Abstract
Acute Lymphoblastic Leukemia (ALL) is the most common childhood neoplasia. Studies have shown that susceptibility to ALL may be modulated by genetic variables. Our study investigated 21 genetic variants in the susceptibility of the population of the Brazilian Amazon region to B-cell ALL. The variants of the genes GGH, CEBPE, ARID5B, MTHFR and MTHFD1 were related to a protective effect against the development of ALL, whereas the variant of the gene ATIC was associated with a risk effect. The results suggest that genetic variants analyzed modulate of the risk of developing ALL in the studied population.
Keywords: ALL, Pediatrics, Polymorphisms, Susceptibility, Admixed
1. Introduction
Acute Lymphoblastic Leukemia (ALL) is the most common type of childhood hematopoietic neoplasia, and is responsible for 30% of all pediatric cancers [1,2], and is one of the principal causes of cancer deaths in children around the world [3]. The etiology of ALL is still poorly understood, although studies have shown that it may be provoked by a combination of environmental exposure and genetic susceptibility [4,5].
Recently, a number of studies of genome-wide association (GWAS) have found single nucleotide polymorphisms (SNPs) in the ARID5B, IKZF1, CEBPE, CDKN2A, and PIP4K2A genes [6], [7], [8], [9], [10] that were related to the risk of developing ALL, specifically, the B cell lineage of the leukemia (B-cell ALL). While each of these variants is responsible for only a minor increase in the risk of developing ALL, they may have a cumulative influence on the genetic susceptibility to the disease [10]. In addition to their influence on genetic susceptibility to ALL, the prevalence of some of these variants may oscillate significantly among patients of different ancestries, which may account for ethnic differences found in the incidence of the disease [10].
In addition to the polymorphisms discovered by the GWAS, one potential genetic pathway in the development of childhood ALL is folate biosynthesis. Folate is involved in the metabolism that plays an essential role in the synthesis, repair, and methylation of the DNA. Reduced ingestion of folate during pregnancy may result in breakage of the DNA molecule, as well as a reduction in repairs and abnormal methylation, which has led to the proposal of a link between polymorphisms in the genes involved in the biosynthesis of folate and the risk of developing ALL [11,12]. These polymorphisms may be associated with genes that codify the central regulator and the transport enzymes (for example, MTHFD1, MTRR, MTHFR, SHMT1, GGH, SLCO1B1 and transporters of the ABC family) involved in the folate transport cycle, as well as the genes involved in the synthesis of purines, such as AMPD1 and ATIC [11,13,14].
Most of the research that identified these genetic variants, either by GWAS or within the scope of the folate metabolism, has focused on European populations. In this case, the patterns of risk associated with these variants in highly admixed populations, such as that of the Brazilian Amazon region are completely unknown. Understanding the potential impact of these risk-associated variants is especially important in the case of variants that are substantially more frequent in non-European populations than in European ones, in order to provide important insights for the prediction of the incidence of the disease in these populations.
The present study investigated the role of 21 polymorphisms in the susceptibility to B-cell ALL in the population of the Brazilian Amazon region. These polymorphisms included five (the ARID5B, IKZF1, CEBPE, CDKN2A, and PIP4K2A genes) selected based on GWAS studies, and 16 (MTHFD1, MTRR, MTHFR, SHMT1, GGH, SLCO1B1, ABCC1, ABCC2, ABCC3, AMPD1, and ATIC genes) related to folate biosynthesis.
2. Patients and methods
2.1. Ethical aspects
The present study was approved by the research committee of the Federal University of Pará (UFPA). Consent for the collection of biological samples and clinical data was obtained personally from each participant prior to the study.
2.2. Cases and controls
The participants in the research were selected based on a case-control study approach. The case group was composed of 121 patients with B-cell ALL diagnosed at two public hospitals (the Ophir Loyola Hospital and the Octavio Lobo Childhood Cancer Hospital) in the city of Belém, Pará (Brazil) that are reference institutions for the treatment of childhood cancer in the Amazon region. The patients were diagnosed between 2006 and 2016, based on the criteria of the French-American-British (FAB) classification systems. The immunophenotyping was determined by flow cytometry [15]. The control group was composed of 155 unrelated individuals from the same socioeconomic level and geographic area as the members of the case group.
2.3. Selection of the genes and polymorphisms
The present study investigated the role of 21 polymorphisms (Supplementary Table S1) in the susceptibility to B-cell ALL. Five of these (the ARID5B, IKZF1, CEBPE, CDKN2A, and PIP4K2A genes) were selected based on GWAS studies. The remaining 16 polymorphisms (the MTHFD1, MTRR, MTHFR, SHMT1, GGH, SLCO1B1, ABCC1, ABCC2, ABCC3, AMPD1, and ATIC genes) are related to folate biosynthesis.
Supplementary Table S1.
