Abstract
Terminally hydroxylated fatty acids or dicarboxylic acids are industrially relevant compounds with broad applications. Here, we present the proof of principle for the de novo biosynthesis of 8-hydroxyoctanoic acid from glucose and ethanol in the yeast Saccharomyces cerevisiae. Toxicity tests with medium-chain length ω-hydroxy fatty acids and dicarboxylic acids revealed little or no growth impairments on yeast cultures even at higher concentrations. The ability of various heterologous cytochrome P450 enzymes in combination with their cognate reductases for ω-hydroxylation of externally fed octanoic acid were compared. Finally, the most efficient P450 enzyme system was expressed in a yeast strain, whose fatty acid synthase was engineered for octanoic acid production, resulting in de novo biosynthesis of 8-hydroxyoctanoic acid up to 3 mg/l. Accumulation of octanoic acid revealed that cytochromes P450 activities were limiting 8-hydroxyoctanoic acid synthesis. The hydroxylation of both externally added and intracellularly produced octanoic acid was strongly dependent on the carbon source used, with ethanol being preferred. We further identified the availability of heme, a cofactor needed for P450 activity, as a limiting factor of 8-hydroxyoctanoic acid biosynthesis.
Keywords: ω-Hydroxy fatty acids; α,ω-dicarboxylic acids; 8-Hydroxyoctanoic acid; Cytochrome P450; Toxicity test; S. cerevisiae; Oleochemicals
Highlights
-
•
Low toxic effects of medium-chain ω-hydroxy fatty acids on yeast cells .
-
•
Systematic comparison of cytochrome P450 enzyme activities on octanoic acid .
-
•
De novo biosynthesis of 8-hydroxyoctanoic acid .
-
•
Improvement of cytochrome P450 activity with ethanol or by addition of hemin .
1. Introduction
The industrial production of fatty acids (FA) and derived compounds is mainly based on extraction from plant oils or animal fats, or on petrochemical syntheses routes (Haupt et al., 1984; Akaike, 1985; Rupilius and Ahmad, 2007). However, environmental concerns regarding the use of fossil resources, deforestation and excessive land and water use for oil plant cultivation (Schmidt, 2015) as well as challenges in chemical synthesis (i.e. ω-hydroxy fatty acids) (Scheps et al., 2013) have increased interest in production through microbial biosynthesis. The engineering of microbial cell factories therefore holds the potential for a “greener” industry through environmentally friendly production conditions and increased sustainability. Hydroxy fatty acids and dicarboxylic acids are fatty acid derived compounds which harbor additional hydroxyl or carboxyl groups. They have compelling chemical properties and are relevant for chemical, pharmaceutical, food and cosmetic industries (Kim and Oh, 2013), are used as additives for lubricants, emulsifiers and can have antibiotic, anti-inflammatory and anticancer properties (Kim and Oh, 2013). Additionally, they can serve as building blocks for polymers and bioplastics (Liu et al., 2011; Scheps et al., 2013).
There is great interest in microbial biosynthesis of ω-hydroxy fatty acids (HyFA) and α,ω-dicarboxylic acids (DCA) (Lu et al., 2010; Clomburg et al., 2015; Durairaj et al., 2015; Han et al., 2017; Haushalter et al., 2017; Liu et al., 2019). In Saccharomyces cerevisiae, a host organism with a long tradition in the biotech industry, the production of medium-chain (C8–C14) HyFA variants by bioconversion (Durairaj et al., 2015; Han et al., 2017) or long-chain (C16–C18) variants by de novo biosynthesis (Liu et al., 2019) has been achieved by terminal hydroxylation of FAs. Subsequent oxidation of the terminal hydroxyl group by endogenous alcohol and aldehyde dehydrogenases has led to formation of DCAs (Han et al., 2017).
Formation of HyFAs from FA precursors requires efficient hydroxylation of the terminal carbon. Such reactions can be catalyzed by enzymes belonging to the superfamily of cytochromes P450 (CYPs), which are active on a large variety of different substrates (Renault et al., 2014). CYPs are hemeproteins which act as monooxygenases by activating molecular oxygen and transferring one atom to its substrate and reducing the other to water (Munro et al., 2013). Two electrons are required for the CYP reaction which are transferred by donor proteins. The membrane anchored eukaryotic CYPs mainly act in a two-component system, in which the electrons are transferred from NADPH to CYP by a cytochrome P450 reductase (CPR) (Munro et al., 2013). Several eukaryotic CYPs have been identified to specifically ω-hydroxylate medium-chain FAs (Fisher et al., 1998; Durairaj et al., 2015; Han et al., 2017).
The aforementioned bioconversions of medium-chain FAs to HyFAs were achieved by supplementation of FAs. In contrast, in this work we aimed to construct a complete pathway from glucose or ethanol to the medium-chain length 8-hydroxyoctanoic acid (C8-HyFA) in S. cerevisiae. This, however, requires direct synthesis of octanoic acid (C8-FA) from those carbon sources. In the yeast cytosol, FAs are synthesized de novo by the type I fatty acids synthase (FAS), a large enzymatic complex (Lomakin et al., 2007), which is encoded by the two genes FAS1 and FAS2. Intermediates of FA biosynthesis are covalently linked to FAS via the ACP domain, and are released in a coenzyme A (CoA) bound form with a length of mainly 16 or 18 carbon atoms (Lomakin et al., 2007). Subsequent cleavage of the CoA-FA-thioester bound by endogenous thioesterases yields free FA. Multiple studies have addressed the enrichment of medium-chain FAs by heterologous expression of medium-chain specific thioesterases (Leber and Da Silva, 2014; Fernandez-Moya et al., 2015; Zhu et al., 2017). An alternative approach for medium-chain FA biosynthesis has been carried out by rational engineering of the FAS (Gajewski et al., 2017). Mutations in relevant domains increased medium-chain FAs production. A single mutation (FAS1R1834K) allowed highly selective production of C8-FA (about 90% of secreted fatty acids) (Gajewski et al., 2017). Thus, we considered the FAS1R1834K/FAS2 system combined with expression of medium-chain FA specific CYPs to be optimal for selective de novo biosynthesis of C8-HyFA.
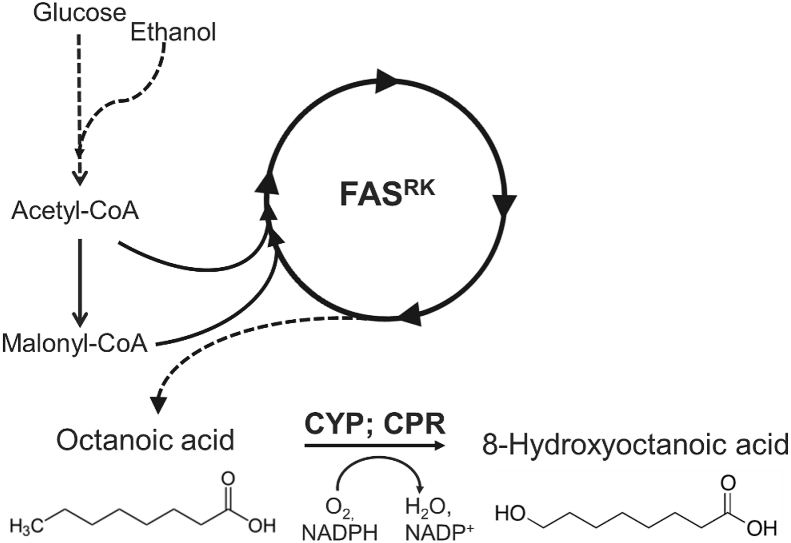
Here, we present for the first time the proof of principle for the de novo biosynthesis of C8-HyFA in S. cerevisiae. We constructed a complete pathway from glucose or ethanol towards C8-HyFA (Fig. 1) by combining C8-FA production via a mutated FAS (FAS1R1834K/FAS2) with direct ω-hydroxylation of the C8-FA by various CYPs.
