Graphical abstract
Abbreviations: RSAT, Regulatory Sequence Analysis Tools; SNP, Single Nucleotide Polymorphism; TF, Transcription Factor; TFBS, Transcription Factor Binding Site; PSSM, Position Specific Scoring Matrix; MPRA, Massively Parallel Reporter Assays: MPRA; LD, Linkage Disequilibrium; rsID, Reference SNP Identifier; SOIs, SNPs of Interest; GWAS, Genome Wide Association Studies; CRM, Cis-Regulatory Module; eQTL, Expression Quantitative Trait Loci; ROC, Receiver Operating Characteristic; CEU, Northern Europeans from Utah
Keywords: Regulatory variants, Transcription factors, Position specific scoring matrix, SNPs, Binding motifs
Abstract
Gene regulatory regions contain short and degenerated DNA binding sites recognized by transcription factors (TFBS). When TFBS harbor SNPs, the DNA binding site may be affected, thereby altering the transcriptional regulation of the target genes. Such regulatory SNPs have been implicated as causal variants in Genome-Wide Association Study (GWAS) studies. In this study, we describe improved versions of the programs Variation-tools designed to predict regulatory variants, and present four case studies to illustrate their usage and applications. In brief, Variation-tools facilitate i) obtaining variation information, ii) interconversion of variation file formats, iii) retrieval of sequences surrounding variants, and iv) calculating the change on predicted transcription factor affinity scores between alleles, using motif scanning approaches. Notably, the tools support the analysis of haplotypes. The tools are included within the well-maintained suite Regulatory Sequence Analysis Tools (RSAT, http://rsat.eu), and accessible through a web interface that currently enables analysis of five metazoa and ten plant genomes. Variation-tools can also be used in command-line with any locally-installed Ensembl genome. Users can input personal collections of variants and motifs, providing flexibility in the analysis.
1. Introduction
Genomic DNA sequence harbors the gene regulatory information necessary spatial and temporal gene expression patterns [38], [31]. Gene regulatory regions encompass short, highly redundant DNA motifs recognized by transcription factors (TF) [36]. These regulatory regions may contain genetic variants, Single Nucleotide Polymorphisms (SNPs) or indels, that alter the DNA TF binding site (TFBS), and thereby the binding of TF [20]. Moreover, it has been reported that 93.7% of variants that have been associated with human traits or diseases have been found to be located in non-coding regions [43], [40], and particularly enriched in open chromatin regions [57], indicating that these variants may affect transcriptional regulatory mechanisms, and thereby explain the observed phenotypes.
The Regulatory Sequence Analysis Tools (RSAT, http://rsat.eu) [47], [26] has established itself in the last 20 years as a major software suite dedicated to the analysis of regulatory regions, with five public servers supporting more than 500 eukaryote and 9,000 prokaryote genomes. With a major focus on usability and accessibility to users with or without formal bioinformatics training, RSAT provides tools to retrieve sequences, perform motifs analysis, evaluate TF motif quality, compare and cluster motifs, convert file formats, etc. Here we describe Variation-tools, a subset of tools included in RSAT that enable users to analyse regulatory variants and assess their putative impact on TF binding sites.
1.1. Current approaches for detecting potential regulatory variants
The fact that many variants are located in non-coding regions triggered the development of bioinformatic tools to identify the regulatory potential of these genetic variants. Starting from a list of SNPs, computational analyses can help formulating hypotheses on which TF may be impacted by a genetic variant. However, there are numerous challenges for in silico analysis to unravel the impact of genetic variations in gene regulatory regions. Several tools and resources have been published, providing alternative methods to tackle this problem (Table 1). Most of them are either based on pattern-matching approaches to evaluate the impact of alleles on TF binding, or on machine learning models built using functional annotations of the regulatory regions, e.g. epigenomics and transcriptomics data. Still, these resources and tools have limitations hampering their usage in several organisms [68], [28], [35], on new annotated variants [5], [63], [54], and/or on analyses with personal collections of TF motifs [68], [28], [41].
Table 1.
Tools similar to variation-scan with available implementation. PM stands for Pattern Matching, ML stands for Machine Learning.
| Name | PMID | Source | Approach | Organism | Input | Output | Matrix flexibility | Type | Last update |
|---|---|---|---|---|---|---|---|---|---|
| deltaSVM | 26075791 | http://www.beerlab.org/deltasvm/ | Gapped k-mer SVM classifier. | Any organism | DNaseI-seq data; putative regulatory regions as positive training set and randomized sequences as negative training set. | deltaSVM, predicted impact of a variant in chromatin accessibility which is measured by adding up the contribution of all 10-mers in which the SNP is present for chromatin accessibility. | It can only be trained for one TF at a time. | ML, non-static. | Last update Sept 2015. |
| DeepSea | 26301843 | http://deepsea.princeton.edu/job/analysis/create/ | Deep convolutional network. | Human | SNPs in VCF format. | Chromatin feature probabilities for reference and alternative alleles, chromatin feature probability log fold changes for each variant, chromatin feature probability differences for each variants, e-values for chromatin feature effects, functional significance score for each variant. There are 919 chromatin features evaluated. | It contains 690 TF binding profiles for 160 different TFs, but does not support the addition of new matrices. | ML, non-static. | Last update May 2017. |
| atSNP | 26092860 | https://github.com/keleslab/atSNP | Importance sampling algorithm for p-value calculation, first-order Markov Model to generate random background sequences. | Any organism whose genome is included in the Bioconductor BSGenome package. | SNP list, motif file. | p-value for binding affinity with alternative and reference allele, p-value for binding affinity change based on log-likelihood ratio and log-rank ratio. It also provides composite logo plots for directly visualizing the SNP effects on motif matches. | It accepts several matrices, and several different formats. It includes a motif library of 2,065 PSSMs from ENCODE and JASPAR, but also allows user-defined motif libraries. | PM, non-static. | Last update Nov 2018. |
| BayesPI-BAR | 26202972 | http://folk.uio.no/junbaiw/BayesPI-BAR/ | Biophysical modeling of protein-DNA interaction, estimation of TF chemical potential (through a bayesian nonlinear regression model) and differential binding affinity. | Any organism | ChIP-seq experiment for TFs to be tested, DNA sequences for selected SNPs,PSSMs for selected TFs. | Given a SNP and a PSSM list, it produces two lists sorted by significance: one composed of binding motifs disrupted by the SNP, and one by sites with an increased affinity to the TF caused by the SNP. | Can use several PSSMs simultaneously. | PM, biophysical modeling.Non-static. | No updates listed, software created July 2015. |
| GWAS4D | 29771388 | http://mulinlab.tmu.edu.cn/gwas4d/gwas4d/gwas4d/gwas4d_server | Variant prioritization method, followed by an integrative analysis of genome-wide association. | Human | Accepts VCF-like, coordinate only, dbSNP ID and PLINK-like formats. | Regulatory variant prioritization table: includes the most likely affected motif by alternative variant effect. | The model includes motifs of 1,480 transcriptional regulators from 13 different resources. It is not possible to upload user-specified matrices. | PM, static | Last update Sept 2018. |
| sTRAP | 20127973 | http://trap.molgen.mpg.de/cgi-bin/home.cgi | Prediction of local binding affinity followed by a normalization of binding affinities to determine difference between reference allele and SNP. | Organisms available in TRANSFAC. | Accepts only two sequences in FASTA format. | List of TFs ranked according to changes induced by the SNP. | There is no option for user-specified matrices, matrices from TRANSFAC versions can be selected. | PM, non-static | No updates listed, software created in 2011. |
| SNP2TFBS | 27899579 | https://ccg.epfl.ch//snp2tfbs/ | Estimation based on PSSM model. | Human. | When working with the code, the input required is the reference genome, a SNP catalogue and a PSSM collection. The web interface accepts SNP IDs and VCF format, as well as a specification of a genomic region through a bed file or by specifying the start and end positions. |
List of affected TFBSs, sorted by the magnitude of the effects. | On the web interface, only matrices from JASPAR can be used. Nonetheless, it is possible to download the code used to generate the database and use a different input. | PM, static. | Last update July 2017. |
| atSNP Search | 30534948 | http://atsnp.biostat.wisc.edu/ | Used atSNP algorithm with dbSNP build 144 for human genome assembly 38 against JASPAR and Encode motifs to create a repository with all the SNP-motif combinations resulting from the previous resources. | Human. | It can receive a set of rsIDs, a rsID and a window size around the SOI, genomic coordinates, a gene symbol and a window size around the gene of interest, or a TF name. | Table including p-values for motif matches for both reference and alternate alleles, as well as the change in the motif matching and the direction of said change. Output includes logo plots, displaying the sequence logos aligned to best motif matches with reference and SNP alleles. | Only JASPAR or ENCODE matrices can be selected, and it is possible to select only one transcription factor at a time. | PM, static. | Last update Jan 2018. |
