L803mt Cy3 to improve MBP mRNA and MBP protein levels correlated with improvements in axonal myelination, as indicated by the decrease of G-ratio of fibres of 0.2 to 0.8 μm diameter in the corpus collosum (Fig. 7E and F).
To evaluate whether this improvement in neuropathology with nanocarrier-mediated treatment also reverses the behavioural deficits observed in adults in this paradigm, spatial learning and memory were assessed starting at P90. First we tested basic behavioural parameters by spontaneous locomotion and rearing using actimetry, and exploratory behaviour using an open field test. This injury model is designed to induce subtle neurobehavioural deficits to mimic the learning problems that become apparent in school age children born preterm (Lindstrom et al., 2011; Spittle et al., 2017). As expected, no changes in these basic parameters was observed between groups of mice, i.e. controls (PBS+3DNA SCR Cy3), injured (IL-1β+SRC Cy3) or the treatment group (IL-1β+3DNA L803mts Cy3) (Supplementary Fig. 7B and C). Spatial learning and memory were tested via the Barnes maze test, which assesses spatial learning over 5 days of probe trials plus short term memory retention at the end of the learning trials and long term memory retention 10 days later. Spatial learning (i.e. distance travelled and number of errors) itself was not modified by IL-1β exposure (Fig. 7G). However, mice exposed to IL-1β had a short-term memory deficit based on a decreased distance travelled in the target sextant during the first 30 s of the probe trial on the final day of testing (Fig. 7H). In addition, IL-1β exposure caused deficits in long term memory recall, as assessed as the increased distance travelled in a single probe trial at 10 days post-learning (Fig. 7I). These short term and long term neuroinflammatory induced memory deficits were both reversed by intraperitoneal treatment by delivery to MG/Mϕ of the Wnt antagonist, L803mts (3DNA L803mts Cy3) by the 3DNA nanocarrier (Fig. 6H and I).
Altogether these data clearly demonstrate that a Wnt agonist therapy can reduce pro-inflammatory MG/Mϕ activation and improve neuropathological and functional outcomes in a model of neuroinflammation mediated white matter injury. These data also highlight a novel 3DNA nanocarrier that has the ability to deliver therapies like this in vivo specifically to microglia.
Discussion
Recent research has highlighted that microglia of the developing brain have a unique phenotype. The aims of this study were to identify in microglia of the developing brain the molecular regulators of injury-related phenotype, and to validate them as therapeutic targets for preventing microglia-mediated injury in the 15 million infants born preterm every year. We undertook a comprehensive set of studies involving pharmacological and genetic manipulations in mouse and zebrafish models, human tissue and patient data. We also introduced an innovative 3DNA nanocarrier that can deliver drugs specifically to microglia in vivo. Initially, we characterized the complex MG/Mϕ phenotype in our mouse model of encephalopathy of prematurity (Favrais et al., 2011; Schang et al., 2014; Krishnan et al., 2017) and demonstrated that MG/Mϕ activation is directly responsible in this model for the clinically relevant myelination defect. Next, using a genome-wide transcriptomic analysis of these activated MG/Mϕ we associated the Wnt pathway with MG/Mϕ activation. We specifically determined that the expression of Wnt/β-catenin pathway receptors, ligands and intracellular signalling components were robustly downregulated in MG/Mϕ isolated from our animal model. A combination of approaches in zebrafish and mouse models in vitro and in vivo allowed us to determine that Wnt/β-catenin pathway inhibition specific to MG/Mϕ is sufficient and necessary to induce in these cells a pro-inflammatory activation, and subsequent hypomyelination in vivo. We highlighted the relevance of the Wnt pathway to the activation state of human primary microglia, and preterm-born infant brain development. Of note, we verified our prediction that genetic variation in the Wnt pathway in preterm infants is related to indices of their cerebral structural connectivity. Although, it is a limitation of these types of human studies that the data are not cell specific and the effects of the WNT SNPs may be on earlier stages of development in other cell types. Finally, we selectively delivered to MG/Mϕ in vivo a Wnt pathway activator using a 3DNA nanocarrier, which we administered via non-invasive intraperitoneal injection. This specific and non-invasive delivery of a Wnt agonist therapeutic prevented a pro-inflammatory MG/Mϕ activation state and the associated white matter damage and cognitive deficit in our mouse model. Together, these data demonstrate that downregulation of the canonical Wnt/β-catenin pathway triggers a hypomyelination-inducing microglia phenotype and validate this pathway as a novel and clinically viable neurotherapeutic target thanks to the MG/Mϕ specificity of 3DNA nanocarriers.