Fig. 1.
Schematic presentation of de novo biosynthesis of 8-hydroxyoctanoic acid. A mutated fatty acid synthase FAS1R1834K/FAS2 (FASRK) produces octanoic acid which is ω-hydroxylated by a cytochrome P450 (CYP) resulting in 8-hydroxyoctanoic acid formation. The electrons required for the CYP reaction are transferred from NADPH to CYP by a cytochrome P450 reductase (CPR).
2. Materials and methods
2.1. Strain construction and transformation
Yeast strains used in this study are listed in Table 1. Yeast strain SHY24 was constructed by deletion of the FAA2 locus in BY4741. Strain SHY34 resulted from the previously described strain RPY21 (Gajewski et al., 2017) by deletion of two kanMX markers which were present in the RPY21 genome as remnants of fas1 and fas2 deletion. RPY21 has a BY background and is based on strain BY.PK1238_1A_KO.
Table 1.
Yeast strains used in this study.
| Strain Name | Relevant features | Reference/Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Brachmann et al. (1998) |
| CEN.PK113-7D | MATa MAL2-8c SUC2 | Euroscarf, Frankfurt am Main, Germany |
| SHY24 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δfaa2 | This work |
| SHY34 | MATα ura3Δ0 his3Δ0 leu2Δ0 TRP1 lys2Δ0 MET15 Δfas1 Δfas2 Δfaa2 | This work |
| LBY3 | CEN.PK113-7D Δpox1 | This work |
RPY21 and SHY34 carry additional deletions in FAA2 (medium chain fatty acyl-CoA synthetase).
Strain LBY3 was constructed from strain CEN.PK113-7D by deletion of POX1. The deletions of kanMX, FAA2 and POX1 were carried out by CRISPR-Cas9 meditated gene deletion as described previously (Generoso et al., 2016). Transformations were performed following the frozen competent cell protocol (Gietz and Schiestl, 2007), whereas SHY34 was transformed by a slightly modified method as previously described (Gajewski et al., 2017). FAS deficient strains are auxotrophic for fatty acids and, to allow growth, their medium was supplemented with 0,5 mM oleic acid and Terigitol-NP-40 (70% Sigma Aldrich Germany) to solubilize it. Transformed yeasts were plated on solid YPD (2% (w/v) peptone, 1% (w/v) yeast extract, 2% (w/v) glucose) containing appropriate antibiotics hygromycin (100 mg/L) or G418 (200 mg/L) for plasmid selection and grown at 30 °C for two to four days.
2.2. Plasmid and strain construction
All plasmids used in this study are listed in Table 2. Plasmids carrying cytochrome P450s (CYP) and cytochrome reductases (CPR) were constructed using the Golden Gate cloning system (Lee et al., 2015) by following the published protocol. Genes of cytochrome P450 and cytochrome reductases were purchased as DNA Strings from Thermo Fischer Scientific, Germany with flanking BsmBI and BasI restriction site overhangs for Golden Gate cloning. The MET25 promoter was included in the Golden Gate toolkit by amplification from BY4741 genomic DNA and cloning into pYTK backbone. CYPs used were: CYP4B1 from Oryctolagus cuniculus, CYP94C1 from Arabidopsis thaliana, CYP539A7 from Fusarium oxysporum f. sp. lycopersici (UniProtKB accession numbers: P15128; Q9ZUX1; A0A0D2X892) and their respective CPRs POR from Oryctolagus cuniculus, ATR1 from Arabidopsis thaliana, cprA from Fusarium oxysporum f. sp. lycopersici (UniProtKB accession numbers: P00389, Q9SB48, A0A0D2XWA9). CYPs and CPRs were cloned into a 2micron backbone with hygromycin resistance marker.
Table 2.
Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| pYTK-CYP4B1-POR | 2micron, KanR, HygR, pTDH3-CYP4B1-tENO1, pMET25-POR-tSSA1 | This study |
| pYTK-CYP94C1-ATR1 | 2micron, KanR, HygR, pTDH3-CYP94C1-tENO1, pMET25-ATR1-tSSA1 | This study |
| pYTK-CYP53A7-cprA | 2micron, KanR, HygR, pTDH3_CYP539A7- tENO1 pMET25-cprA-tSSA1 | This study |
| pYTK- CYP539A7 | 2micron, KanR, HygR, pTDH3-CYP539A7-tENO1 | This study |
| pYTK- CYP539A7-pPGK1-cprA | 2micron, KanR, HygR, pTDH3-CYP539A7-tENO1_pPGK1-cprA-tSSA1 | This study |
| pYTK- CYP539A7-pTEF2-cprA | 2micron, KanR, HygR, pTDH3-CYP539A7-tENO1_pTEF2-cprA-tSSA1 | This study |
| pYTK- CYP539A7-pALD6-cprA | 2micron, KanR, HygR, pTDH3-CYP539A7-tENO1_pALD6-cprA-tSSA1 | This study |
| pYTK- CYP539A7-pRNR1 -cprA | 2micron, KanR, HygR, pTDH3-CYP539A7-tENO1_pRNR1-cprA-tSSA1 | This study |
| pYTK- CYP539A7-pRAD27 -cprA | 2micron, KanR, HygR, pTDH3-CYP539A7-tENO1_pRAD27-cprA-tSSA1 | This study |
| pRS315-FAS1-RK | CEN6/ARS4, AmpR, LEU2, pFAS1-FAS1R1834K-tFAS1 | Gajewski et al. (2017) |
| pRS313-FAS2 | CEN6/ARS4, AmpR, HIS3, pFAS2-FAS2-tFAS2 | Gajewski et al. (2017) |
| pRS315-FAS1RK-FAS2 | CEN6/ARS4, AmpR, kanMX, pFAS1-FAS1R1834K, pFAS2-FAS2-tFAS2 | This study |
| pRS315-FAS1RK-FAS2-2μ | 2micron, AmpR, kanMX, pFAS1-FAS1R1834K, pFAS2-FAS2-tFAS2 | This study |
| pRS62-K | 2micron, AmpR, kanMX, | Farwick et al. (2014) |
KanR and AmpR stand for genes which are responsible for kanamycin or ampicillin resistance for E. coli. HygR and kanMX stand for genes which are responsible for hygromycin or G418 resistance for S. cerevisiae.