Characteristics of the polymorphisms analyzed in the present study and quality control.
| Gene | SNP, ID | Allele | Function | Change in amino acid | Quality Control |
|---|---|---|---|---|---|
| ABCC1 | rs28364006 | A > G | Missense | Thr1337Ala | Genotyping failure and no HWE |
| ABCC2 | rs717620 | C > T | 5′ UTR | – | Genotyping failure |
| ABCC3 | rs9895420 | T > A | 5′ Flanking | – | No HWE |
| AMPD1 | rs17602729 | G > A | Stop Codon | Gln45Ter | No HWE |
| ARID5B | rs10821936 | C > T | Intron | – | Accepted |
| ATIC | rs2372536 | C > G | Missense | Thr116Ser | No HWE |
| ATIC | rs4673993 | T > C | Splice region | – | Accepted |
| CDKN2A | rs3731217 | A > C | Intergenic | – | No HWE |
| CEBPE | rs2239633 | G > A | 5′ UTR | – | Accepted |
| GGH | rs11545078 | G > A | Missense | Thr151Ile | No HWE |
| GGH | rs1800909 | A > G | Missense | Cys6Arg | Accepted |
| GGH | rs3758149 | G > A | 5′ Flanking | – | Accepted |
| IKZF1 | rs4132601 | T > G | UTR '3 | – | No HWE |
| MTHFD1 | rs2236225 | G > A | Missense | Arg653Gln | Accepted |
| MTHFR | rs1801133 | G > A | Missense | Ala222Val | Accepted |
| MTRR | rs1801394 | A > G | Missense | Ile22Met | Genotyping failure and no HWE |
| PIP4K2A | rs7088318 | C > A | Intergenic | – | No HWE |
| SHMT1 | rs1979277 | G > A | Missense | Leu435Phe | No HWE |
| SLCO1B1 | rs2306283 | A > G | Missense | Asn130Asp | No HWE |
| SLCO1B1 | rs4149015 | G > A | 5′ Flanking | – | No HWE |
| SLCO1B1 | rs4149056 | T > C | Missense | Val174Ala | No HWE |
3UTR: 3′UTR regulation; 5UTR: 5′UTR regulation.
2.4. Genotyping of the polymorphisms
The genetic material was extracted from peripheral blood samples of patients in remission, using the commercial Biopur Mini Spin Plus – 250 extraction kit (Biopur, Brazil), based on the manufacturer's instructions, and quantified using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies,Wilmington, DE). The polymorphisms were genotyped using the TaqMan OpenArray Genotyping technology (Applied Biosystems, Life Technologies, Carlsbad, USA) in the QuantStudio™ 12 K Flex Real-Time PCR system (Applied Biosystems, Life Technologies, Carlsbad, USA), using the protocol published by Applied Biosystems. The Taqman Genotyper software was used to analyze the plate data and evaluate the precision of the genotype readings, as well as to control the quality of the genotyping.
2.5. RNA extraction, reverse transcriptase-polymerase chain reaction (RT-PCR)
Cytogenetic data regarding the defining gene fusions of the ALL subtypes: BCR-ABL, ETV6-RUNX1, MLL-AF4, SIL-TAL and TCF3-PBX1 were obtained for 99 B-cell ALL patients investigated. Hyperdiploidy data were not available for most patients and were therefore not included in the study.
Venipuncture and blood collection containing anticoagulant (EDTA) from patients with ALL were performed. The blood was submitted to Ficoll Histopaque® (Sigma-Aldrich, USA) according to the manufacturer's protocol for lymphocyte separation. Subsequently, it was subjected to RNAeasy Mini Kit processing (Qiagen, USA) as standard protocol for total RNA extraction and cDNA conversion using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) according to manufacturer's instructions. For gene fusion analysis, the c-DNA obtained was used to amplify molecular targets by Polymerase Chain Reaction with the GoTaq® Colorless Master Mix kit (Promega, USA), according to the protocol instructions, using primers designed for RT-PCR multiplex reaction for fusions of interest, similar to those described by Galehdari et al. [16] with modifications.
2.6. Analysis of genetic ancestry
The genetic ancestry of the samples was analyzed based on the set of 61 Ancestry Informative Markers (AIMs) described by Santos et al. [17] and Ramos et al. [18]. The proportions of European, African, and Native American ancestries were estimated using STRUCTURE v2.3.3, assuming the existence of three parental populations. To infer the contribution of each parental population, additional data from Native American, Sub-Saharan, and European populations were included and further details on these populations can be found in Amador et al. [19].
2.7. Statistical analysis
The statistical analyses were run in R v.3.4.0 (R Foundation for Statistical Computing). The first step was to verify whether each polymorphism was in Hardy-Weinberg Equilibrium (HWE). The ancestry indices were compared between samples using Mann-Whitney's U test. The influence of the genetic variants on susceptibility to B-cell ALL was evaluated using a Bayesian multivariate logistic regression, which included sex and genetic ancestry to control for potential confounding effects. The effect of each variant was evaluated using recessive, dominant, and log-additive models, with the model being selected based on the lowest AIC (Akaike Information Criterion) value for the association test. A Bayesian logistic regression was used to control for the effects of rare mutations. The significance of the tests was adjusted for multiple comparisons by the Bonferroni correction, with adjusted p values lower than 0.05 being considered statistically significant.
3. Results
3.1. Genotyping
The first step in the analysis of the 21 polymorphisms was to exclude the loci with (i) allele frequencies of less than 1% (MAF≤1%), (ii) for which less than 90% of the samples were genotyped, and (iii) when the control group was not in HWE (Supplementary Table S2). Once these criteria were applied, seven polymorphisms were left to be included in the association analysis.
Supplementary Material Table S2.