| HaploReg | 22064851, 26657631 |
https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php | It contains data from multiple genome annotation resources. PSSMs are scored against reference and alternative alleles, and change in log-odds is calculated. | Human | Users can provide a list of rsIDs or chromosome regions. Users can also select GWAS studies from the NHGRI catalog. | Provides data on allelic frequencies, conservation, chromatin states, and near genes. For each of the regulatory motifs altered by the SNP, it provides the change in log-odds and a logo. | HaploReg contains a library created from literature sources, TRANSFAC, JASPAR and PBM experiments. There is no option for user-specified matrices. | PM, static. | Last update November 2015. |
| RegulomeDB | 22955989 | http://www.regulomedb.org/ | RegulomeDB uses information from several datasets, as well as manual curation and a heuristic method to distinguish between functional and non-functional variants. | Human. | Users can provide a list of dbSNP IDs, hg19 coordinates in BED, VCF or GFF3 format, or hg19 chromosomal regions in the same formats. | Table sorted by likely functionality, containing variant coordinates, score assigned by the algorithm, and evidence of function including protein binding, motifs, chromatin structure, eQTLs and histone modifications. | RegulomeDB includes all PSSMs from TRANSFAC, JASPAR CORE, and UniProbe. There is no option for user-specified matrices. | PM, static. | No updates, listed, software created in Sept 2012. |
| motifbreakR | 26272984 | https://github.com/Simon-Coetzee/MotifBreakR | It has three options of algorithms: the standard sum of log probabilities, weighted sum, and an information content method. | Organisms included in BSgenome. | SNPs can be imported from an R package or provided to the algorithm in BED or VCF format. PSSMs can be selected from the MotifDb package or be user-specified. | Table containing statistics describing the percent of maximum score for a matrix and matrix values for both alleles, as well as the strand. It also reports whether the TFBS is disrupted strongly or weakly. | PSSMs can be imported from the MotifDb package or be user-specified. More than one matrix can be used at a time. | PM, non-static. | Last update Jul 2018. |
| variation-scan | http://rsat.eu | Estimation based on PSSM model. | web interface: installed Ensembl organisms. command-line: any locally installed organism. |
A collection of PSSMs and a set of variants in varSeq format. This format can be obtained using retrieve-variation-seq. | A table with one line per pair of alleles per motif (if there are more than two, there will be one line per possible pair) reporting the position, weight and p-value of each allele, weight difference and p-value ratio. | Users can select for the collections available in RSAT (JASPAR, HOCOMOCO, CisBP), but they can also use personal collections. | PM-non static. | April 2019. |
All tools in the Pattern Matching category, (labeled PM in Table1) use Position-Specific Scoring Matrices (PSSMs) to evaluate the affinity of a TF to a given sequence with an allele. Major differences between these tools can be found in (i) their availability: web pages [5], command line [12] or both [69]; (ii) flexibility for the user to input their own data [61]; (iii) usability: the possibility to use several variant formats [28]; (iv) results representation: figures and/or tables [63]; (v) available organisms: only human [62], or other organisms [61]; and (vi) the possibility to calculate results on-the-fly [41] or access pre-calculated ones [5].
Another set of tools (labeled ML in Table1) aim for the identification of potential regulatory variants by integrating several types of data, beyond taking into account potential disruption of TF binding. Particularly, Lee, et al. [37] integrated DNaseI-seq data with SVM approaches to identify variants that could potentially disrupt TF binding. DeepSea [68] integrates functional genomic data from ChIP-seq, DNaseI-seq, RNA-seq and other functional genomic high-throughput data to assess the potential damage of variants across the human genome. Precalculated results for annotated variants can be accessed on their website.
Both tools can be trained on other organisms, provided that functional genomic data are available. The main limitation of these resources is the required expertise in bioinformatics and/or computational resources for users to analyse their own data sets. Other tools identify potential regulatory effects of a variant by comparing the measured affinity of a TF to the different possible alleles. Our tool, named variation-scan, falls within this category.
1.2. Variation-tools
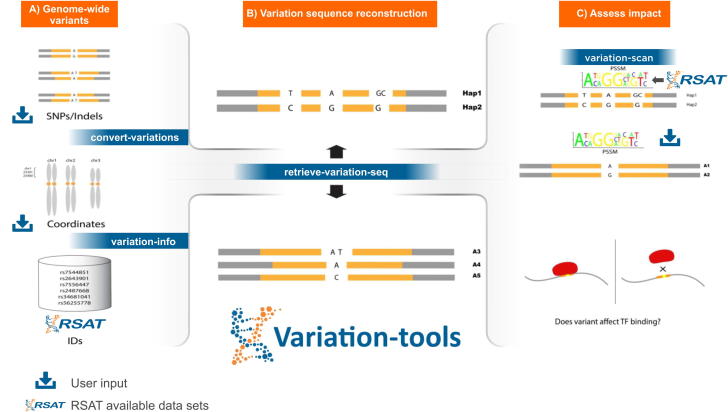
In this context, we have developed Variation-tools to address the main limitations identified in existing programs (Table 1). Variation-tools are composed of four programs that enable (i) retrieval of information of Ensembl annotated variants when available for a given genome in RSAT (variation-info), (ii) conversions between variant file formats (convert-variations), (iii) retrieval of the sequences surrounding variants (retrieve-variation-seq), and (iv) scanning of different alleles of a variant with one or several motifs, comparing the scores and p-values in order to identify affected TFBS (variation-scan) (Fig. 1). Earlier versions of these programs were reported in 2015 as part of a RSAT update article [45], these first versions were developed in perl and were refactored and improved for the 2018 update [47]. In this article we present the latest versions of the tools, with optimized memory usage, and novel support for the inclusion of haplotype information.
Fig. 1.
Schematic representation of Variation-tools: This set of tools, included in the Regulatory Sequence Analysis Tools (RSAT), focuses on assessing the impact of different allelic variants on Transcription factor binding sites. A) convert-variations allows users to input their own variants and convert them to other formats (VCF, GVF and varBed, the latter is the format used in the next step), while variation-info retrieves the annotated information of Ensembl variants installed in RSAT servers. B) The tool retrieve-variation-seq retrieves the surrounding sequence of variants (including possible haplotypes) and generates a text file with one line per allele and per variant or haplotype (varSeq format). C) Users can input their variants in varSeq format and a collection of motifs (direct input by the user or selected from RSAT available collections) to variation-scan; the tool then scans the corresponding sequences with all motifs and perform pairwise comparisons between the binding scores of each transcription factor onto all alleles of a variant or haplotype.
In summary, RSAT Variation-tools provide an accessible resource for experienced and non-expert users to analyze regulatory variants in a web interface for fifteen organisms (five metazoa (http://metazoa.rsat.eu) and ten plants (http://plants.rsat.eu), with flexibility to upload personal variant and PSSM collections. We describe here Variation-tools methodology, along with four case studies demonstrating the flexibility of the tools, enabling the analysis of data sets from different origin (Ensembl variants, Genome-Wide Association Study (GWAS) data, ChIP-seq regions, etc.), complexity, and organisms.
2. Methods
2.1. Variation-tools: from variants to identification of regulatory effects
Variation-tools consist in a subset of four tools within RSAT devoted to the identification of genetic variants putatively affecting TF binding
-
1)
variation-info: this tool relies on the Ensembl genetic variation information [29] annotated and installed on the corresponding server for each particular genome (i.e. human variants are installed in the Metazoa server). It can take two different inputs: 1) variant rsID or 2) genomic loci in bed format. This tool will retrieve the information of the variants matching the IDs or the information of the variants located in the genomic loci. Variants installed in RSAT servers have been processed to remove variants with incomplete annotations (no alleles) or ambiguous coordinates (non matching alleles coordinates). When users have their own variants collections, they can skip this tool and use directly convert-variations.
-
2)
convert-variations: enables the interconversion of variant file formats such as VCF, GVF and varBed. varBed is an internal format of RSAT that facilitates the retrieval of the sequence surrounding the variant (Supplementary Fig. 1A).
-
3)
retrieve-variation-seq: retrieves the sequence surrounding the variant, and produces one sequence for each allele (Supplementary Fig. 1B). The tool can take as input a varBed file (see convert-variations). For organisms with Ensembl annotated variants, it can take a list of IDs or a bed file listing genomic loci. The output is provided in a format named varSeq, with each row giving one allele with its surrounding sequence. Each variant has a specific internal ID to accommodate several variants with various alleles in the same file.
-
4)
variation-scan: performs the scanning of alleles with a PSSM and compares the scores and p-values between alleles to assess the putative effect on TF binding (see details below) (Supplementary Fig. 2). It requires as input a varSeq file (see retrieve-variation-seq), a motif or collection of motifs (over twenty supported file formats), and a background model (for methodological details on background model, refer to [59] Box n°3). Different background models are readily available through the web interface. However, depending on the biological question and related potential biases, we recommend the creation of a dedicated background model, which can be done using the RSAT tool create-background, also available via the RSAT web interface.