Our evidence supports that the Wnt pathway controlling MG/Mϕ phenotype is the canonical Wnt/β-catenin pathway. Our supporting data include that genetic and pharmacological manipulation of the β-catenin degradation complex via AXIN2 robustly effected MG/Mϕ phenotype in vitro and in vivo. In contrast, blockade of non-canonical Wnt signalling with a PKC inhibitor had no effect on microglia activation. Although, we acknowledge that there may still be a role for other facets of non-canonical signalling in the complexity of MG/Mϕ functions. However, MG/Mϕ-specific β-catenin ablation in vivo in-of-itself was sufficient to recapitulate the systemic-inflammation-driven hypomyelination that we see in our animal model of encephalopathy of prematurity. We also found that the WNT genes with variance associated with preterm infant connectivity were predominantly (8/10) from the Wnt/β-catenin pathway. A key role for β-catenin would also begin to explain our observations of Wnt-mediated pro-inflammatory activation via TLR-4 and IL-1 receptor agonists and also conversely Wnt-mediated anti-inflammatory activation via a IL-4 receptor agonist. Downstream signalling of both the TLR/IL-1 and IL-4 receptor families includes activation of the transcription factor NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) (Caamano and Hunter, 2002). The ability of NF-κB to mediate the opposing effects of these receptors on microglia phenotype is conferred by interactions with other regulatory factors, including at multiple levels indirectly and directly by β-catenin (Ma and Hottiger, 2016).
Ours is the first study to demonstrate a robust cell-intrinsic role of the Wnt/β-catenin pathway in microglia activation in vitro and in vivo and validate that this pathway is a viable immunomodulatory neurotherapeutic. Previous work on Wnt and microglia has been limited to in vitro ligand-receptor interactions, specifically WNT3A or WNT5A. However, the specific Wnt pathway activated by a Wnt ligand is dependent on the context of ligand-receptor interaction and not only cell-intrinsic properties (van Amerongen et al., 2008). This fact likely explains the discordance between the data from the previous studies, where in some paradigms Wnt exposure is anti-inflammatory (Halleskog et al., 2012; Yu et al., 2014) or in others the exposure was pro-inflammatory (Halleskog et al., 2011; Hooper et al., 2012; Halleskog and Schulte, 2013) with no relation to the predicted canonical or non-canonical pathway being activated. Altogether these data highlight the difficulty that would be faced in using ligand-receptor interactions to target the Wnt pathway to modulate microglia activity, in contrast with our success of targeting the intracellular cascade of Wnt/β-catenin pathway specifically using our 3DNA-mediated delivery system. We also wish to note that the Wnt pathway has well characterized roles in oligodendrocyte maturation (Fancy et al., 2011; Feigenson et al., 2011; Guo et al., 2015; Zhao et al., 2016), astrocyte activation (Cao et al., 2012) and endothelial cell function (Lengfeld et al., 2017). Our study builds on this understanding of the role of Wnt as a complex cell specific temporally and spatially regulated controller of brain development and response to injury.
The 3DNA nanocarrier is a powerful translational tool as is it can take agents across the blood–brain barrier following intraperitoneal injection, is specifically internalized by MG/Mϕ in the brain and it has no toxicity in vitro or in vivo in our study, or in human retinal tissue in vitro (Gerhart et al., 2017) and in preclinical in vivo targeted delivery of a ovarian cancer therapy (Huang et al., 2016). Wnt activation in OPCs has previously been suggested to block their maturation by a member of our current research team (Fancy et al., 2011). As such it was important that 3DNA nanocarriers allowed us to target this pathway exclusively in MG/Mϕ. 3DNA has a non-injury dependent mechanism of traversing the blood–brain barrier, as it crossed in naïve and inflammation-exposed mice, although the specific mechanism is unknown. We have previously reported that the blood–brain barrier in our mouse model of encephalopathy of prematurity is primarily intact (Krishnan et al., 2017). The chemical and physical attributes of 3DNA likely precludes it from entering the brain via paracellular aqueous routes or transcellular lipophilic pathways. However, investigations of receptor-mediated transcytosis are underway, as similar aptamer-based DNA duplexes can interact with receptors such as nucleolin or transferrin (Reyes-Reyes et al., 2010).