The plasmid pRS315-FAS1RK-FAS2 was constructed by homologous recombination in strain BY4741.
pFAS2-FAS2-tFAS2 and kanMX fragments were amplified with oligonucleotides (Supplementary Table 1) that contained at least 30 bp overhangs to the plasmid backbone for homologous recombination. pFAS2-FAS2-tFAS2 was integrated into XhoI cut site of pRS315-FAS1 backbone and kanMX was integrated into EcoRV cut site (LEU2) with overhangs to replace LEU2 gene. Fragments of pFAS2-FAS2-tFAS2 and kanMX were amplified from pRS313-FAS2 and pRS26-K, respectively. Plasmids containing auxotrophic markers (LEU2 or HIS3) were selected in synthetic media lacking the corresponding compound (leucine or histidine), whereas the plasmids carrying hygromycin (HygR) and G418 (kanMX) resistance genes were selected by adding antibiotics (200 μg/ml) to the media.
2.3. Media and cultivation
Several colonies of yeast strains or transformed yeasts were used to inoculate liquid medium pre-cultures. Liquid cultures were grown in shake flasks at 30 °C and 180 rpm using either SC (0.17% (w/v) yeast nitrogen base without amino acids, 0.5% (w/v) ammonium sulfate, addition of amino acids as previously described (Bruder et al., 2016)) or YP (2% (w/v) peptone, 1% (w/v) yeast extract) medium. 2% (w/v) glucose was used as carbon source for pre-cultures. Main cultures were inoculated from pre-cultures to high cell density (OD600 of 8–10) with either 2% (w/v) glucose, 2% (w/v) galactose, or 2% (v/v) ethanol plus 2% (v/v) glycerol as carbon sources as indicated. Glycerol was added to ethanol containing media with the rationale to provide the cells with C3 carbon skeletons.
High cell density bioconversion experiments were performed by supplementation with 50 mg/L octanoic acid. In bioconversion experiments with SC medium supplementation of 2 mM methionine was done for pre-cultures to repress CPR expression from pMET25. For the bioconversions and de novo biosynthesis with YP medium 100 mM potassium phosphate buffer (pH 6.5) was added. 5-Aminolevulinic acid (ALA) and hemin were purchased from Sigma Aldrich, Germany and dissolved as 100x stock solutions in H20 and DMSO, respectively. They were added to the main cultures with final concentrations of 300 μg/L and 50 μg/L, respectively. Samples for compound extraction were taken at given time points.
2.4. Toxicity assay and degradation analysis
For the toxicity assay a BY4741 pre-culture was inoculated into fresh YPD medium to an OD600 of 0.2 and cultivated for 5–6 h until an OD600 of 0.8–1.0 was reached. The culture was re-diluted in YPD medium to an OD600 of 0.05 and 50 μl was used to inoculate 200 μL of YPD medium in 96-well plates with a dilution series of C8- or C10 fatty acids, ω-hydroxy fatty acids or dicarboxylic acids. The pH values were approximately 5.0 for the media with dicarboxylic acids and approximately 5.9 for the media with ω-hydroxy fatty acids. The compounds were dissolved in YPD medium at their highest concentrations, filtered for sterility (0.2 μm) and subsequently diluted to desired concentrations. Cultivations were performed for 18 h at 30 °C without agitation and cells were mixed thoroughly before OD600 measurement in a plate reader (ClarioStar, BMG Labtech, Germany).
For 8-hydroxyoctanoic acid or octanedioic acid degradation analysis, strains were inoculated from pre-cultures in fresh YPD medium supplemented with 50 mg/L 8-hydroxyoctanoic acid or octanedioic acid to high cell density (OD600 = 10). The pH values were approximately 6.2 for medium with 8-hydroxyoctanoic acid and 5.9 for medium with octanedioic acid. Samples for extraction of compounds were taken at given time points.
2.5. Compound extraction and derivatization
For extraction of fatty acids or hydroxy fatty acids and dicarboxylic acids, yeast cultures were separated from the medium (16 000 rpm, 5 min). Hydroxy fatty acids or dicarboxylic acids were extracted from 1 ml culture supernatant. 0.02 mg heptanoic acid was added as an internal standard followed by acidification with 200 μL 1 M HCl and extraction with equal volumes of ethyl acetate. After phase separation the organic phase was taken and evaporated in a vacuum concentrator (Concentrator 5301, Eppendorf, Germany). The resulting extract was dissolved in 200 μL ethyl acetate and mixed with equal volumes of N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA, Alfa Aesar, Germany) for derivatization. Derivatization was performed for 45 min at 75 °C and samples were analyzed by gas chromatography (GC).
Fatty acids were derivatized to methyl esters for GC analysis and extracted from 10 ml culture supernatant. 0.2 mg heptanoic acid was added as an internal standard followed by acidification with 1 ml 1 M HCl and extraction by 2.5 ml of 1:1 mixture of methanol and chloroform. After centrifugation for phase separation (4000 rpm, 10 min) the organic phase was taken and evaporated in a vacuum concentrator (Concentrator 5301, Eppendorf, Germany). The resulting extract was dissolved in 200 μL toluol and mixed with 10 ml methanol and 300 μL 8% HCl for methylation. Derivatization was performed at 100 °C for 3 h. Fatty acid methyl esters were extracted by addition of 1 ml water and 1 ml hexane. The hexane phase was collected and analyzed by GC.
2.6. Gas chromatography
Analysis was performed on a Clarus 400 gas chromatograph (PerkinElmer, Germany) equipped with a FID detector and an Elite-5MS column (Ø 0.25 mm; length 30 m; film thickness 1.00 μm). 1 μL of sample was injected with 1:10 split and helium was used as carrier gas (90 kPa). 8-Hydroxy fatty acids and dicarboxylic acids– BSTFA derivates were separated with by the following method: run time 25.5 min, start at 50 °C and hold for 5 min; ramp at 20 °C min to 240 °C and hold for 2 min, ramp at 20 °C–300 °C and hold for 5 min. Octanoic acid methyl esters were separated by the following method: run time 42.67 min, start at 50 °C and hold for 5 min; ramp at 10 °C min to 120 °C and hold for 5 min, ramp at 15 °C–220 °C and hold for 10 min, ramp at 20 °C–300 °C and hold for 5 min.
3. Results & discussion
3.1. Evaluating possible toxic effects of medium-chain ω-hydroxy fatty acids and dicarboxylic acids on yeast
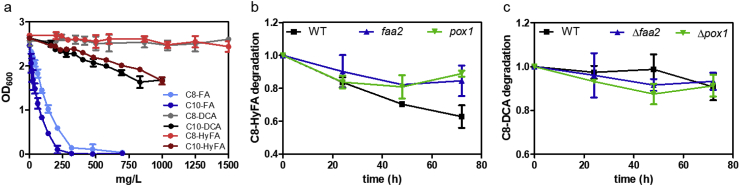
We first evaluated possible toxic or inhibitory effects of HyFAs and DCAs on yeast cells. Medium-chain FAs like C8-FA and decanoic (C10-FA) acid and derivatives like 1-octanol are well known to affect the growth of yeast cells (Viegas et al., 1989; Alexandre et al., 1996; Legras et al., 2010; Borrull et al., 2015; Henritzi et al., 2018). 1-Octanol for instance even causes a stronger growth impairment than C8-FA (Henritzi et al., 2018). As the biosynthesis of products with toxic properties can reduce product yields e.g. by inhibition of cellular processes or even can lead to cell death, we investigated possible toxic effects of HyFAs and DCAs with eight or ten straight carbon atoms. Measurements of cell density (recorded as changes in OD600) were used as proxy to assess the toxicity of extracellularly supplemented compounds to a wild type (BY4741) yeast culture. The toxicity was compared to the well-known toxic compounds C8-FA and C10-FA. As expected, C8 and C10–FAs strongly influenced growth of the yeast strain at rising concentrations (Fig. 2a). At about 500 mg/L and 300 mg/L, respectively, no growth was detectable anymore. In contrast, the C8 or C10-HyFAs and DCAs caused little or no growth impairment even at high concentrations (Fig. 2a).