2.2. Haplotype processing
Genetic variants can be detected using high-throughput techniques. This has enabled the identification of millions of variants in the HapMap [30] and 1000 genomes projects [1]. However, the information on the variants alone is less useful than knowing which groups of alleles are co-located on the same chromosome (haplotype). The process of identifying the variants that belong to each chromosome is known as phasing. Including haplotype phasing information facilitates the identification of relations between variants [6].
VCF files can include haplotype phasing information. The tool convert-variations identifies and retrieves the phasing information of the variants, while the tool retrieve-variation-seq builts the corresponding haplotype with all the SNPs that lay within a defined window (default: 30 bp).
2.3. Computing binding specificity of a transcription factor to a DNA sequence
variation-scan uses PSSMs to assess the binding specificity of a TF to a DNA sequence with different alleles in a given position. The first step of variation-scan (i.e., scanning of the sequences with a given PSSM) is delegated to the RSAT tool matrix-scan. The scoring scheme and p-value calculation are described in detail in [59], Box n°1 and Box n°2, respectively. In brief:
PSSM are used to assess the binding specificity of a TF. This affinity is calculated as a weight score (Ws). The Ws of a site in variation-scan is calculated using [27]:
where S is a sequence segment of the same length of M, M is the PSSM, and B is the background model. Hence, P(S|M) is the probability of the sequence given the PSSM and P(S|B) is the probability of the sequence given the background model. Ws has been related to the affinity of the TF to the sequence, as it assesses similarity of a sequence to a known set of binding sites, providing information about the probability of a sequence to be a new instance of a binding site [56].
Moreover, it is possible to calculate the p-value of a given score as:
where the P-value is calculated as the probability of observing a score of at least W given a background model w|B.
When a sequence is longer than the PSSM, the PSSM is shifted base by base until the full sequence has been scored. This scanning step is performed on the sequences of all reported alleles, so that each allele is compared with all the positions of a given motif.
Background models represent the nucleotide composition of a set of sequences (whole genome, all promoter sequences, etc.). These models are used to estimate the expectancy of a nucleotide being found. Background models can represent dependency between nucleotides in sequences (e.g. taking into account the frequencies of dinucleotides to build a Markov model of order 1 [59] Box n°3). As background models are used to calculate weight scores for a binding site (P(S|B)), it is important to select an appropriate model for each analysis. Examples of selected background models are presented in the different study cases matching each particular biological question.
2.4. Assessment of allele effect on transcription factor binding
In the second step, i.e., evaluating the impact of SNPs, variation-scan compares the obtained Ws (Ws difference = Ws_Allele1 – Ws_Alelle2) and the P-value (P-value ratio = P-value_Allele1/P-value_Allele2) of each of the alleles, position by position throughout the scanning window. To evaluate indels, variation-scan compares the highest Ws and its corresponding P-value for each sequence of the reported alleles. When more than two alleles of a variant are reported, all alleles are compared to all alleles in a pairwise manner.
2.5. variation-scan performance test
2.5.1. Computing efficiency
The tools variation-info and convert-variation are coded in Perl, while retrieve-variation-seq and variation-scan are coded in C, to enable the analysis of large numbers of variants from eukaryotic genomes in a reasonable time. To further improve performance, we reduced the data transfer from the hard drive to memory.
variation-scan performance was assessed by randomly selecting a variant from the 1000 genomes project [1] and a motif from the RSAT non-redundant motifs collection [8]. The randomly selected variant was used to create sets with different numbers of replicates, ranging from one thousand to nine millions, to estimate the relation between running time and the amount of evaluated variants. The processes were run on a Dell PowerEdge C6145 server with 2 AMD Opteron(tm) Processor 6386 SE, 16 cores each, Processor speed of 2.8–3.5 Ghz, RAM 256 Gb and with an operating system CentOS 7 (7.6.1810).
2.5.2. Dataset: experimentally-determined regulatory variants in red blood cells
The regulatory activity of 2,756 red blood cell variants has been systematically measured using Massively Parallel Reporter Assays (MPRA) [60]. MPRA is a high-throughput assay in which a library of putative regulatory elements, each followed by a unique barcode, is inserted into a plasmid, then transfected into a cell, and transcripts are then quantified through the abundance of barcodes. These variants are known to be in strong linkage disequilibrium (LD) with 75 variants associated with common traits of this cell type. Three sliding windows per variant (left, right, and center) were synthesized, barcoded and used to study the effect of slight changes in their genomic context. Following methods described by Ulirsch, et al. [60], for each sequence mRNA/DNA ratio was computed to obtain a quantitative evaluation of the regulatory effect of a sequence variant.
2.5.3. Evaluation of variation-scan
The variant dataset was used as input for variation-scan; the variants assessed in the red blood cell assay were annotated with the Ensembl GRCh37 human genome release, and given as input to convert-variations followed by retrieve-variation-seq. Since three sliding windows were used for each variant in the MPRA, the corresponding windows were merged before computing a background model using the create-background-model tool.
According to the original study [60], binding sites for the following TF were enriched in the sequences of interest: GATA1, KLF1, DHS, TAL1, ETS, FLI1 and AP-1. Therefore, a total of 48 PSSMs annotated as related to these TF were retrieved from the non-redundant RSAT motif collection [8], and given as input to variation-scan.
A negative control set of motifs was created using the RSAT tool permute-matrix [47]; five pemuted motifs were created for each of the 48 motifs, generating a collection of 240 control motifs.
For a variant to be reported in variation-scan as positive, we requested that at least one of the allele sequences was evaluated as a binding sites with a p-value of at most 10−4 (using the parameter -uth pval 1e-4 in the command line), and that the p-value ratio was greater or equal to ten (a change of one order of magnitude between the best and the worst allele p-values) (-lth pval_ratio 10).
We compared variation-scan to two other tools previously used to assess the same set of variants by Ulirsch, et al. [60]: DeepSea [50], [37] deltaSVM. In order to avoid personal biases when calibrating tool parameters, we decided to rely on the published ones [60]. For this analysis variation-scan was run without thresholds to identify the impact of the parameters, particularly the threshold on p-value ratio.
2.6. Case studies
2.6.1. Case study 1: Identification of regulatory variants in the “Platinum” genomes haplotypes
The set of high-confidence variants from the two CEU (Northern Europeans from Utah) human Platinum Genomes NA12877 and NA12878 [23] were downloaded through the Amazon Web Service (AWS) Command Line Interface from the Illumina Platinum Genomes AWS S3 bucket (https://github.com/Illumina/PlatinumGenomes). The downloaded VCF files contained phasing information of each CEU individual haplotype configuration. The genome version used was GRCh37.
We selected SNPs intersecting with the annotated DNAseI-seq clustered peaks V3 from the ENCODE project [4]. The VCF file with the selected SNPs was processed using convert-variations with the option phased and then the haplotype sequences were reconstructed with retrieve-variation-seq.
For a haplotype SNP set or single position variants to be reported in variation-scan, we requested that at least one of the sequences was evaluated as a binding site with a p-value of at most 10−4 (-uth pval 1e-4) and that the p-value ratio between the two alleles was greater or equal to 100 (a change of two orders of magnitude between the best and the worst alleles p-values) (-lth pval_ratio 100). In addition, we require a change of sign between the best and worst score as an additional filter.
We annotated the predicted disrupted TFBS with the TF ChIP-seq non-redundant peak collection and with the Cis-Regulatory Modules (CRM) regions from ReMap [10] using bedtools intersect version 2.27 [49]. We also calculated the enrichment for annotations in the provenance sequence segments of the predicted haplotypes sites.
2.6.2. Case study 2: prediction of regulatory variants associated with susceptibility to Mycobacterium tuberculosis infection
We collected SNPs associated with the phenotypic trait “susceptibility to Mycobacterium tuberculosis infection measurement” (disease ID EFO_0008407) from the 1.0.2 version of the GWAS catalog [40] (https://www.ebi.ac.uk/gwas/). This query returned one study [58] with 67 distinct variants, of which 48 had a valid reference SNP identifier (rsID) and could be further used (denoted hereafter as disease-associated SNPs, or DA-SNPs). To predict the TF binding sites putatively affected by these selected SNPs, we designed an approach combining Variation-tools with different external resources. We further collected from Ensembl REST interface (http://rest.ensembl.org/) 564 SNPs in linkage disequilibrium (LD-SNPs) in the European population [62], with a threshold on the regression coefficient (r2 ≥ 0.8) and a maximal distance of 200 bp.