In preterm infants, it is exposure to inflammatory events that are both difficult to identify or prevent, such as chorioamnionitis and sepsis, which is the leading risk factor for white matter damage. This damage is caused by microglia-mediated neuroinflammatory processes; a link that is supported by post-mortem human studies showing microgliosis in the developing white matter of preterm-born infants (Verney et al., 2010, 2012; Supramaniam et al., 2013) and verified by numerous experimental paradigms (see references in Hagberg et al., 2015). White matter damage in these infants is observed as diffuse white matter signal changes and reduced structural connectivity using MRI analyses (Shah et al., 2008; Spittle et al., 2009). In a large animal model of preterm-born infant brain injury a direct link between diffuse white matter injury and white matter gliosis has been made (Riddle et al., 2011). This reduced structural connectivity in preterm-born infants is in turn strongly associated with poor neurodevelopmental outcomes (Woodward et al., 2006; Ball et al., 2015). As such, the purpose of our imaging-genomics analysis was to link the preclinical observations of microglial-mediated compromise of structural connectivity due to white matter injury to genetic variance that could alter the microglial inflammatory response in our preterm cohort and would affect changes in structural connectivity. Our integrated analysis of imaging and genomics demonstrated that common genetic variation in Wnt pathway genes does indeed influence brain structural connectivity features within our preterm-born infants. We fully acknowledge that the data we derived are not cell-specific and could be due to effects on the white and grey matter. However, it supports in contemporaneous infants the relevance of the Wnt pathway using an imaging modality that is currently used to assess injury and predict outcome. Also, we wish to highlight that changes to the grey matter are also evident in preterm-born infants, including changes in cortical microstructure (Ball et al., 2013), interneuron distribution (Stolp et al., 2019) and degeneration of axons (Back and Miller, 2014). However, the contribution of axonal injury to diffuse white matter is controversial, although it is evident in necrotic foci that are approximated to occur in only 5% of white matter injury cases (Riddle et al., 2011; Buser et al., 2012). Preterm-born infants with apparent focal necrosis were excluded from our imaging-genomics analysis removing a significant contribution of a frank axonopathy from effecting the connectivity phenotype in this cohort. We cannot exclude, however, that effects on the grey matter in these infants are not important determiners of outcome and effected by changes in SNPs in genes of the Wnt pathway. In addition, we found that the Wnt pathway genes containing SNPs associated with white matter tractography phenotype belonged to a human brain-specific gene interaction network. This network further builds a case for a functional effect of WNT SNPs in human preterm infants, with consequences for the development of white matter structure (Greene et al., 2015). Specific prediction of the consequences of our identified SNPs found the majority were intron variants. Previous studies in experimental animals have shown that immune cell intron variants often effect cell function by altering enhancer binding (Farh et al., 2015). Altogether our clinical data build a case that these Wnt pathway SNPs may be a useful way to stratify infants who are at highest risk for white matter damage, and to improve on our currently limited prognostic abilities related to long term outcome.
In conclusion, the activity of the Wnt/β-catenin pathway regulates MG/Mϕ activation, the Wnt pathway is relevant in human brain development, and specific agonism of the Wnt/β-catenin pathway in MG/Mϕ in vivo using 3DNA nanocarrier-mediated drug delivery has a beneficial effect on MG/Mϕ phenotype, myelination and cognitive deficits. This is strong evidence that Wnt signalling modulation may be a promising therapeutic approach in encephalopathy of prematurity and, potentially, in other disorders involving MG/Mϕ-mediated neuroinflammation.
Supplementary Material
Acknowledgements
Our thanks to the children and families who participated in the study, and the nurses, doctors and scientists who supported the project. We also wish to thank Dr Dominique Langui (Institut du Cerveau et de la Moelle épinière, Hôpital Pitié-Salpêtrière, Paris, France) for providing us with access to electron microscopy facilities and Dr Manuela ZinniI INSERM U1141 NeuroDiderot for access to additional molecular biology facilities.
Funding
This study was supported by grants from Inserm, Université Paris Diderot, Université Sorbonne-Paris-Cité, Investissement d'Avenir (ANR-11-INBS-0011, NeurATRIS), ERA-NET Neuron (Micromet), DHU PROTECT, Association Robert Debré, PremUP, Fondation de France, Fondation pour la Recherche sur le Cerveau, Fondation des Gueules Cassées, Roger de Spoelberch Foundation, Grace de Monaco Foundation, Leducq Foundation, Action Medical Research, Cerebral Palsy Alliance Research Foundation Australia, Wellcome Trust (WSCR P32674) and The Swedish Research Council (2015-02493). We wish to acknowledge the support of the Department of Perinatal Imaging and Health, King’s College London. In addition, the authors acknowledge financial support from the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Competing interests
The authors report no competing interests.