Selection criteria for the inclusion of the polymorphisms investigated in the present study.
| Gene | SNP, ID | Quality Control |
|---|---|---|
| ABCC1 | rs28364006 | Genotyping failure and no HWE |
| ABCC2 | rs717620 | Genotyping failure |
| ABCC3 | rs9895420 | No HWE |
| AMPD1 | rs17602729 | No HWE |
| ARID5B | rs10821936 | Accepted |
| ATIC | rs2372536 | No HWE |
| ATIC | rs4673993 | Accepted |
| CDKN2A | rs3731217 | No HWE |
| CEBPE | rs2239633 | Accepted |
| GGH | rs11545078 | No HWE |
| GGH | rs1800909 | Accepted |
| GGH | rs3758149 | Accepted |
| IKZF1 | rs4132601 | No HWE |
| MTHFD1 | rs2236225 | Accepted |
| MTHFR | rs1801133 | Accepted |
| MTRR | rs1801394 | Genotyping failure and no HWE |
| PIP4K2A | rs7088318 | No HWE |
| SHMT1 | rs1979277 | No HWE |
| SLCO1B1 | rs2306283 | No HWE |
| SLCO1B1 | rs4149015 | No HWE |
| SLCO1B1 | rs4149056 | No HWE |
3.2. Demographic characteristics of the patients
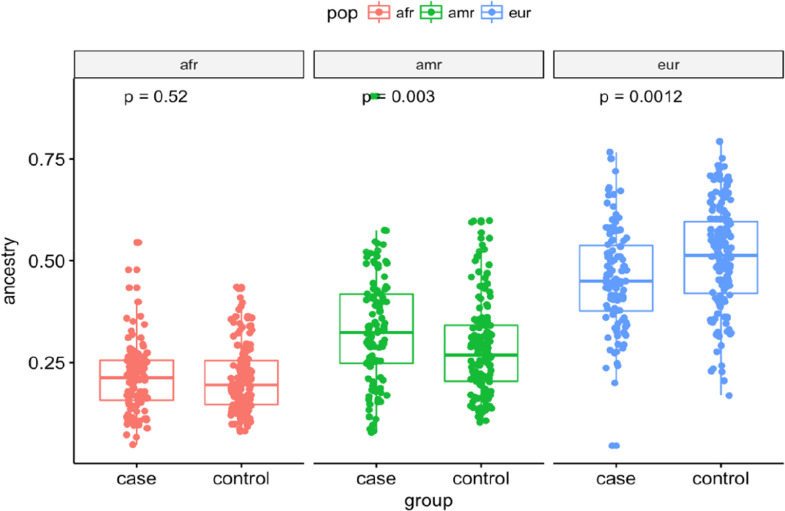
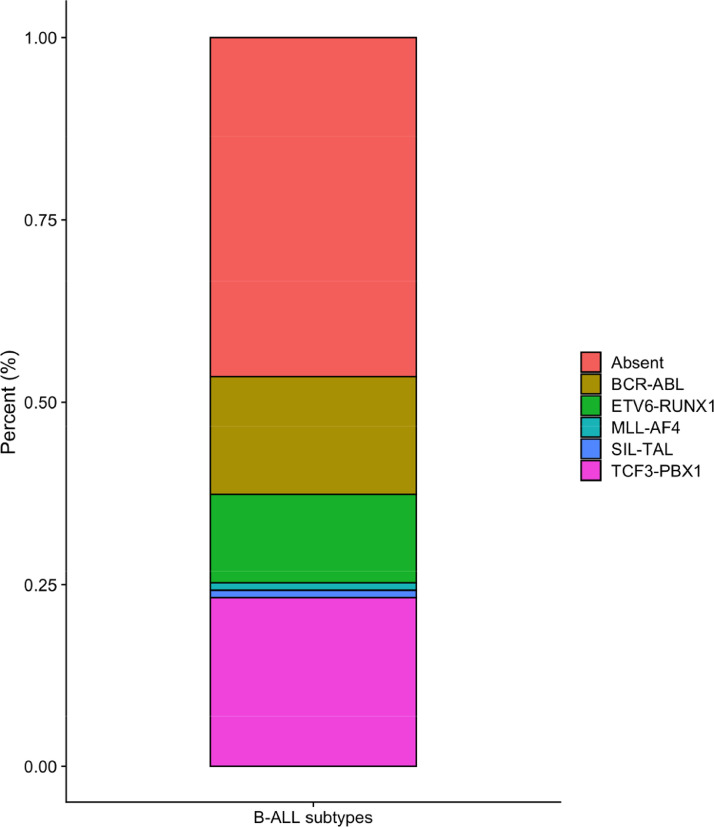
The two groups (case and control) presented distinct demographic characteristics (Table 1). While the case group was predominantly male, the control group had a higher proportion of females. Significant differences were found between the two groups in age (P < 0.001), sex (P = 0.001), and European (P = 0.001) and Amerindian ancestry (P = 0.001). The genetic ancestry of the case group was 45% European, 21% African, and 34% Amerindian, whereas the control group was 50% European 21% African, and 29% Amerindian (Fig. 1). Cytogenetic data for the BCR-ABL, ETV6-RUNX1, MLL-AF4, SIL-TAL and TCF3-PBX1 fusions were available for approximately 82% of the investigated patients (Fig. S1 Supplementary). 46.5% of the patients had none of the translocations studied. The most frequent fusion was TCF3-PBX1, followed by BCR-ABL and ETV6-RUNX1 (Table 1).
Table 1.
Demographic variables of the participants of the present study, by group (case or control).
| Variable | B-cell ALL | Control | P-valor |
|---|---|---|---|
| Number of subjects | 121 | 155 | |
| Age, years* | 5.29±3.32 | 23.77±5.48 | <0.001a |
| Gender (Male/Female) | 72/49 | 61/94 | 0.001 |
| Genetic Ancestry* | |||
| European | 0.451±0.103 | 0.507±0.128 | 0.001b |
| African | 0.213±0.073 | 0.209±0.081 | 0.520b |
| Amerindian | 0.335±0.113 | 0.284±0.113 | 0.003b |
| Chromosomal translocations, n (%) | 99 | – | – |
| Absent | 46 (46.5) | – | – |
| BCR-ABL | 16 (16.2) | – | – |
| ETV6-RUNX1 | 12 (12.1) | – | – |
| MLL-AF4 | 1 (1.0) | – | – |
| TCF3-PBX1 | 23 (23.2) | – | – |
| SIL-TAL | 1 (1.0) | – | – |
Significance determined by Student's t-test.