Fig. 2.
Toxicity assays and degradation analyses of fatty acids, ω-hydroxy fatty acids and dicarboxylic acids. a) BY4741 wild type yeast strain was grown in 96-well plates in YPD medium with different concentrations of C8 or C10 fatty acids (FA), ω-hydroxy fatty acids (HyFA) or dicarboxylic acids (DCA). b) Supplementation of 50 mg/L of 8-hydroxyoctanoic acid (C8-HyFA) or c) supplementation of 50 mg/L octanedioic acid (C8-DCA) to yeast cultures of BY4741 (WT), SHY24 (Δfaa2) or LBY3 (Δpox1). The values in (b) and (c) were normalized to a control sample without yeast cells. Values and error bars represent mean and standard deviation of two biological replicates.
The toxicity of C8–FAs or C10–FAs has been attributed to various mechanisms. S. cerevisiae rapidly acidifies its growth medium and under such conditions weak acids, like medium-chain FAs, are present in their protonated form. In this state, they are more liposoluble and can pass the membrane by passive diffusion (Viegas and Sá-Correia, 1997). In the neutral cytosol, dissociation leads to a decrease of cellular pH and accumulation of acyl-anions, which lead to an inhibition of cellular processes (Cabral et al., 2001). Indeed, the influence of a decreasing pH on increasing toxicity of C8–FAs has been demonstrated (Liu et al., 2013). C8–FAs rapidly enter yeast cells (Henritzi et al., 2018) although the majority is found in cell wall fractions and not inside the cell, which is attributed to energy expensive expulsion by the Pdr12 carrier (Borrull et al., 2015). Additionally, exposure of yeast to C8–FAs is causing membrane stress and disturbs integrity leading to membrane leakage (Liu et al., 2013). For medium-chain FAs, toxicity is increasing with increasing chain length (C10 > C8 > C6) (Liu et al., 2013) possibly because of stronger membrane perturbation due to higher liposolubility. Interestingly, dependency of chain-length on toxicity was also demonstrated here for HyFAs and DCAs, whereby the longer C10 variants cause a growth defect while the C8 variants do not (Fig. 2a). However, due to the additional hydroxyl or carboxyl group HyFAs and DCAs, respectively, are less liposoluble than their corresponding FAs, which likely results in a reduced ability to integrate into or diffuse across the plasma membrane. However, other reasons for lower toxicity cannot be ruled out.
3.2. Degradation of medium-chain ω-hydroxy fatty acids and dicarboxylic acids in yeast
It has been suggested that ω-hydroxy lauric acid can be rapidly degraded via β-oxidation in S. cerevisiae (Durairaj et al., 2015). In order to test the stability of C8-HyFA and C8-DCA in a yeast culture, a wild type (BY4741) strain was inoculated to a high cell density in YPD medium supplemented with either 50 mg/L C8-HyFA or C8-DCA. The amounts of C8-HyFA were reduced by 37% after 72 h (Fig. 2b). To assess if this decrease can be attributed to degradation via the same route as that observed for C8-FA (Leber et al., 2016; Henritzi et al., 2018), FAA2 (encoding the medium-chain fatty acyl CoA synthetase) or POX1 (encoding the acyl-CoA oxidase, the initial enzyme of β-oxidation) were deleted. In these two strains, the degradation of C8-HyFA was reduced by more than 50% compared to the WT (Fig. 2b). This suggests that activation by Faa2 and (subsequent) β-oxidation are the main but not the only reason for reduction of C8-HyFA levels. The remaining decrease of C8-HyFA in faa2 or pox1 deletion strains could be due to competing reactions, e.g. esterification (Borrull et al., 2015). Moreover, the deletions of POX1 and FAA2 could also indirectly (e.g. by altering the lipid composition of the membrane) affect the uptake and intracellular fate of C8-HyFA.
Interestingly, the supplemented C8-DCA showed only small decreases with similar results in all three strains (Fig. 2c). This suggests that C8-DCA either is not degraded via β-oxidation or it does not cross the membrane due to the presence of the additional carboxyl group. The initial pH of the medium supplemented with 50 mg/L C8-HyFA or C8-DCA was around 6.2 and 5.9, respectively. However, cultures of S. cerevisiae acidify the medium rapidly and therefore C8-DCA (pKa: 4.53 and 5.50 (Bretti et al., 2006)) is likely to become increasingly protonated in the course of cultivation, which should facilitate its diffusion across the plasma membrane. This is reasonable to expect, as C8-FA, which exhibits an even lower pKa value (4.89 (Dean, 1987)), is readily taken up by the cells in phosphate-buffered media at starting pH of 6.5 (Henritzi et al., 2018). The results presented here show that medium-chain HyFAs and DCAs have only little or no toxic effects on yeast cells, and degradation of C8 variants either does not take place or can be reduced by blocking of the β-oxidation.
3.3. Bioconversion of octanoic acid results in C8-HyFA production but not of C8-DCA
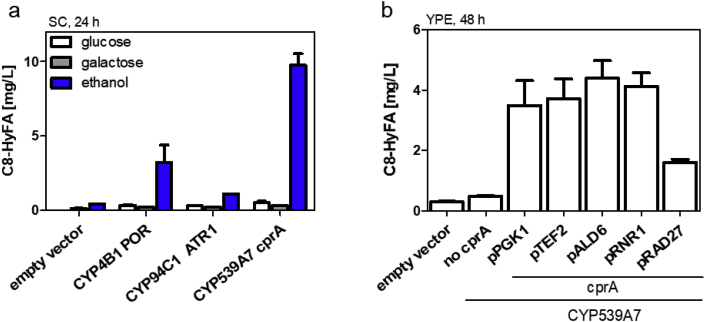
CYPs from various organisms have been shown to ω-hydroxylate medium-chain FAs of eight to twelve carbon atoms by in vitro assays (Fisher et al., 1998) or by heterologous expression in S. cerevisiae and whole-cell bioconversion of externally added FAs (Durairaj et al., 2015; Han et al., 2017). We decided to systematically compare effective ω-hydroxylation of C8–FAs for three CYPs CYP4B1 (Fisher et al., 1998) CYP94C1 (Durairaj et al., 2015) and CYP539A7 (Han et al., 2017) based on their activity towards medium-chain FAs or C8-FA specifically. The hydroxylation reaction consists of a two component system with CYP and CPR. The CYPs were expressed under control of the strong and constitutive TDH3 promoter while CPRs were under control of the methionine-repressible MET25 promoter. The latter was chosen because an excess of CPR over CYP can induce the formation of reactive oxygen species and affect cell viability (Zangar et al., 2004). Therefore, balancing expression between CYP and its reductase can be crucial for cell health and high productivity (Paddon et al., 2013). By controlling expression of the CPRs by the MET25 promoter we prevented its expression in the pre-culture by methionine supplementation, but reached higher expression in the main culture by omitting methionine for high productivity. The cytochromes and their reductases were expressed in strain SHY24 (Δfaa2) so that degradation of C8-HyFA was minimized (see Fig. 2b). Effective ω-hydroxylation was tested in high cell density bioconversions in SC-medium with supplementation of 50 mg/L C8-FA. Initial experiments were performed using 2% glucose as a carbon source. All three tested CYP-CPR variants showed production of C8-HyFA but titers remained below 1 mg/L (Fig. 3a). We additionally tested use of ethanol/glycerol and galactose as carbon sources. With galactose as carbon source C8-HyFA remained mainly below production levels with glucose. Interestingly, in ethanol/glycerol media, all three tested cytochromes displayed production of C8-HyFA. Activity of CYP94C1-ATR1 remained the lowest of the tested CYPs. This is consistent with the finding that its activity is mainly displayed on medium-chain FAs with ten or twelve carbon atoms (Kandel et al., 2007; Han et al., 2017). The best performing enzymes were CYP539A7-cprA which converted 20% of C8-FA to C8-HyFA. Thus, CYP539A7-cprA was chosen for further experiments.