Glossary
Abbreviations
- MACS =
magnetically-activated cell sorting
- MG/Mϕ =
microglia/infiltrating macrophages
- OPC =
oligodendrocyte progenitor cell
- SNP =
single nucleotide polymorphism
References
- Back SA, Luo NL, Mallinson RA, O'Malley JP, Wallen LD, Frei B, et al. Selective vulnerability of preterm white matter to oxidative damage defined by F2-isoprostanes. Ann Neurol 2005; 58: 108–20. [DOI] [PubMed] [Google Scholar]
- Back SA, Miller SP. Brain injury in premature neonates: A primary cerebral dysmaturation disorder? Ann Neurol 2014; 75: 469–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G, Counsell SJ, Anjari M, Merchant N, Arichi T, Doria V, et al. An optimised tract-based spatial statistics protocol for neonates: applications to prematurity and chronic lung disease. Neuroimage 2010; 53: 94–102. [DOI] [PubMed] [Google Scholar]
- Ball G, Pazderova L, Chew A, Tusor N, Merchant N, Arichi T, et al. Thalamocortical Connectivity Predicts Cognition in Children Born Preterm. Cereb Cortex 2015; 25: 4310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G, Srinivasan L, Aljabar P, Counsell SJ, Durighel G, Hajnal JV, et al. Development of cortical microstructure in the preterm human brain. Proc Natl Acad Sci USA 2013; 110: 9541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA 2016; 113: E1738–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiards SS, Haynes RL, Folkerth RD, Borenstein NS, Trachtenberg FL, Rowitch DH, et al. Myelin abnormalities without oligodendrocyte loss in periventricular leukomalacia. Brain Pathol 2008; 18: 153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman JP, Walley A, Ball G, Takousis P, Krishnan ML, Hughes-Carre L, et al. Common genetic variants and risk of brain injury after preterm birth. Pediatrics 2014; 133: e1655–63. [DOI] [PubMed] [Google Scholar]
- Buchanan FG, DuBois RN. Connecting COX-2 and Wnt in cancer. Cancer Cell 2006; 9: 6–8. [DOI] [PubMed] [Google Scholar]
- Buser JR, Maire J, Riddle A, Gong X, Nguyen T, Nelson K, et al. Arrested preoligodendrocyte maturation contributes to myelination failure in premature infants. Ann Neurol 2012; 71: 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci 2014; 17: 131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caamano J, Hunter CA. NF-kappaB family of transcription factors: central regulators of innate and adaptive immune functions. Clin Microbiol Rev 2002; 15: 414–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Yin A, Wen G, Sheikh AM, Tauqeer Z, Malik M, et al. Alteration of astrocytes and Wnt/beta-catenin signaling in the frontal cortex of autistic subjects. J Neuroinflamm 2012; 9: 223. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chhor V, Le Charpentier T, Lebon S, Ore MV, Celador IL, Josserand J, et al. Characterization of phenotype markers and neuronotoxic potential of polarised primary microglia in vitro. Brain Behav Immun 2013; 32: 70–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhor V, Moretti R, Le Charpentier T, Sigaut S, Lebon S, Schwendimann L, et al. Role of microglia in a mouse model of paediatric traumatic brain injury. Brain Behav Immun 2017; 63: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho IH, Hong J, Suh EC, Kim JH, Lee H, Lee JE, et al. Role of microglial IKKbeta in kainic acid-induced hippocampal neuronal cell death. Brain 2008; 131 (Pt 11): 3019–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann O, Leviton A. Inflammatory brain damage in preterm newborns–dry numbers, wet lab, and causal inferences. Early Hum Dev 2004; 79: 1–15. [DOI] [PubMed] [Google Scholar]
- Degos V, Peineau S, Nijboer C, Kaindl AM, Sigaut S, Favrais G, et al. G protein-coupled receptor kinase 2 and group I metabotropic glutamate receptors mediate inflammation-induced sensitization to excitotoxic neurodegeneration. Ann Neurol 2013; 73: 667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delobel-Ayoub M, Arnaud C, White-Koning M, Casper C, Pierrat V, Garel M, et al. Behavioral problems and cognitive performance at 5 years of age after very preterm birth: the EPIPAGE Study. Pediatrics 2009; 123: 1485–92. [DOI] [PubMed] [Google Scholar]
- Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SB, Guyenet PG, et al. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature 2012; 484: 105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Wang P, Yu Y, Chen F, Liu J, Li Y. Gadolinium chloride improves the course of TNBS and DSS-induced colitis through protecting against colonic mucosal inflammation. Sci Rep 2014; 4: 6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature protocols 2009; 4: 1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, et al. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev 2009; 23: 1571–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Harrington EP, Baranzini SE, Silbereis JC, Shiow LR, Yuen TJ, et al. Parallel states of pathological Wnt signaling in neonatal brain injury and colon cancer. Nat Neurosci 2014; 17: 506–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Harrington EP, Yuen TJ, Silbereis JC, Zhao C, Baranzini SE, et al. Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. Nat Neurosci 2011; 14: 1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015; 518: 337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favrais G, van de Looij Y, Fleiss B, Ramanantsoa N, Bonnin P, Stoltenburg-Didinger G, et al. Systemic inflammation disrupts the developmental program of white matter. Ann Neurol 2011; 70: 550–65. [DOI] [PubMed] [Google Scholar]
- Feigenson K, Reid M, See J, Crenshaw EB 3rd, Grinspan JB. Wnt signaling is sufficient to perturb oligodendrocyte maturation. Mol Cell Neurosci 2009; 42: 255–65. [DOI] [PubMed] [Google Scholar]
- Feigenson K, Reid M, See J, Crenshaw IE, Grinspan JB. Canonical Wnt signalling requires the BMP pathway to inhibit oligodendrocyte maturation. ASN Neuro 2011; 3: e00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci G, Dmitrieva N, Gianni D, Fontana EJ, Pan X, Lu Y, et al. Depletion of peripheral macrophages and brain microglia increases brain tumor titers of oncolytic viruses. Cancer Res 2007; 67: 9398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Greenbaum M, Casta L, Clemente A, Mathers K, Getts R, et al. Antibody-Conjugated, DNA-Based Nanocarriers Intercalated with Doxorubicin Eliminate Myofibroblasts in Explants of Human Lens Tissue. J Pharmacol Exp Ther 2017; 361: 60–7. [DOI] [PubMed] [Google Scholar]
- Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet 2015; 47: 569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Lang J, Sohn J, Hammond E, Chang M, Pleasure D. Canonical Wnt signaling in the oligodendroglial lineage–puzzles remain. Glia 2015; 63: 1671–93. [DOI] [PubMed] [Google Scholar]
- Hagberg H, Gressens P, Mallard C. Inflammation during fetal and neonatal life: implications for neurologic and neuropsychiatric disease in children and adults. Ann Neurol 2012; 71: 444–57. [DOI] [PubMed] [Google Scholar]
- Hagberg H, Mallard C. Effect of inflammation on central nervous system development and vulnerability. Curr Opin Neurol 2005; 18: 117–23. [DOI] [PubMed] [Google Scholar]
- Hagberg H, Mallard C, Ferriero DM, Vannucci SJ, Levison SW, Vexler ZS, et al. The role of inflammation in perinatal brain injury. Nat Rev Neurol 2015; 11: 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halleskog C, Dijksterhuis JP, Kilander MB, Becerril-Ortega J, Villaescusa JC, Lindgren E, et al. Heterotrimeric G protein-dependent WNT-5A signaling to ERK1/2 mediates distinct aspects of microglia proinflammatory transformation. J Neuroinflamm 2012; 9: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halleskog C, Mulder J, Dahlstrom J, Mackie K, Hortobagyi T, Tanila H, et al. WNT signaling in activated microglia is proinflammatory. Glia 2011; 59: 119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halleskog C, Schulte G. WNT-3A and WNT-5A counteract lipopolysaccharide-induced pro-inflammatory changes in mouse primary microglia. J Neurochem 2013; 125: 803–8. [DOI] [PubMed] [Google Scholar]
- Hamrick SE, Miller SP, Leonard C, Glidden DV, Goldstein R, Ramaswamy V, et al. Trends in severe brain injury and neurodevelopmental outcome in premature newborn infants: the role of cystic periventricular leukomalacia. J Pediatr 2004; 145: 593–9. [DOI] [PubMed] [Google Scholar]
- Haynes RL, Billiards SS, Borenstein NS, Volpe JJ, Kinney HC. Diffuse axonal injury in Periventricular Leukomalacia as determined by apoptotic marker fractin. Pediatr Res 2008; 63: 656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S, Izzy S, Sen P, Morsett L, El Khoury J. Microglia in neurodegeneration. Nat Neurosci 2018; 21: 1359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier SL, Witkin SS, Krohn MA, Watts DH, Kiviat NB, Eschenbach DA. The relationship of amniotic fluid cytokines and preterm delivery, amniotic fluid infection, histologic chorioamnionitis, and chorioamnion infection. Obstet Gynecol 1993; 81: 941–8. [PubMed] [Google Scholar]