Significance determined by Mann-Whitney test.
Mean (±Standard Deviation).
Fig. 1.
Box plot graph elucidates the difference between case and control groups for Amerindian, African and European ancestries in the study population.
Fig. 1S.
Distribution of cytogenetic subtypes of B-ALL for the patients investigated.
3.3. Analysis of the association with susceptibility
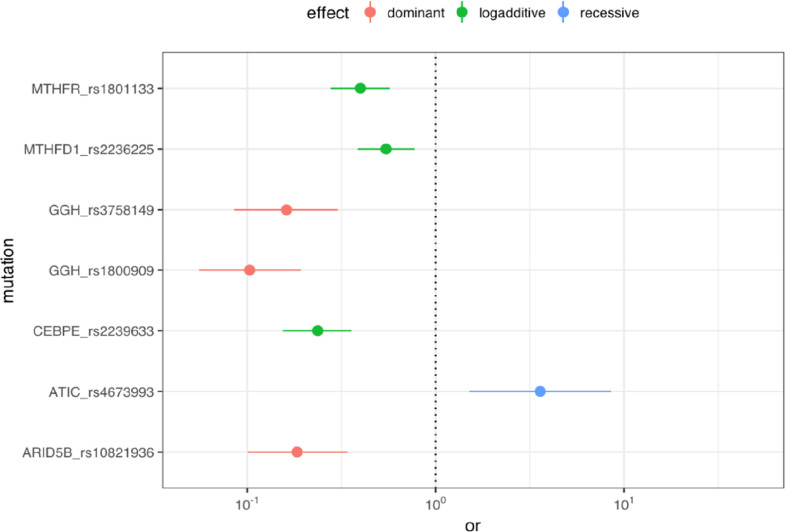
All seven study polymorphisms presented a significant association with susceptibility to B-cell ALL (Table 2). In the case of the GGH (rs1800909, rs3758149), CEBPE (rs2239633), ARID5B (rs10821936), MTHFR (rs1801133), and MTHFD1 (rs2236225) genes, the dominant or log-additive models indicated a significant protective effect in relation to the development of B-cell ALL. In the case of the ATIC (rs4673993) gene, the recessive model indicated a significant association with the risk of development of B-cell ALL in the study population (Fig. 2).
Table 2.
Distribution of the genotypes associated with susceptibility to B-cell ALL.
| Genotype, ID | Model | B-cell ALL (%) | Control (%) | P | OR (95%CI) |
|---|---|---|---|---|---|
| ARID5B_ rs10821936 | Dominant | < 0.001 | CT+TT vs CC: 0.183 (0.100–0.341) |
||
| CC | 57 (47.5) | 20 (13.5) | |||
| CT | 39 (32.5) | 84 (56.8) | |||
| TT | 25 (20.0) | 44 (29.7) | |||
| ATIC_ rs4673993 | Recessive | 0.004 | CC vs TC+TT: 3.594 (1.509–8.518) |
||
| TT | 61 (50.4) | 78 (51.7) | |||
| TC | 40 (33.1) | 65 (43) | |||
| CC | 20 (16.5) | 8 (5.3) | |||
| CEBPE_ rs2239633 | Log-additive | < 0.001 | 0.237 (0.154–0.357) | ||
| GG | 54 (44.6) | 12 (7.8) | |||
| GA | 47 (38.8) | 61 (39.6) | |||
| AA | 20 (16.5) | 81 (52.6) | |||
| GGH_ rs1800909 | Dominant | < 0.001 | AG+GG vs AA: 0.103 (0.056–0.192) |
||
| AA | 97 (80.8) | 48 (32.2) | |||
| AG | 13 (10.8) | 82 (55.0) | |||
| GG | 10 (8.3) | 19 (12.8) | |||
| GGH_ rs3758149 | Dominant | < 0.001 | GA+AA vs GG: 0.162 (0.085–0.303) |
||
| GG | 58 (62.4) | 37 (24.2) | |||
| GA | 11 (11.8) | 81 (52.9) | |||
| AA | 24 (25.8) | 35 (22.9) | |||
| MTHFD1_ rs2236225 | Log-additive | < 0.001 | 0.546 (0.385–0.776) | ||
| GG | 47 (38.8) | 29 (19.0) | |||
| GA | 47 (38.8) | 62 (40.5) | |||
| AA | 27 (22.3) | 62 (40.5) | |||
| MTHFR_ rs1801133 | Log-additive | < 0.001 | 0.399 (0.277–0.571) | ||
| GG | 55 (45.5) | 29 (19.2) | |||
| GA | 42 (34.7) | 61 (40.4) | |||
| AA | 24 (19.8) | 61 (40.4) |
OR: odds ratio; CI: confidential interval. P: Logistic regression adjusted for sex and genetic ancestry.
Fig. 2.
Odds ratio of the polymorphisms investigated between case and control groups. The plot illustrates the polymorphisms odds-ratio and their 95% confidence interval by the point and line ranges, the null effect (OR = 1) is indicated by the vertical dotted line and the genetic effect model is indicated by the colors red, green and blue for dominant, log-additive and recessive models, respectively.