Fig. 3.
Bioconversion of octanoic acid to 8-hydroxyoctanoic acid. Bioconversions were performed with strain SHY24 (Δfaa2) in SC (a) or YP medium (b) supplemented with 50 mg/L C8-FA. Note that ethanol media additionally contained glycerol (see Materials & Methods). a) Expression of different CYPs and CPRs in SHY24 with different carbon sources as indicated. b) WT strain (BY4741) was transformed with the empty vector, CYP539A7 alone (no cprA), or CYP539A7 in combination with cprA under the control of different promoters indicated at the bottom. Samples were taken after 24 h (a) or 48 h (b). Values and error bars represent mean and standard deviation of two biological replicates.
With the aim of a de novo biosynthesis of C8-HyFAs, we wanted to adapt the fermentation conditions to the best conditions previously developed for C8-FA production. Selective C8-FA production was previously achieved by the expression of a mutated fatty acid synthase (FAS1R1834K/FAS2) (Gajewski et al., 2017). However, the C8-FA titers were strongly dependent on the medium. In YPD medium with a potassium phosphate buffer, C8-FA titers were strongly increased (Gajewski et al., 2017). In contrast, production of C8–FAs remained low in SC medium compared to YP medium (Pavlovic, 2016). For a change from SC medium to YP medium the MET25 promoter for controlling CPR expression is not feasible as it would be continuously repressed by methionine in the medium. To balance the expression of cprA we tested expression from five promoters with different strengths (pPGK1 > pTEF2 > pALD6 > pRNR1 > pRAD27 (Lee et al., 2015)) and analyzed production of C8-HyFAs from C8–FAs in buffered YP medium with ethanol as carbon source (Fig. 3b). Because titers remained low after 24 h bioconversion, incubation times were increased to 48 h. Promoter strength of cprA expression had only limited effects on C8-HyFAs production. Four out of five promoters showed comparable C8-HyFA output and only the use of pRAD27 reduced titers by 50% suggesting that expression was too low and cprA became limiting. Indeed, if no CPR was expressed only limited amounts of C8-HyFA were detectable. High expression of cprA i.e. from pPGK1 did not reduce growth of cultures (final OD 26.8 compared to 23.3 for empty vector control) suggesting that there is no toxic effect of high cprA expression. These new bioconversion conditions (promoter, medium, buffering) reduced titers by about 60% compared to the initial experiment in synthetic medium. The best performing expression system with pTDH3-CYP539A7 and pALD6-cprA was chosen for further experiments.
DCAs acids can be formed from HyFAs by two consecutive oxidations catalyzed by alcohol and aldehyde dehydrogenases. It has been reported that expression of a CYP and blocking of the β-oxidation for production of HyFAs alone led to formation of DCAs by endogenous alcohol and aldehyde dehydrogenases (Han et al., 2017). We also considered formation of C8-DCAs from C8-HyFAs under our conditions, but C8-DCA titers always remained below the detection limit. Future strategies for C8-DCA formation should consider and test the overexpression of heterologous alcohol and aldehyde dehydrogenases with specific medium-chain activity.
3.4. Influence of heme on 8-hydroxyoctanoic acid titers in glucose conditions
The reactions of CYPs are dependent on heme in the active center (Munro et al., 2007b). Heme can also serve as a signaling molecule and efforts have been made to understand the regulation of heme biosynthesis in S. cerevisiae (Keng and Guarente, 1987; Zhang and Hach, 1999; Mense and Zhang, 2006). The cellular levels of heme have been found to be dependent on the carbon source and availability of oxygen (Zitromer and Lowrym, 1992). Under glucose/anaerobic conditions heme levels are low but are increased under ethanol/aerobic conditions (Zhang et al., 2017). We hypothesized that - when glucose is used as a carbon source - heme might be limiting the CYP activity, which could result in reduced titers of C8-HyFAs compared to ethanol/glycerol medium (Fig. 3a). Upregulation of known bottleneck reactions of heme biosynthesis HEM2, HEM3 or HEM12 (Michener et al., 2012) or deletion of heme oxygenase HXM1 (Savitskaya et al., 2019), which is involved in heme degradation, have been shown to increase heme levels. It was reported that heme levels can also be increased by supplementation of hemin or by the heme precursor 5-aminolevulinic acid (ALA) (Zhang et al., 2017). Therefore, we tested if supplementation of ALA or hemin influences the production of C8-HyFAs in a bioconversion experiment using buffered YPD medium. ALA did not influence C8-HyFA titers compared to YPD medium without supplement (Fig. 4), which is consistent with the observation that the formation of ALA is not the rate limiting step of heme biosynthesis (Hoffman et al., 2003). However, titers were increased by about 2-fold when hemin was supplemented. Although hemin supplementation increased C8-HyFA titers in glucose medium, titers remained below those measured in ethanol/glycerol medium (about 50% reduced) (Fig. 3, Fig. 4). Therefore, it is unlikely that heme availability is the only bottleneck for C8-FA hydroxylation under glucose conditions. Nevertheless, improvement of heme supply is certainly a target for future studies, and strategies to increase endogenous heme levels (Michener et al., 2012; Savitskaya et al., 2019) will have to be included in production strain engineering.
Fig. 4.