- Hooper C, Sainz-Fuertes R, Lynham S, Hye A, Killick R, Warley A, et al. Wnt3a induces exosome secretion from primary cultured rat microglia. BMC Neurosci 2012; 13: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, et al. Delivery of therapeutics targeting the mRNA-binding protein HuR using 3DNA nanocarriers suppresses ovarian tumor growth. Cancer Res 2016; 76: 1549–59. [DOI] [PubMed] [Google Scholar]
- Johnston MV, Hagberg H. Sex and the pathogenesis of cerebral palsy. Dev Med Child Neurol 2007; 49: 74–8. [DOI] [PubMed] [Google Scholar]
- Kaidanovich-Beilin O, Eldar-Finkelman H. Long-term treatment with novel glycogen synthase kinase-3 inhibitor improves glucose homeostasis in ob/ob mice: molecular characterization in liver and muscle. J Pharmacol Exp Ther 2006; 316: 17–24. [DOI] [PubMed] [Google Scholar]
- Keats EC, Dominguez JM 2nd, Grant MB, Khan ZA. Switch from canonical to noncanonical Wnt signaling mediates high glucose-induced adipogenesis. Stem cells (Dayton, Ohio) 2014; 32: 1649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 2017; 47: 566–81.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan ML, Van Steenwinckel J, Schang AL, Yan J, Arnadottir J, Le Charpentier T, et al. Integrative genomics of microglia implicates DLG4 (PSD95) in the white matter development of preterm infants. Nat Commun 2017; 8: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan ML, Wang Z, Silver M, Boardman JP, Ball G, Counsell SJ, et al. Possible relationship between common genetic variation and white matter development in a pilot study of preterm infants. Brain Behav 2016; 6: e00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengfeld JE, Lutz SE, Smith JR, Diaconu C, Scott C, Kofman SB, et al. Endothelial Wnt/beta-catenin signaling reduces immune cell infiltration in multiple sclerosis. Proc Natl Acad Sci USA 2017; 114: E1168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Adair-Rohani H, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380: 2224–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom K, Lindblad F, Hjern A. Preterm birth and attention-deficit/hyperactivity disorder in schoolchildren. Pediatrics 2011; 127: 858–65. [DOI] [PubMed] [Google Scholar]
- Lips ES, Cornelisse LN, Toonen RF, Min JL, Hultman CM, International Schizophrenia C, et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol Psychiatry 2012; 17: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips ES, Kooyman M, de Leeuw C, Posthuma D. JAG: A computational tool to evaluate the role of gene-sets in complex traits. Genes 2015; 6: 238–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Hottiger MO. Crosstalk between Wnt/beta-Catenin and NF-kappaB Signaling Pathway during Inflammation. Front Immunol 2016; 7: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marret S, Mukendi R, Gadisseux JF, Gressens P, Evrard P. Effect of ibotenate on brain development: an excitotoxic mouse model of microgyria and posthypoxic-like lesions. J Neuropathol Exp Neurol 1995; 54: 358–70. [DOI] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 2016; 353: aad8670. [DOI] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol 1980; 85: 890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melief J, Koning N, Schuurman KG, Van De Garde MD, Smolders J, Hoek RM, et al. Phenotyping primary human microglia: tight regulation of LPS responsiveness. Glia 2012; 60: 1506–17. [DOI] [PubMed] [Google Scholar]
- Michell-Robinson MA, Touil H, Healy LM, Owen DR, Durafourt BA, Bar-Or A, et al. Roles of microglia in brain development, tissue maintenance and repair. Brain 2015; 138 (Pt 5): 1138–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron VE, Boyd A, Zhao JW, Yuen TJ, Ruckh JM, Shadrach JL, et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat Neurosci 2013; 16: 1211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto A, Wake H, Ishikawa AW, Eto K, Shibata K, Murakoshi H, et al. Microglia contact induces synapse formation in developing somatosensory cortex. Nat Commun 2016; 7: 12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Hennessy EM, Myles J, Johnson SJ, Draper ES, Costeloe KL, et al. Neurological and developmental outcome in extremely preterm children born in England in 1995 and 2006: the EPICure studies. BMJ 2012; 345: e7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro S. A DNA Device that Mediates Selective Endosomal Escape and Intracellular Delivery of Drugs and Biologicals. Adv Funct Mater 2014; 24: 2899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI. SNP FAQ Archive [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2005-. The dbSNP Mapping Process. 2005. Available from: https://www.ncbi.nlm.nih.gov/books/NBK44455/ (25 November 2018, date last accessed).