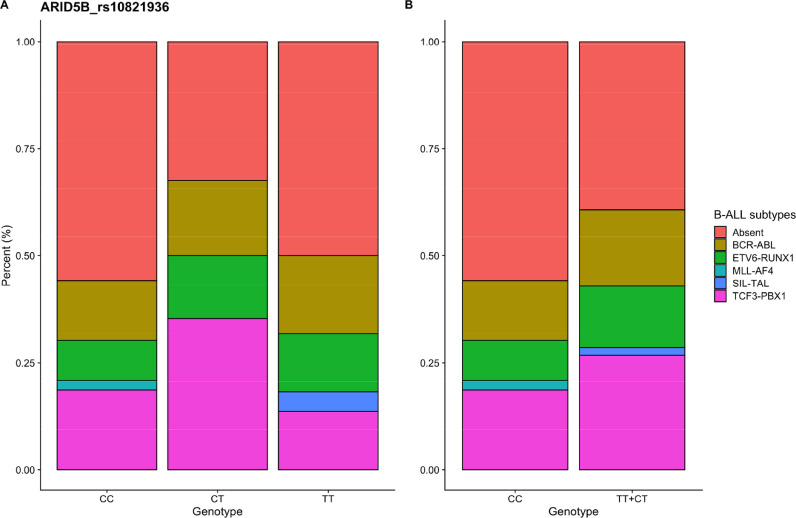
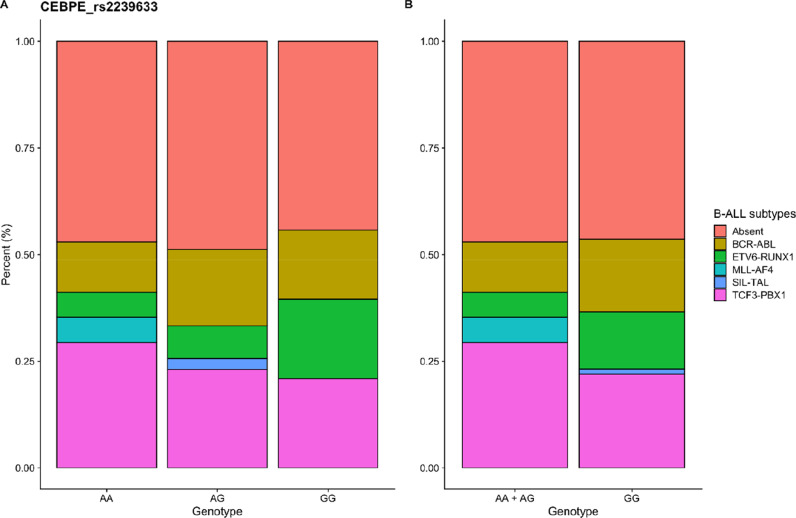
An additional analysis was performed associating the cytogenetic subtypes of B-cell ALL, according to the gene fusion investigated, with the polymorphic variants of the ARID5B (rs10821936) and CEBPE (rs2239633) genes, selected based on GWAS studies. The data are presented in Fig. 3, Fig. 4 and in Supplementary Table S3. The distribution of gene fusions according to the genotypes of the ARID5B and CEBPE gene variants was not statistically significant.
Fig. 3.
Distribution of ARID5B_ rs10821936 gene variants according to B-ALL leukemia subtype.
Fig. 4.
Distribution of CEBPE_ rs2239633 gene variants according to B-ALL leukemia subtype.
Supplementary Table S3.
Associations between genetic variants of ARID5B and CEBPE genes, and leukemia subtype of B-ALL.
| Genotype, ID | Distribution of cytogenetic subtypes of B-ALL | Pa | |||||
|---|---|---|---|---|---|---|---|
| ARID5B_ rs10821936 | Absent n (%) |
BCR-ABL n (%) |
ETV6-RUNX1 n (%) |
MLL-AF4 n (%) |
TCF3-PBX1 n (%) |
SIL-TAL n (%) |
0.337 |
| CC | 24 (52.2) | 6 (37.5) | 4 (33.3) | 1 (100) | 8 (34.6) | 0 | |
| CT | 11 (23.9) | 6 (37.5) | 5 (41.7) | 0 | 12 (52.2) | 0 | |
| TT | 11 (23.9) | 4 (25) | 3 (25) | 0 | 3 (13) | 1 (100) | |
| CT+TT | 22 (47.8) | 10 (62.5) | 8 (66.7) | 0 | 15 (65.2) | 1 (100) | 0.425 |
| CC | 24 (52.2) | 6 (37.5) | 4 (33.3) | 1 (100) | 15 (65.2) | 0 | |
| CEBPE_ rs2239633 | 0.460 | ||||||
| GG | 19 (41.3) | 7 (43.8) | 8 (66.7) | 0 | 9 (39.1) | 0 | |
| GA | 19 (41.3) | 7 (43.8) | 3 (25) | 0 | 9 (39.1) | 1 (100) | |
| AA | 8 (17.4) | 2 (12.5) | 1 (8.3) | 1 (100) | 5 (21.7) | 0 | |
| AA+GA | 27 (58.7) | 9 (56.3) | 4 (33.3) | 1 (100) | 14 (60.9) | 1 (100) | 0.489 |
| GG | 19 (41.3) | 7 (43.8) | 8 (66.7) | 0 | 9 (39.1) | 0 | |
Significance determined by Fisher exact test.
4. Discussion
Demographic factors such as sex and genetic ancestry were controlled for in the analysis of the association between the variants investigated and susceptibility for the development of B-cell ALL. The male sex and Amerindian ancestry were predominant in the case group, whereas the control group had significantly more individuals of the female sex and European ancestry, which is consistent with the findings of a previous study published by our research group [20].