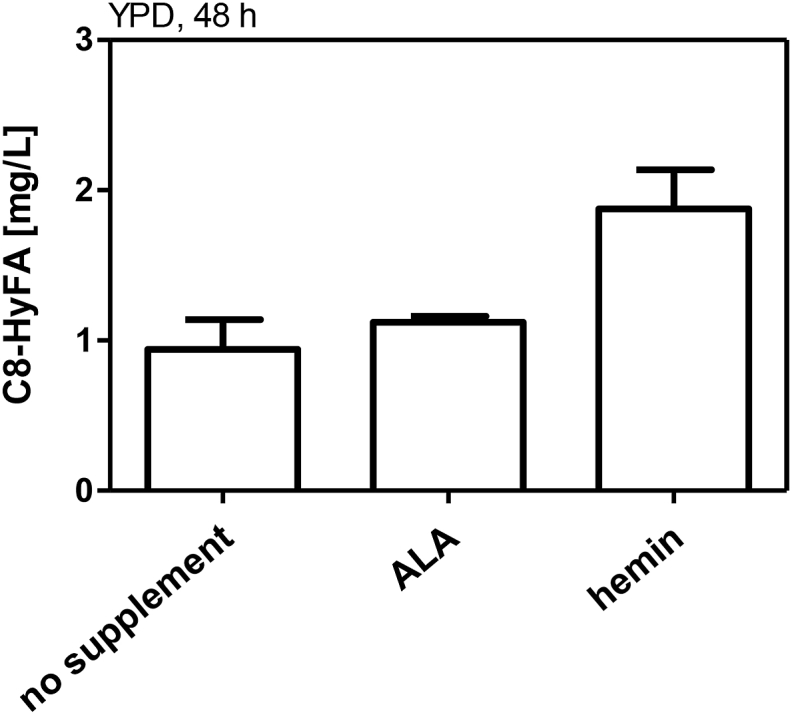
Bioconversion of octanoic acid to 8-hydroxyoctanoic acid with hemin and ALA supplementation. Bioconversions were performed with strain SHY24 (Δfaa2) expressing CYP539A7 pALD6-cprA in buffered YPD medium supplemented with 50 mg/L C8-FA and additional supplementation of 5-aminolevulinic acid (ALA, 300 μg/mL) or hemin (50 μg/mL). Samples were taken after 48 h. Values and error bars represent mean and standard deviation of two biological replicates.
Furthermore, the limited capacity of the yeast cells to express functional membrane proteins such as CYPs must also be considered (Michener et al., 2012). A possible reason for increased hydroxylation activity in ethanol/glycerol media might be attributed to altered lipid composition on non-fermentative carbon sources (Alexandre et al., 1994; Huffer et al., 2011; Henderson and Block, 2014; Cray et al., 2015), which may be beneficial for functional expression of membrane bound CYP539A7-cprA. Additionally, ethanol increases the toxic effect of C8–FAs (Legras et al., 2010) which could trigger higher conversion rates to non-toxic HyFAs. An effect of the carbon source on the transcriptional level may also be considered, since the activity of the TDH3 promoter on ethanol is reduced compared to its activity on glucose and galactose (Peng et al., 2015). A reduced transcript level may be beneficial to avoid possible aggregation caused by the overexpression of a heterologous membrane protein. A very likely explanation for the beneficial effect of ethanol-containing media may also be found in the redox cofactor supply. Ethanol is converted to acetate via acetaldehyde by the consecutive action of the alcohol dehydrogenases and aldehyde dehydrogenases. The involvement of the NADP-dependent isoform Ald6 in ethanol catabolism may provide increased amount of NADPH, which is required for the hydroxylation by CYPs, compared to glucose-grown cells. Thus, redox-cofactor supply is also an important target for future studies.
3.5. De novo biosynthesis of 8-hydroxyoctanoic acid from glucose and ethanol
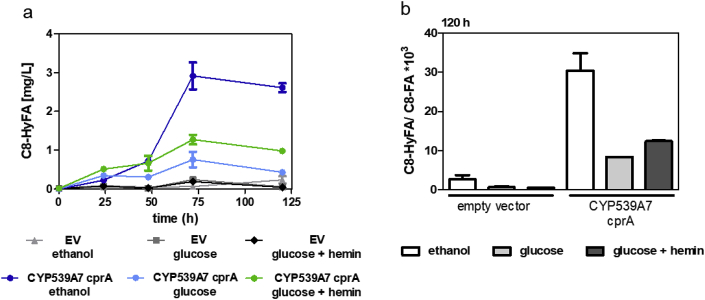
Recently, the biosynthesis of medium-chain FAs was established by rational engineering of the FAS (Gajewski et al., 2017). A mutation in the MPT domain of FAS1 (R1834K) enabled production of C8–FAs with high specificity. We wanted to combine C8-FA synthesis with ω-hydroxylation by CYP. For that, a plasmid expressing mutated fatty acids synthase (FAS1R1834K/FAS2) was transformed together with the best performing CYP (pTHD3-CYP539A7 pALD6-crpA) in a fas1/2 deficient strain with faa2 deletion (SHY34) and fermented in YP medium with ethanol or glucose with or without hemin supplementation. The mutated FAS was expressed from a low copy (CEN6/ARS4) plasmid because a higher copy number (2micron) had no beneficial effect on octanoic acid production (Supplementary Fig. 1). Additionally, an empty vector control was used instead of pTDH3-CYP539A7 pALD6-cprA expression. All three fermentation conditions successfully resulted in de novo formation of C8-HyFA (Fig. 5a). Fermentations in glucose medium resulted in low titers, which did not exceed 1 mg/L. As already shown in the bioconversion experiments, hemin supplementation increased titers by 68%. The use of ethanol/glycerol media additionally increased titers by 2.3-fold. To quantitatively compare the utilization of different carbon sources for C8-HyFA production, we calculated the molar yields (μmol product per mol of carbon source; Supplementary Table 2) at the terminal time point (120 h). In ethanol/glycerol media, no detectable consumption of glycerol has occurred in the course of fermentation (Supplementary Fig. 2), so that the production of C8-HyFA occurred only from ethanol. Interestingly, the molar yields in glucose/hemin media were highest, but when the values were normalized to the cell density (OD600), the values obtained in ethanol/glycerol were comparable to those obtained in glucose/hemin media, while the lowest performance was achieved in glucose media without hemin addition (Supplementary Table 2). This observation underlines the importance of heme for the functionality of CYPs. After 120 h of fermentation in all samples high amounts of C8-FA (52–100 mg/L) remained detectable. Whereas ethanol/glycerol media did not have a significant influence on C8-FA production, they yielded the highest C8-HyFA to C8-FA ratios (Fig. 5b). This again shows that activity of the CYP and not C8-FA production is the bottleneck for C8-HyFA production under all three conditions tested. Efficient electron transfer from CPR to CYP is important for high CYP activity. Artificial fusion proteins of CYP and CPR have been explored in many studies and can improve CYPs catalytic activity by forcing both partners into close proximity (Munro et al., 2007a). Therefore, in order to improve CYP activity, the fusion to CPR should be considered as a future engineering strategy.
Fig. 5.
De novo biosynthesis of 8-hydroxyoctanoic acid. Mutated fatty acid synthase (FAS1R1834K/FAS2) and pTDH3-CYP539A7 with pALD6-cprA were expressed in the strain SHY34 which was grown in YP medium containing indicated carbon sources with or without hemin supplementation. Note that ethanol media additionally contained glycerol (see Materials & Methods). As a negative control, an empty vector was introduced instead of CYP539A7 and cprA. a) extracellular concentrations of C8-HyFAs and (b) the C8-HyFA/C8-FA ratio at 120 h is shown. Values and error bars represent mean and standard deviation of two biological replicates.
4. Conclusions
Here, we present for the first time the proof of concept for de novo biosynthesis of 8-HyFA in S. cerevisiae. Production of 8-HyFA was achieved from glucose or ethanol by combining ω-hydroxylation of C8-FA by cytochrome P450 with C8-FA production via a mutated FAS. We show that the choice of fermentation conditions and of the carbon source are critical and that the ω-hydroxylation is the limiting step in the process. Possible targets for further strain improvement were identified. We show that medium-chain HyFAs and DCAs are little or non-toxic for S. cerevisiae, so that high production titers should be possible in the future.
Authors’ contributions
FW, EB and MO conceived the study. FW conducted the experiments.