- Nosarti C, Giouroukou E, Healy E, Rifkin L, Walshe M, Reichenberg A, et al. Grey and white matter distribution in very preterm adolescents mediates neurodevelopmental outcome. Brain 2008; 131 (Pt 1): 205–17. [DOI] [PubMed] [Google Scholar]
- Nosarti C, Nam KW, Walshe M, Murray RM, Cuddy M, Rifkin L, et al. Preterm birth and structural brain alterations in early adulthood. Neuroimage Clin 2014; 6: 180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opgen-Rhein R, Strimmer K. From correlation to causation networks: a simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Syst Biol 2007; 1: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit AS, Ball G, Edwards AD, Counsell SJ. Diffusion magnetic resonance imaging in preterm brain injury. Neuroradiology 2013; 55 (Suppl 2): 65–95. [DOI] [PubMed] [Google Scholar]
- Perry VH, Nicoll JA, Holmes C. Microglia in neurodegenerative disease. Nat Rev Neurol 2010; 6: 193–201. [DOI] [PubMed] [Google Scholar]
- Rangon CM, Schang AL, Van Steenwinckel J, Schwendimann L, Lebon S, Fu T, et al. Myelination induction by a histamine H3 receptor antagonist in a mouse model of preterm white matter injury. Brain Behav Immun 2018; 74: 265–76. [DOI] [PubMed] [Google Scholar]
- Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res 2010; 70: 8617–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle A, Dean J, Buser JR, Gong X, Maire J, Chen K, et al. Histopathological correlates of magnetic resonance imaging-defined chronic perinatal white matter injury. Ann Neurol 2011; 70: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 2003; 52: 588–95. [DOI] [PubMed] [Google Scholar]
- Rivest S. Regulation of innate immune responses in the brain. Nat Rev Immunol 2009; 9: 429–39. [DOI] [PubMed] [Google Scholar]
- Robinson EC, Valstar M, Hammers A, Ericsson A, Edwards AD, Rueckert D. Multivariate statistical analysis of whole brain structural networks obtained using probabilistic tractography. Med Image Computing Comput-Assist Intervent 2008; 11 (Pt 1): 486–93. [DOI] [PubMed] [Google Scholar]
- Schafer J, Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics 2005; 21: 754–64. [DOI] [PubMed] [Google Scholar]
- Schang AL, Van Steenwinckel J, Chevenne D, Alkmark M, Hagberg H, Gressens P, et al. Failure of thyroid hormone treatment to prevent inflammation-induced white matter injury in the immature brain. Brain Behav Immun 2014; 37: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah DK, Doyle LW, Anderson PJ, Bear M, Daley AJ, Hunt RW, et al. Adverse neurodevelopment in preterm infants with postnatal sepsis or necrotizing enterocolitis is mediated by white matter abnormalities on magnetic resonance imaging at term. J Pediatr 2008; 153: 170–5, 175.e1. [DOI] [PubMed] [Google Scholar]
- Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, et al. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One 2011; 6: e18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Kagawa T, Wada T, Muroyama Y, Takada S, Ikenaka K. Wnt signaling controls the timing of oligodendrocyte development in the spinal cord. Dev Biol 2005; 282: 397–410. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Smits R, Ikenaka K. Microglia-induced activation of non-canonical Wnt signaling aggravates neurodegeneration in demyelinating disorders. Mol Cell Biol 2016; 36: 2728–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiow LR, Favrais G, Schirmer L, Schang AL, Cipriani S, Andres C, et al. Reactive astrocyte COX2-PGE2 production inhibits oligodendrocyte maturation in neonatal white matter injury. Glia 2017; 65: 2024–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieger D, Moritz C, Ziegenhals T, Prykhozhij S, Peri F. Long-range Ca2+ waves transmit brain-damage signals to microglia. Dev Cell 2012; 22: 1138–48. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002; 17: 143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article3. [DOI] [PubMed] [Google Scholar]
- Sokol SY. Spatial and temporal aspects of Wnt signaling and planar cell polarity during vertebrate embryonic development. Semin Cell Dev Biol 2015; 42: 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittle AJ, Boyd RN, Inder TE, Doyle LW. Predicting motor development in very preterm infants at 12 months' corrected age: the role of qualitative magnetic resonance imaging and general movements assessments. Pediatrics 2009; 123: 512–7. [DOI] [PubMed] [Google Scholar]
- Spittle AJ, Walsh JM, Potter C, McInnes E, Olsen JE, Lee KJ, et al. Neurobehaviour at term-equivalent age and neurodevelopmental outcomes at 2 years in infants born moderate-to-late preterm. Dev Med Child Neurol 2017; 59: 207–15. [DOI] [PubMed] [Google Scholar]
- Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, et al. Microglia modulate wiring of the embryonic forebrain. Cell reports 2014; 8: 1271–9. [DOI] [PubMed] [Google Scholar]
- Stolp BH, Fleiss B, Arai Y, Supramaniam V, Vontell R, Birtles S, et al. Interneuron development is disrupted in preterm brains with diffuse white matter injury: observations in mouse and human. Frontiers in physiology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supramaniam V, Vontell R, Srinivasan L, Wyatt-Ashmead J, Hagberg H, Rutherford M. Microglia activation in the extremely preterm human brain. Pediatr Res 2013; 73: 301–9. [DOI] [PubMed] [Google Scholar]
- Thion MS, Ginhoux F, Garel S. Microglia and early brain development: An intimate journey. Science 2018; 362: 185–9. [DOI] [PubMed] [Google Scholar]
- Tran FH, Zheng JJ. Modulating the wnt signaling pathway with small molecules. Protein science : a publication of the Protein Society 2017; 26: 650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twilhaar ES, Wade RM, de Kieviet JF, van Goudoever JB, van Elburg RM, Oosterlaan J. Cognitive outcomes of children born extremely or very preterm since the 1990s and associated risk factors a meta-analysis and meta-regression. JAMA Pediatr 2018; 172: 361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R. Alternative wnt signaling is initiated by distinct receptors. Sci Signal 2008; 1: re9. [DOI] [PubMed] [Google Scholar]
- van der Sar AM, Stockhammer OW, van der Laan C, Spaink HP, Bitter W, Meijer AH. MyD88 innate immune function in a zebrafish embryo infection model. Infect Immun 2006; 74: 2436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haastert IC, Groenendaal F, Uiterwaal CS, Termote JU, van der Heide-Jalving M, Eijsermans MJ, et al. Decreasing incidence and severity of cerebral palsy in prematurely born children. J Pediatr 2011; 159: 86–91.e1. [DOI] [PubMed] [Google Scholar]
- Verdonk F, Roux P, Flamant P, Fiette L, Bozza FA, Simard S, et al. Phenotypic clustering: a novel method for microglial morphology analysis. J Neuroinflamm 2016; 13: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verney C, Monier A, Fallet-Bianco C, Gressens P. Early microglial colonization of the human forebrain and possible involvement in periventricular white-matter injury of preterm infants. J Anat 2010; 217: 436–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verney C, Pogledic I, Biran V, Adle-Biassette H, Fallet-Bianco C, Gressens P. Microglial reaction in axonal crossroads is a hallmark of noncystic periventricular white matter injury in very preterm infants. J Neuropathol Exp Neurol 2012; 71: 251–64. [DOI] [PubMed] [Google Scholar]
- Volpe JJ. Brain injury in premature infants: a complex amalgam of destructive and developmental disturbances. Lancet neurology 2009a; 8: 110–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe JJ. The encephalopathy of prematurity–brain injury and impaired brain development inextricably intertwined. Semin Pediatr Neurol 2009b; 16: 167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010; 38(Web Server issue): W214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SA, Boddeke HW, Kettenmann H. Microglia in Physiology and Disease. Annu Rev Physiol 2017; 79: 619–43. [DOI] [PubMed] [Google Scholar]
- Wood NS, Marlow N, Costeloe K, Gibson AT, Wilkinson AR. Neurologic and developmental disability after extremely preterm birth. EPICure Study Group. N Engl J Med 2000; 343: 378–84. [DOI] [PubMed] [Google Scholar]
- Woodward LJ, Anderson PJ, Austin NC, Howard K, Inder TE. Neonatal MRI to predict neurodevelopmental outcomes in preterm infants. N Engl J Med 2006; 355: 685–94. [DOI] [PubMed] [Google Scholar]
- Wu HC, Shen CM, Wu YY, Yuh YS, Kua KE. Subclinical histologic chorioamnionitis and related clinical and laboratory parameters in preterm deliveries. Pediatr Neonatol 2009; 50: 217–21. [DOI] [PubMed] [Google Scholar]
- Yu CH, Nguyen TT, Irvine KM, Sweet MJ, Frazer IH, Blumenthal A. Recombinant Wnt3a and Wnt5a elicit macrophage cytokine production and tolerization to microbial stimulation via Toll-like receptor 4. Eur J Immunol 2014; 44: 1480–90. [DOI] [PubMed] [Google Scholar]
- Yuen TJ, Silbereis JC, Griveau A, Chang SM, Daneman R, Fancy SP, et al. Oligodendrocyte-encoded HIF function couples postnatal myelination and white matter angiogenesis. Cell 2014; 158: 383–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017; 36: 1461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng Y, Liu L, Yu K, Zhang L, Wang H, et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat Commun 2016; 7: 10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All new data are available from the authors on request, subject to ethical restrictions related to the human studies.