Variations in the genes that regulate the folate cycle (for example, GGH, MTHFR, MTHFD1, ATIC) may influence susceptibility to ALL [11,12]. The γ-glutamyl hydrolase (GGH) gene plays an important role in folate homeostasis, catalyzing the hydrolysis of the active polyglutamates of the natural folates into monoglutamates for the biosynthesis of nucleotides. This means that variations in the GGH gene may alter the amount of folate available in the cell and influence susceptibility to ALL [14,21].
Variants of the GGH gene have previously been associated with altered susceptibility for ALL [13], although the present study was the first to show that the rs1800909 and rs3758149 variants of the GGH gene were associated with the modulation of the risk of developing B-cell ALL. Here, the homozygous genotypes of the wild variants rs1800909 and rs3758149 of the GGH gene were associated with a protective effect against the development of ALL in the study population (rs1800909: OR = 0.103; 95%CI = 0.056–0.192; P < 0.001; rs3758149: OR = 0.162; 95%CI=0.085- 0.303; P < 0.001).
Polymorphisms of the methylenotetrahydrofolate reductase (MTHFR) and methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) genes are known to reduce the activity of the enzyme that is essential for folate bio-availability and metabolism [11,12]. The rs1801133 variant of the MTHFR gene and the rs2236225 variant of MTHFD1 have been widely investigated in relation to the modulation of the risk of ALL, in a number of different populations [12,13,22,23]. In the present study, the log-additive association models of both genes were associated with a protective effect for the development of B-cell ALL (MTHFR: OR = 0.399; 95%CI = 0.277–0.571; P < 0.001; MTHFD1: OR = 0.546; 95%CI = 0.385–0.776; P < 0.001).
The ATIC gene codifies a bifuncional protein (5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase and IMP cyclohydrolase) that catalyzes the last two stages of the synthesis of Inosine monophosphate [24]. The rs4673993 variant of the ATIC gene has been widely shown to be associated with toxicity to treatment with methotrexate, an inhibitor of the folate pathway [25], [26], [27]. The present study represents the first report of the role of the rs4673993 variant of the ATIC gene as a risk factor for ALL. The homozygous mutant genotype of the rs4673993 variant of the ATIC gene was associated with a 360% increase in the risk of developing ALL in the study population (OR = 3.594; 95%CI = 1.509–8.518; P = 0.004).
A number of GWAS studies have found an over-representation of polymorphisms of the ARID5B and CEBPE genes in ALL patients in comparison with individuals that do not have the disease [[6], [7], [8],10]. The ARID5B gene plays a fundamental role in the regulation of the development of the embryo, and cell growth and differentiation through the repression of the expression of specific differentiation genes. Deficiencies in the expression of the ARID5B gene in the developing fetus may interrupt the maturation of the B lymphocytes and contribute to leukemogenesis [28,29]. The rs10821936 variant of the ARID5B gene has been associated with susceptibility to ALL in children in a number of different populations [7,6,10,30,31]. In the present study, however, the homozygous genotype of the wild rs10821936 variant was associated with a protective effect for the development of B-cell ALL (OR = 0.183; 95%CI = 0.100–0.341; P < 0.001).
The CCAAT ligation/potentializing proteins (CEBPs) are transcription factors involved in the development of hematopoietic cells, including granulopoiesis [32]. A number of studies [7,8,30,33,34,35] have demonstrated a strong association between the rs2239633 variant of the CEBPE gene and susceptibility to childhood ALL. In the present study, the log-additive model of the rs2239633 variant of the CEBPE gene indicated a protective effect against the development of B-cell ALL (OR = 0.237; 95%CI = 0.154–0.357; P < 0.001).
Variants of the GWAS-identified ARID5B and CEBPE genes are reported in the literature associated with certain ALL subtypes. For example the variant rs10821936 of the ARID5B gene is strongly associated with hyperdiploid B-ALL and MLL-germline. And the rs2239633 variant of the CEBPE gene is largely null in children with and without MLL rearrangements. Most of these association studies were performed in poorly mixed populations [36]. Furthermore, the association between ALL cytogenetic subtypes and ethnic variation is poorly verified in the literature. In order to better explore the relationship of variants identified in GWAS studies with ALL cytogenetic subtypes in the population of the Brazilian Amazon region, we performed an additional analysis comparing the BCR-ABL, ETV6-RUNX1, MLL-AF4, SIL-TAL and TCF3-PBX1 gene fusions with the variants of the ARID5B and CEBPE genes studied. Most patients did not present any of the investigated translocations. The distribution of gene fusions according to the ARID5B and CEBPE gene polymorphism genotypes was not statistically significant.
5. Conclusion
The results of the present study indicate that the variants of the genes GGH (rs1800909, rs3758149), MTHFR (rs1801133), MTHFD1 (rs2236225), ATIC (rs4673993), ARID5B (rs10821936), and CEBPE (rs2239633) play a fundamental role in the risk of developing B-cell ALL in the study population. This is the first study to describe the influence of these variants in the development of B-cell ALL in the population of the Brazilian Amazon region.
Author contributions statement
DC, PA and N.S designed the study. DC conducted the molecular genetic study, participated in the statistical analyses, and wrote the manuscript. AW, FA, AC, LL, JC and TS participated in the molecular genetic studies, the collection of the clinical data, and the organization of the study. AS and LL participated in the design of the study and conducted the statistical analysis. AK, SS, PA and NS participated in the inception and coordination of the project. All the authors have read and approved the final manuscript.