Annotations (chromosomal location, type of genomic region) of the resulting 612 SNPs (48 DA + 564 LD) were collected from Ensembl BioMart [22], [21]. We then restricted the selection to SNPs in non-coding regions, resulting in a set of 572 SNPs of interest (SOIs) for the detection of regulatory variants. Using SNPs in LD, we determined LD-Block regions. These were then annotated based on overlaps with ChIP-seq peaks collected from the ReMap database [10]. We also calculated enrichment for disease annotations using the R XGR package [24].
Finally, we used retrieve-variation-seq to retrieve the sequence variants around each SOI, and predicted the impact of the variation on TF binding for each motif of the JASPAR non-redundant RSAT motif collection [8] using variation-scan, with the thresholds of 1e-4 on the p-value and 100 on the p-value ratio.
2.6.3. Case study 3: Assessment of the regulatory effect of GWAS reported variants in promoters with enhancer function
The STARR-seq assay [2] is in its principle similar to the MPRA, and helps identify self-transcribing active regulatory regions that have enhancer potential. Using this approach Dao et al. [17], analysed the enhancer potential of annotated RefSeq promoters [48]. In the two cell lines K562 and HELA, they identified 632 and 493 promoters with enhancer potential (ePromoters), respectively. Moreover, the authors identified enrichment of eQTL variants reported by GTEx [25].
To identify ePromoters variants that could be affecting TF binding, we retrieved the GWAS catalog version 1.0 (downloaded on 7/01/19) [40]. Using bedtools overlap version 2.26.0 [49], we computed the overlap between SNPs and the ePromoter coordinates reported in [17]. PSSMs representing TF enriched in ePromoters were also obtained from [17], corresponding to SMRC1, JUN, FOS, ATF:MAF:NEF2, YY1, ETS family, Creb and USF1/2.
Using the selected GWAS variants that fall within ePromoters and the TF motifs enriched in these regions, we applied variation-scan to assess the potential regulatory effect of these variants. variation-scan was run with the parameters – lth w_diff 1 – lth pval_ratio 10, with a background model built using create-background with all RefSeq promoter sequences. In order to filtrate variants with the highest putative regulatory disruption, we further selected variants that showed a change of sign in the weight score between alleles.
2.6.4. Case study 4: identification of regulatory variants affecting VRN1 binding in barley
The latest version of Hordeum vulgare (barley) reference genome [42] and a panel of mapped genetic variants were imported from Ensembl Genomes release 42 [34] and installed in the RSAT Plants server (http://plants.rsat.eu). We obtained experimentally determined binding sites (ChIP-seq) for VRN1 from [19]. Since these peaks were originally positioned within contigs of the 2012 genome assembly [14], they had to be matched to the corresponding regions of the current assembly with BLAST + v2.9.0 (blastn) local alignments against the repeat-masked genome sequence (perfect matches) [7]. Using bedtools overlap version 2.26.0 [49], we selected variants falling within the VRN1 reported binding peaks. The selected variants in VCF format were then processed using convert-variations and retrieve-variation-seq to obtain the sequences with the alternative alleles.
The VRN1 DNA motif used to scan the variants was obtained from the footprintDB plant collection [16] version: 2018-06 (http://floresta.eead.csic.es/footprintdb/index.php?motif=AY750993:VRN1:EEADannot). variation-scan was used with a pre-computed background Markov model (order 1) for barley to assess the effect of variants in TF binding, with the following parameters: – lth score 1 – lth w_diff 1 – lth pval_ratio 10 – uth pval 1e-3.
2.7. Availability
Variation-tools are available on the web (Metazoa: http://metazoa.rsat.eu/, Plants: http://plants.rsat.eu/, Teaching: http://teaching.rsat.eu/). The tools can be also installed for command-line usage with the RSAT suite (http://download.rsat.eu/).
The code and material to reproduce the results presented in the article can be accessed through GitHub (https://github.com/RSAT-doc/supp-material-publications.git).
3. Results
The Variation-tools provide complementary programs enabling the retrieval of variants (variation-info) and of their surrounding sequences (retrieve-variation-seq), as well as interconversion between file formats (convert-variation). The main predictive program is variation-scan, which can be used with any set of variants provided by the user (in VCF or GVF formats) or annotated in Ensembl (from a list of rsIDs or a bed file to identify overlapping variants in genome coordinates), with any set of motifs selected from the collections available in RSAT, or provided by the user.
3.1. variation-scan accurately assesses the effect of experimentally validated regulatory variants
The original version of variation-scan [45] required approximately five hours to assess the allele effect of nine millions variants. The novel version [47] significantly reduces the processing time to about one hour (Supplementary Fig. 3).
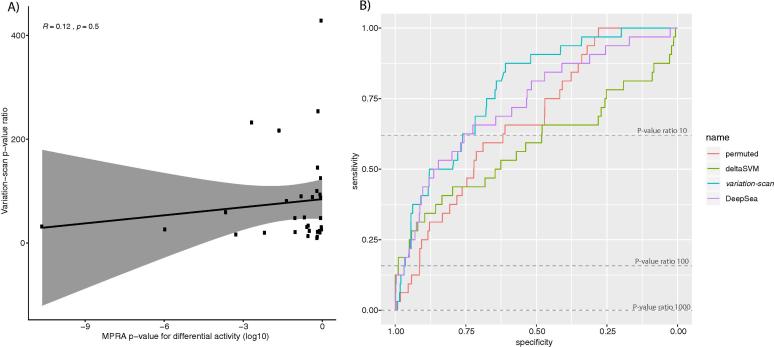
To evaluate the performance of variation-scan, we used an experimentally validated regulatory variant set obtained from a MPRA experiment [60]. For all of the assessed allele pairs, we compared the weight score differences computed with variation-scan with the mRNA/DNA ratio of the MPRA (see methods). As shown in Supplementary Fig. 4A, we are able to recover only 9.37% of the experimentally validated variants with variation-scan, as we requested at least one of the alleles to have a binding site of high confidence (p-value ≤ 10−4). Focusing on the variants reported as positive in the MPRA data set, we observed a weak correlation between the weight difference and the MPRA mRNA/DNA ratio in positive variants. However, this correlation is not significant, as MPRA values do not scale with the variation-scan weight differences. Nevertheless, all variants show a p-value ratio indicative of allele binding effects, showing that variation-scan gives accurate measurements of the impact of regulatory variants (Fig. 2A).
Fig. 2.
Identification of experimentally validated regulatory variants using variation-scan. A) Correlation of the Massively Parallel Reporter Assays (MPRA) p-value of the mRNA/DNA ratio of positive variants and the variation-scan weight difference for the MPRA variants with significant change. B) Receiver Operating Characteristic (ROC) curve comparing the performance when aiming to classify MPRA experimentally analyzed variants using variation-scan (turquoise), DeepSea (purple), deltaSVM (green), and a negative control which consists of permuted motifs scored with variation-scan (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
With the proposed thresholds, we can confidently reject 96.35% of MPRA negative sequences, which could be improved using more restrictive parameter, with a concomitant reduction in true positives. Noteworthy, as any high-throughput assay, MPRA has its limitations [51] and sequencing biases could increase the number of false negatives.
We performed a negative control, consisting of 240 permuted matrices (five permuted versions of the 48 motifs). With this collection, it was still possible to recover a group of variants, but it only represented 31.2% of the MPRA positive variants (Supplementary Fig. 4B).
We compared the performance of variation-scan to two other tools that had been previously used by Ulirsch, et al [60] to assess the same set of MPRA variants: DeepSea [68] and deltaSVM [37]. We decided to use the same parameters in order to avoid personal biases when calibrating the tools. Therefore, training weights for DNAse I hypersensitivity sites were used in the deltaSVM analysis. As for DeepSea, the web implementation of the tool was used, with the metric Functional Significance Score.
Tools were compared based on ROC curves (Fig. 2B), we additionally ran variation-scan using a set of permuted matrices as negative control (Fig. 2B, red line). The three tools show very similar sensitivity vs specificity at the beginning of the curves, but only variation-scan and DeepSea further remain separated from the negative control. As expected DeepSea performs slightly better than variation-scan at the beginning of the curve, nevertheless this tool requires training using epigenetic data, while variation-scan requires only a motif and a set of variants.
3.2. Variation-tools case studies
To illustrate the diverse applications of Variation-tools to tackle various biological questions, we designed four different case studies:
-
1.
Impact of regulatory variants in the same haplotype on TF binding sites.
-
2.
Identification of the regulatory potential of variants reported in GWAS.
-
3.
Assessment of the regulatory potential of GWAS variants within experimentally determined regulatory regions.
-
4.
Determination of regulatory variants within TF binding regions identified using ChIP-seq [19].
3.2.1. Genome-wide haplotype variant information can be used to identify sets of regulatory variants affecting the same TFBS
The lowering costs in sequencing have made it possible to obtain whole genome sequences of more individuals, opening the possibility of knowing, not only the variants of a genome, but also the haplotypes, and determining which variants are passed linked within the same chromosome. This enables the assessment of the regulatory effects of sets of variants within the same haplotype in a given TFBS.