FW, EB, and MO analyzed the data and wrote the paper. All authors read and approved the manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 720824.
Declaration of competing interest
E.B. is co-inventor of EP patent application No. 15 162 192.7 filed on April 1, 2015, and of EP patent application No. 15 174 342.4 filed on June 26, 2015, by Goethe-University Frankfurt, concerning short-chain acyl-CoA producing FAS variants. There are no other competing interests.
Acknowledgements
We thank Renata Pavlovic for providing yeast strain RPY21, Sandra Henritzi for strains SHY24, SHY34 as well as providing Fig. S1, and Leonie Baumann for strain LBY3.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2019.e00111.
Abbreviations
- OD600
optical density at 600 nm
- CoA
coenzyme A
- FAS
fatty acid synthase
- CYP
cytochrome P450
- CPR
cytochrome P450 reductase
- ALA
5-Aminolevulinic acid
- FA
fatty acid
- HyFA
ω-hydroxy fatty acid
- DCA
α,ω-dicarboxylic acid
- SC
synthetic complete
- YP
yeast extract and peptone
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Akaike Y. Other oleochemical uses: palm oil products. JAOCS (J. Am. Oil Chem. Soc.) 1985;62:335–340. [Google Scholar]
- Alexandre H., Mathieu B., Charpentier C. Alteration in membrane fluidity and lipid composition, and modulation of H+-ATPase activity in Saccharomyces cerevisiae caused by decanoic acid. Microbiology. 1996;142:469–475. doi: 10.1099/13500872-142-3-469. [DOI] [PubMed] [Google Scholar]
- Alexandre H., Rousseaux I., Charpentier C. Relationship between ethanol tolerance, lipid composition and plasma membrane fluidity in Saccharomyces cerevisiae and Kloeckera apiculata. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 1994;124:17–22. doi: 10.1111/j.1574-6968.1994.tb07255.x. [DOI] [PubMed] [Google Scholar]
- Borrull A., Lopez-Martinez G., Poblet M., Cordero-Otero R., Rozes N. New insights into the toxicity mechanism of octanoic and decanoic acids on Saccharomyces cerevisiae. Yeast. 2015;32:451–460. doi: 10.1002/yea.3071. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bretti C., Crea F., Foti C., Sammartano S. Solubility and activity coefficients of acidic and basic nonelectrolytes in aqueous salt solutions. 2. Solubility and activity coefficients of suberic, azelaic, and sebacic acids in NaCl(aq), (CH3)4NCl(aq), and (C2H5)4NI(aq) at different ionic strengths and at t = 25 °C. J. Chem. Eng. Data. 2006;51:1660–1667. [Google Scholar]
- Bruder S., Reifenrath M., Thomik T., Boles E., Herzog K. Parallelised online biomass monitoring in shake flasks enables efficient strain and carbon source dependent growth characterisation of Saccharomyces cerevisiae. Microb. Cell Factories. 2016;15:127. doi: 10.1186/s12934-016-0526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral M.G., Viegas C.A., Sá-Correia I. Mechanisms underlying the acquisition of resistance to octanoic-acid-induced-death following exposure of Saccharomyces cerevisiae to mild stress imposed by octanoic acid or ethanol. Arch. Microbiol. 2001;175:301–307. doi: 10.1007/s002030100269. [DOI] [PubMed] [Google Scholar]
- Clomburg J.M., Blankschien M.D., Vick J.E., Chou A., Kim S., Gonzalez R. Integrated engineering of β-oxidation reversal and ω-oxidation pathways for the synthesis of medium chain ω-functionalized carboxylic acids. Metab. Eng. 2015;28:202–212. doi: 10.1016/j.ymben.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Cray J.A., Stevenson A., Ball P., Bankar S.B., Eleutherio E.C.A., Ezeji T.C., Singhal R.S., Thevelein J.M., Timson D.J., Hallsworth J.E. Chaotropicity: a key factor in product tolerance of biofuel-producing microorganisms. Curr. Opin. Biotechnol. 2015;33:228–259. doi: 10.1016/j.copbio.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Dean J.A. McGraw-Hill Book Company; 1987. Handbook of Organic Chemistry. [Google Scholar]
- Durairaj P., Malla S., Nadarajan S.P., Lee P.-G., Jung E., Park H.H., Kim B.-G., Yun H. Fungal cytochrome P450 monooxygenases of Fusarium oxysporum for the synthesis of ω-hydroxy fatty acids in engineered Saccharomyces cerevisiae. Microb. Cell Factories. 2015;14:45. doi: 10.1186/s12934-015-0228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farwick A., Bruder S., Schadeweg V., Oreb M., Boles E. Proceedings of the National Academy of Sciences of the United States of America. vol. 111. 2014. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose; pp. 5159–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moya R., Leber C., Cardenas J., Da Silva N.A. Functional replacement of the Saccharomyces cerevisiae fatty acid synthase with a bacterial type II system allows flexible product profiles. Biotechnol. Bioeng. 2015;112:2618–2623. doi: 10.1002/bit.25679. [DOI] [PubMed] [Google Scholar]
- Fisher M.B., Zheng Y.-M., Rettie A.E. Positional specificity of rabbit CYP4B1 for ω-hydroxylation of short-medium chain fatty acids and hydrocarbons. Biochem. Biophys. Res. Commun. 1998;248:352–355. doi: 10.1006/bbrc.1998.8842. [DOI] [PubMed] [Google Scholar]
- Gajewski J., Pavlovic R., Fischer M., Boles E., Grininger M. Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat. Commun. 2017;8 doi: 10.1038/ncomms14650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generoso W.C., Gottardi M., Oreb M., Boles E. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microbiol. Methods. 2016;127:203–205. doi: 10.1016/j.mimet.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:1–4. doi: 10.1038/nprot.2007.17. [DOI] [PubMed] [Google Scholar]
- Han L., Peng Y., Zhang Y., Chen W., Lin Y., Wang Q. Designing and creating a synthetic omega oxidation pathway in Saccharomyces cerevisiae enables production of medium-chain α,ω-dicarboxylic acids. Front. Microbiol. 2017;8:2184. doi: 10.3389/fmicb.2017.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt D.E., Drinkard G., Pierce H.F. Future of petrochemical raw materials in oleochemical markets. JAOCS (J. Am. Oil Chem. Soc.) 1984;61:276–281. [Google Scholar]
- Haushalter R.W., Phelan R.M., Hoh K.M., Su C., Wang G., Baidoo E.E.K., Keasling J.D. Production of odd-carbon dicarboxylic acids in Escherichia coli using an engineered biotin-fatty acid biosynthetic pathway. J. Am. Chem. Soc. 2017;139:4615–4618. doi: 10.1021/jacs.6b11895. [DOI] [PubMed] [Google Scholar]
- Henderson C.M., Block D.E. Examining the role of membrane lipid composition in determining the ethanol tolerance of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2014;80:2966–2972. doi: 10.1128/AEM.04151-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henritzi S., Fischer M., Grininger M., Oreb M., Boles E. An engineered fatty acid synthase combined with a carboxylic acid reductase enables de novo production of 1-octanol in Saccharomyces cerevisiae. Biotechnol. Biofuels. 2018;11:150. doi: 10.1186/s13068-018-1149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M., Góra M., Rytka J. Identification of rate-limiting steps in yeast heme biosynthesis. Biochem. Biophys. Res. Commun. 2003;310:1247–1253. doi: 10.1016/j.bbrc.2003.09.151. [DOI] [PubMed] [Google Scholar]