Funding source
This work was supported by the Brazilian National Council for Scientific and Technological Development (CNPQ) and Pro-rector of Research and Post-graduation (Propesp) of Federal University of Para.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgments
The authors would like to thank all the patients and their families and the clinical staff for their skilled assistance. We also thank Antonio A.C. Modesto, Fernando A.R. Mello and Marcos A.T. Amador for technical assistance.
References
- 1.Pui C.-H., Carroll W.L., Meshinchi S., Arceci R.J. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J. Clin. Oncol. 2011;29:551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward E., Desantis C., Robbins A., Kohler B., Jemal A. Childhood and adolescent cancer statistics. Ca. Cancer J. Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 3.Lo Nigro L. Biology of childhood acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 2013;35(4):245–252. doi: 10.1097/MPH.0b013e31828f8746. [DOI] [PubMed] [Google Scholar]
- 4.Chang J.S. Parental smoking and childhood leukemia. Methods Mol. Biol. 2009;472:103–137. doi: 10.1007/978-1-60327-492-0_5. [DOI] [PubMed] [Google Scholar]
- 5.Gharbi H., Ben Hassine I., Soltani I., Safra I., Ouerhani S., Bel Haj Othmen H. Association of genetic variation in IKZF1, ARID5B, CDKN2A, and CEBPE with the risk of acute lymphoblastic leukemia in Tunisian children and their contribution to racial differences in leukemia incidence. Pediatr. Hematol. Oncol. 2016;33(3):157–167. doi: 10.3109/08880018.2016.1161685. [DOI] [PubMed] [Google Scholar]
- 6.Treviño L.R., Shimasaki N., Yang W., Panetta J.C., Cheng C., Pei D. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J. Clin. Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papaemmanuil E., Hosking F.J., Vijayakrishnan J., Price A., Olver B., Sheridan E. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherborne A.L., Hosking F.J., Prasad R.B., Kumar R., Koehler R., Vijayakrishnan J. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat. Genet. 2010;42:492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orsi L., Rudant J., Bonaventure A., Goujon-Bellec S., Corda E., Evans T.-J. Genetic polymorphisms and childhood acute lymphoblastic leukemia: GWAS of the ESCALE study (SFCE) Leukemia. 2010;26:2561–2564. doi: 10.1038/leu.2012.148. [DOI] [PubMed] [Google Scholar]
- 10.Xu H., Yang W., Perez-Andreu V., Devidas M., Fan Y., Cheng C. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J. Natl. Cancer Inst. 2013;105:733–742. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne E., Greenop K.R., Scott R.J., Haber M., Norris M.D., Attia J. Folate pathway gene polymorphisms, maternal folic acid use, and risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol. Biomark. Prev. 2015;24(1):48–56. doi: 10.1158/1055-9965.EPI-14-0680. [DOI] [PubMed] [Google Scholar]
- 12.Metayer C., Scélo G., Chokkalingam A.P., Barcellos L.F., Aldrich M.C., Chang J.S. Genetic variants in the folate pathway and risk of childhood acute lymphoblastic leukemia. Cancer Causes Control. 2011;22(9):1243–1258. doi: 10.1007/s10552-011-9795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lautner-Csorba O., Gézsi A., Erdélyi D.J., Hullám G., Antal P., ÁF S. Roles of genetic polymorphisms in the folate pathway in childhood acute lymphoblastic leukemia evaluated by Bayesian relevance and effect size analysis. PLoS ONE. 2013;8(8):e69843. doi: 10.1371/journal.pone.0069843. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Liu S., Wang H., Mai H., Yuan X., Li C. Methylation level of CPG islands in GGH gene promoter in pediatric acute leukemia. PLoS ONE. 2017;12(3) doi: 10.1371/journal.pone.0173472. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikoma M.R., Sandes A.F., Thiago L.S., Cavalcanti Júnior G.B., Lorand-Metze I.G., Costa E.S. GBCFLUX. First proposed panels on acute leukemia for four-color immunophenotyping by flow cytometry from the Brazilian group of flow cytometry-GBCFLUX. Cytomet. B Clin. Cytom. 2015;88(3):194–203. doi: 10.1002/cyto.b.21175. [DOI] [PubMed] [Google Scholar]
- 16.Galehdari H., Abedini N., Kazeminezhad R., Pedram M. Evaluation of RT-PCR to detect translocations in children diagnosed with acute lymphoblastic leukemia. Iran. J. Publ. Health. 2009;38(4):117–121. 2009. [Google Scholar]
- 17.Santos N.P., Ribeiro-Rodrigues E.M., Ribeiro-Dos-Santos A.K., Pereira R., Gusmão L., Amorim A. Assessing individual interethnic admixture and population substructure using a 48 – insertion-deletion. Hum. Mutat. 2010;31(2):184–190. doi: 10.1002/humu.21159. [DOI] [PubMed] [Google Scholar]
- 18.Ramos B.R., D’Elia M.P., Amador M.A., Santos N.P., Santos S.E., da Cruz Castelli E. Neither self-reported ethnicity nor declared family origin are reliable indicators of genomic ancestry. Genetica. 2016;144(3):259–265. doi: 10.1007/s10709-016-9894-1. [DOI] [PubMed] [Google Scholar]