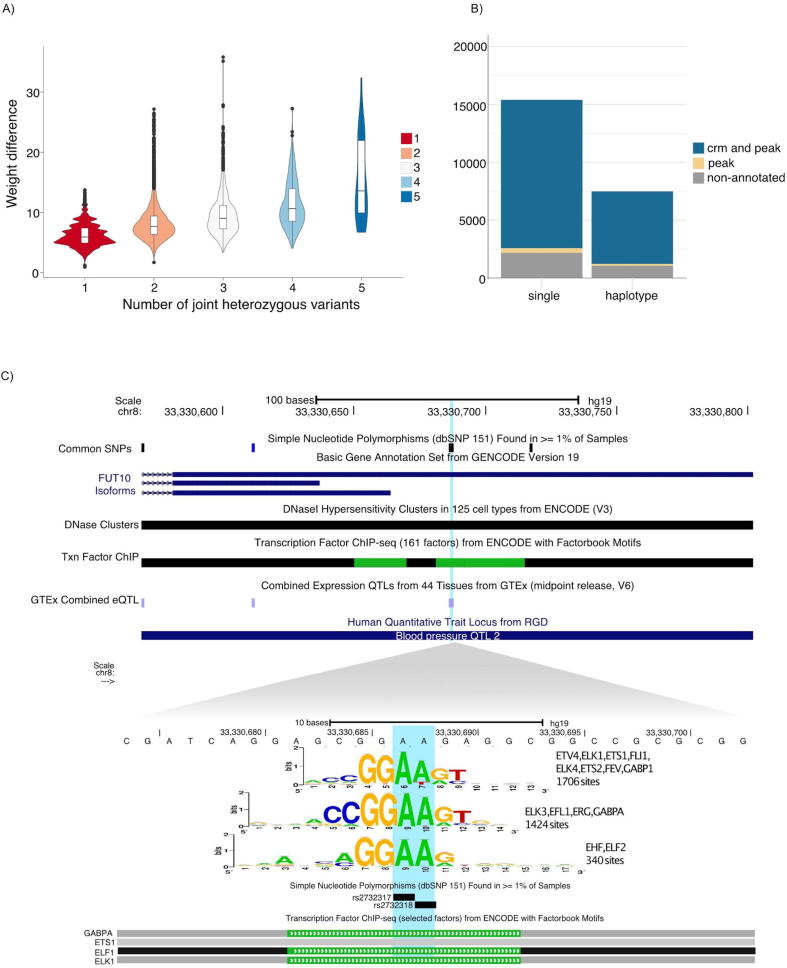
Using the high-confidence SNPs from two “Platinum” Genomes [23], we determined haplotype variants that are likely to affect one TFBS. We selected variants 30bps apart, located in open chromatin, to be analysed with variation-scan using the non-redundant motif collection at RSAT [8]. We detected 7,406 haplotype sites with at least two heterozygous variants and a probable effect in binding of 361 TFs. Overall the number of heterozygous variants within a haplotype increases the measured weight difference. This is expected as more changes in the binding sites are more likely to change TF affinity (Fig. 3A).
Fig. 3.
Haplotype analysis in high-quality human genomes. A) The number of heterozygous variants (X-axis) within the same putative binding site tend to have a greater impact on the TF binding probability. This is expected as the increase of weight difference observed on the violin plot corresponds to the expected cumulated impact of variations affecting different positions of the same binding site. B) Number of predicted disrupted Transcription Factor Binding Sites (TFBSs) with Cis-Regulatory Modules (CRMs) and TF ChIP-seq peak annotation (blue), with only peak annotation (yellow), and non-annotated predictions (grey). C) University of California Santa Cruz (UCSC) browser [48] screen shot, showing a locus encompassing two SNPs that compose an heterozygous haplotype in one of the Northern Europeans from Utah (CEU) individuals. The figure shows the reference genome haplotype. The variants are located in the FUT10 promoter (top). variation-scan predicts an effect in three motifs that represent binding sites for GABPA, ETS1 and ELF2, factors that have been proven to have binding sites in this region by the ENCODE project. The variant rs2732317 has been associated with effects in gene expression by the GTEx project. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To assess the biological relevance of all the putative disrupted TFBS predictions, we annotated 7,485 predicted haplotypes sites containing two or more variants with at least one heterozygous variant and 15,396 predicted sites containing a SNP (singletons) with the TF ChIP-seq peaks and the Cis-Regulatory Modules (CRM) regions from ReMap [10]. We found that almost all the predicted disrupted TFBS (~85%) contain a CRM or peak annotation or both (Fig. 3B). Interestingly, we found enrichment of CRM and peak annotations in the provenance sequence segments of the 7,485 predicted haplotypes sites compared to the provenance sequence segments of the single variants (Fisher exact test, p-value < 2.2e-16).
One of these annotated haplotypes is composed of the minor alleles of two SNPs (rs2732317 and rs2732318), where we observed a potential regulatory effect likely affecting three binding motifs, for EHF/ELF2, ETV4/ELK1/ETS1/FLI1/ELK4/ETS2/FEV/GABP1, and ELK3/ELF1/ERG/GABPA (Fig. 3C).
3.2.2. Genetic variants associated with Mycobacterium tuberculosis infection show potential regulatory effects
The second case study illustrates a knowledge-free use of Variation-tools to identify regulatory variants from GWAS studies for a user-specified disease, without prior indication about the potentially involved transcription factors or binding motifs. The approach is based on the prediction of regulatory variants with RSAT Variation-tools, narrowed down by selecting the regulatory SNPs that overlap ChIP-seq peaks in ReMap [10], in order to identify convergent indications for a potential impact of the variants on the binding of a TF.
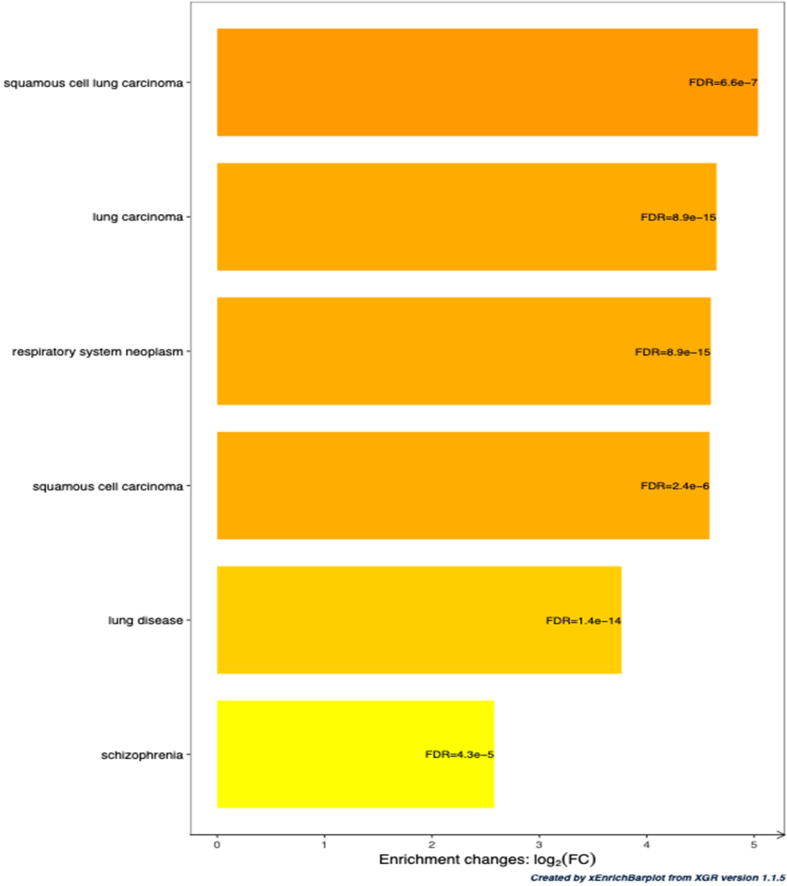
Interestingly, the 572 SNPs of interest (SOIs, see methods) show a significant enrichment for diseases related to respiratory functions (lung carcinoma, respiratory neoplasm, squamous cell carcinoma, lung disease), as well as for schizophrenia (Fig. 4), confirming the relevance of the collection of SNPs.
Fig. 4.
Enrichment of the set of SNPs of Interest (SOIs) for diseases. The SNPs of interest includes SNPs reported by a GWAS to be associated with resistance to Mycobacterium tuberculosis infection and the SNPs in linkage disequilibrium with those. The genes associated with these SNPs were compared to each term of a catalogue of diseases.