- Huffer S., Clark M.E., Ning J.C., Blanch H.W., Clark D.S. Role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Biotechnol. Biofuels. 2011;77:6400–6408. doi: 10.1128/AEM.00694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel S., Sauveplane V., Compagnon V., Franke R., Millet Y., Schreiber L., Werck-Reichhart D., Pinot F. Characterization of a methyl jasmonate and wounding-responsive cytochrome P450 of Arabidopsis thaliana catalyzing dicarboxylic fatty acid formation in vitro. FEBS J. 2007;274:5116–5127. doi: 10.1111/j.1742-4658.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- Keng T., Guarente L. Proceedings of the National Academy of Sciences of the United States of America. vol. 84. 1987. Constitutive expression of the yeast HEMJ gene is actually a composite of activation and repression; pp. 9113–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-R., Oh D.-K. Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 2013;31:1473–1485. doi: 10.1016/j.biotechadv.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Leber C., Choi J.W., Polson B., Da Silva N.A. Disrupted short chain specific beta-oxidation and improved synthase expression increase synthesis of short chain fatty acids in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2016;113:895–900. doi: 10.1002/bit.25839. [DOI] [PubMed] [Google Scholar]
- Leber C., Da Silva N.A. Engineering of Saccharomyces cerevisiae for the synthesis of short chain fatty acids. Biotechnol. Bioeng. 2014;111:347–358. doi: 10.1002/bit.25021. [DOI] [PubMed] [Google Scholar]
- Lee M.E., DeLoache W.C., Cervantes B., Dueber J.E. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 2015;4:975–986. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- Legras J.L., Erny C., Le Jeune C., Lollier M., Adolphe Y., Demuyter C., Delobel P., Blondin B., Karst F. Activation of two different resistance mechanisms in Saccharomyces cerevisiae upon exposure to octanoic and decanoic acids. Appl. Environ. Microbiol. 2010;76:7526–7535. doi: 10.1128/AEM.01280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Liu F., Cai J., Xie W., Long T.E., Turner S.R., Lyons A., Gross R.A. Polymers from fatty acids: poly(ω-hydroxyl tetradecanoic acid) synthesis and physico-mechanical studies. Biomacromolecules. 2011;12:3291–3298. doi: 10.1021/bm2007554. [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang C., Lu W. Biosynthesis of long-chain ω-hydroxy fatty acids by engineered Saccharomyces cerevisiae. J. Agric. Food Chem. 2019;67:4545–4552. doi: 10.1021/acs.jafc.9b00109. [DOI] [PubMed] [Google Scholar]
- Liu P., Chernyshov A., Najdi T., Fu Y., Dickerson J., Sandmeyer S., Jarboe L. Membrane stress caused by octanoic acid in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2013;97:3239–3251. doi: 10.1007/s00253-013-4773-5. [DOI] [PubMed] [Google Scholar]
- Lomakin I.B., Xiong Y., Steitz T.A. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell. 2007;129:319–332. doi: 10.1016/j.cell.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Lu W., Ness J.E., Xie W., Zhang X., Minshull J., Gross R.A. Biosynthesis of monomers for plastics from renewable oils. J. Am. Chem. Soc. 2010;132:15451–15455. doi: 10.1021/ja107707v. [DOI] [PubMed] [Google Scholar]
- Mense S.M., Zhang L. Heme: a versatile signaling molecule controlling the activities of. Cell Res. 2006:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- Michener J.K., Nielsen J., Smolke C.D. Proceedings of the National Academy of Sciences of the United States of America. vol. 109. 2012. Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases; pp. 19504–19509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro A.W., Girvan H.M., Mason A.E., Dunford A.J., McLean K.J. What makes a P450 tick? Trends Biochem. Sci. 2013;38:140–150. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Munro A.W., Girvan H.M., McLean K.J. Cytochrome P450--redox partner fusion enzymes. Biochim. Biophys. Acta. 2007;1770:345–359. doi: 10.1016/j.bbagen.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Munro A.W., Girvan H.M., McLean K.J. Variations on a (t)heme—novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat. Prod. Rep. 2007;24:585–609. doi: 10.1039/b604190f. [DOI] [PubMed] [Google Scholar]
- Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., Leavell M.D., Tai A., Main A., Eng D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Pavlovic R. Doctoral dissertation (Goehte University Frankfurt); 2016. Genetic Modification of Enzymes and Metabolic Pathways for the Improvement of Fatty Acid Synthesis in the Yeast Saccharomyces cerevisiae. [Google Scholar]
- Peng B., Williams T.C., Henry M., Nielsen L.K., Vickers C.E. Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microb. Cell Factories. 2015;14:91. doi: 10.1186/s12934-015-0278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H., Bassard J.-E., Hamberger B., Werck-Reichhart D. Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Curr. Opin. Plant Biol. 2014;19:27–34. doi: 10.1016/j.pbi.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Rupilius W., Ahmad S. Palm oil and palm kernel oil as raw materials for basic oleochemicals and biodiesel. Eur. J. Lipid Sci. Technol. 2007;109:433–439. [Google Scholar]
- Savitskaya J., Protzko R.J., Li F.-Z., Arkin A.P., Dueber J.E. Iterative screening methodology enables isolation of strains with improved properties for a FACS-based screen and increased L-DOPA production. Sci. Rep. 2019;9:5815. doi: 10.1038/s41598-019-41759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheps D., Honda Malca S., Richter S.M., Marisch K., Nestl B.M., Hauer B. Synthesis of ω-hydroxy dodecanoic acid based on an engineered CYP153A fusion construct. Microb. Biotechnol. 2013;6:694–707. doi: 10.1111/1751-7915.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.H. Life cycle assessment of five vegetable oils. J. Clean. Prod. 2015;87:130–138. [Google Scholar]
- Viegas C.A., Rosa M.F., Sà-Correia I., Novais J.M. Inhibition of yeast growth by octanoic and decanoic acids produced during ethanolic fermentation. Appl. Environ. Microbiol. 1989;55:21–28. doi: 10.1128/aem.55.1.21-28.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas C.A., Sá-Correia I. Effects of low temperatures (9-33°C) and pH (3.3-5.7) in the loss of Saccharomyces cerevisiae viability by combining lethal concentrations of ethanol with octanoic and decanoic acids. Int. J. Food Microbiol. 1997;34:267–277. doi: 10.1016/s0168-1605(96)01200-7. [DOI] [PubMed] [Google Scholar]
- Zangar R.C., Davydov D.R., Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol. Appl. Pharmacol. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Zhang L., Hach A. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 1999;56:415–426. doi: 10.1007/s000180050442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Bu P., Zeng J., Vancura A. Increased heme synthesis in yeast induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J. Biol. Chem. 2017;292:16942–16954. doi: 10.1074/jbc.M117.790923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Zhou Y.J., Krivoruchko A., Grininger M., Zhao Z.K., Nielsen J. Expanding the product portfolio of fungal type I fatty acid synthases. Nat. Chem. Biol. 2017;13:360–362. doi: 10.1038/nchembio.2301. [DOI] [PubMed] [Google Scholar]
- Zitromer R.S., Lowrym C.V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.