- 19.Amador M.A., Cavalcante G.C., Santos N.P., Gusmão L., Guerreiro J.F., Ribeiro-dos-Santos Â. Distribution of allelic and genotypic variants in native American, African, European and Brazilian populations. BMC Res. Notes BioMed Central. 2016;9:101. doi: 10.1186/s13104-016-1906-9. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho D.C., Wanderley A.V., Amador M.A., Fernandes M.R., Cavalcante G.C., Pantoja K.B. Amerindian genetic ancestry and INDEL polymorphisms associated with susceptibility of childhood B-cell Leukemia in an admixed population from the Brazilian Amazon. Leuk. Res. 2015 doi: 10.1016/j.leukres.2015.08.008. pii: S0145-2126(15)30361-1. [DOI] [PubMed] [Google Scholar]
- 21.Galivan J., Ryan T.J., Chave K., Rhee M., Yao R., Yin D. Glutamyl hydrolase. pharmacological role and enzymatic characterization. Pharmacol. Ther. 2000;85(3):207–215. doi: 10.1016/s0163-7258(99)00063-7. [DOI] [PubMed] [Google Scholar]
- 22.Pei J.S., Hsu C.M., Tsai C.W., Chang W.S., Ji H.X., Hsiao C.L. The association of methylenetetrahydrofolate reductase genotypes with the risk of childhood leukemia in Taiwan. PLoS ONE. 2015;10(3) doi: 10.1371/journal.pone.0119776. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia X., Duan Y., Cui J., Jiang J., Lin L., Peng X. Association of methylenetetrahydrofolate reductase gene-gene interaction and haplotype with susceptibility to acute lymphoblastic leukemia in Chinese children. Leuk. Lymphoma. 2017;58(8):1887–1892. doi: 10.1080/10428194.2016.1265117. [DOI] [PubMed] [Google Scholar]
- 24.Rayl E.A., Moroson B.A., Beardsley G.P. The human purH gene product, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase. Cloning, sequencing, expression, purification, kinetic analysis, and domain mapping. J. Biol. Chem. 1996;271:2225–2233. doi: 10.1074/jbc.271.4.2225. [DOI] [PubMed] [Google Scholar]
- 25.Halilova K.I., Brown E.E., Morgan S.L., Bridges S.L., Jr., Hwang M.H., Arnett D.K. Markers of treatment response to methotrexate in rheumatoid arthritis: where do we stand? Int. J. Rheumatol. 2012;2012 doi: 10.1155/2012/978396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lima A., Monteiro J., Bernardes M., Sousa H., Azevedo R., Seabra V. Prediction of methotrexate clinical response in Portuguese rheumatoid arthritis patients: implication of MTHFR rs1801133 and ATIC rs4673993 polymorphisms. Biomed. Res. Int. 2014;2014 doi: 10.1155/2014/368681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima A., Bernardes M., Azevedo R., Seabra V., Medeiros R. Moving toward personalized medicine in rheumatoid arthritis: sNPs in methotrexate intracellular pathways are associated with methotrexate therapeutic outcome. Pharmacogenomics. 2016;17(15):1649–1674. doi: 10.2217/pgs-2016-0067. [DOI] [PubMed] [Google Scholar]
- 28.Huang T.H., Oka T., Asai T., Okada T., Merrills B.W., Gertson P.N. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 1996;24:1695–1701. doi: 10.1093/nar/24.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilsker D., Patsialou A., Dallas P.B., Moran E. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 2002;13:95–106. [PubMed] [Google Scholar]
- 30.Bekker-Méndez V.C., Núñez-Enríquez J.C., Torres Escalante J.L., Alvarez-Olmos E., González-Montalvoc P.M., Jiménez-Hernández E. ARID5B, CEBPE and PIP4K2A germline genetic polymorphisms and risk of childhood acute lymphoblastic leukemia in Mexican patients: a MIGICCL study. Arch. Med. Res. 2016;47(8):623–628. doi: 10.1016/j.arcmed.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Archer N.P., Perez-Andreu V., Stoltze U., Scheurer M.E., Wilkinson A.V., Lin T.N. Family-based exome-wide association study of childhood acute lymphoblastic leukemia among Hispanics confirms role of ARID5B in susceptibility. PLoS ONE. 2017;12(8) doi: 10.1371/journal.pone.0180488. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akasaka T., Balasas T., Russell L.J., Sugimoto K.J., Majid A., Walewska R. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) Blood. 2007;109(8):3451–3461. doi: 10.1182/blood-2006-08-041012. [DOI] [PubMed] [Google Scholar]
- 33.Sun J., Zheng J., Tang L., Healy J., Sinnett D., Dai Y. Association between CEBPE variant and childhood acute leukemia risk: evidence from a meta-analysis of 22 studies. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0125657. e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C., Chen J., Sun H., Sun L., Liu Y. CEBPE polymorphism confers an increased risk of childhood acute lymphoblastic leukemia: a meta-analysis of 11 case-control studies with 5,639 cases and 10,036 controls. Ann. Hematol. 2015;94(2):181–185. doi: 10.1007/s00277-014-2186-x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X.-X., Du Y.-F., Zhai Y.-J., Gao F., Yang Y.-J., Ma X.-C. A common genetic variation in CEBPE and acute lymphoblastic leukemia: a meta-analysis of the available evidence. Onco. Targets Ther. 2015;8:2443–2451. doi: 10.2147/OTT.S89661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams L.A., Yang J.J., Hirsch B.A., Marcotte E.L., Spector L.G. Is there etiologic heterogeneity between subtypes of childhood acute lymphoblastic leukemia? A review of variation in risk by subtype. Cancer Epidemiol. Biomark. Prev. 2019;28(5):846–856. doi: 10.1158/1055-9965.EPI-18-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]