The scanning of the SOIs with the 579 matrices of Jaspar core non-redundant Vertebrate collection predicted 107 modifications of TF binding, covering 66 distinct SNPs and 181 distinct motifs. There are 4,847 overlaps between the 80 million ChIP-seq peaks of the ReMap catalog and 263 of the 572 initial SOIs, but only two of them show a match between the TF of the ChIP-seq peak and that corresponding to the motif returned by variation-scan: CEBPB for reference SNP rs3131071 and ELF1 for rs3132397. Noticeably, CEBPB has been reported as the main regulator for genes differentially expressed between tuberculosis patients and control cases [39]. CEBPB has also been associated with the pathogenesis of tuberculosis. This factor is involved in the differentiation and activation of macrophages and in the regulation of the immune and inflammatory response. It also plays a crucial role in the stimulation of IgG immune compounds [39]. In summary, the convergence between ChIP-seq and motif scanning results enabled the identification of two promising candidates among the 66 candidate regulatory SNPs. The same approach can be applied to other association studies in order to predict regulatory variations potentially involved in user-specified diseases.
3.2.3. Assessment of the regulatory effect of GWAS reported variants in promoters with enhancer function
ePromoters are regulatory regions with dual functions: as promoters, they regulate the gene downstream, but they also show enhancer potential according to [17]. ePromoters have been described to be enriched for eQTLs, suggesting that their function could be affected by genetic variants. We thus set out to investigate if GWAS reported variants could be affecting TF binding in ePromoters.
We identified five and twelve GWAS reported variants falling within the reported coordinates of ePromoters corresponding to the human cell lines HELA and K562, respectively. Using variation-scan with the motifs of TF reported as enriched in ePromoters, we were able to detect two variants (rs3771180, rs3822259) affecting two TF binding motifs in HELA (MAF::NFE2, FLI1/FEV/ETS2/ELK4/ELK4/GABP1/Gabpa), and two variants (rs147997200, rs62229372) affecting three binding motifs in K562 (Creb3l2, Atf3/MAFG::NFE2L1/MAFG/NF2L1, FOS::JUN).
Noteworthy, in HELA ePromoters, we found the SNP rs3771180, which is also described as an eQTL in the whole blood dataset by the GTEx project. This variant has been associated with asthma and is upstream the Interleukin Receptor 1 gene, which is consistent with ePromoters related to inflammatory response in HELA [17].
3.2.4. Population genetic variations in barley can potentially affect VRN1 binding
Variation in traits in crops can be due to changes in TF binding that likely affect gene regulation. As a proof of concept that Variation-tools can be used for this purpose, we set out to identify reported variants in Hordeum vulgare (barley) that can putatively affect binding of VRN1, a TF involved in vernalization response. We focused this analysis on VRN1 ChIP-seq reported regions [19], selecting only variants that overlapped them (n = 1604).
Using the VRN1 motif annotated in footprintDB and the barley variants annotated in Ensembl Plants, we identified a total of thirteen variants likely to affect VRN1 binding. Of these, two are proximal to genes MLOC_73196 and MLOC_79452, which belong to a set of 38 genes known to change their expression level upon VRN1 binding in RNA-seq experiments [19].
3.3. Limitations and parameter selection
One issue arising from the analysis of big data sets is the number of false positives [44], due to the weak information content of TF binding motifs relative to genome sizes. This is one of the limitations of any bioinformatics approach to predict TF binding sites, and thus also affects the performances of variation-scan (Supplementary Fig. 4A and B).
Taking advantage of specific biological insights (e.g. identification of relevant TF, reduction of genomic regions using functional genomic information) can significantly improve results, reducing the number of false positives [9], [55]. In this respect, Case Studies 3 and 4 focused on the analysis on TF known to bind on the regions of interest, which enabled us to consistently assess the performance of the tool in the evaluation set, and further helped us to identify biologically relevant regulatory variants, affecting ePromoters function in Case Study 3, or affecting VRN1 binding in barley in Case Study 4. Regarding Case Study 2, the usage of ChIP-seq information enabled the identification of potentially relevant variants related to tuberculosis.
Furthermore, the selection of adequate thresholds to select variants with variation-scan has an impact on reducing the number of false positives:
-
•
P-value (-uth pval): This option refers to the upper threshold set on the p-value; this criteria has to be valid for the binding site prediction associated to at least one of the alleles; this means that at least one of the alleles allows for the prediction of a reliable binding site.
-
•
Weight difference (-w_diff): This option determines minimal allowed weight differences between the predicted binding sites of two alleles (see methods for a description of how the weight difference is calculated).
-
•
P-value ratio (-lth pval_ratio): This option determines the lower threshold for the p-value ratio between the predicted binding sites of two alleles (see methods for a description of how the weight difference is calculated).
Depending on the biological question, users should decide to use more or less restrictive thresholds. As shown in Case Studies 3 and 4, when the biological hypothesis is well defined, lower thresholds return manageable numbers of predictions with interesting biological insights. For more general biological questions, as in Case Studies 1 and 2, using a larger number of data, we recommend to select more stringent thresholds to reduce the number of false positives, and thereby focus the analysis on the best predictions.
Regarding the selection of a particular set of motifs, there are multiple databases installed within RSAT, which provide easy access to several reference collections (i.e. JASPAR, HOCOMOCO, etc.). The selection will depend on the biological question. For some TF and TF families there are structural descriptions of the protein-DNA interfaces. In some cases these structures can be used to map TF residues to particular bases within the DNA motif. Motifs from 3D-footprint [15], which are part of the footprintDB collection (http://floresta.eead.csic.es/footprintdb), allow users to further investigate the effect of variants in the light of structural information.
4. Discussion
The lowering costs of sequencing technologies has facilitated the identification of genetic variants associated with traits and diseases in humans and other species [64]. For this reason, the identification of variants affecting TF binding sites has become mainstream [3], [53], [32], calling for efficient computational approaches to analyse large sets of variants [18].
The case studies presented here demonstrate the application of RSAT Variation-tools to a diverse selection of real-world problems. Current genotype information facilitates the characterization of haplotypes, but this requires tools designed to take advantage of this information [13], [52], [11]. In Case Study 1, we show how the tool convert-variations facilitates the usage of this information, by enabling users to analyse the impact of combinations of several variants located within a 30 bp window of a chromosome. Indeed, a specific combination of variations in the same haplotype may have a synergic impact on a given TF binding site, whilst the analysis of individual variations may fail to reveal some actual regulatory impact. In the absence of information enabling TF preselection, we decided to use the complete collection of motifs. Nevertheless, by requiring more than one SNP affecting one TF binding site, we were able to identify haplotypes with potential regulatory effects. In the advent of new genome-wide characterization in population studies, this function will facilitate the integration of phasing information in the search of regulatory variants.
The identification of causal regulatory variants is a real challenge, for several reasons: (i) GWAS, which typically cover one million SNPs (“tag SNPs”), only represent a small fraction of the actual variants (150 millions currently known); (ii) the information content of a TF binding motif is relatively small, so that testing the potential impact of hundreds – or thousands – of candidate SNPs on the binding of hundreds of TF will unavoidably return an important number of false positives. A strategy to circumvent this intrinsic limitation is to take into account TF binding regions evidenced by ChIP-seq peak experiments, in order to prioritize the predictions of regulatory variants. Case Study 2 shows that this approach ranks first SNPs highly relevant for susceptibility to Mycobacterium tuberculosis infection. It has to be noted that the absence of ChIP-seq peak does not preclude a predicted regulatory variant from being valid. Indeed, ChIP-seq data are only available for a subset of transcription factors, and only indicate the TF binding locations for the specific cell types or tissues in which the experiments have been performed, which may differ from those involved in the aetiology of the considered disease. The consistency between ChIP-seq peaks and predicted regulatory variants should thus be considered as a way to identify the most promising candidates rather than as a strict requirement to consider a prediction as valid.
The third case study focuses on ePromoters, defined as are regulatory regions with the capacity to act both as promoters and as enhancers, and thus the potential to affect more than one gene. Hence, regulatory variants associated with human diseases in these loci can have complex effects on gene regulation. We were able to identify four variants affecting TF binding within ePromoters in HELA and K562, ePromoters function has been linked to quick response gene expression related to inflammation and stress [46]. We were able to identify four SNPs (rs3771180, rs3822259, rs147997200 and rs62229372) putatively affecting TF binding, which are associated with traits related to inflammation and stress, supporting the relevance of ePromoters in inflammatory response.
The fourth case study takes published barley data and enlights natural variations in two regulatory regions bound by transcription factor VRN1 that are predicted to have an effect on the expression of two downstream genes. One of them is annotated as an amino acid permease (MLOC_73196), but the other one (MLOC_79452) is a protein-coding gene of unknown function. Further work would be required to confirm whether these natural variants display relevant phenotypes.
Finally, while Variation-tools provide a flexible framework to assess the effect of variants in TF gene regulation, there are other factors affecting regulatory mechanisms that may be taken into account, such as i) DNA accessibility [33], ii) DNA shape [66], iii) DNA methylation (Xuan [65] and iv) TF protein availability [67].
5. Conclusions
Variation-tools enables the prediction of the effect of sequence variants on TF binding. In addition to reasonable computing time, the focus is put on usability and high flexibility: annotated variants can be retrieved from specific genomic loci, as well as from personal collections of variants, motifs (provided as PSSMs) can be chosen from the collections available in RSAT (JASPAR, HOCOMOCO, CisBP, etc.), as well as from user-provided PSSM sets. The tools supports various organisms in selected RSAT servers: currently Metazoa, Plants and Teaching. In addition to the web interface, Variation-tools can also be used on the command line to facilitate analysis of custom data sets. Variation-tools can be used in combination with external databases, as exemplified with the study of GWAS data. Finally, as part of the long-lasting RSAT suite, Variation-tools programs are continuously maintained and updated.
Acknowledgments
Acknowledgements
We thank the persons contributing to the maintenance of the RSAT servers, in particular Laboratorio Nacional de Visualización Científica Avanzada (Mexico) specially Luis Alberto Aguilar Bautista and Jair Garcia Sotelo, the ABims platform in Roscoff, France, Pierre Vincens at the ENS, Paris and Aurora Martín Cotaina from EEAD-CSIC for her help on managing the Plants server. We acknowledge Salvatore Spicuglia for useful comments during the development of the tools. We thank Lambert Moyon and Swann Floc’hlay for providing feedback on the use of Variation-tools. We thank Alejandra Castillo and Carina Uribe for technical assistance. We thank Mauricio Guzman for styling the figures.
Funding
A.M.-R.’s laboratory is supported by a Consejo Nacional de Ciencia y Tecnologia (CONACYT) grant [269449]; Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica – Universidad Nacional Autónoma de México (PAPIIT-UNAM) grant [IA206517-IA201119]; M.T.-C., A.M.-R and D.T. further acknowledge SEP-CONACYT-ECOS-ANUIES [291235] support. M. T.-C. and W. S.-G. are supported by the Institut Universitaire de France. W. S.-G. benefits from a Master fellowship of the Institut de Convergences Q-life of PSL. B.C.M. was supported by Gobierno de Aragón grant A08_17R (“Genética, genómica, biotecnología y mejora de cultivos”).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.09.009.
Contributor Information
Jacques van Helden, Email: Jacques.van-Helden@univ-amu.fr.
Alejandra Medina-Rivera, Email: amedina@liigh.unam.mx.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.1000 Genomes Project Consortium, Adam Auton, Lisa D. Brooks, Richard M. Durbin, Erik P. Garrison, Hyun Min Kang, Jan O. Korbel, et al. 2015. A Global Reference for Human Genetic Variation. Nature 526 (7571): 68–74. [DOI] [PMC free article] [PubMed]
- 2.Arnold, Cosmas D., Daniel Gerlach, Christoph Stelzer, Łukasz M. Boryń, Martina Rath, Alexander Stark. 2013. Genome-Wide quantitative enhancer activity maps identified by STARR-Seq. Science, March. https://doi.org/10.1126/science.1232542. [DOI] [PubMed]
- 3.Behera Vivek, Evans Perry, Face Carolyne J., Hamagami Nicole, Sankaranarayanan Laavanya, Keller Cheryl A. Exploiting genetic variation to uncover rules of transcription factor binding and chromatin accessibility. Nat Commun. 2018;9(1):782. doi: 10.1038/s41467-018-03082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein Bradley E., Birney Ewan, Dunham Ian, Green Eric D., Gunter Chris, Snyder Michael. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle, Alan P., Eurie L. Hong, Manoj Hariharan, Yong Cheng, Marc a. Schaub, Maya Kasowski, Konrad J. Karczewski, et al. 2012. Annotation of functional variation in personal genomes using RegulomeDB. Genome Research 22 (9): 1790–97. [DOI] [PMC free article] [PubMed]
- 6.Browning Sharon R., Browning Brian L. Haplotype phasing: existing methods and new developments. Nat Rev Genet. 2011;12(10):703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho Christiam, Coulouris George, Avagyan Vahram, Ma Ning, Papadopoulos Jason, Bealer Kevin. BLAST+: architecture and applications. BMC Bioinf. 2009;10(December):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro-Mondragon, Jaime Abraham, Sébastien Jaeger, Denis Thieffry, Morgane Thomas-Chollier, Jacques Van Helden. 2017. RSAT Matrix-clustering: dynamic exploration and redundancy reduction of transcription factor binding motif collections. Nucleic Acids Res 45 (13). https://doi.org/10.1093/nar/gkx314. [DOI] [PMC free article] [PubMed]
- 9.Chen, Chih-Yu, I-Shou Chang, Chao A. Hsiung, and Wyeth W. Wasserman. 2014. On the identification of potential regulatory variants within genome wide association candidate SNP sets. BMC Med Genom 7 (June): 34. [DOI] [PMC free article] [PubMed]
- 10.Chèneby Jeanne, Gheorghe Marius, Artufel Marie, Mathelier Anthony, Ballester Benoit. ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-Seq experiments. Nucleic Acids Res. 2018;46(D1):D267–D275. doi: 10.1093/nar/gkx1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi Yongwook, Chan Agnes P., Kirkness Ewen, Telenti Amalio, Schork Nicholas J. Comparison of phasing strategies for whole human genomes. PLoS Genet. 2018;14(4) doi: 10.1371/journal.pgen.1007308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coetzee Simon G., Coetzee Gerhard A., Hazelett Dennis J. motifbreakR: an R/bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31(23):3847–3849. doi: 10.1093/bioinformatics/btv470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International Hapmap Consortium A haplotype map of the human genome. Nature. 2005;437(7063):1299. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The International Barley Genome Sequencing Consortium, The International Barley Genome Sequencing Consortium A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012 doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 15.Contreras-Moreira, Bruno. 2010. 3D-Footprint: A database for the structural analysis of protein-DNA complexes. Nucleic Acids Res 38 (Database issue): D91–97. [DOI] [PMC free article] [PubMed]
- 16.Contreras-Moreira Bruno, Sebastian Alvaro. FootprintDB: analysis of plant cis-regulatory elements, transcription factors, and binding interfaces. Methods Mol Biol. 2016;1482:259–277. doi: 10.1007/978-1-4939-6396-6_17. [DOI] [PubMed] [Google Scholar]
- 17.Dao, Lan T. M., Ariel O. Galindo-Albarrán, Jaime A. Castro-Mondragon, Charlotte Andrieu-Soler, Alejandra Medina-Rivera, Charbel Souaid, Guillaume Charbonnier, et al. 2017. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat Genetics 49 (7): 1073–81. [DOI] [PubMed]
- 18.D’Argenio, Valeria. 2018. The high-throughput analyses era: are we ready for the data struggle? High-Throughput 7 (1). https://doi.org/10.3390/ht7010008. [DOI] [PMC free article] [PubMed]
- 19.Deng Weiwei, Cristina Casao M., Wang Penghao, Sato Kazuhiro, Hayes Patrick M., Jean Finnegan E. Direct links between the vernalization response and other key traits of cereal crops. Nat Commun. 2015;6(January):5882. doi: 10.1038/ncomms6882. [DOI] [PubMed] [Google Scholar]
- 20.Deplancke Bart, Alpern Daniel, Gardeux Vincent. The genetics of transcription factor DNA binding variation. Cell. 2016;166(3):538–554. doi: 10.1016/j.cell.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Durinck Steffen, Moreau Yves, Kasprzyk Arek, Davis Sean, De Moor Bart, Brazma Alvis. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21(16):3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 22.Durinck Steffen, Spellman Paul T., Birney Ewan, Huber Wolfgang. Mapping identifiers for the integration of genomic datasets with the R/bioconductor package biomaRt. Nat Protoc. 2009;4(8):1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberle Michael A., Fritzilas Epameinondas, Krusche Peter, Källberg Morten, Moore Benjamin L., Bekritsky Mitchell A. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. 2017;27(1):157–164. doi: 10.1101/gr.210500.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang Hai, Knezevic Bogdan, Burnham Katie L., Knight Julian C. XGR software for enhanced interpretation of genomic summary data, illustrated by application to immunological traits. Genome Med. 2016;8(1):129. doi: 10.1186/s13073-016-0384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI, et al. 2017. Genetic Effects on Gene Expression across Human Tissues. Nature 550 (7675): 204–13. [DOI] [PMC free article] [PubMed]
- 26.van Helden Jacques. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31(13):3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hertz G.Z., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15(7–8):563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 28.Huang Dandan, Yi Xianfu, Zhang Shijie, Zheng Zhanye, Wang Panwen, Xuan Chenghao. GWAS4D: multidimensional analysis of context-specific regulatory variant for human complex diseases and traits. Nucleic Acids Res. 2018;46(W1):W114–W120. doi: 10.1093/nar/gky407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt Sarah E, William McLaren, Laurent Gil, Anja Thormann, Helen Schuilenburg, Dan Sheppard, Andrew Parton, et al. 2018. Ensembl variation resources.Database: J Biol Databases Curat 2018 (January). 10.1093/database/bay119. [DOI] [PMC free article] [PubMed]
- 30.International HapMap Consortium The international HapMap project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 31.Inukai Sachi, Kock Kian Hong, Bulyk Martha L. Transcription factor–DNA binding: beyond binding site motifs. Curr Opin Genet Dev. 2017;43(April):110–119. doi: 10.1016/j.gde.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalita Cynthia A., Brown Christopher D., Freiman Andrew, Isherwood Jenna, Wen Xiaoquan, Pique-Regi Roger. High-throughput characterization of genetic effects on DNA–protein binding and gene transcription. Genome Res. 2018;28(11):1701–1708. doi: 10.1101/gr.237354.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan Tommy, Li Xiao-Yong, Sabo Peter J., Thomas Sean, Stamatoyannopoulos John A., Biggin Mark D. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early drosophila development. PLoS Genet. 2011 doi: 10.1371/journal.pgen.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kersey Paul Julian, Allen James E., Allot Alexis, Barba Matthieu, Boddu Sanjay, Bolt Bruce J. Ensembl genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018;46(D1):D802–D808. doi: 10.1093/nar/gkx1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar Sunil, Ambrosini Giovanna, Bucher Philipp. SNP2TFBS-a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 2017;45(D1):D139–D144. doi: 10.1093/nar/gkw1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert Samuel A., Jolma Arttu, Campitelli Laura F., Das Pratyush K., Yin Yimeng, Albu Mihai. The human transcription factors. Cell. 2018;175(2):598–599. doi: 10.1016/j.cell.2018.09.045. [DOI] [PubMed] [Google Scholar]
- 37.Lee Dongwon, Gorkin David U., Baker Maggie, Strober Benjamin J., Asoni Alessandro L., McCallion Andrew S. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet. 2015;47(8):955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lelli Katherine M., Slattery Matthew, Mann Richard S. Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet. 2012;46(August):43–68. doi: 10.1146/annurev-genet-110711-155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Yan, Duan Zipeng, Feng Xu., Zhang Jiayuan, Shulgina Marina V., Li Fan. Construction and analysis of the transcription factor-microRNA co-regulatory network response to mycobacterium tuberculosis: a view from the blood. Am J Transl Res. 2017;9(4):1962–1976. [PMC free article] [PubMed] [Google Scholar]
- 40.MacArthur Jacqueline, Bowler Emily, Cerezo Maria, Gil Laurent, Hall Peggy, Hastings Emma. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manke Thomas, Heinig Matthias, Vingron Martin. Quantifying the effect of sequence variation on regulatory interactions. Hum Mutat. 2010;31:477–483. doi: 10.1002/humu.21209. [DOI] [PubMed] [Google Scholar]
- 42.Mascher Martin, Gundlach Heidrun, Himmelbach Axel, Beier Sebastian, Twardziok Sven O., Wicker Thomas. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544(7651):427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 43.Maurano, Matthew T., Hao Wang, Tanya Kutyavin, John A. Stamatoyannopoulos. 2012. Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genetics 8 (3): e1002599. [DOI] [PMC free article] [PubMed]
- 44.Medina-Rivera Alejandra, Abreu-Goodger Cei, Thomas-Chollier Morgane, Salgado Heladia, Collado-Vides Julio, van Helden Jacques. Theoretical and empirical quality assessment of transcription factor-binding motifs. Nucleic Acids Res. 2011;39(3):808–824. doi: 10.1093/nar/gkq710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medina-Rivera Alejandra, Defrance Matthieu, Sand Olivier, Herrmann Carl, Castro-Mondragon Jaime A., Delerce Jeremy. RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 2015;43(W1):W50–W56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medina-Rivera Alejandra, Santiago-Algarra David, Puthier Denis, Spicuglia Salvatore. Widespread enhancer activity from core promoters. Trends Biochem Sci. 2018;43(6):452–468. doi: 10.1016/j.tibs.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen Nga Thi, Thuy Bruno Contreras-Moreira, Castro-Mondragon Jaime A., Santana-Garcia Walter, Ossio Raul, Robles-Espinoza Carla Daniela. RSAT 2018: regulatory sequence analysis tools 20th anniversary. Nucleic Acids Res. 2018;46(W1):W209–W214. doi: 10.1093/nar/gky317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Leary Nuala A., Wright Mathew W., Rodney Brister J., Ciufo Stacy, Haddad Diana, McVeigh Rich. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan Aaron R., Hall Ira M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramirez Fidel, Ryan Devon P., Gruning Bjorn, Bhardwaj Vivek, Kilpert Fabian, Richter Andreas S. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(April):160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santiago-Algarra, David, Lan T. M. Dao, Lydie Pradel, Alexandre España, Salvatore Spicuglia. 2017. Recent advances in high-throughput approaches to dissect enhancer function. F1000Research 6 (June): 939. [DOI] [PMC free article] [PubMed]
- 52.Seo Jeong-Sun, Rhie Arang, Kim Junsoo, Lee Sangjin, Sohn Min-Hwan, Kim Chang-Uk. De Novo assembly and phasing of a Korean human genome. Nature. 2016;538(7624):243–247. doi: 10.1038/nature20098. [DOI] [PubMed] [Google Scholar]
- 53.Sewell, Jared Allan, Shaleen Shrestha, Clarissa Stephanie Santoso, Elena Forchielli, Sebastian Carrasco Pro, Melissa Martinez, and Juan Ignacio Fuxman Bass. 2018. Uncovering human transcription factor interactions associated with genetic variants, Novel DNA motifs, and repetitive elements using enhanced yeast one-hybrid assays. bioRxiv. 10.1101/459305. [DOI] [PMC free article] [PubMed]
- 54.Shin Sunyoung, Hudson Rebecca, Harrison Christopher, Craven Mark, Keles Sündüz. atSNP search: a web resource for statistically evaluating influence of human genetic variation on transcription factor binding. Bioinformatics. 2018;(December) doi: 10.1093/bioinformatics/bty1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi Wenqiang, Fornes Oriol, Mathelier Anthony, Wasserman Wyeth W. Evaluating the impact of single nucleotide variants on transcription factor binding. Nucleic Acids Res. 2016;44(21):10106–10116. doi: 10.1093/nar/gkw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stormo Gary D. Modeling the specificity of protein-DNA interactions. Quantitative Biology (Beijing, China) 2013;1(2):115–130. doi: 10.1007/s40484-013-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E. The accessible chromatin landscape of the human genome. Nature. 2012;488(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian Chao, Hromatka Bethann S., Kiefer Amy K., Eriksson Nicholas, Noble Suzanne M., Tung Joyce Y. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat Commun. 2017;8(1):599. doi: 10.1038/s41467-017-00257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turatsinze Jean-Valery, Thomas-Chollier Morgane, Defrance Matthieu, Van Helden Jacques. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc. 2008;3(10):1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- 60.Ulirsch Jacob C., Nandakumar Satish K., Wang Li, Giani Felix C., Zhang Xiaolan, Rogov Peter. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell. 2016;165(6):1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Junbai, Batmanov Kirill. BayesPI-BAR: a new biophysical model for characterization of regulatory sequence variations. Nucleic Acids Res. 2015;43(21) doi: 10.1093/nar/gkv733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward Lucas D., Kellis Manolis. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward Lucas D., Kellis Manolis. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wetterstrand KA. 2019. DNA Sequencing Costs: Data. Genome.gov. July 23, 2019. https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data.
- 65.Lin Xuan, Xiao Quy, Sian Stephanie, An Omer, Thieffry Denis, Jha Sudhakar. MethMotif: an integrative cell specific database of transcription factor binding motifs coupled with DNA methylation profiles. Nucleic Acids Res. 2019;47(D1):D145–D154. doi: 10.1093/nar/gky1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, Lin, Yaron Orenstein, Arttu Jolma, Yimeng Yin, Jussi Taipale, Ron Shamir, Remo Rohs. 2017. Transcription factor family‐specific DNA shape readout revealed by quantitative specificity models. Mol Syst Biol 13 (2). 10.15252/msb.20167238. [DOI] [PMC free article] [PubMed]
- 67.Zabet Nicolae Radu, Adryan Boris. Estimating binding properties of transcription factors from genome-wide binding profiles. Nucleic Acids Res. 2015 doi: 10.1093/nar/gku1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou Jian, Troyanskaya Olga G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12(10):931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zuo Chandler, Shin Sunyoung, Keleş Sündüz. atSNP: transcription factor binding affinity testing for regulatory SNP detection. Bioinformatics. 2015;31(20):3353–3355. doi: 10.1093/bioinformatics/btv328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.